Abstract
Fbh1 (F-box DNA helicase 1) orthologues are conserved from Schizosaccharomyces pombe to chickens and humans. Here, we report the disruption of the FBH1 gene in DT40 cells. Although the yeast fbh1 mutant shows an increase in sensitivity to DNA damaging agents, FBH1−/− DT40 clones show no prominent sensitivity, suggesting that the loss of FBH1 might be compensated by other genes. However, FBH1−/− cells exhibit increases in both sister chromatid exchange and the formation of radial structures between homologous chromosomes without showing a defect in homologous recombination. This phenotype is reminiscent of BLM−/− cells and suggests that Fbh1 may be involved in preventing extensive strand exchange during homologous recombination. In addition, disruption of RAD54, a major homologous recombination factor in FBH1−/− cells, results in a marked increase in chromosome-type breaks (breaks on both sister chromatids at the same place) following replication fork arrest. Further, FBH1BLM cells showed additive increases in both sister chromatid exchange and the formation of radial chromosomes. These data suggest that Fbh1 acts in parallel with Bloom helicase to control recombination-mediated double-strand-break repair at replication blocks and to reduce the frequency of crossover.
DNA damage is continuously generated by cellular metabolic products as well as by environmental factors (23) and poses a serious threat to the cells. In cycling cells, it may inhibit the progression of replication forks, resulting in gaps and occasionally double-strand breaks (DSBs) in the daughter strands (4, 34). To release replication blocks and thereby prevent DSBs, cells have evolved two major repair pathways, translesion DNA synthesis and homologous recombination (HR) (15). Translesion synthesis is carried out by a number of specialized translesion synthesis DNA polymerases, which are able to synthesize directly across damaged DNA bases. As an alternative to translesion synthesis, bypass can be effected by a form of HR involving a transient template switch from damaged strand to the newly synthesized daughter strand on the sister chromatid (16). HR is also required for repair of DSBs arising at collapsed replication forks, caused by DNA-damaging agents such as the DNA topoisomerase I inhibitor camptothecin (1, 14). This chemotherapeutic agent covalently attaches to topoisomerase I, allowing it to cleave but not religate DNA. Upon encounter with a replication fork, the topoisomerase I-cleaved DNA complex induces a single-end break (4, 8, 37). Restart of replication from these single-end breaks requires HR with the intact sister chromatid.
The initial step of recombination-mediated DSB repair involves processing of the DSBs to produce a 3′ single-strand overhang, followed by polymerization of Rad51 on the single-stranded DNA. In concert with Rad54 (2, 31, 38), the resulting nucleoprotein filament facilitates the homology search and pairing with the intact duplex DNA donor to form a D-loop structure. Following DNA synthesis from the invading 3′ end, the strand is dissociated from the D loop and rehybridizes with the 3′ single-strand tail of the other side of the DSB. This type of HR-dependent DSB repair is called synthesis-dependent strand annealing, which is thought to account for a majority of mitotic HR (18, 30). In contrast to synthesis-dependent strand annealing, which does not result in the formation of crossovers, strand exchange following D-loop formation results in the formation of a Holliday junction. Holliday junctions can be resolved by incisions at the junction, which can result in crossover formation (24). It has also been proposed that double Holliday junctions are dissolved by the concerted action of the RecQ helicase Blm and topoisomerase III α (42).
The BLM helicase is mutated in Bloom's syndrome. This rare autosomal recessive genetic disease features an elevated rate of sister chromatid exchange (SCE), frequent recombination between allelic genes, increased genome instability, and a high incidence of tumorigenesis (9). The Bloom helicase is one of five human RecQ helicases, which share 3′-to-5′ DNA helicase activity as well as structural homology. Yeast species, on the other hand, have only a single orthologous gene, Sgs1 in Saccharomyces cerevisiae and Rqh1 in Schizosaccharomyces pombe (12). Although these RecQ family helicases have been implicated in the initiation of replication restart without crossover and resolution of recombination intermediates (32, 40, 42), their function remains poorly understood even in yeast due to the complexity of repair reactions following replication block and the lack of an appropriate phenotypic assay to monitor the late steps of HR in vivo. Likewise, the detailed molecular function of the mammalian RecQ helicase remains elusive.
The FBH1 (F-box DNA helicase 1) gene was recently identified in Schizosaccharomyces pombe, and its orthologous genes are conserved in chickens, mice, and humans but not in budding yeast (22). Fbh1 belongs to the superfamily 1 helicase and has unwinding activity with a 3′-to-5′ direction. Fbh1 is structurally similar to the Srs2 DNA helicase family, which includes the PcrA, Rep, and UvrD helicases (43). Fbh1 also contains an F box toward its N terminus, suggesting that Fbh1 acts as a ubiquitin ligase (E3) in the SCF complex, although the substrate of the ubiquitin ligase has not been determined (21). The S. pombe fbh1 mutant is moderately sensitive to UV; methylmethane sulfonate (MMS), an alkylating agent; and γ-rays. Genetic studies show that Fbh1 is essential for viability in the absence of Rqh1 and that this lethality is suppressed by additional inactivation of Rad51 paralogs, which promote an early step of HR. Thus, Fbh1 may affect recombination-dependent repair downstream of Rhp51 (S. pombe Rad51 ortholog), probably by promoting the processing of recombination intermediates. This notion is supported by the finding that spontaneous Rad51 focus formation was enhanced in an fbh1-deficient S. pombe mutant (27, 29). Therefore, Fbh1 may share a redundant function with Srs2 in S. pombe.
In this study, we established FBH1-deficient DT40 cells to investigate Fbh1 function in vertebrate cells. The resulting FBH1−/− cells exhibited essentially a normal phenotype. The functional overlap between yeast Fbh1 and Rqh1 led us to investigate whether other genes compensate for the loss of Fbh1 in DT40 cells. To this end, we disrupted the FBH1 gene in DT40 lines mutant for the FANCC−, RAD18−/−, RAD54−/−, and BLM−/− genes (5, 13, 17, 44). We show that although inactivation of FBH1 did not reduce the efficiency of HR, Fbh1 acts in parallel with Blm and restricts the extent of strand exchange during recombination initiated by stalled replication, thereby reducing the frequency of crossover formation.
MATERIALS AND METHODS
Cloning of the chicken FBH1 gene.
A chicken FBH1 cDNA was partially amplified from chicken testis cDNA by reverse transcriptase PCR (RT-PCR) with the primer pair 5′-AAATCATGGCTTTTGCNGGNACTGG-3′ and 5′-CACCTCGGAAGGAATAGATCTGCTG-3′, the design of which was based on conserved sequences between the human and xenopus FBH1 homologs. Rapid amplification of cDNA ends was carried out to isolate the 3′ and 5′ termini of the chicken FBH1 transcript. Full-length cDNA of chicken FBH1 (GdFBH1) was amplified from chicken testis cDNA by RT-PCR with a primer pair, 5′-AGAAAATGCACCTTACAGCTG-3′ and 5′-GCAATGGCTCTGGGAGTTGG-3′. An amplified 3-kb fragment was subcloned into a pCR2.1-TOPO vector (Invitrogen) and sequenced.
Construction of targeting vectors.
The positions of exons and introns were determined by base sequencing. Genomic DNA sequences were amplified with two sets of primers, 5′-CTGCCACAGATATGCTAGAG-3′ and 5′-TTTCCCATGTACTCATCCTC-3′ plus 5′-GACTTTGTGGATGTACCGTG-3′ and 5′-ATGGAGCCAAAACGCTTCTG-3′, for the left and right arms, respectively, of the disruption construct. Amplified PCR products (1.5 kb for the left arm and 3.5 kb for the right arm) were cloned into pCR2.1-TOPO vector (Invitrogen). The 3.5-kb KpnI (blunt ended)-NotI fragment was cloned into the EcoRV-NotI site of pCR2.1 containing the 1.5-kb left arm sequence. The BamHI site was used to insert marker gene cassettes, hisD and bsr selection marker genes flanked by loxP sequences, to generate the FBH1-hisD and FBH1-bsr gene disruption constructs, respectively (3). The puro marker gene was inserted into the BamHI site to generate the FBH1-puro gene disruption construct. The 0.5-kb fragment generated by PCR amplification of genomic DNA using the primers 5′-TTCAGGAAGAGCCCATCGTG-3′ and 5′-AATGTCCCCACTGAAACAGG-3′ was used as a probe for Southern blot analysis to screen gene-targeting events. Plasmids were linearized prior to transfection into DT40 cells; FBH1-hisD and FBH1-bsr were digested with XhoI, and FBH1-puro was digested with KpnI.
Generation of gene-disrupted cells.
Wild-type DT40 cells that stably express the CreER chimeric recombination enzyme were transfected sequentially with FBH1-bsr and FBH1-hisD targeting constructs to obtain FBH1+/− and FBH1−/− cells (Fig. 1A) (10, 25). Stable expression of CreER did not affect the viability of the cells or their sensitivity to DNA damaging agents (data not shown). Targeting events were verified by the appearance of a 6-kb band and the disappearance of a 9-kb band in Southern blot analysis of EcoRI-digested genomic DNA (Fig. 1B). Gene disruption of each transfected clone was confirmed by RT-PCR analysis of FBH1 gene transcription. The primers used for RT-PCR were the forward primer 5′-TTGAGCGTGGTCAGATAGTG-3′ and the reverse primer 5′-CACCTCGGAAGGAATAGATCTGCTG-3′ (Fig. 1D). β-Actin transcripts were analyzed as positive controls for RT-PCR analysis. Identified FBH1−/− clones were treated with 100 nM tamoxifen to exclude bsr and hisD marker cassettes from chromosomal DNA (44). The resulting FBH1−/− cells were sequentially transfected with BLM-hisD and BLM-bsr targeting constructs to obtain FBH1−/− BLM+/− and FBH1−/− BLM−/− clones, respectively (41). Likewise, RAD54-hisD and RAD54-puro targeting constructs were used for generating FBH1−/− RAD54−/− cells (5). The FANCC gene, which is encoded in a single sex chromosome, was disrupted in FBH1−/− cells by using the FANCC-puro plasmid (13). RAD18−/− cells were sequentially transfected with FBH1-bsr and FBH1-puro targeting constructs to obtain FBH1−/− RAD18+/− and FBH1−/− RAD18−/− clones, respectively (44). To express chicken Fbh1, its cDNA was inserted into the p176 expression vector containing the Neo selection marker gene (10). The expression plasmid was linearized with PvuI prior to transfection into cells. The conditions for cell culture, selection, and DNA transfections have been described previously (35).
FIG. 1.
Gene targeting of the FBH1 locus. (A) Schematic representation of a part of the FBH1 locus and the targeting constructs. The FBH1 replacement constructs, which contain a bsr or hisD selection marker gene, were used to disrupt the FBH1 locus. The filled box represents the FBH1 exons. RI, EcoRI. (B) Southern blot analysis of EcoRI-digested DNA using the probe indicated in panel A. (C) RT-PCR analysis of total RNA using the primers shown in panel A. The coding region of the chicken β-actin gene was amplified as a control. (D) Normal growth kinetics of FBH1−/− cells. (E) Cell cycle distribution of the indicated cell lines. Cells were stained with FITC-anti-BrdU antibody (y axis, log scale) to detect BrdU uptake and with propidium iodide to measure the total DNA (x axis, linear scale). The upper gate identifies cells incorporating BrdU (∼S phase), the lower left gate identifies G1 cells, and the lower right gate displays G2/M cells. The sub-G1 fraction reflects dead cells. The numbers given in the boxes indicate the percentages of gated events.
Proliferation analysis and colony formation assay.
A colony formation assay was performed as described previously (28). To measure the sensitivity to camptothecin, cells were plated in methylcellulose medium containing camptothecin. The experimental methods for cell counting and cell cycle analysis were described previously (39).
Measurement of SCE levels.
Measurement of SCE levels was carried out as described previously (28, 44). To measure camptothecin-induced SCE, cells were labeled with bromodeoxyuridine (BrdU) for two cell cycle periods (∼20 h) and treated with 5 nM camptothecin for the last 9 h. To measure SCE induced by UV, cells were treated with 0.25 J/m2 of UV and incubated with 10 uM BrdU for 20 h. The cells were incubated with Colcemid to enrich metaphase cells for the last 3 h before harvest.
Chromosomal aberration analysis.
Preparation of chromosome spreads and karyotype analysis were performed as described previously (35, 44). To measure camptothecin-induced chromosomal aberrations, cells were incubated with 10 nM camptothecin for 9 h before harvest and fixation. To analyze UV-induced chromosomal aberrations, cells were suspended in 1 ml of phosphate-buffered saline, spread onto six-well plates, and exposed to 1 J/m2 UVC, followed by the addition of 5 ml of the complete medium. Cells were harvested every 3 h for 15 h. To enrich mitotic cells, cells were exposed to Colcemid for the last 3 h.
Measurement of targeted integration frequencies.
HR was evaluated by the following three methods. First, the frequency of I-SceI restriction enzyme-induced HR was measured as described previously (11, 19). The assay plasmid was inserted into the OVALBUMIN locus. Second, to analyze targeted integration events at the OVALBUMIN, RAD54, and CENP-H loci, each disruption construct was transfected into cells. Gene targeting events were identified by either Southern blot analysis or FACS analysis of EGFP-CENP-H fusion protein expression (20). Lastly, HR-mediated immunoglobulin V (IgV) diversification (Ig gene conversion) was monitored, as described previously (6). To this end, FBH1−/− clones were generated from wild-type DT40 cells that carry a specific frameshift mutation. The rate of Ig gene conversion was assessed by measuring the gain of surface IgM expression during the 3-week period with clonal expansion.
Nucleotide sequence accession number.
The chicken FBH1 cDNA sequences have been submitted to the GenBank database under accession number EF066526.
RESULTS
No apparent defect in HR in the absence of Fbh1.
The chicken FBH1 encodes a polypeptide comprised of 1,012 amino acids that has 55%, 50%, and 26% identity at the amino acid level to the human, mouse, and S. pombe orthologs, respectively. Gene-targeting constructs were generated to delete amino acids 118 to 875, including both the F-box domain and five helicase motifs (Ia, Ib, II, III, IV) of the FBH1 gene (Fig. 1A; also see Fig. S1 in the supplemental material). Two independent FBH1−/− clones were identified by Southern blot analysis (Fig. 1B). Gene disruption of these clones was verified by RT-PCR (Fig. 1C). FBH1−/− cells were able to proliferate with normal kinetics (Fig. 1D). Flow cytometric analysis of BrdU pulse-labeled cells showed a normal cell cycle distribution (Fig. 1E).
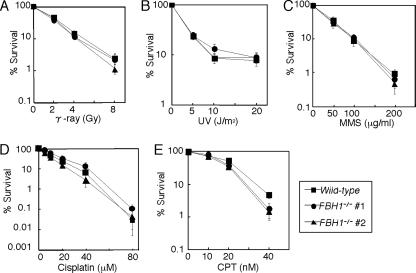
We evaluated the capability of HR in FBH1−/− cells by using the following phenotypic assays: sensitivity to DNA damaging agents (Fig. 2), gene-targeting frequency (Table 1), HR-mediated repair of DSBs induced by the I-SceI restriction enzyme, HR-dependent diversification of the Ig gene (Ig gene conversion), and the kinetics of Rad51 subnuclear focus formation. FBH1−/− cells were tolerant to γ-rays, MMS, UV, and cisplatin, as were wild-type cells (Fig. 2A to D), while exhibiting modest sensitivity to camptothecin (Fig. 2E). The efficiency of both gene targeting (Table 1) and HR-dependent DSB repair following I-SceI expression (Fig. 3A) was indistinguishable between wild-type and FBH1−/− cells. Likewise, FBH1−/− cells appear to undergo Ig gene conversion with normal kinetics (data not shown). The kinetics of subnuclear Rad51 focus formation following γ-irradiation were the same between wild-type and FBH1−/− cells during the cell cycle and at 3 h, 6 h (Fig. 3B), and 24 h (data not shown) after γ-irradiation. Collectively, FBH1−/− cells can perform HR reactions with normal kinetics, which agrees with the previous finding of no effect on spontaneous and DNA damage-induced recombination in Fbh1-deficient yeast (29).
FIG. 2.
Sensitivities of the FBH1 mutant to genotoxic stresses. The indicated genotypes of cells were exposed to γ-rays (A) and UV (B), after a 1-h treatment with MMS (C) and cisplatin (D) and after continuous exposure to camptothecin (CPT) (E). The doses of genotoxic agents are displayed on the x axes on a linear scale, while the fractions of surviving colonies are displayed on the y axes on a logarithmic scale. Error bars show the standard error of the mean for at least three independent experiments.
TABLE 1.
Targeted integration frequenciesa
| Genotype | No. of targeted clones/no. of drug-resistant clones analyzed (%) at the indicated locus
|
|||
|---|---|---|---|---|
| OVALBUMIN | RAD54 |
CENP-H
|
||
| Expt 1 | Expt 2 | |||
| Wild type | 46/47 (97.8) | 21/52 (40.4) | 19/22 (86.4) | 16/29 (55.2) |
| FBH1−/− (1) | 44/46 (95.6) | ND | 32/60 (53.3) | 14/27 (51.8) |
| FBH1−/− (2) | 35/36 (97.2) | 6/36 (16.7) | 46/73 (63.0) | 28/45 (62.2) |
Wild-type and FBH1−/− cells were transfected with targeting constructs of the indicated loci. ND, not determined.
FIG. 3.
No significant defect in HR in FBH1−/− cells. (A) I-SceI-induced gene conversion assay with the indicated genotypes. The frequency of HR-dependent double-strand-break repair is shown as the number of G418-resistant colonies per 5 × 106 cells (white bars). The experiments were done at least three times. (B) Rad51 focus formation with the indicated genotypes before and at 3 and 6 h after 4 Gy γ-irradiation. (C) Spontaneous and 5 nM camptothecin (CPT)-induced SCE events in the macrochromosomes of 50 metaphase cells. Means ± standard errors are shown in the upper right corner. Underlined numbers indicate subtraction of spontaneous SCE from SCE following CPT treatment. The level of spontaneous SCE in FBH1−/− cells significantly differs from that in wild-type cells (P < 0.0001). The level of CPT-induced SCE was indistinguishable between wild-type and FBH1−/− cells (P < 0.0001). Statistical significance was calculated by the Mann-Whitney nonparametric U test (36). (D) UV-induced SCE (0.25 J/m2) is shown as in panel C. The level of UV-induced SCE was indistinguishable between wild-type and FBH1−/− cells (P < 0.0004).
Deletion of the FBH1 gene has no significant effect on FANCC- or RAD18-deficient cells.
Despite apparently normal HR, FBH1−/− cells exhibited significant increases in the level of spontaneously arising SCE (Fig. 3C and D). This increased SCE was reversed by reconstitution of FBH1−/− cells with chicken FBH1 cDNA (data not shown). Taking the normal HR efficiency of FBH1−/− cells into account, the increased spontaneous SCE suggests either an increase in the amount of endogenous DNA damage that stimulates SCE or an elevated ratio of crossover-type HR relative to non-crossover-type HR. To gain an insight into the increased spontaneous SCE, we measured SCE induced by camptothecin and UV (Fig. 3C and D), which result in DSBs and single-strand gaps, respectively, when replication forks encounter the damaged sites. Subtraction of spontaneous SCE from camptothecin-induced SCE indicates that camptothecin-induced SCE was at the same level in wild-type and FBH1−/− cells (Fig. 3C). In contrast, UV induced a moderately higher level of SCE in FBH1−/− cells than in wild-type cells (Fig. 3D). Thus, we suggest that single-strand gaps spontaneously formed at stalled replication forks stimulate crossover-type HR more frequently in the absence of Fbh1. Further, camptothecin-induced DSBs may not be repaired adequately by HR and may lead to cell death in FBH1−/− cells (Fig. 2E).
The elevation of spontaneous SCE led us to explore the functional interactions between Fbh1 and either FancC or Rad18, because cells deficient in these proteins exhibit increased levels of spontaneous SCE (13, 44). To this end, we made FBH1−/− FANCC− and FBH1−/− RAD18−/− DT40 cells. Inactivation of FBH1 had no significant impact on FANCC− and RAD18−/− cells in terms of cellular sensitivity to cisplatin and MMS (see Fig. S2A and C and Fig. S3A in the supplemental material; also data not shown). On the other hand, FBH1−/− RAD18−/− cells exhibited slightly higher UV sensitivity than did RAD18−/− cells (see Fig. S2D in the supplemental material). In addition, camptothecin sensitivity and spontaneous SCE were increased in FBH1−/− RAD18−/− and FBH1−/− FANCC− cells compared with each single mutant (see Fig. S2B and S3B in the supplemental material; also data not shown). These observations indicate no significant functional overlap between Fbh1 and either Rad18 or FancC in suppression of SCE.
Synergistic effect of defective FBH1 and RAD54 on genome instability and hypersensitivity to camptothecin.
Given that the SCE phenotype of FBH1−/− cells suggested that recombination events more frequently resulted in crossover, we sought to test the relationship between Fbh1 and the core recombination factor Rad54 (5). FBH1−/− RAD54−/− cells grew more slowly, displaying a significant fraction of dying cells compared to each single mutant (Fig. 1E and 4A). Likewise, in FBH1−/− RAD54−/− cells, higher fractions in the sub-G1 and G2 phases were elevated in comparison to the FBH1−/− and RAD54−/− single mutants (Fig. 1E). As expected, the level of spontaneous SCE was reduced by deletion of the RAD54 gene in FBH1−/− cells (data not shown). Collectively, deletion of FBH1 in a RAD54−/− background compromises genome stability, leading to stimulation of a G2 damage checkpoint and apoptosis.
FIG. 4.
FBH1−/− RAD54−/− cells display genome instability and hypersensitivity against camptothecin (CPT). (A) Growth curves of cells of the indicated genotypes. Results were obtained from at least three independent experiments. (B and C) Colony survival assay after treatment with CPT and cisplatin performed as described for Fig. 2. Error bars show the standard error of the mean for at least three independent experiments. (D) Chromosome aberrations before and following 10 nM CPT treatment for 9 h with the indicated genotypes.
FBH1−/− RAD54−/− cells were more sensitive to camptothecin than was either single mutant (Fig. 4B). On the other hand, cellular sensitivity to cisplatin (Fig. 4C), γ-rays, and MMS (data not shown) was indistinguishable between RAD54−/− and FBH1−/− RAD54−/− cells. To assess the cause of cellular hypersensitivity to camptothecin, chromosomal aberrations were monitored following exposure of cells to camptothecin (Fig. 4D). In agreement with colony survival data, the FBH1−/− RAD54−/− cells exhibited a marked increase in camptothecin-induced chromosomal aberrations compared to RAD54−/− cells (Fig. 4D). The number of chromosomal aberrations, as well as camptothecin sensitivity, was reversed by ectopic expression of chicken FBH1 cDNA (GdFBH1) in FBH1−/− RAD54−/− cells (Fig. 4B and D). It should be noted that camptothecin-induced chromosome-type breaks (DSBs at the same position on two sister chromatids) were significantly increased in FBH1−/− RAD54−/− cells in comparison with RAD54−/− cells. Since camptothecin induces DSBs in one of the two sister chromatids and subsequently stimulates HR with the other intact sister, DSBs at the same position in two sister chromatids, i.e., chromosome-type breaks, indicate that sister recombination is not efficiently completed in FBH1−/− RAD54−/− cells, leading to DSBs in both sister chromatids during the S and G2 phases (35).
This raises two hypotheses concerning the function of Fbh1 in DSB repair. First, both Fbh1 and Rad54 may be involved in the same stage of HR, with Fbh1 acting as a backup for Rad54-dependent HR. Alternatively, Fbh1 may prevent the formation of abnormal recombination intermediates, which lead to chromosome-type breaks in the absence of Rad54. To test the former hypothesis, we assayed HR in RAD54−/− and FBH1−/− RAD54−/− cells by monitoring the repair of I-SceI induced DSBs. Deletion of FBH1 did not further diminish the capability of HR in RAD54−/− cells (Fig. 3A). In addition, Rad51 focus formation following γ-irradiation and camptothecin was comparable in RAD54−/− and FBH1−/− RAD54−/− clones (data not shown). These data are consistent with disruption of FBH1 resulting in the formation of recombination intermediates that are preferentially resolved with crossover and which lead to chromosome-type breaks in the absence of Rad54.
Complementary role of Fbh1 and Blm in releasing the DNA replication block.
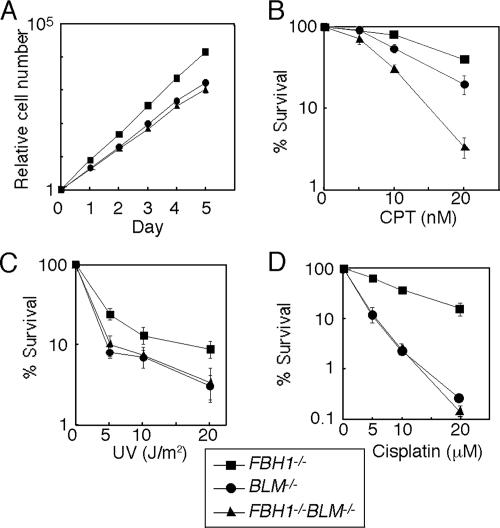
The synthetic lethality of the fbh1 and rqh1 mutations in yeast, as well as an increase in the level of spontaneous SCE in Fbh1-deficient DT40 clones, prompted us to disrupt the BLM gene in FBH1−/− cells (9, 17, 41). Unlike in S. pombe, FBH1−/− BLM−/− DT40 cells were able to proliferate with the same kinetics as BLM−/− cells (Fig. 5A), but FBH1−/− BLM−/− cells were considerably more sensitive to camptothecin than was either single mutant (Fig. 5B). Conversely, the sensitivity of FBH1−/− BLM−/− cells to UV, MMS, and cisplatin was the same as that of BLM−/− cells (Fig. 5C and D and data not shown). Thus, Fbh1 may substitute for the loss of Blm, particularly when DSBs occur as a consequence of a replication block.
FIG. 5.
Combined mutations of the FBH1 and BLM genes synergistically elevate sensitivity to camptothecin (CPT). (A) Growth curves of cells of the indicated genotypes. (B to D) Colony survival assay after treatment with CPT, UV, and cisplatin performed as described for Fig. 2. Error bars show the standard error of the mean for at least three independent experiments.
To further explore the functional overlap between Fbh1 and Blm, we examined SCE and mitotic exchanges between homologous chromosomes, because a significant increase in these two types of crossovers is the characteristic feature of BLM-deficient cells (17, 41). FBH1−/− and BLM−/− DT40 cells showed 1.8- and 9.4-fold-higher numbers of spontaneous SCE events than did wild-type cells, respectively (Fig. 6A). The FBH1−/− BLM−/− cells displayed further increases in the level of spontaneous SCE. We next examined the level of UV-induced SCE (Fig. 6B). The induction of SCE tended to be higher in FBH1−/− and FBH1−/− BLM−/− cells than in wild-type and BLM−/− cells, respectively (Fig. 6B); the number of SCEs in a UV-irradiated BLM−/− cell is likely to be saturated, making definitive quantitative measurements unreliable.
FIG. 6.
Increased genome instability of FBH1−/− BLM−/− cells observed in both UV-induced SCE and exchange between homologous chromosomes. Cells were labeled with BrdU during two cell cycle periods with (B) or without (A) prior UV treatment (0.25 J/m2). Spontaneous and UV-induced SCEs in the macrochromosomes of 50 metaphase cells were counted. Underlined numbers indicate subtraction of spontaneous SCE from SCE following UV. Histograms show the distribution of cells with the indicated numbers of SCEs per cell. The mean number of SCEs per cell ± standard error is shown in each histogram. The level of spontaneous SCE in BLM−/− cells significantly differs from that in FBH1−/− BLM−/− cells (P < 0.02). The level of UV-induced SCE was distinguishable between BLM−/− and FBH1−/− BLM−/− cells (P < 0.03), though it may have reached a plateau level. Statistical significance was calculated by the Mann-Whitney nonparametric U test. (C) Representative homologous chromosome exchange in FBH1−/− BLM−/− cells. Arrowheads indicate chromosome 1. (D) Example of nonhomologous exchange events between chromosomes 1 and 2 and between chromosomes 2 and 4 shown by arrowheads. (E) Frequency of chromosomal aberrations in mitotic cells harvested at indicated times after exposure of asynchronous cell populations to 1 J/m2 UV. Chromosome aberrations were scored for 100 metaphase cells. All samples were treated with Colcemid for the last 3 h prior to harvest of cells.
We monitored mitotic chromosome aberrations with time following UV irradiation (Fig. 6C to E). Mitotic cells were collected every 3 h from 0 to 15 h after 1 J/m2 UVC irradiation of asynchronous cells. Cells entering metaphase at 0 to 6 h and 9 to 12 h after UV irradiation reflect cells that received UV irradiation in the late S to G2 phase and the G1 to early S phase, respectively (17). UV irradiation increased exchange between homologous chromosomes, i.e., symmetrical chromosomal quadriradials (Fig. 6C), but did not increase exchange between different chromosomes (Fig. 6D) 9 to 12 h after UV irradiation in asynchronous wild-type cells (i.e., cells that were exposed to UV in G1 to early S phase). These chromosomal quadriradials may result from persisting Holliday junctions formed at sites of replication stalled by UV-induced DNA damage. As expected, BLM−/− cells exhibited a marked increase in the number of homologous chromosome exchanges in comparison to wild-type cells (Fig. 6E). This type of chromosomal aberration depends on HR, because deletion of HR factors such as RAD54 and XRCC3 in BLM−/− cells reduced the formation of symmetrical chromosomal quadriradials by ∼10-fold (M. Seki, personal communication). Like BLM−/− cells, FBH1−/− cells displayed an increase in homologous chromosome exchange. Furthermore, concurrent deletion of FBH1 and BLM had an additive effect on UV-induced quadriradial formation compared to FBH1−/− and BLM−/− single mutant cells. The total numbers of chromosomal quadriradials were 15 (wild type), 23 (FBH1−/−), 56 (BLM−/−), and 72 (FBH1−/− BLM−/−) per 600 metaphase cells in each genotype. These results suggest a possible involvement of the Fbh1 helicase, in parallel with the Blm helicase, in resolving otherwise toxic recombination intermediates produced at the sites of stalled replication forks.
DISCUSSION
Role for vertebrate Fbh1 during DNA replication.
As shown in Fig. 7, there are some processes to restrict the extent of strand exchange during recombination produced at the sites of stalled replication forks, resulting in a reduced frequency of crossover formation. Although several DNA helicases have been involved in these processes, the precise mechanism of this model remains to be determined. Several lines of evidence point to Fbh1 being involved in preventing the formation of recombination intermediates following replication blockage. The formation of such recombination intermediates (Fig. 7; transition from C to E) could explain the phenotypes associated with the inactivation of Fbh1. None of the experimental data presented here indicate that non-crossover-type HR, which accounts for the majority of HR during somatic cell division in vertebrates (18), is increased in the absence of FBH1. Neither gene targeting, HR-dependent repair of I-SceI-induced DSBs, nor Ig gene conversion was decreased in frequency in FBH1−/− cells. Deletion of FBH1 had no effect on sensitivity to UV, MMS, and cisplatin, which block replication and induce recombination, even in the absence of BLM. Thus, it does not seem likely that Fbh1 processes putative recombination intermediates (such as the chicken foot or double Holliday junction) directly. Despite a lack of evidence for a general increase in HR events in FBH1−/− cells, FBH1 deficiency leads to a significant increase in the number of crossovers between homologous chromosomes (radial structures) after UV irradiation. This, coupled with an increased level of camptothecin-induced chromosome-type breaks in FBH1−/− RAD54−/− cells, indicates that sister-mediated HR-dependent repair is blocked after physical interactions of two sister chromatids in the absence of Fbh1. Such a defect in the suppression of processing recombination intermediates in FBH1−/− cells after DNA damage accords with the phenotype of S. pombe Fbh1 cells (27). In conclusion, Fbh1 is likely to prevent extensive exchange between two homologous sequences, thereby facilitating HR-dependent repair of single-ended breaks formed during replication.
FIG. 7.
Model for the function of Fbh1 and Blm at stalled replication forks. A replication fork encounters damages on the template (oval) and is blocked. This block can be bypassed by translesion synthesis (transition from A to G), or the fork can collapse (transition from A to B). HR to restart the collapsed fork is usually carried out by synthesis-dependent strand annealing (SDSA), which is not associated with crossover (transition from C to D). More extensive strand exchange between sister chromatids (E) leads to the formation of a double Holliday junction, which may be resolved with crossover (F). Blm may play a number of roles that reduce crossovers. It may suppress D-loop formation (transition from B to C) (40), reverse Holliday junction formation (transition from C to E) (26) and, with topoisomerase IIIα, promote the dissolution of double Holliday junctions, thereby avoiding resolution by incision (42). The present data support a role for Fbh1 acting alongside Blm in the second of these possibilities, the avoidance of extensive strand exchange and Holliday junction formation (transition from C to E).
Fbh1 and Blm act in parallel in the repair of DSBs that occur as a consequence of replication block.
The present study reveals phenotypic similarities as well as differences in the mutant phenotype of the FBH1 orthologs of DT40 and S. pombe. In both species, deletion of the FBH1 gene augments the phenotype of BLM−/−/rqh1 mutants. On the other hand, FBH1−/− cells did not show a prominent phenotype, while S. pombe fbh1 mutants exhibit sensitivity to variety of DNA damaging agents and enhanced Rad51 focus formation. Thus, the prominent role of Fbh1 in S. pombe is not seen in DT40, suggesting that either vertebrate cells have a reduced requirement for this helicase or the functions of Fbh1 can be covered by other helicases as well as Blm. BLM-deficient cells are characterized by hyper-recombination between sister chromatids and also between homologous chromosomes (7). These phenotypes were also observed, although to a lesser degree, in FBH1−/− cells (Fig. 3C and D and 6E). It should be noted that the FBH1 mutation increased the sensitivity of BLM−/− cells to camptothecin but not to any other damaging agents (Fig. 5 B to D). Spontaneous single-strand nicks are likely to collapse replication forks to produce single-ended breaks similar to those induced by camptothecin (Fig. 7B) (33). These single-ended breaks trigger recombination-mediated replication restart, hence a requirement for Rad54, explaining the additive compromise in genomic stability and sensitivity to camptothecin in the FBH1−/− RAD54−/− double mutant. Thus, the absence of Fbh1 or Blm may cause an accumulation of recombination intermediates that normally require these DNA helicases for their resolution. The synergistic effect of disruption of RAD54 in both FBH1−/− and BLM−/− cells provides further support for a functional similarity between Fbh1 and Blm (41).
Taking our results together, we conclude that the Fbh1-dependent pathway is likely to act in parallel to Blm in cellular tolerance of single-ended breaks resulting from replication blocks. In humans, Fbh1 may play a role in suppression of the loss of heterozygosity, like Blm, thereby preventing tumor formation. No human FBH1 mutants have been reported, possibly because FBH1 disruption alone does not result in a clinically detectable syndrome or, although less likely given the lack of a phenotype in DT40 cells, is lethal. However, it will be interesting to determine whether a defect in this helicase function is associated with cancer predisposition or even a genetic disease akin to Bloom's and Werner's syndromes.
Supplementary Material
Acknowledgments
We thank the members of the Takeda laboratories for help and support. Special thanks go to Y. Sato, M. Nakaoka, and R. Ohta for technical assistance. We also thank M. Seki (Tohoku University) for helpful discussions and M. C. Whitby (University of Oxford) for critical reading and discussion.
Financial support was provided in part by a grant from Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology Corporation; by a Center of Excellence (COE) grant to S.T.; by a Grant-In-Aid for Priority Areas to H.S. for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan; and by grants from The Uehara Memorial Foundation and The Naito Foundation.
Footnotes
Published ahead of print on 5 February 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adachi, N., S. So, and H. Koyama. 2004. Loss of nonhomologous end joining confers camptothecin resistance in DT40 cells. Implications for the repair of topoisomerase I-mediated DNA damage. J. Biol. Chem. 279:37343-37348. [DOI] [PubMed] [Google Scholar]
- 2.Alexeev, A., A. Mazin, and S. C. Kowalczykowski. 2003. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 10:182-186. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa, H., D. Lodygin, and J. M. Buerstedde. 2001. Mutant loxP vectors for selectable marker recycle and conditional knock-outs. BMC Biotechnol. 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnaudeau, C., C. Lundin, and T. Helleday. 2001. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 307:1235-1245. [DOI] [PubMed] [Google Scholar]
- 5.Bezzubova, O., A. Silbergleit, Y. Yamaguchi-Iwai, S. Takeda, and J. M. Buerstedde. 1997. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell 89:185-193. [DOI] [PubMed] [Google Scholar]
- 6.Buerstedde, J. M., C. A. Reynaud, E. H. Humphries, W. Olson, D. L. Ewert, and J. C. Weill. 1990. Light chain gene conversion continues at high rate in an ALV-induced cell line. EMBO J. 9:921-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraverty, R. K., and I. D. Hickson. 1999. Defending genome integrity during DNA replication: a proposed role for RecQ family helicases. Bioessays 21:286-294. [DOI] [PubMed] [Google Scholar]
- 8.Doe, C. L., and M. C. Whitby. 2004. The involvement of Srs2 in post-replication repair and homologous recombination in fission yeast. Nucleic Acids Res. 32:1480-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis, N. A., and J. German. 1996. Molecular genetics of Bloom's syndrome. Hum. Mol. Genet. 5:1457-1463. [DOI] [PubMed] [Google Scholar]
- 10.Fujimori, A., S. Tachiiri, E. Sonoda, L. H. Thompson, P. K. Dhar, M. Hiraoka, S. Takeda, Y. Zhang, M. Reth, and M. Takata. 2001. Rad52 partially substitutes for the Rad51 paralog XRCC3 in maintaining chromosomal integrity in vertebrate cells. EMBO J. 20:5513-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukushima, T., M. Takata, C. Morrison, R. Araki, A. Fujimori, M. Abe, K. Tatsumi, M. Jasin, P. K. Dhar, E. Sonoda, T. Chiba, and S. Takeda. 2001. Genetic analysis of the DNA-dependent protein kinase reveals an inhibitory role of Ku in late S-G2 phase DNA double-strand break repair. J. Biol. Chem. 276:44413-44418. [DOI] [PubMed] [Google Scholar]
- 12.Hickson, I. D. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3:169-178. [DOI] [PubMed] [Google Scholar]
- 13.Hirano, S., K. Yamamoto, M. Ishiai, M. Yamazoe, M. Seki, N. Matsushita, M. Ohzeki, Y. M. Yamashita, H. Arakawa, J. M. Buerstedde, T. Enomoto, S. Takeda, L. H. Thompson, and M. Takata. 2005. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J. 24:418-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hochegger, H., D. Dejsuphong, T. Fukushima, C. Morrison, E. Sonoda, V. Schreiber, G. Y. Zhao, A. Saberi, M. Masutani, N. Adachi, H. Koyama, G. de Murcia, and S. Takeda. 2006. Parp-1 protects homologous recombination from interference by Ku and ligase IV in vertebrate cells. EMBO J. 25:1305-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hochegger, H., E. Sonoda, and S. Takeda. 2004. Post-replication repair in DT40 cells: translesion polymerases versus recombinases. Bioessays 26:151-158. [DOI] [PubMed] [Google Scholar]
- 16.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 17.Imamura, O., K. Fujita, C. Itoh, S. Takeda, Y. Furuichi, and T. Matsumoto. 2002. Werner and Bloom helicases are involved in DNA repair in a complementary fashion. Oncogene 21:954-963. [DOI] [PubMed] [Google Scholar]
- 18.Johnson, R. D., and M. Jasin. 2000. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 19:3398-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, R. D., N. Liu, and M. Jasin. 1999. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature 401:397-399. [DOI] [PubMed] [Google Scholar]
- 20.Kikuchi, K., Y. Taniguchi, A. Hatanaka, E. Sonoda, H. Hochegger, N. Adachi, Y. Matsuzaki, H. Koyama, D. C. van Gent, M. Jasin, and S. Takeda. 2005. Fen-1 facilitates homologous recombination by removing divergent sequences at DNA break ends. Mol. Cell. Biol. 25:6948-6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, J., J. H. Kim, S. H. Lee, D. H. Kim, H. Y. Kang, S. H. Bae, Z. Q. Pan, and Y. S. Seo. 2002. The novel human DNA helicase hFBH1 is an F-box protein. J. Biol. Chem. 277:24530-24537. [DOI] [PubMed] [Google Scholar]
- 22.Kim, J. H., J. Kim, D. H. Kim, G. H. Ryu, S. H. Bae, and Y. S. Seo. 2004. SCFhFBH1 can act as helicase and E3 ubiquitin ligase. Nucleic Acids Res. 32:2287-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindahl, T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709-715. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Y., and S. C. West. 2004. Happy Hollidays: 40th anniversary of the Holliday junction. Nat. Rev. Mol. Cell Biol. 5:937-944. [DOI] [PubMed] [Google Scholar]
- 25.Metzger, D., J. Clifford, H. Chiba, and P. Chambon. 1995. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl. Acad. Sci. USA 92:6991-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohaghegh, P., J. K. Karow, R. M. Brosh Jr., V. A. Bohr, and I. D. Hickson. 2001. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 29:2843-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morishita, T., F. Furukawa, C. Sakaguchi, T. Toda, A. M. Carr, H. Iwasaki, and H. Shinagawa. 2005. Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates. Mol. Cell. Biol. 25:8074-8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okada, T., E. Sonoda, Y. M. Yamashita, S. Koyoshi, S. Tateishi, M. Yamaizumi, M. Takata, O. Ogawa, and S. Takeda. 2002. Involvement of vertebrate polkappa in Rad18-independent postreplication repair of UV damage. J. Biol. Chem. 277:48690-48695. [DOI] [PubMed] [Google Scholar]
- 29.Osman, F., J. Dixon, A. R. Barr, and M. C. Whitby. 2005. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol. Cell. Biol. 25:8084-8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petukhova, G., S. Stratton, and P. Sung. 1998. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature 393:91-94. [DOI] [PubMed] [Google Scholar]
- 32.Ralf, C., I. D. Hickson, and L. Wu. 2006. The Bloom's syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 281:22839-22846. [DOI] [PubMed] [Google Scholar]
- 33.Saleh-Gohari, N., H. E. Bryant, N. Schultz, K. M. Parker, T. N. Cassel, and T. Helleday. 2005. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol. Cell. Biol. 25:7158-7169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599-602. [DOI] [PubMed] [Google Scholar]
- 35.Sonoda, E., M. S. Sasaki, J. M. Buerstedde, O. Bezzubova, A. Shinohara, H. Ogawa, M. Takata, Y. Yamaguchi-Iwai, and S. Takeda. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17:598-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sonoda, E., M. S. Sasaki, C. Morrison, Y. Yamaguchi-Iwai, M. Takata, and S. Takeda. 1999. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 19:5166-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strumberg, D., A. A. Pilon, M. Smith, R. Hickey, L. Malkas, and Y. Pommier. 2000. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol. Cell. Biol. 20:3977-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugawara, N., X. Wang, and J. E. Haber. 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12:209-219. [DOI] [PubMed] [Google Scholar]
- 39.Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17:5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Brabant, A. J., T. Ye, M. Sanz, I. J. German, N. A. Ellis, and W. K. Holloman. 2000. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry 39:14617-14625. [DOI] [PubMed] [Google Scholar]
- 41.Wang, W., M. Seki, Y. Narita, E. Sonoda, S. Takeda, K. Yamada, T. Masuko, T. Katada, and T. Enomoto. 2000. Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J. 19:3428-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, L., and I. D. Hickson. 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426:870-874. [DOI] [PubMed] [Google Scholar]
- 43.Wu, L., and I. D. Hickson. 2006. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 40:279-306. [DOI] [PubMed] [Google Scholar]
- 44.Yamashita, Y. M., T. Okada, T. Matsusaka, E. Sonoda, G. Y. Zhao, K. Araki, S. Tateishi, M. Yamaizumi, and S. Takeda. 2002. RAD18 and RAD54 cooperatively contribute to maintenance of genomic stability in vertebrate cells. EMBO J. 21:5558-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.