Abstract
Ets1 is a member of the Ets transcription factor family. Alternative splicing of exon VII results in two naturally occurring protein isoforms: full-length Ets1 (p51-Ets1) and Ets1ΔVII (p42-Ets1). These isoforms bear key distinctions regarding protein-protein interactions, DNA binding kinetics, and transcriptional target specificity. Disruption of both Ets1 isoforms in mice results in the loss of detectable NK and NKT cell activity and defects in B and T lymphocytes. We generated mice that express only the Ets1ΔVII isoform. Ets1ΔVII homozygous mice express no p51-Ets1 and elevated levels of the p42-Ets1 protein relative to the wild type and display increased perinatal lethality, thymomegaly, and peripheral lymphopenia. Proliferation was increased in both the thymus and the spleen, while apoptosis was decreased in the thymus and increased in the spleen of homozygotes. Significant elevations of CD8+ and CD8+CD4+ thymocytes were observed. Lymphoid cell (CD19+, CD4+, and CD8+) reductions were predominantly responsible for diminished spleen cellularity, with fewer memory cells and a failure of homeostatic proliferation to maintain peripheral lymphocytes. Collectively, the Ets1ΔVII mutants demonstrate lymphocyte maturation defects associated with misregulation of p16Ink4a, p27Kip1, and CD44. Thus, a balance in the differential regulation of Ets1 isoforms represents a potential mechanism in the control of lymphoid maturation and homeostasis.
Ets1 is a member of the Ets winged helix-turn-helix transcription factor family. Ets transcription factors are activators and repressors of transcription, important in the development of organisms throughout the Metazoa (9, 10, 22, 38, 48, 50). Previous Ets1 knockout mice show partial perinatal lethality. Surviving mice and chimeras derived from Ets1-targeted cells exhibit pan-lymphoid defects. These defects include reduced numbers of thymocytes, lymphoid cells in the spleen, and NK and NKT cells; loss of detectable NK and NKT cell activity; decreased proliferation and elevated apoptosis in mature T cells (4, 6, 40, 58); increased differentiation of B cells to plasma cells with elevated serum immunoglobulin M; B-cell receptor signaling defects (12); abnormalities in T-cell receptor (TCR) β-selection in thymopoiesis; and an ineffective Th1 response (11, 19). Complicating the assessment of Ets1 function is the fact that Ets1-null mice have been studied not only on a mixed genetic background (C57BL/6 × 129Sv) predisposed to autoimmunity (53) but also with a line of Ets1-null mice that expresses a very low level of a neomorphic protein (∼1 to 5%) lacking the pointed domain of Ets1 (7, 59). These Ets1-null mice present with a deficiency in CD8+ thymocyte development and an elevated proportion of single-positive effector memory (CD62L− CD44+) cells in both CD4+ and CD8+ peripheral T-cell populations (7).
The mammalian Ets1 gene produces two major protein isoforms, full-length p51-Ets1 and p42-Ets1, which arise from alternative splicing of exon VII of Ets1 hnRNA with no disruption in the reading frame (5, 24, 29). The p51-Ets1 and p42-Ets1 isoforms have common and distinctive physical properties that allow for both functional redundancy and isoform-specific activities. Shared domains include the Pointed (PNT), transactivation, and ETS DNA binding domains. The domain encoded by exon VII has been identified as a negative regulator and modulator of DNA binding, a mediator of homo- and heterotypic protein-protein interactions, and a target of calcium-mediated phosphorylation (2, 8, 14, 28, 46). Collectively, these distinctions contribute to the Ets1 isoform-specific modulation of particular Ets1 target genes. Exon VII-mediated homotypic and AML1 (Runx1)-Ets1 heterotypic protein-protein interactions augment transcription of MMP-3 and granulocyte-macrophage colony-stimulating factor, respectively (2, 35, 36). Conversely, other target genes, such as the VE-cadherin gene, are transcriptionally activated more strongly by the p42-Ets1 variant in a cell context-specific manner (33). These distinctions are physiologically relevant; ectopic expression of the p42-Ets1 isoform but not the full-length p51-Ets1 isoform in DLD-1 colon cancer cells restores Fas-mediated apoptosis (32). A similar proapoptotic role for the p42-Ets1 isoform has been suggested for MDA-MB-231 invasive breast cancer cells (3). While much work characterizing the functions of Ets1 has been done, relatively little is known about the interplay between the two protein isoforms in mediating these functions in vivo. To extend the functional assessment of Ets1 protein isoforms in vivo, we have generated Ets1ΔVII gene-targeted mice, which express only the p42-Ets1 isoform (lacking the full-length p51-Ets1 isoform). Analyses of Ets1ΔVII mice have demonstrated a requirement for the full-length p51-Ets1 isoform in the regulation of lymphopoiesis and homeostasis. Genes important in these processes and misexpressed in Ets1ΔVII homozygotes include those for p16Ink4a, p27Kip1, and CD44 and provide possible mechanisms for the phenotypes observed.
MATERIALS AND METHODS
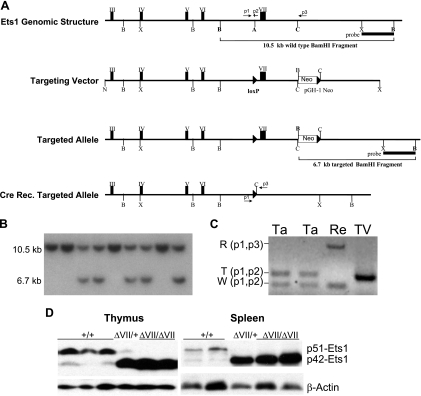
Mice.
Ets1 genomic clones from an isogenic library (Stratagene; strain 129/SvJ) were isolated using a murine Ets1 cDNA probe. The genomic structure of isolated clones was determined by restriction mapping, hybridization, and sequencing. The targeting vector was generated by inserting an ApaI adapter containing a loxP element into a unique ApaI site in intron VI immediately 5′ to exon VII. A positive selectable, single loxP-flanked Neor cassette (54) was inserted in reverse orientation into a unique ClaI site 1.4 kb 3′ to exon VII, allowing for excision of exon VII upon Cre-mediated recombination. Approximately 10 kb and 5 kb of Ets1 genomic sequences flank the Neor cassette. TC1-10 embryonic stem (ES) cells were electroporated, positively selected with G418 (230 μg/ml), and expanded, as described previously (54). Screening for targeted ES cell lines by Southern blotting involved BamHI digestion of genomic DNA and use of a 3′ flanking probe which recognizes a 10.5-kb wild-type fragment and a 6.7-kb targeted fragment (introduction of a BamHI site in the Neor cassette). Cre recombination for excision of exon VII was accomplished by adenoviral delivery of Cre recombinase into ES cells, as described previously (25). Chimeric mice were generated by ES cell-morula aggregation, as described previously (54). Analyses were conducted with 8- to 16-week-old 129Sv coisogenic littermates from heterozygous intercrosses. Genotyping was performed with DNA from tail biopsies by PCR using the following primers: p1, 5′-CTAGTGTAGACTCAGGACTTCTG-3′; p2, 5′-TAGGAAGAGGGCAGGGAAGAG-3′; and p3, 5′-GTCTTATCCACACCCAGTTGGTG-3′, with p1-p2 amplifying the wild-type sequence (167 bp) and p1-p3 amplifying exon VII-deleted sequences (550 bp) (Fig. 1A). Coisogenic lines were established by two successive crosses to the 129Sv strain from which ES cell lines were derived. Mice were housed under specific pathogen-free conditions, and experiments were performed in accordance with the institutional guidelines for animal care at the Medical University of South Carolina under approved protocols.
FIG. 1.
Generation and molecular analysis of Ets1ΔVII mice. (A) Targeting strategy. The top row shows Ets1 genomic structure and restriction map of an Ets1 genomic clone isolated from a 129/Sv mouse genomic library. Restriction sites: A, ApaI; B, BamHI; C, ClaI; N, NotI; and X, XhoI. Primer binding sites are indicated by numbered arrows (p1, p2, p3). Three-prime flanking probe and 10.5-kb wild-type BamHI fragment are indicated. The second row from the top shows the structure of the Ets1ΔVII targeting vector. A loxP element (black triangle) was cloned into a unique ApaI site 5′ to exon VII, and a pGH-1 G418 resistance cassette (Neor) and loxP element were cloned into a unique ClaI site 3′ to exon VII. The third row from the top shows the structure of the Cre conditional Ets1ΔVII-targeted allele. The reduction in size of the 3′ BamHI fragment is indicated. The bottom row depicts a Cre recombined allele lacking exon VII. Genotyping primers are indicated. (B) Confirmation of gene targeting in ES cells by Southern blotting using a 3′ BamHI-XhoI probe to show 10.5-kb wild-type and 6.7-kb targeted alleles. (C) PCR verification of Cre recombination. Recombinant (R), targeted (T), and wild-type (W) alleles are indicated with primer pairs. Heterozygous targeted (Ta) and recombined (Re) cells and the targeting vector (TV) control are shown. (D) Western blot confirmation of protein expression from the targeted allele in isolated thymocytes and splenocytes. Twofold-greater total spleen protein versus total thymus protein was loaded due to reduced Ets1 expression in the spleen. Ets1 genotypes are given above each blot.
Western blot analysis.
Single-cell suspensions were obtained by mechanical disruption of thymus or spleen and passage through 40-μm cell strainers (352340; BD Biosciences). Thymocytes and splenocytes were resuspended in modified radioimmunoprecipitation assay buffer with protease inhibitors (1836153; Roche) and quantitated by the bicinchoninic acid method (23225; Pierce). Seventy micrograms of total spleen protein or 25 to 40 μg of total thymus protein was separated on 12.5% sodium dodecyl sulfate-polyacrylamide gels and transferred to membranes. The following antibodies were used: β-actin (sc-47778; Santa Cruz Biotechnology), Ets1 c-20 (sc-350; Santa Cruz Biotechnology), Runx1 (PC284L; Calbiochem), p27Kip1 (sc-528; Santa Cruz Biotechnology), p16Ink4a (sc-1207; Santa Cruz Biotechnology), anti-mouse antibody-horseradish peroxidase (HRP) (M30007; Caltag), and anti-rabbit antibody-HRP (L42007; Caltag). Membranes were developed using a West Pico ECL kit (34079; Pierce).
Flow cytometry and cell sorting.
Thymocytes and splenocytes were prepared as described above. Briefly, 2 × 105 cells were blocked with anti-Fc receptor (553141; BD Biosciences) and stained with combinations of antibodies listed. The antibodies used in these experiments included CD94 (550773; Pharmingen), CD4 (557307; Pharmingen), CD8 (553032, 553034; Pharmingen), CD19 (557398; Pharmingen), CD44 (553134; Pharmingen), Gr-1 (553128; Pharmingen), and Mac-1 (557395; Pharmingen). For annexin V staining, cells were stained according to the manufacturer's instructions (556547; BD Biosciences). Two- and three-color flow cytometric analyses were performed by using a FACScalibur (Becton Dickinson). Each dot plot or histogram represents an analysis of 104 events using WinMDI software version 2.8.
BrdU labeling, histology, and IHC.
For detection of 5-bromodeoxyuridine (BrdU) incorporation in vivo by splenocytes and thymocytes, 1.5 mg BrdU (B5002; Sigma) in 150 μl of sterile phosphate-buffered saline was injected intraperitoneally 4 h before organ isolation. Organs were fixed in Amsterdam's fixative, paraffin embedded, and serially sectioned in 7-μm sections. For histological evaluation, the sections were stained with hematoxylin and eosin. For immunohistochemical (IHC) detection of BrdU incorporation into the DNA of dividing cells, sections were deparaffinized and hydrated. Endogenous peroxidase was inhibited by incubation with freshly prepared 3% H2O2 with 0.1% sodium azide. DNA was denatured by acid (4 N HCl) treatment, and nonspecific binding was blocked with 2% bovine serum albumin in phosphate-buffered saline. Sections were incubated with monoclonal anti-BrdU antibody (347580; BD Pharmingen) at a 1:150 dilution overnight at 4°C followed by anti-mouse antibody-HRP (K1015; Dako). After incubation with primary antibody, tissue sections were sequentially incubated with goat anti-mouse antibody-HRP (K1016; Dako) at a dilution of 1:100. Staining was developed with diaminobenzidine (SK-4100; Vector) substrate, and sections were counterstained with hematoxylin.
RT-PCR.
Total RNA was isolated from flow-sorted thymocyte subpopulations using RNA Stat 60 reagent (Cs-111; Tel-Test) according to the manufacturer's protocol. Isolated RNA was DNase I treated to remove genomic DNA. The RNA was reverse transcribed using an Advantage RT-for-PCR kit (639505; Clontech) with oligo(dT) primers. Real-time quantitative PCR (qPCR) reactions were performed in triplicate using Platinum SYBR Green qPCR SuperMix-UDG (11733-038; Invitrogen) and an ABI Prism 5700 real-time PCR machine (Applera). Relative transcript expression was determined by the ΔΔCT method: relative expression is equal to 2−(ΔCTsample − ΔCTβActin), in which CT is the threshold cycle and 40 cycles is the limit of detectable product. Primers were designed to span intron sequences, when possible, and were tested by nonquantitative reverse transcription (RT)-PCR to verify that only one amplicon was produced. The full-length Ets1-specific primer set includes one primer that binds to exon VII sequences. The alternate spliced Ets1ΔVII-specific primer set includes one primer that spans the exon VI-exon VIII junction. Primers used were as follows: β-Actin CCGGGACCTGACAGACTACC (forward) and TGCCACAGGATTCCATACCC (reverse); p42Ets1, AGAGCCAGTCGTGGAAGTGG and CGCACGGCTCAGTTTCTCAT; p51Ets1, TAGTTGTGACCGCCTCACCC and TCGGCCCACTTCCTGTGTAG; Cdkn2a, CAGGTGATGATGATGGGCAAC and CAGCGTGTCCAGGAAGCCT; Cdkn1b, CAGGCAAACTCTGAGGACCG and CCTTTTGTTTTGCGAAGAAGAATC; and CD44, CCGCACTGTGACTCATGGAT and CTTCTGCCCACACCTTCTCC.
Statistics.
Data are reported as means ± standard errors of the means. Differences in means were analyzed by unpaired t test or analysis of variance (ANOVA), as appropriate, depending upon the number of groups in the analysis. Unpaired t tests were used for post hoc testing. Calculated cell mass was determined as the product of the relative frequency of cells (number of cells/104 events) by total organ weight. This approach was taken as a proxy for the conventional method of taking the product of the estimated total cell number and multiplying it by the relative frequency of cells to determine the absolute cell number.
RESULTS
Generation and molecular analysis of Ets1ΔVII-targeted mice.
Homologous recombination in murine ES cells was used to generate Cre-conditional Ets1ΔVII cell lines, with loxP sites flanking the alternatively spliced exon VII (Fig. 1A, targeted allele). Homologous recombination was observed in 22 of 97 G418-resistant ES cell colonies by Southern blot analysis (a representative example is shown in Fig. 1B). Excision of the floxed exon VII targeted allele was accomplished by adenoviral delivery of Cre recombinase to Ets1ΔVIIfloxed/+ ES cells and verified in subclones by PCR (Fig. 1C). After in vitro Cre recombination-mediated exon VII deletion, targeted ES cell lines were used to generate aggregation chimeras. Germ line transmission of the targeted allele in chimeric mice was verified by Southern blot analysis of DNA from tail biopsies of agouti offspring (data not shown). Heterozygous offspring were used to establish Ets1ΔVII 129-strain coisogenic mice, which were interbred for analysis. Loss of the full-length transcript and expression of the Ets1ΔVII transcript were verified by real-time qPCR (Table 1). Loss of the full-length p51-Ets1 protein and expression of the p42-Ets1 protein was confirmed by Western blotting (Fig. 1D). In the thymus and spleen of wild-type mice, full-length p51-Ets1 is the most abundantly expressed isoform; however, in heterozygotes, there is a marked increase in the expression of the targeted allele (p42-Ets1) and a concomitant decrease in p51-Ets1 expression (Fig. 1D). This result suggests that relative Ets1 isoform levels are maintained in normal lymphoid tissues via a feedback autoregulatory mechanism involving the autoinhibition domain of p51-Ets1 or by changes in posttranscriptional processing. While variable expression levels were reported in other studies (4), we observe that wild-type thymocytes and splenocytes have a reproducibly low proportion of p42-Ets1 protein relative to p51-Ets1 (Fig. 1D, lower band). Homozygote expression of Ets1ΔVII transcripts ranges from a 12% reduction to a twofold elevation relative to Ets1ΔVII transcript levels in wild-type mice, depending upon the thymocyte subpopulation analyzed (Table 1). Consistent with other published studies (1), the highest expression of all Ets1 transcripts is in the CD4+CD8+ population of thymocytes in Ets1ΔVII homozygotes and wild-type littermates (Table 1).
TABLE 1.
Relative quantification of Ets1 transcripts in wild-type and targeted micea
| Thymocyte subpopulation | Result for indicated genotype
|
ΔΔCT | Relative expression | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ets1+/+
|
Ets1ΔVII/ΔVII
|
|||||||||
| Expt 1 | Expt 2 | Mean | ΔCT | Expt 1 | Expt 2 | Mean | ΔCT | |||
| ETS1full | ||||||||||
| DN | 36.71 | 37.14 | 36.93 | 11.85 | 40.00 | 40.00 | 40.00 | |||
| DP | 36.16 | 36.23 | 36.20 | 9.79 | 40.00 | 40.00 | 40.00 | |||
| CD8 | 37.18 | 38.19 | 37.69 | 11.91 | 40.00 | 40.00 | 40.00 | |||
| CD4 | 36.45 | 37.41 | 36.93 | 11.55 | 40.00 | 40.00 | 40.00 | |||
| ETS1ΔVII | ||||||||||
| DN | 31.69 | 31.85 | 31.77 | 6.70 | 36.16 | 34.63 | 35.40 | 5.96 | −0.74 | 1.670 |
| DP | 31.36 | 31.30 | 31.33 | 4.93 | 31.18 | 32.19 | 31.69 | 5.05 | 0.12 | 0.920 |
| CD8 | 32.36 | 32.61 | 32.49 | 6.71 | 31.06 | 30.82 | 30.94 | 5.70 | −1.02 | 2.028 |
| CD4 | 31.67 | 32.02 | 31.85 | 6.07 | 31.71 | 31.28 | 31.50 | 6.25 | 0.18 | 0.883 |
DN, double negative; DP, double positive; CD8, single-positive CD8; CD4, single-positive CD4 thymocytes.
Thymomegaly in Ets1ΔVII homozygous mice.
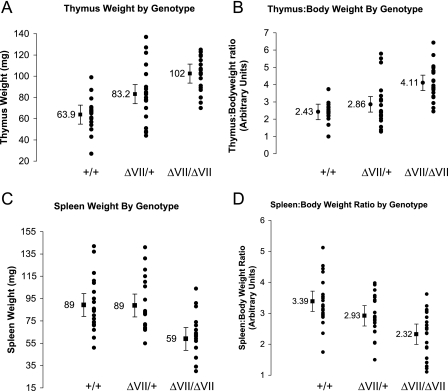
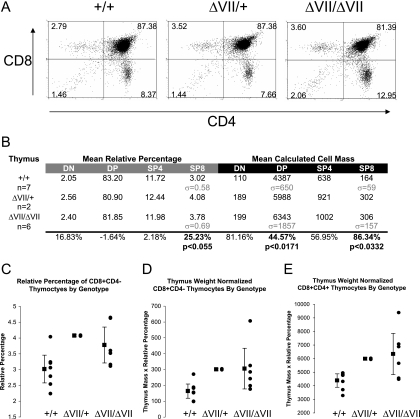
Ets1ΔVII homozygous mice are born at less than the expected Mendelian ratio from heterozygous intercrosses of 129-strain coisogenic mice, suggesting a low penetrant pre- or perinatal lethality (1:1.7:0.57 +/+:+/−:−/− survival, n = 242). Surviving mice mature to adulthood, are fertile, and are not overtly distinguishable from wild-type littermates. However, gross anatomic examinations of mice revealed pronounced thymic enlargement (thymomegaly) in homozygotes and more moderate thymomegaly in heterozygotes. The mean thymic weight was 60% greater in homozygotes and 30% greater for heterozygotes than for wild-type littermates (P < 0.000001 by ANOVA) (Fig. 2A). This difference was not attributable to alterations in total body mass between genotypes, as homozygotes have a 69% higher thymus:body mass ratio than wild-type mice (P < 0.000005 by ANOVA) (Fig. 2B). The thymomegaly of heterozygotes is more modest relative to total body mass (Fig. 2B). Thymomegaly in homozygotes was not associated with a histologically identifiable alteration in thymic structure other than increased organ size and more intense hematoxylin staining in the medulla, potentially indicating increased numbers of mature thymocytes (data not shown). Flow cytometric analyses of isolated thymocytes revealed statistically significant differences in relative thymocyte subpopulations, showing increased relative frequency of CD4−CD8+ cells (25% increase; P < 0.055) (Fig. 3A to C). To normalize these data to total cell numbers, we multiplied organ weight by relative frequency of each cell type to determine the calculated cell mass for each population. This revealed a trend toward increased mean cell number in all thymocyte subtypes, with a 45% increase in CD4+CD8+ cells and an 86% increase in CD4−CD8+ cells, results which achieved statistical significance (P < 0.05) (Fig. 3A, D, and E). Thus, the CD4−CD8+ compartment showed both the greatest increase in mean normalized cell number and the highest relative increase in Ets1ΔVII transcripts. In summary, all thymocyte populations are expanded in the mutant when measured on the basis of normalized cell number and compared to the pool size of these cells in the wild-type mice. Statistical significance was achieved for this difference in the CD4+CD8+ and CD4−CD8+ cells but not in the CD4−CD8− and CD4+CD8− cells. Also, the normalized cell number increase in CD4−CD8+ cells is not accompanied by a decrease in CD4+CD8− cells.
FIG. 2.
Thymomegaly and microsplenia in Ets1ΔVII homozygotes are independent of total body mass. (A) Wet thymus mass of adult mice plotted by genotype shows thymomegaly in Ets1ΔVII homozygotes (P < 0.000001 by ANOVA). (B) Thymus mass-to-body mass ratio plotted by genotype shows an increase in homozygotes comparable to that in wild-type mice (P < 0.000005 by ANOVA). (C) Wet spleen mass of adult mice plotted by genotype reveals microsplenia in homozygote mice (P = 0.00006 by ANOVA). (D) Spleen mass-to-body mass ratio plotted by genotype reveals that variance in spleen mass does not result from alterations in body mass (P < 0.00013 by ANOVA). Black circles indicate individuals; the black square indicates the mean value for the group, with 95% confidence intervals indicated by error bars. Mutant versus wild type, n = 20 for each group.
FIG. 3.
Loss of full-length p51-Ets1 alters thymopoiesis for mice with indicated Ets1 genotypes. (A) Representative flow cytometry of CD4/CD8 double-labeled thymocytes from Ets1ΔVII and wild-type mice. Relative percentages are indicated in each quadrant. (B) Table summarizing results of two-color flow cytometry of CD4 and CD8 labeling of thymocytes from 8- to 16-week-old Ets1ΔVII-targeted mice and wild-type littermates. Left half (gray bar) shows average relative percentages of each population, based upon 104 cell counts/mouse. Right half (black bar) shows “mean calculated cell mass” for each population, which was used to normalize data to total cell numbers (calculated by multiplying relative percentages of each population by organ weight). Relative increase or decrease of mean Ets1ΔVII values compared to wild-type values is indicated at the bottom of the chart. (C) Summary data showing relative increases in CD8+ thymocytes in Ets1ΔVII mice (P < 0.055). (D) Summary data showing increased absolute number of CD8+ thymocytes in Ets1ΔVII mice (P < 0.05). (E) Summary data showing increased absolute number of DP thymocytes in Ets1ΔVII mice (P < 0.01). Black circles indicate individuals; the black square indicates the mean value for the group, with 95% confidence intervals indicated by error bars. Mutant versus wild type. DN, double negative; DP, double positive; CD8, single-positive CD8; CD4, single-positive CD4 thymocytes; σ, standard deviation.
Microsplenia in Ets1ΔVII homozygous mice.
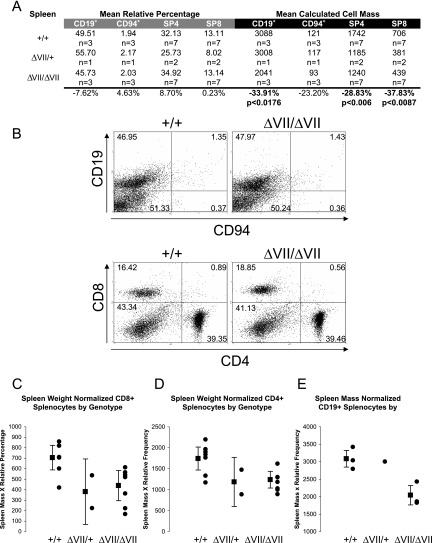
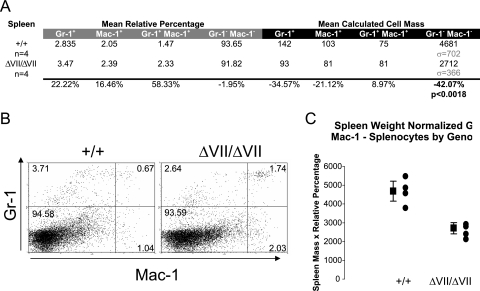
Phenotypic differences detected in the peripheral lymphoid compartment of Ets1ΔVII homozygotes include reduced spleen size (microsplenia) and peripheral blood lymphopenia. A statistically significant 34% decrease in the spleen weight of Ets1ΔVII homozygous mice was confirmed by ANOVA (P = 0.00006) (Fig. 2C). This phenotype was independent of changes in total body mass. Mean spleen:body mass of homozygotes was 31.5% reduced from the wild-type mean (P < 0.00013 by ANOVA) (Fig. 2D). No statistically significant alteration in the relative frequency of lymphoid-derived cells was observed for homozygotes relative to wild-type littermates (CD19+ B cells, CD94+NK cells, CD4+ T cells, CD8+ T cells) (Fig. 4A and B). However, based on a normalized cell number, statistically significant reductions in both B- and T-lymphocyte populations were observed. B cells were reduced 34% relative to the wild type (P < 0.05), and CD4+ and CD8+ T cells were reduced 29% and 38%, respectively (P < 0.01) (Fig. 4A, right half; Fig. 4C to E). This result is similar to the 22% and 44% reductions reported for Ets1 knockout mice (4). As in the thymus, the CD8+ population is the most severely affected, though the expression level of Ets1ΔVII transcripts in sorted splenocytes has yet to be determined. In the myeloid compartment, there were no statistically significant alterations in the relative percentages of splenocytes; however, there was a trend toward an increased proportion of myeloid cells (Gr-1+ and/or Mac-1+) (Fig. 5A and B). Consistent with these results, there was a 42% decrease in nonmyeloid cells based on normalized cell numbers (Gr-1−Mac-1−, P < 0.01) (Fig. 5A, right half; Fig. 5C). Taken together, these results suggest that the underlying microsplenia is likely due to a decrease in lymphoid-derived cells.
FIG. 4.
Microsplenia in Ets1ΔVII mice results principally from reduced lymphocyte numbers. (A) Table summarizing results of two-color flow cytometry of lymphoid cells of the spleen by CD4/CD8 or CD19/CD94 labeling of splenocytes from 8- to 16-week-old Ets1ΔVII-targeted mice and wild-type littermates. The left half (gray bar) shows average relative percentages of each population, based upon 104 cell counts/mouse. The right half (black bar) shows mean calculated cell mass for each population, as described for Fig. 3B. Relative increase or decrease of mean Ets1ΔVII values compared to wild-type values are indicated at the bottom of the chart. (B) Representative flow cytometry of either CD4/CD8 or CD19/CD94 double-labeled splenocytes from Ets1ΔVII and wild-type mice. Relative percentages are indicated in each quadrant. (C to E) Summary data showing absolute decreases in CD8+, CD4+, and CD19+ splenocytes, respectively, in Ets1ΔVII mice. Black circles indicate individuals; the black square indicates the mean value for the group, with 95% confidence intervals indicated by error bars. Mutant versus wild type (P < 0.01).
FIG. 5.
Reduced numbers of nonmyeloid cells in Ets1ΔVII spleens. (A) Table summarizing results of two-color flow cytometry of myeloid cells of the spleen by Gr-1/Mac-1 labeling of splenocytes from 8- to 16-week-old Ets1ΔVII-targeted mice and wild-type littermates. Left half (gray bar) shows average relative percentages of each population, based upon 104 cell counts/mouse. Right half (black bar) shows mean calculated cell mass for each population, as described for Fig. 3B. Relative increase or decrease of mean Ets1ΔVII values compared to wild-type values is indicated at the bottom of the chart. (B) Representative flow cytometry of either Gr-1/Mac-1 double-labeled splenocytes from Ets1ΔVII and wild-type mice. Relative percentages are indicated in each quadrant. (C) Summary data showing absolute decreases in nonmyeloid splenocytes in Ets1ΔVII mice. Black circles indicate individuals; the black square indicates the mean value for the group, with 95% confidence intervals indicated by error bars. Mutant versus wild type (P < 0.01).
Variable peripheral lymphopenia in Ets1ΔVII homozygous mice.
In addition to microsplenia, an incompletely penetrant lymphopenia in the peripheral blood of three out of seven homozygous mice tested was observed. Affected mice had a 50% reduction in the number of leukocytes per volume of blood and a twofold elevation in the relative percentage of circulating Gr-1hi Mac-1+ cells (data not shown). Collectively, these results suggest that the size of the peripheral lymphocyte pool is dependent upon the expression of proper ratios of both the full-length p51-Ets1 and the p42-Ets1 isoforms.
Increased cell proliferation in lymphoid organs of Ets1ΔVII homozygous mice.
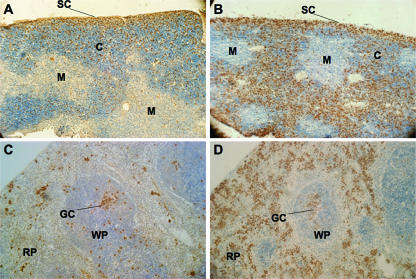
In order to establish the etiology of the thymomegaly, microsplenia, and peripheral lymphopenia in Ets1ΔVII mice, we assessed cell size, proliferation, and apoptosis. No significant differences in cell size were detected by comparisons of mean forward scatter of freshly isolated splenocytes and thymocytes, irrespective of genotype (data not shown). To determine the role of cell proliferation in the thymomegaly and microsplenia of Ets1ΔVII mutants, BrdU was administered to mice intraperitoneally, and S-phase cells were measured by IHC. In contrast to reports of Ets1-null mice having normal cell cycles for unchallenged thymocytes and splenocytes, Ets1ΔVII homozygotes showed a marked increase in the numbers of proliferative cells of both the spleen and the thymus relative to those of wild-type littermates. In the wild-type thymus, the vast majority of proliferating cells are found in the subcapsular region, with more diffuse BrdU staining in the cortex, and few proliferating cells are detected in the medulla (Fig. 6A). By contrast, thymuses of homozygotes show a marked increase in the levels of proliferating cortical thymocytes, levels comparable to those seen in the subcapsular region (Fig. 6B). The zone of proliferating cells in the subcapsular region of homozygotes appears less organized and less distinct than that observed for wild-type mice. This altered distribution may result from a loss of regional specificity of subcapsular CD4−CD8− thymocytes and/or increased proliferation of thymocytes normally found in the cortex. Though our data did not achieve statistical significance, the trend toward an increased relative proportion of CD4−CD8− cells suggests the former. These data collectively implicate cell cycle misregulation as contributing to thymomegaly. Spleens of homozygotes were found to have a marked increase in the proliferation of splenocytes in the red pulp (Fig. 6C and D), perhaps as a compensatory response to the marked reduction in the lymphocyte compartment of the spleen (Fig. 4). No obvious change in proliferation was observed for the white pulp splenocytes, with germinal centers being observed for both wild types and homozygotes.
FIG. 6.
Marked increase in cell proliferation in the thymic cortex and splenic red pulp of Ets1ΔVII mice. BrdU staining of 7-μm thymus sections from wild-type (A) and Ets1ΔVII (B) homozygotes demonstrates increased proliferation in the cortex of homozygote thymuses. BrdU staining of 7-μm spleen sections from wild-type (C) and Ets1ΔVII (D) homozygotes demonstrates marked increase in proliferation in the red pulp of the spleen of homozygotes. Hematoxylin counterstain. SC, subcapsular; C, cortex; M, medulla; GC, germinal center; WP, white pulp; RP, red pulp.
Altered apoptosis in splenocytes and thymocytes of Ets1ΔVII mutant mice.
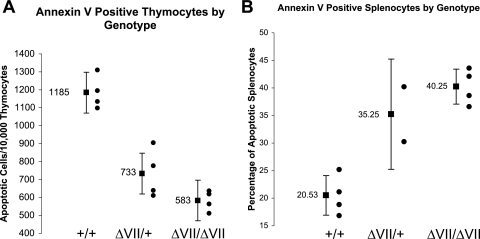
To determine the contribution of apoptosis to the thymomegaly and microsplenia in Ets1ΔVII mice, primary thymocytes and splenocytes were stained with annexin V and subjected to flow cytometric quantification. Annexin V detects the early membrane asymmetry of phosphatidylserine characteristic of cells initiating apoptosis. Consistent with the observed thymomegaly, thymocytes of both Ets1ΔVII homozygotes and heterozygotes had a 50% mean reduction in the frequency of apoptosis compared to wild-type littermates (P < 0.001) (Fig. 7A).
FIG. 7.
Altered apoptosis in Ets1ΔVII mice. (A) Marked reduction in the apoptotic rate of Ets1ΔVII thymocytes. Summary results of annexin V staining of thymocytes indicate a reduction in the average rate of apoptosis in Ets1ΔVII thymocytes. Black circles indicate individuals; the black square indicates the mean value for the group, with 95% confidence intervals indicated by error bars (P < 0.001). (B) Pronounced increase in the apoptotic rate in Ets1ΔVII spleens. Summary results of annexin V staining of splenocytes indicates a reduction in the average rate of apoptosis in Ets1ΔVII splenocytes (P < 0.001). Black circles indicate individuals; the black square indicates the mean value for the group, with 95% confidence intervals indicated by error bars.
Apoptosis is also critical to the regulation of the peripheral lymphocyte homeostasis, both in limiting the numbers of peripheral lymphocytes and in the regulation of the affinity/avidity and diversity of the antigen receptor pool (18). To study this, splenocytes were stained with annexin V and subjected to flow cytometric quantification. Annexin V staining of primary splenocytes from littermates showed a twofold mean elevation in the frequency of apoptosis in the spleens of homozygotes relative to those of heterozygotes and wild-type mice (P < 0.001) (Fig. 7B). Given the marked increase in numbers of apoptotic cells in homozygotes, it can be argued that apoptosis significantly contributes to the microsplenic phenotype. Furthermore, given the 30 to 40% reduction in lymphoid mean splenocyte mass in homozygotes, it is possible that this increased rate of apoptosis corresponds to T- and B-cell apoptosis. Collectively, these results suggest a role specific to the p51-Ets1 isoform in the regulation of lymphocyte survival, since heterozygotes and homozygotes have comparable levels of p42-Ets1 overexpression but different apoptotic rates.
Reduced expression of key cell cycle regulators p16Ink4a and p27Kip1 in thymuses of Ets1ΔVII homozygous mice.
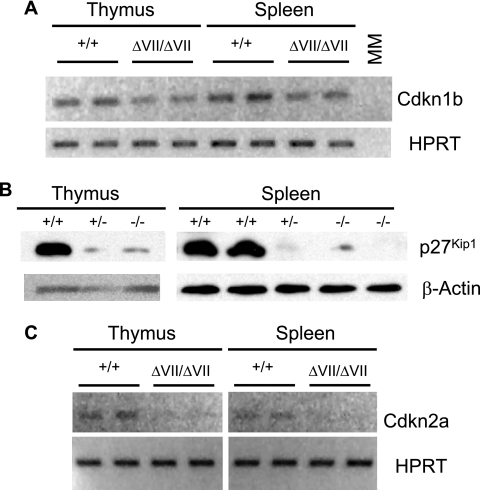
To investigate the molecular mechanism underlying the increased proliferation in Ets1ΔVII thymuses, we studied the expression of p16Ink4a (Cdkn2a) and p27Kip1 (Cdkn1b). These two proteins are reported to be expressed throughout the thymus, at levels consistent with roles as important negative regulators of thymocyte proliferation in mice (26). Furthermore, alterations in the expression of these proteins have been shown to be important in lymphoid organ size. RT-PCR analyses of littermates demonstrate a slightly diminished expression of Cdkn1b transcripts in homozygotes relative to wild types (Fig. 8A). Western blot analyses of protein from Ets1ΔVII homozygotes and heterozygotes show profound decreases in p27Kip1 protein expression relative to those for wild-type littermates (Fig. 8B). Ets1 has been implicated as a direct regulator of Cdkn2a transcription (23, 43). RT-PCR analyses of littermates also demonstrate a diminished expression of Cdkn2a transcripts in homozygotes relative to wild types (Fig. 8C). These results suggest that full-length p51-Ets1 may positively regulate the expression of these genes to limit proliferation in the thymus and spleen of wild-type mice.
FIG. 8.
Decreased expression of cell cycle regulators CDKN2a and CDKN1b in Ets1ΔVII mice is associated with increased proliferation. (A) RT-PCR analysis of thymocyte and splenocyte mRNA show diminished expression of CDKN1b transcript levels in Ets1ΔVII mice, hypoxanthine phosphoribosyltransferase (HPRT) loading control (30 cycles). (B) Western blot analysis of total protein from isolated thymocytes and splenocytes shows diminished expression of p27Kip1 (CDKN1b) protein in Ets1ΔVII thymocytes. β-Actin loading control. (C) RT-PCR analysis of thymocyte and splenocyte mRNA show diminished expression of CDKN2a transcript levels in Ets1ΔVII mice. HPRT loading control (30 cycles). Ets1 genotypes are given above each blot.
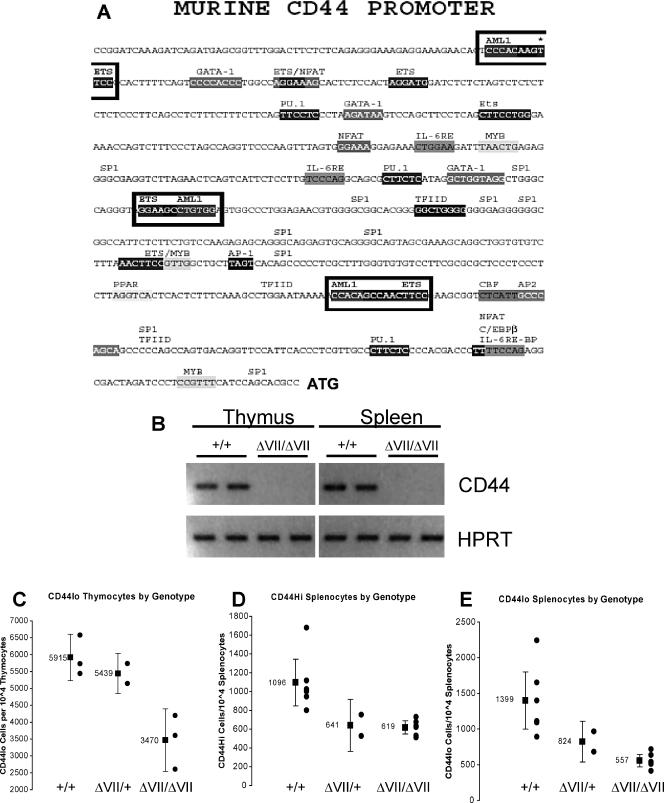
Reduced expression of CD44 suggests memory cell defects in Ets1ΔVII homozygous mice.
Full-length p51-Ets1 participates in promoter regulation via cooperative binding. The murine CD44 gene has three putative heterodimeric Ets/Runx1 (AML1) binding sites, which may be occupied by p51-Ets1 and partner AML1 in cooperative binding (Fig. 9A). RT-PCR analyses of thymocyte and splenocyte mRNA demonstrated greatly diminished expression of CD44 transcripts in Ets1ΔVII mice (Fig. 9B), even when cycle number was increased and other CD44-specific primer pairs were used (data not shown). Cell-surface expression of CD44 is important for homing of thymic progenitors from the bone marrow to the thymus and also as a definitive differentiation antigen of subtypes of CD4−CD8− thymocytes. Flow cytometric analysis of thymocyte cell-surface CD44 expression demonstrated a reduced frequency of CD44-expressing cells compared to wild types; a slight reduction in heterozygotes and an additional 40% reduction in homozygotes was observed (Fig. 9C).
FIG. 9.
Reduced CD44 expression in Ets1ΔVII mice. (A) Murine CD44 promoter sequence (GenBank no. AF262063) contains three putative AML1 (Runx1)/Ets1 coordinated binding sites, indicated by boxes. (B) RT-PCR analysis of thymocyte and splenocyte mRNA shows diminished expression of CD44 transcript levels in Ets1ΔVII mice. HPRT loading control (all 30 cycles). Note that transcripts were detectable, albeit reduced, in Ets1ΔVII mice when increased cycle numbers were used with other CD44-specific primer sets. CD44+ cell-surface expression in Ets1ΔVII mice is displayed in panels C to E. (C) Loss of CD44+ thymocytes in Ets1ΔVII mice. Summary results demonstrate the diminished average representation of CD44 in thymocytes of Ets1ΔVII mice. Black circles indicate individuals; the black square indicates the mean value for the group, with 95% confidence intervals indicated by error bars. (D and E) Loss of CD44hi splenocytes suggests defective activation or memory cell generation in Ets1ΔVII mice. Summary results demonstrate diminished representation of CD44hi (D) and CD44lo (E) splenocytes in Ets1ΔVII mice. Black circles indicate individuals; the black square indicates the mean value for the group, with 95% confidence intervals indicated by error bars.
In the periphery, high-level CD44 expression is a marker of memory cells or cells undergoing homeostatic proliferation. In wild-type mice, CD44hi memory cells account for 10 to 20% of peripheral cells (52). Consistent with this, flow cytometric analysis of splenocytes from wild-type mice revealed 8 to 18% of cells to be CD44hi (Fig. 9D and E). In marked contrast, heterozygotes and homozygotes had a 50% reduction in the mean relative frequency of these cells (Fig. 9D and E). This reduction is even more pronounced when considering that affected mice have microsplenia; thus, this relative reduction corresponds to a much lower absolute number of cells.
DISCUSSION
A preliminary characterization of mice engineered to express only the p42-Ets1 isoform has revealed a potential role for Ets1 isoforms in the regulation of lymphopoiesis and peripheral lymphoid homeostasis. A summary comparing and contrasting the phenotypes observed for the Ets1ΔVII mutant to those previously published for Ets1 knockout mice is presented in Table 2. An analysis of Ets1ΔVII mutants demonstrates that the absence of full-length p51-Ets1 is associated with superphysiological expression of p42-Ets1, increased proliferation, and altered apoptosis. Increased proliferation in the thymus and spleen of homozygotes was observed, with associated reductions in the expression of p16Ink4a and p27Kip1, whereas results for heterozygotes were more comparable to those for wild types (Fig. 8). Thymocytes of both Ets1ΔVII homozygotes and heterozygotes had a 50% mean reduction in apoptosis, compared to that of wild-type littermates (Fig. 7A). This contrasts with the diminished survival reported for Ets1 knockout mice and complemented Rag2−/− chimeras (12, 40). These results suggest that dosage of Ets1 isoforms may be a critical modulator of apoptosis, as Ets1ΔVII heterozygous mice exhibit an intermediate effect on apoptosis.
TABLE 2.
Comparison of Ets1ΔVII and Ets1 knockout mouse phenotypes
| Parameter | Result for indicated genotype
|
Relationshipa | Reference(s) | |
|---|---|---|---|---|
| Ets1ΔVII | Ets1 knockout | |||
| ETS1 expression | ||||
| Full-length p51-Ets1 protein | None | Very low neomorphb | S | 7 |
| ΔVII p42-Ets1 protein | High | Very low neomorphb | D | 7 |
| Thymus | ||||
| Cellularity | 60% increased | 65% reduction | D | 4, 7 |
| CD4−CD8+ thymocytes | Expanded | Diminished | D | 7 |
| Proliferation | Increased | Undocumented | ||
| Apoptosis | Decreased | Increased | D | 6, 11, 12, 40 |
| Spleen | ||||
| Size | Decreased | Undocumented | ||
| Proliferation | Increased | Decreased (T cells) | D | 40 |
| Apoptosis | Increased | Increased | S | 6, 40 |
| NK cells | Present | Absent | D | 4 |
| CD44hi splenocytes | Diminished | Expanded | D | 7 |
| Systemic | ||||
| Lethality | 33% pre- or perinatal | 50% pre- or perinatal | S | 6, 7, 12 |
| Rapid IFN-γ secretion | Diminished | Diminished | S | 6, 7, 12 |
| Peripheral lymphopenia | Yes | Yes, but abnormal | D | 7, 12 |
S, similar phenotypes for Ets1ΔVII mice and (previously reported) Ets1-targeted mice; D, divergent phenotypes for these two strains.
The Ets1-targeted allele has been demonstrated to generate protein from in-frame splicing from exon II to exon V at ∼1 to 5% of wild-type expression levels. Also, this variant undergoes alternative splicing of exon VII to generate a neomorphic p42-Ets1 (p29).
Cell survival is a key regulatory mechanism of thymopoiesis, allowing the elimination of defective thymocytes. Underscoring this point, less than 5% of all thymocytes normally survive the process to become mature T cells (37). Early thymocytes are dependent upon stem cell factor and interleukin 7 for survival. These signals are replaced by signals from the pre-TCR that allow for proliferation, survival, differentiation, and allelic exclusion. Previously described Ets1 knockout mice have defective allelic exclusion of TCR-β, supporting a model where Ets1 is a downstream effector of pre-TCR signaling (11). While we have shown trends in the relative frequency of thymocyte subtypes, it is clear that expansion of all thymocyte populations occurs in Ets1ΔVII homozygotes, suggesting that this phenotype begins at the earliest (CD4−CD8−) stage of thymopoiesis. At this stage, Ets1ΔVII may act as a downstream amplifier of pre-TCR signals. This is plausible mechanistically, given that the pre-TCR proliferative signal is transduced by Ras/Erk1/2, which may impinge on Ets1. ERK1/2 phosphorylation of Thr38 residue on both Ets1 isoforms has been demonstrated to augment Ets1 transcriptional activity (49). Also, CD4−CD8− cells have a lower threshold for ligand-independent pre-TCR signaling, based in part on increased Ca2+ capacitive entry (20, 45). This is significant, considering that such Ca2+ flux-mediated signaling normally represses full-length p51-Ets1, but not p42-Ets1, transcriptional activity. Thus, in Ets1ΔVII mutants, p42-Ets1 not only is expressed at higher levels than normal but also may represent a more active transcriptional regulator based on a lack of responsiveness to the inhibitory Ca2+ signal. We have shown that p16Ink4a levels are diminished in the Ets1ΔVII homozygote thymus. p16Ink4a is known to antagonize pre-TCR-mediated expansion and/or survival of differentiating thymocytes (30). Collectively, these data may implicate either hyperactivation of Ets1 downstream effector functions in response to pre-TCR signal or transduction of a spurious signal (57). Alterations in the expression of p16Ink4a or p27Kip1 have also been shown to be important in lymphoid organ size. Mice transgenic for either p16Ink4a or p27Kip1 show partial developmental arrest at the CD4−CD8− stage and reduced proliferation and function of lymphocytes (30, 56). Gene-targeted mice deficient in either p16Ink4a or p27Kip1 develop thymic hyperplasia (41, 13, 51). Mice deficient in p27Kip1 also develop splenic hyperplasia. Chromatin immunoprecipitation demonstrates that Ets1 binds to the Cdkn2a promoter in vivo, supporting the notion that it is a direct Ets1 target (43). Our results argue that full-length Ets1 positively regulates these genes in the thymus and spleen of wild-type mice; therefore, in Ets1ΔVII thymuses, these genes are not activated and cause increased proliferation. Thus, the thymomegaly phenotype could be due to elevated expression of the p42-Ets1 isoform and/or the loss of the full-length p51-Ets1 isoform, with a balance of isoforms being more important in the regulation of apoptosis and the presence of p51-Ets1 (even at low levels) being more important in the suppression of proliferation. Thymomegaly in Ets1ΔVII mutants contrasts with the 65% reduction in thymic cellularity reported for Ets1-null animals (Table 2; 4, 7). Furthermore, the loss of both Ets1 isoforms results in the absence of proliferative phenotypes (reduced thymic cellularity), arguing that the thymomegaly phenotype results from elevated expression of p42-Ets1 (40). Nevertheless, it can also be argued that the presence of low levels of p51-Ets1 counteracts the effects of elevated p42-Ets1 levels, as observed in Ets1ΔVII heterozygotes. In addition, the continued expression of the Ets1ΔVII allele may block a compensatory response that is active in Ets1 knockout mice. This could explain the relatively large increase in p42-Ets1 protein expression shown on immunoblots compared to the twofold elevation at the transcript level, which may suggest the involvement of posttranscriptional regulation of Ets1 expression (Fig. 1D; Table 1).
Diminished spleen mass with no disruption in the organization of red pulp, white pulp, and germinal centers is a disorder generally referred to as microsplenia and a phenotype observed for Ets1ΔVII mutant mice. No change in proliferation was observed in the white pulp splenocytes, with germinal centers being observed for both wild-type and Ets1ΔVII homozygotes (Fig. 4 and 6). Since the majority of splenic lymphoid cells are located in the white pulp region, these results indicate that the observed increase in splenic red pulp proliferation is due to either nonlymphoid cells or ectopic lymphocytes. Since myeloid cell numbers in Ets1ΔVII mutants were normal, this result would also suggest either that there is either (i) a global induction of apoptosis across splenocyte lineages and myeloid cells but not lymphoid cells (which are able to mount proliferative compensation) or (ii) a mislocalization of lymphocytes that are undergoing proliferative expansion (e.g., homeostatic expansion). Such a mislocalization may result in the loss of appropriate costimulation and contribute to the elevated apoptosis that is observed with these animals. The presence of organized white pulp and germinal centers suggests that lymphocyte migration and immune response may not be completely lost in Ets1ΔVII homozygotes.
We have also demonstrated a role for Ets1 isoforms in maintaining peripheral lymphoid homeostasis. There are many models that can be proposed to explain how Ets1 may exert these effects. Peripheral lymphopenia in the Ets1ΔVII homozygotes may be due to defects in primary thymopoiesis. Alternatively, the reverse could be the case, i.e., peripheral lymphopenia may drive defects in primary thymopoiesis. One model is that Ets1 functions in primary thymopoiesis as a survival signal. Elevated p42-Ets1 expression in Ets1ΔVII mutants results in abnormally high-level survival of T cells that would normally have been lost during negative selection. These defective T cells have persistently high-level Ets1 expression, which normally diminishes with migration to the periphery (Fig. 1D), and they may have abnormal TCR signaling. Upon entry into the peripheral compartment, however, such defects would render these surviving mutant T cells more susceptible to marked apoptosis, as shown by the Ets1ΔVII mutants.
In the thymus and peripheral lymphoid compartment, cell-surface receptor expression, cell division, and apoptosis exert strict controls on lymphocyte type and number. Lymphocyte homeostasis can control the balance between an insufficient number of cells to mount an immune response or the excessive proliferation and/or overstimulation of an immune response associated with pathological conditions, such as cancer and autoimmune disorders (16, 42). Studies using antibodies to block CD44 function have demonstrated a role in lymphocyte activation, differentiation, homing/migration, and proliferation (15, 21, 31, 44, 47). We have shown that CD44 expression is diminished in the Ets1ΔVII mutants. As CD44 is important in the homing of thymic progenitors to the subcapsular region of the thymus, the disorganization of this region in Ets1ΔVII homozygotes, as shown by BrdU IHC data, suggests the possibility that diminished CD44 expression may lead to ectopic localization of these cells throughout the cortex (Fig. 6; data not shown). In experimental lymphopenia, antigen-independent proliferation of cells expressing memory cell markers (CD44Hi, CD122+, Ly6C+) leads to recovery of normal or near-normal lymphocyte numbers (39). Such homeostatic proliferation occurs from the expansion of preexisting memory cells or from naïve cells that acquire memory cell characteristics (27, 55). The process requires normal thymopoiesis to establish a competent naïve pool that expresses the required cytokines (e.g., interleukin 7) and has the ability to upregulate the genetic program(s) required for the memory cell phenotype.
Flow cytometric analysis of splenocytes from wild-type mice revealed 8 to 18% of cells to be CD44hi (Fig. 9D). In marked contrast, heterozygotes and homozygotes had a 50% reduction in the mean relative frequency of these cells (Fig. 9D). This difference is even more pronounced when considering the fact that affected mice have microsplenia and that the relative frequency corresponds to a much lower absolute number of cells. Thus, the resulting naïve cell pool for the Ets1ΔVII mutants is expected to be defective in effecting an immune response and in establishing a memory cell phenotype. While it is not clear the relative contributions of memory cells or cells undergoing homeostatic proliferation accounting for the remaining CD44hi splenocytes in homozygotes, two things are clear: (i) that there are fewer cells with the memory phenotype than would be expected for wild-type mice and (ii) that there is a failure of homeostatic proliferation to maintain proper numbers of peripheral lymphocytes. Moreover, CD44 has been shown to affect cell proliferation via posttranslational control of p27Kip1 expression (17) and, thus, could contribute to the diminished expression of p27Kip1 protein in Ets1ΔVII heterozygotes and homozygotes (Fig. 8B). Supporting this possibility is the observed dramatic loss of CD44 expression in the thymus and spleen of Ets1ΔVII homozygotes, as shown by RT-PCR (Fig. 9B).
Alterations in the splenic microenvironment, for example, by deregulation of Fas or TNF expression, could also contribute to the peripheral lymphopenia phenotype. A marked increase in proliferation in spleens of Ets1ΔVII homozygotes was observed, indicating the possibility that homeostatic proliferation is initiated but with defective lymphocytes undergoing apoptosis at an accelerated rate. In the periphery, CD44 expression is a marker of activated cells, memory cells, and cells undergoing homeostatic proliferation. In unpublished observations, we have confirmed the loss of memory cell function in these mice by vaccination with the MVA strain of vaccinia virus, followed by a challenge one week later (data not shown). The day following challenge, gamma interferon (IFN-γ) production was elevated in wild-type mice, while IFN-γ levels in homozygotes was equivalent to levels in baseline unchallenged mice (data not shown). This rapid IFN-γ response is a measure of memory cell activity (34).
Collectively, these results indicate isoform-specific requirements for Ets1 in the control of normal thymopoiesis, T-cell homeostasis, and peripheral lymphoid homeostasis. Ongoing studies are focused on the impact of compensatory mechanisms induced by persistent or transient imbalances in Ets1 isoform expression. Such compensatory mechanisms can be pursued in vivo and in vitro using inducible, tissue-specific, Cre excision-mediated disruption of the conditional Ets1VII allele described here.
Acknowledgments
T.H., F.O.B., D.K.W., and D.D.S. were supported by NIH grant P01 CA78582. F.O.B., R.C.M.-H., M.J.K. and D.D.S. were supported by grant SC COBRE P20 RR16434. R.L. was supported by grant K22CA109577.
We acknowledge Takis S. Papas for his role in inspiring the initiation of this project, Makio Ogawa for helpful suggestions and guidance during the course of this work, Yong Z. Gong and the MUSC Gene Targeting & Knockout Mouse Facility for their assistance in the generation of the gene-targeted mouse lines described, HaiQun Zeng and the Hollings Cancer Center Cell Sorting Facility for cellular analyses, and E. Starr Hazard and the Biomolecular Computing Resource for assistance with bioinformatic computations.
Footnotes
Published ahead of print on 5 March 2007.
REFERENCES
- 1.Anderson, M. K., G. Hernandez-Hoyos, R. A. Diamond, and E. V. Rothenberg. 1999. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development 126:3131-3148. [DOI] [PubMed] [Google Scholar]
- 2.Baillat, D., A. Begue, D. Stehelin, and M. Aumercier. 2002. ETS-1 transcription factor binds cooperatively to the palindromic head to head ETS-binding sites of the stromelysin-1 promoter by counteracting autoinhibition. J. Biol. Chem. 277:29386-29398. [DOI] [PubMed] [Google Scholar]
- 3.Ballschmieter, P., M. Braig, R. K. Lindemann, A. Nordheim, and J. Dittmer. 2003. Splicing variant DeltaVII-Ets1 is downregulated in invasive Ets1-expressing breast cancer cells. Int. J. Oncol. 22:849-853. [PubMed] [Google Scholar]
- 4.Barton, K., N. Muthusamy, C. Fischer, C. N. Ting, T. L. Walunas, L. L. Lanier, and J. M. Leiden. 1998. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity 9:555-563. [DOI] [PubMed] [Google Scholar]
- 5.Bellacosa, A., K. Datta, S. E. Bear, C. Patriotis, P. A. Lazo, N. G. Copeland, N. A. Jenkins, and P. N. Tsichlis. 1994. Effects of provirus integration in the Tpl-1/Ets-1 locus in Moloney murine leukemia virus-induced rat T-cell lymphomas: levels of expression, polyadenylation, transcriptional initiation, and differential splicing of the Ets-1 mRNA. J. Virol. 68:2320-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bories, J. C., D. M. Willerford, D. Grevin, L. Davidson, A. Camus, P. Martin, D. Stehelin, and F. W. Alt. 1995. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature 377:635-638. [DOI] [PubMed] [Google Scholar]
- 7.Clements, J. L., S. A. John, and L. A. Garrett-Sinha. 2006. Impaired generation of CD8+ thymocytes in Ets-1-deficient mice. J. Immunol. 177:905-912. [DOI] [PubMed] [Google Scholar]
- 8.Cowley, D. O., and B. J. Graves. 2000. Phosphorylation represses Ets-1 DNA binding by reinforcing autoinhibition. Genes Dev. 14:366-376. [PMC free article] [PubMed] [Google Scholar]
- 9.Degnan, B. M., S. M. Degnan, T. Naganuma, and D. E. Morse. 1993. The ets multigene family is conserved throughout the Metazoa. Nucleic Acids Res. 21:3479-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dittmer, J. 2003. The biology of the Ets1 proto-oncogene. Mol. Cancer 2:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eyquem, S., K. Chemin, M. Fasseu, and J. C. Bories. 2004. The Ets-1 transcription factor is required for complete pre-T cell receptor function and allelic exclusion at the T cell receptor beta locus. Proc. Natl. Acad. Sci. USA 101:15712-15717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eyquem, S., K. Chemin, M. Fasseu, M. Chopin, F. Sigaux, A. Cumano, and J. C. Bories. 2004. The development of early and mature B cells is impaired in mice deficient for the Ets-1 transcription factor. Eur. J. Immunol. 34:3187-3196. [DOI] [PubMed] [Google Scholar]
- 13.Fero, M. L., M. Rivkin, M. Tasch, P. Porter, C. E. Carow, E. Firpo, K. Polyak, L. H. Tsai, V. Broudy, R. M. Perlmutter, K. Kaushansky, and J. M. Roberts. 1996. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell 85:733-744. [DOI] [PubMed] [Google Scholar]
- 14.Fisher, R. J., M. Fivash, J. Casas-Finet, J. W. Erickson, A. Kondoh, S. V. Bladen, C. Fisher, D. K. Watson, and T. Papas. 1994. Real-time DNA binding measurements of the ETS1 recombinant oncoproteins reveal significant kinetic differences between the p42 and p51 isoforms. Protein Sci. 3:257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foger, N., R. Marhaba, and M. Zoller. 2000. CD44 supports T cell proliferation and apoptosis by apposition of protein kinases. Eur. J. Immunol. 30:2888-2899. [DOI] [PubMed] [Google Scholar]
- 16.Fry, T. J., B. L. Christensen, K. L. Komschlies, R. E. Gress, and C. L. Mackall. 2001. Interleukin-7 restores immunity in athymic T-cell-depleted hosts. Blood 97:1525-1533. [DOI] [PubMed] [Google Scholar]
- 17.Gadhoum, Z., M. P. Leibovitch, J. Qi, D. Dumenil, L. Durand, S. Leibovitch, and F. Smadja-Joffe. 2004. CD44: a new means to inhibit acute myeloid leukemia cell proliferation via p27Kip1. Blood 103:1059-1068. [DOI] [PubMed] [Google Scholar]
- 18.Green, D. R., N. Droin, and M. Pinkoski. 2003. Activation-induced cell death in T cells. Immunol. Rev. 193:70-81. [DOI] [PubMed] [Google Scholar]
- 19.Grenningloh, R., B. Y. Kang, and I. C. Ho. 2005. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J. Exp. Med. 201:615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haks, M. C., S. M. Belkowski, M. Ciofani, M. Rhodes, J. M. Lefebvre, S. Trop, P. Hugo, J. C. Zuniga-Pflucker, and D. L. Wiest. 2003. Low activation threshold as a mechanism for ligand-independent signaling in pre-T cells. J. Immunol. 170:2853-2861. [DOI] [PubMed] [Google Scholar]
- 21.Hogerkorp, C. M., S. Bilke, T. Breslin, S. Ingvarsson, and C. A. Borrebaeck. 2003. CD44-stimulated human B cells express transcripts specifically involved in immunomodulation and inflammation as analyzed by DNA microarrays. Blood 101:2307-2313. [DOI] [PubMed] [Google Scholar]
- 22.Hsu, T., and R. A. Schulz. 2000. Sequence and functional properties of Ets genes in the model organism Drosophila. Oncogene 19:6409-6416. [DOI] [PubMed] [Google Scholar]
- 23.Huot, T. J., J. Rowe, M. Harland, S. Drayton, S. Brookes, C. Gooptu, P. Purkis, M. Fried, V. Bataille, E. Hara, J. Newton-Bishop, and G. Peters. 2002. Biallelic mutations in p16INK4a confer resistance to Ras- and Ets-induced senescence in human diploid fibroblasts. Mol. Cell. Biol. 22:8135-8143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jorcyk, C. L., D. K. Watson, G. J. Mavrothalassitis, and T. S. Papas. 1991. The human ETS1 gene: genomic structure, promoter characterization and alternative splicing. Oncogene 6:523-532. [PubMed] [Google Scholar]
- 25.Kaartinen, V., and A. Nagy. 2001. Removal of the floxed neo gene from a conditional knockout allele by the adenoviral Cre recombinase in vivo. Genesis 31:126-129. [DOI] [PubMed] [Google Scholar]
- 26.Kanavaros, P., K. Stefanaki, D. Rontogianni, D. Papalazarou, M. Sgantzos, D. Arvanitis, C. Vamvouka, V. Gorgoulis, I. Siatitsas, N. J. Agnantis, and M. Bai. 2001. Immunohistochemical expression of p53, p21/waf1, rb, p16, cyclin D1, p27, Ki67, cyclin A, cyclin B1, bcl2, bax and bak proteins and apoptotic index in normal thymus. Histol. Histopathol. 16:1005-1012. [DOI] [PubMed] [Google Scholar]
- 27.Kieper, W. C., and S. C. Jameson. 1999. Homeostatic expansion and phenotypic conversion of naive T cells in response to self peptide/MHC ligands. Proc. Natl. Acad. Sci. USA 96:13306-13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, W. Y., M. Sieweke, E. Ogawa, H. J. Wee, U. Englmeier, T. Graf, and Y. Ito. 1999. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 18:1609-1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koizumi, S., R. J. Fisher, S. Fujiwara, C. Jorcyk, N. K. Bhat, A. Seth, and T. S. Papas. 1990. Isoforms of the human ets-1 protein: generation by alternative splicing and differential phosphorylation. Oncogene 5:675-681. [PubMed] [Google Scholar]
- 30.Lagresle, C., B. Gardie, S. Eyquem, M. Fasseu, J. C. Vieville, M. Pla, F. Sigaux, and J. C. Bories. 2002. Transgenic expression of the p16INK4a cyclin-dependent kinase inhibitor leads to enhanced apoptosis and differentiation arrest of CD4−CD8− immature thymocytes. J. Immunol. 168:2325-2331. [DOI] [PubMed] [Google Scholar]
- 31.Lesley, J., R. Hyman, and P. W. Kincade. 1993. CD44 and its interaction with extracellular matrix. Adv. Immunol. 54:271-335. [DOI] [PubMed] [Google Scholar]
- 32.Li, R., H. Pei, and T. Papas. 1999. The p42 variant of ETS1 protein rescues defective Fas-induced apoptosis in colon carcinoma cells. Proc. Natl. Acad. Sci. USA 96:3876-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lionneton, F., E. Lelievre, D. Baillat, D. Stehelin, and F. Soncin. 2003. Characterization and functional analysis of the p42Ets-1 variant of the mouse Ets-1 transcription factor. Oncogene 22:9156-9164. [DOI] [PubMed] [Google Scholar]
- 34.Liu, F., and J. L. Whitton. 2005. Cutting edge: re-evaluating the in vivo cytokine responses of CD8+ T cells during primary and secondary viral infections. J. Immunol. 174:5936-5940. [DOI] [PubMed] [Google Scholar]
- 35.Liu, H., and T. Grundstrom. 2002. Calcium regulation of GM-CSF by calmodulin-dependent kinase II phosphorylation of Ets1. Mol. Biol. Cell 13:4497-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, H., M. Holm, X. Q. Xie, M. Wolf-Watz, and T. Grundstrom. 2004. AML1/Runx1 recruits calcineurin to regulate granulocyte macrophage colony-stimulating factor by Ets1 activation. J. Biol. Chem. 279:29398-29408. [DOI] [PubMed] [Google Scholar]
- 37.Lucas, B., F. Vasseur, and C. Penit. 1993. Normal sequence of phenotypic transitions in one cohort of 5-bromo-2′-deoxyuridine-pulse-labeled thymocytes. Correlation with T cell receptor expression. J. Immunol. 151:4574-4582. [PubMed] [Google Scholar]
- 38.Maroulakou, I. G., and D. B. Bowe. 2000. Expression and function of Ets transcription factors in mammalian development: a regulatory network. Oncogene 19:6432-6442. [DOI] [PubMed] [Google Scholar]
- 39.Murali-Krishna, K., and R. Ahmed. 2000. Cutting edge: naive T cells masquerading as memory cells. J. Immunol. 165:1733-1737. [DOI] [PubMed] [Google Scholar]
- 40.Muthusamy, N., K. Barton, and J. M. Leiden. 1995. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature 377:639-642. [DOI] [PubMed] [Google Scholar]
- 41.Nakayama, K., N. Ishida, M. Shirane, A. Inomata, T. Inoue, N. Shishido, I. Horii, D. Y. Loh, and K. Nakayama. 1996. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85:707-720. [DOI] [PubMed] [Google Scholar]
- 42.O'Flaherty, E., W. K. Wong, S. J. Pettit, K. Seymour, S. Ali, and J. A. Kirby. 2000. Regulation of T-cell apoptosis: a mixed lymphocyte reaction model. Immunology 100:289-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohtani, N., Z. Zebedee, T. J. Huot, J. A. Stinson, M. Sugimoto, Y. Ohashi, A. D. Sharrocks, G. Peters, and E. Hara. 2001. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409:1067-1070. [DOI] [PubMed] [Google Scholar]
- 44.Ponta, H., L. Sherman, and P. A. Herrlich. 2003. CD44: from adhesion molecules to signalling regulators. Nat Rev. Mol. Cell. Biol. 4:33-45. [DOI] [PubMed] [Google Scholar]
- 45.Puthier, D., F. Joly, M. Irla, M. Saade, G. Victorero, B. Loriod, and C. Nguyen. 2004. A general survey of thymocyte differentiation by transcriptional analysis of knockout mouse models. J. Immunol. 173:6109-6118. [DOI] [PubMed] [Google Scholar]
- 46.Rabault, B., and J. Ghysdael. 1994. Calcium-induced phosphorylation of ETS1 inhibits its specific DNA binding activity. J. Biol. Chem. 269:28143-28151. [PubMed] [Google Scholar]
- 47.Rafi, A., M. Nagarkatti, and P. S. Nagarkatti. 1997. Hyaluronate-CD44 interactions can induce murine B-cell activation. Blood 89:2901-2908. [PubMed] [Google Scholar]
- 48.Remy, P., and M. Baltzinger. 2000. The Ets-transcription factor family in embryonic development: lessons from the amphibian and bird. Oncogene 19:6417-6431. [DOI] [PubMed] [Google Scholar]
- 49.Seidel, J. J., and B. J. Graves. 2002. An ERK2 docking site in the Pointed domain distinguishes a subset of ETS transcription factors. Genes Dev. 16:127-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seth, A., and D. K. Watson. 2005. ETS transcription factors and their emerging roles in human cancer. Eur. J. Cancer 41:2462-2478. [DOI] [PubMed] [Google Scholar]
- 51.Sharpless, N. E., N. Bardeesy, K. H. Lee, D. Carrasco, D. H. Castrillon, A. J. Aguirre, E. A. Wu, J. W. Horner, and R. A. DePinho. 2001. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature 413:86-91. [DOI] [PubMed] [Google Scholar]
- 52.Sprent, J. 2003. Turnover of memory-phenotype CD8+ T cells. Microbes Infect. 5:227-231. [DOI] [PubMed] [Google Scholar]
- 53.Spyropoulos, D. D., F. O. Bartel, T. Higuchi, T. Deguchi, M. Ogawa, and D. K. Watson. 2003. Marker-assisted study of genetic background and gene-targeted locus modifiers in lymphopoietic phenotypes. Anticancer Res. 23:2015-2026. [PubMed] [Google Scholar]
- 54.Spyropoulos, D. D., P. N. Pharr, K. R. Lavenburg, P. Jackers, T. S. Papas, M. Ogawa, and D. K. Watson. 2000. Hemorrhage, impaired hematopoiesis, and lethality in mouse embryos carrying a targeted disruption of the Fli1 transcription factor. Mol. Cell. Biol. 20:5643-5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tanchot, C., F. A. Lemonnier, B. Perarnau, A. A. Freitas, and B. Rocha. 1997. Differential requirements for survival and proliferation of CD8 naive or memory T cells. Science 276:2057-2062. [DOI] [PubMed] [Google Scholar]
- 56.Tsukiyama, T., N. Ishida, M. Shirane, Y. A. Minamishima, S. Hatakeyama, M. Kitagawa, K. Nakayama, and K. Nakayama. 2001. Down-regulation of p27Kip1 expression is required for development and function of T cells. J. Immunol. 166:304-312. [DOI] [PubMed] [Google Scholar]
- 57.Venanzoni, M. C., L. R. Robinson, D. R. Hodge, I. Kola, and A. Seth. 1996. ETS1 and ETS2 in p53 regulation: spatial separation of ETS binding sites (EBS) modulate protein: DNA interaction. Oncogene 12:1199-1204. [PubMed] [Google Scholar]
- 58.Walunas, T. L., B. Wang, C. R. Wang, and J. M. Leiden. 2000. Cutting edge: the Ets1 transcription factor is required for the development of NK T cells in mice. J. Immunol. 164:2857-2860. [DOI] [PubMed] [Google Scholar]
- 59.Wang, D., S. A. John, J. L. Clements, D. H. Percy, K. P. Barton, and L. A. Garrett-Sinha. 2005. Ets-1 deficiency leads to altered B cell differentiation, hyperresponsiveness to TLR9 and autoimmune disease. Int. Immunol. 17:1179-1191. [DOI] [PubMed] [Google Scholar]