Abstract
Phosphorylation of a yeast histone H2A at C-terminal serine 129 has a central role in double-strand break repair. Mimicking H2A phosphorylation by replacement of serine 129 with glutamic acid (hta1-S129E) suggested that phosphorylation destabilizes chromatin structures and thereby facilitates the access of repair proteins. Here we have tested chromatin structures in hta1-S129 mutants and in a C-terminal tail deletion strain. We show that hta1-S129E affects neither nucleosome positioning in minichromosomes and genomic loci nor supercoiling of minichromosomes. Moreover, hta1-S129E has no effect on chromatin stability measured by conventional nuclease digestion, nor does it affect DNA accessibility and repair of UV-induced DNA lesions by nucleotide excision repair and photolyase in vivo. Similarly, deletion of the C-terminal tail has no effect on nucleosome positioning and stability. These data argue against a general role for the C-terminal tail in chromatin organization and suggest that phosphorylated H2A, γ-H2AX in higher eukaryotes, acts by recruitment of repair components rather than by destabilizing chromatin structures.
Eukaryotic DNA is packaged in nucleosomes and higher-order chromatin structures. The nucleosome core consists of 147 bp of DNA that is wrapped in ∼1.7 negative superhelical turns around a histone octamer composed of two H2A-H2B dimers and an (H3-H4)2 tetramer. The (H3-H4)2 tetramer binds to the central 60 bp of DNA, and the two H2A-H2B dimers organize 30 bp toward either end of the nucleosomal DNA, reflecting a modular assembly of the nucleosome. The histones are folded in histone fold domains in the center of the nucleosome core and have flexible N-terminal tails that protrude from the nucleosome core particle (29). The tails are thought to interact with the DNA or histones of adjacent nucleosomes, thereby contributing to higher-order structural organization of chromatin (30). The C-terminal tail of H2A is located close to the site where the DNA enters and exits the nucleosome, and in crystal structures obtained with yeast histones, it is brought into close proximity to the N-terminal tail of H2A in the neighboring nucleosome (64). On the other hand, cross-linking data showed that it binds to linker DNA but rearranges to bind the central part of the nucleosome core DNA when the linker is removed (59). Thus, it is conceivable that the C-terminal tail and its modification by phosphorylation as it occurs in DNA repair impact the stability of individual nucleosomes, nucleosome positioning, and nucleosome-nucleosome contacts in higher-order structures.
Nucleosomes control the accessibility of DNA to proteins involved in transcription, replication, recombination, and DNA repair. Occluded sites may become exposed and accessible to proteins by dynamic transitions involving transient dissociation of histones, unwrapping of DNA, or changing the position of histone octamers in the DNA sequence (nucleosome mobility). Those intrinsic properties may be modulated by chromatin remodeling activities that chemically modify the histones, exchange histone variants, and/or alter the structure and position of nucleosomes. Posttranslational modifications of histones include acetylation, methylation, phosphorylation, ubiquitylation, sumoylation, and poly-ADP-ribosylation (10, 39, 62). Histone modifications can act as binding interfaces for the recruitment of chromatin-associated proteins (19, 50). On the other hand, modifications that alter the charge of histones, such as histone acetylation and phosphorylation, may alter the stability of nucleosomes and higher-order structures (17, 47, 66).
Increasing evidence supports a role for chromatin remodeling in DNA repair pathways. In nucleotide excision repair (NER) of UV lesions, remodeling includes rearrangement and/or reassembly of nucleosomes after DNA repair synthesis, acetylation of histones after UV damage induction, and monoubiquitination of histone H2A (15, 16, 21, 67). Double-strand break (DSB) repair involves phosphorylation of H2AX, acetylation of histones in the vicinity of the lesion, and local chromatin expansion immediately after DNA damage induction (25, 57, 61). The C-terminal region of the H2A variant H2AX of mammals and the bulk H2A (Hta1 and Hta2) of Saccharomyces cerevisiae contain a serine 4 residues from the carboxyl terminus in an invariant SQ motif. This serine residue is phosphorylated in response to DSBs by the ataxia-telangiectasia mutated and ataxia-telangiectasia mutated- and Rad3-related checkpoint kinases (Tel1 and Mec1 in S. cerevisiae) (9, 43). In mammals, phosphorylated H2AX (γ-H2AX) spreads over megabase chromatin domains and is required for the stable accumulation of repair proteins into nuclear foci (4, 38, 42). The major mediator of γ-H2AX recognition is likely MDC1, which binds γ-H2AX through its tandem BRCT domain (52). In S. cerevisiae, γ-H2A spreads ∼50 kb in either direction from a defined DSB and repair factors (Mre11 and Rad51) are recruited near the break (48). In addition, several chromatin-remodeling complexes, including INO80 (35, 60), NuA4, and SWR1 (8), and cohesins (51, 58) assemble on chromatin in a γ-H2A-dependent manner. But phosphorylation of H2A and nucleosome loss at the break seem to occur independently (57). After completion of DSB repair, γ-H2A(X) becomes dephosphorylated, which is necessary for efficient recovery from the DNA damage checkpoint (5, 22).
Downs et al. (9) showed that mutation of serine 129 in the SQ motif of H2A to alanine (hta1-S129A) results in sensitivity to methyl methanesulfonate (MMS) to a similar degree as when the last four C-terminal amino acids (SQEL) are missing. In contrast, if serine 129 of histone H2A is replaced with glutamic acid (hta1-S129E), yeast cells survive in the presence of MMS almost as well as the H2A (HTA1) wild-type strain, indicating that glutamic acid 129, at least in part, mimics constitutive phosphorylation. Since hta1-S129E showed enhanced nuclease sensitivity of chromatin and changes in supercoiling of a 2μm plasmid, it was suggested that the C-terminal motif of H2A has a role in higher-order chromatin structure and that phosphorylation of the SQ motif may modulate chromatin structure in the vicinity of a DSB.
Here we used the hta1-S129E, hta1-S129A, and hta1-S129*/hta2-S129* mutants to investigate the influence of the H2A C-terminal tail on chromatin organization and stability in detail. Minichromosomes and genomic loci were analyzed with respect to nucleosome positioning, nucleosome-nucleosome contacts, and nucleosome stability. The accessibility of DNA was tested by micrococcal nuclease (MNase) digestion of nuclei and, as an alternative in vivo approach, by DNA repair of UV lesions. We found that in hta1-S129E strains, the stability of the chromatin is not compromised and it is indistinguishable from the chromatin of HTA1 wild-type cells and the hta1-S129A mutant. The data support a role for S129 phosphorylation in the recruitment of repair proteins rather than in chromatin destabilization. Moreover, we show that deletion of the C-terminal tail has no obvious effect on nucleosome stability and positioning.
MATERIALS AND METHODS
Yeast strains and media.
The yeast strains used in this study are described in Table 1. The following S. cerevisiae strains were provided by J. A. Downs. FY406, JHY2, JHY3, and JHY8 are all isogenic to S288C, except that all are GAL2+ (18, 65). JDY22 (hta1-S129*/hta2-S129*) is in the W303α background (9). MFY1, MFY2, and MFY3 were constructed by disruption of TRP1 in JHY2, JHY3, and JHY8 (hta2-htb2Δ::trp1::URA3) by using an EcoRI-XhoI disruption fragment of pTU10 (7). MFY5, MFY6, MFY36, RHY1, and RHY2 were derived from MFY2, MFY3, MFY1, W303α, and JDY22, respectively, by transformation with minichromosome YRpFT35 [TRP1 ARS1 UmidA UmidB], which was obtained from pBT2UmidR-YRpFT35 as a 2,527-bp EcoRI fragment and circularized by ligation prior to transformation. MFY13, MFY14, MFY37, RHY5, and RHY6 were derived from JHY3, JHY8, JHY2, W303α, and JDY22, respectively, by transformation with minichromosome YRpFT68 [trp1::URA3 ARS1 UmidA UmidB], obtained from pFT54-YRpFT68 as a 3,693-bp EcoRI fragment and circularized by ligation (56). All transformations were verified by Southern blot hybridization. The following media were as previously described (46): YPD (1% Bacto yeast extract, 2% Bacto peptone, 2% dextrose), SD (0.67% yeast nitrogen base without amino acids, 2% dextrose), and SD minimal medium (SD supplemented with adenine, uracil, histidine, leucine, lysine, and tryptophan as required).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source (reference) |
|---|---|---|
| FY406 | MATahis3Δ200 leu2Δ1 lys2-128Δ trp1Δ63 ura3-52 (hta1-htb1)Δ::LEU2 (hta2-htb2)Δ::TRP1 YCpSAB6 [pRS316-HTA1-HTB1] | J. Downs (18) |
| JHY2 | Same as FY406, except YCpJH2 [pRS313-HTA1-HTB1] instead of YCpSAB6 | J. A. Downs |
| JHY3 | Same as FY406, except YCpJH3 [pRS313-hta1-S129A-HTB1] instead of YCpSAB6 | J. A. Downs |
| JHY8 | Same as FY406, except YCpJH8 [pRS313-hta1-S129E-HTB1] instead of YCpSAB6 | J. A. Downs |
| MFY1 | Same as JHY2, but (hta2-htb2)Δ::trp1::URA3 | This study |
| MFY2 | Same as JHY3, but (hta2-htb2)Δ::trp1::URA3 | This study |
| MFY3 | Same as JHY8, but (hta2-htb2)Δ::trp1::URA3 | This study |
| MFY5 | Same as MFY2, plus YRpFT35 [TRP1] | This study |
| MFY6 | Same as MFY3, plus YRpFT35 [TRP1] | This study |
| MFY36 | Same as MFY1, plus YRpFT35 [TRP1] | This study |
| MFY13 | Same as JHY3, plus YRpFT68 [URA3] | This study |
| MFY14 | Same as JHY8, plus YRpFT68 [URA3] | This study |
| MFY37 | Same as JHY2, plus YRpFT68 [URA3] | This study |
| W303α | MATα ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1-100 | J. A. Downs |
| JDY22 | Same as W303α, but hta1-S129* hta2-S129* | J. Downs (9) |
| RHY1 | Same as W303α, plus YRpFT35 [TRP1] | This study |
| RHY2 | Same as JDY22, plus YRpFT35 [TRP1] | This study |
| RHY5 | Same as W303α, plus YRpFT68 [URA3] | This study |
| RHY6 | Same as JDY22, plus YRpFT68 [URA3] | This study |
Chromatin analysis by MNase.
Yeast cells were grown at 30°C in 5 liters of SD minimal medium to an A600 of about 0.8 to 1.2, and crude nuclear extract was prepared essentially as described previously (27). Two-milliliter aliquots of the crude nuclear extract were digested with MNase (1 to 60 U/ml; Roche Diagnostics) at 37°C for 5 min. The reactions were terminated by addition of 3 ml of 2.5× buffer G2 (2 M guanidine HCl, 75 mM Tris [pH 8.0], 75 mM EDTA, 12.5% Tween 20, 1.25% Triton X-100, 200 mg/ml RNase A [Sigma], 300 mg/ml proteinase K [Roche Diagnostics]) and incubation for 2 h at 50°C. Genomic DNA was isolated with Genomic-tips 100/G (QIAGEN) and dissolved in 10 mM Tris (pH 8.0)-1 mM EDTA.
Mapping of MNase cutting sites by indirect end labeling.
DNA was cut with XbaI (YRpFT35 and YRpFT68), EcoRV (GAL1-10), or StuI (PHO3-5); electrophoresed on 1% agarose-TBE (89 mM Tris-borate, 2 mM EDTA [pH 8.3]) gels; transferred to Zeta-Probe GT membranes; and hybridized with 32P-labeled DNA probes. Probes were generated by random hexanucleotide-primed DNA synthesis with a HexaLabel DNA labeling kit (Fermentas), [α-32P]CTP (Amersham Biosciences), and short DNA templates as indicated in the figure legends. MNase digestion patterns were analyzed with a PhosphorImager screen (Amersham Biosciences) and ImageQuant software (Molecular Dynamics).
Plasmid DNA topology analysis.
The superhelical density of minichromosomes YRpFT35 and YRpFT68 and the 2μm plasmid was determined as previously described (11). Briefly, yeast cells were grown at 30°C in SD minimal medium to an A600 of about 1 to 1.5 or in YPD to an A600 of about 2.5 to 3.0 and then mixed with an equal volume of buffered ethanol-toluene (20 mM Tris [pH 8.0], 95% ethanol, 3% toluene) prechilled to −20°C; this was followed immediately by addition of 0.5 M EDTA to a final concentration of 10 mM (20). Spheroplasts were prepared by incubation with Zymolyase 100T (Seikagaku Kogyo Co., Ltd.) for 30 min at 30°C. The DNA was isolated with Genomic-tips 100/G (QIAGEN), electrophoresed in a 1% agarose gel (for YRpFT35 and YRpFT68) or a 0.75% agarose gel (for the 2μm plasmid) containing TBE (89 mM Tris [pH 8], 89 mM boric acid, 2 mM EDTA, 1 μg/ml chloroquine), and transferred to Zeta-Probe GT membranes. YRpFT35 and YRpFT68 topoisomers were probed with a 32P-labeled EcoRI-XbaI fragment of the TRP1 gene. 2μm plasmid topoisomers were probed with a 32P-labeled 339-bp SnabI-XbaI 2μm plasmid fragment containing the 2μm origin of replication. Topoisomers were visualized by PhosphorImager analysis.
Two-dimensional chloroquine gels were prepared as described by Kim and Clark (24). Two-dimensional gels were 0.8% agarose in 40 mM Tris-30 mM NaH2PO4-1 mM EDTA (pH 8.2). The first dimension was 10 μg/ml chloroquine (55 V, 15 h), and the second dimension was 20 μg/ml chloroquine (35 V, 15 h). The DNA was transferred to Zeta-Probe GT membranes, probed, and analyzed as described above.
UV irradiation and repair by photolyase and NER.
UV irradiation of yeast cultures and repair of UV lesions were done as described previously (53). Briefly, yeast cultures of MFY6 and MFY36 strains were grown in 6 liters of SD minimal medium at 30°C to a density of about 1 × 107 cells/ml and resuspended in SD to about 3 × 107 cells/ml. Suspensions were irradiated with UV light by use of Sylvania G15T8 germicidal lamps (predominantly 254 nm) at a dose of 150 J/m2 (measured by a UVX radiometer; UVP Inc., Upland, CA), and the medium was supplemented with the appropriate ingredients. For photoreactivation (PR) and NER, the cell suspension was exposed to photoreactivating light (Sylvania type F15 T8/BLB bulbs, peak emission at 375 nm) at ∼1.3 mW/cm2 (measured by a UVX radiometer with a 365-nm photocell) and 25 to 27°C for 30 to 120 min. For NER alone, aliquots were incubated at room temperature for 30 to 240 min. Samples were chilled on ice, and genomic DNA was isolated with Genomic-tips 100/G (QIAGEN) and dissolved in 10 mM Tris (pH 8.0)-1 mM EDTA. All steps until lysis of cells were done in yellow light (Sylvania GE Gold fluorescent light) to prevent undesired PR.
Cyclobutane pyrimidine dimer (CPD) analysis by indirect end labeling.
DNA was digested with XbaI (YRpFT35 and YRpFT68) or EcoRV (GAL1-10) and nicked at CPDs with T4 endonuclease V (Epicenter) in 50 mM Tris (pH 7.5)-5 mM EDTA or mock treated with the same buffer. The DNA was electrophoresed on 1.5% alkaline agarose gels, transferred to Zeta-Probe GT membranes, and hybridized with 32P-labeled strand-specific DNA probes. Strand-specific probes were generated by primer extension by using the DNA fragments as described in the figure legends, [α-32P]CTP (Amersham Biosciences), and Taq DNA polymerase (Fermentas) for 20 cycles. The membranes were analyzed with PhosphorImager screens (Amersham Biosciences), and CPDs were quantified with ImageQuant software (Molecular Dynamics). The CPD content (CPDs/top strand and CPDs/bottom strand) was calculated with the Poisson expression −ln(RFa/RFb), where RFa and RFb represent the signal intensity of the intact restriction fragment of the T4 endonuclease V and mock-treated DNA, respectively (32). CPD repair was expressed as a percentage with respect to the initial damage (0 min = 100% damage).
RESULTS
Mutations of phosphorylation site S129 of H2A do not affect nucleosome positioning and stability on minichromosomes.
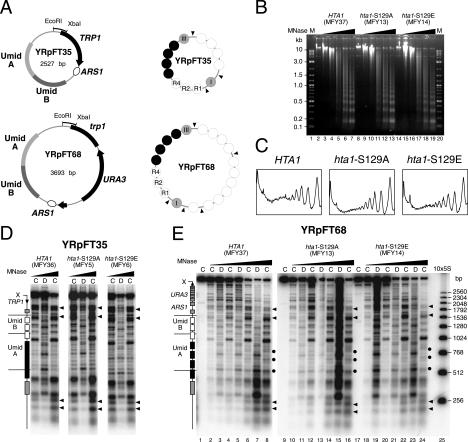
To test whether the mutations S129A and/or S129E of histone H2A affect the stability of nucleosomes, nucleosome positioning, and nucleosome-nucleosome contacts, we used two minichromosomes with well-characterized nucleosome positions (YRpFT35 and YRpFT68; Fig. 1A) (56). YRpFT35 shows nuclease-sensitive regions at the TRP1 promoter (EcoRI site) and at the origin of replication (ARS1), four nucleosomes on TRP1, and nine nucleosomes in a region between EcoRI and ARS1 that contains a tandem repeat of Umid sequences (UmidA and UmidB; Umid corresponds to 3.5 nucleosomes of the URA3 coding region). Four of these nucleosomes are in close contact, forming a nuclease-resistant “tetranucleosome.” In YRpFT68, which was generated by insertion of the URA3 gene into YRpFT35, most of the structural features of YRpFT35 were maintained, except that the linker DNA in the tetranucleosome became accessible to MNase. The different structures on UmidA and UmidB demonstrate that the DNA sequence does not determine nucleosome positioning in these regions (56). The presence of positioned nucleosomes, tightly packed nucleosomes, and nuclease-sensitive regions makes those minichromosomes suitable substrates to test nucleosome positioning, stability, and nucleosome-nucleosome contacts.
FIG. 1.
Chromatin structures of minichromosomes in hta1-S129 mutants. (A) Schematic representation of minichromosomes YRpFT35 and YRpFT68. YRpFT35 contains the TRP1 gene and the autonomously replicating ARS1 sequence. UmidA and UmidB are fragments of the URA3 gene inserted in the TRP1/ARS1 circle. YRpFT68 was generated by insertion of URA3 in TRP1 of YRpFT35. Indicated are the EcoRI-XbaI fragments used as probes for indirect end labeling and nucleosome positions (circles) determined by MNase digestion. Four nucleosomes are tightly packed in YRpFT35, forming a tetranucleosome (dark circles). In YRpFT68, the linker in the tetranucleosome is accessible (56). (B) Analysis of bulk chromatin by MNase digestion. Nuclei from HTA1 (MFY37), hta1-S129A (MFY13), and hta1-S129E (MFY14) cells were digested with increasing amounts of MNase (0, 1, 2, 5, 10, and 15 U/ml). Purified DNA was separated on a 1% agarose gel containing ethidium bromide. M is a 2-log DNA ladder (New England BioLabs). (C) Scans of the lanes corresponding to 15 U of MNase/ml shown in panel B. The values were normalized with respect to the band intensities of the mononucleosomes. (D) Nucleosome footprints on YRpFT35 obtained by MNase digestion. Chromatin (C) and DNA (D) isolated from yeast strains expressing wild-type HTA1 (MFY36) or mutant allele hta1-S129A (MFY5) or hta1-S129E (MFY6) were digested with MNase, and the DNA was purified, digested with XbaI, run on 1% agarose gels, blotted to Zeta-Probe GT membranes, and hybridized to radioactive probes (indicated in panel A). Wedges above of the lanes denote increasing MNase concentrations. Rectangles on the left of the blot mark the positions of nucleosomes in the sequences of UmidA and UmidB and flanking sequences of the minichromosome. The arrow denotes the direction of TRP1 transcription. X indicates the XbaI restriction site. Arrowheads indicate open, nonnucleosomal regions. (E) Nucleosome footprints on YRpFT68. Chromatin and DNA isolated from HTA1 (MFY37), hta1-S129A (MFY13), and hta1-S129E (MFY14) strains were analyzed as described for panel C. 10×5S is a molecular size marker indicating multiples of 256 bp (55). Dots indicate cleavage between nucleosomes at elevated levels of digestion.
The minichromosomes were transfected into yeast strains in which both genomic loci coding for histones H2A and H2B (HTA1-HTB1 and HTA2-HTB2) were disrupted and which carry a wild-type or a mutant allele on a centromeric plasmid (9, 18) (Table 1). Figure 1B shows chromatin analysis by MNase of several strains that contained wild-type HTA1 (MFY37) or mutant allele hta1-S129A (MFY13) or hta1-S129E (MFY14). Both the DNA patterns after MNase digestion of nuclei (nucleosomal repeats) and the digestion kinetics were indistinguishable in those strains. Scans of the lanes with clearly visible nucleosomal repeats (Fig. 1B, lanes 7, 13, and 19) confirmed that there was no obvious difference in global nucleosomal organization among HTA1, hta1-S129A, and hta1-S129E cells (Fig. 1C). Thus, the mutations in HTA1 did not affect the general stability and organization of bulk genomic chromatin.
Figure 1D and E reveal details of the chromatin structures of the minichromosomes with respect to nucleosome positioning and stability. Cleavage sites in chromatin are compared with those in naked DNA. Regions that are protected from cleavage in chromatin and encompass 140 to 160 bp are interpreted as positioned nucleosomes (55). Most clearly, the cutting patterns were similar in all of the strains, in both YRpFT35 (Fig. 1D) and YRpFT68 (Fig. 1E). Nucleosome footprints (boxes in Fig. 1D and E) are readily identified, as well as a long footprint characteristic of the tetranucleosome (dark box in UmidA) and the nuclease-sensitive regions around the EcoRI site and ARS1 (arrowheads). Thus, these structures were maintained in the hta1-S129E and hta1-S129A mutants, demonstrating that the S129 mutations affected the positions of neither spaced nucleosomes nor tightly packed nucleosomes. Moreover, the strong footprints indicate that nucleosomes were not destabilized. In addition, all nuclease-sensitive regions were maintained in the different genetic backgrounds, indicating that the mutations in the H2A C-terminal tail does not affect the ARS1 origin of replication and TRP1 promoter regions.
On the basis of our previous observations, we expected enhanced cleavage between nucleosomes in UmidA of YRpFT68. Instead, the four nucleosomes remained quite resistant and enhanced cutting was evident only at higher levels of digestion (Fig. 1E, dots). This is not an effect of the S129 mutants, since it is also observed in MFY37 containing a wild-type HTA1 allele. The variation in DNA accessibility in the tetranucleosome can be attributed to the use of different chromatin preparations in these studies (see Discussion).
Taken together, these data suggested that neither of the hta1 mutants destabilized nucleosomes in minichromosomes or in bulk chromatin but maintained the structural and dynamic characteristics of wild-type chromatin.
H2A-S129E maintains chromatin stability.
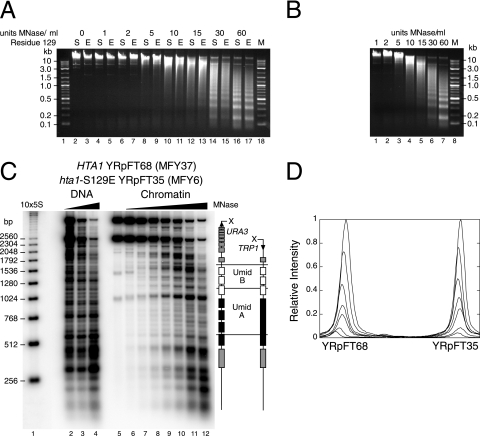
Since the kinetics of digestion with MNase are sensitive to variations in nuclear preparations, we performed codigestion experiments with hta1-S129E and HTA1 wild-type cells. We prepared in parallel nuclei from HTA1 cells containing YRpFT68 (MFY37) and nuclei from hta1-S129E cells containing YRpFT35 (MFY6) from cultures of equal cell densities. The nuclear suspensions were split into two equal fractions. One fraction of MFY37 nuclei was mixed with an equal fraction of MFY6 nuclei for codigestion with MNase. The remaining nuclear fractions were digested separately. Visualized by ethidium bromide staining, bulk chromatins from both HTA1 and hta1-S129E cells were similarly accessible to MNase and had indistinguishable nucleosomal repeat patterns (Fig. 2A). The codigestion of mixed nuclei also showed a similar kinetic, and the distinct bands reflecting the nucleosomal repeat manifested no difference in the nucleosomal repeat length of HTA1 and hta1-S129E chromatin (Fig. 2B). Indirect end labeling confirmed the arrangement of nucleosomes in YRpFT35 and YRpFT68 in the individual digests (data not shown; Fig. 1D and E). Probing the codigested samples with a TRP1 fragment detected both minichromosomes and allowed us to assess digestion kinetics quantitatively (Fig. 2C and D). The signals corresponding to full-length linearized YRpFT68 and YRpFT35 (top bands) decreased similarly with increasing amounts of MNase. From scanning of the top bands of the minichromosomes, it became evident that hta1-S129E chromatin (YRpFT35) was not more susceptible to degradation by MNase than HTA1 chromatin (YRpFT68) (Fig. 2D). Thus, both genomic chromatin and minichromosomes maintain stable nucleosomes and nucleosome positions in the hta1-S129E mutant.
FIG. 2.
Comparative MNase analyses of Hta1 and Hta1-S129E chromatin. Nuclei isolated from HTA1 cells containing minichromosome YRpFT68 (MFY37) and nuclei from hta1-S129E cells containing minichromosome YRpFT35 (MFY6) were separately digested or mixed and codigested with increasing amounts of MNase (0 to 60 U/ml). (A) Analysis of bulk chromatin of separately digested nuclei. Purified genomic DNA was analyzed on a 1% agarose gel containing ethidium bromide. S (serine) or E (glutamic acid) indicates the residue at position 129 of histone H2A that was present in the digested nuclei. M (lanes 1 and 18) is a 2-log DNA ladder (New England BioLabs). (B) Equal amounts of nuclei of both strains were mixed and codigested. DNA was analyzed as described for panel A. (C) Nucleosome footprints of codigested YRpFT35 and YRpFT68 were done as described in the legend to Fig. 1. 10×5S is a molecular size marker indicating multiples of 256 bp. (D) PhosphorImager scans of the top two bands in lanes 6 to 12 of the blot shown in panel C. The values were normalized with respect to the band intensities of lane 6.
Deletion of the C-terminal SQEL motif of H2A does not destabilize chromatin.
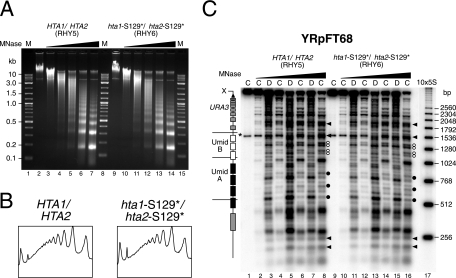
The C-terminal SQEL motif of H2A might stabilize nucleosomes or higher-order structures, irrespective of the presence or absence of a negative charge at position 129. We therefore examined the chromatin structures of YRpFT35 and YRpFT68 in strains RHY2 and RHY6, respectively, in which serine 129 of both genomic copies of H2A was mutated to a stop codon (hta1-S129*/hta2-S129*) (9). Chromatin from RHY6 showed an accessibility to MNase similar to that of chromatin isolated from an isogenic wild-type strain (RHY5) (Fig. 3A and B). The hta1-S129*/hta2-S129* mutant maintained the nucleosome positions in YRpFT68 (Fig. 3C) and YRpFT35 (not shown), demonstrating that the loss of the last four C-terminal amino acids of H2A does not destabilize chromatin.
FIG. 3.
Comparative MNase analyses of Hta1/Hta2 and hta1-S129*/hta2-S129* chromatin. Nuclei isolated from W303α (RHY5) and hta1-S129*/hta2-S129* (RHY6) cells containing minichromosome YRpFT68 were digested with increasing amounts of MNase (0, 5, 10, 15, 30, and 60 U/ml). (A) Analysis of bulk chromatin of digested nuclei. Purified genomic DNA was analyzed on a 1% agarose gel containing ethidium bromide. M (lanes 1, 8, and 15) is a 2-log DNA ladder (New England BioLabs). (B) Scans of the lanes corresponding to 30 U of MNase/ml shown in panel A. The values were normalized with respect to the maximal band intensities (dinucleosomes). (C) Nucleosome footprints on YRpFT68. The DNA shown in panel A was analyzed as described in the legend to Fig. 1. Solid dots indicate cleavage between nucleosomes on YRpFT68 at elevated levels of digestion. White dots represent doublets possibly originating from alternative nucleosome positions. In the W303 genetic background, the EcoRI-XbaI fragment cross-hybridizes with the genomic TRP1 locus (asterisk). 10×5S is a molecular size marker indicating multiples of 256 bp.
However, we noticed subtle differences between different sets of strains in the area of nucleosomes R1, R2, and R4 of YRpFT68, irrespective of the C-terminal deletions in H2A. RHY5 and RHY6 (Fig. 3C, white dots) showed two double bands of similar intensities. In MFY13, MFY14, and MFY37, the upper bands of those doublets were more pronounced (Fig. 1 and 2). These differences in nucleosome positions R1, R2, and R4 may be related to altered histone expression in different genetic backgrounds (see Discussion).
The H2A-S129E mutation maintains nucleosome positions in chromosomal loci.
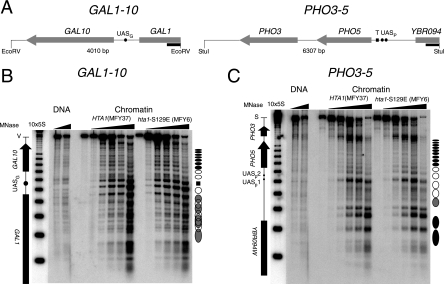
To test whether maintaining nucleosome positioning in hta1-S129E mutant cells is a unique property of minichromosomes, chromatin structures of the chromosomal GAL1-10 and PHO3-5 loci were analyzed (Fig. 4). Under repressing conditions, intergenic regions between YBR094W and PHO5, between PHO5 and PHO3, and between GAL1 and GAL10 contain the histone variant H2AZ (Htz1) (40) but hemagglutinin-tagged Hta1 was relatively uniformly distributed across these loci (45). Cells were grown under conditions in which GAL1, GAL10, and PHO5 were not transcribed. Nucleosome footprinting by MNase revealed the characteristic chromatin structure in the GAL1-10 region (3), namely, positioned nucleosomes in GAL10, three nucleosomes in the promoter region flanking UASG, and less precisely and partially overlapping nucleosome positions in GAL1 (Fig. 4B). Similarly, the PHO3-5 locus showed the characteristic chromatin structure, in particular, the positioned nucleosomes in the PHO5 gene and the PHO5 promoter (Fig. 4C) (1). The chromatin structures in both loci were indistinguishable in HTA1 and hta1-S129E cells.
FIG. 4.
Nucleosome positions in the repressed genomic loci GAL1-10 and PHO3-5 revealed by MNase footprinting. (A) Maps of the genomic loci GAL1-10 and PHO3-5 on chromosome II. Shown are the EcoRV (GAL1-10) and StuI (PHO3-5) restriction fragments with the GAL1, GAL10, PHO3, PHO5, and YBR094W coding regions, respectively; the upstream activating sequences (UASG, UASp1, and UASp2); and the TATA box (T) in the PHO5 promoter. Black rectangles indicate the probes used for Southern blotting. (B) Nucleosome footprints on GAL1-10. DNA (shown in Fig. 2A) was digested with EcoRV (V) and separated on a 1% agarose gel. The Southern blot probed with a fragment of GAL1 shows positioned nucleosomes (circles) and delocalized nucleosomes (overlapping circles). The black rectangle indicates the Gal4 binding site on UASG. 10×5S is a molecular size marker indicating multiples of 256 bp. (C) Nucleosome footprints on PHO3-5. DNA (shown in Fig. 2A) was digested with StuI (S) and separated on a 1% agarose gel. The Southern blot probed with a fragment of YBR094W shows the nucleosomal arrangement in the PHO3-5 locus as in panel B.
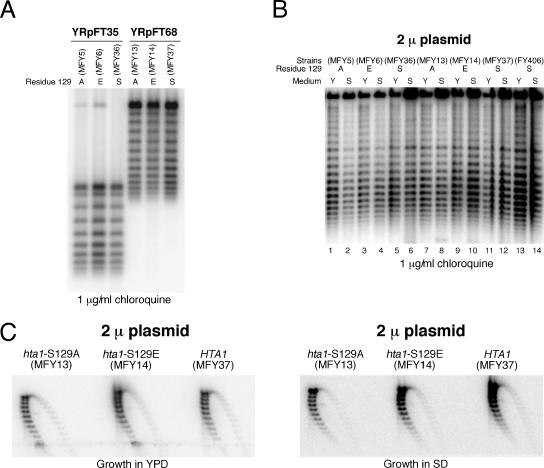
H2A-S129 mutations have no effect on the superhelical density of minichromosomes and the 2μm plasmid.
Studying the superhelical density of circular minichromosomes can be used to characterize chromatin in vivo. Previous studies have reported that histone depletion (23) and histone mutations (including hta1-S129E) (9, 31, 63) affect supercoiling of the yeast 2μm plasmid. Since the MNase assay did not detect substantial chromatin changes in the hta1 mutants, we first tested the supercoiling of minichromosomes YRpFT35 and YRpFT68. Strains with wild-type and mutant histone H2A were grown in SD minimal medium, and DNA was purified in parallel under conditions that prevent changes in plasmid DNA topology during cell lysis (20). Plasmid supercoils were resolved in chloroquine-agarose gels and analyzed by Southern blotting (Fig. 5A). The topoisomer distribution was indistinguishable in strains containing wild-type HTA1 or the mutated allele (hta1-S129A or hta1-S129E). Thus, the mutations did not obviously affect the average number of nucleosomes or their stability, which is consistent with the MNase digestion results. Since the hta1-S129E mutation was reported to change supercoiling in the 2μm plasmid (9) and since the culture medium might influence transcription and thereby supercoiling, we analyzed 2μm DNA of cells grown in YPD and SD minimal medium. No obvious variation in supercoiling of the 2μm plasmid was found in the different strains in either SD minimal medium or YPD (Fig. 5B). Moreover, similar supercoil distributions in 2μm plasmids were observed by using two-dimensional gels (Fig. 5C). Thus, the supercoiling assays confirm that neither hta1-S129A nor hta1-S129E destabilized nucleosomes.
FIG. 5.
Superhelical densities of the minichromosomes isolated from strains expressing wild-type (HTA1) or mutated histone H2A (hta1-S129A and hta1-S129E). (A) Topoisomer distribution of YRpFT35 and YRpFT68. DNAs isolated from the strains indicated were electrophoresed on 1% agarose gels containing 1 μg/ml chloroquine and transferred to a Zeta-Probe GT membrane. YRpFT35 and YRpFT68 superhelical density was analyzed by hybridization with a radiolabeled EcoRI-XbaI fragment from the TRP1 gene. A (alanine), E (glutamic acid), or S (serine) indicates the residue at position 129 of histone H2A. (B) Topoisomer distribution of the 2μm plasmid. The yeast strains indicated were grown in either YPD (Y) or SD minimal medium (S). DNAs isolated from these strains were separated on a 0.75% agarose gel containing 1 μg/ml chloroquine and transferred to a Zeta-Probe GT membrane. The superhelical density of the 2μm plasmid was analyzed by hybridization with a SnaBI-XbaI probe of the 2μm origin of replication. (C) Topoisomer distribution of the 2μm plasmid in two dimensions. The same DNA as shown in panel B was separated in 0.8% agarose gels containing 10 and 20 μg/ml chloroquine in the first and second dimensions, respectively. The 2μm plasmid was detected as described for panel B.
Repair of UV lesions is not affected by H2A-S129E.
As an alternative way to study chromatin structure and accessibility of DNA to proteins in living cells, we measured repair of UV lesions by photolyase and NER. CPDs and pyrimidine(6-4)pyrimidone photoproducts (6-4PPs) are two major classes of DNA lesions generated by UV light (13). Both CPDs and 6-4PPs are repaired by NER, a multistep pathway that includes damage recognition, excision of an oligonucleotide containing the DNA lesion, DNA repair synthesis, and chromatin remodeling to reestablish chromatin structures (15, 54). Alternatively, many organisms, including yeast, have a CPD-photolyase that selectively binds CPDs and reverses the damage in a light-dependent reaction (PR) (12). NER and PR are modulated in chromatin by protein-DNA interactions, positioned nucleosomes, and heterochromatin (54).
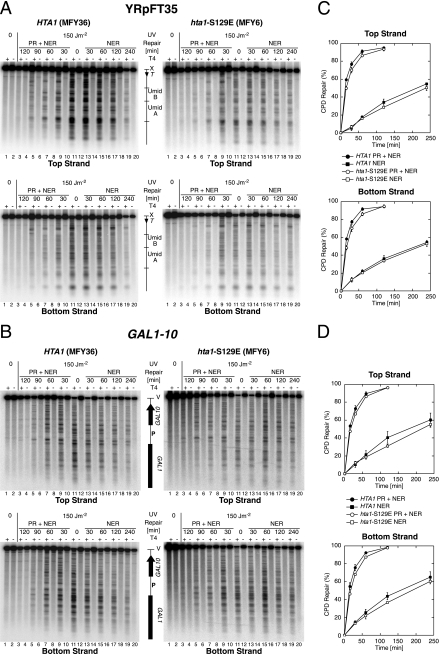
Budding yeast hta1-S129A mutants are sensitive to MMS and camptothecin but insensitive to UV (9, 33, 41), indicating that H2A-S129 phosphorylation is not important for the repair of UV lesions. We confirmed that none of the hta1-S129 mutant strains was hypersensitive to UV irradiation, but we noticed a very weak UV sensitivity of hta1-S129A and hta1-S129E cells, compared to HTA1 cells, at the relatively high dose used for repair experiments (150 J/m2) (data not shown). DNA repair of UV lesions by photolyase (PR) and NER was investigated in MFY36 (HTA1, YRpFT35) and MFY6 (hta1-S129E, YRpFT35). Cells were UV irradiated with 150 J/m2 to generate about 0.3 CPD/kb and either exposed to light for PR or incubated in the dark for NER. To analyze the distribution of CPDs in YRpFT35 and GAL1-10, the DNA was purified, cut at CPDs with T4 endonuclease V, and digested with XbaI or EcoRV, respectively. The DNA fragments were separated on alkaline gels, transferred to nylon membranes, and hybridized to short, strand-specific probes.
DNA of nonirradiated cells showed the intact restriction fragment (top band), irrespective of T4 endonuclease V treatment (Fig. 6A and B, lanes 1 and 2). DNA of irradiated cells showed the intact restriction fragment in the mock-treated lanes (lanes 12) but a specific band pattern and a weaker top band when cut at CPDs with T4 endonuclease V (lanes 11). The bands represent the yields and distribution of CPDs along the DNA fragment. The top and bottom strands revealed different patterns, demonstrating strand specificity. The CPD patterns disappeared when cells were exposed to photoreactivating light (PR + NER, lanes 3 to 9) or incubated in the dark (NER, lanes 13 to 19). Repair of CPDs was quantified as a time-dependent increase in the intact restriction fragment (Fig. 6C and D). The CPDs were slowly removed by NER alone (about 50 to 70% in 4 h for YRpFT35 and GAL1-10). However, CPDs were efficiently removed by PR and NER on GAL1-10 and YRpFT35 (>80% in one h), indicating that under those conditions photolyase was the predominant pathway used to repair CPDs. Most clearly, there was no dramatic difference between the repair curves of MFY36 (HTA1) and MFY6 (hta1-S129E). Thus, the accessibility and repair of UV lesions are largely independent of the hta1-S129E mutation, which is consistent with the results of MNase digestion and supercoiling. We noticed, however, that CPDs in the hta1-S129E strain seemed to be repaired more slowly, which is consistent with the subtle UV sensitivity of the hta1-S129E mutant. However, whether this very subtle effect reflects a less accessible, and hence a more stable, chromatin structure in the context of repair or a lower repair capacity in the mutant remains speculative.
FIG. 6.
Repair of CPDs by photolyase and NER. HTA1 (MFY36) and hta1-S129E (MFY6) cells containing minichromosome YRpFT35 were irradiated with UV light at 150 J/m2 (lanes 3 to 20), exposed to photoreactivating light (PR + NER) for 30 to 120 min (lanes 3 to 10), or kept in the dark for 0 to 240 min for NER (lanes 11 to 20). The DNA was purified and cut at CPDs with T4 endonuclease V (T4). (A) To analyze CPD repair on minichromosome YRpFT35, the DNA was digested with XbaI (X), separated on 1.5% alkaline agarose gels, transferred to Zeta-Probe GT membranes, and hybridized with strand-specific probes generated by primer extension with the RIX DNA fragment as the template (Fig. 1). PhosphorImager screens are shown for the top and bottom strands. (B) To analyze CPD repair at the GAL1-10 locus, the DNA was digested with EcoRV (V) and further processed as described for panel A. The GAL1-10 locus was hybridized with strand-specific probes generated by primer extension with the GAL1-SS1 DNA fragment as the template (Fig. 3). (C and D) Quantitative analysis of CPD repair on GAL1-10 and YRpFT35. The data are means with standard deviations from three independent experiments.
DISCUSSION
Driven by the hypothesis that phosphorylation of yeast H2A might disturb chromatin structures to facilitate repair of strand breaks, we tested chromatin structures in an hta1-S129E mutant mimicking phosphorylation and the corresponding wild-type strain (HTA1). In contrast to a previous observation (9), we did not observe a difference between these two strains with respect to nuclease digestion or in supercoiling assays. In addition, we show that the hta1-S129E mutation affects neither the stability nor the positioning of nucleosomes. Finally, repair of UV lesions, as an alternative way to assess chromatin accessibility in vivo, revealed similar repair rates in both strains. We cannot explain the different observations made by us and by Downs et al. (9); however, we strongly emphasize that the discrepancy in chromatin accessibility does not affect the major conclusions about DNA repair drawn by Downs et al.
We can only speculate that the different observations reflect subtle variations in the nuclease digestion and supercoiling assays. Indeed, since supercoiling depends not only on the number and stability of nucleosomes but also on the transcriptional activity, temperature, and topoisomerase activity during isolation, variations in growth or preparation conditions might impact supercoiling (e.g., see references 14, 23, 44, and 49). We therefore investigated two different artificial minichromosomes with one active gene each (TRP1 and URA3, respectively) and the natural 2μm plasmid, and we used YPD and SD minimal medium. Moreover, we checked the strains with respect to UV sensitivity and MMS sensitivity and verified the hta1 mutations in our strains by sequencing (data not shown). Thus, we are confident that under our conditions, there is no substantial difference in supercoiling in the different histone mutants. It should, however, be mentioned that other histone mutants were reported to impair the ability of nucleosomes to constrain supercoils in vivo, i.e., a histone H3 residue (hht1-K56Q) that makes water-mediated DNA contacts at the entry-and-exit site of the nucleosome core (31) and Sin mutations in histone H4 that define a discrete domain on the nucleosome surface (63).
There is also variability in chromatin analysis by MNase digestion. First, it must be considered that MNase cuts DNA and RNA (2). Hence, variations in the RNA content of nuclear preparations might affect digestion kinetics. Second, disruption of either histone locus HTA1-HTB1 or HTA2-HTB2 was shown to result in different phenotypes (6, 37), including alterations of chromatin structure (36). (hta1-htb1)Δ strains can compensate histone levels by amplification of the HTA2-HTB2 gene pair (26), whereas (hta2-htb2)Δ strains increase transcription of HTA1-HTB1 (34, 37). Thus, it is conceivable that, depending on the genetic background, the strains might have different histone levels, which could affect chromatin structure. Third, MNase digestions might be compromised by variations in nuclear and chromatin preparations that affect the composition and stability of chromatin. Different chromatin preparations (e.g., partial purification of minichromosomes versus crude nuclear preparations) might explain the different extents of linker cleavage in the tetranucleosome region of YRpFT68. Here we compared chromatin results by using the same chromatin preparation technique and found no major differences between hta mutants and wild-type strains in the same genetic background. Interestingly, we observed subtle differences in nucleosome positions R1, R2, and R4 of YRpFT68, depending on the genetic background of the strains. Strains expressing H2A and H2B from endogenous alleles (RHY5 and RHY6) showed two doublets of similar intensities (Fig. 3C, white dots). However, in strains which express the HTA1-HTB1 allele from a centromeric plasmid and lack the HTA2-HTB2 allele, the upper bands of those doublets were more pronounced (Fig. 1 and 2). It is possible that the differences in nucleosome positions R1, R2, and R4 are related to the different genetic background expressing all histone loci and of strains expressing only one subtype or mutation from a centromeric plasmid.
Contacts of the C-terminal tail of H2A with the N-terminal tail of H2A of a neighboring particle in crystallized yeast nucleosome core particles (64) and cross-linking of the C-terminal tail to linker DNA (59) indicated that this tail might affect both the stability of nucleosomes and potential interactions with flanking nucleosomes and linker DNA in higher-order structures. Phosphorylation at H2A-S129, as it occurs in DNA repair, as well as the hta1-S129E mutation, alters the charge of an amino acid residue and thereby might mediate changes in chromatin by disruption of histone-DNA contacts or nucleosome-nucleosome interactions. However, our experiments with yeast do not support that hypothesis. Nucleosomes remained stable with respect to nuclease sensitivity and positioning in all of the regions analyzed. Moreover, no effect was obvious in the region of tightly packed nucleosomes. In addition, we show that PR and NER, two different DNA repair pathways which are known to be modulated by chromatin structures (54), did not reveal major effects of the hta1-S129E mutant on chromatin structure and stability. Thus, we take this as a strong indication that a negative charge at S129 of the C-terminal tail has no direct role in the organization and stability of nucleosomes and nucleosome-nucleosome contacts in yeast.
γ-H2AX might act in DSB repair by disruption of chromatin and/or by recruitment of chromatin remodeling activities and repair components. Tsukuda et al. (57) showed that no histone loss occurred in the first 30 min after induction of a DSB, although γ-H2A accumulated rapidly and extensively on either side of the break. Furthermore, an H2A mutant lacking the SQ motif lost histones at the break to the same extent as a wild-type strain. While those experiments do not address the stability of nucleosomes, they indicate that nucleosome displacement at the break depends on the recruitment of remodeling factors. These data are consistent with our observation that the negative charge at H2A-S129E does not destabilize chromatin structure.
Studies with mammalian cells revealed that the presence of H2AX is not a prerequisite for chromatin decondensation around a DSB. Rather, decondensation depends on ATP, indicating the action of remodeling factors near the break, also in the absence of H2AX (25). Phosphorylated H2AX appears to regulate cellular responses to DSBs by binding effector proteins, as shown for MDC1 (52). In yeast, a specific phosphorylated H2A protein interacts with Nhp10, a subunit of the INO80 remodeling complex (35) and the NuA4 subunit Arp4 binds phospho-H2A peptides (8, 35). Thus, phosphorylation of H2A(X) may act as a binding interface for chromatin-associated proteins involved in DSB repair rather than directly alter the level of chromatin compaction. In support of this suggestion, recent data show that loss of MDC1-γ-H2AX interaction or ablation of H2AX produces the same phenotype in mammalian cells and in mice as the H2AX deletion (28, 52), thus supporting the idea that, in mammals, the main function of γ-H2AX is to interact with MDC1.
Acknowledgments
We thank U. Suter for continuous support, J. Downs and S. Jackson for strains and discussions, D. Clark for help with the supercoiling assays, and A. Bucceri, C. Capiaghi, R. Hahn, K. Kapitza, M. Lopes, and J. Sogo for comments.
This research was supported by grant 3100AO-102152 from the Swiss National Science Foundation and ETH research grant TH41/02-3 (to F.T.).
Footnotes
Published ahead of print on 12 March 2007.
REFERENCES
- 1.Almer, A., and W. Horz. 1986. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 5:2681-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anfinsen, C., P. Cuatrecasas, and H. Taniuchi. 1971. Staphylococcal nuclease, chemical properties and catalysis, p. 177-204. In P. D. Boyer (ed.), The enzymes, vol. 4, 3rd ed. Academic Press, Inc., New York, NY. [Google Scholar]
- 3.Cavalli, G., and F. Thoma. 1993. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J. 12:4603-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Celeste, A., O. Fernandez-Capetillo, M. J. Kruhlak, D. R. Pilch, D. W. Staudt, A. Lee, R. F. Bonner, W. M. Bonner, and A. Nussenzweig. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675-679. [DOI] [PubMed] [Google Scholar]
- 5.Chowdhury, D., M. C. Keogh, H. Ishii, C. L. Peterson, S. Buratowski, and J. Lieberman. 2005. γ-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell 20:801-809. [DOI] [PubMed] [Google Scholar]
- 6.Clark-Adams, C. D., D. Norris, M. A. Osley, J. S. Fassler, and F. Winston. 1988. Changes in histone gene dosage alter transcription in yeast. Genes Dev. 2:150-159. [DOI] [PubMed] [Google Scholar]
- 7.Cross, F. R. 1997. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast 13:647-653. [DOI] [PubMed] [Google Scholar]
- 8.Downs, J. A., S. Allard, O. Jobin-Robitaille, A. Javaheri, A. Auger, N. Bouchard, S. J. Kron, S. P. Jackson, and J. Cote. 2004. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16:979-990. [DOI] [PubMed] [Google Scholar]
- 9.Downs, J. A., N. F. Lowndes, and S. P. Jackson. 2000. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408:1001-1004. [DOI] [PubMed] [Google Scholar]
- 10.Felsenfeld, G., and M. Groudine. 2003. Controlling the double helix. Nature 421:448-453. [DOI] [PubMed] [Google Scholar]
- 11.Fertala, J., J. R. Vance, P. Pourquier, Y. Pommier, and M. A. Bjornsti. 2000. Substitutions of Asn-726 in the active site of yeast DNA topoisomerase I define novel mechanisms of stabilizing the covalent enzyme-DNA intermediate. J. Biol. Chem. 275:15246-15253. [DOI] [PubMed] [Google Scholar]
- 12.Friedberg, E. C. 2003. DNA damage and repair. Nature 421:436-440. [DOI] [PubMed] [Google Scholar]
- 13.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, DC.
- 14.Giaever, G. N., and J. C. Wang. 1988. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell 55:849-856. [DOI] [PubMed] [Google Scholar]
- 15.Gong, F., Y. Kwon, and M. J. Smerdon. 2005. Nucleotide excision repair in chromatin and the right of entry. DNA Repair 4:884-896. [DOI] [PubMed] [Google Scholar]
- 16.Gontijo, A. M., C. M. Green, and G. Almouzni. 2003. Repairing DNA damage in chromatin. Biochimie 85:1133-1147. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, J. C., C. Tse, and A. P. Wolffe. 1998. Structure and function of the core histone N-termini: more than meets the eye. Biochemistry 37:17637-17641. [DOI] [PubMed] [Google Scholar]
- 18.Hirschhorn, J. N., A. L. Bortvin, S. L. Ricupero-Hovasse, and F. Winston. 1995. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol. 15:1999-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 20.Johnston, L. H., and D. H. Williamson. 1978. An alkaline sucrose gradient analysis of the mechanism of nuclear DNA synthesis in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 164:217-225. [DOI] [PubMed] [Google Scholar]
- 21.Kapetanaki, M. G., J. Guerrero-Santoro, D. C. Bisi, C. L. Hsieh, V. Rapic-Otrin, and A. S. Levine. 2006. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. USA 103:2588-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keogh, M. C., J. A. Kim, M. Downey, J. Fillingham, D. Chowdhury, J. C. Harrison, M. Onishi, N. Datta, S. Galicia, A. Emili, J. Lieberman, X. Shen, S. Buratowski, J. E. Haber, D. Durocher, J. F. Greenblatt, and N. J. Krogan. 2006. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature 439:497-501. [DOI] [PubMed] [Google Scholar]
- 23.Kim, U. J., M. Han, P. Kayne, and M. Grunstein. 1988. Effects of histone H4 depletion on the cell cycle and transcription of Saccharomyces cerevisiae. EMBO J. 7:2211-2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, Y., and D. J. Clark. 2002. SWI/SNF-dependent long-range remodeling of yeast HIS3 chromatin. Proc. Natl. Acad. Sci. USA 99:15381-15386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruhlak, M. J., A. Celeste, G. Dellaire, O. Fernandez-Capetillo, W. G. Muller, J. G. McNally, D. P. Bazett-Jones, and A. Nussenzweig. 2006. Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J. Cell Biol. 172:823-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Libuda, D. E., and F. Winston. 2006. Amplification of histone genes by circular chromosome formation in Saccharomyces cerevisiae. Nature 443:1003-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livingstone-Zatchej, M., and F. Thoma. 1999. Mapping of nucleosome positions in yeast. Methods Mol. Biol. 119:363-378. [DOI] [PubMed] [Google Scholar]
- 28.Lou, Z., K. Minter-Dykhouse, S. Franco, M. Gostissa, M. A. Rivera, A. Celeste, J. P. Manis, J. van Deursen, A. Nussenzweig, T. T. Paull, F. W. Alt, and J. Chen. 2006. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 21:187-200. [DOI] [PubMed] [Google Scholar]
- 29.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 30.Luger, K., and T. J. Richmond. 1998. The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 8:140-146. [DOI] [PubMed] [Google Scholar]
- 31.Masumoto, H., D. Hawke, R. Kobayashi, and A. Verreault. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436:294-298. [DOI] [PubMed] [Google Scholar]
- 32.Mellon, I., G. Spivak, and P. C. Hanawalt. 1987. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell 51:241-249. [DOI] [PubMed] [Google Scholar]
- 33.Moore, J. D., O. Yazgan, Y. Ataian, and J. E. Krebs. 8 October 2006. Diverse roles for histone H2A modifications in DNA damage response pathways in yeast. Genetics [Epub ahead of print.] 10.1534/genetics.106.063792. [DOI] [PMC free article] [PubMed]
- 34.Moran, L., D. Norris, and M. A. Osley. 1990. A yeast H2A-H2B promoter can be regulated by changes in histone gene copy number. Genes Dev. 4:752-763. [DOI] [PubMed] [Google Scholar]
- 35.Morrison, A. J., J. Highland, N. J. Krogan, A. Arbel-Eden, J. F. Greenblatt, J. E. Haber, and X. Shen. 2004. INO80 and γ-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell 119:767-775. [DOI] [PubMed] [Google Scholar]
- 36.Norris, D., B. Dunn, and M. A. Osley. 1988. The effect of histone gene deletions on chromatin structure in Saccharomyces cerevisiae. Science 242:759-761. [DOI] [PubMed] [Google Scholar]
- 37.Norris, D., and M. A. Osley. 1987. The two gene pairs encoding H2A and H2B play different roles in the Saccharomyces cerevisiae life cycle. Mol. Cell. Biol. 7:3473-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 39.Peterson, C. L., and M. A. Laniel. 2004. Histones and histone modifications. Curr. Biol. 14:R546-R551. [DOI] [PubMed] [Google Scholar]
- 40.Raisner, R. M., and H. D. Madhani. 2006. Patterning chromatin: form and function for H2A.Z variant nucleosomes. Curr. Opin. Genet. Dev. 16:119-124. [DOI] [PubMed] [Google Scholar]
- 41.Redon, C., D. R. Pilch, E. P. Rogakou, A. H. Orr, N. F. Lowndes, and W. M. Bonner. 2003. Yeast histone 2A serine 129 is essential for the efficient repair of checkpoint-blind DNA damage. EMBO Rep. 4:678-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 44.Saavedra, R. A., and J. A. Huberman. 1986. Both DNA topoisomerases I and II relax 2 micron plasmid DNA in living yeast cells. Cell 45:65-70. [DOI] [PubMed] [Google Scholar]
- 45.Santisteban, M. S., T. Kalashnikova, and M. M. Smith. 2000. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103:411-422. [DOI] [PubMed] [Google Scholar]
- 46.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 47.Shogren-Knaak, M., H. Ishii, J. M. Sun, M. J. Pazin, J. R. Davie, and C. L. Peterson. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311:844-847. [DOI] [PubMed] [Google Scholar]
- 48.Shroff, R., A. Arbel-Eden, D. Pilch, G. Ira, W. M. Bonner, J. H. Petrini, J. E. Haber, and M. Lichten. 2004. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 14:1703-1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simpson, R. T., F. Thoma, and J. M. Brubaker. 1985. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell 42:799-808. [DOI] [PubMed] [Google Scholar]
- 50.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 51.Ström, L., H. B. Lindroos, K. Shirahige, and C. Sjogren. 2004. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol. Cell 16:1003-1015. [DOI] [PubMed] [Google Scholar]
- 52.Stucki, M., J. A. Clapperton, D. Mohammad, M. B. Yaffe, S. J. Smerdon, and S. P. Jackson. 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123:1213-1226. [DOI] [PubMed] [Google Scholar]
- 53.Suter, B., M. Livingstone-Zatchej, and F. Thoma. 1999. Mapping cyclobutane-pyrimidine dimers in DNA and using DNA-repair by photolyase for chromatin analysis in yeast. Methods Enzymol. 304:447-461. [DOI] [PubMed] [Google Scholar]
- 54.Thoma, F. 2005. Repair of UV lesions in nucleosomes—intrinsic properties and remodeling. DNA Repair 4:855-869. [DOI] [PubMed] [Google Scholar]
- 55.Thoma, F., L. W. Bergman, and R. T. Simpson. 1984. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J. Mol. Biol. 177:715-733. [DOI] [PubMed] [Google Scholar]
- 56.Thoma, F., and M. Zatchej. 1988. Chromatin folding modulates nucleosome positioning in yeast minichromosomes. Cell 55:945-953. [DOI] [PubMed] [Google Scholar]
- 57.Tsukuda, T., A. B. Fleming, J. A. Nickoloff, and M. A. Osley. 2005. Chromatin remodelling at a DNA double-strand break site in Saccharomyces cerevisiae. Nature 438:379-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Unal, E., A. Arbel-Eden, U. Sattler, R. Shroff, M. Lichten, J. E. Haber, and D. Koshland. 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16:991-1002. [DOI] [PubMed] [Google Scholar]
- 59.Usachenko, S. I., S. G. Bavykin, I. M. Gavin, and E. M. Bradbury. 1994. Rearrangement of the histone H2A C-terminal domain in the nucleosome. Proc. Natl. Acad. Sci. USA 91:6845-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Attikum, H., O. Fritsch, B. Hohn, and S. M. Gasser. 2004. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119:777-788. [DOI] [PubMed] [Google Scholar]
- 61.van Attikum, H., and S. M. Gasser. 2005. The histone code at DNA breaks: a guide to repair? Nat. Rev. Mol. Cell Biol. 6:757-765. [DOI] [PubMed] [Google Scholar]
- 62.Varga-Weisz, P. D., and P. B. Becker. 2006. Regulation of higher-order chromatin structures by nucleosome-remodelling factors. Curr. Opin. Genet. Dev. 16:151-156. [DOI] [PubMed] [Google Scholar]
- 63.Wechser, M. A., M. P. Kladde, J. A. Alfieri, and C. L. Peterson. 1997. Effects of Sin− versions of histone H4 on yeast chromatin structure and function. EMBO J. 16:2086-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White, C. L., R. K. Suto, and K. Luger. 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20:5207-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]
- 66.Wolffe, A. P., and J. J. Hayes. 1999. Chromatin disruption and modification. Nucleic Acids Res. 27:711-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu, Y., Y. Teng, H. Liu, S. H. Reed, and R. Waters. 2005. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc. Natl. Acad. Sci. USA 102:8650-8655. [DOI] [PMC free article] [PubMed] [Google Scholar]