Abstract
Transcriptional insulators are specialized cis-acting elements that protect promoters from inappropriate activation by distal enhancers. The H19 imprinting control region (ICR) functions as a CTCF-dependent, methylation-sensitive transcriptional insulator. We analyzed several insertional mutations and demonstrate that the ICR can function as a methylation-regulated maternal chromosome-specific insulator in novel chromosomal contexts. We used chromosome conformation capture and chromatin immunoprecipitation assays to investigate the configuration of cis-acting elements at these several insertion sites. By comparing maternal and paternal organizations on wild-type and mutant chromosomes, we hoped to identify mechanisms for ICR insulator function. We found that promoter and enhancer elements invariably associate to form DNA loop domains at transcriptionally active loci. Conversely, active insulators always prevent these promoter-enhancer interactions. Instead, the ICR insulator forms novel loop domains by associating with the blocked promoters and enhancers. We propose that these associations are fundamental to insulator function.
For all multicellular organisms, the reliable establishment and maintenance of complex patterns of gene expression are fundamental to the development of a normal, healthy individual. Not only loss of expression but also inappropriate or promiscuous gene transcription can lead to disease and developmental defects. Transcriptional insulators are specialized cis-acting DNA elements that act as barriers to protect genes from both positive and negative influences of their genomic or chromatin environment and thus maintain the accurate temporal and spatial transcriptional patterns critical to normal development.
Two types of insulators have been defined (for recent reviews, see references 9, 19, 33, and 67). Barrier insulators protect genes from chromosomal position effects by preventing the spread of heterochromatin-mediated silencing. Enhancer-blocking insulators, the subject of this study, protect promoters from activation by a distal enhancer. Since enhancers are indiscriminate in their choice of promoters and can activate promoters over very long distances (even hundreds of kilobases), they have the potential to activate many genes, and it is therefore critical to restrict their action to the appropriate target promoter. Enhancer blocking is completely position dependent: blocking occurs only when the insulator is inserted between the promoter and the enhancer element. Enhancer blocking occurs without actual inactivation of either the promoter or the enhancer (7, 21, 49).
Insulators were first identified and have been best characterized for Drosophila melanogaster by use of the gypsy retrotransposon and the scs/scs′ paired elements flanking the Hsp70 (heat shock protein 70) gene (7, 21, 28, 29, 49). The minimal DNA sequence essential for enhancer blocking by gypsy contains a cluster of binding sites for the Suppressor of Hairy wing [Su(Hw)] (50). Su(Hw) protein interacts with CP190 and with mod(mdg4) proteins and then, through interactions with topoisomerase I-interacting protein, is localized to the nuclear lamina (43). By these interactions, gypsy insulator elements come together to form clusters called insulator bodies, which are localized to the nuclear periphery. The loop domains created by these clusters are proposed to isolate the enhancer and promoters separated by the gypsy insulators and somehow prevent their productive interactions. Molecular and structural analysis of scs/scs′ provides some support for the importance of loop domains in insulator function (28, 29). However, several transcriptional studies indicate that the mechanisms for enhancer blocking by scs/scs′ may be distinct from those used by gypsy (8, 32, 39).
Insulators have also been identified in invertebrates. Best characterized is the cHS4 (constitutive DNase I hypersensitive site 4) element at the 5′ end of the chicken β-globin locus (46, 47). The enhancer-blocking activity of cHS4 is associated with strong binding sites for CTCF (5), a very interesting multitalented zinc finger protein (30, 41). The ability of CTCF proteins to interact with each other and their association with nucleoplasmin suggest that CTCF might organize the genome into insulator bodies analogous to those suggested for Su(Hw) (66).
In this study, we focus on a CTCF-dependent insulator at the imprinted mouse Igf2-H19 locus (Fig. 1A). Igf2 and H19 are about 80 kb apart. Their extensive and complex expression patterns are essentially identical, and in fact the two genes share enhancer elements located around kb +8 and around kb +25 that drive expression in endodermal and mesodermal tissues, respectively (Fig. 1A) (25, 37). (Note that all sequences are referenced relative to the start site for H19 transcription, which is set at +1 bp). While sharing temporal and spatial specificities, the two genes are reciprocally imprinted. Igf2 is expressed from the paternal chromosome, while only the maternal H19 allele is transcribed (2, 14). The imprinting of Igf2 and H19 is dependent upon a shared cis-acting element called the imprinting control region (ICR) (59). The 2.4-kb ICR is located 2 kb upstream of the H19 promoter and thus separates the Igf2 promoters but not the H19 promoter from the shared enhancers (Fig. 1A). This element was originally identified molecularly because its CpGs were methylated specifically on the paternal chromosome (1, 18, 61, 62). At the same time, the ICR was highlighted genetically because H19 transgenes were expressed specifically upon maternal inheritance only when they included ICR sequences (13, 16, 45).
FIG. 1.
Long-range interactions at the Igf2 locus on wild-type (WT) and ΔICR chromosomes. (A) Schematic depiction of the 100-kb Igf2-H19 locus includes the three Igf2 promoters (P1 at kb −78, P2 at kb −76, and P3 at kb −74), the shared ICR (at kb −4.4 to −2), the H19 promoter (H19P at bp 0), and the shared endodermal (open circle at kb +8) and mesodermal (filled circle at kb +25) enhancers. DMR1 and DMR2, flanking the Igf2 promoters, become methylated on the paternal chromosome in the postimplantation embryo and play a role in the activation of paternal Igf2 in liver cells and in the repression of maternal Igf2 in muscle cells, respectively. The ΔICR chromosome carries a 5-kb deletion from kb −6 to −1 that removes the ICR. The vertical bars above and below the map indicate BamHI and BglII restriction sites, respectively. Arrowheads depict the orientations and locations of PCR primers used for 3C analysis. Asterisks indicate RFLPs that distinguish between wild-type M. castaneus alleles and M. domesticus alleles. (B to K) 3C analysis of long-range interactions at the Igf2 locus was carried out on using the primers indicated. Animal genotypes C/D, D/C, C/ΔICR, and ΔICR/C are indicated (maternal allele listed first). The top panels for each experiment represent the 3C PCR product. The bottom panels, when included, depict the banding patterns after digestion with enzymes distinguishing between the M. castaneus (C-labeled arrowheads)- and M. domesticus (D-labeled arrowheads)-derived DNAs. Note that the ΔICR mutation is on an M. domesticus chromosome. (B and C) In wild-type muscle cells (B) and liver cells (C), the Igf2 promoters associate with the mesodermal and endodermal enhancers, respectively, only on the active paternal chromosome. That is, the C/D and D/C extracts yield only PCR products that are all M. domesticus (D-labeled arrowheads) and all M. castaneus (C-labeled arrowheads) alleles, respectively. (D and E) The maternally inherited ICR insulator is necessary to prevent maternal promoter-enhancer interactions in both muscle (D) and liver (E) cells. Maternal inheritance of the ICR deletion mutant results in biallelic interactions between the Igf2 promoters and the enhancers, as indicated by the presence of both M. domesticus and M. castaneus bands in the 3C products of ΔICR/C extracts. (F and G) Inactive maternal Igf2 promoters associate with the ICR insulator in both muscle (F) and liver (G) cells. (H and I) Igf2 promoters interact only with a maternally inherited ICR in both muscle (H) and liver (I) cells. (J) The maternal ICR interacts with blocked downstream enhancers. (K) Only the unmethylated maternal ICR interacts with downstream mesodermal and endodermal enhancers. NS, nonspecific PCR product; C+D, digestion products indicative of both M. castaneus and M. domesticus DNAs comigrate.
The critical importance of the ICR was demonstrated by in vivo deletion experiments which indicated that it has at least three distinct functions (26, 54, 59). First, the ICR is the imprinting box that carries the actual mark distinguishing the parental identity of each allele. Second, paternal inheritance of the (methylated) ICR results in repression of the paternal H19 allele through a developmentally programmed silencing of the promoter (53, 54). Finally, the maternal ICR functions as a CTCF-dependent enhancer-blocking insulator that prevents expression of the maternal Igf2 allele (4, 24, 26, 27, 48, 56). The enhancer blocking is maternal chromosome specific because CTCF binding is methylation sensitive. Thus, H19ICR represents a case where insulator activity can be regulated by the organism to alter gene expression patterns.
We have recently established several novel mouse models to investigate the ability of the ICR to function autonomously in a context-independent manner (44). In one line (the H19R line), we inserted the 2.4-kb BglII fragment encompassing the ICR at the kb +10 position upstream of the H19 gene, thereby separating the H19 promoter from the mesodermal but not from the endodermal enhancers (Fig. 2A). In a second line (the AfpICR line), we inserted the ICR at the kb −0.8 position at the nonimprinted Afp (alpha fetoprotein) locus on chromosome 5 (Fig. 3A). This insertion thus separates the Afp promoter from enhancers that drive its expression in liver (51). We have already shown that the ICR can function as an imprinting box at both loci (44). That is, the ICR becomes methylated in somatic cells only when paternally inherited. In this study, we investigate the ability of the unmethylated maternally inherited ICR to function as an enhancer blocker. We find that these maternally inherited ICR insertions, H19R and AfpICR, efficiently bind CTCF and block mesodermal H19 and liver Afp expression, respectively. Thus, the ability of a single ICR insertion to function as an enhancer blocker is context independent.
FIG. 2.
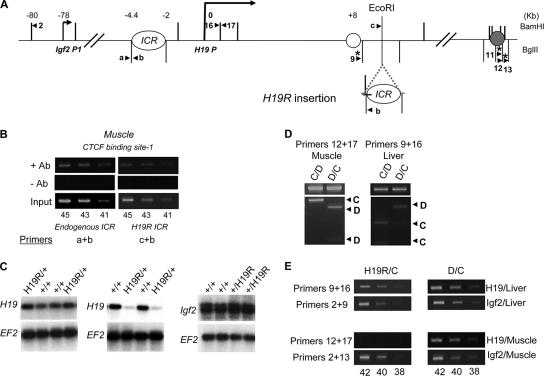
Long-range interactions at the H19 locus on wild-type and H19R chromosomes. (A) Schematic representation of the Igf2-H19 locus, including the Igf2 promoter 1 (Igf2 P1 at kb −78), the ICR (at kb −4.4 to −2), the H19 promoter (H19 P at bp 0), and the shared endodermal (open circle at kb 8) and mesodermal (filled circle at kb 25) enhancers. The H19R mutation, depicted on the lower line, is an insertion of the 2.4-kb ICR fragment at the kb +10 EcoRI site. The vertical bars above and below the maps indicate BamHI and BglII restriction sites, respectively. Arrowheads depict the orientations and locations of PCR primers used for 3C and ChIP analysis. Asterisks indicate RFLPs that distinguish between M. castaneus and M. domesticus alleles. (B) ChIP analyses demonstrate that CTCF proteins can bind in vivo to the ICR insertion on H19R. After preparing cross-linked protein-DNA extracts, the presence of the endogenous and H19R ICR sequences was detected by PCR amplification for 45, 43, or 41 cycles with the primers indicated. Ab, antisera. (C) The 2.4-kb ICR element is a transcriptional insulator at a heterologous location. RNAs prepared from liver (left panel) and muscle (middle and right panels) of P2 littermates were analyzed by Northern blotting using probes specific to H19 or Igf2, as indicated. Subsequently, blots were stripped and hybridized with probes to Elongation Factor 2 (EF2). +/+, wild-type maternal and paternal chromosomes; H19R/+, maternal inheritance of the H19R chromosome; +/H19R, paternal inheritance of the H19R chromosome. (D) In wild-type muscle cells and liver cells, the H19 promoter associates with the mesodermal and endodermal enhancers, respectively, only on the active maternal chromosome. 3C analysis was performed using the primers indicated and extracts from wild-type C/D and D/C pups. Top panels depict the 3C PCR product. Bottom panels depict the banding patterns after digestion with enzymes distinguishing the M. castaneus (C-labeled arrowheads)- and M. domesticus (D-labeled arrowheads)-derived DNAs. (E) The H19R ICR insertion blocks H19 promoter-enhancer associations in muscle but not in liver. 3C analysis was performed on extracts prepared from liver and muscle cells from H19R/C and D/C animals. Primers 9 plus 16 test for H19 promoter-endodermal enhancer interactions, while primers 12 plus 17 test for H19 promoter-mesodermal enhancer interactions. Primer pairs 2 plus 9 and 2 plus 13 identify paternal Igf2 promoter-enhancer interactions (endodermal and mesodermal, respectively) and control for the integrity of the extracts. Each PCR was analyzed at 42, 40, and 38 cycles.
FIG. 3.
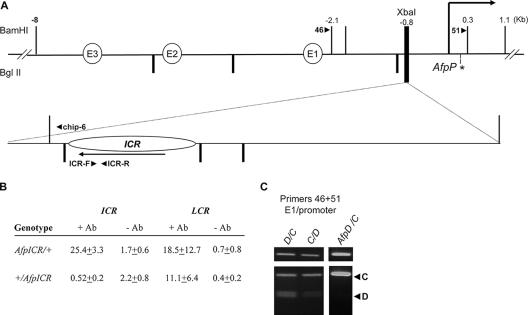
Long-range interactions on the wild-type and AfpICR chromosomes. (A) Schematic representation of the wild-type Afp locus (top line) and the ICR insertion mutation on AfpICR (bottom line). The three enhancer elements (E3, E2, and E1) and the Afp promoter (AfpP, horizontal arrow) are depicted. Vertical lines above and below the maps represent BamHI and BglII sites, respectively. Arrowheads depict the orientations and locations of PCR primers used for 3C and ChIP. Asterisks indicate RFLPs that distinguish between M. castaneus and M. domesticus alleles. Note that the AfpICR insertion is on an M. domesticus chromosome. (B) Maternal chromosome-specific binding of CTCF to the AfpICR insertion. After cross-linked protein-DNA extracts were prepared, the presence of ICR sequences was detected by real-time quantitative PCR using the primers ICR-F and ICR-R. The amount of ICR DNA was determined for samples treated with (+ Ab) or without (− Ab) antisera to CTCF and then compared to input (genomic) DNA, and that ratio is reported. For comparison, all samples were also analyzed using primers specific for the CTCF binding sites at the β-globin locus (LCR). (C) 3C analysis was performed using the primers 46 plus 51 and liver extracts prepared from D/C, C/D, and AfpICR/C animals. C-labeled arrowhead, M. castaneus allele product; D-labeled arrowhead, M. domesticus allele product.
Next, we used chromosome conformation capture (3C) (15) and chromatin immunoprecipitation (ChIP) (34) to identify long-range interactions of the promoters, enhancers, and ICR elements at these loci. We thus sought to identify interactions that are associated with active gene expression as well as interactions associated with insulator-mediated repression. Imprinted loci are particularly conducive to these sorts of analysis because each cell carries both an active and inactive copy of each allele for comparison. By comparing long-range interactions on maternal and paternal chromosomes, structures essential to gene activation and gene repression can be highlighted. In fact, two exciting reports describing some of the facets of genome organization at this locus have recently been published (35, 40). Our current study expands on this earlier work in several ways. First, we examined several novel interactions at Igf2 promoters 2 and 3 and at the shared mesodermal enhancers. More importantly, we utilized the array of mouse mutations that we have generated over the last several years, comparing wild-type and mutant chromosomes that had been either maternally or paternally inherited and in endodermal and mesodermal cells. We show that interactions between promoter and enhancer elements are a hallmark of transcriptionally active chromosomes but are never associated with inactive chromosomes. Rather, the presence of a transcriptional insulator prevents these interactions and replaces them with a specific alternative organization of the chromosome into loops that include the promoter/ICR elements. These associations occur independently of the specific chromosomal context of the ICR. Finally, we discuss these results in terms of current models for insulator and enhancer function. Specifically, our results indicate that enhancer-blocking insulators can function by directly interacting with the regulated promoter and enhancer elements.
MATERIALS AND METHODS
Animal husbandry.
All animal work was approved the NICHD Animal Care and Use Committee. The H19R line and the AfpICR line (previously called the AfpD line) (44) and Cast7 line (22) have been described previously. Cast5 mice were derived by crossing Mus castaneus females with FVB males and then backcrossing female progeny for four additional generations to FVB males, each time selecting for M. castaneus markers on chromosome 5.
RNA analysis.
RNA for Northern analyses was obtained and analyzed as previously described (26). RNase analysis was done using an RPA II RNase protection assay kit (Ambion) with probes specific for each isoform. cDNAs were generated using an iScript cDNA synthesis kit (Bio-Rad). By exploiting single-nucleotide polymorphisms, the expression of Afp mRNA from the AfpICR allele (on an SvJ129 chromosome) relative to expression from the wild-type allele (FVB) was determined by analyzing the melting property of hybridization probes (Afp5-S, Afp-A) spanning the single-nucleotide polymorphism region (S. Jeong and K. Pfeifer, unpublished data). Quantitative competitive PCR for Igf2 expression analysis was performed on the cDNAs in the presence of the appropriate competitor pair. Competitors for promoter-specific cDNA were generated by PCR using one of the promoter-specific forward primers, i.e., e1f, e2f, and e3f for exons 1, 2 and 3, respectively, and a mismatch-carrying reverse primer, e4snpr, which spans the 3′ part of exon 4 and the 5′ part of exon 5. Each of these competitors was then combined in a 1:1 molar ratio with a second competitor to a shared region of the Igf2 cDNA, which was generated using PCR primers e6f and e6snpr. A mix containing the muscle and liver cDNA samples and the competitor pairs was subjected to two independent PCRs, one for the amplification of the promoter-specific region and one for the common cDNA part. The relative amount of each Igf2 cDNA to corresponding competitor was determined by melting analysis using hybridization probes e4-A and e4-S for the promoter-specific region and e6-A and e6-S for the common region, yielding the relative amount of promoter-specific cDNA part to the common cDNA part. These values were than utilized to generate the relative promoter usages between three promoters. Primer sequences for these assays are shown in Table 1.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′ → 3′)a |
|---|---|
| 1 | GTGAACAGAACAAATGCTGACCGA |
| 2 | GGACCACAGAGAACTAGAGCTGA |
| 3 | GGACCACAGAGAACTAGAGCTGA |
| 4 | CCCAAAGGCTGCTAGGAGATCCCA |
| 5 | CCTCTAGCTCAAAGCCTGCG |
| 6 | GCCATTCTCCTGGGATTAGG |
| 7 | GTGATTCGGGAACTGTAGGCAATGGCTA |
| 8 | GCTATGTTCCTCCTGTATGGTCA |
| 9 | GGCAGTGCTAGAGATATGTGGGCC |
| 10 | GACAGGCATAGAAAGAGCCAAGA |
| 11 | GGGAATGCTGTCCTCTGAATTAATAG |
| 12 | CTAAGACACAGAGACCTCTAAAGGGGAA |
| 13 | CCTAATGAGCTGTTTCCAAGCCCTTTGAT |
| 14 | GTGATTCGGGAACTGTAGGCAATGGCTA |
| 15 | AGCTCCACCATGTACCTCACTG |
| 16 | CTCTGGAGTCCGATACCTGC |
| 17 | CAGGTGGAAAGAGCTCTTAGAGA |
| 46 | AACCCAGTCGCCATATGTTC |
| 51 | GCTGTCCTGGAGCTCACTTT |
| a | AGCTTACTGCCCTCATTGTACTTTC |
| b | CTTGGGTGACCCACAGCATT |
| c | GAACTCCTATTCGTCCTGCTTCTA |
| d | AGGCTGCTAGGAGATCCCAG |
| e | CGAGGTCCCATGTCATGTTTCC |
| f | TACCTCAGGGGGGTCACAAATG |
| g | GGGATCATAGATGGTGATAGGG |
| h | CTTGACAGGCATAGAAAGAGCCAA |
| i | GGGACTCACAGGCCTGTATG |
| j | GTGGACTAGGATGAAGGCAGC |
| k | CCAGCTCATCCCAATTCTAAGCAA |
| CD3-F | TCCCCAGACAGATGACCTTC |
| CD3-R | AGGACACTCTGGGACACCAC |
| ICR-F | GACCATGCCCTATTCTTGGA |
| ICR-R | TGCAGAGAGTAAGCCGACCT |
| LCR-F | CACTTAAGCAGACTCCTTCCAG |
| LCR-R | GGATTTCTAGGACGAAAGCCAC |
| e1f | GGCCTTGTGGTACCAATGG |
| e2f | GGCTTCCAGGTACCAATGG |
| e3f | CCCAACTTCAGGTACCAATGG |
| e4snpr | GAAGGCCTGCTGAAGTAGAAGCCGCGTTCCG |
| e5snpr | GAAGGCCTGCTGAAGTAGAAGCCGCGTTCCG |
| e5r | GAAGGCCTGCTGAAGTAGAAG |
| e6f | CCATCGGGCAAGGGGATC |
| e6snpr | CACCATCGGGCAAGGGGATCTCAGCAGTTCTAAAAAAGCAAATTTG |
| e6r | TGGGTTCTGGGATCCAAGTC |
| Afp-5A | ACATCTCCAGAAGGAAGAGTGGACAA-FITC |
| Afp-5S | Red640-AAATGTGTTGACGCTTTGGTGTGAG |
| e4A | GGGGAGCTTGTTGACACGCT-FITC |
| e4S | Red640-CAGTTTGTCTGTTCGGACCGC |
| e6A | Red640-AAACAACCCAATTGACACCCCCCAAA |
| e6S | CAGCAGTTCTAAAAAACCAAATTTGATTGGC-FITC |
FITC, fluorescein isothiocyanate.
3C.
Single-cell suspensions of fetal liver or skeletal muscle tissue were obtaining by sieving pooled material from four or five animals through a 100-μm nylon cell strainer into PBS. The 3C procedure was then performed as described previously (15). The specific restriction enzymes and primer pairs used to analyze long-range interactions are described below and in the figure legends. Table 1 includes the sequences for primers used in this study. A detailed description of the PCR products, including the restriction maps with restriction fragment length polymorphisms (RFLPs) used to distinguish between M. castaneus and M. domesticus DNAs, is given elsewhere (unpublished).
ChIP.
Single cells from fetal or neonatal liver and muscle tissue were isolated and fixed with formaldehyde using the procedures described above for 3C. Nuclei were harvested using a ChIP assay kit (Upstate, Lake Placid, NY). DNAs were sonicated to an average size of 500 bp (range of 200 to 1,000 bp) and incubated with or without anti-CTCF polyclonal antisera (Upstate, Lake Placid, NY) overnight at 4°C. After a second overnight incubation with Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Santa Cruz, CA), agarose beads were pelleted and DNAs prepared according to the Upstate ChIP assay kit instructions. Samples were analyzed by either real-time PCR using a Roche Light Cycler or by conventional PCR, in which case multiple reactions of different cycle numbers were assayed and referenced to input DNAs also assayed using multiple cycle numbers.
RESULTS
cis-acting elements essential for expression of H19 and Igf2 in fetal liver and muscle cells.
In vivo analyses have identified enhancer elements essential for the expression of both Igf2 and H19 in fetal liver and muscle (Fig. 1A). While the H19 promoter is relatively simple, multiple Igf2 isoforms that are due to differential splicing and differential promoter use have been identified. To determine which of these promoters are actually used in the tissues analyzed in this study, we performed RNase protection assays and also developed a novel quantitative reverse transcription-PCR assay in which we compared levels of exons 1, 2, and 3 to that of a shared internal exon (data not shown). These studies confirm that while promoters 1, 2, and 3 each contribute to fetal expression, most Igf2 mRNA (>90%) is from promoters 2 and 3. Thus, we focused our attentions accordingly.
Interactions between Igf2 promoters and enhancers in skeletal muscle and in liver tissues are specific to the active paternal chromosome.
Considerable experimental evidence now supports the notion that physical interactions between promoter and enhancer elements are essential for transcriptional activation (10, 11, 35, 52, 60). To test whether this is always true at the Igf2 locus, we looked for interactions between Igf2 promoters and enhancers by use of 3C technology. In brief, single cells were isolated from embryonic liver or muscle tissues and treated with formaldehyde to cross-link proteins and DNA with the goal of trapping the native chromatin organization. The DNA-protein complex was then subjected to restriction enzyme digestion using enzymes that isolate the cis elements of interest. Next, the resultant digested DNAs were diluted and then incubated with T4 DNA ligase. Under these conditions, only intramolecular ligations are expected. Finally, ligation products were detected by PCR, choosing primers to test systematically for a physical association of the three Igf2 promoters with the shared endodermal and mesodermal enhancers.
In these experiments, fetal samples were generated by intercrossing FVB mice (Mus domesticus) with Cast7 mice. Cast7 mice are FVB congenics that carry an M. castaneus version of the mouse distal chromosome 7 (22). Pups resulting from such reciprocal crosses are referred to as C/D (for maternal M. castaneus and paternal M. domesticus Igf2-H19 locus) or D/C (for maternal M. domesticus and paternal M. castaneus Igf2-H19 locus). M. castaneus and M. domesticus DNAs carry multiple RFLPs that allowed us to identify the parental origin of the PCR products and thus deduce the parental origin of the promoter-enhancer interactions.
To identify promoter-enhancer interactions in muscle cells, we digested skeletal muscle nuclei with both BamHI and BglII and used PCR primers shown in Fig. 1A. We identified mesodermal enhancer interactions with each of the three active Igf2 promoters (Fig. 1B, upper panels). Most critically, RFLP analysis of each of these PCR products shows that they all are specific to the transcriptionally active paternal chromosome (Fig. 1B, lower panels). That is, upon digestion with enzymes that distinguish between M. castaneus- and M. domesticus-derived PCR products, we noted an M. domesticus pattern in C/D pups but an M. castaneus pattern in D/C pups. We obtained similar results when we tested fetal liver cells for interactions between the three Igf2 promoters and the endodermal enhancer (Fig. 1C). That is, we noted associations between the Igf2 promoters and the endodermal enhancers in all pups; these associations were specific to the active paternal allele. Thus, we conclude that Igf2 promoters on the paternal chromosome become physically proximal to enhancers located over 100 kb downstream. Our data suggest an importance to these physical interactions, because we see them on active but not on inactive chromosomes.
The tissue specificity of these interactions supports the idea that they are of functional importance. In muscle extracts, Igf2 promoter-mesodermal enhancer interactions are about eightfold enriched compared to promoter-endoderm enhancer interactions. In liver extracts, Igf2 promoter-endoderm enhancer interactions are about 16-fold enriched compared to promoter-mesoderm enhancer interactions (data not shown).
The maternal ICR insulator is required to prevent maternal promoter-enhancer associations.
It is now well established that the unmethylated maternal ICR binds CTCF protein at four sites and acts as a transcriptional insulator to prevent expression of the maternal Igf2 (3, 24, 27, 56). We used a ΔICR mutant mouse (54) to test the effect of ICR deletion on the physical associations we observed between the Igf2 promoters and tissue-specific enhancers. The ΔICR mouse carries an M. domesticus chromosome with a 5-kb deletion that encompasses the ICR insulator region (Fig. 1A). Paternal deletion of the ICR (i.e., in C/ΔICR pups) has no significant phenotype in regards to Igf2 transcription (26, 54, 58, 59). That is, Igf2 expression remains robust and paternal chromosome specific. Thus, it was not surprising that 3C analyses of these pups show that the promoter-enhancer interactions remain paternal chromosome specific and are indistinguishable from the 3C patterns noted for wild-type animals (Fig. 1D [for muscle] and E [for liver]). In contrast, maternal deletion of the ICR insulator (i.e., in ΔICR/C pups) results in an overall increase in Igf2 expression in fetal muscle with approximately equal contributions by the paternal and maternal alleles (26, 54, 58, 59). 3C analyses of these pups demonstrate robust interactions between the Igf2 promoter and enhancer elements, while RFLP analysis demonstrates that these promoter-enhancer interactions in ΔICR/C pups occur equally well on both the maternal and paternal chromosomes (Fig. 1D). Identical results were obtained using fetal liver cells (Fig. 1E). Thus, the ICR insulator is necessary in cis to prevent both transcription and promoter-enhancer association. In sum, analyses of both wild-type and ΔICR chromosomes in two tissue types indicate that active transcription at Igf2 correlates strictly with the physical association of the Igf2 promoters and enhancers. These data suggest that the ICR insulator blocks expression by preventing this association of the promoter and enhancer elements.
The ICR insulator functions are not promoter specific.
We next wished to test whether it is a generalized property of the ICR insulator to reorganize promoter-enhancer interactions or if this mechanism was specific to the Igf2 locus. For example, several specialized cis-acting transcriptional elements, including Igf2-Differentially methylated Region 1 (DMR1) and Igf2-DMR2, have been identified proximal to the Igf2 promoter region (Fig. 1A), and these might be essential for the ability of the ICR insulator to regulate and organize long-range interactions at Igf2 (12, 17, 40). We recently generated mutant mice in which the 2.4-kb ICR element was inserted into heterologous positions in the genome (44). At that time, we noted that wherever it was inserted, the insert was methylated in liver and muscle cells only on the paternal chromosome. Since the maternal ICR insertion remains unmethylated, we reasoned that it had the potential to bind CTCF and act as a transcriptional insulator. Thus, these insertion mutations offered the promise of model systems to examine the context dependence of the ICR's insulator activity.
We first examined an insertion of the ICR at the EcoRI site 10 kb downstream of the H19 promoter (Fig. 2A). The ICR insertion on this chromosome, called H19R, separates the H19 promoter from its muscle-specific but not from its liver-specific enhancers. We tested for in vivo binding of CTCF in fetal muscle by use of ChIP. The results shown in Fig. 2B directly demonstrate in vivo binding at CTCF site 1 both of the endogenous ICR and of the inserted ICR on the maternal H19R chromosome. We also noted CTCF binding at site 4 (data not shown). In contrast, but as expected given that CTCF cannot recognize methylated DNA, we could not detect any in vivo binding to the ICR insert on the paternally inherited H19R chromosome (data not shown).
To directly measure the transcriptional insulator activity of the maternally inherited, nonmethylated ICR insert, we used Northern blots to measure liver and muscle H19 RNAs isolated from H19R/+ embryos and their wild-type (+/+) littermates. A maternally inherited H19R insertion has no effect on H19 expression in liver, consistent with the location of the potential insulator distal to both the H19 promoter and the endodermal enhancers (Fig. 2C, left panels). However, H19 expression in muscle was reduced eightfold (average from three litters), consistent with the location of the ICR between the promoter and mesodermal enhancer (Fig. 2C, middle panels). This degree of reduction in H19 RNA is actually quite similar to that caused by a complete deletion of the mesodermal enhancers (25), indicating that the insertion is highly effective in enhancer blocking. The ICR insertion phenotype is restricted to the maternal (unmethylated) inherited chromosome, because Igf2 expression in muscle is unaffected when the H19R mutation is paternally inherited. That is, +/H19R pups and their wild-type littermates show identical levels of Igf2 expression in muscle tissue (Fig. 2C, right panels). Thus, we conclude that the 2.4-kb BglII fragment can act very effectively as a CTCF-based methylation-sensitive insulator even in a heterologous chromosomal context.
We next used 3C technology to compare H19 promoter and enhancer interactions on wild-type and H19R chromosomes. For wild-type cells (C/D and D/C), we readily identified interactions of the H19 promoter with the mesodermal and endodermal enhancers in muscle and liver cells, respectively (Fig. 2D, upper panels). RFLP analyses demonstrate that these promoter interactions are specific to the transcriptionally active maternal chromosome (Fig. 2D, lower panels). (Note that the paternal H19 allele is silenced not by transcriptional insulation but by a developmentally programmed silencing mechanism that is dependent upon the presence of a methylated ICR. Presumably, the heterochromatinization of the H19 promoter associated with this silencing [1, 18, 23, 57] disrupts or prevents its interactions with the enhancers.) Thus, as with Igf2, promoter-enhancer interactions are a signpost of an actively transcribing allele.
We then prepared extracts from H19R/C animals to examine the promoter-enhancer interactions on the maternal H19R chromosome. In liver cells, where there is no phenotype associated with the H19R insertion, interactions between the H19 promoter and the endodermal enhancers were readily detected using our routine PCR conditions in both mutant (H19R/C) and wild-type (D/C) animals (Fig. 2E, top panels, primers 9 plus 16). As expected, these interactions were all from the maternal allele (data not shown). In contrast, we could not detect H19 promoter-mesodermal enhancer interactions in muscle cells isolated from H19R/C animals (Fig. 2E, bottom panels, primers 12 plus 17). As a reference, we used primers 2 plus 13 to quantitate Igf2 promoter and mesodermal enhancer interactions and thus demonstrated that other long-range interactions were readily identified in these same extracts (Fig. 2E, bottom panels). Thus, we conclude that the ICR insertion on the H19R chromosome permits maternal H19 promoter association with the endodermal enhancer in liver cells but prevents associations with the mesodermal enhancer in muscle cells.
Functional analysis of an ICR insertion at the Afp locus.
We reasoned that the ICR insertions we generated at the Afp locus on chromosome 5 actually represented a more stringent assessment of the generality of ICR's insulator function at a heterologous locus. Afp is not near any known imprinted locus, its expression is fully biallelic (see below), and we have not been able to identify any DMRs at this locus that might confound our ability to draw general conclusions about the ability of the ICR insulator to function autonomously. As with the ICR insertion on the H19R chromosome, we have already reported that ICR insertions at Afp remain unmethylated specifically on the maternal chromosome (44). Thus, we focused our attention on the maternal inheritance of the AfpICR chromosome depicted in Fig. 3A.
We first used ChIP to test for in vivo binding of CTCF. Normalizing for input DNA, we noted that precipitation with antisera to CTCF enriched for the ICR insertion (about 15-fold) specifically upon maternal inheritance (Fig. 3B). As a control, we tested for enrichment of CTCF binding sequences from the well-characterized insulator at the β-globin locus. To directly test whether this ICR insertion acted as a transcriptional insulator, we analyzed Afp RNA isolated from livers of wild-type mice and mice carrying the AfpICR allele. We developed a novel quantitative reverse transcription-PCR assay that uses differential DNA melting analysis to distinguish between FVB and SvJ129 Afp cDNAs, which differ at a single base pair (Jeong and Pfeifer, unpublished data). (Note that the ICR insertion is on an SvJ129 chromosome.) Our assay directly measured the ratio of SvJ129 to FVB RNAs, which we converted to a percentage by multiplying by 100. Thus, if Afp were transcribed equally from both chromosomes, we would expect a value for the SvJ129 allele of 100%. For livers isolated from wild-type SvJ129/FVB and FVB/SvJ129 neonatal pups, the actual values were 105% and 106%, respectively, thus confirming that Afp is transcribed without any measurable parental or allele bias. In contrast, in AfpICR/FVB animals, expression from the maternally inherited AfpICR chromosome is less than 2% ± 1% (n = 3) of that from the wild-type paternal chromosome. Thus, the maternally inherited ICR insertion can completely block Afp expression. We reasoned that an analysis of paternal inheritance of the ICR insertion would allow us to understand which part of this repression was due to insulation and which represented side effects of the insertion and concomitant lengthening of the distance between the Afp promoters and enhancers. The paternal AfpICR allele in FVB/AfpICR pups expresses at 30% ± 10% (n = 6). Together, these results demonstrate that ICR insertion between the Afp promoter and enhancers acts as a strong transcriptional insulator that blocks expression around 15-fold.
Figure 3C, top panels, includes examples of analyses that demonstrate that Afp promoter-enhancer interactions are readily detected both in wild-type (D/C and C/D) pups and in pups carrying a maternal ICR insertion and a paternal wild-type chromosome of M. castaneus origin (AfpICR/C). However, RFLP analysis shows that these interactions are biallelic in wild-type cells (D/C and C/D) but are M. castaneus allelic (i.e., paternal) in AfpICR/C livers, indicating that the maternally inherited ICR insertion prevents association of the Afp promoter and enhancer elements just as it prevents transcription from the maternal chromosome.
The maternally inherited ICR insulator physically associates with the blocked promoter and enhancers.
The results of our analysis of the natural Igf2 locus and of the two artificially generated loci, H19R and AfpICR, are all quite consistent. First, we have observed that promoter-enhancer interactions are limited to and a hallmark of actively transcribing genes. Second, we have noted that CTCF-dependent insulation is associated with loss of these interactions.
We supposed that one way the insulator might abolish promoter-enhancer associations was through induction of alternative interactions (20, 31). Specifically, we conjectured that the ICR insulator itself formed associations with the blocked promoters or enhancers. To test these hypotheses, we first looked for interactions between the ICR and the Igf2 promoters in wild-type animals. We found that the ICR does become associated with the Igf2 promoters and that these interactions are from the maternal chromosome (Fig. 1F and G). To confirm these results, we prepared extracts from ΔICR/C and C/ΔICR animals. We were able to identify promoter-ICR associations in tissues isolated from C/ΔICR animals but not from ΔICR/C animals, which lack a maternally inherited ICR insulator (Fig. 1H and I).
Likewise, we identified an association of the H19R ICR insulator insert with the H19 promoter specifically in muscle cells upon maternal inheritance, and we also noted an association of the AfpICR insulator insert with the Afp promoter in liver cells upon maternal inheritance (data not shown).
We next looked for an association of the maternal ICRs with the blocked maternal enhancers. We first looked for interactions between the ICR and the shared enhancer elements on wild-type chromosomes. We readily identified such interactions, and RFLP analyses demonstrate that these associations are from the maternal chromosome (Fig. 1J). Analysis of extracts from ΔICR/C and C/ΔICR animals confirms these results in that we identify ICR association with the two enhancers only where there is a maternally inherited ICR (Fig. 1K). Likewise, maternal chromosome-specific interactions between the ICR insert on H19R and the mesoderm enhancer and also between the ICR insert on AfpICR and the Afp enhancers were identified (data not shown).
ChIP with antisera to CTCF enriches for the maternal ICR and the maternal Igf2 promoter and enhancer sequences.
Our 3C results indicate that the active insulator associates with the promoters and enhancers it regulates on the maternal chromosome. The reproducibility of this phenomenon at several insulated loci suggests that this might be a critical mechanism to prevent promoter-enhancer interactions that would induce transcription. To confirm this initial observation, we performed ChIP assays. Single cells isolated from neonates were treated with formaldehyde, sonicated to reduce the average DNA size to about 500 bp, and immunoprecipitated with anti-CTCF polyclonal antibodies, and DNA sequences were detected by PCR using primers specific to the ICR, to the Igf2 promoters, or to the shared enhancer elements (Fig. 4A). As expected, anti-CTCF-precipitated DNAs were enriched for ICR sequences by use of either muscle (Fig. 4B) or liver (Fig. 4C) cells. We also noted an enrichment of enhancer and Igf2 promoter sequences. However, antibody treatment did not enrich for random sequences such as the CD3 intergenic region. Finally, RFLP analyses show that enriched DNAs at the Igf2 promoter and enhancer regions are largely maternal in origin (data not shown).
FIG. 4.
ChIP assays confirm the proximity of the insulator and promoter and enhancer elements on the maternal chromosome. (A) Schematic representation of the Igf2-H19 locus including the three Igf2 promoters, the ICR, the H19 promoter, and the shared endodermal (open circle) and mesodermal (closed circle) enhancers. Numbers above the line indicate the relative positions of the corresponding elements. Arrowheads indicate the locations and orientations of primers used the ChIP analysis. (B and C) Cross-linked extracts were prepared from muscle (B) and liver (C) cells and analyzed by ChIP using polyclonal antisera specific to CTCF protein. ICR, enhancer, and Igf2 promoter 2 sequences were identified using the primers indicated and quantitated by testing PCR products after 41, 43, and 45 cycles. In addition, primers specific to the CD3 locus were tested as a nonspecific control. + Ab, with antisera; − Ab, without antisera.
DISCUSSION
In all organisms, regulated gene expression is fundamental in establishing cell identity and function. Enhancer-blocking insulators can play a critical role in maintaining appropriate gene expression patterns by circumscribing the promiscuous activity of enhancers and thus targeting enhancer activation to the appropriate promoters (9, 19, 33, 67). Two key properties appear to be common to all enhancer-blocking insulators. First, they are position dependent and block gene expression only when placed between the enhancer and promoter and not from a flanking position. Second, enhancer blockers prevent expression without actually inactivating either the promoter or the enhancer.
We have focused on the enhancer-blocking activity associated with the ICR element at the Igf2-H19 locus. The ICR element separates the Igf2 promoters but not the H19 promoter from shared enhancer elements. Thus, binding of the CTCF protein to the ICR blocks expression of Igf2 but not of H19. Enhancer blocking at this locus is parental origin specific because the paternal chromosome-specific methylation of the ICR prevents CTCF binding and thus prevents enhancer blocking. In this work, we first used 3C technology to examine long-range interactions among the Igf2 promoters, enhancers, and the ICR, comparing maternal and paternal wild-type chromosomes and also wild-type and ΔICR chromosomes. To generalize our findings, we also examined two novel model systems where the ICR element had been inserted into heterologous positions in the mouse genome. The H19R chromosome carries an ICR element inserted to separate the H19 promoter from its mesoderm enhancers but not from its endoderm enhancers. The AfpICR chromosome carries an ICR insertion that separates the Afp promoter from its liver enhancers. We demonstrate CTCF binding and enhancer-blocking function for the ICR insertion at each of these loci, thus demonstrating that ICR's insulator function is context independent. Moreover, we also characterize the effects of these insertions on long-range interactions among cis-acting elements. Imprinted loci are particularly well suited for characterization by 3C because each cell carries an active and an inactive allele whose confirmations can thus readily be compared in a highly controlled experiment.
Our first major finding is that active transcription is always coupled with physical association of the transcribed promoter and its enhancer. We noted these interactions in multiple cases examining five promoters and three enhancers in two tissues. Specifically, we observed an association of paternal Igf2 promoters 1, 2, and 3 and of the maternal H19 promoter with the mesodermal and endodermal enhancers (located up to 100 kb away) in skeletal muscle and in liver cells, respectively. We also identify promoter-enhancer interactions from maternal and paternal chromosomes of the nonimprinted Afp locus. The identification of these interactions is consistent with several recent studies using 3C technology and supports the importance of direct interactions between the enhancer binding proteins and the promoter elements in gene activation, as proposed by DNA looping models (for reviews, see references 63 and 67.
Our second finding is that enhancer-blocking insulators regulate promoter-enhancer interactions. Again, this conclusion is based on several comparisons. First, Igf2 promoter-enhancer associations are seen only when the ICR insulator is either inactivated by methylation (as when paternally inherited) or deleted by mutagenesis (as on ΔICR chromosomes). Second, H19 promoter-enhancer interactions are blocked by the H19R insertion specifically in muscle cells and upon maternal inheritance. Third, Afp promoter-enhancer interactions are blocked by maternal inheritance of the AfpICR insertion mutation. In sum, we examined the ICR insulator in three contexts, testing six promoter regions (Igf2P1, Igf2P2, Igf2P3, H19P, and AfpP) and three enhancers (Igf2-H19 endoderm, Igf2-H19 mesoderm, and Afp liver) and saw a consistent effect on promoter-enhancer physical association.
Our third finding is that the insulator actively organizes the locus by itself forming associations with the blocked enhancer and promoter elements. The maternal endogenous ICR associates with the maternal Igf2 promoters and the maternal enhancers, the maternal H19R insertion associates with the maternal H19 promoter and mesoderm enhancers, and the maternal AfpICR insertion associates with the maternal Afp promoter and liver enhancer. Using an alternative molecular genetics approach, physical interaction between the Fab-7 insulator and the Abd-B promoter has recently been identified for Drosophila (11).
Two general mechanisms to explain enhancer-blocking function have been proposed. One mechanism emphasizes a large-scale structural role for the insulator in organizing the chromosome into separate DNA loop domains that isolate regulatory elements so that they cannot productively interact (36, 64). As described in the introduction, several analyses of both Drosophila and vertebrate insulators identify loop structures consistent with this model. However, this model is unable to explain the ability to detect enhancer-blocking activity even in transient transfection assays where constructs do not integrate into chromosomes (47). A second alternative mechanism emphasizes a transcriptional role for insulators and proposes that insulators behave as cis-acting sites that act locally to control gene regulation by interacting with the gene's enhancer or promoter elements. (For example, one specific model of this class proposes that the insulator might act as a decoy and competes with promoters for enhancer interactions [20].) These two mechanisms are not necessarily mutually exclusive but may differ only in their emphasis on different aspects of insulator function (63).
Consistent with studies of other systems, we note that the ICR insulator is involved in organizing the chromosome across large distances. That is, the association of the ICR with elements across a >100-kb region supports the notion that insulators function by generating loop domains. However, perhaps more interesting is the specific composition of these DNA loops. We specifically note interactions with promoter and enhancer elements, a finding more consistent with transcription-based models for enhancer blocking. Thus, we propose that the insulator functions by interacting with promoter and enhancer elements in ways that do not favor the productive association of promoters and enhancers with each other and that these alternative structures prevent gene activation.
One significant limitation of this analysis is that only binary interactions are tested. For example, for muscle cells, we noted that on the maternal chromosomes, the H19R insertion is associated with the H19 promoter and also with the H19 enhancer. However, we cannot distinguish whether this represents an association of all three elements into one large complex or if there are two populations of chromosomes and the insulator is tying up the enhancer in some cases and the promoter in others. Likewise, on wild-type maternal chromosomes, we have established that the ICR interacts with the Igf2 promoters and enhancers. However, these same maternal enhancers also associate with the maternal H19 promoter. It would obviously be very valuable if we could understand whether these interactions are occurring by localization of all these elements or whether in distinct cell populations the enhancer is either activating H19 or, alternatively, being tied up in nonproductive associations with the ICR insulator.
Two studies of chromosome conformation at the Igf2-H19 locus in fetal liver cells have been reported (35, 40). Our studies extend on these earlier reports in the several ways. First, we characterized Igf2 promoter use in fetal cells and noted that promoters 2 and 3 account for >90% of Igf2 mRNA and thus focused on these elements as well as on promoter 1. We also examined expression and chromosome organization in both endodermal and mesodermal tissues and thus characterized novel enhancer elements. Most importantly, we compared ICR functions in several deletion and insertion mutations in order to understand what is general and what is locus specific about CTCF-mediated insulator function. We have found that the ability of the ICR to regulate gene expression and to organize chromosome conformation is entirely context independent. That is, the ICR insertions downstream of H19 and at the Afp locus on chromosome 5 appear as efficacious as the endogenous ICR in parent-of-origin-dependent insulation. Thus, we suggest that locus-specific interactions such as those noted between the maternal ICR and DMR1 (reference 40 and data not shown) are likely not obligatory to enhancer blocking. Rather, a key feature of insulators is likely to be their promiscuous ability to interact with enhancer and promoters (55), just as enhancers and promoters can promiscuously interact with each other. This is a particularly intriguing question, given the recent findings that the H19ICR can interact quite promiscuously not only with sequences quite distant on chromosome 7 but also with sequences on other chromosomes (38, 68). Our results certainly do not support the notion that any of these interactions are direct but rather support that they all can occur through other proteins or transcriptional structures (42).
In one major regard, our results appear to be contradictory with previous studies of Igf2-H19 organization. We note interactions of the H19 promoter and its enhancer elements only on the active maternal chromosome. Likewise, as discussed above, we note that in wild-type cells, ICR-enhancer interactions are restricted to the maternal chromosomes where the insulator is functional. In contrast, Kurukuti et al. (35) concluded that these enhancer interactions are biallelic. We suggest that this apparent discrepancy can be explained by the choice of restriction enzymes in the two studies. Previous studies used EcoRI, which is particularly useful for analyzing cis elements near the Igf2 transcription unit but is less optimal at the H19 gene. First, EcoRI digestion leaves the H19 promoter and the H19ICR on a single DNA fragment. Second, in vivo and in vitro analyses both indicate that the EcoRI fragment analyzed actually does not contain any enhancer sequences (references 6 and 65 and M. Miller and K. Pfeifer, unpublished observations). In contrast, the enhancers are entirely localized to the BamHI-BglII fragment used in this study. Thus, we conclude that the H19 promoter-endodermal enhancer interactions are maternal chromosome specific. Our results for the endodermal enhancer are entirely consistent with the results we saw for the mesodermal enhancer. Our results showing a lack of interaction of the H19 promoter and enhancers on the paternal chromosome are also consistent with early studies demonstrating that the paternal H19 promoter is in a highly condensed inaccessible chromatin state (18).
Instead, we believe that the biallelic interactions noted for the EcoRI fragment are indicative of the general compaction of the whole locus, a finding that we also noted when we initially scanned for interactions across the region (data not shown). In fact, this compacted feature of the locus focused our attention on the parent-of-origin-specific interactions, as we reasoned that this was a good way to identify associations that were likely to be of functional significance.
As discussed above, our results are in one sense fully consistent with the idea that long-range interactions between enhancers and promoters (via DNA looping) are essential for transcriptional activation. That is, we consistently note promoter-enhancer association at active but never at inactive loci. However, our results also clearly demonstrate the phenomenon that most compellingly challenges the DNA looping/direct interaction models. Like all other enhancer blockers characterized to date, the H19ICR insulator is position dependent. That is, the endogenous ICR blocks maternal Igf2 but not maternal H19. Likewise, the ICR inserted on the H19R chromosome blocks H19 expression in muscle but not in liver cells. This position dependence is not predicted or explained by DNA looping models for enhancer activation of promoter transcription. Rather, position dependence is better explained by tracking models, which suggest that a positive vector originates from the enhancer and travels linearly along the chromosome until it reaches the relevant promoter (67). By this model, an enhancer-blocking element prevents further transmission of the insulator signal, perhaps by conferring directionality on the enhancer (31). One way to explain this apparent paradox is to emphasize the dynamic nature of the enhancer interactions with other cis sites. The loop between the enhancer and the promoter is presumably particularly stable and thus can be identified in the 3C assays. (Likewise, interactions of the enhancer with the active maternal insulator must be relatively stable to be identified by the 3C assay.) However, the many transitory interactions formed as the enhancer scans for appropriate partners are each not stable enough and so are not present in sufficient quantity to be identified in these assays.
In sum, we present analyses of the transcriptional activation and long-range DNA loop structures at the Igf2, H19, and Afp loci, comparing chromosomes with and without the H19ICR insulator element. In each case, we also compare maternal chromosomes, where the ICR functions as an enhancer blocker, with paternal chromosomes, where methylation of the ICR prevents CTCF binding and therefore blocks insulator function. Our results suggest that insulators are highly promiscuous in their ability to block activation of promoters and enhancers and that the ICR functions by disrupting the promoter-enhancer connections invariably associated with transcriptional activation. Instead, the ICR promotes alternative long-range interactions between itself and the blocked enhancer and promoter. The physical structures noted in this report are consistent with DNA looping and long-range interactions of promoters and enhancers being critical to gene activation. However, the position dependence of the enhancer-blocking activity suggests that this model is not adequate on its own to explain transcriptional activation. Rather, some sort of tracking model remains required to account for insulator function.
Acknowledgments
We thank Judy Kassis, Michael Miller, and Claudia Gebert for helpful discussions and advice.
This work was funded by the NICHD Intramural Research Program.
Footnotes
Published ahead of print on 5 March 2007.
REFERENCES
- 1.Bartolomei, M. S., A. L. Webber, M. E. Brunkow, and S. M. Tilghman. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev. 7:1663-1673. [DOI] [PubMed] [Google Scholar]
- 2.Bartolomei, M. S., S. Zemel, and S. M. Tilghman. 1991. Parental imprinting of the mouse H19 gene. Nature 351:153-155. [DOI] [PubMed] [Google Scholar]
- 3.Bell, A., and G. Felsenfeld. 1999. Stopped at the border: boundaries and insulators. Curr. Opin. Genet. Dev. 9:191-198. [DOI] [PubMed] [Google Scholar]
- 4.Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. [DOI] [PubMed] [Google Scholar]
- 5.Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. [DOI] [PubMed] [Google Scholar]
- 6.Brunkow, M. E., and S. M. Tilghman. 1991. Ectopic expression of the H19 gene in mice causes prenatal lethality. Genes Dev. 5:1092-1101. [DOI] [PubMed] [Google Scholar]
- 7.Cai, H., and M. Levine. 1995. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature 376:533-536. [DOI] [PubMed] [Google Scholar]
- 8.Cai, H., and P. Shen. 2001. Effects of cis arrangements of chromatin insulators on enhancer-blocking activity. Science 291:493-495. [DOI] [PubMed] [Google Scholar]
- 9.Capelson, M., and V. Corces. 2004. Boundary elements and nuclear organization. Biol. Cell 96:617-629. [DOI] [PubMed] [Google Scholar]
- 10.Carter, D. 2002. Long range chromatin regulatory interactions in vivo. Nat. Genet. 32:623-626. [DOI] [PubMed] [Google Scholar]
- 11.Cleard, F., Y. Moshkin, F. Karch, and R. Maeda. 2006. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat. Genet. 38:931-935. [DOI] [PubMed] [Google Scholar]
- 12.Constancia, M., W. Dean, S. Lopes, T. Moore, G. Kelsey, and W. Reik. 2000. Deletion of a silencer element in Igf2 results in loss of imprinting independent of H19. Nat. Genet. 26:203-206. [DOI] [PubMed] [Google Scholar]
- 13.Cranston, M., T. Spinka, D. Elson, and M. Bartolomei. 2001. Elucidation of the minimal sequence required to imprint H19 transgenes. Genomics 73:98-107. [DOI] [PubMed] [Google Scholar]
- 14.DeChiara, T. M., E. J. Robertson, and A. Efstratiadis. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64:849-859. [DOI] [PubMed] [Google Scholar]
- 15.Dekker, J. 2002. Capturing chromosome conformation. Science 295:1306-1311. [DOI] [PubMed] [Google Scholar]
- 16.Elson, D. A., and M. S. Bartolomei. 1997. A 5′ differentially methylated sequence and the 3′-flanking region are necessary for H19 transgene imprinting. Mol. Cell. Biol. 17:309-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feil, R., J. Walter, N. D. Allen, and W. Reik. 1994. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development 120:2933-2943. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson-Smith, A. C., H. Sasaki, B. M. Cattanach, and M. A. Surani. 1993. Parental-origin-specific epigenetic modifications of the mouse H19 gene. Nature 362:751-755. [DOI] [PubMed] [Google Scholar]
- 19.Gaszner, M., and G. Felsenfeld. 2005. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 7:703-713. [DOI] [PubMed] [Google Scholar]
- 20.Geyer, P., and I. Clark. 2002. Protecting against promiscuity: the regulatory role of insulators. Cell. Mol. Life Sci. 59:2112-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geyer, P., and V. G. Corces. 1992. DNA position-specific repression of transcription by a Drosophila zinc-finger protein. Genes Dev. 6:1865-1873. [DOI] [PubMed] [Google Scholar]
- 22.Gould, T. D., and K. Pfeifer. 1998. Imprinting of mouse Kvlqt1 is developmentally regulated. Hum. Mol. Gen. 7:483-487. [DOI] [PubMed] [Google Scholar]
- 23.Grandjean, V., L. O'Neill, T. Sado, B. Turner, and A. Ferguson-Smith. 2001. Relationship between DNA methylation, histone H4 acetylation and gene expression in the mouse imprinted Igf2-H19 domain. FEBS Lett. 488:165-169. [DOI] [PubMed] [Google Scholar]
- 24.Hark, A. T., C. J. Schoenherr, D. J. Katz, R. S. Ingram, J. M. Levorse, and S. M. Tilghman. 2000. CTCF mediates methylation-sensitive enhancer blocking activity at the H19/Igf2 locus. Nature 405:486-489. [DOI] [PubMed] [Google Scholar]
- 25.Kaffer, C., A. Grinberg, and K. Pfeifer. 2001. Regulatory mechanisms at the mouse Igf2/H19 locus. Mol. Cell. Biol. 21:8189-8196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaffer, C. R., M. Srivastava, K. Park, E. Ives, S. Hsieh, J. Batlle, A. Grinberg, S. P. Huang, and K. Pfeifer. 2000. A transcriptional insulator at the imprinted H19/Igf2 locus. Genes Dev. 14:1908-1919. [PMC free article] [PubMed] [Google Scholar]
- 27.Kanduri, C., V. Pant, D. Loukinov, E. Pugacheva, C. Qi, A. Wolffe, R. Ohlsson, and V. Lobanenkov. 2000. Functional association of CTCF with the insulator upstream of the H19 gene is parent-of-origin specific and methylation-sensitive. Curr. Biol. 10:853-856. [DOI] [PubMed] [Google Scholar]
- 28.Kellum, R., and P. Schedl. 1992. A group of scs elements function as domain boundaries in an enhancer-blocking assay. Mol. Cell. Biol. 12:2424-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kellum, R., and P. Schedl. 1991. A position-effect assay for boundaries of higher order chromosomal domains. Cell 64:941-950. [DOI] [PubMed] [Google Scholar]
- 30.Klenova, E. M., H. Morse, R. Ohlsson, and V. V. Lobanenkov. 2002. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin. Cancer Biol. 12:399-414. [DOI] [PubMed] [Google Scholar]
- 31.Krebs, J., and M. Dunaway. 1998. The scs and scs′ insulator elements impart a cis requirement on enhancer-promoter interactions. Mol. Cell 1:301-308. [DOI] [PubMed] [Google Scholar]
- 32.Kuhn, E., M. Viering, K. Rhodes, and P. Geyer. 2003. A test of insulator interactions in Drosophila. EMBO J. 22:2463-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhn, E. J., and P. Geyer. 2003. Genomic insulators: connecting properties to mechanisms. Curr. Opin. Cell Biol. 15:259-265. [DOI] [PubMed] [Google Scholar]
- 34.Kuo, M., and C. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 35.Kurukuti, S., V. Tiwari, G. Tavoosidana, E. Pugacheva, A. Murrell, Z. Zhao, V. Lobanenkov, W. Reik, and R. Ohlsson. 2006. CTCF binding at the H19 imprinting control region mediates maternally inherited higher-order chromatin conformation to restrict enhancer access to Igf2. Proc. Natl. Acad. Sci. USA 103:10684-10689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labrador, M., and V. Corces. 2002. Setting the boundaries of chromatin domains and nuclear organization. Cell 111:151-154. [DOI] [PubMed] [Google Scholar]
- 37.Leighton, P. A., J. R. Saam, R. S. Ingram, C. L. Stewart, and S. M. Tilghman. 1995. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev. 9:2079-2089. [DOI] [PubMed] [Google Scholar]
- 38.Ling, J., T. Li, J. Hu, T. Vu, H. Chen, X. Qiu, A. Cherry, and A. R. Hoffman. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312:269-272. [DOI] [PubMed] [Google Scholar]
- 39.Muravyova, E., A. Golovnin, E. Gracheva, A. Parshikov, T. Belenkaya, V. Pirrotta, and P. Georgiev. 2001. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science 291:495-498. [DOI] [PubMed] [Google Scholar]
- 40.Murrell, A., S. Heeson, and W. Reik. 2004. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat. Genet. 36:889-893. [DOI] [PubMed] [Google Scholar]
- 41.Ohlsson, R., R. Renkawitz, and V. Lobanenkov. 2001. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 17:520-527. [DOI] [PubMed] [Google Scholar]
- 42.Osborne, C., L. Chakalova, K. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065-1071. [DOI] [PubMed] [Google Scholar]
- 43.Pai, C.-Y., E. Lei, D. Ghosh, and V. Corces. 2004. The centrosomal protein CP190 is a component of the gypsy insulator. Mol. Cell 16:737-748. [DOI] [PubMed] [Google Scholar]
- 44.Park, K., E. Sellars, A. Grinber, S. Huang, and K. Pfeifer. 2004. The H19 differentially methylated region marks the parental origin of a heterologous locus without gametic DNA methylation. Mol. Cell. Biol. 24:3588-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfeifer, K., P. A. Leighton, and S. M. Tilghman. 1996. The structural H19 gene is required for transgene imprinting. Proc. Natl. Acad. Sci. USA 93:13876-13883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prioleau, M., P. Nony, M. Simpson, and G. Felsenfeld. 1999. An insulator element and condensed chromatin region separate the chicken Β-globin locus from an independently regulated erythroid-specific folate receptor gene. EMBO J. 18:4035-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Recillas-Targa, F., A. Bell, and G. Felsenfeld. 1999. Positional enhancer-blocking activity of the chicken Β-globin insulator in transiently transfected cells. Proc. Natl. Acad. Sci. USA 96:14354-14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoenherr, C., J. Levorse, and S. M. Tilghman. 2003. CTCF maintains differential methylation at the Igf2/H19 locus. Nat. Genet. 33:66-69. [DOI] [PubMed] [Google Scholar]
- 49.Scott, K., and P. Geyer. 1995. Effects of the Su(Hw) insulator protein on the expression of the divergently transcribed Drosophila yolk sac protein genes. EMBO J. 14:6258-6267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spana, C., D. Harrison, and V. Corces. 1988. The Drosophila melanogaster suppressor of Hairy-wing protein binds to specific sequences of the gypsy retrotransposon. Genes Dev. 2:1414-1423. [DOI] [PubMed] [Google Scholar]
- 51.Spear, B. 1999. Alpha-fetoprotein gene regulation: lessons from transgenic mice. Semin. Cancer Biol. 9:109-116. [DOI] [PubMed] [Google Scholar]
- 52.Spilianakis, C., and R. Flavell. 2004. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 5:1017-1027. [DOI] [PubMed] [Google Scholar]
- 53.Srivastava, M., E. Frolova, B. Rottinghaus, S. Boe, A. Grinberg, E. Lee, P. Lover, and K. Pfeifer. 2003. Imprint control element mediated secondary methylation imprints at the Igf2/H19 locus. J. Biol. Chem. 278:5977-5983. [DOI] [PubMed] [Google Scholar]
- 54.Srivastava, M., S. Hsieh, A. Grinberg, L. Williams-Simon, S.-P. Huang, and K. Pfeifer. 2000. H19 and Igf2 monoallelic expression is regulated in two distinct ways by a shared cis acting element. Genes Dev. 14:1186-1195. [PMC free article] [PubMed] [Google Scholar]
- 55.Szabo, P., S. Tang, M. Reed, F. Silva, W. Tsark, and J. Mann. 2002. The chicken beta-globin insulator conveys chromatin boundary activity but not imprinting at the mouse Igf2/H19 domain. Development 129:897-904. [DOI] [PubMed] [Google Scholar]
- 56.Szabo, P., S. Tang, A. Rentsendorj, G. Pfeifer, and J. R. Mann. 2000. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 10:607-610. [DOI] [PubMed] [Google Scholar]
- 57.Szabo, P. A., G. P. Pfeifer, and J. R. Mann. 1998. Characterization of novel parent-specific epigenetic modifications upstream of the imprinted mouse H19 gene. Mol. Cell. Biol. 18:6767-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thorvaldsen, J., M. Mann, O. Nwoko, K. Duran, and M. S. Bartolomei. 2002. Analysis of sequence upstream of the endogenous H19 reveals elements both essential and dispensable for imprinting. Mol. Cell. Biol. 22:2450-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thorvaldsen, J. L., K. L. Duran, and M. S. Bartolomei. 1998. Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev. 12:3693-3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tolhuis, B. 2002. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol. Cell 10:1453-1465. [DOI] [PubMed] [Google Scholar]
- 61.Tremblay, K. D., K. L. Duran, and M. S. Bartolomei. 1997. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 17:4322-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tremblay, K. D., J. R. Saam, R. S. Ingram, S. M. Tilghman, and M. S. Bartolomei. 1995. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat. Genet. 9:407-413. [DOI] [PubMed] [Google Scholar]
- 63.West, A., and P. Fraser. 2005. Remote control of gene transcription. Hum. Mol. Genet. 14:R101-R111. [DOI] [PubMed] [Google Scholar]
- 64.West, A., M. Gaszner, and G. Felsenfeld. 2002. Insulators: many functions, many mechanisms. Genes Dev. 16:271-288. [DOI] [PubMed] [Google Scholar]
- 65.Yoo-Warren, H., V. Pachnis, R. S. Ingram, and S. M. Tilghman. 1988. Two regulatory domains flank the mouse H19 gene. Mol. Cell. Biol. 8:4707-4715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yusufzai, T., H. Tagami, Y. Nakatani, and G. Felsenfeld. 2004. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13:291-298. [DOI] [PubMed] [Google Scholar]
- 67.Zhao, H., and A. Dean. 2005. Organizing the genome: enhancers and insulators. Biochem. Cell Biol. 83:516-524. [DOI] [PubMed] [Google Scholar]
- 68.Zhao, Z., G. Tavoosidana, M. Sjoelinder, A. Goendoer, P. Mariano, S. Wang, C. Kanduri, M. Lezcano, K. Sandhu, W. Singh, V. Pant, V. Tiwari, S. Kurukuti, and R. Ohlsson. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat. Genet. 38:1341-1347. [DOI] [PubMed] [Google Scholar]