Abstract
Histone acetyltransferases are associated with the elongating RNA polymerase II (Pol II) complex, supporting the idea that histone acetylation and transcription are intertwined mechanistically in gene coding sequences. Here, we studied the establishment and function of histone acetylation and transcription in noncoding sequences by using a model locus linking the β-globin HS2 enhancer and the embryonic ɛ-globin gene in chromatin. An intact HS2 enhancer that recruits RNA Pol II is required for intergenic transcription and histone H3 acetylation and K4 methylation between the enhancer and target gene. RNA Pol II recruitment to the target gene TATA box is not required for the intergenic transcription or intergenic histone modifications, strongly implying that they are properties conferred by the enhancer. However, Pol II recruitment at HS2, intergenic transcription, and intergenic histone modification are not sufficient for transcription or modification of the target gene: these changes require initiation at the TATA box of the gene. The results suggest that intergenic and genic transcription complexes are independent and possibly differ from one another.
The role of intergenic antisense transcription in heterochromatin formation and silencing is well established (29). Intergenic transcription is also associated with activation in several developmentally regulated mammalian gene loci, and the transcribed regions often colocalize with regions of histone H3 acetylation and H3 K4 dimethylation (reviewed in reference 8). Acetylation of the amino-terminal tails of the core histones H3 and H4 and methylation of H3 lysine 4 are modifications associated with active genes and are thought to render the chromatin more “open” or permissive to the process of transcription (4, 19). The association of histone acetyltransferases and methyltransferases with RNA polymerase II (Pol II) provides a rationale for the establishment of these modifications in transcribed coding sequences (47) and possibly across significant noncoding domains of active gene loci (46).
In the human major histocompatibility complex class II locus, intergenic transcripts are detected both up- and downstream of an enhancer and overlap with a domain of H3 acetylation; both are lost along with HLA-DRA expression in cells null for either of two enhancer binding activator proteins, indicating that they are enhancer dependent and suggesting that one or the other or both is required for HLA-DRA expression (34). Intergenic transcription and histone acetylation also correlate in the TH2 cytokine locus in tissues that express or will express the interleukin 4 (IL-4)/IL-13 cytokines (39). However, intergenic transcription is not necessary for histone acetylation or transcriptional permissiveness of the IL-4/IL-13 genes (3). In the human growth hormone (hGH) locus, intergenic sense and antisense transcription can also be disrupted without affecting histone acetylation encompassing the locus control region (LCR) and the hGH gene (18). Both intergenic transcription and histone acetylation are LCR dependent in this locus (17, 18). However, unlike with the TH2 locus, intergenic transcription is required for hGH expression. Thus, the importance of intergenic transcription to target gene activity can be variable. The possibility of separating intergenic transcription and histone acetylation in the TH2 and growth hormone loci argues that their establishment is not intertwined as it is in coding sequences.
The human β-globin locus contains five globin genes that are sequentially activated during development: embryonic ɛ, fetal Aγ and Gγ, and adult δ and β (42). All of the genes depend on a far-upstream LCR comprised of DNase I hypersensitive sites (HS1 to HS4) for high-level expression in erythroid cells. Extensive intergenic transcription occurs across this locus (2, 13), including antisense transcription (16; also A. Kim, C. M. Kiefer, and A. Dean, unpublished data). The transcribed intergenic regions of the locus include the LCR and the region surrounding each actively transcribed gene, which switches during development (10, 13). Histone acetylation and H3 K4 dimethylation profiles correlate well with the intergenic transcribed regions (10, 26). There is an LCR dependence of histone acetylation flanking the LCR and at the active human β-globin promoter (41), but there has been no description of locus-wide histone modification or intergenic transcription in a situation wherein the LCR has been deleted. Inhibition of intergenic transcription by DRB does not affect intergenic histone acetylation, suggesting that they are established independently in this locus and raising the question of how the intergenic histone modification is targeted (16, 23).
A domain of modified histones and intergenic transcripts encompasses the β-globin LCR and the most proximal gene, ɛ-globin, in human K562 cells that mimic the embryonic erythroid stage when this gene is active (16, 25). Salient features of this domain are faithfully recapitulated on manipulable, chromatinized episomes, where HS2 activates the ɛ-globin gene (24). Using this system, we asked whether intergenic transcription and H3 acetylation and K4 methylation depended on the HS2 enhancer or the target ɛ-globin gene and to what extent the intergenic transcription and histone modification contributed to gene activation. Inactivation of HS2 by mutation of the NF-E2 sites, which results in the loss of gene transcription and loss of the domain of modified histones, also resulted in the loss of intergenic transcripts, indicating that the enhancer mediates all of these processes. Inhibition of Pol II initiation by mutation of the TATA box in the ɛ-globin promoter reduced transcription and histone modifications within the coding region. However, intergenic transcription and histone acetylation were unaffected, indicating that they are not a consequence of gene transcription and do not result per se in transcription of the target gene. Instead, target gene transcription and histone modification depend on a functional TATA box.
MATERIALS AND METHODS
Minichromosome construction, cell culture conditions, and transfection.
Minichromosomes containing HS2 and the ɛ-globin gene have been described previously (24). The tandem NF-E2 binding motifs in HS2 and the ɛ-globin TATA box in the promoter were mutated by site-directed mutagenesis in minichromosomes (11, 35). Minichromosomes were transfected into K562 cells, and multiple individual clones were selected and maintained as described previously (11).
RNA/DNA FISH.
Cells were spotted onto poly-l-lysine-coated slides and fixed in 4% formaldehyde containing 5% acetic acid, and RNA fluorescence in situ hybridization (FISH) was carried out as described previously (12). The ɛ-globin probe (pBSɛ458) was labeled by nick translation with digoxigenin-11-dUTP (Dig-Nick Translation Mix; Roche Diagnostics). Antibody detection of the digoxigenin label was carried out with sheep anti-Dig antibody (Roche Diagnostics) followed by rabbit anti-sheep fluorescein isothiocyanate (FITC) and goat anti-rabbit FITC antibodies (both from Calbiochem). After digoxigenin detection, the slides were washed, dehydrated in ethanol, air dried, coverslipped with Vectashield mounting medium containing 0.1 μg/ml DAPI (4′,6′-diamidino-2-phenylindole) (Vector Laboratories, Burlingame, CA), and imaged. The same slides were used for DNA FISH, as previously described (44). The coverslips were removed and hybridization was carried out with a CEP 11 alpha satellite probe (chromosome enumeration DNA FISH probe for chromosome 11 specific tandem-repeat DNA sequences; Vysis, Inc., Des Plaines, IL) labeled with spectrum orange. The slides were then washed and mounted in Vectashield, as described above. Digital images of fluorescent signals were acquired with a Zeiss Axioskop fluorescence microscope equipped with a Hamamatsu charge-coupled-device camera. Images were analyzed with IPlab, version 3.9, software.
RNase protection assay.
RNA was prepared from 5 × 106 K562 cells carrying minichromosomes by use of the PUREscript kit (Gentra). The episomal copy of the ɛ-globin gene is marked by a distinguishing mutation in the 5′ untranslated region (11). RNase digestion and gel analyses were performed according to the protocol of the manufacturer of the reagents (RPA II kit; Ambion). RNase protection assay results were normalized to the actin control signal and corrected for the minichromosome copy number. Clones used in these studies carried between 8 and 30 copies of the minichromosome. The endogenous ɛ-globin mRNA signal varied among K562 cell clones but was unrelated to minichromosome transcription.
Reverse transcription reaction.
RNA was prepared as described above and treated with RNase-free DNase I. One microgram of RNA was then reverse transcribed with random hexamers using the Superscript II First Strand synthesis kit, as suggested by the manufacturer (Invitrogen). cDNA was diluted to 400 μl, and 5 μl of cDNA was amplified in a 25-μl reaction volume by real-time PCR. The amount of cDNA was compared with a genomic DNA standard and then corrected by the amount of actin cDNA compared with genomic DNA.
ChIP.
Histone modification was analyzed by chromatin immunoprecipitation (ChIP), as described previously (24, 33). Briefly, nuclei were prepared from 5 × 107 K562 cells and digested with different micrococcal nuclease (MNase) concentrations (0.0025 unit/μl, 0.01 unit/μl, and 0.04 unit/μl) for 10 min at 37°C. Digested chromatin was combined and then fractionated on a sucrose gradient (5 to 30%). Mono- and dinucleosomes were pooled and then reacted with antibodies after preclearing with protein A agarose. Alternatively, cells were cross-linked with 1% formaldehyde and chromatin was fragmented by both MNase digestion and sonication, which produced primarily mononucleosome-sized fragments (36). Similar results were obtained with both protocols. ChIP for analysis of Pol II was performed with cells that were cross-linked with 0.4% formaldehyde. Chromatin was fragmented to 200 to 500 bp by sonication and reacted with antibodies as described previously (20, 25). Analysis of CBP and p300 by ChIP was carried out after 1% cross-linking, and chromatin was fragmented by both MNase digestion and sonication, as described above.
Anti-diacetylated (K9, K14) histone H3 and anti-dimethylated H3 (K4) were purchased from Upstate Biotechnology, Lake Placid, NY. Anti-Pol II (N20), anti-CBP, anti-p300, and normal rabbit immunoglobulin G were purchased from Santa Cruz Biotechnology, Santa Cruz, CA.
Quantitative real-time PCR analysis, primers, and TaqMan probes.
Immunoprecipitated DNA was analyzed by quantitative real-time PCR (ABI Prism 7900) using TaqMan probes and primers (Primer Express 1.0; PE Applied Biosystems). For non-cross-linked ChIP samples, DNA was quantified by using Pico green, and 1 ng of DNA was amplified in a 25-μl reaction volume (24). Data were collected at the threshold at which amplification was linear. The difference for each primer pair was determined by comparing the amount of target sequence in immunoprecipitated DNA to the amount of target sequence in input DNA by the comparative threshold cycle method (33). For cross-linked ChIP samples, the relative intensity was determined by comparing the amount of target sequence in 2.5% of the immunoprecipitated DNA to the amount of target sequence in 0.2% of the input DNA. Sequences of primers and TaqMan probes have been described previously (24).
RESULTS
Intergenic transcription requires a functional enhancer.
Minichromosomes, maintained at stable, moderate copy numbers in K562 cells, contained the strong HS2 enhancer of the β-globin LCR and the ɛ-globin gene (Fig. 1) in the form of a 5.2-kb minilocus in which the enhancer and gene reside about 2.5 kb apart (24). HS2 contains a previously mapped intergenic initiation site (40) and activates the linked ɛ-globin gene to a high level. The minichromosomal mRNA transcripts can be distinguished from endogenous ɛ-globin transcripts in K562 cells because the 5′ untranslated region has been marked with a 2-bp mismatch (11). However, in experiments to measure intergenic transcription using reverse transcriptase (RT) PCR and in ChIP experiments, both the endogenous and minichromosomal templates are recognized. Since multiple copies of the minichromosomes are present, the signal in these chromatin-based experiments should be primarily attributable to the minichromosomes; however, this is the case only if all templates behave similarly, i.e., if all are being activated by HS2 and transcribe ɛ-globin. To ascertain whether this is the case, we employed DNA/RNA FISH.
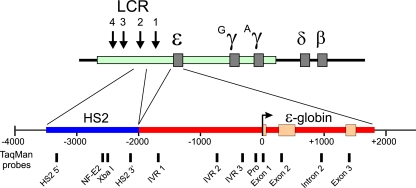
FIG. 1.
Structure of the human β-globin locus and minichromosomal locus containing the ɛ-globin gene and LCR HS2. The human β-globin locus is diagrammed across the top of the figure. A domain of histone modification mapped over the LCR and ɛ- and γ-globin genes in K562 cells is indicated by the colored rectangle. LCR HS2 (1.46 kb) was fused to the ɛ-globin gene (3.7 kb) in the minichromosome locus (see Materials and Methods). Two mutant loci were created in which either the HS2 NF-E2 site or the ɛ-globin promoter TATA box was destroyed by clustered point mutations. The positions of TaqMan probes used for real-time PCR are indicted at the bottom.
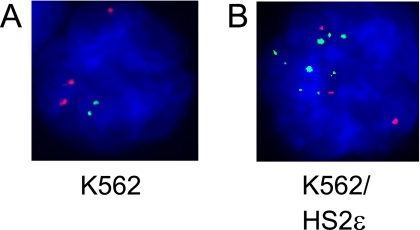
K562 cells were fixed and permeabilized on a slide, followed by sequential hybridization to RNA and DNA FISH probes. In K562 cells, three copies of chromosome 11, which is the location of the β-globin locus, were detected in most nuclei by hybridizing a DNA probe corresponding to the centromeric α-satellite repeat region (Fig. 2A). RNA FISH performed on the same slides revealed that only two chromosomes actively transcribe ɛ-globin. When a K562 clone carrying HS2-ɛ-globin minichromosomes was examined, many more foci of transcription were observed (Fig. 2B). The number of additional foci observed, in this case eight, corresponded well with the minichromosome copy number determined by Southern blotting (not shown). We conclude that essentially all copies of the minichromosomes actively transcribe ɛ-globin and that intergenic transcript measurement and chromatin-based experiments will predominantly reflect the state of these templates.
FIG. 2.
RNA FISH indicates active transcription on the multiple copies of minichromosomes in K562 cells. RNA FISH was carried out as described in Materials and Methods with K562 cells and a K562 clone carrying HS2ɛ minichromosomes. The ɛ-globin transcripts were labeled with FITC (green). The slides were then fixed, and DNA FISH was carried out with a chromosome 11-specific centromeric probe labeled with Spectrum Orange. One hundred random nuclei from each cell line on each of three slides were counted to determine the number of transcribing endogenous and minichromosomal templates. (A) DAPI-stained K562 nucleus containing three copies of chromosome 11 (red). Two ɛ-globin transcriptional foci are observed at some distance from each centromere, as expected (green). (B) A nucleus from a cell clone with HS2ɛ minichromosomes has many additional foci (8.5 ± 0.9) of ɛ-globin transcription, in good agreement with the Southern blot assessment of the number of minichromosomal templates (not shown).
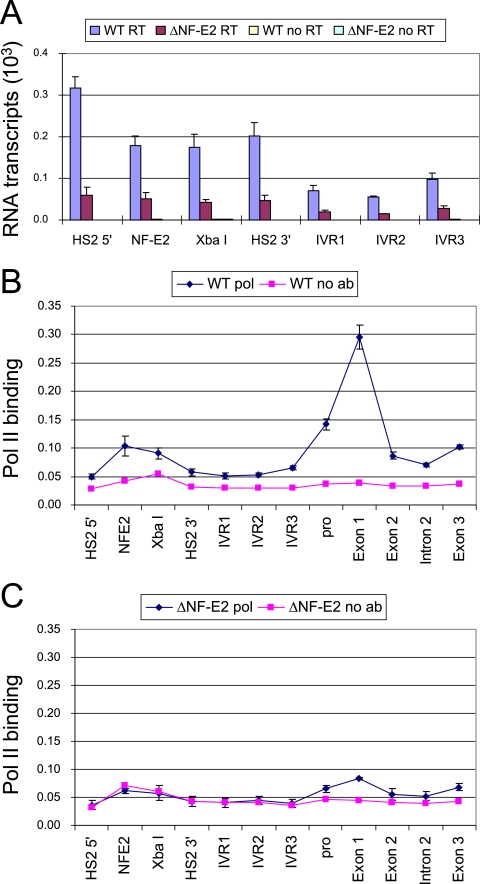
Tandem NF-E2 motifs in HS2 are required for ɛ-globin transcription and for H3 acetylation and K4 dimethylation across the minilocus on minichromosomes (11, 24). We asked whether intergenic transcription similarly requires NF-E2 interaction at HS2. Enhancer dependence of globin locus intergenic transcription has not been demonstrated. Random primed cDNA was generated from RNA of K562 cells carrying minichromosomes. Quantitative real-time PCR was used to survey intergenic sites across the minilocus with TaqMan probes (Fig. 1). Relative levels of transcripts were determined for multiple RNA preparations (see Materials and Methods). Transcripts were detected across the locus from HS2 to the linked ɛ-globin gene (Fig. 3A) at levels several-hundred-fold to 1,000-fold lower than spliced ɛ-globin mRNA (not shown), comparable to other data (16, 26). When the NF-E2 motifs in HS2 were eliminated, transcripts were greatly reduced at all positions. This decrease can be attributed to changes in minichromosome template transcription, since endogenous intergenic transcription should be unaffected. These results indicate that intergenic transcription is highly HS2 enhancer dependent, similar to H3 acetylation and K4 dimethylation.
FIG. 3.
Intergenic transcription and Pol II association are dependent on NF-E2 binding in HS2. (A) cDNA was prepared by reverse transcription using 1 μg of RNA isolated from K562 cells containing minichromosomes and then amplified by real-time PCR using the indicated probes. The cDNA signal was compared with that of genomic DNA purified from K562 cells containing minichromosomes and was corrected by the amount of actin cDNA compared with genomic DNA. No signal was produced without RT. The results of three independent experiments ± standard errors of the means are graphed. (B) Chromatin was prepared from 0.4% formaldehyde-cross-linked K562 cells carrying wild-type minichromosomes. Chromatin was sonicated to 100- to 500-bp fragments, and immunoprecipitation was performed with antibodies to Pol II. ChIP DNA was amplified by real-time PCR using the indicated probes. Control samples were incubated without antibody (ab). The relative intensity was determined as described in Materials and Methods. The results of three independent experiments ± standard errors of the means are graphed. (C) Chromatin was prepared from K562 cells carrying NF-E2 mutant minichromosomes, and ChIP was carried out as described for panel B.
Considering that Pol II is recruited to LCR HS core regions (21, 22, 25, 26) and that at least some nongenic transcripts originate within HS2, we were interested in whether the loss of NF-E2 sites in HS2 affected the localization of Pol II in the model locus. ChIP was carried out with antibodies to the large subunit of RNA Pol II. Quantitative real-time PCR was performed with the TaqMan probes shown in Fig. 1. Pol II detection was maximal at the start site of the highly transcribed ɛ-globin gene but was considerably weaker in more 3′ coding sequences (Fig. 3B). Within HS2, Pol II was maximal at the core NF-E2 sites, where it is recruited by transcription activators, but levels above background were detected 5′ of the core and between HS2 and ɛ-globin. When the NF-E2 motifs in HS2 were eliminated, Pol II was almost completely lost from the gene, consistent with the loss of HS2 enhancer activity and loss of gene transcription (Fig. 3C). Furthermore, Pol II was not detected above the background signal at the nongenic sequences or at HS2, consistent with transcript reduction locus-wide.
Transcription initiation-dependent histone alterations at gene coding sequences.
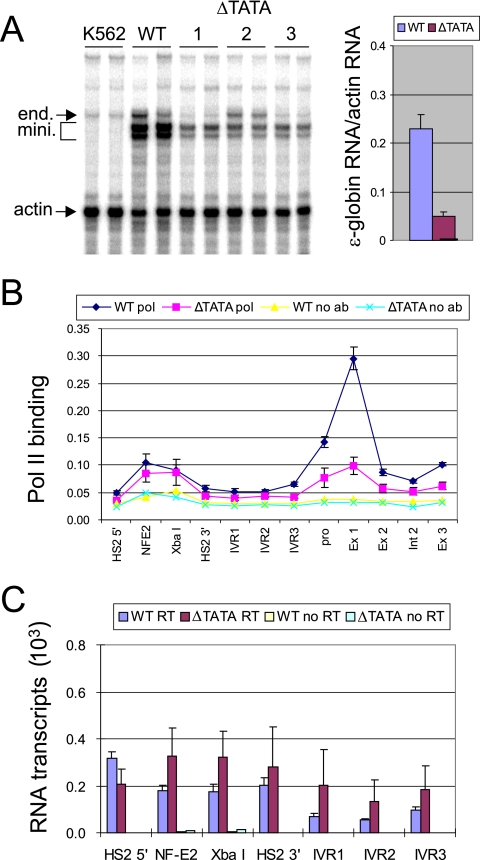
The above results do not allow us to distinguish whether or not the nongenic transcripts depend on Pol II entry at HS2, because when the enhancer is inactivated Pol II is also lost from the gene promoter. Therefore, we compromised Pol II initiation at the ɛ-globin promoter by mutating the TATA box. The TATA mutation reduced the transcription of the ɛ-globin gene to less than 20% of the wild-type level (Fig. 4A), and detection of Pol II was greatly reduced at the ɛ-globin start site (Fig. 4B). Notably, mutation of the TATA box had little effect on Pol II binding at HS2 or between the enhancer and the gene, nor did it interrupt intergenic transcription, which tended to be slightly increased in the presence of the TATA mutation (Fig. 4C).
FIG. 4.
TATA box mutation does not alter intergenic transcription or Pol II recruitment at HS2. (A) RNase protection analysis was performed with RNA isolated from K562 cells containing minichromosomes with a mutated ɛ-globin TATA box. Two RNA preparations for each of three ΔTATA clones are shown. The bands protected by endogenous (end) and minichromosomal ɛ-globin (mini) RNA are indicated by arrows. Controls include K562 cell RNA and RNA from K562 cells carrying wild-type minichromosomes. Actin served as the loading control, and the results were corrected for minichromosome copy numbers. The results for multiple clones ± standard errors of the means are depicted graphically. (B) ChIP was carried out as described in the legend to Fig. 3B. The relative intensity of detection at each position was determined as described in Materials and Methods. The results of three independent experiments ± standard errors of the means are presented. (C) cDNA was prepared by reverse transcription using 1 μg of RNA isolated from K562 cells containing minichromosomes and then amplified by real-time PCR using the indicated probes. No signal was produced without RT. The results of three independent experiments ± standard errors of the means are graphed.
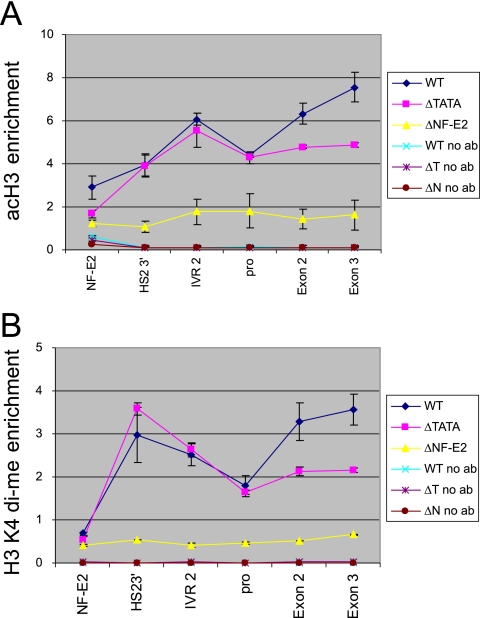
Inhibition of initiation reduced histone H3 acetylation and H3 K4 dimethylation across the gene coding sequences, indicating that the high level of histone modification there relates to active transcription. (Fig. 5). However, mutation of the TATA box did not affect the pattern of histone acetylation and K4 dimethylation in the intergenic region or K4 dimethylation in HS2, although H3 acetylation at HS2 was reduced somewhat. The effect of HS2 NF-E2 mutation, which essentially eliminates H3 acetylation and K4 dimethylation, is shown for comparison (24). Thus, histone modification and transcription in the intergenic region is separable from gene transcription. We conclude that the promoter TATA box is required for a high level of histone modifications in the coding region of the ɛ-globin gene, as it is for high-level transcription, consistent with transcription-coupled histone acetyltransferase (HAT) activity in coding sequences. Histone modification and transcription in the enhancer and intergenic region are independent of promoter function.
FIG. 5.
The TATA box mutation affects coding sequence histone modifications. Mono- and dinucleosomes were prepared from non-cross-linked chromatin by MNase digestion (see Materials and Methods) and reacted with antibodies to diacetylated H3 K9 and K14. Input and immunoprecipitated DNAs were analyzed by real-time PCR using the indicated probes. The enrichment was determined by comparing the amount of target sequence in immunoprecipitated DNA to the amount of target sequence in input DNA. Three independent experiments were carried out, and the average results ± standard errors of the means are shown. (B) ChIP was carried out using 1% formaldehyde cross-linking (see Materials and Methods) before immunoprecipitation with antibodies to dimethylated H3 K4. The relative intensity of detection at each position was determined as described in Materials and Methods. The results of three independent experiments ± standard errors of the means are presented.
Localized recruitment of HATs to the HS2 enhancer.
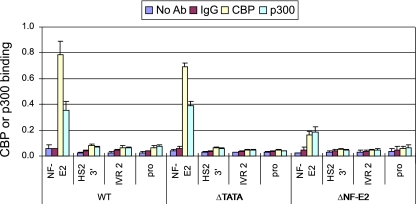
From the above results, the TATA box/active promoter is not required to establish histone acetylation in the nongenic regions. To examine the distribution of HATs through the locus under various conditions, chromatin was immunoprecipitated with antibodies against CBP and p300 and analyzed by PCR. CBP and p300 are known to play a role in the expression of globin genes (5), and CBP interacts at LCR HS3 (30). We had observed recruitment of these HATs to a small core HS2 fragment (48).
The results of ChIP experiments shown in Fig. 6 indicate that both CBP and p300 are strongly detected in the core region of the 1.5-kb HS2 enhancer, as expected, but not in the extended HS2 flanking sequences or at the gene promoter. Mutation of the NF-E2 sites reduced recruitment of the HATs to HS2, consistent with the loss of histone modifications throughout the model globin locus (24). In contrast, inhibition of Pol II initiation by mutation of the ɛ-globin promoter TATA box did not affect HAT recruitment at HS2. Thus, the change in acetylation status within the ɛ-globin gene lacking its TATA box does not relate to impairment of CBP/p300 recruitment to HS2. We conclude that gene transcription and histone acetylation of coding sequences require entry of Pol II and an associated HAT(s) at the TATA box.
FIG. 6.
NF-E2 mutation inhibits recruitment of CBP and p300 to HS2. Chromatin was prepared from 1% formaldehyde-cross-linked K562 cells carrying minichromosomes and fragmented by MNase digestion and sonication (see Materials and Methods). Chromatin was subjected to immunoprecipitation using antibodies to CBP or p300 followed by amplification using real-time PCR with the indicated probes. The difference for each primer pair was determined by comparing antibody-precipitated DNA with input DNA. The results for three chromatin preparations ± standard errors of the means are presented.
DISCUSSION
The results presented in this study show that positive histone modifications and intergenic transcription between the HS2 enhancer and ɛ-globin gene in a model chromatinized locus depend on an intact enhancer. Furthermore, when Pol II initiation at the gene is prevented by mutation of the TATA box, the intergenic histone modification and intergenic transcription persist, indicating that they occur independently of gene transcription. Transcription in the intergenic region is insufficient by itself to activate gene transcription, suggesting that while some polymerases may reach the vicinity of the target gene through transcribing across intergenic sequences (32), high-level gene transcription depends on initiation at the gene promoter. This result suggests that the Pol II complexes involved in intergenic versus coding region transcription may be different. Interestingly, the RNA products produced in the two cases have been shown to be quite different (32).
Role of the enhancer in histone modification and intergenic transcription.
In our model locus, inactivation of the HS2 enhancer by mutation of the NF-E2 sites resulted in loss of gene transcripts and intergenic transcripts and loss of acetylated histones locus-wide, establishing a critical role for NF-E2. NF-E2 may have a direct role in recruiting CBP/p300 (7) or may function primarily to disrupt the chromatin structure of HS2 (1, 11), after which other enhancer binding activators such as GATA-1 might mediate recruitment (30, 49). Our data also implicate NF-E2 in recruitment of Pol II to HS2. This function could also be mediated by disruption of HS2 chromatin structure to allow activator binding, since it is known that cells null for GATA-1 fail to recruit Pol II to the LCR (22). Recruitment of Pol II to HS2 in CB3 MEL cells which are null for NF-E2 (21) argues against this possibility. However, it is plausible that an NF-E2-related factor occupies HS2 in CB3 cells, disrupts chromatin structure, and permits GATA-1 binding, since it is known that NF-E2 is not required to maintain hypersensitivity at HS2 in CB3 cells (27).
Although the above results are consistent with HS2 being the site from which the domain of modified histones begins to form, they do not prove that this is the case. The observations that LCRs recruit HATs (9, 30) and are often the peaks of histone modifications in endogenous loci (17, 34) are consistent with this idea, as is the diminution of widespread histone acetylation after interposition of a chromatin insulator between HS2 and the ɛ-globin gene (48). Unidirectional spread of histone acetylation between an enhancer and a gene as a function of time after induction has been reported at the HNF-4 locus (15). Our data also suggest that Pol II may transcribe from HS2, where it is recruited across intergenic sequences to the linked gene, consistent with other data (32). However, we propose that high-level transcription activation of ɛ-globin is not likely to be accomplished in this manner because it requires initiation at the gene TATA box. Although Pol II tracking is required for activation of the PSA and hGH genes (18, 45), intergenic transcription in the growth hormone locus terminates upstream of the gene itself, indicating that it has a function separate from Pol II delivery to the gene (18).
Role of the promoter in intergenic transcription and domain-wide histone modifications.
Inhibition of Pol II initiation by mutation of the TATA box of the ɛ-globin gene reduced histone acetylation and K4 dimethylation within the coding region without affecting intergenic transcription or histone modification, indicating that they are independent of promoter activity. It appears that CBP and p300, even though present at HS2, cannot mediate a high level of histone acetylation in coding sequences in the absence of a TATA box at the promoter. Possibly, a different HAT acetylates coding sequences during active gene transcription. Both CBP and PCAF are involved in transcript elongation via association with the elongating Pol II complex, and interestingly, their association is gene specific (28). Unfortunately, we were unable to detect reliable signals with antibodies to PCAF in ChIP assays.
Another possible reason for the inability of HATs recruited to HS2 to modify the linked gene could be that direct promoter-enhancer interaction is compromised by the TATA box mutation. In the endogenous β-globin locus, loops between the LCR HSs and active gene promoters have been demonstrated (6, 43). Loop formation between HS2 and ɛ-globin on minichromosomes has not been documented; however, we consider it to be highly likely. The formation of loops in minichromosomes would not be impeded, since 2.5 kb of native sequence separate the enhancer and promoter (38). The TATA box may be important for such interactions in the model locus (14). Looping interactions in chromosomes between the LCR HSs and the β-globin gene are not compromised by deletion of the promoter of the gene, arguing against this possibility (37). However, on chromosomes, more-extensive sequences than are present in the minilocus might contribute to stabilizing looping interactions.
Function of domains of modified histones and intergenic transcription.
A chromatin remodeling and transcription model in which intergenic transcription demarcates differentially regulated chromatin subdomains of the β-globin locus has been proposed (13). However, establishing whether the intergenic transcripts are a byproduct of gene transcription or occur independently has been problematic. On minichromosomes, the intergenic region between the HS2 enhancer and target globin gene is transcribed and the histones are modified in the absence of gene transcription. Consistent with these observations, transcription can be initiated in HS2 without a cis-linked globin promoter on an integrated plasmid (32) and intergenic transcription occurs in the TH2 cytokine locus in vivo before the onset of cytokine gene transcription (39). Thus, intergenic transcription and histone domains are not simply by-products of gene transcription, and they may have an independent function. Other data obtained with either a chromatin insulator or a transcription terminator also provide support for the functional importance of domains of histone acetylation and intergenic transcription to enhancer function at a distant target gene (18, 31, 48). It will be important to conduct further tests of these concepts with endogenous loci.
Acknowledgments
We thank Peter Fraser and Cameron Osborne for advice on carrying out RNA FISH. We are especially grateful to Amalia Dutra and Evgenia Pak for generous help with DNA FISH and kind permission to use their facility for the imaging and analysis of the FISH experiments.
This research was supported by the Intramural Research Program of the NIH, NIDDK (A.D.).
Footnotes
Published ahead of print on 5 February 2007.
REFERENCES
- 1.Armstrong, J. A., and B. M. Emerson. 1996. NF-E2 disrupts chromatin structure at human β-globin locus control region hypersensitive site 2 in vitro. Mol. Cell. Biol. 16:5634-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashe, H. L., J. Monks, M. Wijgerde, P. Fraser, and N. J. Proudfoot. 1997. Intergenic transcription and transinduction of the human β-globin locus. Genes Dev. 11:2494-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baguet, A., X. Sun, T. Arroll, A. Krumm, and M. Bix. 2005. Intergenic transcription is not required in Th2 cells to maintain histone acetylation and transcriptional permissiveness at the Il4-Il13 locus. J. Immunol. 175:8146-8153. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 5.Blobel, G. A. 2000. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood 95:745-755. [PubMed] [Google Scholar]
- 6.Carter, D., L. Chakalova, C. S. Osborne, Y. Dai, and P. Fraser. 2002. Long-range chromatin regulatory interactions in vivo. Nat. Genet. 32:623-626. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, X., M. J. Reginato, N. C. Andrews, and M. A. Lazar. 1997. The transcriptional integrator CREB-binding protein mediates positive cross talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol. Cell. Biol. 17:1407-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean, A. 2006. On a chromosome far, far away: LCRs and gene regulation. Trends Genet. 22:38-45. [DOI] [PubMed] [Google Scholar]
- 9.Elefant, F., N. E. Cooke, and S. A. Liebhaber. 2000. Targeted recruitment of histone acetyltransferase activity to a locus control region. J. Biol. Chem. 275:13827-13834. [DOI] [PubMed] [Google Scholar]
- 10.Forsberg, E. C., K. M. Downs, H. M. Christensen, H. Im, P. A. Nuzzi, and E. H. Bresnick. 2000. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 97:14494-14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong, Q. H., J. C. McDowell, and A. Dean. 1996. Essential role of NF-E2 in remodeling of chromatin structure and transcriptional activation of the ɛ-globin gene in vivo by 5′ hypersensitive site 2 of the β-globin locus control region. Mol. Cell. Biol. 16:6055-6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gribnau, J., E. de Boer, T. Trimborn, M. Wijgerde, E. Milot, F. Grosveld, and P. Fraser. 1998. Chromatin interaction mechanism of transcriptional control in vivo. EMBO J. 17:6020-6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gribnau, J., K. Diderich, S. Pruzina, R. Calzolari, and P. Fraser. 2000. Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell 5:377-386. [DOI] [PubMed] [Google Scholar]
- 14.Gui, C. Y., and A. Dean. 2003. A major role for the TATA box in recruitment of chromatin modifying complexes to a globin gene promoter. Proc. Natl. Acad. Sci. USA 100:7009-7014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatzis, P., and I. Talianidis. 2002. Dynamics of enhancer-promoter communication during differentiation-induced gene activation. Mol. Cell 10:1467-1477. [DOI] [PubMed] [Google Scholar]
- 16.Haussecker, D., and N. J. Proudfoot. 2005. Dicer-dependent turnover of intergenic transcripts from the human beta-globin gene cluster. Mol. Cell. Biol. 25:9724-9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho, Y., F. Elefant, N. Cooke, and S. Liebhaber. 2002. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell 9:291-302. [DOI] [PubMed] [Google Scholar]
- 18.Ho, Y., F. Elefant, S. A. Liebhaber, and N. E. Cooke. 2006. Locus control region transcription plays an active role in long-range gene activation. Mol. Cell 23:365-375. [DOI] [PubMed] [Google Scholar]
- 19.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, K. D., and E. H. Bresnick. 2002. Dissecting long-range transcriptional mechanisms by chromatin immunoprecipitation. Methods 26:27-36. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, K. D., H. M. Christensen, B. Zhao, and E. H. Bresnick. 2001. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell 8:465-471. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, K. D., J. A. Grass, M. E. Boyer, C. M. Kiekhaefer, G. A. Blobel, M. J. Weiss, and E. H. Bresnick. 2002. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. USA 99:11760-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, K. D., J. A. Grass, C. Park, H. Im, K. Choi, and E. H. Bresnick. 2003. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol. Cell. Biol. 23:6484-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, A., and A. Dean. 2003. A human globin enhancer causes both discrete and widespread alterations in chromatin structure. Mol. Cell. Biol. 23:8099-8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, A., and A. Dean. 2004. Developmental stage differences in chromatin sub-domains of the β-globin locus. Proc. Natl. Acad. Sci. USA 101:7028-7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, A., C. M. Kiefer, and A. Dean. 2007. Distinctive signatures of histone methylation in transcribed coding and noncoding human β-globin sequences Mol. Cell. Biol. 27:1271-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotkow, K. J., and S. H. Orkin. 1995. Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol. Cell. Biol. 15:4640-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouskouti, A., and I. Talianidis. 2005. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 24:347-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lachner, M., R. J. O'Sullivan, and T. Jenuwein. 2003. An epigenetic road map for histone lysine methylation. J. Cell Sci. 116:2117-2124. [DOI] [PubMed] [Google Scholar]
- 30.Letting, D. L., C. Rakowski, M. J. Weiss, and G. A. Blobel. 2003. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol. Cell. Biol. 23:1334-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ling, J., L. Ainol, L. Zhang, X. Yu, W. Pi, and D. Tuan. 2004. HS2 enhancer function is blocked by a transcriptional terminator inserted between the enhancer and the promoter. J. Biol. Chem. 279:51704-51713. [DOI] [PubMed] [Google Scholar]
- 32.Ling, J., B. Baibakov, W. Pi, B. M. Emerson, and D. Tuan. 2005. The HS2 enhancer of the beta-globin locus control region initiates synthesis of non-coding, polyadenylated RNAs independent of a cis-linked globin promoter. J. Mol. Biol. 350:883-896. [DOI] [PubMed] [Google Scholar]
- 33.Litt, M. D., M. Simpson, M. Gaszner, C. D. Allis, and G. Felsenfeld. 2001. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science 293:2453-2455. [DOI] [PubMed] [Google Scholar]
- 34.Masternak, K., N. Peyraud, M. Krawczyk, E. Barras, and W. Reith. 2003. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat. Immunol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 35.McDowell, J. C., and A. Dean. 1999. Structural and functional cross-talk between a distant enhancer and the ɛ-globin gene promoter shows interdependence of the two elements in chromatin. Mol. Cell. Biol. 19:7600-7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morshead, K. B., D. N. Ciccone, S. D. Taverna, C. D. Allis, and M. A. Oettinger. 2003. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc. Natl. Acad. Sci. USA 100:11577-11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patrinos, G. P., M. de Krom, E. de Boer, A. Langeveld, A. M. Imam, J. Strouboulis, W. de Laat, and F. G. Grosveld. 2004. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 18:1495-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rippe, K., P. H. von Hippel, and J. Langowski. 1995. Action at a distance: DNA-looping and initiation of transcription. Trends Biochem. Sci. 20:500-506. [DOI] [PubMed] [Google Scholar]
- 39.Rogan, D. F., D. J. Cousins, S. Santangelo, P. A. Ioannou, M. Antoniou, T. H. Lee, and D. Z. Staynov. 2004. Analysis of intergenic transcription in the human IL-4/IL-13 gene cluster. Proc. Natl. Acad. Sci. USA 101:2446-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Routledge, S. J., and N. J. Proudfoot. 2002. Definition of transcriptional promoters in the human β-globin locus control region. J. Mol. Biol. 323:601-611. [DOI] [PubMed] [Google Scholar]
- 41.Schubeler, D., C. Francastel, D. M. Cimbora, A. Reik, D. I. Martin, and M. Groudine. 2000. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human β-globin locus. Genes Dev. 14:940-950. [PMC free article] [PubMed] [Google Scholar]
- 42.Stamatoyannopoulos, G. 2005. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 33:259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tolhuis, B., R. J. Palstra, E. Splinter, F. Grosveld, and W. de Laat. 2002. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10:1453-1465. [DOI] [PubMed] [Google Scholar]
- 44.van Raamsdonk, C. D., and S. M. Tilghman. 2001. Optimizing the detection of nascent transcripts by RNA fluorescence in situ hybridization. Nucleic Acids Res. 29:E42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, Q., J. S. Carroll, and M. Brown. 2005. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell 19:631-642. [DOI] [PubMed] [Google Scholar]
- 46.West, A. G., and P. Fraser. 2005. Remote control of gene transcription. Hum. Mol. Genet. 14:R101-R111. [DOI] [PubMed] [Google Scholar]
- 47.Workman, J. L. 2006. Nucleosome displacement in transcription. Genes Dev. 20:2009-2017. [DOI] [PubMed] [Google Scholar]
- 48.Zhao, H., and A. Dean. 2004. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 32:4903-4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao, H., A. Kim, S. H. Song, and A. Dean. 2006. Enhancer blocking by chicken beta-globin 5′ HS4: role of enhancer strength and insulator nucleosome depletion. J. Biol. Chem. 281:30573-30580. [DOI] [PubMed] [Google Scholar]