Abstract
HOXA10 is necessary for embryonic patterning of skeletal elements, but its function in bone formation beyond this early developmental stage is unknown. Here we show that HOXA10 contributes to osteogenic lineage determination through activation of Runx2 and directly regulates osteoblastic phenotypic genes. In response to bone morphogenic protein BMP2, Hoxa10 is rapidly induced and functions to activate the Runx2 transcription factor essential for bone formation. A functional element with the Hox core motif was characterized for the bone-related Runx2 P1 promoter. HOXA10 also activates other osteogenic genes, including the alkaline phosphatase, osteocalcin, and bone sialoprotein genes, and temporally associates with these target gene promoters during stages of osteoblast differentiation prior to the recruitment of RUNX2. Exogenous expression and small interfering RNA knockdown studies establish that HOXA10 mediates chromatin hyperacetylation and trimethyl histone K4 (H3K4) methylation of these genes, correlating to active transcription. HOXA10 therefore contributes to early expression of osteogenic genes through chromatin remodeling. Importantly, HOXA10 can induce osteoblast genes in Runx2 null cells, providing evidence for a direct role in mediating osteoblast differentiation independent of RUNX2. We propose that HOXA10 activates RUNX2 in mesenchymal cells, contributing to the onset of osteogenesis, and that HOXA10 subsequently supports bone formation by direct regulation of osteoblast phenotypic genes.
Patterning and development of the skeleton are complex processes involving signaling proteins and transcription factors that function as determinants for bone formation. Among the principal regulatory cascades for the development of the skeleton are Hox genes that determine the position and shape of a tissue element (19, 54) and the bone morphogenetic proteins (BMPs), which induce the differentiation of mesenchymal cells to osteoblast and chondroblast lineages (18, 84). BMP2 signaling leads to the induction of a number of transcription factors, including RUNX2 (28) and OSTERIX (42, 56), which are essential for bone development (34, 45, 69). Several HOX and homeodomain proteins have been identified as molecular targets of BMP-mediated gene transcription during early stages of bone formation by microarray gene expression profiling studies (3, 33). The present study was aimed at characterizing a BMP2-inducible gene, the Homeobox a10 (HOXA10) gene, as a candidate for contributing to commitment and development of the osteoblast phenotype.
Mammalian Hox genes (homologues of the Drosophila homeotic genes) encode transcription factors crucial in regional development along the anterior-posterior axis during embryogenesis (44, 64). The mouse and human genomes contain 39 Hox genes, which are grouped into four clusters, Hoxa, Hoxb, Hoxc, and Hoxd, positioned on four separate chromosomes in 13 paralogs (38, 44). Hoxa10 is a member of the Abdominal B (Abd B) class of homeobox protein-encoding genes, representing the most 5′ genes in the cluster consisting of paralog genes from Hoxa 9 to Hoxa 13 (4, 44, 50). The corresponding class of homeobox proteins binds preferentially to a consensus core of TTAT or TTAC, which is distinct from the TAAT homeodomain consensus core binding site recognized by MSX and DLX proteins (4). HOXA10 DNA binding is influenced by flanking sequences and the formation of complexes with HOXA10-interacting proteins of the MEIS and PBX classes of transcription factors, as well as other coregulatory proteins, such as histone deacetylase 2 (13, 51, 60, 72, 74, 81).
Several of the Hox genes are essential for normal skeletal development (31, 64). The nonparalogous Hoxa10 and Hoxd11 genes cooperate in the development of the forelimbs and axial skeleton and are required to globally pattern the mammalian skeleton (6, 7, 26, 68, 80). Inactivation of the paralogous Hoxa10 and Hoxd10 genes results in alterations in the formation of the forelimbs and hind limbs. Hoxa10−/− mice revealed an active role for the gene in modeling the femur, tibia, and fibula (12, 79). Transgenic expression of HOXA10 in presomitic mesoderm of the mouse resulted in vertebrae without ribs (11). HOXA10 is expressed in the presomitic mesoderm, which develops into the axial skeleton and cooperates with other Hox genes (e.g., Hoxd11) for normal skeletal development (11, 26, 31, 64). Despite the considerable genetic evidence that HOXA10 has critical skeletal functions, target genes of HOXA10 in bone have not been identified.
In this study we have characterized HOXA10 regulation of the key osteogenic factor Runx2, as well as RUNX2 target genes, identifying Hoxa10-specific regulatory elements in promoters of the Runx2, osteocalcin (OC), alkaline phosphatase, and bone sialoprotein osteoblast-related genes. This discovery of a Hox regulatory factor in activating Runx2 provides novel insights for a mechanism that regulates a transcription factor essential for bone formation (43, 58). Although Runx2 is rapidly induced in response to BMP2 and present in developing limbs, somites, and mesenchymal condensations prior to chondrogenic and osteoblast differentiation (23, 46, 76), a Smad-responsive element has not been defined. Thus, BMP2-induced HOXA10 represents a key regulator of Runx2 transcription during embryogenesis. Our studies also show that HOXA10 can regulate osteoblast genes independent of RUNX2. We have thus identified an additional role for HOXA10 in postnatal bone formation and maintenance of the osteoblast phenotype. We propose that HOXA10 functions in two capacities: as a component of a BMP2 signaling cascade prior to RUNX2 to mediate the developmental induction of osteogenesis and during osteoblast differentiation to regulate the temporal expression of bone phenotypic genes to drive osteoblast maturation through mechanisms involving chromatin remodeling of gene promoters.
(Brian Weiner's contribution to this paper is in fulfillment of a Worcester Polytechnic Institute undergraduate thesis project.)
MATERIALS AND METHODS
Cell cultures.
C3H10T1/2 and NIH 3T3 cells were maintained in Dulbecco's minimal essential medium (MEM) (GIBCO) supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Georgia). C3H10T1/2 cells were induced to osteogenesis by BMP2 (300 ng/ml). MC3T3 cells were maintained in α-MEM supplemented with 10% FBS. Primary rat osteoblast cells were isolated from calvaria according to the procedures described previously (34). The rat osteosarcoma cell line ROS 17/2.8, representing a mature osteoblast phenotype, was maintained in F-12 medium supplemented with 5% FBS (70). Cells were cultured under osteogenic conditions with MEM (GIBCO-BRL) supplemented with 10% FBS, 50 μg of ascorbic acid/ml, and 10 mM β-glycerol phosphate. Bone marrow stromal cells were isolated from 8-week-old C57BL6 mice and cultured in MEM containing 10% FBS (29).
Runx2 null cells were isolated from calvarial tissues of mouse embryos (17.5 days postcoitum) of the Runx2 null mouse and immortalized using mouse telomerase (TERT). Characterization of this cell line has been described previously (1). The BMP2 used in these studies was a kind gift from John Wozney (Wyeth Research, Women's Health and Musculoskeletal Biology).
Transfection and reporter assays.
The Hoxa10 expression clone containing the mouse cDNA (1.2 kb) of the Hoxa10-1 variant (55 kDa) containing the transactivation domain was kindly provided by Richard L. Maas (Harvard Medical School, Boston, MA) (4). Transient transfections were performed in six-well plates at 50 to 70% confluence with 5 μl of FuGENE6 transfection reagent with wild-type (WT) and deleted promoter reporter DNA according to the manufacturer's instructions (Roche, Indianapolis, IN). A tagged Hoxa10 expression vector (pcDNA3.1-Xpress-Hoxa10) was constructed was used along with a Runx2 expression vector (pcDNA3.1-HA-Runx2) (85) in this study. For control of expression, vector pcDNA3.1 was transfected according to the experimental condition. The following Runx2 WT and deletion promoter reporter plasmids (21) were used: the WT Runx2 0.6-kb fragment and the deletion series (−490, −458, −351, −288, −128) fragments. These fragments were cloned in pGL3 basic luciferase vector (Promega, Madison WI). Cells transfected with either Runx2 promoter luciferase or −208 OC promoter chloramphenicol acetyltransferase (CAT) reporter constructs along with Hoxa10, Runx2, or control vector were harvested 24 to 36 h after transfection, and all lysates were assayed for luciferase or CAT activity according to the manufacturer's instructions (Promega, Madison, WI). All results were normalized to the luciferase activity resulting from transfection of the promoterless pGL3 luciferase construct (Promega, Madison, WI). The OC promoter activity was assayed by using the −208-bp promoter DNA fragment from rat OC genes cloned in the pCAT basic vector (Promega, Madison, WI) (32). The percent CAT conversion was the average of values for six similar transfection samples.
cDNA synthesis and QPCR.
RNA was isolated from cultures of MC3T3, NIH 3T3, and C3H10T1/2 cells by use of TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. cDNAs were synthesized with oligo(dT) primers by use of a SuperScript first-strand cDNA synthesis kit (Invitrogen) according to the manufacturer's protocol. Gene expression was assessed by real-time quantitative PCR (RT-QPCR) using Power SYBR green PCR master mix (Applied Biosystems, California). Primers used for PCRs are listed in Table 1.
TABLE 1.
Primers for real-time PCR assays
| Primer for indicated gene | Sequence |
|---|---|
| Bone marker primers (mouse) | |
| BSP | |
| Forward | 5′-GCA CTC CAA CTG CCC AAG A-3′ |
| Reverse | 5′-TTT TGG AGC CCT GCT TTC TG-3′ |
| ALP | |
| Forward | 5′-TTG TGC CAG AGA AAG AGA GAG A-3′ |
| Reverse | 5′-GTT TCA GGG CAT TTT TCA AGG T-3′ |
| Runx2 | |
| Forward | 5′-CGG CCC TCC CTG AAC TCT-3′ |
| Reverse | 5′-TGC CTG CCT GGG ATC TGT A-3′ |
| OC | |
| Forward | 5′-CTG ACA AAG CCT TCA TGT CCA A-3′ |
| Reverse | 5′-GCG GGC GAG TCT GTT CAC TA-3′ |
| Hoxa10 | |
| Forward | 5′-TTC GCC GGA GAA GGA CTC-3′ |
| Reverse | 5′-TCT TTG CTG TGA GCC AGT TG-3′ |
| Primers for ChIP (rat) | |
| Runx2 | |
| Forward | 5′-TCA GCA TTT GTA TTC TAT CCA AAT CC-3′ |
| Reverse | 5′-TGG CAT TCA GAA GGT TAT AGC TTT T-3′ |
| ALP | |
| Forward | 5′-CCT GTG CAT TTC CCA ACA CGG CGG-3′ |
| Reverse | 5′-CCA CTT CCC AGG CAG TGG AGA CAG-3′ |
| BSP | |
| Forward | 5′-GTT TAA ATG CTT AAG TCG TTT GC-3′ |
| Reverse | 5′-GGC TGT GGG TTC TCA CCA GAA A-3′ |
| OC | |
| Forward | 5′-GGC AGC CTC TGA TTG TGT CC-3′ |
| Reverse | 5′-TAT ATC CAC TGC CTG AGC GG-3′ |
| Control 3′ UTR primers for ChIP (mouse) | |
| OC | |
| Forward | 5′-GAT CCC ATA TCA GCC AGC AC-3′ |
| Reverse | 5′-GAC TGC CCT GGA TCA CAA GT-3′ |
| Runx2 | |
| Forward | 5′-CGT CCA CCT GTT CCA AAG TT-3′ |
| Reverse | 5′-GGC ATT GCC ATT TTC AGT TT-3′ |
| BSP | |
| Forward | 5′-CCT TTT CGG TGA TTG CAG TT-3′ |
| Reverse | 5′-AAG GTT GAG GGT GTC AGT GG-3′ |
| ALP | |
| Forward | 5′-TTG TTC CTC TTG CCT CAG GT-3′ |
| Reverse | 5′-TGA CAA TCA CAT GGC CTC TC-3′ |
| Control 3′ UTR primers for ChIP (rat) | |
| OC | |
| Forward | 5′-GCA CTG CAC AGA TGT GGA AC-3′ |
| Reverse | 5′-CAG GTT TTC CCT TTC TCA GG-3′ |
| Runx2 | |
| Forward | 5′-TGC TTT GCA ACC AAA TCA AG-3′ |
| Reverse | 5′-TCT GAA GGG AAG CTT TGG AA-3′ |
| BSP | |
| Forward | 5′-TGG AAG ATG CTT GAT GAC CA-3′ |
| Reverse | 5′-AAG GGG TCA GAG GAC AAG GT-3′ |
| ALP | |
| Forward | 5′-CCA GTG TGA TCC CCA GAA CT-3′ |
| Reverse | 5′-TGT CTG TAG CAA TCC CAC CA-3′ |
Electrophoretic mobility shift analysis (EMSA).
Recombinant HOXA10 protein was translated using Promega TNT coupled with a rabbit reticulocyte lysate system (Promega). WT and mutant oligonucleotides containing the Hoxa10 binding site derived from the Runx2 P1 promoter site 1 (WT, 5′G CAT TCA GAA GGT TAT AGC TTT 3′; mutant, 5′G CAT TCA GAA GGC GAT AGC TTT 3′ [underlining indicates the HOXA10 binding site, and boldface indicates the mutation]) were end labeled with [γ-32P]ATP by use of T4 polynucleotide kinase (New England Biolabs, Massachusetts). The detailed procedures have been described previously (34). Anti-HOXA10 (N20) or a nonspecific antibody (either α-actin or normal immunoglobulin G [IgG] as indicated) (Santa Cruz) were used for the immunoshift studies. Complexes were visualized by autoradiography of a 6.5% acrylamide gel.
Immunoblotting.
Each well of a six-well plate was lysed in 50 μl lysis buffer (2% sodium dodecyl sulfate [SDS], 10 mM dithiothreitol, 10% glycerol, 12% urea, 10 mM Tris-HCl [pH 7.5], 1 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail [Roche], 25 μM MG132 [proteosome inhibitor from Calbiochem]) and boiled for 5 min. Equal amounts of total protein were analyzed by SDS-polyacrylamide gel electrophoresis and probed with suitable antibodies. Immunocomplexes were detected using Western Lightning chemiluminescence reagent (Perkin Elmer, Boston, MA).
Antibodies.
The following antibodies were purchased from Santa Cruz Biotechnology. HOXA10 N20 (SC-17158) was for chromatin immunoprecipitation (ChIP) and EMSA, and A20 (5C-17159) was for Western blotting. RUNX2 antibodies were PEBP2αA (M-70 [SC-10758]), PEBP2αA (C-19 [SC-8566]), and actin (I-19 [SC-1616]). Mouse monoclonal RUNX2 antibody was a generous gift from Yoshi Ito and Kosei Ito (National University, Singapore, Republic of Singapore). Anti-hyperacetylated histone H4 (Penta) and anti-trimethyl histone H3 (Lys4), clone MC315, were purchased from Upstate Cell Signaling Solutions (Charlottesville, VA). The mouse monoclonal anti-Xpress antibody was obtained from Invitrogen (Carlsbad, CA). Mouse monoclonal antibody against RNA polymerase II (Pol II) (clone 8WG16) was obtained from Covance (Princeton, NJ) and used in ChIP studies.
ChIP assays.
The procedure for ChIP in primary rat osteoblasts has already been described (34). Control primer pairs from 3′ untranslated regions (UTR) of the genes were used to verify specific and nonspecific binding of DNA fragments (Table 1). IgG antibody was used as a control for nonspecific pull-down of immunocomplexes. Sequential ChIP studies were performed using the primary pull-down from one antibody, which was divided into equal aliquots for the second pull-down with antibodies specific for coregulatory molecules. Instead of being eluted in 1% SDS and 100 mM Na2HCO3 after cross-linking and washing, immunocomplexes were eluted in 10 mM dithiothreitol. The eluate was further diluted 1:40 in ChIP dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 167 mM Tris-HCl [pH 8.1], 167 mM NaCl) and used for the second immunoprecipitations. Aliquots (2 to 3 μl) of DNA samples from different pull-downs were assayed by either radioactive labeling or RT-QPCR using Power SYBR green PCR master mix (Applied Biosystems, California) for the detection of specific DNA fragments with primers in the proximal promoters of bone-related genes that encompass the Hoxa10 binding sites (Table 1).
Immunohistochemistry and immunofluorescence.
Long bones from normal newborn mice were fixed with 4% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.4, for 48 h, dehydrated, and embedded in paraffin by standard procedures. Paraffin-embedded tissues (5-μm sections) were immunoperoxidase labeled and blocked by 0.3% H2O2 in absolute methanol for 30 min at room temperature. To reveal antigens, sections were put in a 1 mM Tris solution, pH 9.0, supplemented with 0.5 mM EGTA. The sections for RUNX2 immunohistochemistry were heated in a microwave oven for 10 min after EGTA treatment, while a normal steaming antigen retrieval method was used for HOXA10. Nonspecific immunoglobulin binding was prevented by incubating the sections in 50 mM NH4Cl for 30 min followed by blocking with phosphate-buffered saline supplemented with 1% bovine serum albumin, 0.05% saponin, and 0.2% gelatin. Serial sections were incubated overnight at 4°C with anti-rabbit RUNX2 (M-70 and C-19) or anti-goat HOXA10 (N20) antibody diluted (1:100) in phosphate-buffered saline supplemented with 0.1% bovine serum albumin and 0.3% Triton X-100. Equal amounts of the respective blocking peptides (2 μg/ml) for HOXA10 and RUNX2 were also used. Normal IgG was used as a nonspecific control. Immunolabeling controls were performed by using antibodies preabsorbed with immunizing peptides. Labeling was visualized with the horseradish peroxidase-conjugated secondary antibody (P448, 1:200; Dako).
MC3T3 cells were plated at a density of 0.6 × 105 cells/well on gelatin-coated coverslips in six-well plates. Cells were processed for in situ immunofluorescence analyses, which were carried out as described previously (39). HOXA10 was detected by a goat polyclonal antibody at a dilution of 1:100 (Santa Cruz Biotechnology). The secondary antibody used was Alexa 488-anti-goat antibody (Molecular Probes) at a dilution of 1:800. For overexpressed HOXA10 protein, HeLa cells at 0.5 × 105cells/well on gelatin-coated coverslips were transfected with 0.5 μg of Hoxa10 expression construct, and 24 h after transfection the coverslip was processed for immunofluorescence study (39). Xpress-HOXA10 was detected by a mouse monoclonal antibody against the Xpress tag at a dilution of 1:3,000 (Invitrogen). The secondary antibody used was Alexa 568-anti-mouse antibody (Molecular Probes, Eugene, OR) at a dilution of 1:800. Slides were examined on a Zeiss Axioplan 2 microscope fitted with epifluorescence (Carl Zeiss, Jena, Germany) attached to a charge-coupled-device camera. Images were saved and processed using Metamorph imaging software, version 6.1 (Universal Imaging, Downingtown, PA).
RNA interference (RNAi) of Hoxa10.
The mouse MC3T3-E1 osteoblastic cells at 30 to 50% confluence were transfected using Oligofectamine (Invitrogen Life Technologies) with small interfering RNA (siRNA) duplexes specific for murine Hoxa10 r(CCA AAU UAU CCC ACA ACA A)dTdT and r(UUG UUG UGG GAU AAU UUG G)dCdG obtained from QIAGEN Inc. (Stanford, CA). Six different sets of siRNA duplexes at different concentrations were used to evaluate the target specificity and knockdown efficiency. The cells were also transfected with control siRNA duplexes specific for green fluorescent protein by use of the same concentrations to check the transfection efficiency. The siRNA experiment was carried out for 72 h. Total RNA and proteins from the specific siRNA oligonucleotide-treated, untreated, Oligofectamine-treated and nonspecific-oligonucleotide-treated cells were analyzed by Western blotting and RT-QPCR. To study chromatin modifications after Hoxa10 siRNA treatment, cells were harvested 72 h posttransfection and used for immunoprecipitation with trimethyl histone H3K4 and hyperacetylated H4 antibody for ChIP analyses.
RESULTS
Hoxa10 is a BMP2-responsive gene correlating to the induction of Runx2-mediated osteogenesis.
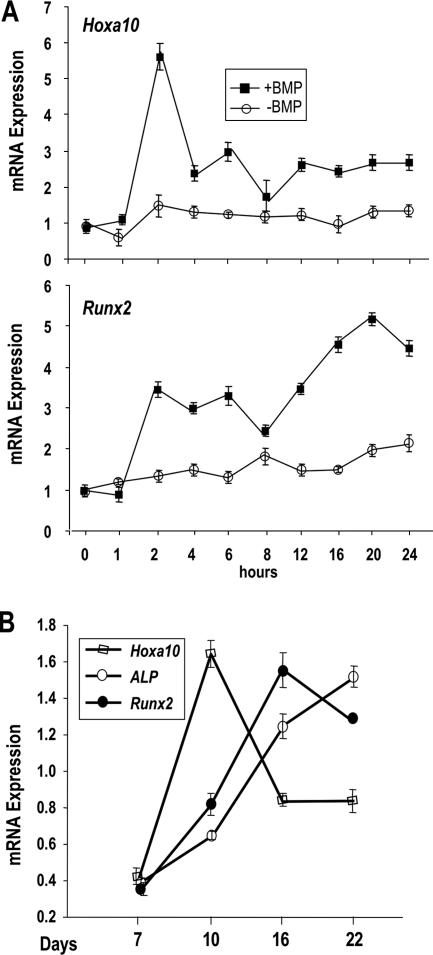
Hoxa10 was identified as an early BMP2-responsive gene in an Affymetrix cDNA profiling study of BMP2-mediated osteogenic induction of premyogenic C2C12 cells (3). During the 24-h time course, Hoxa10 expression clustered into a group of genes in which the osteogenic transcription factor Runx2 was also present. We carried out independent experiments in this study and quantitated gene expression by RT-QPCR to validate the microarray report (Fig. 1A). Both Hoxa10 and Runx2 mRNAs are present at very low levels in untreated cells, immediately upregulated by BMP2 within 2 h, and then partly downregulated, but they exhibit a second spike at 6 h. After the 8-h commitment point to osteogenesis, when bone phenotypic markers (alkaline phosphatase and OC) are expressed (3), Hoxa10 mRNA remains at a steady state, while Runx2 continuously increases.
FIG. 1.
Hoxa10 expression in relation to osteogenic differentiation. (A) Premyogenic C2C12 cells were treated with 100 ng/ml BMP2 for 24 h. Total RNA was isolated at different time points (0, 1, 2, 4, 6, 8, 12, 16, 20, and 24 h). Five micrograms of total RNA was reverse transcribed with oligo(dT) primer and amplified (RT-QPCR) by gene-specific primers. (Top) Hoxa10 expression (+BMP, −BMP) was normalized to Gapdh expression (+BMP, −BMP), and relative transcript levels were plotted; (bottom) the induction of Runx2 expression with BMP2 treatment is shown for the same time course. Error bars represent triplicate analyses of each sample from two independent experiments. (B) Temporal expression of Hoxa10 and the osteogenic markers Runx2 and ALP during MC3T3 cell growth and differentiation. cDNAs from different time points were amplified using bone-specific gene primers (Table 1). Expression values from RT-QPCR were normalized to Gapdh values. Error bars represent triplicate sample analyses from one experiment. Independent experiments exhibited similar temporal expressions (data not shown).
To further evaluate the expression of Hoxa10 and Runx2 during the development of the osteoblast phenotype, we examined the temporal expression of Hoxa10 during the differentiation of osteoprogenitor MC3T3 cells. Hoxa10 is endogenously expressed at low levels in the proliferating cells (day 7) and is induced four- to fivefold upon the maturation of the phenotype, concurrent with cellular multilayering (day 10 in this experiment) (Fig. 1B). Peak mRNA expression levels of Hoxa10 are transient and downregulated by 50% by day 16, the onset of mineral deposition in MC3T3 cells. Expression of Runx2 and alkaline phosphatase phenotypic genes continuously increases during differentiation. These observations suggest that Hoxa10 expression is important at the early stages of osteoblast differentiation. We further showed that either treatment of pluripotent C3H10T1/2 cells with BMP2 or growth of adherent bone marrow cells in osteogenic media results in a twofold increase in Hoxa10 concomitant with the expression of the osteoblast phenotype (data not shown). These studies implicate HOXA10 in establishing early events in the development of the osteoblast phenotype.
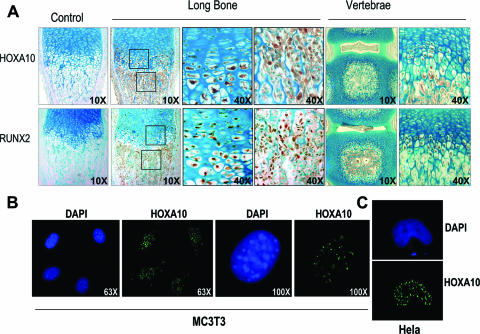
To appreciate the role of HOXA10 in osteogenesis in vivo, we examined its expression in bone cell populations in situ in relation to endochondral bone formation and skeletal development. Sections of long bones and vertebrae from newborn mice were treated with specific antibodies to detect RUNX2 and HOXA10 (Fig. 2). We found strong expression of HOXA10 in the periosteum, the hypertrophic chondrocyte zone of the growth plate, and osteoblasts on all bone surfaces. RUNX2 was also expressed in these cell populations. Weak expressions of HOXA10 and RUNX2 were found in the flattened cells that represent the prehypertrophic chondrocyte phenotype. It is noteworthy that both HOXA10 and RUNX2 were absent from the epiphyseal chondrocytes that represent permanent hyaline cartilage. Similar observations were made for vertebrae tissue. We conclude from these studies that RUNX2 and HOXA10 are expressed in osteogenic lineage cells and maturing chondrocytes at the growth plate in both the axial and appendicular skeletons.
FIG. 2.
Coexpression of HOXA10 and RUNX2 in vivo and in vitro. (A) HOXA10 and RUNX2 are visualized in hypertrophic chondrocytes and osteoblast lineage cells in long bones and vertebrae as indicated. Immunohistochemistry was performed using anti-HOXA10 antibody and anti-RUNX2 antibody. For nonspecific controls, normal rabbit IgG was substituted for primary antibody, and blocking peptides specific for the HOXA10 and RUNX2 antibodies were used. Bone and vertebra sections are shown at ×10 and ×40 magnification as indicated for the boxed areas at the growth plate and the primary spongiosa. Trabeculae show robust expression of both proteins in surface osteoblasts. The first column shows peptide blocking controls (magnification, ×10). (B) In situ immunofluorescence examination of endogenous HOXA10 protein in MC3T3 cells to demonstrate nuclear staining with few cytoplasmic foci using the Santa Cruz antibody. (C) Nuclear localization of transfected Xpress-tagged HOXA10 is also observed for HeLa cells detected with anti-Xpress antibody (see Materials and Methods). DAPI, 4′,6′-diamidino-2-phenylindole.
While most immunohistochemical staining is nuclear, some hypertrophic chondrocytes and osteoblasts have cytoplasmic staining representing either background or specific HOXA10 staining. To address a potential artifact of cytoplasmic staining, we examined the endogenous localization of HOXA10 by in situ immunofluorescence in isolated osteoblasts. Punctate nuclear staining was observed, with a few foci detected in the cytoplasms of MC3T3 cells (Fig. 2B). However, 100% nuclear localization was observed with exogenously expressed HOXA10 protein in HeLa cells by use of the Xpress tag antibody (Fig. 2C). These findings indicate that the localization of the HOXA10 isoform with transactivation activity is nuclear.
The coordinate BMP2 induction of Hoxa10 and Runx2 in response to BMP2-induced osteogenesis in several cell models, taken together with the profiles of expression of these genes during the maturation of MC3T3 cells as well as their coexpression in osteogenic lineage cells, suggests a functional relationship between RUNX2 and HOXA10.
The P1 bone-related promoter of Runx2 and the endogenous Runx2 gene are regulated by HOXA10.
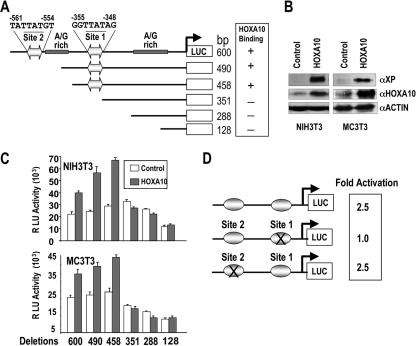
As activating signals on the Runx2 promoter during early stages of embryonic development are limited, we pursued the characterization of a Hoxa10 regulatory element in the bone-related Runx2 P1 promoter. Examination of the Runx2 0.6-kb promoter, which has been studied for its regulation in osseous and nonosseous cells, revealed the presence of three putative Hox consensus sequences. Two TTAT-containing core sequences (4, 24) and one sequence with a TTAC core (72), which binds HOXA9 well but HOXA10 with a low frequency (4, 73), are present in the proximal promoter (Fig. 3A). These sequences differ from the TAAT binding site for homeodomain proteins of the MSX and DLX classes (34, 45). To identify the HOXA10-responsive regulatory element(s) in the Runx2 gene, deletion mutation analyses were performed with both NIH 3T3 (nonosseous) and MC3T3 (osseous) cells cotransfected with Runx2 promoter deletion fragments and HOXA10. The endogenous protein levels of HOXA10 are slightly higher in the MC3T3 preosteoblasts than in the NIH 3T3 fibroblasts (Fig. 3B, control lanes) which may be contributing to the basal expression of the promoter constructs in MC3T3 cells, where the −351 fragment is reduced 20% (Fig. 3C). In the presence of exogenous HOXA10, a 2- to 2.5-fold increase in promoter activity of the −600, −490, and −458 fragments is observed, suggesting that the proximal TTAT site 1 is sufficient for HOXA10 regulation of Runx2 (Fig. 3C). Furthermore, complete loss of HOXA10 responsiveness occurs with the −351 Runx2-Luc fragment, which lacks the putative site 1 for HOXA10 binding. Furthermore, mutation of site 1 blocked HOXA10-mediated transcriptional activation (Fig. 3D). The site 2 mutation had no effect on HOXA10 induction of Runx2, i.e., the construct retained the same 2.5-fold activation level as the WT. The TTAC core site, located at −420/−417, cannot be functional, as the site 1 mutation showed no HOXA10-induced change in promoter activity. Taken together, these results are consistent with the location of a functional Hoxa10 site in the proximal promoter fragment at −353/−351, a regulatory element mediating HOXA10 activation of the Runx2 P1 promoter.
FIG. 3.
HOXA10 directly regulates Runx2 promoter activity and transcription. (A) Schematic representation of the deletions of the 0.6-kb Runx2 promoter with the firefly luciferase reporter gene. The full-length 0.6-kb promoter contains two Hoxa10 putative binding sites (site 1 and site 2) having the core TTAT motifs. Deletion clones −490- and −458-Luc contain only site 1, whereas clones −351-, −288-, and −108-Luc are devoid of Hoxa10 binding sites. (B) Western blots of endogenous and exogenous HOXA10 protein levels in nonosseous (NIH 3T3) and osseous (MC3T3) cells. For detection of endogenous and exogenous HOXA10 protein, anti-HOXA10 (N20; Santa Cruz Biotech Inc.) was used. α, anti-. (C) A functional Hoxa10 resides in the proximal Runx2 promoter. Runx2 promoter deletion mutant constructs were cotransfected with 400 ng of either backbone vector (control representing basal promoter activity [open bars]) or Hoxa10 expression construct (treated [solid bars]). Runx2 promoter activities are calculated from relative luciferase values for both NIH 3T3 and MC3T3 cells. All transfections were performed in triplicate. RLU, relative light units. (D) Two nucleotide mutations (TTAT to CGAT) were constructed in the distal (site 2) and proximal (site 1) Hoxa10 binding motifs in the 0.6-kb Runx2 P1 promoter as illustrated. Hoxa10-mediated activation for WT/mutant is indicated.
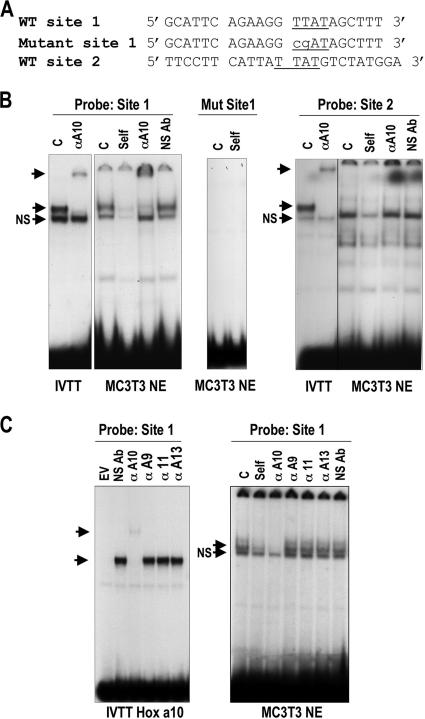
Sequence-specific binding of HOXA10 to the proximal regulatory motif was validated in EMSA by WT and mutant oligonucleotide binding studies (Fig. 4A and B). Confirming the mutation studies, site 2 does not form a HOX protein-DNA complex with osteoblast nuclear extracts (Fig. 4B). In contrast, site 1 binds in vitro-transcribed and -translated (IVTT) HOXA10 protein and HOXA10 from nuclear extracts of osteoblasts, as indicated by antibody supershifts (Fig. 4B). To rule out the possibility of redundant HOX protein binding to site 1 because other HOX factors are expressed in osteoblasts (20, 30), we further examined the interaction of other paralogous HOX proteins expressed in MC3T3 cells with Runx2 site 1. Antibody specificities were established by testing IVTT HOXA10 protein bound to probe with supershift analyses using antibodies specific to other Abd B class proteins. None of these antibodies cross-reacted with HOXA10 (Fig. 4C, left panel), nor did they supershift/block shift the HOXA10 complex from nuclear extracts bound to Runx2 site 1 (Fig. 4C, right panel). In conclusion, the deletion and site-directed mutagenesis of putative Hox elements in the 0.6-kb Runx2 promoter demonstrates that HOXA10 protein binds preferentially to the Runx2 TTAT site 1.
FIG. 4.
Specificity of the Hoxa10 site 1 for HOXA10 interactions. (A) Oligonucleotide sequences are shown for the WT and the Hoxa10 site mutations and compared to that for the mouse consensus optimal Hoxa10 binding site (4). Underlining indicates the core binding site; lowercase indicates mutant nucleotides. (B) DNA binding activity of HOXA10 is demonstrated by EMSA. Ten femtomoles of labeled double-stranded oligonucleotides derived from the WT or mutant (Mut) probe as indicated was incubated with 2 μl of IVTT HOXA10 or 5 μg of MC3T3 nuclear extract (NE). Self-competition (Self), antibody supershift, and nonspecific antibody (NS Ab) controls are shown. The mutant double-stranded oligonucleotide for Hoxa10 does not bind nuclear proteins. The top unlabeled arrows indicate the supershifted band; the lower unlabeled arrows show the position of the HOXA10-specific band, below which is a nonspecific complex (arrows labeled NS). α, anti-; C, control. (C) IVTT of indicated HOX proteins is complexed with the Hox site 1 in Runx2 to show the specificity of the HOXA10 antibody. Nuclear extracts of MC3T3 osteoblasts were used in EMSA studies to show the specificity of the HOXA10 interaction (arrow) with the site 1 Runx2 probe. GFP, green fluorescent protein; EV, empty vector.
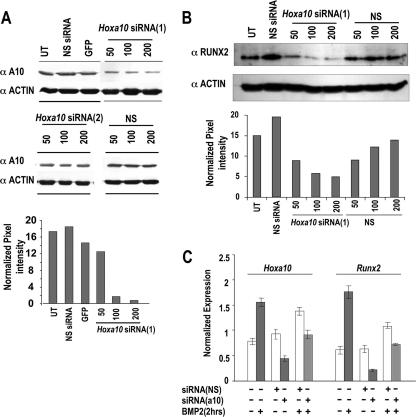
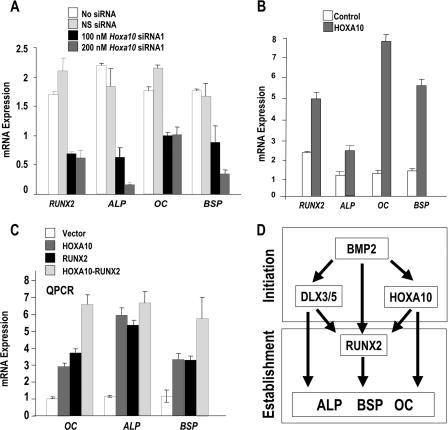
To address the biological significance of the HOXA10-mediated positive regulation of the Runx2 gene at the onset of osteoblast differentiation, cellular levels of HOXA10 were inhibited by siRNA knockdown in two osteogenic cell models. Preosteoblastic MC3T3 cells were treated with several Hoxa10 siRNA duplex oligonucleotides for 72 h, which resulted in the inhibition of HOXA10 cellular protein levels compared with the nonsilencing duplex control. Figure 5A shows results for two siRNAs, one of which only modestly decreased HOXA10 protein (siRNA2; decrease of 10 to 15% from densitometry measurements), while siRNA1 exhibited a robust dose-dependent inhibitory effect on HOXA10 and also caused a significant dose-related decrease in RUNX2 protein (Fig. 5B).
FIG. 5.
RNAi of Hoxa10 in MC3T3 and C2C12 cells inhibits Runx2 and osteogenic gene expression. (A) Knockdown of Hoxa10 in MC3T3-E1 cells by two sets of Hoxa10 siRNA duplexes and nonspecific controls (NS). The lower panel shows densitometric quantitation of HOXA10 knockdown when cells were isolated 72 h after siRNA treatment. (B) Preosteoblast MC3T3 cells at 30 to 50% confluence were transfected with siRNA1 specific for murine Hoxa10 at different concentrations (50, 100, and 200 nM). (Top) The Western blot shows the knockdown of RUNX2 protein upon treatment with Hoxa10-specific siRNA duplexes. UT, untransfected; NS, nonsilencing duplexes (used as controls). Densitometry shows reductions of RUNX2 protein upon knockdown of Hoxa10. (C) C2C12 cells were plated on day 0 at 30 to 40% confluence and treated with Hoxa10 siRNA or nonspecific siRNA [siRNA(NS)] (100 nM) for 48 h. After the medium was changed, BMP2 (100 ng/ml) was added to induce Hoxa10 and Runx2 for 2 h. mRNA levels were assayed by RT-QPCR. White bars represent the control for the treatment as indicated. GFP, green fluorescent protein; α, anti-.
The potency of HOXA10 in regulating Runx2 at the onset of BMP2-induced osteogenesis was examined with C2C12 cells (Fig. 5C). Cells were first treated with siRNA for 48 h, which was followed by BMP2 for 2 h. Hoxa10 siRNA resulted in a 50% knockdown of Hoxa10, a decrease which prevented the twofold BMP2 induction of Hoxa10 mRNA in the control (absence of Hoxa10 siRNA). Runx2 was reduced nearly 75% by Hoxa10 siRNA in the absence of BMP2. In the presence of BMP2 and Hoxa10 siRNA, Runx2 was not induced to a level over that seen for the control. However, the inhibition was less (33% decrease) than in the absence of BMP2 (>50% decrease). This finding is likely the consequence of BMP2 induction of other transcription factors that can upregulate Runx2. Thus, we were prompted to examine levels of Dlx3, a BMP2 early response gene in this cell model (35). Dlx3 was induced over the control in the presence of Hoxa10 siRNA and BMP2 (data not shown). These studies define HOXA10 as a significant regulator of Runx2 transcriptional activity and gene expression.
Functional recruitment of HOXA10 to promoters of Runx2 and other osteoblast-related genes.
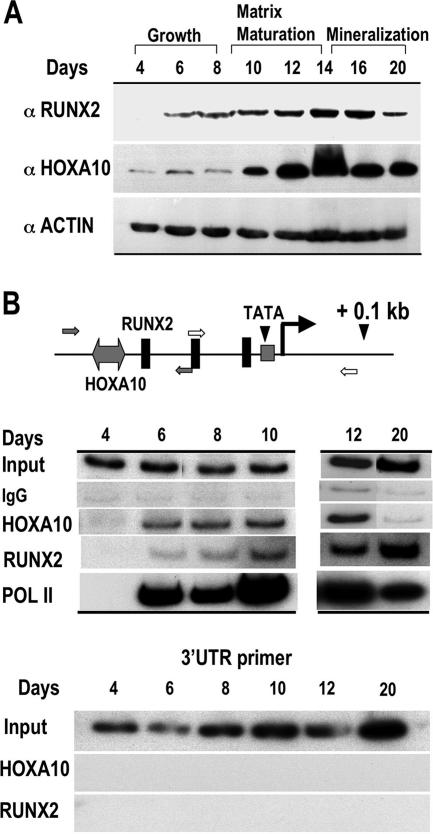
To gain further insight into the in vivo regulation of Runx2 transcription by HOXA10 during the development of the osteoblast phenotype, we performed ChIP assays with primary rat calvarial osteoblasts, which undergo a well-documented sequence of maturation stages (59). Western blot analysis for RUNX2 and HOXA10 protein levels during the differentiation time course shows that a low basal level of HOXA10 protein is present in proliferative osteoblasts (day 4), while RUNX2 protein is not observed until day 6 (Fig. 6A). When bone-like nodules form on day 10, HOXA10 is upregulated twofold, increases again on day 12, and remains at a constitutive level into the mineralization stage (day 20). RUNX2 protein levels increase continuously during osteoblast maturation but are reduced on day 20 (heavily mineralized cultures). These findings emphasize that HOXA10 is present in osteoprogenitors prior to RUNX2 protein appearance and that HOXA10 may function throughout differentiation.
FIG. 6.
HOXA10 protein and recruitment to the Runx2 promoter in primary calvarial cells during osteoblast growth and differentiation. (A) Western blot analyses for RUNX2 and HOXA10 protein expression are shown during stages of growth and differentiation of isolated primary calvarial osteoblasts as indicated (antibody information is provided in Materials and Methods). Protein profiles of actin demonstrate equivalent amounts of total protein loaded in the gel. (B) ChIP studies of the Runx2 proximal promoter locus during growth and differentiation. The top panel illustrates the positions of regulatory elements and primers used to amplify the Runx2 promoter-specific DNA fragments. Open arrow, RNA Pol II; solid arrow, HOXA10 and RUNX2 occupancy. For the middle panel, ChIP analysis was performed on the indicated days with antibodies for HOXA10, RUNX2, or RNA Pol II (see Materials and Methods). One percent of the soluble chromatin fraction was taken as the input fraction. IgG is shown as an antibody control. The lower panel shows the control for ChIP primer specificity.
The timing of recruitment of HOXA10 to the Runx2 gene in relation to transcription was determined by ChIP assays at the indicated days during differentiation (Fig. 6B). Trace levels of HOXA10 above those for the IgG control were observed on days 4 and 5 in different experiments. Consistently, we found that RNA Pol II and HOXA10 were robustly associated with the promoter, beginning on day 6, when RUNX2 protein was first detected in this experiment (Fig. 6B). This coordinate occupancy was reproducible over multiple time courses (data not shown). HOXA10 remained bound to the Runx2 promoter but dissociated on day 20, the late mineralization stage, a finding consistent with decreased RUNX2 protein levels in the cells (Fig. 6A) and decreased RNA Pol II association with the promoter (Fig. 6B). On the other hand, RUNX2 association with its own promoter gradually increased from day 6 to day 20 in late mineralization, reflecting its role in maintaining physiologic transcription by autoregulation (21). Thus, HOXA10 contributes both to the significant activation of Runx2 expression throughout the early stages of osteoblastogenesis and to the regulation of physiologic levels in mature osteoblasts by dissociation from the Runx2 promoter.
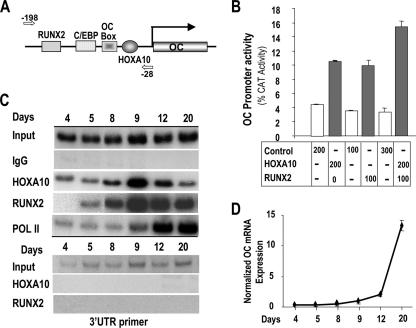
ChIP studies allow for assays of transcription factors associated with different promoters in the same sample. We therefore examined the promoters of several osteoblast genes for potential Hoxa10 binding sites and designed primers to amplify the DNA of the appropriate promoter domain. Table 2 shows the putative sequences. We examined the direct regulation of the osteoblast-specific OC gene, which is highly regulated by RUNX2 (40). A Hox binding site was located in the proximal −208 promoter segment, which exhibits tissue-restricted expression (37) (Fig. 7A). Using a −208 OC-CAT reporter construct, we found a two- to threefold increase in promoter activity in response to HOXA10 in multiple experiments (Fig. 7B). By ChIP analysis (Fig. 7C), we found that HOXA10 is associated with OC chromatin 24 h prior to RUNX2 association. During osteoblast differentiation, both HOXA10 and RUNX2 binding were enhanced on days 8 and 9, when OC mRNA first began to accumulate (Fig. 7D), consistent with increased Pol II binding on day 9 (Fig. 7C). After this initial induction of OC, HOXA10 association with the OC promoter diminished in the matrix maturation and mineralization stages, while RUNX2 remained present on OC chromatin. The coordinate increase in HOXA10 and RUNX2 binding to OC prompted us to examine the combined effects of HOXA10 and RUNX2 on OC promoter activity. We found an additive effect on promoter activity as a result of the coexpression of the factors (Fig. 7B), suggesting that the Hoxa10 and Runx2 regulatory elements function independently on the OC promoter, not synergistically. Taken together, these results demonstrate HOXA10 supports the regulation of OC expression earlier than RUNX2 and significantly contributes to OC-induced mRNA.
TABLE 2.
HOXA10 binding motifs in bone-specific promoters
| Promoter | Nucleotide at indicated positiona
|
Binding | Functional activity | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −4 | −3 | −2 | −1 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| Hoxa10 consensus | A | A | T | T | T | T | A | T | T | A | C | ||
| Runx2 (mouse)e | A | A | G | G | T | T | A | T | A | G | Cb | ++ | + |
| Runx2 (rat)e | T | T | C | A | T | T | A | T | T | A | Tc | + | − |
| A | A | C | C | T | T | A | C | A | G | Gd | − | − | |
| OC (mouse) | C | T | G | C | T | T | A | C | A | T | C | ||
| OC (rat) | C | T | G | C | T | T | A | C | A | T | T | ||
| BSP (mouse) | T | A | T | T | T | T | A | T | T | T | G | ||
| BSP (rat) | T | A | G | T | T | T | A | T | T | T | T | ||
| T | T | A | T | T | T | A | T | A | G | A | |||
| ALP (mouse) | C | T | G | C | T | T | A | T | G | A | A | ||
| ALP (rat) | G | T | C | A | T | T | A | T | A | A | C | ||
| T | C | G | T | T | T | A | T | G | C | A | |||
| T | T | A | C | T | T | A | T | A | G | G | |||
Osteoblast gene sequences were obtained from the NCBI database. HOXA10-allowed nucleotides (−4 to −1 and 5 to 7) outside the core (1, T; 2, T; 3, A; 4, T or C) are as follows for the indicated positions, as detailed in the work of Benson et al. (4): −1, T>G; −2, T>G; −3, A>T>G; −4, A>G = T; 5, T>G>A; 6, A>G>C; 7, C>A = G. HOXA9 can bind to TTAC as well as to TTAT sites, as shown by Shen et al. (72-74).
HOXA10 binds IVTT protein but not in nuclear extracts and is nonresponsive to Runx2 site 2 (Fig. 3 and 4).
Uncharacterized Runx2 binding site with TTAC motif (−419/−416).
The corresponding HOXA10 binding site indicated in nucleotides across species and obtained for different bone marker genes (Runx2, OC, BSP, and ALP).
FIG. 7.
HOXA10 is recruited to the OC promoter to directly regulate gene expression. (A) Activation of OC gene transcription by HOXA10. Schematic representation of rat proximal OC promoter segment illustrating the location of the only Hoxa10 consensus motif in the −208-bp 5′ OC fragment. Arrows are the forward (−198) and reverse (−28) OC primers used in ChIP analysis. (B) The mouse preosteoblast cell line MC3T3-E1 was cotransfected with either empty vector (control) or Hoxa10 and Runx2 expression plasmids as indicated and with the proximal OC promoter (−208 OC-CAT) by use of FuGENE6 (Roche Molecular Biologicals). Control cells contained the indicated amount of backbone plasmid (pGL3). Cells were harvested 24 h posttransfection for CAT assay. Promoter activity was calculated from values equivalent to those for six samples and expressed as percent CAT activity. (C) In vivo occupancy of HOXA10 on the OC gene. Cross-linked chromatin samples from primary rat calvarium-derived osteoblast cultures at the indicated stages of differentiation (days 4, 5, 8, 9, 12, and 20) were used for immunoprecipitation reactions with 2 μg of HOXA10, RUNX2, Pol II, and nonspecific antibody (IgG). The pull-down DNA fragments were purified and assayed by PCR. Input represents 1% of each chromatin fraction used for immunoprecipitation. The ChIP data presented are representative of multiple experiments in which all the time point data were derived from the same osteoblast preparation. The lower panel shows the control for ChIP primer specificity. (D) Expression of OC mRNA (QPCR) during differentiation of primary calvarial osteoblasts at the indicated days. C/EBP, CCAAT/enhancer-binding proteins.
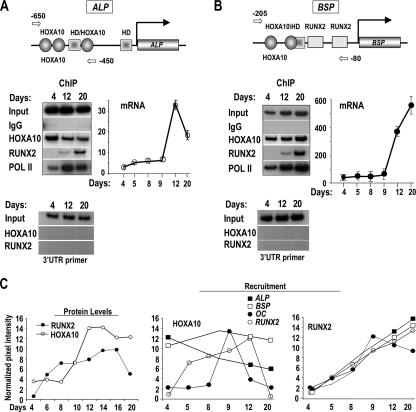
We also examined the promoters for alkaline phosphatase (ALP) and bone sialoprotein (BSP) and found Hoxa10-regulatory elements (Table 2 and Fig. 8). In proliferating primary osteoblasts, ALP (Fig. 8A) and BSP (Fig. 8B) are expressed, but at low levels. Association of HOXA10 with the ALP and BSP promoters was confirmed by ChIP assay with the osteoprogenitor cells (day 4) as well as with postproliferative mature osteoblasts (days 12 and 20). Interestingly, HOXA10 association with the promoter of ALP, the early marker of osteoblasts, was most prominent on day 4 and reduced on day 20, when ALP mRNA is downregulated. In contrast, both HOXA10 binding to the BSP promoter and BSP expression (mRNA) increased during differentiation. These observations suggest that HOXA10 directly regulates genes independent of RUNX2 binding.
FIG. 8.
Hoxa10 regulatory elements are present in the ALP (A) and BSP (B) genes. ChIP assay was performed with primary rat calvarial osteoblasts by use of the indicated antibodies at the proliferation stage (day 4) and the differentiation stages (days 12 and 20, early and mature osteoblasts in mineralized matrix). HD, homeodomain. (Top) Illustration of Hoxa10 sites and locations of primers (arrows) used to detect the association of HOXA10 with the ALP and BSP promoters; (bottom) ChIP quantitated by radioactivity with control primers shown below (left) and expression of ALP and BSP genes by RT-QPCR (right). (C) Summary profiles of cell protein levels and recruitment of HOXA10 and RUNX2 to four bone promoters. The data represent densitometric values obtained from Western blot and ChIP experiments normalized to actin and IgG values, respectively.
These ChIP studies reveal that the recruitment of RUNX2 to most gene promoters continuously increases during differentiation, with similar profiles in each case (Fig. 8C). Figure 8C summarizes the profiles (from densitometry measurements of the ChIPs) of RUNX2 and HOXA10 association with four gene promoters. In contrast to RUNX2, HOXA10 has distinct profiles of association/dissociation with individual gene promoters during differentiation. It is notable that HOXA10 association to the gene promoters is critical to gene regulation independent of HOXA10 cellular levels (Fig. 8C, left panel). These differences may reflect very dynamic functional characteristics of HOXA10 promoter binding in relation to the regulation of the transcription of osteoblast genes at stages of osteoblast phenotype development. We conclude from these studies that Hoxa10 functional elements are present in multiple osteoblast genes, including the OC, bone sialoprotein, and alkaline phosphatase genes, contributing to their regulated expression.
HOXA10 regulates osteoblast genes both dependent on (in MC3T3 cells) and independent of (in Runx2 null cells) RUNX2.
The ChIP studies (Fig. 6, 7, and 8) suggest that HOXA10 may regulate osteoblast genes that are significant functional components of bone, either synergistically, by increasing Runx2 expression, or directly, through Hox regulatory motifs. We therefore examined the consequences of the depletion of Hoxa10 on the endogenous expression of these skeletal markers of osteoblast differentiation by RT-QPCR (Fig. 9A). MC3T3 cells were treated with Hoxa10 siRNA for 48 h, which reduced Runx2 gene expression threefold. Dose-dependent decreases of the early osteoblast markers ALP and BSP were also observed, as was a twofold reduction in OC mRNA. Exogenous expression of HOXA10 in MC3T3 cells (Fig. 9B) shows that Runx2 and ALP expression is increased twofold, while the mature osteoblast-related genes OC and BSP are induced between five- and eightfold.
FIG. 9.
HOXA10 regulation of osteoblast phenotypic genes dependent on and independent of RUNX2. (A) Transcript levels of bone phenotypic markers are decreased by Hoxa10 siRNA treatment at the indicated doses. RT-QPCR mRNA expression profiles in Hoxa10 siRNA-treated (100 and 200 nM) MC3T3 cells. The relative mRNA expression profiles of the endogenous marker genes were normalized to Gapdh expression profiles. Error bars are means ± standard deviations from triplicate samples. (B) Forced expression of Hoxa10 (200 ng) for 24 h induces expression of bone phenotypic genes (RT-QPCR analyses of six samples). (C) A Runx2−/− TERT-immortalized stable cell line was transfected with 400 ng cytomegalovirus-driven Hoxa10, 200 ng Runx2, or both expression plasmids for 24 to 36 h (six samples). Cell layers were harvested for total cellular RNA and analyzed for expression of indicated genes by RT-QPCR. (D) Schematic illustration of the BMP2-induced HOXA10, homeodomain proteins DLX3/DLX5, and RUNX2 gene expression representing the initiation phase of osteogenesis. Together these genes establish the osteoblast phenotype by direct and Runx2-dependent activities on target genes.
To address the possibility of the direct regulation of osteoblast genes, we tested the competency of HOXA10 to induce their expression independent of RUNX2 by using a TERT-immortalized preosteoblastic cell line derived from Runx2 null mice (1) (Fig. 9C). Cells were transfected with Hoxa10 and/or Runx2 expression vector, and total cellular RNA was analyzed 24 h later. Expression profiling by real-time PCR revealed that HOXA10 induced OC, ALP, and BSP to an extent the same as that seen for their induction by RUNX2 (three- to fourfold for OC or BSP and five- to sixfold for ALP). The presence of HOXA10 and RUNX2 together resulted in an additive induction for OC and BSP, genes whose expression continues to increase during differentiation (Fig. 7 and 8), but not ALP (Fig. 8).
HOXA10 activation of osteogenic genes OC, ALP, and BSP in Runx2 null cells, taken together with the association of HOXA10 with binding elements in these promoters, supports a RUNX2-independent direct role for HOXA10 in promoting osteoblast differentiation. However, these promoters are also Runx2 responsive, and HOXA10 directly induces Runx2, thereby contributing to HOXA10 RUNX2-dependent transcription of osteoblast genes. The BMP2-initiated induction of Hoxa10, Runx2, and other transcriptional factors (e.g., DLX3 and DLX5) indicates that these factors all contribute specific regulatory functions to support bone formation. This combinatorial control is schematically illustrated in Fig. 9D.
HOXA10 contributes to chromatin modification of osteoblast target genes for induced transcription.
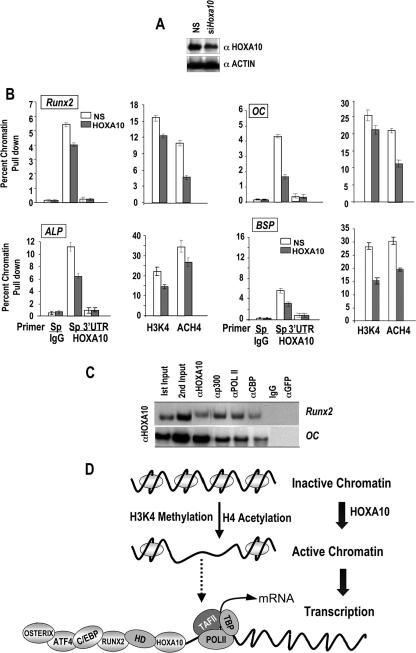
The early recruitment of HOXA10, prior to RUNX2, on the OC, ALP, and BSP gene promoters suggests that HOXA10 may contribute to remodeling the chromatin of these phenotypic genes. The remodeling of chromatin structure is mediated in part by enzymes that topologically alter DNA interactions with histones or that covalently modify the core histone proteins H3 and H4 (41). Acetylation of histone H4 and H3K4 methylation are modifications that strongly correlate with transcriptionally active chromatin (10, 75, 77). To test this hypothesis, we examined the effects of Hoxa10 depletion on histone acetylation and H3K4 methylation of gene promoters, which reflect transcriptionally active chromatin. Cells were treated with Hoxa10-specific siRNA for 72 h (Fig. 10A), and these chromatin modifications were assayed by ChIP analysis for the four bone phenotypic genes (Fig. 10B). We confirmed a 50% knockdown of Hoxa10 by siRNA and a 50% reduction in HOXA10 binding to OC, ALP, and BSP promoters but a modest decrease in the recruitment of HOXA10 to the RUNX2 promoter (Fig. 10B, left panels). We observed a significant 50% reduction in the acetylation of OC, BSP, and Runx2 chromatin but a modest decrease in ALP chromatin acetylation in cells treated with Hoxa10 siRNA relative to control levels (nonsilencing siRNA) (Fig. 10B). On the other hand, we found in multiple studies (n = 3) a significant decrease in the H3K4 methylation of ALP and BSP, while the methylation of OC and Runx2 chromatin was less decreased.
FIG. 10.
siRNA knockdown of Hoxa10 decreases histone acetylation and H3K4 methylation of osteogenic genes. (A) MC3T3 cells were treated with Hoxa10 siRNA and nonspecific control (NS) for 72 h. The knockdown effect is shown by Western blotting. (B) DNA samples from ChIP with HOXA10, nonspecific IgG, methylated histone K4 (H3K4), and acetylated histone H4 (ACH4) were amplified by gene-specific (Sp) and 3′ control UTR primers (Table 1) for the indicated gene promoters. The left panel for each gene shows the effect of Hoxa10 siRNA on HOXA10 recruitment in ChIP assay of the indicated promoter. The right panel shows the status of H3K4 methylation or H4 acetylation of the chromatin modification by Hoxa10-specific knockdown. (C) ChIP-reChIP assays were performed to identify the association of coregulatory factors p300 and CBP with HOXA10 on the Runx2 and OC promoters. A HOXA10 ChIP was performed with HOXA10 antibody, and the resulting chromatin immunoprecipitate (second input) was subjected to reChIP with the indicated antibodies (α) (secondary pull-down). Normal IgG and green fluorescent protein (GFP) antibodies provided controls. The DNA fragments for Runx2 and OC were amplified as described in Materials and Methods. (D) Schematic illustration to show how HOXA10 belongs to an epigenetic coregulatory complex for remodeling chromatin to induce transcription of osteogenic genes. We propose that HOXA10 may be among the earliest factors recruited to bone promoters. HOXA10 recruitment is followed by the coordinated occupancy of other bone-related transcription factors for maximal expression of individual genes during stages of differentiation. HD, homeodomain.
Upon RNAi-mediated depletion of HOXA10, we found an impairment of histone modifications in the chromatin of the Runx2, OC, ALP, and BSP genes, suggesting that coregulatory proteins with histone acetyltransferase (HAT) activity may be present in a HOXA10 complex associated with chromatin. To address this mechanism of chromatin remodeling by HOXA10 binding to target genes, we examined the association of coregulatory proteins having HAT activity, p300 and CBP, with HOXA10 (Fig. 10C). We performed sequential chromatin immunoprecipitation studies (ChIP-reChIP) to determine the in vivo interactions of HOXA10 with coactivator proteins on the Runx2 and OC promoters in ROS 17/2.8 cells. These cells robustly express endogenous bone-related factors. A first ChIP was performed with HOXA10 antibody, followed by pull-down with antibodies specific to each coregulatory protein (Fig. 10C). Although the interaction of HOXA10 is greater with the OC gene than with the Runx2 promoter, p300 and CBP are found in the HOXA10 complex associated with both genes. These results demonstrate that the presence of HOXA10 on bone target genes can contribute to epigenetic alterations in chromatin that favor transcription (summarized in Fig. 10D).
DISCUSSION
In this study we have defined a principal role for HOXA10 in contributing to osteoblast cell fate and differentiation. Hoxa10, which is essential for skeletal patterning, is characterized in our studies as an activator of Runx2 and three phenotypic genes (ALP, BSP, and OC) that represent progressive stages of osteoblastogenesis. Our data support the concept that HOXA10 functions in initiating osteogenic gene expression but that the bone phenotype is not fully established until Runx2 is expressed. The evidence includes the following: (i) the finding that Hoxa10 and Runx2 are immediate-response genes to the BMP2 morphogenetic signal and exhibit similar expression profiles during the induction of the osteoblast phenotype in C2C12 cells; (ii) the exhibition by both genes of robust expression in osteoprogenitors, growth plate chondrocytes, and osteoblasts in vivo, which further demonstrates the coexpression of HOXA10 and RUNX2 in osteogenic lineage cells in vivo; (iii) the activation of Runx2 transcription by HOXA10 in mesenchymal cells; (iv) the early recruitment of HOXA10 to target gene chromatin during induction of the osteoblast phenotype, which has consequential effects on acetylation and H3K4 methylation, a signature of active chromatin; and (v) RNAi-directed depletion and exogenous expression of HOXA10, which confirm a coordinate change in endogenous expression of Runx2 and osteoblast genes. Further, the demonstration that HOXA10 can directly regulate osteogenic genes in the absence of Runx2 (studies performed with Runx2−/− cells) identifies HOXA10 as a contributor to bone formation independent of Runx2. Together, these results indicate that HOXA10 plays a pivotal role at the onset of osteoblast commitment and may be a significant factor in contributing to osteoblastogenesis by dynamic regulation of osteoblast genes throughout stages of differentiation. Thus, Hoxa10 has a key role in promoting bone tissue formation beyond its requirement for patterning the embryonic skeleton.
Our findings suggest HOXA10 functions in a linear pathway with RUNX2 to promote bone formation as well as independently of RUNX2. Our model (Fig. 9D) proposes that HOXA10 functions as an immediate early response gene to the BMP2 signal for the initiation of osteogenic gene regulation and provides an amplification step for RUNX2-induced expression to establish the osteoblast phenotype. As other BMP2-induced transcription factors regulate Runx2 and osteoblastic genes (e.g., homeodomain proteins [MSX2, DLX3, and DLX5], TCF/LEF1, AP-1, OSTERIX, CCAAT/enhancer-binding proteins [encoded by C/EBP], and ATF4) (22, 28, 45, 82, 83, 86), a compelling question is that of the hierarchy of HOXA10, RUNX2, and these other factors in regulating gene expression for bone formation. Null mouse models have identified OSTERIX function later than RUNX2 (56). From a developmental perspective, BMP/TGFβ, Wnt, and HOX signaling interactions are documented (5, 62), and all of these proteins contribute to bone development. From a gene regulation perspective, a hierarchy of transcriptional control can be better deduced by understanding the ordered recruitment of factors to gene promoters and the temporal levels of the regulatory factors during osteoblast differentiation. Abd B class Hoxa10, Msx2, and the distal-less Dlx3 genes are induced within a few hours, while other factors (e.g., Dlx5 and C/EBPβ) are induced later (after 8 h) in the C2C12 model. Recent studies provide evidence from ChIP studies for a homeodomain protein regulatory network for osteoblast commitment and differentiation (35). Here we show HOXA10 to be associated with bone promoters prior to RUNX2, suggesting that it contributes to the initiation of the osteoblast phenotype. However, increased HOXA10 binding appears to be coordinated with increased recruitment of Runx2, homeodomain proteins, and other factors postproliferatively for accumulation of mRNA, thereby establishing the bone phenotype (35). The selective temporal binding of HOXA10 to promoters reflects binding and dissociation during differentiation. This is in contrast to the continual increase in RUNX2 binding to promoters, suggesting that HOXA10 has a very dynamic regulatory function for regulating osteogenic gene expression during osteoblast differentiation.
We found modifications of histones at the level of chromatin in response to altered HOXA10 expression, indicating a remodeling of the transcriptional machinery that is necessary for in vivo gene regulation. HOXA10 recruitment to bone promoters prior to a significant accumulation of mRNA but after Pol II association emphasizes a functional role for HOXA10 increasing transcription through chromatin remodeling. This is supported by knockdown of HOXA10 reducing histone acetylation and H3K4 methylation, which are associated with active transcription and by HOXA10 and p300/CBP coassociation on the Runx2 and OC promoters (ChIP-reChIP studies). Earlier studies have reported that HOX factors can modify transcriptional activation and repression by interacting with p300/CBP (14, 71). Our studies have focused on HOXA10 in supporting chromatin remodeling for the activation of osteoblast-related genes, as was shown for p21 (9). Equally important is the fact that HOXA10 can promote histone deacetylation by recruiting histone deacetylase 2 and SIRT2 histone deacetylases to repress gene transcription (67). Histone deacetylase-HOXA10 interactions would reverse HAT activity and could be operative in osteoblasts for HOXA10 to mediate either activation or attenuation of gene expression. Such mechanisms are important for HOXA10 suppressor function in undifferentiated myeloid cells and HOXA9 regulation of neovascularization (2, 51, 65, 78). In addition to coregulatory factors that modify chromatin, the other well-characterized partner proteins for the Abd B HOX proteins, PBX1 and MEIS, both having homeodomain protein modules, form complexes with HOX proteins and also bind to DNA to regulate repressor and enhancer activities of Hox genes. HOXA9, HOXA10, HOXA11, HOXD12, and HOXB13 each have different properties of interaction with these coregulators (52, 61, 72-74). For example, high-affinity DNA binding can be achieved when HOXB9 and HOXA10 proteins are dimerized with PBX1. While our forced-expression and siRNA knockdown studies show that HOXA10 has anabolic activity on osteoblast gene expression, how these endogenous partner proteins influence the HOXA10-mediated expression of osteoblast genes is a provocative question.
The specificity of HOXA10 for regulating osteoblast differentiation should be considered in relation to Hox nonparalogous and paralogous genes, which are known to overlap in expression in some tissues but can have preferential activities (15, 17, 27). For example, HOXA10 and HOXD10 regulate the skeleton but may exhibit specific gene regulation properties (36, 49, 79). Also, HOXA10 and HOX11 homeobox proteins are equivalent for axial but not appendicular skeletal development (87). The TTAT regulatory element is recognized by several HOX proteins; however, protein-DNA interactions as well as protein-protein interactions are dependent on the contextual sequence of the core TTAT motif (8, 13, 57). Our analysis of the functional specificity of the Runx2 Hoxa10 site 1 and no other core motifs in the 0.6-kb Runx2 promoter is consistent with other studies evaluating the specificities of Hoxa10 sites (4, 72, 74). Although several HOX proteins are likely present in osteoblast nuclear extracts (e.g., HOXA9), our findings revealed a supershifted/block shifted complex by the HOXA10 antibody. However, it is possible that other HOX protein interactions may occur at this and potentially other HOX motifs within the Runx2 gene. Our data do support the concept that the Runx2 site 1 (TTAT) has a high degree of specificity for HOXA10 binding. Interestingly, the OC gene is characterized by a TTAC core motif, implicating this motif in the formation of complexes that support a high degree of OC expression in mature osteoblasts. It is also noteworthy that the flanking nucleotides are distinct among the four target genes studied here (Table 2). This finding implies that their sequences may hold information for the dynamic association of HOXA10 with an individual gene throughout differentiation.
HOXA10 is expressed in many different tissues in which its functions have been addressed and may be related to its activities in bone tissue. Progesterone and 17β-estradiol, hormones that regulate turnover of adult bone (53, 63), increase Hoxa10 expression (16, 48). HOXA10 regulates hematopoietic differentiation in part via activation of p21 (WAF1/CIP1), the cyclin-dependent kinase inhibitor (9, 25). In the spinal cord, expression of Hoxa10 is confined to the postmitotic cell population (15). We find that HOXA10 is expressed in proliferating cells and upregulated in postproliferative mature osteoblasts in vitro. HOXA10 is also a target of induction by vitamin D3 (66), a hormone that increases the expression of osteoblast-related genes. Thus, HOXA10 appears to have a broad role in gene regulation for contributing to cell differentiation. Our studies have added to the growing recognition that Hox genes have important functions in the adult, including now the skeleton (55).
In conclusion, our characterization of HOXA10 positive regulation of genes that represent major functional components for bone formation, including those encoding RUNX2 (a bone essential transcription factor), alkaline phosphatase (required for matrix mineralization), bone sialoprotein (an important cell matrix-binding phosphoprotein with hydroxyapatite nucleation capabilities), and OC (the calcium binding bone-specific protein) (47), solidifies the concept that HOXA10 is an important regulator of gene expression throughout the progression of bone formation. These novel functions for HOXA10 in regulating target genes for osteoblast differentiation and bone formation in the postnatal skeleton have broad implications for HOXA10 functions in normal bone metabolism and bone-related disorders.
Acknowledgments
Studies reported were supported in part by grants from the National Institutes of Health (DE12528, AR39588, AR48818, and P30 DK32520).
The contents of this work are solely our responsibility and do not necessarily represent the official views of the National Institutes of Health.
We thank Richard L. Maas (Department of Medicine, Brigham and Women's Hospital, Harvard Medical School, Boston, MA) for reagents, Anthony Imbalzano (University of Massachusetts Medical School) for helpful discussions, Dana Fredericks for technical assistance, and Judy Rask for manuscript preparation.
Footnotes
Published ahead of print on 26 February 2007.
REFERENCES
- 1.Bae, J.-S., S. Gutierrez, R. Narla, J. Pratap, R. Devados, J. L. Stein, J. B. Lian, G. S. Stein, and A. Javed. 2007. Reconstitution of Runx2/Cbfa1 null cells identifies Runx2 functional domains required for osteoblast differentiation and responsiveness to osteogenic regulators BMP2, TGFα and 1,25(OH)2D3. J. Cell. Biochem. 100:434-449. [DOI] [PubMed] [Google Scholar]
- 2.Bae, N. S., M. J. Swanson, A. Vassilev, and B. H. Howard. 2004. Human histone deacetylase SIRT2 interacts with the homeobox transcription factor HOXA10. J. Biochem. (Tokyo) 135:695-700. [DOI] [PubMed] [Google Scholar]
- 3.Balint, E., D. Lapointe, H. Drissi, C. van der Meijden, D. W. Young, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 2003. Phenotype discovery by gene expression profiling: mapping of biological processes linked to BMP-2-mediated osteoblast differentiation. J. Cell. Biochem. 89:401-426. [DOI] [PubMed] [Google Scholar]
- 4.Benson, G. V., T. H. Nguyen, and R. L. Maas. 1995. The expression pattern of the murine Hoxa-10 gene and the sequence recognition of its homeodomain reveal specific properties of Abdominal B-like genes. Mol. Cell. Biol. 15:1591-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bondos, S. 2006. Variations on a theme: Hox and Wnt combinatorial regulation during animal development. Sci. STKE 2006:e38. [DOI] [PubMed] [Google Scholar]
- 6.Boulet, A. M., and M. R. Capecchi. 2002. Duplication of the Hoxd11 gene causes alterations in the axial and appendicular skeleton of the mouse. Dev. Biol. 249:96-107. [DOI] [PubMed] [Google Scholar]
- 7.Boulet, A. M., and M. R. Capecchi. 2004. Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forelimb zeugopod. Development 131:299-309. [DOI] [PubMed] [Google Scholar]
- 8.Brake, R. L., U. R. Kees, and P. M. Watt. 2002. A complex containing PBX2 contributes to activation of the proto-oncogene HOX11. Biochem. Biophys. Res. Commun. 294:23-34. [DOI] [PubMed] [Google Scholar]
- 9.Bromleigh, V. C., and L. P. Freedman. 2000. p21 is a transcriptional target of HOXA10 in differentiating myelomonocytic cells. Genes Dev. 14:2581-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulger, M. 2005. Hyperacetylated chromatin domains: lessons from heterochromatin. J. Biol. Chem. 280:21689-21692. [DOI] [PubMed] [Google Scholar]
- 11.Carapuco, M., A. Novoa, N. Bobola, and M. Mallo. 2005. Hox genes specify vertebral types in the presomitic mesoderm. Genes Dev. 19:2116-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carpenter, E. M., J. M. Goddard, A. P. Davis, T. P. Nguyen, and M. R. Capecchi. 1997. Targeted disruption of Hoxd-10 affects mouse hindlimb development. Development 124:4505-4514. [DOI] [PubMed] [Google Scholar]
- 13.Chang, C. P., L. Brocchieri, W. F. Shen, C. Largman, and M. L. Cleary. 1996. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol. Cell. Biol. 16:1734-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chariot, A., C. van Lint, M. Chapelier, J. Gielen, M. P. Merville, and V. Bours. 1999. CBP and histone deacetylase inhibition enhance the transactivation potential of the HOXB7 homeodomain-containing protein. Oncogene 18:4007-4014. [DOI] [PubMed] [Google Scholar]
- 15.Choe, A., H. Q. Phun, D. D. Tieu, Y. H. Hu, and E. M. Carpenter. 2006. Expression patterns of Hox10 paralogous genes during lumbar spinal cord development. Gene Expr. Patterns 6:730-737. [DOI] [PubMed] [Google Scholar]
- 16.Daftary, G. S., and H. S. Taylor. 2006. Endocrine regulation of HOX genes. Endocr. Rev. 27:331-355. [DOI] [PubMed] [Google Scholar]
- 17.Davis, A. P., D. P. Witte, H. M. Hsieh-Li, S. S. Potter, and M. R. Capecchi. 1995. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature 375:791-795. [DOI] [PubMed] [Google Scholar]
- 18.de la Fuente, L., and J. A. Helms. 2005. Head, shoulders, knees, and toes. Dev. Biol. 282:294-306. [DOI] [PubMed] [Google Scholar]
- 19.Depew, M. J., C. A. Simpson, M. Morasso, and J. L. Rubenstein. 2005. Reassessing the Dlx code: the genetic regulation of branchial arch skeletal pattern and development. J. Anat. 207:501-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobreva, G., M. Chahrour, M. Dautzenberg, L. Chirivella, B. Kanzler, I. Farinas, G. Karsenty, and R. Grosschedl. 2006. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell 125:971-986. [DOI] [PubMed] [Google Scholar]
- 21.Drissi, H., Q. Luc, R. Shakoori, S. Chuva de Sousa Lopes, J.-Y. Choi, A. Terry, M. Hu, S. Jones, J. C. Neil, J. B. Lian, J. L. Stein, A. J. van Wijnen, and G. S. Stein. 2000. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J. Cell. Physiol. 184:341-350. [DOI] [PubMed] [Google Scholar]
- 22.Drissi, H., A. Pouliot, J. L. Stein, A. J. van Wijnen, G. S. Stein, and J. B. Lian. 2002. Identification of novel protein/DNA interactions within the promoter of the bone-related transcription factor Runx2/Cbfa1. J. Cell. Biochem. 86:403-412. [DOI] [PubMed] [Google Scholar]
- 23.Ducy, P., R. Zhang, V. Geoffroy, A. L. Ridall, and G. Karsenty. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747-754. [DOI] [PubMed] [Google Scholar]
- 24.Ekker, S. C., D. G. Jackson, D. P. von Kessler, B. I. Sun, K. E. Young, and P. A. Beachy. 1994. The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. EMBO J. 13:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eklund, E. A. 2006. The role of HOX genes in myeloid leukemogenesis. Curr. Opin. Hematol. 13:67-73. [DOI] [PubMed] [Google Scholar]
- 26.Favier, B., F. M. Rijli, C. Fromental-Ramain, V. Fraulob, P. Chambon, and P. Dolle. 1996. Functional cooperation between the non-paralogous genes Hoxa-10 and Hoxd-11 in the developing forelimb and axial skeleton. Development 122:449-460. [DOI] [PubMed] [Google Scholar]
- 27.Fromental-Ramain, C., X. Warot, S. Lakkaraju, B. Favier, H. Haack, C. Birling, A. Dierich, P. Dollé, and P. Chambon. 1996. Specific and redundant functions of the paralogous Hoxa-9 and Hoxd-9 genes in forelimb and axial skeleton patterning. Development 122:461-472. [DOI] [PubMed] [Google Scholar]
- 28.Gaur, T., C. J. Lengner, H. Hovhannisyan, R. A. Bhat, P. V. N. Bodine, B. S. Komm, A. Javed, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 2005. Canonical WNT signaling promotes osteogenesis by directly stimulating RUNX2 gene expression. J. Biol. Chem. 280:33132-33140. [DOI] [PubMed] [Google Scholar]
- 29.Gaur, T., C. J. Lengner, S. Hussain, B. Trevant, D. Ayers, J. L. Stein, P. V. N. Bodine, B. S. Komm, G. S. Stein, and J. B. Lian. 2006. Secreted frizzled protein 1 regulates Wnt signaling for BMP induced chondrocyte differentiation. J. Cell. Physiol. 208:87-96. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 30.Gersch, R. P., F. Lombardo, S. C. McGovern, and M. Hadjiargyrou. 2005. Reactivation of Hox gene expression during bone regeneration. J. Orthop. Res. 23:882-890. [DOI] [PubMed] [Google Scholar]
- 31.Goodman, F. R. 2002. Limb malformations and the human HOX genes. Am. J. Med. Genet. 112:256-265. [DOI] [PubMed] [Google Scholar]
- 32.Gutierrez, S., A. Javed, D. Tennant, M. van Rees, M. Montecino, G. S. Stein, J. L. Stein, and J. B. Lian. 2002. CCAAT/enhancer-binding proteins (C/EBP) β and δ activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J. Biol. Chem. 277:1316-1323. [DOI] [PubMed] [Google Scholar]
- 33.Harris, S. E., D. Guo, M. A. Harris, A. Krishnaswamy, and A. Lichtler. 2003. Transcriptional regulation of BMP-2 activated genes in osteoblasts using gene expression microarray analysis: role of Dlx2 and Dlx5 transcription factors. Front. Biosci. 8:s1249-s1265. [DOI] [PubMed] [Google Scholar]
- 34.Hassan, M. Q., A. Javed, M. I. Morasso, J. Karlin, M. Montecino, A. J. van Wijnen, G. S. Stein, J. L. Stein, and J. B. Lian. 2004. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol. Cell. Biol. 24:9248-9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hassan, M. Q., R. S. Tare, S. Lee, M. Mandeville, M. I. Morasso, A. Javed, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 2006. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by Dlx3 and a homeodomain transcriptional network. J. Biol. Chem. 281:40515-40526. [DOI] [PubMed] [Google Scholar]
- 36.Hedlund, E., S. L. Karsten, L. Kudo, D. H. Geschwind, and E. M. Carpenter. 2004. Identification of a Hoxd10-regulated transcriptional network and combinatorial interactions with Hoxa10 during spinal cord development. J. Neurosci. Res. 75:307-319. [DOI] [PubMed] [Google Scholar]
- 37.Hoffmann, H. M., T. L. Beumer, S. Rahman, L. R. McCabe, C. Banerjee, F. Aslam, J. A. Tiro, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 1996. Bone tissue-specific transcription of the osteocalcin gene: role of an activator osteoblast-specific complex and suppressor hox proteins that bind the OC box. J. Cell. Biochem. 61:310-324. [DOI] [PubMed] [Google Scholar]
- 38.Izpisua-Belmonte, J. C., H. Falkenstein, P. Dolle, A. Renucci, and D. Duboule. 1991. Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 10:2279-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Javed, A., B. Guo, S. Hiebert, J.-Y. Choi, J. Green, S.-C. Zhao, M. A. Osborne, S. Stifani, J. L. Stein, J. B. Lian, A. J. van Wijnen, and G. S. Stein. 2000. Groucho/TLE/R-Esp proteins associate with the nuclear matrix and repress RUNX (CBFα/AML/PEBP2α) dependent activation of tissue-specific gene transcription. J. Cell Sci. 113:2221-2231. [DOI] [PubMed] [Google Scholar]
- 40.Javed, A., S. Gutierrez, M. Montecino, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 1999. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D responsive transcription and contribute to chromatin organization. Mol. Cell. Biol. 19:7491-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 42.Kim, Y. J., H. N. Kim, E. K. Park, B. H. Lee, H. M. Ryoo, S. Y. Kim, I. S. Kim, J. L. Stein, J. B. Lian, G. S. Stein, A. J. van Wijnen, and J. Y. Choi. 2006. The bone-related Zn finger transcription factor Osterix promotes proliferation of mesenchymal cells. Gene 366:145-151. [DOI] [PubMed] [Google Scholar]
- 43.Komori, T., H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimizu, R. T. Bronson, Y.-H. Gao, M. Inada, M. Sato, R. Okamoto, Y. Kitamura, S. Yoshiki, and T. Kishimoto. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755-764. [DOI] [PubMed] [Google Scholar]
- 44.Krumlauf, R. 1994. Hox genes in vertebrate development. Cell 78:191-201. [DOI] [PubMed] [Google Scholar]
- 45.Lee, M. H., Y. J. Kim, W. J. Yoon, J. I. Kim, B. G. Kim, Y. S. Hwang, J. M. Wozney, X. Z. Chi, S. C. Bae, K. Y. Choi, J. Y. Cho, J. Y. Choi, and H. M. Ryoo. 2005. Dlx5 specifically regulates Runx2-II expression by binding to homeodomain response elements in the Runx2 distal promoter. J. Biol. Chem. 280:35579-35587. [DOI] [PubMed] [Google Scholar]
- 46.Lengner, C. J., M. Q. Hassan, R. W. Serra, C. Lepper, A. J. van Wijnen, J. L. Stein, J. B. Lian, and G. S. Stein. 2005. Nkx3.2 mediated repression of RUNX2 promotes chondrogenic differentiation. J. Biol. Chem. 280:15872-15879. [DOI] [PubMed] [Google Scholar]
- 47.Lian, J. B., A. Javed, S. K. Zaidi, C. Lengner, M. Montecino, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 2004. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit. Rev. Eukaryot. Gene Expr. 14:1-41. [PubMed] [Google Scholar]
- 48.Lim, H., L. Ma, W. G. Ma, R. L. Maas, and S. K. Dey. 1999. Hoxa-10 regulates uterine stromal cell responsiveness to progesterone during implantation and decidualization in the mouse. Mol. Endocrinol. 13:1005-1017. [DOI] [PubMed] [Google Scholar]
- 49.Lin, A. W., and E. M. Carpenter. 2003. Hoxa10 and Hoxd10 coordinately regulate lumbar motor neuron patterning. J. Neurobiol. 56:328-337. [DOI] [PubMed] [Google Scholar]
- 50.Lowney, P., J. Corral, K. Detmer, M. M. LeBeau, L. Deaven, H. J. Lawrence, and C. Largman. 1991. A human Hox 1 homeobox gene exhibits myeloid-specific expression of alternative transcripts in human hematopoietic cells. Nucleic Acids Res. 19:3443-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lu, Y., I. Goldenberg, L. Bei, J. Andrejic, and E. A. Eklund. 2003. HoxA10 represses gene transcription in undifferentiated myeloid cells by interaction with histone deacetylase 2. J. Biol. Chem. 278:47792-47802. [DOI] [PubMed] [Google Scholar]
- 52.Mann, R. S., and M. Affolter. 1998. Hox proteins meet more partners. Curr. Opin. Genet. Dev. 8:423-429. [DOI] [PubMed] [Google Scholar]
- 53.Manolagas, S. C., S. Kousteni, and R. L. Jilka. 2002. Sex steroids and bone. Recent Prog. Horm. Res. 57:385-409. [DOI] [PubMed] [Google Scholar]
- 54.Merabet, S., J. Pradel, and Y. Graba. 2005. Getting a molecular grasp on Hox contextual activity. Trends Genet. 21:477-480. [DOI] [PubMed] [Google Scholar]
- 55.Morgan, R. 2006. Hox genes: a continuation of embryonic patterning? Trends Genet. 22:67-69. [DOI] [PubMed] [Google Scholar]
- 56.Nakashima, K., X. Zhou, G. Kunkel, Z. Zhang, J. M. Deng, R. R. Behringer, and B. de Crombrugghe. 2002. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108:17-29. [DOI] [PubMed] [Google Scholar]
- 57.Neuteboom, S. T., and C. Murre. 1997. Pbx raises the DNA binding specificity but not the selectivity of antennapedia Hox proteins. Mol. Cell. Biol. 17:4696-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Otto, F., A. P. Thornell, T. Crompton, A. Denzel, K. C. Gilmour, I. R. Rosewell, G. W. H. Stamp, R. S. P. Beddington, S. Mundlos, B. R. Olsen, P. B. Selby, and M. J. Owen. 1997. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89:765-771. [DOI] [PubMed] [Google Scholar]
- 59.Owen, T. A., M. Aronow, V. Shalhoub, L. M. Barone, L. Wilming, M. S. Tassinari, M. B. Kennedy, S. Pockwinse, J. B. Lian, and G. S. Stein. 1990. Progressive development of the rat osteoblast phenotype in vitro: reciprocal relationships in expression of genes associated with osteoblast proliferation and differentiation during formation of the bone extracellular matrix. J. Cell. Physiol. 143:420-430. [DOI] [PubMed] [Google Scholar]
- 60.Pineault, N., C. Abramovich, H. Ohta, and R. K. Humphries. 2004. Differential and common leukemogenic potentials of multiple NUP98-Hox fusion proteins alone or with Meis1. Mol. Cell. Biol. 24:1907-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pineault, N., C. D. Helgason, H. J. Lawrence, and R. K. Humphries. 2002. Differential expression of Hox, Meis1, and Pbx1 genes in primitive cells throughout murine hematopoietic ontogeny. Exp. Hematol. 30:49-57. [DOI] [PubMed] [Google Scholar]
- 62.Pockwinse, S. M., J. B. Lawrence, R. H. Singer, J. L. Stein, J. B. Lian, and G. S. Stein. 1993. Gene expression at single cell resolution associated with development of the bone cell phenotype: ultrastructural and in situ hybridization analysis. Bone 14:347-352. [DOI] [PubMed] [Google Scholar]
- 63.Riggs, B. L., S. Khosla, and L. J. Melton III. 2002. Sex steroids and the construction and conservation of the adult skeleton. Endocr. Rev. 23:279-302. [DOI] [PubMed] [Google Scholar]
- 64.Rijli, F. M., and P. Chambon. 1997. Genetic interactions of Hox genes in limb development: learning from compound mutants. Curr. Opin. Genet. Dev. 7:481-487. [DOI] [PubMed] [Google Scholar]
- 65.Rossig, L., C. Urbich, T. Bruhl, E. Dernbach, C. Heeschen, E. Chavakis, K. Sasaki, D. Aicher, F. Diehl, F. Seeger, M. Potente, A. Aicher, L. Zanetta, E. Dejana, A. M. Zeiher, and S. Dimmeler. 2005. Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. J. Exp. Med. 201:1825-1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rots, N. Y., M. Liu, E. C. Anderson, and L. P. Freedman. 1998. A differential screen for ligand-regulated genes: identification of HoxA10 as a target of vitamin D3 induction in myeloid leukemic cells. Mol. Cell. Biol. 18:1911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saleh, M., I. Rambaldi, X. J. Yang, and M. S. Featherstone. 2000. Cell signaling switches HOX-PBX complexes from repressors to activators of transcription mediated by histone deacetylases and histone acetyltransferases. Mol. Cell. Biol. 20:8623-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salsi, V., and V. Zappavigna. 2006. Hoxd13 and Hoxa13 directly control the expression of the EphA7 ephrin tyrosine kinase receptor in developing limbs. J. Biol. Chem. 281:1992-1999. [DOI] [PubMed] [Google Scholar]
- 69.Shao, J. S., S. L. Cheng, J. M. Pingsterhaus, N. Charlton-Kachigian, A. P. Loewy, and D. A. Towler. 2005. Msx2 promotes cardiovascular calcification by activating paracrine Wnt signals. J. Clin. Investig. 115:1210-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shen, J., M. A. Montecino, J. B. Lian, G. S. Stein, A. J. van Wijnen, and J. L. Stein. 2002. Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin D-dependent, bone tissue-specific transcription. J. Biol. Chem. 277:20284-20292. [DOI] [PubMed] [Google Scholar]
- 71.Shen, W. F., K. Krishnan, H. J. Lawrence, and C. Largman. 2001. The HOX homeodomain proteins block CBP histone acetyltransferase activity. Mol. Cell. Biol. 21:7509-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen, W. F., J. C. Montgomery, S. Rozenfeld, J. J. Moskow, H. J. Lawrence, A. M. Buchberg, and C. Largman. 1997. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol. Cell. Biol. 17:6448-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen, W. F., S. Rozenfeld, A. Kwong, L. G. Kömüves, H. J. Lawrence, and C. Largman. 1999. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol. Cell. Biol. 19:3051-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen, W. F., S. Rozenfeld, H. J. Lawrence, and C. Largman. 1997. The Abd-B-like Hox homeodomain proteins can be subdivided by the ability to form complexes with Pbx1a on a novel DNA target. J. Biol. Chem. 272:8198-8206. [DOI] [PubMed] [Google Scholar]
- 75.Sims, R. J., III, and D. Reinberg. 2006. Histone H3 Lys 4 methylation: caught in a bind? Genes Dev. 20:2779-2786. [DOI] [PubMed] [Google Scholar]
- 76.Smith, N., Y. Dong, J. Pratap, J. B. Lian, P. Kingsley, A. J. van Wijnen, J. L. Stein, E. M. Schwarz, R. J. O'Keefe, G. S. Stein, and M. H. Drissi. 2005. Overlapping expression of Runx1(Cbfa2) and Runx2(Cbfa1) transcription factors supports cooperative induction of skeletal development. J. Cell. Physiol. 203:133-143. [DOI] [PubMed] [Google Scholar]
- 77.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Thorsteinsdottir, U., G. Sauvageau, M. R. Hough, W. Dragowska, P. M. Lansdorp, H. J. Lawrence, C. Largman, and R. K. Humphries. 1997. Overexpression of HOXA10 in murine hematopoietic cells perturbs both myeloid and lymphoid differentiation and leads to acute myeloid leukemia. Mol. Cell. Biol. 17:495-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wahba, G. M., S. L. Hostikka, and E. M. Carpenter. 2001. The paralogous Hox genes Hoxa10 and Hoxd10 interact to pattern the mouse hindlimb peripheral nervous system and skeleton. Dev. Biol. 231:87-102. [DOI] [PubMed] [Google Scholar]
- 80.Wellik, D. M., and M. R. Capecchi. 2003. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science 301:363-367. [DOI] [PubMed] [Google Scholar]
- 81.Williams, T. M., M. E. Williams, and J. W. Innis. 2005. Range of HOX/TALE superclass associations and protein domain requirements for HOXA13:MEIS interaction. Dev. Biol. 277:457-471. [DOI] [PubMed] [Google Scholar]
- 82.Xiao, G., D. Jiang, C. Ge, Z. Zhao, Y. Lai, H. Boules, M. Phimphilai, X. Yang, G. Karsenty, and R. T. Franceschi. 2005. Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression. J. Biol. Chem. 280:30689-30696. [DOI] [PubMed] [Google Scholar]
- 83.Yang, X., and G. Karsenty. 2004. ATF4, the osteoblast accumulation of which is determined post-translationally, can induce osteoblast-specific gene expression in non-osteoblastic cells. J. Biol. Chem. 279:47109-47114. [DOI] [PubMed] [Google Scholar]
- 84.Yoon, B. S., and K. M. Lyons. 2004. Multiple functions of BMPs in chondrogenesis. J. Cell. Biochem. 93:93-103. [DOI] [PubMed] [Google Scholar]
- 85.Zaidi, S. K., A. Javed, J.-Y. Choi, A. J. van Wijnen, J. L. Stein, J. B. Lian, and G. S. Stein. 2001. A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J. Cell Sci. 114:3093-3102. [DOI] [PubMed] [Google Scholar]
- 86.Zambotti, A., H. Makhluf, J. Shen, and P. Ducy. 2002. Characterization of an osteoblast-specific enhancer element in the CBFA1 gene. J. Biol. Chem. 277:41497-41506. [DOI] [PubMed] [Google Scholar]
- 87.Zhao, Y., and S. S. Potter. 2002. Functional comparison of the Hoxa 4, Hoxa 10, and Hoxa 11 homeoboxes. Dev. Biol. 244:21-36. [DOI] [PubMed] [Google Scholar]