Abstract
The CDP-ethanolamine pathway is responsible for the de novo biosynthesis of ethanolamine phospholipids, where CDP-ethanolamine is coupled with diacylglycerols to form phosphatidylethanolamine. We have disrupted the mouse gene encoding CTP:phosphoethanolamine cytidylyltransferase, Pcyt2, the main regulatory enzyme in this pathway. Intercrossings of Pcyt2+/− animals resulted in small litter sizes and unexpected Mendelian frequencies, with no null mice genotyped. The Pcyt2−/− embryos die after implantation, prior to embryonic day 8.5. Examination of mRNA expression, protein content, and enzyme activity in Pcyt2+/− animals revealed the anticipated 50% decrease due to the gene dosage effect but rather a 20 to 35% decrease. [14C]ethanolamine radiolabeling of hepatocytes, liver, heart, and brain corroborated Pcyt2 gene expression and activity data and showed a decreased rate of phosphatidylethanolamine biosynthesis in heterozygotes. Total phospholipid content was maintained in Pcyt2+/− tissues; however, this was not due to compensatory increases in the decarboxylation of phosphatidylserine. These results establish the necessity of Pcyt2 for murine development and demonstrate that a single Pcyt2 allele in heterozygotes can maintain phospholipid homeostasis.
Phosphatidylethanolamine (PE) is a dominant inner-leaflet phospholipid in cell membranes (9), where it plays a role in membrane function by structurally stabilizing membrane-anchored proteins (10, 13, 26), and participates in important cellular processes such as cell division (10), cell fusion (24, 25), blood coagulation (7, 11, 12), and apoptosis (11). In Drosophila, where PE is the major membrane phospholipid, the release of sterol regulatory element-binding proteins is controlled by PE, as opposed to sterols in mammalian cells (8). Biosynthesis of PE occurs de novo via the Kennedy or CDP-ethanolamine pathway (17), which is terminated in the endoplasmic reticulum, and can also be produced by the decarboxylation of phosphatidylserine (PS) in mitochondria or by base-exchange mechanisms (38). Studies using rat liver hepatocytes and hamster heart have shown that, quantitatively, PE is mainly produced via the de novo pathway (33, 44). In vivo radiolabeling experiments have also shown that the CDP-ethanolamine pathway is the major route of PE production in rat liver, heart, and kidney (1), which corroborates previous work on isolated hepatocytes and human Y79 retinoblastoma cells (33, 43). In vitro evidence, however, showed that in hamster CHO and BHK cell lines, PS decarboxylation could become the major contributing pathway for PE production (18, 40).
CTP:phosphoethanolamine cytidylyltransferase (Pcyt2) is the main regulatory enzyme in the CDP-ethanolamine pathway and catalyzes the formation of CDP-ethanolamine from phosphoethanolamine and CTP (3, 17, 30). Pcyt2 is a single gene that is alternatively spliced at exon 7 to produce two transcripts, Pcyt2α and Pcyt2β (23), which are differentially expressed among tissues. This splicing event is evolutionarily conserved among mammals, birds, and amphibians. The analogous rate-limiting enzyme in the CDP-choline pathway, CTP:phosphocholine cytidylyltransferase (Pcyt1), has been extensively investigated, while the understanding of Pcyt2 remains in its infancy.
Various knockout models have been generated to investigate phospholipid metabolism (37). The liver-specific PE-N-methyltransferase (PEMT), which converts PE to phosphatidylcholine (PC), was disrupted in mice, and Pemt−/− mice were viable due to the compensation of the CDP-choline pathway. When fed a choline-deficient diet, however, Pemt−/− mice experienced steatohepatitis and died within 5 days (19). Various models have also addressed the isoforms of Pcyt1, showing that the deletion of Pcyt1a results in embryonic lethality preimplantation (42) and that Pcyt1b2−/− mice are viable, but both male and female mice experience gonadal dysfunction (16). The necessity of the genes responsible for PS synthesis, PS synthase 1 and 2 (Pss1/2), has been evaluated in a Pss2 knockout model (2, 28). Pss2−/− mice develop normally due to an increase in Pss1 expression and a decrease in PS degradation (28); however, a percentage of male mice experience testicular abnormalities and infertility, as Pss2 was determined to be abundant in the testes (2). Most recently, an isoform of ethanolamine kinase, EKI2, was targeted. Although there was no reduction in the rate of PE synthesis via the CDP-ethanolamine pathway, likely due to the presence of both EKI1 and choline kinase, EKI2−/− female mice experience placental thrombosis (32).
There is evidence supporting the formation of two distinct pools of PE, via the CDP-ethanolamine and PS decarboxylation pathways, which are utilized for distinct PE functions (35); however, until recently the necessity for each individual pathway had not been fully appreciated. The rate-controlling enzyme in the decarboxylation pathway, PS decarboxylase (Pisd), has been disrupted in mice, resulting in early embryonic lethality around embryonic day 9.0 (E9.0), likely due to malformed and dysfunctional mitochondria (27). In addition, although Pisd+/− mice experienced approximately half of the wild-type Pisd expression, they were phenotypically indistinguishable from controls. In Pisd heterozygous animals, Pcyt2 protein expression as well as the production of PE via the CDP-ethanolamine pathway was increased to compensate for the loss of production via the PS decarboxylation pathway (27). We have created a Pcyt2 knockout model to investigate the importance of Pcyt2 and therefore the CDP-ethanolamine pathway in mammalian phospholipid biosynthesis. Here, we report that the homozygous disruption of this gene results in embryonic lethality prior to E8.5, while heterozygotes have increased expression of the remaining Pcyt2 allele required for maintaining necessary levels of PE for phospholipid homeostasis. At the same time, there is no change in the level of other phospholipids, nor is there compensation from the alternative PE synthetic pathway via PS decarboxylation.
MATERIALS AND METHODS
Targeted disruption of the Pcyt2 gene.
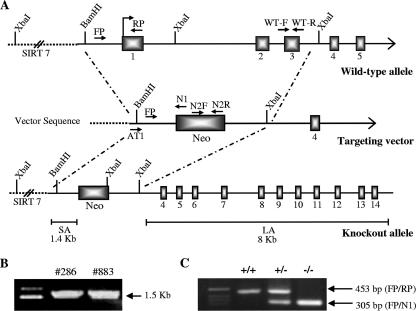
As a first step toward generating Pcyt2−/− mice, we characterized the mouse Pcyt2 gene and promoter (23), and a 12.5-kb genomic region used to construct the targeting vector was first subcloned from a positively identified bacterial artificial chromosome clone (AY189524). The targeting vector was created by homologous recombination and was designed such that the short homology arm extends ∼1.4 kb 5′ to exon 1. The long homology arm starts at the 3′ side of exon 3 and is ∼8 kb long. The neomycin cassette is trapped inside exon 1, replacing 2.8 kb of the gene including the promoter, ATG, and exons 2 and 3. The targeting vector was linearized by digestion with NotI and then transfected by electroporation of 129 SvEviTL embryonic stem (ES) cells (InGenious Targeting, New York). After selection in G418, surviving clones were screened by PCR using the primers AT1 (ATGATTTCTTCATGGTGTAGTCTG) and N1 (TGCGAGGCCAGAGGCCACTTGTGTAGC) to identify recombinant ES clones containing the neomycin cassette. Selection screening for homologous recombination identified two positive ES clones, 286 and 883. These clones were injected into mouse blastocysts and implanted into pseudopregnant females. Agouti, chimeric male offspring were then crossed back to C57BL/6 mice to achieve germ line transmission.
Animals.
All procedures were approved by the University of Guelph's Animal Care Committee and were in accordance with guidelines of the Canadian Council on Animal Care. Mice were exposed to a 12-h light/12-h dark cycle beginning with light at 7:00 a.m. Male and female mice were fed a standardized diet (Harlan Teklad S-2335) ad libitum and had free access to water. Mice described are of mixed genetic background (C57BL/6 × 129/Sv).
Genotyping.
Pcyt2 genotypes were identified from genomic DNA isolated from mouse tails, embryo yolk sacs, and total embryos. Tissues were digested with 200 μg of proteinase K at 55°C in a buffer (10 mM Tris-HCl pH 8.0, 0.1 M EDTA pH 8.0, 0.5% sodium dodecyl sulfate [SDS], 0.1 M NaCl). To eliminate RNA, 10 μg of RNase A was added. Purified genomic DNA was amplified using a common upstream primer, FP (CCTGGAACTCATGAGATCCTCCTG), in combination with either a downstream primer, RP (ATCGCACCACACCCGCACGA), specific for the wild-type allele or primer N1 (TGCGAGGCCAGAGGCCACTTGTGTAGC), specific for the knockout allele. Confirmatory screenings were performed with separate sets of primers specific to the neomycin cassette (N2F, GCACCGCTGAGCAATGGAAG; N2R, CGATTGTCTGTTGTGCCCAGTC) or the wild-type allele (ET-FP, CCTAGAGGAGATTGCCAAGC; ET-RP, CTGCCGTGAACAGAGAAGTC). PCRs utilized 0.3 units of REDtaq genomic DNA polymerase (Sigma). Initial denaturation was at 94°C for 5 min, followed by 32 cycles at 94°C for 1 min, 60°C for 30 s, and 72°C for 1 min, with a final extension at 72°C for 10 min. The primer pairs FP/RP, FP/N1, N2F/N2R, and ET-FP/ET-RP yielded products of 450, 305, 339, and 167 bp, respectively.
RNA analyses.
Tissues were harvested, immediately frozen in liquid nitrogen, and stored at −80°C. A total of 50 to 100 mg of tissue was homogenized in Trizol reagent (Invitrogen), and total RNA was extracted according to the manufacturer's protocol. cDNA was synthesized from 2 μg of total RNA using a poly(dT) primer and Superscript II reverse transcriptase (Invitrogen). Pcyt2 cDNA was amplified using the upstream primer F11 (ACCATACTCCGTGACAGCGG) and the downstream primer R13 (GGTGGGCACAGGGCAAGGGC), corresponding to positions in exons 11 and 13, respectively. The two splice variants of Pcyt2, α and β, were amplified using F6 (GGAGATGTCCTCTGAGTACCG) and R7 (GGCACCAGCCACATAGATGAC) primers, which flank the spliced region. Pisd was amplified using primers previously described (27). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified using the upstream primer ACCACAGTCCATGCCATCAC and the downstream primer TCCACCACCCTGTTGCTGTA. PCRs for Pcyt2 total, Pcyt2α and β, Pisd, and GAPDH were carried out using the following cycle parameters: 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s for 28, 32, 32, and 24 cycles, respectively, which were predetermined as the given phases proportioned to the product concentration. Pcyt2 total, Pcyt2α, Pcyt2β, Pisd, and GAPDH PCRs yielded products of 342 bp, 225 bp, 172 bp, 133 bp, and 451 bp, respectively, which were resolved on a 1.5% agarose gel, visualized by ethidium bromide staining, and quantified using Image J densitometry software (NIH).
Production of Pcyt2-specific antibody.
A polyclonal Pcyt2 antibody that recognizes both α and β splice variants was generated against the peptide VTKAHHSSQEMSSEYRE, situated at the end of the first catalytic motif and directly before the spliced peptide (23). Rabbits were immunized with the hemocyanine-conjugated peptide, and immune serum was collected after four successive immunizations. The antibody (V-5497) was purified on an affinity column containing immobilized peptide antigen busing 0.2 M glycine/HCl (pH 2.2) and 250 mM NaCl. The pure antibody was preserved by adding 0.02% thimerosal and was then stored at −20°C.
Histology and immunohistochemistry.
For histological staining, the uterine tissue was dissected from pregnant females at various stages of pregnancy and fixed in 10% formalin in phosphate-buffered saline (PBS). Tissues were then embedded in paraffin wax, dried with ethanol, and sectioned into 20-μm sections. Slides were then stained with hematoxylin and eosin. For immunohistochemistry (IHC) analysis, embryos were dissected from pregnant females at various stages of pregnancy and fixed for 2 h in 4% paraformaldehyde at 4°C and then in 10% formalin in PBS. Tissues were then embedded in paraffin wax, dried with ethanol, and sectioned into 20-μm sections and stored at 4°C. Slides were first exposed to xylene to remove the paraffin and then to 100% ethanol, then to 3% H2O2 in methanol, and successively decreasing concentrations of ethanol for hydration. Slides were then incubated in a citrate buffer (9.4 mM citric acid and 40 mM sodium citrate, pH 6.0) for 15 min at 94°C for antigen retrieval and then blocked for 30 min in PBS with 0.2% bovine serum albumin (BSA), 0.5% Triton X-100, and 1% goat serum. The Pcyt2 antibody was diluted in PBS with 1% BSA (1:50), and slides were incubated at 4°C overnight. After washes with PBS with 0.1% Tween 20, slides were introduced to a goat anti-rabbit immunoglobulin G (IgG) secondary conjugated to horseradish peroxidase (HRP) at a dilution of 1:200 for 30 min. Antigen visualization was performed for 5 min with a diaminobenzidine substrate kit (Vector Laboratories), and samples were subsequently counterstained using Harris's hematoxylin and dehydrated.
Pcyt2 immunoblotting.
Frozen tissue samples were homogenized in a cold lysis buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, and 10 mM NaF) containing protease (1/10) and phosphatase (1/100) inhibitor cocktails (Sigma). The lysate was centrifuged at 2,000 × g for 20 min at 4°C to remove cell debris. Proteins (10 μg for liver and 25 μg for heart and brain) were separated by 10% SDS-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes. Prior to blotting, red die (Ponceau S) staining was used to ensure proper protein transfer and loading. Membranes were blocked with 5% milk in 20 mM Tris-HCl (pH 7.5), 500 mM NaCl, and 0.05% Tween-20 (TBS-T) for 2 h at room temperature, followed by brief washing in TBS-T alone. Membranes were then incubated with the Pcyt2-specific antibody at a 1:2,000 dilution in TBS-T at 4°C overnight. Membranes were then washed three times for 15 min each in TBS-T. Membranes were incubated with the secondary antibody, an HRP-conjugated goat anti-rabbit IgG, diluted 1:20,000, for 1 h at room temperature, and then washed five times for 15 min in TBS-T and visualized with enhanced chemiluminescence (Amersham). For the control β-actin protein, anti-β-actin antibody (Biovision) was used at a dilution of 1:10,000 (1% BSA in TBS-T), and an anti-mouse HRP-conjugated IgG at 1:10,000 dilution was used.
Pcyt2 enzymatic activities.
Tissue homogenates were twice frozen in liquid nitrogen, thawed on ice, and then centrifuged for 2 min at 13,000 × g to pellet cellular debris. The supernatants were then removed, and the enzymatic activity was assayed using [14C]phosphoethanolamine (American Radiolabeled Chemical) as a substrate (34). Briefly, a mixture of 20 mM Tris-HCl (pH 7.8), 10 mM MgCl2, 5 mM dithiothreitol, 2 mM CTP, 1 mM unlabeled phosphoethanolamine, and 3.6 μM [14C]phosphoethanolamine (55 μCi/μmol) was incubated with tissue homogenate (50 μg of protein) at 37°C for 15 min. Reactions were terminated by boiling for 2 min. A total of 25 μl was then loaded onto silica G plates (Analtech) with CDP-ethanolamine and phosphoethanolamine standards and separated in a solvent system of methanol-0.5% NaCl-ammonia (50:50:5). Plates were then sprayed with 1% ninhydrin and heated for 10 min at 80°C, and the CDP-ethanolamine band was scraped for liquid scintillation counting. The Pcyt2 activity (nmol/min/mg of protein) was determined from the number of disintegrations per minute using a known amount of [14C]phosphoethanolamine as a standard. This assay is linear for the time performed and up to ∼150 μg of protein as determined previously (34). Protein content was determined with bicinchoninic acid protein assay (Pierce).
Phospholipid content.
Tissues were homogenized in PBS, and lipids were extracted from homogenates according to the method of Bligh and Dyer (4). The lower organic phase from the lipid extraction was dried under a stream of nitrogen gas, and lipids were resuspended in a constant volume of chloroform. Lipids were then separated by thin-layer chromatography on silica gel 60 plates (VWR) in a system of chloroform-methanol-acetic acid-water (40:12:2:0.75, vol/vol/vol/vol). The fluorescent probe 1,6-diphenylhexatriene was added to a final fluorophore concentration of 100 μM (15). Plates were dried, and phospholipids were first visualized by excitation at 302 nm and documented. Plates were then sprayed with 15% sulfuric acid with 0.5% K2CrO7 and heated for 20 min at 110°C. Plates were then scanned, and phospholipids were quantified using standard curves generated for each lipid using reflection densitometry. Phosphatidylinositol, PC, and PS standards were from Sigma, and the PE standard was from Avanti Polar Lipids, Inc.
Hepatocyte isolation and metabolic labeling.
Hepatocytes were isolated from mice as previously described (32, 42). Briefly, livers were removed and rinsed briefly in a buffer containing 66.7 mM NaCl, 6.7 mM KCl, 100 mM HEPES (pH 7.6), and 36 mM glucose. Livers were then minced with a Mcllwain tissue chopper (Vibratome) and digested in the same buffer plus 5 mM CaCl2·2H2O, collagenase (0.5 mg/ml) and DNase (6 μg/ml) for 20 min at 37°C three times successively. After each step the supernatant containing the cell suspension was collected, and the combined cell suspension was filtered through 41-μm-pore-size Spectra/Mesh nylon (Fisher Scientific). The hepatocytes in the filtrate were collected by centrifugation at 100 × g for 2 min and washed three times with buffer containing 137 mM NaCl, 5 mM KCl, 0.65 mM Mg2SO4·7H2O, 1.2 mM CaCl2·2H2O, 10 mM HEPES (pH 7.4), and BSA (15g/ml). The cells were resuspended in 10 ml of Dulbecco's PBS containing 5 mM glucose and 3.3 mM pyruvate plus 0.5% BSA. Cells were then plated on 60-mm collagen-coated plates (BD Biosciences) and incubated at 37°C with 5% CO2 for 2 h. The medium was then removed, and fresh medium containing 0.2 μCi of [1,2-14C]ethanolamine or 2 μCi of l-[3-3H]serine (American Radiolabeled Chemicals) was added for up to 3 h. After incubation, cells were transferred to ice, washed with PBS, and collected for lipid extraction as described above. Phospholipids and water-soluble intermediates were resolved as described above, and the radioactivity associated with each was determined by liquid scintillation counting. Duplicate plates were used for bicinchoninic acid protein determination.
Mitochondrial staining.
Hepatocytes were isolated and plated as described above. After 20 h the growth medium was replaced with fresh prewarmed medium (Dulbecco's modified Eagle's medium plus 17% fetal bovine serum) containing 100 nM MitoTracker Red (Molecular Probes). The cells were additionally incubated for 30 min at 37°C, and the medium was than replaced with freshly prepared, prewarmed medium containing 4% formaldehyde. Plates were incubated for 15 min at 37°C, and the fixed cells were washed three times with PBS before being viewed with a fluorescent microscope.
In vivo labeling.
Experiments were conducted as previously described (1) with some alterations. Mice were weighed and injected with the radiolabeled substrate via intraperitoneal injection. The quantity of radioactive precursor used was 0.5 μCi of [1,2-14C]ethanolamine and 5μCi of l-[3-3H]serine. Mice were allowed to recover for 3 h and were sacrificed using CO2. Brain, heart, kidney, and liver were then dissected, weighed, and immediately homogenized in PBS. Total lipids were extracted as described above, and an aliquot of the total extract as well as the lipid-containing organic phase was counted for total radioactivity. PE, PC, and PS were resolved via thin-layer chromatography as described below and individually counted.
Phospholipid fatty acid composition.
Liver lipid samples extracted as described above were subsequently transmethylated to allow the determination of fatty acid side chain composition, as previously described (6). Briefly an aliquot of hepatic lipid extract was dried under a stream of nitrogen and transmethylated with 6% (by volume) H2SO4 in methanol at 80°C for 3 h. After being cooled to room temperature, the methylated fatty acids were collected by the addition of petroleum ether and deionized H2O followed by centrifugation at 1,000 × g for 15 min at 4°C. Samples were then stored at −20°C until analysis by gas-liquid chromatography (HP 5890 Series II; Hewlett Packard).
Statistical analysis.
All significant differences were determined with a Student's t test using GraphPad Prism 4.0 software; significance was determined as a P value of <0.05.
RESULTS
Disruption of Pcyt2 results in embryonic lethality.
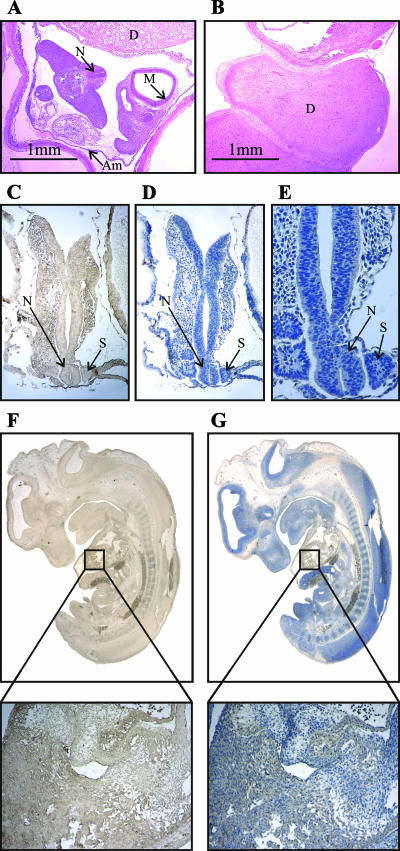
Chimeric mice were generated as described in Materials and Methods, chosen for agouti coloration, and subsequently bred to C57BL/6 mice to determine if germ line transmission was achieved. Eight Pcyt2+/− mice were generated as founders and crossed to generate Pcyt2−/− mice. Two individual mouse lines were generated from positive ES clones 286 and 883; however, no differences were established between the two. Although Pcyt2+/− mice were fertile, initial intercrosses yielded small litter sizes. PCR genotyping (Fig. 1A and C) revealed no homozygous mutants from a total of 119 mice (Table 1), which led to the hypothesis that Pcyt2−/− mice die in utero. To further investigate this, embryos were collected from timed matings and genotyped. Embryos from E13.5 were dissected, and the yolk sac was genotyped; however, at this stage, no Pcyt2−/− embryos were identified. There were, however, identifiable resorption sites on the uterus. E10.5 embryos were then dissected and genotyped with a similar distribution of genotypes and similar pattern of resorption (Fig. 2A and B). Finally, embryos were isolated at E8.0 to E9.0. At this stage of development, the entire embryo was used for genotyping, and from a total of 56 embryos, only three Pcyt2−/− embryos were identified. Upon dissection, it was noticeable that the embryos that had been genotyped as Pcyt2 null were necrotic. Unlike the later stages of embryonic development, no distinguishable resorption sites were observed on the uterus, and certain implantation sites were void of recognizable embryos, indicating that these embryos were being resorbed at earlier stages of development and had mostly completed the resorption process. These results force the conclusion that Pcyt2−/− embryos implant and are lethal prior to E8.5. The pattern of expression of Pcyt2 in wild-type embryos at this stage was established using IHC. As shown in Fig. 2C to G, the protein was ubiquitously detected throughout the embryo at gestational stages E8.5 and E12, demonstrating that Pcyt2 is expressed in the early stages of embryogenesis. IHC utilizing only the HRP-conjugated secondary antibody was used as a negative control to test for endogenous peroxidase activity and for nonspecific staining (not shown). It has also been demonstrated previously that Pcyt2 is expressed in rat liver between days E17 and E22 (birth), showing a gradual increase in Pcyt2 activity which then remained constant during the adult stage (3). The rat liver Pcyt2 protein had a profile similar to that of Pcyt2 activity (3). Thus, the embryonic lethality of Pcyt2 homozygotes led us to investigate further the consequences of the single Pcyt2 allele in the heterozygous state.
FIG. 1.
Targeted disruption of the Pcyt2 gene. (A) The targeting vector was created by replacing a 2.8-kb fragment, including exons 1 to 3 and the translational start site, with a neomycin (Neo) cassette. Short and long homologous arms of 1.4 and 8 kb, respectively, flank the neomycin cassette. Genotyping primer locations are indicated. (B) PCR confirmation of two recombinant ES clones, 286 and 883, using primers AT1 and N1 (1.5 kb). (C) Genotyping of mouse genomic DNA using a common forward primer, FP, and two specific reverse primers, RP and N1, for the wild-type and knockout alleles, respectively.
TABLE 1.
Genotypes of pups and embryos from Pcyt2+/− intercrosses
| Agea | Total no. of pups or embryos | Genotype
|
No. of resorbed embryosb | ||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | |||
| 3 wks | 119 | 49 | 70 | 0 | NA |
| E13.5 | 21 | 4 | 12 | 0 | 5 |
| E10.5 | 41 | 10 | 22 | 0 | 9 |
| E8.5c | 56 | 15 | 30 | 3 | 8 |
Heterozygous mice were intercrossed, and embryos and pups were collected at the specified developmental stage and were genotyped as described in Materials and Methods.
The uterus was examined for evidence of resorbed embryos. NA, not applicable.
Genomic DNA from E8.5 embryos was isolated using whole embryos.
FIG. 2.
Embryo histology and developmental Pcyt2 protein expression. Hematoxylin and eosin staining of a wild-type E10.5 embryo (A) and an implantation site where the embryo has been completely resorbed (B). The ubiquitous expression of Pcyt2 is shown by IHC in wild-type E8.5 embryos (magnification, ×5) in which Pcyt2 stained (C) and counterstained (D) with hematoxylin is shown along with a twofold magnification of expression in the neural tube and somites of E8.5 embryos (E). For a later developmental stage, E12 (magnification, ×1.5) Pcyt2 stained (F) and counterstained (G) embryos are shown, each with a magnified view of expression in the developing ventricle (below panels). Figures are representative of at least four embryos at each developmental stage: D, decidua; M, mesencephalon; Am, amnion; N, neural tube; S, somites. Pcyt2 staining is shown in brown (C and F), and counterstaining is shown in blue (D, E, and G).
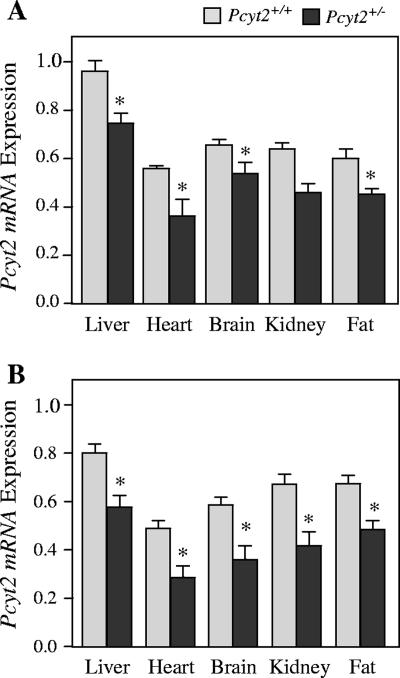
Heterozygosity decreases Pcyt2 mRNA expression.
We next investigated the transcriptional consequences of a single Pcyt2 allele by measuring Pcyt2 mRNA in various tissues. As shown in Fig. 3, mRNA levels were reduced 20 to 35% in liver, heart, brain, kidney, and adipose tissue of both male and female Pcyt2+/− mice. It is firmly established that Pcyt2 is encoded by a single gene; however, it can exist as two distinct transcripts due to the alternative splicing of exon 7 (23), for which there is a tissue-specific pattern of expression. We investigated whether the disruption of a single Pcyt2 allele had effects on the mRNA expression of the two splice variants in the various tissues and found that there were no differences between genotype and gender in heterozygous mice (data not shown). The levels of Pcyt2 mRNA expression did not decrease to the expected level in the heterozygous state, as predicted by the gene dosage effect. Therefore, to further investigate we next assessed the characteristics of Pcyt2 protein expression and activity.
FIG. 3.
Pcyt2 mRNA expression. Primers are common to both Pcyt2 isoforms, and the amplified product is shown relative to GAPDH expression for females (A) and males (B). Data shown represent means ± standard errors of the means for four mice of each gender and genotype quantified in triplicate. *, P < 0.05.
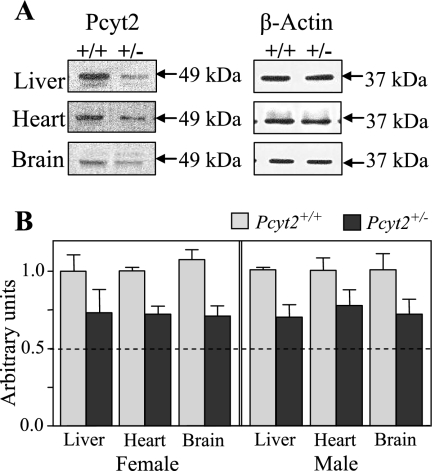
Pcyt2+/− protein expression and enzymatic activity are also diminished.
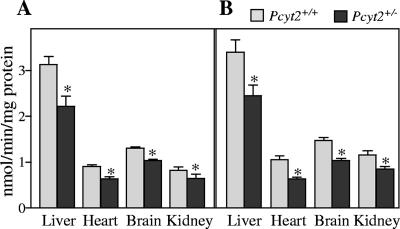
To establish Pcyt2 protein content in tissues of Pcyt2+/+ and Pcyt2+/− mice, immunoblotting was conducted using a polyclonal Pcyt2 antibody (Fig. 4). Although the single Pcyt2 gene gives rise to two splice variants, the molecular mass differs by less than 2 kDa; and, therefore, since the antibody recognizes a common peptide on both splice variants, they are indistinguishable when resolved by SDS-polyacrylamide gel electrophoresis mini-gels. Thus, we have designated the band corresponding to ∼49 kDa to be total Pcyt2. We established that total Pcyt2 protein content in Pcyt2+/− liver, heart, and brain was reduced similarly for both genders and to the approximate level corresponding to mRNA levels, using β-actin as a control. We next examined the enzyme activity of Pcyt2 in the liver, heart, brain, and kidney (Fig. 5). Importantly, there were significant decreases (20 to 35%) in Pcyt2+/− animals relative to littermate controls in all tissues and for both genders. These data demonstrate that although Pcyt2 activity is decreased in heterozygous animals, the actual values do not decrease to 50% of the wild-type levels. This suggests that an up-regulation of the remaining functional allele occurs in heterozygous animals, which is consistently observed at various levels of gene expression.
FIG. 4.
Pcyt2 protein expression in liver, heart, and brain tissues between Pcyt2+/+ and Pcyt2+/− mice. (A) The left panel shows total Pcyt2 (∼49 kDa), and the right panel shows β-actin (∼37 kDa) as an internal control. Blots are representative of four mice of each gender and genotype. (B) Densitometry plots for Pcyt2 expression were normalized to β-actin expression and compared to wild-type levels (set at 1). Data shown represent means ± standard errors of the means for four mice of each gender and genotype quantified in duplicate.
FIG. 5.
Pcyt2 in vitro enzyme activity was assessed in tissue homogenates from Pcyt2+/+ and Pcyt2+/− mice using [14C]phosphoethanolamine as a substrate for females (A) and males (B). Data represent means ± standard errors of the means for at least four mice from each gender and genotype quantified in triplicate. *, P < 0.05.
Phospholipid content does not change in Pcyt2+/− mice.
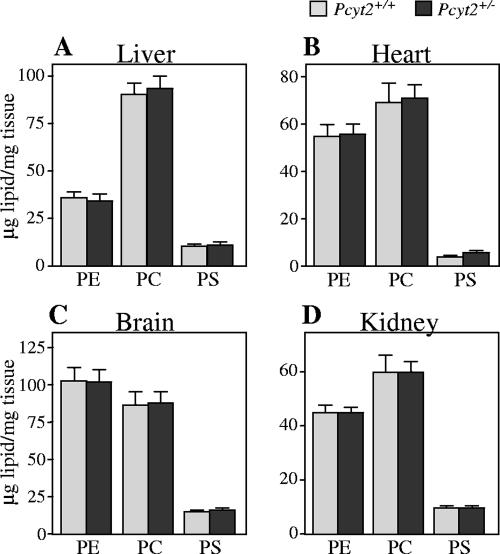
We next investigated the tissue content of the major phospholipids, PE, PC, and PS, in liver, heart, brain, and kidney of Pcyt2+/− and control mice from both genders (Fig. 6). Tissues from a total of seven mice of both genders were analyzed; however, since no difference was observed between genders, results were combined for analyses. In the heterozygous state, the level of PE did not differ between genotypes, and the amounts of PC and PS were also consistent. Unaltered levels of PE, regardless of decreased expression and activity of Pcyt2 in heterozygous animals, suggested the possibility that there could be a compensatory increase in PE production via PS decarboxylation or that the single Pcyt2 allele may be sufficient for PE production via the CDP-ethanolamine pathway. To address these questions, we first investigated the limiting enzyme in the decarboxylation of PS in the mitochondria.
FIG. 6.
Comparison of phospholipid content between Pcyt2+/+ and Pcyt2+/− mice. Phospholipids were isolated from tissues of liver, heart, brain, and kidney as described in Materials and Methods. Data represent means ± standard errors of the means of results from at least seven mice from each gender and genotype.
PS decarboxylase and mitochondria are unaltered.
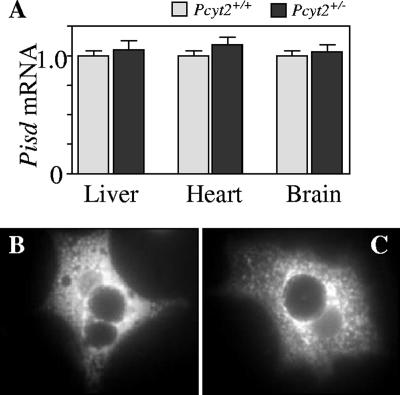
Pcyt2 heterozygous mice maintain PE homeostasis in spite of diminished Pcyt2 expression; therefore, we asked whether the decarboxylation of PS in the mitochondria would be up-regulated as a means of maintaining PE levels. The mRNA expression of Pisd, however, was not altered in the liver, heart, or brain of Pcyt2+/− mice (Fig. 7A). The results were first normalized to GAPDH and expressed relative to wild-type levels, so that each tissue is compared relative to a value of 1. Due to the possibility of an altered state of PS decarboxylation in the mitochondria, we addressed the relative state of the mitochondria by introducing MitoTracker Red to isolated hepatocytes (Fig. 7B and C). It was hypothesized that the mitochondria of Pcyt2+/− mice would be unaffected in relation to their wild-type controls, and this is shown to be the case, as both Pcyt2+/+ and Pcyt2+/− binuclear hepatocytes show similar distributions of mitochondria around the nucleus. Although there were no increases in Pisd mRNA or changes in mitochondrial distribution, we additionally investigated the contribution of the PS decarboxylation pathway to the synthesis of PE in isolated hepatocytes.
FIG. 7.
Pisd mRNA expression and mitochondrial staining of hepatocytes. (A) The amplified product is normalized to GAPDH mRNA and expressed relative to the wild-type value (set at 1). Data shown represent mean ± standard error of the mean for four mice of each gender and genotype quantified in triplicate. Fluorescence micrographs of 20-h cultured primary hepatocytes from Pcyt2+/+ (B) and Pcyt2+/− (C) mice are shown stained with the mitochondrial probe MitoTracker Red.
In vitro and in vivo PE synthesis.
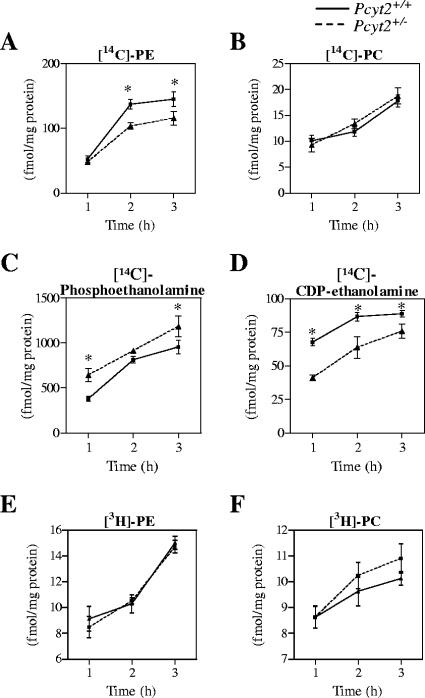
To investigate the contributions of the CDP-ethanolamine and PS decarboxylation pathways in vitro, [14C]ethanolamine and [3H]serine were introduced to hepatocytes isolated from the livers of Pcyt2+/+ and Pcyt2+/− animals. Hepatocytes were incubated for a total of 3 h, with samples taken at 1-, 2-, and 3-h time points. [14C]ethanolamine labeling of hepatocytes revealed a decreased synthesis of PE via the CDP-ethanolamine pathway in Pcyt2+/− hepatocytes (Fig. 8A). The diminished rate of PE synthesis is due to lower levels of Pcyt2 activity, as demonstrated by corresponding differences in the water-soluble intermediates which show an increase in [14C]-phosphoethanolamine (Fig. 8C) and a decreased level of [14C]-CDP-ethanolamine (Fig. 8D). The results were consistent with the hypothesis that there is a decreased rate of PE synthesis by the CDP-ethanolamine pathway that was limited by lowered activity of Pcyt2 in heterozygous animals.
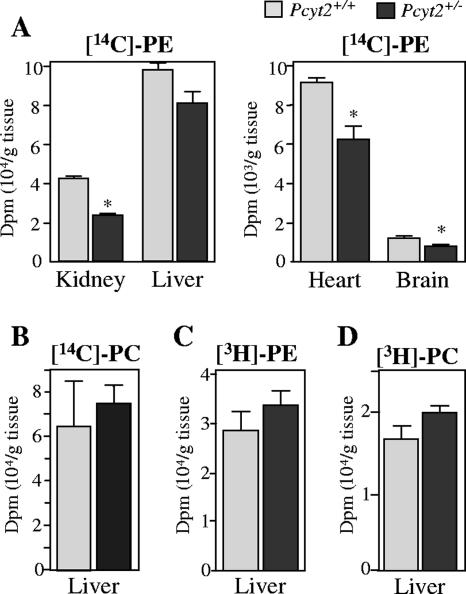
FIG. 8.
In vitro contributions of PE biosynthetic pathways. Primary hepatocytes were incubated with [14C]ethanolamine or [3H]serine as described in Materials and Methods. Data shown for the synthesis of [14C]PE, [14C]PC, [14C]phosphoethanolamine, and [14C]CDP-ethanolamine (A to D) are values after ethanolamine incubation. Data shown for the synthesis of [3H]PE and [3H]PC (E and F) are values after [3H]serine incubation. All data are means ± standard errors of the means and are representative of at least two livers for experiments performed in triplicate. *, P < 0.01.
PE conversion to PC by the methylation pathway was measured by the amount of PC produced after [14C]ethanolamine labeling in hepatocytes. Data show that the methylation of PE derived via the CDP-ethanolamine pathway is not altered between genotypes (Fig. 8B). Finally, hepatocytes were radiolabeled with [3H]serine to reveal the PE produced from PS. These experiments showed that there were no differences between genotypes in the decarboxylation of PS to form PE (Fig. 8E), as determined by the amount of [3H]PE produced; there were also no differences between genotypes in the amount of [3H]PC formed from [3H]serine (Fig. 8F).
To investigate the contributions of the CDP-ethanolamine and PS decarboxylation pathways in vivo, [14C]ethanolamine and [3H]serine were injected into heterozygous and wild-type animals, and incorporation into PE, PC, and PS was measured. Upon intraperitoneal injections, 14C-radiolabeled products were quickly found in all tissues examined, although in different quantities. After 3 h, the incorporation of [14C]ethanolamine into PE in the liver, kidney, heart, and brain (Fig. 9A) was reduced (20 to 35%) in Pcyt2+/− mice compared to wild-type littermates. The amount of PC formed from [14C]ethanolamine via PE methylation was also determined (Fig. 9C); however, no differences between genotypes were observed. Finally, after [3H]serine injections, there were no differences in the amount of PE formed in liver after 3 h (Fig. 9B), again suggesting that the production of PE by decarboxylation was not affected by Pcyt2 deletion. The in vivo labeling experiments also indicated that in the liver, serine radiolabeling of PC was also not altered in Pcyt2+/− mice (Fig. 9D).
FIG. 9.
In vivo contributions of PE biosynthetic pathways. Data represent the incorporation of [14C]ethanolamine into PE (A) in the indicated tissues and the incorporation of [3H]serine into PE in liver (B). The amounts of PC derived from the incorporation of [14C]PE (CDP-ethanolamine pathway) (C) and of PC derived from [3H]PE (PS decarboxylation pathway) (D) via the PEMT pathway in the liver are shown. Data represent means ± standard errors of the means of four mice quantified in duplicate. dpm, disintegrations per min; *, P < 0.05.
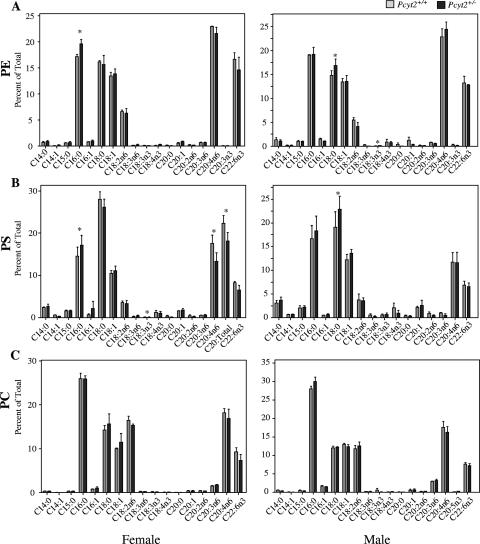
Hepatic phospholipid fatty acid composition is remodeled.
We next investigated whether there was a remodeling of fatty acid side chains in the major phospholipid classes in liver. PE from male and female heterozygous mice revealed slightly different profiles (Fig. 10A). Pcyt2+/− liver PE generally had a higher saturated fatty acid (SFA) content (palmitic and stearic acids) and lower unsaturated fatty acid content compared to controls. Females showed a general decrease in polyunsaturated fatty acids (PUFA), while males had significantly lower levels of ω-3 fatty acids only. Although PE was expected to be the most affected by the disruption of Pcyt2, the composition of PC and PS was also examined. The fatty acid composition of liver PS also demonstrated a significant increase in SFAs (palmitic and stearic acid) and decreased PUFA content in Pcyt2+/− mice, specifically, arachidonic acid and docosahexaenoic acid (Fig. 10B). Although there were fluctuations in the fatty acid composition of PE and PS, PC composition remained relatively undisturbed in heterozygotes of both genders (Fig. 10C).
FIG. 10.
Comparison of hepatic phospholipid compositions. Analyses of individual fatty acid side chains of phospholipids for Pcyt2+/+ and Pcyt2+/− livers expressed as a percentage of total. Results are for three mice of each gender and genotype. Data for female and male analyses for PE, PS, and PC are shown. *, P < 0.05.
DISCUSSION
In the current investigation, we describe the phenotype resulting from the complete and partial disruption of the Pcyt2 gene in mice. Pcyt2 is a single gene that can be alternatively spliced to code for two distinct isoforms. The importance of Pcyt2 within the CDP-ethanolamine pathway, as well as the importance of the entire pathway to mammalian PE biosynthesis, had not been fully elucidated previously. However, by targeting the Pcyt2 gene, we show that its complete disruption in mice results in early embryonic lethality prior to stage E8.5, which makes the Pcyt2 gene and the de novo CDP-ethanolamine pathway for PE biosynthesis indispensable for murine development.
Moreover, heterozygous embryos show no overt phenotypic differences during embryonal development, and pups are indistinguishable from the wild-type littermate controls. Pcyt2 mRNA and protein expression decreased in heterozygous mice relative to their littermate controls, although not to the expected levels that are associated with a true gene dosage effect as seen in other heterozygous knockout models (27, 42). Most importantly, no variations were detected in the amount of total PE and other phospholipids in heterozygous mice, and we demonstrate that the single Pcyt2 allele is able to maintain normal phospholipid homeostasis.
The effect of Pcyt2 deletion on the CDP-ethanolamine pathway was established at multiple levels in Pcyt2+/− and control mice. Radiolabeling of primary hepatocytes demonstrated that the de novo pathway was diminished in Pcyt2+/− cells, as indicated by a decreased rate of PE production, increased levels of the Pcyt2 substrate phosphoethanolamine, and decreased levels of the Pcyt2 product CDP-ethanolamine (Fig. 8). In support of the in vitro experiments, the whole-animal incorporation of [14C]ethanolamine into PE in the liver, kidney, heart, and brain closely resembled the expression pattern and the in vitro activity of Pcyt2, with an overall reduction. Our results show a reduced production rate of PE in Pcyt2+/− hepatocytes and mice as well as consistent phospholipid levels between genotypes in all of the tissues examined. Therefore, we addressed the possibility that an alternative PE synthetic pathway may have been up-regulated in Pcyt2+/− animals to compensate for the diminished rate of PE synthesis. It has been previously demonstrated that phospholipid content could still be maintained regardless of the disruption of certain phospholipid synthetic pathways. In mice, liver PC production is maintained by the CDP-choline pathway when the PE methylation pathway is targeted by the deletion of the Pemt gene (41); in Pss2−/− mice, phospholipid levels were preserved by the presence of Pss1 (28), and in mice heterozygous for Pisd, PE levels were compensated by up-regulation of Pcyt2 and an associated increase in PE production via the CDP-ethanolamine pathway (27). To determine if the reverse phenomenon occurs in Pcyt2+/− mice, we investigated the expression of Pisd as well as the radiolabeling of the decarboxylation pathway by [3H]serine, both in hepatocytes and in whole animals. Our results indicate that there is no significant change in the decarboxylation of PS to compensate for PE production in Pcyt2+/− animals. An alternative fate for PE in the liver is the specific role for the conversion to PC by PEMT. It has been shown that PC produced by PE methylation is destined for bile and lipoprotein secretion (36, 39) and that PE produced by decarboxylation is preferable for methylation (39). Our results, however, demonstrate that PS decarboxylation to PE and PC production are not altered in the heterozygous liver.
Pcyt2 was up-regulated from the anticipated levels due to gene dosage at all of the stages of gene expression analyzed. It is tempting to speculate, therefore, that since the levels of Pcyt2 expression effectively mimicked that of the transcriptional output, the increase in Pcyt2 regulation is initiated at the transcriptional level. In spite of the up-regulation of the remaining Pcyt2 allele, data demonstrate a slower production of PE in Pcyt2+/− mice compared to controls (Fig. 8). The important question that remains, however, is how this occurs without a change in total PE content. To account for the diminished rate of synthesis, it may occur that PE turnover is also slowed to effectively maintain the PE levels in heterozygous animals. We establish that complete Pcyt2 disruption results in embryonic lethality and that these embryos ultimately begin resorption at an early developmental stage, likely due to the absence of sustainable amounts of PE. However, in Pcyt2+/− embryos and mature mice, the critical level of membrane phospholipids is maintained by Pcyt2 protein expression from the single allele.
As part of our investigation into the overall phospholipid phenotype of Pcyt2+/− and control mice, we analyzed the fatty acid composition of PE, PS, and PC in liver. Although there were no changes in total phospholipid content, the SFA content of PE and PS, but not PC, increased in both male and female Pcyt+/− mice, while their total PUFA content decreased. The ramifications and/or significance of these alterations in PE and PS composition have yet to be established; however, numerous other studies and models have demonstrated that fatty acid side chains could affect membrane fluidity and prostanoid production (21) and that PUFA deficiency is strongly associated with fetal development (31) and numerous chronic disorders such as mood disorders (22), neurodegenerative conditions (14, 29), and cardiovascular disease (5).
Finally, our results suggest that the consequence of single-nucleotide polymorphisms (SNPs) in the human Pcyt2 gene may have a similar outcome as in heterozygous mice. The Human dbSNP BLAST database shows that 12 different Pcyt2 polymorphisms have been identified. Specifically for translated regions, an A/G transition at position 244 would alter the existing His to a Tyr (rs17850615), which is the first residue in the putative catalytic sequence (HXGH) (20); a C/T transition exists in the spliced exon 7 (rs2230472), and a C/G transversion occurs in exon 13 (rs2230474). Our results suggest that if such mutations reduce Pcyt2 function, the consequences may not be severe in the heterozygous state since the remaining allele could become up-regulated in response and could also have an effect on PE turnover and fatty acid composition. However, since this gene is essential, homozygous SNPs that completely compromise Pcyt2 function and therefore PE production via the CDP-ethanolamine pathway would likely not exist.
Regardless of past investigations where the necessity of the CDP-ethanolamine pathway in mammalian cells has been questioned, lethality of Pcyt2 null embryos demonstrates that both Pcyt2 and therefore the CDP-ethanolamine pathway are essential for murine development. The presence of a single Pcyt2 allele in heterozygous animals can maintain normal levels of PE for cellular functions, although the remaining allele is not at the level of a true gene dosage effect. The single Pcyt2 allele in heterozygous animals is therefore sufficient, since no compensatory increases in PE production via the mitochondrial PS decarboxylation pathway were established.
Acknowledgments
This work was supported by an operating grant from the Canadian Institute of Health Research held by M.B.
We thank Craig Park for technical assistance with immunohistochemistry.
Footnotes
Published ahead of print on 26 February 2007.
REFERENCES
- 1.Arthur, G., and L. Page. 1991. Synthesis of phosphatidylethanolamine and ethanolamine plasmalogen by the CDP-ethanolamine and decarboxylase pathways in rat heart, kidney and liver. Biochem. J. 273:121-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergo, M. O., B. J. Gavino, R. Steenbergen, B. Sturbois, A. F. Parlow, D. A. Sanan, W. C. Skarnes, J. E. Vance, and S. G. Young. 2002. Defining the importance of phosphatidylserine synthase 2 in mice. J. Biol. Chem. 277:47701-47708. [DOI] [PubMed] [Google Scholar]
- 3.Bleijerveld, O. B., W. Klein, A. B. Vaandrager, J. B. Helms, and M. Houweling. 2004. Control of the CDPethanolamine pathway in mammalian cells: effect of CTP:phosphoethanolamine cytidylyltransferase overexpression and the amount of intracellular diacylglycerol. Biochem. J. 379:711-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 5.Breslow, J. L. 2006. n-3 fatty acids and cardiovascular disease. Am. J. Clin. Nutr. 83:1477S-1482S. [DOI] [PubMed] [Google Scholar]
- 6.Denomme, J., K. D. Stark, and B. J. Holub. 2005. Directly quantitated dietary (n-3) fatty acid intakes of pregnant Canadian women are lower than current dietary recommendations. J. Nutr. 135:206-211. [DOI] [PubMed] [Google Scholar]
- 7.Dobner, P., E. Koller, and B. Engelmann. 1999. Platelet high affinity low density lipoprotein binding and import of lipoprotein derived phospholipids. FEBS Lett. 444:270-274. [DOI] [PubMed] [Google Scholar]
- 8.Dobrosotskaya, I. Y., A. C. Seegmiller, M. S. Brown, J. L. Goldstein, and R. B. Rawson. 2002. Regulation of SREBP processing and membrane lipid production by phospholipids in Drosophila. Science 296:879-883. [DOI] [PubMed] [Google Scholar]
- 9.Dowhan, W. 1997. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem. 66:199-232. [DOI] [PubMed] [Google Scholar]
- 10.Emoto, K., T. Kobayashi, A. Yamaji, H. Aizawa, I. Yahara, K. Inoue, and M. Umeda. 1996. Redistribution of phosphatidylethanolamine at the cleavage furrow of dividing cells during cytokinesis. Proc. Natl. Acad. Sci. USA 93:12867-12872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emoto, K., N. Toyama-Sorimachi, H. Karasuyama, K. Inoue, and M. Umeda. 1997. Exposure of phosphatidylethanolamine on the surface of apoptotic cells. Exp. Cell Res. 232:430-434. [DOI] [PubMed] [Google Scholar]
- 12.Engelmann, B., B. Schaipp, P. Dobner, M. Stoeckelhuber, C. Kogl, W. Siess, and A. Hermetter. 1998. Platelet agonists enhance the import of phosphatidylethanolamine into human platelets. J. Biol. Chem. 273:27800-27808. [DOI] [PubMed] [Google Scholar]
- 13.Escriba, P. V., A. Ozaita, C. Ribas, A. Miralles, E. Fodor, T. Farkas, and J. A. Garcia-Sevilla. 1997. Role of lipid polymorphism in G protein-membrane interactions: nonlamellar-prone phospholipids and peripheral protein binding to membranes. Proc. Natl. Acad. Sci. USA 94:11375-11380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farooqui, A. A., W. Y. Ong, and L. A. Horrocks. 2003. Plasmalogens, docosahexaenoic acid and neurological disorders. Adv. Exp. Med. Biol. 544:335-354. [DOI] [PubMed] [Google Scholar]
- 15.Igal, R. A., M. C. Pallanza de Stringa, and I. N. Tacconi de Gómez Dumm. 1994. Simple method for quantitation of neutral and polar lipids by TLC and fluorescence emission. Int. J. Biochromatogr. 1:137-142. [Google Scholar]
- 16.Jackowski, S., J. E. Rehg, Y. M. Zhang, J. Wang, K. Miller, P. Jackson, and M. A. Karim. 2004. Disruption of CCTβ2 expression leads to gonadal dysfunction. Mol. Cell. Biol. 24:4720-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy, E. P., and S. B. Weiss. 1956. The function of cytidine coenzymes in the biosynthesis of phospholipids. J. Biol. Chem. 222:193-214. [PubMed] [Google Scholar]
- 18.Kuge, O., M. Nishijima, and Y. Akamatsu. 1986. Phosphatidylserine biosynthesis in cultured Chinese hamster ovary cells. II. Isolation and characterization of phosphatidylserine auxotrophs. J. Biol. Chem. 261:5790-5794. [PubMed] [Google Scholar]
- 19.Li, Z., L. B. Agellon, and D. E. Vance. 2005. Phosphatidylcholine homeostasis and liver failure. J. Biol. Chem. 280:37798-37802. [DOI] [PubMed] [Google Scholar]
- 20.Min-Seok, R., Y. Kawamata, H. Nakamura, A. Ohta, and M. Takagi. 1996. Isolation and characterization of ECT1 gene encoding CTP: phosphoethanolamine cytidylyltransferase of Saccharomyces cerevisiae. J. Biochem. (Tokyo) 120:1040-1047. [DOI] [PubMed] [Google Scholar]
- 21.Onuki, Y., M. Morishita, Y. Chiba, S. Tokiwa, and K. Takayama. 2006. Docosahexaenoic acid and eicosapentaenoic acid induce changes in the physical properties of a lipid bilayer model membrane. Chem. Pharm. Bull. 54:68-71. [DOI] [PubMed] [Google Scholar]
- 22.Parker, G., N. A. Gibson, H. Brotchie, G. Heruc, A. M. Rees, and D. Hadzi-Pavlovic. 2006. Omega-3 fatty acids and mood disorders. Am. J. Psychiatry 163:969-978. [DOI] [PubMed] [Google Scholar]
- 23.Poloumienko, A., A. Cote, A. T. Quee, L. Zhu, and M. Bakovic. 2004. Genomic organization and differential splicing of the mouse and human Pcyt2 genes. Gene 325:145-155. [DOI] [PubMed] [Google Scholar]
- 24.Santini, M. T., P. L. Indovina, A. Cantafora, and I. Blotta. 1990. The cesium-induced delay in myoblast membrane fusion is accompanied by changes in isolated membrane lipids. Biochim. Biophys. Acta 1023:298-304. [DOI] [PubMed] [Google Scholar]
- 25.Sessions, A., and A. F. Horwitz. 1981. Myoblast aminophospholipid asymmetry differs from that of fibroblasts. FEBS Lett. 134:75-78. [DOI] [PubMed] [Google Scholar]
- 26.Soulages, J. L., Z. Salamon, M. A. Wells, and G. Tollin. 1995. Low concentrations of diacylglycerol promote the binding of apolipophorin III to a phospholipid bilayer: a surface plasmon resonance spectroscopy study. Proc. Natl. Acad. Sci. USA 92:5650-5654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steenbergen, R., T. S. Nanowski, A. Beigneux, A. Kulinski, S. G. Young, and J. E. Vance. 2005. Disruption of the phosphatidylserine decarboxylase gene in mice causes embryonic lethality and mitochondrial defects. J. Biol. Chem. 280:40032-40040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steenbergen, R., T. S. Nanowski, R. Nelson, S. G. Young, and J. E. Vance. 2006. Phospholipid homeostasis in phosphatidylserine synthase-2-deficient mice. Biochim. Biophys. Acta 1761:313-323. [DOI] [PubMed] [Google Scholar]
- 29.Stillwell, W., S. R. Shaikh, M. Zerouga, R. Siddiqui, and S. R. Wassall. 2005. Docosahexaenoic acid affects cell signaling by altering lipid rafts. Reprod. Nutr. Dev. 45:559-579. [DOI] [PubMed] [Google Scholar]
- 30.Sundler, R., and B. Akesson. 1975. Regulation of phospholipid biosynthesis in isolated rat hepatocytes. Effect of different substrates. J. Biol. Chem. 250:3359-3367. [PubMed] [Google Scholar]
- 31.Tam, O., and S. M. Innis. 2006. Dietary polyunsaturated fatty acids in gestation alter fetal cortical phospholipids, fatty acids and phosphatidylserine synthesis. Dev. Neurosci. 28:222-229. [DOI] [PubMed] [Google Scholar]
- 32.Tian, Y., P. Jackson, C. Gunter, J. Wang, C. O. Rock, and S. Jackowski. 2006. Placental thrombosis and spontaneous fetal death in mice deficient in ethanolamine kinase 2. J. Biol. Chem. 281:28438-28449. [DOI] [PubMed] [Google Scholar]
- 33.Tijburg, L. B., M. J. Geelen, and L. M. Van Golde. 1989. Biosynthesis of phosphatidylethanolamine via the CDP-ethanolamine route is an important pathway in isolated rat hepatocytes. Biochem. Biophys. Res. Commun. 160:1275-1280. [DOI] [PubMed] [Google Scholar]
- 34.Tijburg, L. B., P. S. Vermeulen, and L. M. van Golde. 1992. Ethanolamine-phosphate cytidylyltransferase. Methods Enzymol. 209:258-263. [DOI] [PubMed] [Google Scholar]
- 35.Vance, J. E., E. J. Aasman, and R. Szarka. 1991. Brefeldin A does not inhibit the movement of phosphatidylethanolamine from its sites for synthesis to the cell surface. J. Biol. Chem. 266:8241-8247. [PubMed] [Google Scholar]
- 36.Vance, J. E., T. M. Nguyen, and D. E. Vance. 1986. The biosynthesis of phosphatidylcholine by methylation of phosphatidylethanolamine derived from ethanolamine is not required for lipoprotein secretion by cultured rat hepatocytes. Biochim. Biophys. Acta 875:501-509. [DOI] [PubMed] [Google Scholar]
- 37.Vance, J. E., and D. E. Vance. 2005. Metabolic insights into phospholipid function using gene-targeted mice. J. Biol. Chem. 280:10877-10880. [DOI] [PubMed] [Google Scholar]
- 38.Vance, J. E., and D. E. Vance. 2004. Phospholipid biosynthesis in mammalian cells. Biochem. Cell Biol. 82:113-128. [DOI] [PubMed] [Google Scholar]
- 39.Vance, J. E., and D. E. Vance. 1986. Specific pools of phospholipids are used for lipoprotein secretion by cultured rat hepatocytes. J. Biol. Chem. 261:4486-4491. [PubMed] [Google Scholar]
- 40.Voelker, D. R. 1984. Phosphatidylserine functions as the major precursor of phosphatidylethanolamine in cultured BHK-21 cells. Proc. Natl. Acad. Sci. USA 81:2669-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walkey, C. J., L. R. Donohue, R. Bronson, L. B. Agellon, and D. E. Vance. 1997. Disruption of the murine gene encoding phosphatidylethanolamine N-methyltransferase. Proc. Natl. Acad. Sci. USA 94:12880-12885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, L., S. Magdaleno, I. Tabas, and S. Jackowski. 2005. Early embryonic lethality in mice with targeted deletion of the CTP:phosphocholine cytidylyltransferase α gene (Pcyt1a). Mol. Cell. Biol. 25:3357-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yorek, M. A., R. T. Rosario, D. T. Dudley, and A. A. Spector. 1985. The utilization of ethanolamine and serine for ethanolamine phosphoglyceride synthesis by human Y79 retinoblastoma cells. J. Biol. Chem. 260:2930-2936. [PubMed] [Google Scholar]
- 44.Zelinski, T. A., and P. C. Choy. 1982. Phosphatidylethanolamine biosynthesis in isolated hamster heart. Can. J. Biochem. 60:817-823. [DOI] [PubMed] [Google Scholar]