Abstract
La is an RNA-processing-associated phosphoprotein so highly conserved that the human La protein (hLa) can replace the tRNA-processing function of the fission yeast La protein (Sla1p) in vivo. La proteins contain multiple trafficking elements that support interactions with RNAs in different subcellular locations. Prior data indicate that deletion of a nuclear retention element (NRE) causes nuclear export of La and dysfunctional processing of associated pre-tRNAs that are spliced but 5′ and 3′ unprocessed, with an accompanying decrease in tRNA-mediated suppression, in fission yeast. To further pursue these observations, we first identified conserved residues in the NREs of hLa and Sla1p that when substituted mimic the NRE deletion phenotype. NRE-defective La proteins then deleted of other motifs indicated that RNA recognition motif 1 (RRM1) is required for nuclear export. Mutations of conserved RRM1 residues restored nuclear accumulation of NRE-defective La proteins. Some RRM1 mutations restored nuclear accumulation, prevented disordered pre-tRNA processing, and restored suppression, indicating that the tRNA-related activity of RRM1 and its nuclear export activity could be functionally separated. When mapped onto an hLa structure, the export-sensitive residues comprised surfaces distinct from the RNA-binding surface of RRM1. The data indicate that the NRE has been conserved to mask or functionally override an equally conserved nuclear export activity of RRM1. The data suggest that conserved elements mediate nuclear retention, nuclear export, and RNA-binding activities of the multifunctional La protein and that their interrelationship contributes to the ability of La to engage its different classes of RNA ligands in different cellular locations.
La is an abundant protein whose capacity to bind a variety of noncoding RNAs and mRNAs lends itself to numerous activities (26). La proteins from yeast to human have been implicated in the production of tRNAs, rRNAs, ribosomal proteins, and other components of the translational machinery (13, 16, 19, 20). In human cells, most La is phosphorylated on serine-366 by protein kinase CK2, resides in the nucleoplasm, and is associated with nascent pre-tRNAs (17, 32). Nonphosphorylated La is most concentrated in the nucleolus (15, 16) and was independently found at tRNA and other RNA polymerase III-transcribed genes (4), but it also resides in the cytoplasm associated with 5′TOP mRNAs that encode ribosomal proteins and translation factors (16). Trafficking signals in human La include a nuclear localization signal (NLS), a nuclear retention element (NRE) first identified by microinjection of Xenopus oocytes and later confirmed in fission yeast (14, 35), and a nucleolar localization signal (10, 15). Although trafficking may be important for its different activities (19), knowledge of the functional significance of alterations in La trafficking in specific RNA pathways is limited.
Sequence-specific binding to 3′ UUU-OH, the termination motif found on nascent pre-tRNAs and other transcripts synthesized by RNA polymerase III, is the activity responsible for the best-established function of La proteins, protection of RNA ligands from 3′ exonucleolytic digestion (11, 12; reviewed in references 27 and 41). Nascent pre-tRNAs require 5′ and 3′ end processing, numerous modifications, CCA addition, aminoacylation, nuclear export, and splicing, if necessary, prior to the appearance of a mature functional tRNA in the cytoplasm (9). Recent findings have revealed that the tRNA production pathway is highly complex in biochemistry, spatial organization, and sequential order (9, 29). In what order are the 5′ leader, intron, and 3′ trailer normally removed from a pre-tRNA, and in which cellular compartments do these reactions occur?
In yeast, removal of the 5′ leader by RNase P appears to be the earliest processing step for most pre-tRNAs, occurring at or near tRNA transcription sites, in the nucleolus (2; reviewed in references 6, 9, and 30). Consistent with this and the idea that La is the first protein to interact with nascent pre-tRNAs, La directs 5′ processing to precede 3′ processing, since this order is reversed in its absence (17, 43). Removal of the 3′ trailer is believed to occur in the nucleus, as do some modifications, CCA addition, and aminoacylation (9, 24, 42). The discovery that tRNA splicing occurs in the cytoplasm in yeast (44) is consistent with splicing occurring after 5′ end processing (9, 29). An equally surprising discovery which revealed further complexity was that tRNAs can move in a “retrograde” manner, from the cytoplasm to the nucleus (29, 33, 36). Retrograde tRNA transport may also occur in mammals (45). Although more data are needed, this raises the possibility that nuclear enzymes may have more than one chance to process pre-tRNA, once prior to export and again following retrograde import. The recent advances raise new questions as to the order and transport aspects of tRNA processing.
In contrast to the disordering of 5′ and 3′ end processing that occurs in yeast mutants lacking La (43), removal of the NRE from either human La (hLa) or Schizosaccharomyces pombe La (Sla1p) (hLaΔNRE or Sla1pΔNRE, respectively) leads to disorder in pre-tRNA splicing relative to end processing (hLaΔNRE = hLaΔ316-332 and Sla1pΔNRE = Sla1pΔ234-255) (14). In the presence of an NLS but absence of the NRE, La enters nuclei and associates with pre-tRNAs but is inappropriately exported to the cytoplasm without the pre-tRNAs undergoing end processing (14). Thus, deletion of the NRE promotes the nuclear export of La, which causes associated pre-tRNAs to bypass nuclear 5′ and 3′ end processing and produce spliced pre-tRNAs that retain their 5′ and 3′ extensions, which do not support tRNA-mediated suppression (14). Since, as reviewed above, the current understanding is that 5′ end processing is an early step followed by nuclear export and cytoplasmic splicing, we will refer to the processing defect caused by NRE-deficient La proteins as disordered because it causes splicing to occur prior to end processing.
Both the S. pombe La and human La proteins deleted of their NREs produce indistinguishable effects (14), suggesting that the NRE has been conserved to mask or functionally override an equally conserved nuclear export element that affects pre-tRNA processing. These findings coupled with discoveries of cytoplasmic splicing and retrograde transport led to a suggestion that La may be involved in tRNA transport (29).
Our laboratory uses a suppressor tRNASerUGA that suppresses a nonsense codon in ade6-704 to study the role of La in tRNA biogenesis in the fission yeast, S. pombe (7, 11, 12, 14, 17). Unsuppressed ade6-704 results in red colonies, and suppression results in pink to white colonies. Substitutions in pre-tRNASerUGA that cause dependency on Sla1p or hLa for maturation have been described (11). In this system, La-dependent processing is a primary determinant of mature tRNASerUGA levels, and suppression is dependent on accumulation of mature tRNASerUGA (7, 12, 14, 17). We used this system to investigate determinants of the nuclear retention and export of Sla1p and hLa and the effects of altered localization on tRNA processing. We first attempted to distinguish between two possible models: in the first, the La NRE would contribute directly to pre-tRNA processing and its deletion would secondarily lead to export of La-associated pre-tRNAs that bypassed end processing in the nucleus. In the second model, the NRE does not contribute directly to processing but functions only to prevent export of La and associated pre-tRNAs. We present data supporting the second model, in which normal pre-tRNA processing by NRE-defective La protein was rescued by mutation of conserved residues in hLa and Sla1p that are important for the nuclear export of LaΔNRE. The observed rescue could also be obtained through inhibition of LaΔNRE export by leptomycin B (LMB) treatment. We characterized highly conserved residues both in the NRE and in RNA recognition motif 1 (RRM1) that are required for nuclear retention and export, respectively, of Sla1p and hLa in fission yeast. We found mutations of conserved RRM1 residues that block nuclear export of NRE-defective La proteins and restore normal pre-tRNA processing and tRNA-mediated suppression and modeled these onto available high-resolution hLa structures. Our data indicate that nuclear export of La is detrimental to tRNA maturation. The data are consistent with a model in which conserved structural features of La protein mediate La nuclear export and retention and the interrelationship of these trafficking elements determines the ability of La to engage RNAs residing in different cellular locations.
MATERIALS AND METHODS
Northern blotting.
Northern blotting using LysCUU-int (40) and U1 probes was as described previously (17).
Site-specific mutagenesis.
Site-specific mutagenesis was performed with QuikChange XL (Stratagene) using pREP4-La and pRep4-Sla1 or their ΔNRE or KK derivatives as templates (17). All constructs were verified by sequencing. Deletions of the La motif (LM), RRM1, and RRM2 were designed based on the domain boundaries indicated for the published structures (1, 18); details are in Table 1.
TABLE 1.
hLa and Sla1p constructs
| Protein
|
Localizationa | Suppressionb | Processingc | |
|---|---|---|---|---|
| No. | Name | |||
| 1 | hLa (wild type) | N | +++ | N |
| 2 | hLaΔNRE (hLaΔ316-332)d | C | − | D |
| 3 | hLaK316A/K317A (hLaKK) | C | − | D |
| 4 | hLaΔNREΔLM (hLaΔ7-107 Δ316-332) | C | − | |
| 5 | hLaKKΔLM (hLaK316A/K317A Δ7-107) | C | − | |
| 6 | hLaΔNREΔRRM1 (hLaΔ316-33 Δ101-194) | N | − | |
| 7 | hLaKKΔRRM1 (hLaK316A/K317A Δ101-194) | N | − | |
| 8 | hLaΔNREΔRRM2 (hLaΔ223-332) | C | − | |
| 9 | hLaKK-E103A/Y104A/K105A/V108A/R111A (A2) | N | − | N |
| 10 | hLaKK-V137A/I140A/M142A/R143A/R144A (A5) | N | − | N |
| 11 | hLaKK-K148A/F150A/K151A/S153A (A6) | N | − | N |
| 12 | hLaKK-E132A/D133A (A4.5) | N | + | N |
| 13 | hLaKK-Y175A/K176A/E177A/D179A/L180A/L181A/I182A/I183A (A8) | C | − | |
| 14 | hLaKK-Y114A | C | − | |
| 15 | hLaKK-F155A | C | − | |
| 16 | hLaKK-E162A | C | − | |
| 17 | hLaKK-D187A | C | − | |
| 18 | hLaKK-L124A/D125A | C | ||
| 19 | hLaΔNRE-V137A/I140A/M142A/R143A/R144A (A5) | N | − | |
| 20 | hLaΔNRE-K148A/F150A/K151A/S153A (A6) | N | − | |
| 21 | hLaΔNRE-E132A/D133A (A4.5) | N | ++ | N |
| 22 | hLaΔNRE-F150A | N | + | N |
| 23 | hLaΔNRE-I140A/M142A | C | − | |
| 24 | hLa-E103A/Y104A/K105A/V108A/R111A (A2) | N | ||
| 25 | hLa-E132A/D133A (A4.5) | N | +++ | |
| 26 | hLa-V137A/I140A/M142A/R143A/R144A (A5) | N | − | |
| 27 | hLa-K148A/F150A/K151A/S153A (A6) | N | − | |
| 28 | hLa-F150A | N | ++ | |
| 29 | Sla1p (wild type) | N | +++ | |
| 30 | Sla1pΔNRE | C | − | D |
| 31 | Sla1pKK (K234A/K235A) | C | − | D |
| 32 | Sla1pKK-E177A/E178A | N | + | |
| 33 | Sla1pKK-F196A | N | + | |
| 34 | Sla1pKK-V186A/M188A | C | − | |
| 35 | Sla1pΔNRE-E177A/E178A | N | ++ | N |
| 36 | Sla1pΔNRE-F196A | N | + | N |
| 37 | Sla1pΔNRE-V186A/M188A | C | − | |
| 38 | Sla1p-V186A/M188A | − | ||
| 39 | Sla1p-F196A | +++ | ||
| 40 | Sla1p-E177A/E178A | ++ | ||
Localization determined by immunofluorescence. N, nuclear; C, cytoplasmic; blank space, not determined.
tRNA-mediated suppression. +++, full suppression; −, no suppression; blank space, not determined.
D, disordered spliced pre-tRNA; N, no disordered spliced pre-tRNA; blank space, not determined.
From reference 12.
IF.
Immunofluorescence (IF) using primary antibody (“Go” anti-hLa) or anti-Sla1p, each at a 1:1,000 dilution, followed by fluorescein isothiocyanate (FITC)-conjugated goat anti-human or anti-rabbit immunoglobulin G (Jackson Immunoresearch) at 1:200, was as described previously (14).
tRNA-mediated suppression assay.
The tRNA-mediated suppression was as described previously (17), using 10 mg/liter adenine. LMB was obtained from Sigma.
Molecular modeling.
Molecular modeling was done using MacPyMOL (DeLano Scientific LLC, Palo Alto, CA).
RESULTS
Conserved residues in the NREs of Sla1p and hLa that are important for function.
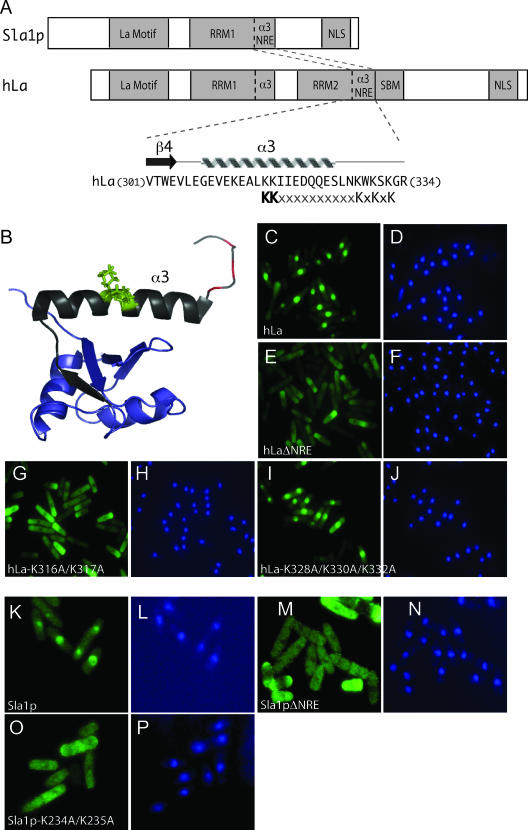
Although the architectures of hLa and Sla1p differ, their NREs are each located adjacent to an RRM (Fig. 1A and B). Attempts to decipher the determinants of the hLa NRE should consider the consensus bipartite NLS [KKx(10)KxKxK] (Fig. 1A) at amino acids 316 to 332 (34). The upstream KK residues of the NRE are highly conserved in the predicted NREs of several La proteins (Fig. 1A), whereas the downstream basic residues are less conserved (14). In a prior investigation that used deletion mutagenesis, no distinction was made between the upstream and downstream basic residues (14). Here, we examined full-length hLa proteins harboring either a double substitution (hLa-K316A/K317A) or a triple substitution (hLa-K328A/K330A/K332A) and compared them to the NRE deletion mutant (hLaΔNRE), and wild-type hLa by IF (Fig. 1C to J). hLa and hLaΔNRE were nuclear and cytoplasmic, respectively, as expected (14) (Fig. 1C versus E), whereas hLa-K316A/K317A was cytoplasmic (Fig. 1G) and hLa-K328A/K330A/K332A was nuclear (Fig. 1I). These data suggest that the upstream, more highly conserved lysines at positions 316 to 317 of hLa are important for NRE function, while the more divergent basic residues at 328 to 332 are not. Substitution of the Sla1p homologous NRE upstream lysines also led to cytoplasmic accumulation, since Sla1p-K234A/K235A was indistinguishable from Sla1pΔNRE (Fig. 1K to P). As will be seen below, the KK substitutions inactivate hLa and Sla1p for tRNA-mediated suppression and cause disordered pre-tRNA splicing indistinguishable from that of the NRE-deleted proteins. We conclude that one or both of the conserved NRE lysines, hLa K316/K317 and Sla1p K234A/K235A, are important determinants of nuclear retention in fission yeast. Hereafter, we also refer to the NRE-defective hLa-K316A/K317A and Sla1p-K234A/K235A as hLaKK and Sla1pKK, respectively.
FIG. 1.
Point mutations of conserved residues in the NRE mimic an NRE deletion. (A) Schematic of human and S. pombe La proteins. The β4 strand of hLa RRM2 and adjacent α3 helix comprising the NRE are represented, with corresponding sequence below (18). A potential consensus bipartite NLS at positions 316 to 332, whose upstream KK residues are in bold, is indicated below (see text). (B) NMR structure model of hLa RRM2 (18). The RRM is blue, except for β4 and the α3 helix in black. The side chains of the conserved lysines at positions 316 and 317 are green, and the backbone chain of the lysines in the KXKXK sequence is shown in red. (C to P) Anti-La indirect IF using fluorescence-conjugated secondary antibody (FITC, green) and DAPI (4′,6′-diamidino-2-phenylindole) (blue). (C) hLa, FITC. (D) hLa, DAPI. (E) hLaΔΝRE, FITC. (F) ΔΝRE DAPI. (G) hLa-K316A/K317A, FITC. (H) hLa-K316A/K317A, DAPI. (I) hLa-K328A/K330A/K332A, FITC. (J) hLa-K328A/K330A/K332A, DAPI. (K) Sla1p, FITC. (L) Sla1p, DAPI. (M) Sla1pΔΝRE, FITC. (N) Sla1pΔΝRE, DAPI. (O) Sla1p-K234A/K235A FITC. (P) Sla1p-K234A/K235A DAPI. Panels K to P are at a slightly higher magnification than panels C to J.
LMB rescues the tRNA-mediated suppression activity of hLaΔNRE.
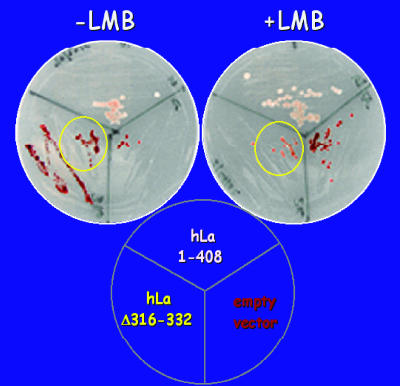
Nuclear accumulation of NRE-deleted La proteins was restored in S. pombe by LMB, an inhibitor of Crm1-mediated nuclear export (14). We reasoned that if inactivity of tRNA-mediated suppression by hLaΔNRE was due solely to nuclear export, LMB might restore its suppression activity. Alternatively, if the NRE was more directly required for tRNA maturation, hLaΔNRE may remain inactive even if restored to the nucleus. S. pombe cells harboring hLa, hLaΔNRE, or empty vector were grown in the presence of LMB. LMB had to be used at a lower concentration than typically used for an IF assay to minimize toxicity during growth (data not shown). Even so, LMB led to significant recovery of suppression for hLaΔNRE but not the control cells (Fig. 2). This suggested that hLaΔNRE can support tRNA-mediated suppression if retained in the nucleus and therefore that we might be able to identify second-site mutations in NRE-defective La proteins that restore nuclear accumulation and tRNA-mediated suppression.
FIG. 2.
LMB restores tRNA-mediated suppression activity to hLaΔNRE. tRNA-mediated suppression of ade6-704 in the presence or absence of LMB is shown. Cells were transformed with empty vector, full-length hLa, or hLaΔNRE. Lighter color indicates that LMB increases suppression by hLaΔNRE but not hLa or empty vector.
Deletion of RRM1 restores nuclear accumulation of NRE-defective La proteins.
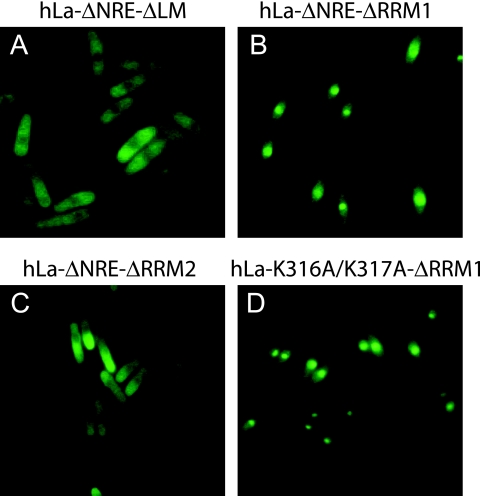
In an attempt to restore nuclear accumulation of hLaΔNRE, we separately deleted the LM, RRM1 and RRM2 (Fig. 3), based on the domain boundaries indicated for the published structures (1, 18). Because of the physical proximity of NRE and RRM2 we anticipated that RRM2 harbored a nuclear export determinant and that its deletion would block export and restore nuclear accumulation. However, deletion of RRM2 did not restore nuclear accumulation (Fig. 3C). Moreover, cytoplasmic accumulation of the resulting protein, hLaΔNREΔRRM2, was fully reversed to a nuclear pattern by LMB, indicating the presence of a nuclear export determinant (data not shown). Likewise, deletion of the LM did not restore nuclear accumulation (Fig. 3A). By contrast to deletion of the LM and RRM2, deletion of RRM1 did restore nuclear accumulation, in the contexts of both hLaΔNRE and hLaKK (Fig. 3B and D). We conclude that RRM1 mediates nuclear export of NRE-defective La proteins. The conservation of RRM1 fits with this conclusion, since hLa and Sla1p responded indistinguishably, by multiple functional criteria, to deletion of their NREs in fission yeast (14).
FIG. 3.
RRM1 is required for nuclear export of hLaΔΝRE. IF of S. pombe cells transformed with various hLa NRE-defective constructs that lack the LM (hLaΔΝRE-ΔLM) (A), RRM1 (hLaΔΝRE-ΔRRM1) (B), RRM2 (hLaΔΝRE-ΔRRM2) (C), or RRM1 in the context of K316A/K317A (hLaK316A/K317A-ΔRRM1) (D) is shown.
Conserved RRM1 residues direct nuclear export of NRE-defective hLa and Sla1p.
While most proteins that undergo Crm1-dependent export contain a nuclear export sequence (NES) that is recognized by Crm1p, others do not (21, 38, 39). We were unable to identify a functional NES of the consensus sequence, L-X(2,3)-(LIVFM)-X(2,3)-L-X-(LI) (22), in hLaΔNRE and Sla1pΔNRE, despite examinations of many mutations (not shown).
Mapping of the nuclear export activity to RRM1 raised the issue of whether this activity would overlap with or interfere with the principal activity attributed to an RRM, RNA binding, which maps to the β-sheet surface of the RRM and adjacent loops (28). We showed that the β-sheet surface of RRM1 is required for normal tRNA maturation and tRNA-mediated suppression (11). Accordingly, LaΔNREΔRRM1, although nuclear, was inactive for suppression (Table 1). We therefore sought to isolate site-specific mutations in RRM1 that would inactivate nuclear export of NRE-defective La proteins and restore functional pre-tRNA processing and tRNA-mediated suppression. This would indicate that the nuclear export activity and the tRNA maturation activity of La RRM1 might be distinct and/or separable.
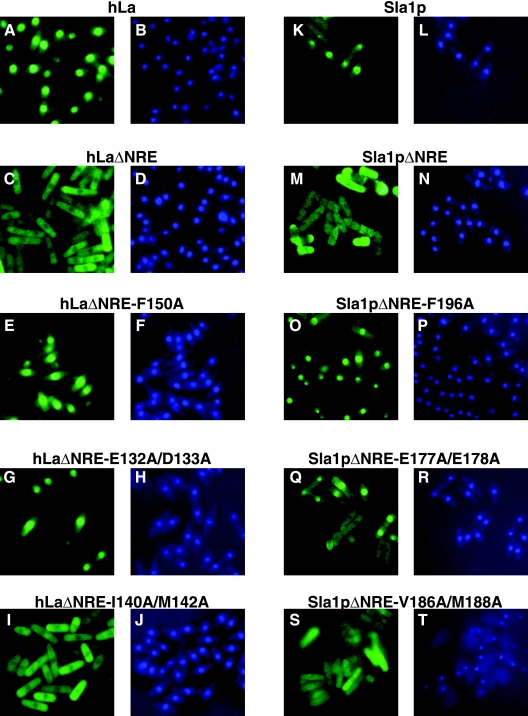
A series of alanine substitutions of conserved surface side chains in RRM1 of hLa were introduced into hLaKK and hLaΔNRE (Table 1, proteins 9 to 23) as well as Sla1pKK and Sla1pΔNRE (Table 1, proteins 31 to 37) proteins. Some of these substitutions restored nuclear accumulation, while others did not (Fig. 4; Table 1). Especially noteworthy are the hLa E132/D133 and homologous Sla1p E177A/E178A substitutions in ΔNRE and NRE-substituted (KK) proteins, as well as hLa F150 and homologous Sla1p F196 substitutions, which largely restored nuclear accumulation (Fig. 4; Table 1). We note that Sla1pΔNRE-E177A/E178A was nuclear in the majority of the cells, while it was cytoplasmic in others (Fig. 4Q). A greater majority of hLaΔNRE-E132/D133 was nuclear (Fig. 4G). Although these observations might be explained by cell cycle-dependent localization, we note that nuclear staining was in general less complete with anti-Sla1p antibodies, even in wild-type cells, than with anti-hLa (data not shown).
FIG. 4.
Conserved RRM1 residues important for nuclear export of hLaΔΝRE and Sla1pΔNRE. IF of S. pombe cells transformed with various hLa NRE-defective constructs, as described for Fig. 1, is shown. (A) hLa, FITC. (B) hLa, DAPI. (C) hL,aΔΝRE FITC. (D) hLaΔΝRE, DAPI. (E) hLaΔΝRE-F150A, FITC. (F) hLaΔΝRE-F150A, DAPI. (G) hLaΔΝRE-E132A/D133A, FITC. (H) hLaΔΝRE-E132A/D133A, DAPI. (I) hLaΔΝRE-I140A/M142A, FITC. (J) hLaΔΝRE-I140A/M142A, DAPI. (K) Sla1p, FITC. (L) Sla1p, DAPI. (M) Sla1pΔNRE, FITC. (N) Sla1pΔΝRE, DAPI. (O) Sla1pΔΝRE-F196A, FITC. (P) Sla1pΔΝRE-F196A, DAPI. (Q) Sla1pΔΝRE-E177A/E178A, FITC. (R) Sla1pΔΝRE-E177A/E178A, DAPI. (S) Sla1pΔΝRE-V186A/M188A, FITC. (T) Sla1pΔΝRE-V186A/M188A, DAPI.
Mutations that restore nuclear accumulation of NRE-defective La proteins also reverse accumulation of disordered spliced pre-tRNA.
Northern analysis was performed to address two issues: whether hLaKK causes disordered spliced pre-tRNA, similar to the case for hLaΔNRE, and whether the RRM1 mutations that restore nuclear accumulation of NRE-defective proteins also reverse the accumulation of the disordered spliced pre-tRNA.
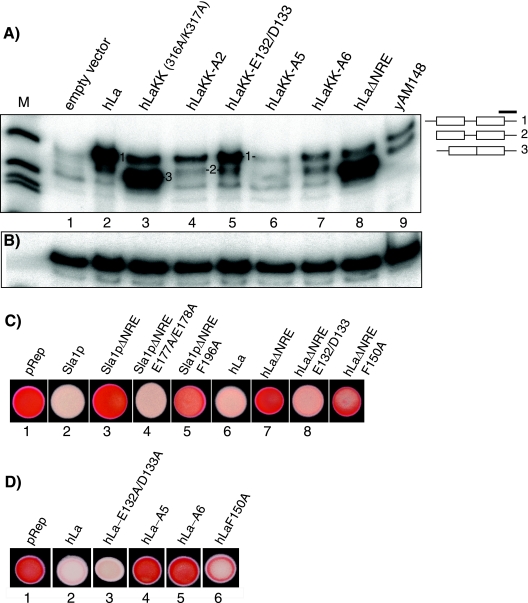
Probes directed to distinct regions of tRNALysCUU precursors have characterized multiple pre-tRNA intermediates in Sla1Δ cells expressing ectopic La proteins (11, 14, 17, 40). hLa produces a pattern in which nascent pre-tRNALysCUU is the most prominent species (Fig. 5A, lane 2, band 1), as noted previously (14). In sharp contrast to the pattern with hLa, hLaKK led to the disordered spliced pre-tRNA (Fig. 5A, lane 3, band 3) that is characteristic of hLaΔNRE (lane 8) (14).
FIG. 5.
Second-site mutations of NRE-defective La proteins that restore nuclear accumulation, prevent accumulation of disordered spliced pre-tRNA, and rescue tRNA-mediated suppression. (A) Northern blot of RNA from control cells (lanes 1 and 9) or cells expressing the La proteins indicated above lanes 2 to 8. Bands corresponding to the pre-tRNALysCUU intermediates as previously characterized are numbered and schematically represented on the right, band 3 reflects the disordered spliced pre-tRNA species (14), and the thick horizontal line represents the 3′ trailer probe used for detection. (B) The same blot as in panel A probed for U1 snRNA. (C and D) Suppression data for selected mutants, as indicated above the spots.
RRM1 mutations that restored nuclear accumulation of hLaKK-derived proteins led to much less, if any, of the disordered spliced pre-tRNA (Fig. 5A, lanes 4 to 7). In addition, some of the RRM1 mutants led to more of the nascent pre-tRNA (band 1) than others (Fig. 5A, lanes 4 to 7), a reproducible characteristic more similar to the pattern for hLa and also observed for hLaΔNRE RRM1 mutated proteins (Table 1 and data not shown). Immunoblot analysis indicated that the wild-type and mutated proteins were expressed at comparable levels (data not shown). Sla1pΔNRE-homologous RRM1 mutants that restored nuclear accumulation also lost the disordered spliced pre-tRNA (e.g., proteins 35 and 36 in Table 1).
A subset of RRM1 mutations restore nuclear accumulation and tRNA-mediated suppression to NRE-defective La proteins.
Some RRM1 mutations that restored nuclear accumulation also restored tRNA-mediated suppression, while others did not (Fig. 5C; Table 1). hLaΔNRE-E132A/D133A restored nuclear accumulation and suppression, as did the homolog, Sla1pΔNRE-E177A/E178A (Fig. 5C). It is noteworthy that hLaKK-E132/D133, which led to more of band 1 than the other hLaKK-RRM1 mutants (Fig. 5A), as also observed for hLaΔNRE-E132/D133 (not shown), also showed the highest suppression activity among the hLa RRM1 mutants (Fig. 5C). hLaΔNRE-F150A and its homolog Sla1pΔNRE-F196A also restored some suppression (Fig. 5C). These RRM1 mutants corroborate the LMB data which suggested that nuclear accumulation of NRE-defective La proteins can enable them for suppression.
Multiple simultaneous substitutions are referred to in abbreviated form as A2, A5, A6, etc. (Table 1). We also examined some of the RRM1 mutations that restored nuclear retention, but not suppression, in the context of otherwise wild-type La proteins. hLa carrying only the A5 or A6 mutations was nuclear but inactive for suppression (Fig. 5D, spots 4 and 5; Table 1, proteins 26 and 27), indicating these to be previously unknown residues that are important for tRNA-mediated suppression.
DISCUSSION
La proteins are associated with a variety of RNAs in different cellular compartments. In this work, we have attempted to detail the different trafficking elements important for La nucleocytoplasmic transport by using a fission yeast system that can assess function in tRNA maturation. We uncovered conserved determinants of the nuclear export and nuclear retention activities of hLa and Sla1p proteins and demonstrated opposing effects of these trafficking elements on La-dependent tRNA maturation. Substitution of two highly conserved lysines in the NRE produced phenocopies of NRE deletions, which are characterized by nuclear exclusion of La, disordered pre-tRNA splicing, and failure to support tRNA-mediated suppression. These substitution data validate the NRE as a functional element that maintains La in the nucleus, where it promotes tRNA maturation. We then found that deletion of RRM1 from hLaΔNRE and hLaKK returned these La proteins to the nucleus (Fig. 3). The data indicate that the NRE has been conserved to functionally mask or override an equally conserved nuclear export activity of the RRM1s of hLa and Sla1p.
However, since RRM1 is required for tRNA-mediated suppression activity of La (11), its complete deletion could not tell us if its nuclear export and tRNA-related activities were distinct or overlapping. We therefore attempted to identify site-specific mutations in RRM1 that could restore nuclear accumulation and tRNA-mediated suppression activity to the NRE-defective La proteins. Mutation of certain conserved RRM1 residues inactivated nuclear export of both the Sla1p and hLa NRE-defective proteins. These results led to two relevant conclusions. First, hLaΔNRE and Sla1pΔNRE proteins lacking the NRE could be made competent for tRNA-mediated suppression by RRM1 mutations that prevented nuclear export (e.g., hLaΔNRE-E132A/D133A and Sla1pΔNRE-E177A/E178A). This further suggested that the NRE does not contribute directly to tRNA maturation but rather serves to prevent untimely nuclear export of La and dysfunctional processing of associated pre-tRNAs. Second, the tRNA-related activity of RRM1 and its nuclear export activity could be functionally separated.
Residues in RRM1 can direct nuclear export but are otherwise masked or overridden by the NRE.
It is noteworthy that RRM1 of the Saccharomyces cerevisiae La protein, Lhp1p, was suspected of having nuclear export activity (31), suggesting that the conservation of nuclear export activity by RRM1 may extend to budding yeast.
Mutation of some RRM1 residues restored nuclear accumulation of NRE-defective La proteins, prevented disordered pre-tRNA splicing, and restored a more orderly pathway of processing and tRNA-mediated suppression. RRM1 mutations that restored nuclear accumulation and suppression activity to the NRE-defective proteins (e.g., E132A/D133A) did not impair nuclear accumulation or suppression when introduced into wild-type La (Fig. 5D, lane 3, and Table 1). These data suggest that the conserved nuclear export activity of RRM1 is not required for the functional maturation of intron-containing pre-tRNAs. Rather, functional unmasking or overriding of the nuclear export activity of La by the NRE appears to be detrimental to tRNA maturation. Thus, the data in this report do not support a positive role for La in tRNA nucleocytoplasmic transport, as speculated previously (29). The fact that we could identify mutations of conserved residues that block export without impairing tRNA maturation (e.g., hLa E132/D133 and Sla1p E177/E178) suggests that these residues were conserved to function in an important process other than tRNA maturation that involves nuclear export.
Although we have not identified a nuclear export factor for hLa or Sla1p, we have addressed the possibility that pre-tRNA binding may affect export. Examination of hLa26-408 and other (point) mutants that are severely impaired for UUU-OH binding and tRNA-mediated suppression revealed no localization defects in the context of either wild-type La or La containing other trafficking mutations (data not shown). While other data argue for a RNA-binding-dependent effect on localization in human cells (10), our analysis with fission yeast does not suggest that the capacity for pre-tRNA binding is a significant determinant of the nuclear export activity of La.
Some RRM1 mutations restored nuclear accumulation but not suppression (see, e.g., proteins 9 to 11, 19, and 20 in Table 1). These would appear to reflect residues that contribute to the RRM1-mediated tRNA maturation activity of La, since these same mutations inactivated wild-type La (Fig. 5D and Table 1, proteins 26 and 27). Other site-specific La RRM1 mutations that impair the maturation of some pre-tRNAs have been described (11).
Functional determinants of the NRE α-helix of hLa.
Our analysis, which included IF, tRNA-mediated suppression, and Northern blotting, showed that the more highly conserved basic residues at hLa positions 316 and 317, but not the less conserved basic residues at positions 328 to 332, are important for NRE function.
The hLa NRE corresponds to a well-ordered α-helix (Fig. 1B, α3) whose underside contacts the β-sheet surface of RRM2 (18). It was therefore anticipated that RRM2 harbored a nuclear export determinant that was masked by the overlying NRE and that deletion of RRM2 would block export and restore nuclear accumulation of the NRE-defective hLa proteins. However, we could not identify a functional NES anywhere in RRM2, and hLaΔNREΔRRM2 accumulated in the cytoplasm in an LMB-sensitive manner (data not shown), indicating a nuclear export determinant elsewhere in hLa. We are left with the conclusion that the NRE masks or functionally overrides the nuclear export potential of RRM1.
In addition to K316 and K317, two other highly conserved residues of the NRE are E320, which is on the same face of α3 as K316, and E324, which is on the same face as K317 (14, 18). These side chains are not directed toward the RRM2 β-sheet surface and would be potentially available for other interactions (18).
Conserved and diverged features of La trafficking and tRNA processing.
While La proteins from yeast to human reside in the nucleoplasm, nucleolus, and cytoplasm (23, 25, 31), some of the trafficking mechanisms appear to have been conserved while others have diverged. The endogenous NLS of S. cerevisiae Lhp1p does not coincide with the C-terminal location of hLa and Sla1p NLSs, and Lhp1p uses a different karyopherin for nuclear import than that used by hLa and Sla1p (31). Implications of these differences have been discussed previously (31).
Other species-specific differences in La trafficking have also been noted. Nuclear export of Sla1pΔNRE and hLaΔNRE is sensitive to the Crm1p export inhibitor LMB in fission yeast (with total reversal of the nuclear exclusion pattern to complete nuclear accumulation by 2 h) (14). By contrast to fission yeast, LMB only partially restores nuclear accumulation of Sla1pΔNRE and hLaΔNRE in primate cells (14) and is ineffective in preventing nuclear export of hLa during nucleo-cytoplasmic shuttling in primate cells (5). It is unknown whether these differences reflect differences in the kinetics or in the transport machineries used for La transport in the mammalian and yeast cells. In any case, an outstanding issue is whether the nuclear export activity of RRM1 elucidated here contributes to nucleo-cytoplasmic shuttling in HeLa cells (5).
Because tRNA splicing occurs in the cytoplasm in yeasts but in the nucleus in vertebrates (reviewed in reference 8), we should expect that while NRE-deficient La mutants undergo nuclear export in yeast and vertebrates (14), the effects on disordered splicing may be different.
We note that although phospho-hLa is nucleoplasmic whereas non-phospho-hLa is nucleolar and cytoplasmic, prior results indicate that phosphorylation is not a determinant of localization, since nonphosphorylatable La mutants or phosphomemetic La mutants do not mislocalize in human cells (3, 15, 16) or yeast cells (16). Moreover, examination of hLa proteins expressed in yeast revealed that the NRE deletion did not affect S366 phosphorylation status (14). In addition, examination of a variety of hLa trafficking mutants both in the S366 native form and containing nonphosphorylatable or phosphomemetic residues failed to support a causative relationship between phosphorylation and localization (reference 15 and data not shown). Although we have not examined the S366 phosphorylation status of the hLa NES mutants characterized here, the cumulative data suggest that any difference in S366 phosphorylation would be a consequence of localization rather than a functional determinant of localization.
Structural modeling of residues involved in nuclear export onto the hLa RRM1 structure.
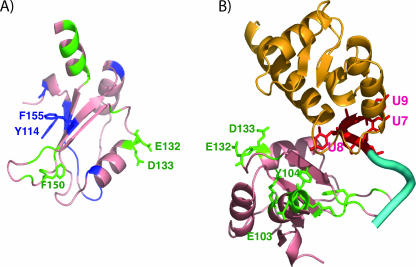
Many RRM1 substitutions did not restore nuclear accumulation (Table 1, proteins 13 to 18, 23, 34, and 37). We examined the spatial arrangement of mutated residues on the hLa structure (Fig. 6). In general, residues that restored nuclear accumulation appear on a side of the RRM facing away from the canonical RNA-binding surface, whereas those that did not face in the same general direction as the RNA-binding surface (Fig. 6A). Mutated residues that restored nuclear accumulation are also shown in the LM-RRM1-UUU-OH RNA structure; all are quite distant from the LM and the RNA (Fig. 6B), consistent with the idea that they may be recognized by a transport carrier. These data support the idea that the nuclear export and tRNA-related activities of RRM1 are separable.
FIG. 6.
Views of the RRM1 residues important for nuclear export that are masked or functionally overridden by the NRE in the isolated RRM1 (A) and in the LM-RRM1-UUU-OH RNA complex (B). (A) The mutated RRM1 residues that restored nuclear accumulation to the NRE-defective hLa proteins are shown in green; the mutated residues that restored nuclear accumulation and suppression (E132, D133, and F150) are shown as side chains. Mutated residues that did not restore nuclear accumulation are shown in blue, including Y114 and F155, the highly conserved aromatic side chains on the β-sheet surface. (B) The LM is shown in gold. The 3′ UUU-OH (U7 U8 U9 [37]) is shown in red, with the rest of the RNA chain in blue.
Acknowledgments
We thank M. Blum for medium preparation, Laboratory of Molecular Growth Regulation members for discussion and/or comments, and Vera Cherkasova and the reviewers for comments on the manuscript.
This research was supported by the Intramural Research Program of the NICHD, NIH. R.J.M. is a commissioned officer in the U.S. Public Health Service.
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Alfano, C., D. Sanfelice, J. Babon, G. Kelly, A. Jacks, S. Curry, and M. R. Conte. 2004. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 11:323-329. [DOI] [PubMed] [Google Scholar]
- 2.Bertrand, E., F. Houser-Scott, A. Kendall, R. H. Singer, and D. R. Engelke. 1998. Nucleolar localization of early tRNA processing. Genes Dev. 12:2463-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broekhuis, C. H., G. Neubauer, A. van der Heijden, M. Mann, C. G. Proud, W. J. van Venrooij, and G. J. Pruijn. 2000. Detailed analysis of the phosphorylation of human La (SS-B) autoantigen. (De)phosphorylation does not affect subcellular distribution. Biochemistry 39:3023-3033. [DOI] [PubMed] [Google Scholar]
- 4.Fairley, J. A., T. Kantidakis, N. S. Kenneth, R. V. Intine, R. J. Maraia, and R. J. White. 2005. Human La is found at RNA polymerase III-transcribed genes in vivo. Proc. Natl. Acad. Sci. USA 102:18350-18355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fok, V., K. Friend, and J. A. Steitz. 2006. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J. Cell Biol. 173:319-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haeusler, R. A., and D. R. Engelke. 2006. Spatial organization of transcription by RNA polymerase III. Nucleic Acids Res. 34:4826-4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamada, M., A. L. Sakulich, S. B. Koduru, and R. Maraia. 2000. Transcription termination by RNA polymerase III in fission yeast: a genetic and biochemical model system. J. Biol. Chem. 275:29076-29081. [DOI] [PubMed] [Google Scholar]
- 8.Hopper, A. K. 2006. Cellular dynamics of small RNAs. Crit. Rev. Biochem. Mol. Biol. 41:3-19. [DOI] [PubMed] [Google Scholar]
- 9.Hopper, A. K., and E. M. Phizicky. 2003. tRNA transfers to the limelight. Genes Dev. 17:162-180. [DOI] [PubMed] [Google Scholar]
- 10.Horke, S., K. Reumann, M. Schweizer, H. Will, and T. Heise. 2004. Nuclear trafficking of La protein depends on a newly identified NoLS and the ability to bind RNA. J. Biol. Chem. 279:26563-26570. [DOI] [PubMed] [Google Scholar]
- 11.Huang, Y., M. A. Bayfield, R. V. Intine, and R. J. Maraia. 2006. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat. Struct. Mol. Biol. 13:611-618. [DOI] [PubMed] [Google Scholar]
- 12.Huang, Y., R. V. Intine, A. Mozlin, S. Hasson, and R. J. Maraia. 2005. Mutations in the RNA polymerase III subunit Rpc11p that decrease RNA 3′ cleavage activity increase 3′-terminal oligo(U) length and La-dependent tRNA processing. Mol. Cell. Biol. 25:621-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inada, M., and C. Guthrie. 2004. Identification of Lhp1p-associated RNAs by microarray analysis in Saccharomyces cerevisiae reveals association with coding and noncoding RNAs. Proc. Natl. Acad. Sci. USA 101:434-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Intine, R. V., M. Dundr, T. Misteli, and R. J. Maraia. 2002. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Mol. Cell 9:1113-1123. [DOI] [PubMed] [Google Scholar]
- 15.Intine, R. V., M. Dundr, A. Vassilev, E. Schwartz, Y. Zhao, M. L. Depamphilis, and R. J. Maraia. 2004. Nonphosphorylated human La antigen interacts with nucleolin at nucleolar sites involved in rRNA biogenesis. Mol. Cell. Biol. 24:10894-10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Intine, R. V., S. A. Tenenbaum, A. S. Sakulich, J. D. Keene, and R. J. Maraia. 2003. Differential phosphorylation and subcellular localization of La RNPs associated with precursor tRNAs and translation-related mRNAs. Mol. Cell 12:1301-1307. [DOI] [PubMed] [Google Scholar]
- 17.Intine, R. V. A., A. L. Sakulich, S. B. Koduru, Y. Huang, E. Pierstorrf, J. L. Goodier, L. Phan, and R. J. Maraia. 2000. Transfer RNA maturation is controlled by phosphorylation of the human La antigen on serine 366. Mol. Cell 6:339-348. [DOI] [PubMed] [Google Scholar]
- 18.Jacks, A., J. Babon, G. Kelly, I. Manolaridis, P. D. Cary, S. Curry, and M. R. Conte. 2003. Structure of the C-terminal domain of human La protein reveals a novel RNA recognition motif coupled to a helical nuclear retention element. Structure (Cambridge) 11:833-843. [DOI] [PubMed] [Google Scholar]
- 19.Kenan, D. J., and J. D. Keene. 2004. La gets its wings. Nat. Struct. Mol. Biol. 11:303-305. [DOI] [PubMed] [Google Scholar]
- 20.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 21.Kutay, U., and S. Guttinger. 2005. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 15:121-124. [DOI] [PubMed] [Google Scholar]
- 22.la Cour, T., R. Gupta, K. Rapacki, K. Skriver, F. M. Poulsen, and S. Brunak. 2003. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 31:393-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long, K. S., T. Cedervall, C. Walch-Solimena, D. A. Noe, M. J. Huddleston, R. S. Annan, and S. L. Wolin. 2001. Phosphorylation of the Saccharomyces cerevisiae La protein does not appear to be required for its functions in tRNA maturation and nascent RNA stabilization. RNA 7:1589-1602. [PMC free article] [PubMed] [Google Scholar]
- 24.Lund, E., and J. E. Dahlberg. 1998. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science 282:2082-2085. [DOI] [PubMed] [Google Scholar]
- 25.Maraia, R. J. 2001. La protein and the trafficking of nascent RNA polymerase III transcripts. J. Cell Biol. 153:F13-F17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maraia, R. J., and M. A. Bayfield. 2006. The La protein-RNA complex surfaces. Mol. Cell 21:149-152. [DOI] [PubMed] [Google Scholar]
- 27.Maraia, R. J., and R. V. Intine. 2001. Recognition of nascent RNA by the human La antigen: conserved and diverged features of structure and function. Mol. Cell. Biol. 21:367-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maris, C., C. Dominguez, and F. H. Allain. 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 272:2118-2131. [DOI] [PubMed] [Google Scholar]
- 29.Phizicky, E. M. 2005. Have tRNA, will travel. Proc. Natl. Acad. Sci. USA 102:11127-11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reiner, R., Y. Ben-Asouli, I. Krilovetzky, and N. Jarrous. 2006. A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes Dev. 20:1621-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosenblum, J. S., L. F. Pemberton, N. Bonifaci, and G. Blobel. 1998. Nuclear import and the evolution of a multifunctional RNA-binding protein. J. Cell Biol. 143:887-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz, E., R. V. Intine, and R. J. Maraia. 2004. CK2 is responsible for phosphorylation of human La protein serine-366 and can modulate 5′TOP mRNA metabolism. Mol. Cell. Biol. 24:9580-9591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaheen, H. H., and A. K. Hopper. 2005. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 102:11290-11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simons, F. H., F. J. Broers, W. J. Van Venrooij, and G. J. Pruijn. 1996. Characterization of cis-acting signals for nuclear import and retention of the La (SS-B) autoantigen. Exp. Cell Res. 224:224-236. [DOI] [PubMed] [Google Scholar]
- 35.Simons, F. H., G. J. Pruijn, and W. J. van Venrooij. 1994. Analysis of the intracellular localization and assembly of Ro ribonucleoprotein particles by microinjection into Xenopus laevis oocytes. J. Cell Biol. 125:981-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takano, A., T. Endo, and T. Yoshihisa. 2005. tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309:140-142. [DOI] [PubMed] [Google Scholar]
- 37.Teplova, M., Y.-R. Yuan, S. Ilin, L. Malinina, A. T. Phan, A. Teplov, and D. J. Patel. 2006. Structural basis for recognition and sequestration of UUU-OH 3′-termini of nascent RNA pol III transcripts by La, a rheumatic disease autoantigen. Mol. Cell 21:75-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas, F., and U. Kutay. 2003. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 116:2409-2419. [DOI] [PubMed] [Google Scholar]
- 39.Trotta, C. R., E. Lund, L. Kahan, A. W. Johnson, and J. E. Dahlberg. 2003. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 22:2841-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Horn, D. J., C. J. Yoo, D. Xue, H. Shi, and S. L. Wolin. 1997. The La protein in Schizosaccharomyces pombe: a conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA 3:1434-1443. [PMC free article] [PubMed] [Google Scholar]
- 41.Wolin, S. L., and T. Cedervall. 2002. The La protein. Annu. Rev. Biochem. 71:375-403. [DOI] [PubMed] [Google Scholar]
- 42.Wolin, S. L., and A. G. Matera. 1999. The trials and travels of tRNA. Genes Dev. 13:1-10. [DOI] [PubMed] [Google Scholar]
- 43.Yoo, C. J., and S. L. Wolin. 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89:393-402. [DOI] [PubMed] [Google Scholar]
- 44.Yoshihisa, T., K. Yunoki-Esaki., N. Tanaka, and T. Endo. 2003. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol. Biol. Cell 14:3266-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaitseva, L., R. Myers, and A. Fassati. 2006. tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol. 4:e332. [DOI] [PMC free article] [PubMed] [Google Scholar]