Abstract
As a multifunctional protein, Yin Yang 1 (YY1) has been demonstrated to regulate both gene expression and protein posttranslational modifications. However, gaps still exist in our knowledge of how YY1 can be modified and what the consequences of its modifications are. Here we report that YY1 protein can be sumoylated both in vivo and in vitro. We have identified lysine 288 as the major sumoylation site of YY1. We also discovered that PIASy, a SUMO E3 ligase, is a novel YY1-interacting protein and can stimulate the sumoylation of YY1 both in vitro and in vivo. Importantly, the effects of PIASy mutants on in vivo YY1 sumoylation correlate with the YY1-PIASy interaction but do not depend on the RING finger domain of PIASy. This regulation is unique to YY1 sumoylation because PIASy-mediated p53 sumoylation still relies on the integrity of PIASy, which is also true of all of the previously identified substrates of PIASy. In addition, PIASy colocalizes with YY1 in the nucleus, stabilizes YY1 in vivo, and differentially regulates YY1 transcriptional activity on different target promoters. This study demonstrates that YY1 is a target of SUMOs and reveals a novel feature of a SUMO E3 ligase in the PIAS family that selectively stimulates protein sumoylation independent of the RING finger domain.
Transcriptional control and posttranslational modifications are two major types of epigenetic regulation in mammalian cells. The dysregulation of either part may lead to severe physiological disorders in mammals. For instance, the repression of tumor suppressor genes and stabilization of oncogene products play important roles in tumorigenesis (35). Studies of the posttranslational modifications of essential regulatory proteins will likely shed light on the mechanisms that determine the cell fate. In this regard, Yin Yang 1 (YY1) is an excellent example due to its regulation of both gene expression and protein posttranslational modifications (54, 62).
YY1 is ubiquitously expressed in all tissues and highly conserved from Xenopus to humans. As a transcription factor, YY1 is able to activate, repress, or initiate gene expression (55). Many reported target genes of YY1 encode proteins that play essential roles in cell proliferation and differentiation, such as c-myc, c-fos, erbb2, p53, cdc6, and the histone H3.2 and H4 genes (6, 12, 31, 33, 52, 65, 69). The functional role of YY1 has been well characterized in developmental studies of Drosophila melanogaster using Pleiohomeotic (pho) and pho like (phol), which are the two orthologs of YY1 (9). Pho and Phol recruit the polycomb group (PcG) silencing complexes to chromatin and establish gene repression (64). YY1 could compensate for the loss of pho protein and rescue the defects in the pho mutant flies (1). Importantly, as a member of the PcG protein family, YY1 possesses two unique features that other PcG proteins do not have: YY1 directly binds a DNA consensus binding site, and YY1 can both establish and maintain gene repression (10).
Many reported YY1-associated proteins are involved in gene regulation and can be divided into four categories: (i) tumor suppressors, such as p53, p14ARF (58), and pRb (45); (ii) oncogene products, such as E1A (55), Mdm2 (58), and c-Myc (56); (iii) posttranslational modification enzymes, such as p300/CBP (32) (acetylation), histone deacetylases 1 to 3 (HDAC1 to -3) (66) (deacetylation), Ezh2 (43), Ezh1 (64), PRMT1 (47) (methylation), and Mdm2 (58) (ubiquitination); and (iv) transcriptional and chromatin remodeling proteins, such as RNA polymerase II (63), Sp1 (53), ATF/CREB (68), nucleophosmin (B23) (21), CtBP (57), and RYBP (13). The association of YY1 with these proteins determines the three layers of its regulatory function in gene expression. First, YY1 directly binds to a consensus element in the promoters of its target genes to physically exert its regulation (54). Second, YY1 recruits different transcriptional factors and cofactors to regulate gene expression, which leads to a greatly extended regulation on YY1-mediated gene expression. Third, YY1 recruits the protein modifiers to mediate the posttranslational modifications of histone and nonhistone proteins associated with YY1 to the YY1-targeted promoters. This will consequently determine the status of the local chromatin environment and subsequently impact target gene expression. For example, YY1-mediated histone acetylation (via recruitment of p300) and deacetylation (via recruitment of HDAC1) contribute to the YY1-regulated gene activation and repression, respectively (10, 32). In addition, YY1 was also involved in histone methylation by recruiting Ezh2 (on H3-K27) (10) and PRMT1 (on H4-R3) (47). The YY1-mediated protein modifications largely increase the complexity of gene expression in eukaryotic cells. These three layers of YY1-mediated gene expression may exert the differential regulation of YY1 on genes under different physiological conditions. These multiple functions and unique properties endow YY1 with a pivotal role in epigenetic regulations, including genomic imprinting and chromatin remodeling (16).
As stated above, YY1 associates with various protein modifiers that mediate different types of protein posttranslational modifications. However, the modifications of YY1 itself have not been extensively studied. To date, only phosphorylation and acetylation of YY1 have been reported. The phosphorylation of YY1 results in differential effects on its DNA binding ability (5). The acetylation of YY1 was regulated by p300 and PCAF, while its deacetylation is mediated by HDACs (67). The acetylation status of YY1 alters its transcriptional activity. Actually, lysine is the target residue of various posttranslational modifications, including acetylation, sumoylation, ubiquitination, methylation, and neddylation. YY1 has great potential to possess other posttranslational modifications besides acetylation due to the presence of 32 lysine residues in its total of 414 amino acids. As a comparison, p53 contains 20 lysines in 393 amino acids and has been reported to have all of the five different lysine-based modifications mentioned above (8). Thus, the high lysine content and its association with many protein modifiers make YY1 a vulnerable target for different modifications. It is also conceivable to predict that the modifications of YY1 itself may play an essential role in its association with other proteins and accordingly determine the gene expression and chromatin structure.
SUMOs (small ubiquitin-related modifiers) modify many proteins by forming an isopeptide bond between the C-terminal portion of a mature SUMO protein and the ɛ-amino group of a lysine residue in the target protein (41). SUMO conjugation, like the process of ubiquitination, is carried out by the sequential action of three enzymes: an activating enzyme (E1), a conjugating enzyme (E2), and a ligase (E3) (23). Unlike the multiple E2s in ubiquitination pathway, Ubc9 is the only E2 that has been identified for sumoylation. Four different SUMO proteins have been reported in mammals: SUMO-1, -2, -3, and -4 (15). These SUMOs possess less than 19% homology to ubiquitin protein in their primary sequences, but their three-dimensional structures share large similarity with that of ubiquitin (4). Among the four SUMOs, SUMO-2, -3, and -4 have over 83% homology, but they only have less than 48% homology with SUMO-1 (15). These SUMO homologs may conjugate to either the common or different target proteins, mostly by recognizing the consensus sequence ψKXD/E (ψ is a hydrophobic residue, and X is any amino acid) (37). The functional outcomes of SUMO and ubiquitin modifications are likely to be very distinct (15). The majority of SUMO conjugations occur in specific regions that are responsible for protein-protein interactions, and, therefore, sumoylation may alter the function of a protein by changing its binding partners (14). Meanwhile, the reported sumoylated proteins are mostly involved in various processes, including transcriptional regulation, chromatin remodeling, and DNA repair (15). A number of tumor suppressors and oncogene products have been identified as targets of SUMOs (3), such as p53 (27, 30), Mdm2 (40), and NF-κB (29).
The E3 ligases of sumoylation promote the conjugation of SUMOs to target proteins. Three different types of SUMO E3 ligases have been described (38). The first type of E3 ligases consists of the proteins in the PIAS family. At least five members of the mammalian PIAS family possess SUMO E3 activity, including PIAS1 (27), PIAS3 (61), PIASxα and PIASxβ (28), and PIASy (50). These proteins have a common RING finger structure. They directly bind to the E2 enzyme, Ubc9, and some substrate proteins. The second type of SUMO E3 ligase is RanBP2 (also known as Nup358) (46). Although RanBP2 does not have a RING finger motif and may not interact with target proteins, it binds to Ubc9 with a 1:1 stoichiometry. The third type of Sumo E3 ligase is Pc2 (26). Pc2 is a member of the Polycomb protein family and has been reported to stimulate the sumoylation of CtBP1, CtBP2, and SIP1 (25, 26, 34).
In the present study, we demonstrate that YY1 can be sumoylated both in vivo and in vitro and we have mapped the major sumoylation site to lysine 288. Consistently, YY1 directly interacts with Ubc9 and a SUMO E3 ligase, PIASy. Importantly, PIASy stimulates YY1 sumoylation in a manner independent of the RING finger domain, but correlating with the PIASy-YY1 interaction.
MATERIALS AND METHODS
Expression vectors, reporter genes, siRNA constructs, and oligonucleotides.
The YY1 mutant K288R was created by PCR-based site-directed mutagenesis, followed by insertion into a pcDNA3-HA vector. pcDNA3/Flag-PIASy, glutathione S-transferase (GST)-tagged PIASy, and His-tagged PIASy were constructed based on the human PIASy cDNA clone kindly provided by Ke Shuai. The PIASy mutants of the RING finger (amino acids [aa] 324 to 368) deletion, aa 1 to 323, 1 to 202, 1 to 100, and 324 to 511 were also generated based on this PIASy cDNA and subcloned into a pcDNA3-Flag vector. pcDNA3/YY1-EGFP and pcDNA3/PIASy-RFP were made by fusing the enhanced green fluorescent protein (EGFP) and red fluorescent protein (RFP) cDNAs to the 3′ ends of human yy1 cDNA (nucleotides 1 to 1242) and human piasy cDNA (nucleotides 1 to 1530), respectively. The p53 wild type (wt) and K386R mutant were generously provided by Jiandong Chen (11). The reporter cdc6-luciferase plasmid was provided by Joseph Nevins. The control plasmid for secreted alkaline phosphatase (SEAP) was a gift from Kazushi Inoue. The small interfering RNA (siRNA) constructs with a scrambled sequence and yy1 cDNA have been described previously (58). The siRNA plasmids, pBS/U6/piasy and pBS/U6/ubc9, were constructed based on the methodology in our previously published papers (59). The target sequences of piasy and ubc9 are 5′-GTGCTCTACGGAAAGTACTT-3′ and 5′-GGGAAGGAGGCTTGTTTAAAC-3′, respectively. The lentiviral vector LTV1 was used to deliver the siRNAs into cells by lentiviral infection. It was constructed based on a previously reported vector (49) with the cytomegalovirus promoter replaced with a ubiquitin C promoter to drive EGFP expression. The siRNA expression cassettes for a scrambled control, yy1, ubc9, and piasy, were subcloned into the LTV1 vector. The packaging of lentivirus followed a standard protocol (49). The sequences of the oligonucleotides used in semiquantitative reverse transcription-PCR (RT-PCR) are as follows: yy1, 5′-CAGAATATATGACAGGAAAGAAAC-3′ and 5′-ACAAAAGCTTTGCCACATTCTGC-3′; cdc6, 5′-CTGGAGTTTGCTGCTGCCGCT-3′ and 5′-GAGCACCAGAAAGGTAAAGGC-3′; piasy, 5′-CTGAAGCCCACCGAATTAGTCC-3′ and 5′-GTGGTGGCGATCTCGCTGTCAGG-3′; and β-actin, 5′-CTGGCACCACACCTTCTACAATGAG-3′ and 5′-CAGACAGCACTGTGTTGGCGTACAG-3′.
Antibodies.
All antibodies used in this study were purchased from Santa Cruz Biotechnology unless otherwise indicated.
Cell culture and transient transfection.
Cos-7 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Transient transfection was carried out using Lipofectamine 2000 (Invitrogen) based on the protocol provided by the manufacturer. In the reporter assay, luciferase reporter plasmid (25 ng of cdc6, 100 ng of c-myc, or 100 ng of ezh2), 80 ng of plasmid expressing the SEAP driven by a β-actin promoter, and other expression plasmids if needed were transfected into Cos-7 cells in 12-well plate with empty vectors to keep the total amount of DNA equal in each well. The transfected cells were harvested 48 h posttransfection, and the luciferase activity assay was performed following a standard protocol using luciferin (Fisher).
In vivo sumoylation.
The in vivo sumoylation experiment was performed essentially as previously described (11). Cos-7 cells in six-well plates were transfected with 0.2 μg of pcDNA3/HA-YY1 wt or mutant plasmid and 0.4 μg each of His-SUMO-1, SUMO-2, or SUMO-3 expression plasmid. Forty-eight hours posttransfection, cells were scraped down into ice-cold phosphate-buffered saline buffer containing 10 mM N-ethylmaleimide (NEM) and were divided into two aliquots. One aliquot was analyzed by Western blotting to confirm the expression of transfected proteins. The other aliquot was used for purification of His-tagged proteins by Ni-nitrilotriacetic acid (NTA) beads (QIAGEN). The cell pellet was lysed in buffer A (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 0.01 M Tris-Cl [pH 8.0], 5 mM imidazole, 0.1% β-mercaptoethanol, 0.1% NP-40) and incubated with Ni-NTA beads for 4 h at room temperature. The beads were washed twice with each of buffer A, buffer B (8 M urea, 0.1 M Na2PO4/NaH2PO4, 0.01 M Tris-Cl [pH 8.0], 0.1% β-mercaptoethanol, 0.1% NP-40), and buffer C (8 M urea, 0.1 M Na2PO4/NaH2PO4, 0.01 M Tris-Cl [pH 6.3], 0.1% β-mercaptoethanol, 0.1% NP-40). The beads were finally treated with buffer D (200 mM imidazole, 0.15 M Tris-Cl [pH 6.7], 30% glycerol, 0.2 M dithiothreitol, 5% sodium dodecyl sulfate). The eluted proteins were examined by Western blotting using hemagglutinin (HA) antibody to detect the modified HA-YY1.
To detect the endogenous YY1 sumoylation, 1.5 mg of Cos-7 cell lysates was incubated with 2 mg of YY1 antibody (C-20; Santa Cruz) overnight at 4°C in a binding buffer (0.05 M Tris-HCl [pH 7.5], 0.15 M NaCl, 5 mM EDTA, 0.1% NP-40) containing 10 mM NEM and 1× protease inhibitor cocktail (Roche Applied Science), followed by incubation with protein G-agarose (Invitrogen) for 1 h. After extensive washing with the binding buffer, the samples were analyzed by Western blotting using the YY1 antibody (H-414).
In vitro sumoylation.
The in vitro sumoylation was carried out as described previously (46). Bacterially expressed His-tagged YY1 wt and K288R mutant, His-Ubc9, His-Aos1/Uba2 (SUMO E1), and GST-SUMO-1 were purified by Ni-NTA beads (QIAGEN) or glutathione-agarose (Sigma). Each in vitro sumoylation reaction mixture contained 250 ng of His-YY1 wt or K288R mutant, 25 ng of His-Ubc9, 100 ng of GST-SUMO-1, 50 ng of SUMO E1, and 5 μl of 10× reaction buffer (50 mM Tris-HCl [pH 7.5], 5 mM MgCl2, 20 mM ATP, 1 mM dithiothreitol, and 2.5 mM ATP). Water was added to make the final volume of 50 μl. The reaction was conducted at 30°C for 30 min and stopped by adding 50 μl of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer and incubating this mixture at 95°C for 5 min. Twenty microliters of each reaction mixture was resolved by Western blotting, and His-YY1 was detected by immunoblotting with an anti-YY1 monoclonal antibody.
Immunofluorescence and confocal microscopy.
Cos-7 cells grown on sterilized glass coverslips overnight were fixed with 4% paraformaldehyde and permeabilized with 0.5% Triton X-100. After being blocked with 1% bovine serum albumin (catalog no. 001-000-161; Jackson ImmunoResearch Laboratory), the cells were incubated either individually or simultaneously with mouse anti-YY1 (H-10; Santa Cruz) and goat anti-PIASy (F-20; Santa Cruz) antibodies diluted in the blocking solution. After an extensive wash, the cells were simultaneously incubated with Cy3-conjugated rabbit anti-mouse and fluorescein isothiocyanate (FITC)-conjugated donkey anti-goat secondary antibodies (Jackson ImmunoResearch Laboratory). Images were captured with a Zeiss confocal microscope (Oberkochen, Germany).
YY1 stability determination.
pcDNA3/HA-YY1 was cotransfected with either pcDNA3 vector or pcDNA3/Flag-PIASy into Cos-7 cells grown in 10-cm dishes. After 24 h, the cells in each dish were trypsinized and replated onto four 6-cm dishes. After another 24 h of growth, one dish was harvested at “time zero.” Cycloheximide was added to a final concentration of 15 μg/ml to the remaining dishes. Cells from one dish were collected at each of the following time points: 4 h, 8 h, and 12 h. The collected cells from each time point were washed with cold phosphate-buffered saline and resuspended in lysis buffer (50 mM Tris [pH 7.5], 5 mM EDTA, 0.1% NP-40, 300 mM NaCl, with freshly added 0.5 mM phenylmethylsulfonyl fluoride and 1× protease inhibitor). After brief sonication and centrifugation, the cell lysates were normalized and analyzed by Western blotting to detect and compare the levels of HA-YY1 at each time point.
RESULTS
YY1 can be sumoylated both in vivo and in vitro.
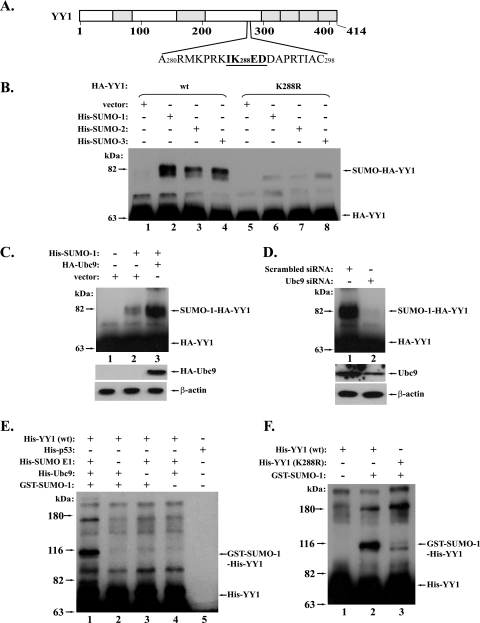
The inspection of YY1 protein indicated the presence of a sequence (IK288ED) that fits the conserved motif (ψKXD/E) of SUMO conjugation (Fig. 1A). This motif and the adjacent residues (Fig. 1A) are completely conserved in YY1 proteins from different species, including humans, mice, rats, chickens, and Xenopus laevis. We thereby used site-directed mutagenesis to replace the lysine at position 288 with an arginine to investigate whether YY1 could be modified by SUMOs through this putative sumoylation site. In the in vivo sumoylation study using Cos-7 cells, HA epitope-tagged wt YY1 (HA-YY1) and its K288R mutant were respectively cotransfected with His-tagged SUMO-1, -2, or -3. Ni-NTA agarose was used to isolate His-tagged proteins, followed by immunoblotting with HA antibody. As shown in Fig. 1B, when wt HA-YY1 was cotransfected, the Ni-NTA agarose could pull down a slower-migrating HA-YY1 species that exhibits a molecular weight (MW) similar to that of the SUMO-conjugated HA-YY1 proteins (compare lanes 2 to 4 with lane 1). However, when the HA-YY1 K288R mutant was used, the majority of this slower-migrating form of YY1 disappeared, suggesting that lysine 288 is a primary sumoylation site of YY1 (Fig. 1B, compare lanes 6 to 8 with lanes 2 to 4). The residues 70 to 80 of human YY1 protein consist of an 11-histidine cluster that can be recognized by the Ni-NTA beads. Therefore, HA-YY1 without SUMO conjugation could also be pulled down by the beads and exhibits a strong band when immunoblotted with HA antibody (Fig. 1B). Although we believe that the conjugation of different SUMO homologs may exert different effects on YY1 function, in the present study, we will focus mostly on YY1 sumoylation by SUMO-1.
FIG. 1.
YY1 can be sumoylated both in vivo and in vitro. (A) The putative sumoylation site of YY1. Sequence analysis of YY1 protein by SUMOPlot (http://www.abgent.com/doc/sumoplot) revealed the presence of a sumoylation motif (the underlined sequence) with lysine 288 as a putative SUMO conjugation site. (B) Sumoylation of wt YY1, but not the YY1 (K288R) mutant, by SUMO-1, -2, and -3. In Cos-7 cells, wt HA-YY1 (lanes 1 to 4) or its K288R mutant (lanes 5 to 8) was cotransfected with the plasmids indicated at the top of the figure. His-SUMO-conjugated proteins were pulled down by Ni-NTA agarose (QIAGEN) and then analyzed in a Western blot using an anti-HA antibody (F-7; Santa Cruz) (see Materials and Methods for details). The sumoylated and free YY1 species are marked on the right (same in the following figures). (C) Overexpression of Ubc9 enhances the sumoylation of YY1. HA-YY1 plasmid was cotransfected with the indicated plasmids into Cos-7 cells. The sumoylated HA-YY1 proteins were analyzed as described above and shown in the top panel. The direct Western blots of HA-Ubc9 and β-actin are shown in the two lower panels. (D) Depletion of Ubc9 decreases the sumoylation of YY1. Plasmids expressing HA-YY1 and His-SUMO-1 were cotransfected with pBS/U6 siRNA plasmids against either a scrambled sequence or ubc9. The sumoylated HA-YY1 was analyzed as described above and shown in the top panel. The direct Western blots of Ubc9 and β-actin are shown in the two lower panels. (E) In vitro sumoylation of YY1. The sumoylation system was reconstituted in vitro with the purified components expressed in bacteria as described in Materials and Methods. His-YY1 was analyzed in this in vitro sumoylation system (lane 1). The controls consist of dropping of each component individually (lanes 2 to 4) and replacement of His-YY1 with His-p53 (lane 5). The reaction mixtures were analyzed by Western blotting using an anti-YY1 (H-10; Santa Cruz). (F) YY1 K288R mutant lost the capability of in vitro sumoylation. The purified wt His-YY1 and its K288R mutant were analyzed in an in vitro sumoylation assay (lanes 2 and 3) with the removal of GST-SUMO-1 as a control (lane 1).
To further confirm that these slower-migrating bands are SUMO-modified YY1, we tested if the fluctuation in Ubc9 (SUMO E2) could affect the formation of these higher-MW species of YY1. Therefore, we studied the YY1 sumoylation by either providing exogenous Ubc9 or knocking down the endogenous Ubc9 using siRNA. Indeed, we observed that the slower-migrating YY1 species significantly increased when Ubc9 was cotransfected (Fig. 1C), but became diminished when Ubc9 was depleted by its siRNA (Fig. 1D). These results further support our prediction that the slower-migrating bands are the sumoylated form of YY1.
In order to determine if YY1 could be sumoylated in vitro, YY1 sumoylation was studied in a reconstituted system using purified proteins from bacteria. As shown in Fig. 1E, the SUMO-1 conjugation of YY1 was only observed in the presence of SUMO E1, Ubc9, and SUMO-1, but disappeared when any of these components was individually removed from the reaction or when His-YY1 was replaced with His-p53 (compare lanes 2 to 5 with lane 1). In addition, the YY1 mutant K288R, which showed a deficiency in the in vivo sumoylation assay, also lacked the major sumoylated form in the in vitro sumoylation experiment, further verifying that the K288 is indeed the primary sumoylation site of YY1 (Fig. 1F).
YY1 directly interacts with Ubc9.
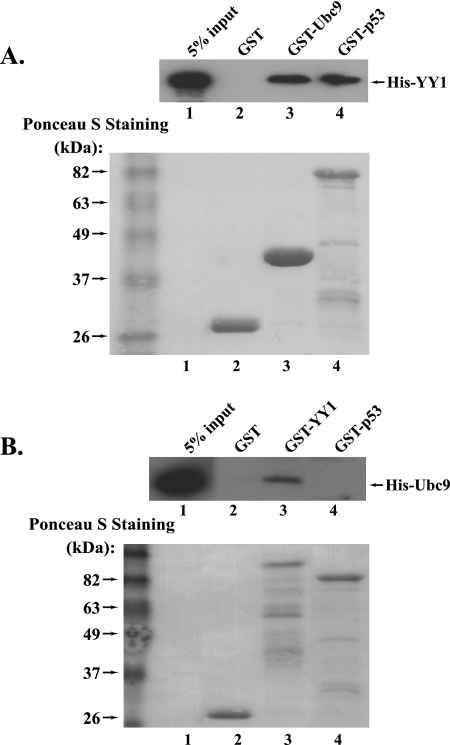
Ubc9 is the only SUMO E2-conjugating enzyme and plays an indispensable role in the sumoylation process. The association of Ubc9 with target proteins could facilitate the SUMO conjugation of target proteins. Thereby, we asked whether YY1 could directly interact with Ubc9. In the in vitro protein binding experiment, we examined the interaction of the purified GST-Ubc9 and His-YY1 with GST and GST-p53, respectively, as negative and positive controls. Indeed, we observed that purified GST-Ubc9 could pull down His-YY1 (Fig. 2A), suggesting a direct interaction between YY1 and Ubc9. In the reciprocal pull down, GST-YY1 could also pull down His-Ubc9, while GST and GST-p53 could not (Fig. 2B). The data from the direct interaction of YY1 and Ubc9 strongly support our observation that YY1 could be sumoylated.
FIG. 2.
YY1 interacts with Ubc9 in vitro. (A) Equal amounts (3 μg) of purified GST, GST-Ubc9, and GST-p53 binding to glutathione-agarose beads (Sigma) were individually incubated with purified His-YY1 (1.5 μg) at 4°C for 4 h. After extensive washing, the samples were analyzed by Western blotting using the anti-YY1 antibody (H-10). (B) In the reciprocal in vitro protein binding experiment, 3 μg each of purified GST, GST-YY1, or GST-p53 was individually incubated with 1.5 μg of purified His-Ubc9. The samples were subjected to Western blot analysis using an anti-Ubc9 antibody (H-81; Santa Cruz).
PIASy is a SUMO E3 ligase of YY1.
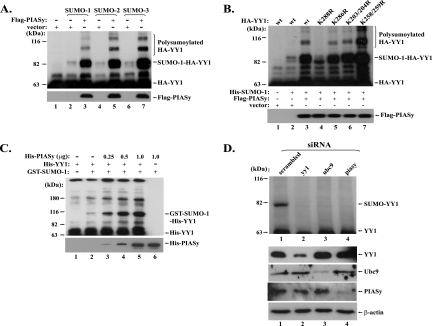
In the sumoylation process, the SUMO E3 ligases can greatly facilitate the sumoylation process, although they are not definitely required for the SUMO conjugation. We asked if any known SUMO E3 ligase could enhance the sumoylation of YY1. In the in vivo sumoylation experiments, when PIASy, a SUMO E3 ligase, was cotransfected with HA-YY1 and His-SUMO-1, -2, or -3, we observed significantly increased YY1 sumoylation and polysumoylation (Fig. 3A), suggesting that PIASy could stimulate YY1 sumoylation in vivo. We also studied the effect of PIASy on the sumoylation-deficient YY1 mutant K288R and other lysine-to-arginine YY1 mutants (K286R, K203/204R, and K258/259R) that could be sumoylated to a similar extent as wt YY1 (data not shown). As shown in Fig. 3B, except for the K288R mutant (lane 4), the sumoylation of all other YY1 mutants (lanes 5 to 7) could be greatly stimulated when PIASy was cotransfected. We have noticed the presence of a YY1 species with a slightly smaller MW than that of the K288-derived SUMO-YY1, which became especially obvious in the YY1 K288R mutant when cotransfected with PIASy (lane 3, Fig. 3B). It is unclear if this resulted from the SUMO conjugation to another site of YY1 or from another type of YY1 modification. However, provided there is another sumoylation site, we believe that K288 is the primary sumoylation target in YY1, since the intensity of this lower band is significantly weaker than that of the K288-derived SUMO-YY1, especially in the in vitro sumoylation studies (Fig. 1F). Nevertheless, to search for a potential secondary sumoylation site, we used arginine to individually or combinatorially replace K173 and K229/K230, which showed a higher probability of being conjugated by SUMO than other 29 lysines in YY1, except K288, when analyzed by the SUMOPlot. Especially, the K229/K230 residues reside in a “DEKK” motif that had previously revealed as a SUMO conjugation site (51). However, the in vitro sumoylation study of these YY1 mutants indicated that the lower band is still present (see Fig. S1 in the supplemental material), suggesting that these lysines are not the secondary sumoylation site that may exist. We predict that this YY1 species may be derived from the SUMO conjugation to one or more recessive lysine residues, which become more apparent when SUMO and SUMO E3 ligase are overexpressed. Consistent with this prediction, we only detected a single band of sumoylated endogenous YY1 (see below). Despite these speculations, the identity of this YY1 species requires further investigation.
FIG. 3.
PIASy is a SUMO E3 ligase of YY1. (A) PIASy enhances the conjugation of SUMO-1, -2, and -3 to YY1 in vivo. Plasmids expressing HA-YY1 and His-SUMO-1, -2, and -3 were individually cotransfected with either an empty vector or Flag-PIASy (as labeled at the top of lanes 2 to 7). His-SUMO-conjugated YY1 was analyzed as described above and immunoblotted with the anti-HA antibody (F-7). Polysumoylated HA-YY1 is indicated on the right. Immunoblotting of Flag-PIASy using an anti-Flag antibody is shown in the lower panel. (B) PIASy stimulates the sumoylation and polysumoylation of wt YY1 and its lysine mutants, except YY1 K288R. Plasmids expressing wt YY1 and its lysine mutants, K288R, K286R, K203/204R, and K258/259R (as labeled at the top), were individually cotransfected with the plasmids indicated in the middle panel. The sumoylated HA-YY1 proteins were analyzed as described above. The expressed Flag-PIASy detected by an anti-Flag antibody (D-8; Santa Cruz) is shown in the lower panel. (C) Dose-dependent effect of PIASy on YY1 sumoylation in vitro. Increased amounts of purified PIASy (0, 0.25, 0.5, and 1.0 μg) were added to the reconstituted in vitro sumoylation system with purified His-YY1 (lanes 2 to 5), followed by Western blotting using the anti-YY1 antibody (H-10). Reactions without GST-SUMO-1 or His-YY1 were used as controls (lanes 1 and 6). His-PIASy in these reactions was immunoblotted by the anti-PIASy antibody (D-8) and is shown in the lower panel. (D) PIASy depletion leads to decreased sumoylation of endogenous YY1. Cos-7 cells were infected by lentiviruses carrying siRNA expression cassettes for a scrambled control, yy1, ubc9, and piasy. The samples were individually immunoprecipitated with YY1 C-20 antibody (Santa Cruz) and analyzed by Western blotting using YY1 H-414 antibody. The sumoylated endogenous YY1 is indicated in the top panel. The direct Western blots of YY1, Ubc9, PIASy, and β-actin are shown in the four lower panels.
To confirm the SUMO E3 activity of PIASy on YY1, we carried out the sumoylation study with a reconstituted in vitro sumoylation system. As shown in Fig. 3C, the PIASy-mediated YY1 sumoylation could be well recapitulated in vitro. With increased amounts of His-PIASy (0.25, 0.5, and 1.0 μg) added in the in vitro sumoylation reaction, the sumoylated form of His-YY1 significantly increased, providing unequivocal evidence for PIASy as a SUMO E3 ligase of YY1.
We next investigated whether decreased PIASy could hamper the sumoylation of the endogenous YY1. Cos-7 cells were infected by four different lentiviruses carrying different siRNA cassettes that target a control (scrambled) sequence, yy1, ubc9, and piasy, respectively. The inhibition of the expression of YY1, Ubc9, and PIASy proteins is shown in the lower panel of Fig. 3D. YY1 antibody C-20 was used to pull down all species of YY1 protein from the cell lysates, followed by Western blot analysis using YY1 antibody H-414. As shown in the upper panel of Fig. 3D, the sample from the cells infected by the scrambled siRNA exhibited a slower-migrating YY1 species with a molecular weight corresponding to that of the SUMO-conjugated YY1 (lane 1 in the upper panel of Fig. 3D). This high-molecular-weight YY1 species is very likely to be the sumoylated form of endogenous YY1 because the band became significantly decreased when either yy1 or ubc9 was knocked down by the siRNAs (compare lanes 2 and 3 with lane 1). Importantly, the depletion of PIASy resulted in a decrease in the sumoylated form of YY1 (compare lane 4 with lane 1), equivalently to the effect of the depletion of SUMO E2 Ubc9 or YY1 itself, suggesting that PIASy is a SUMO E3 ligase of endogenous YY1.
YY1 and PIASy interact and colocalize in vivo.
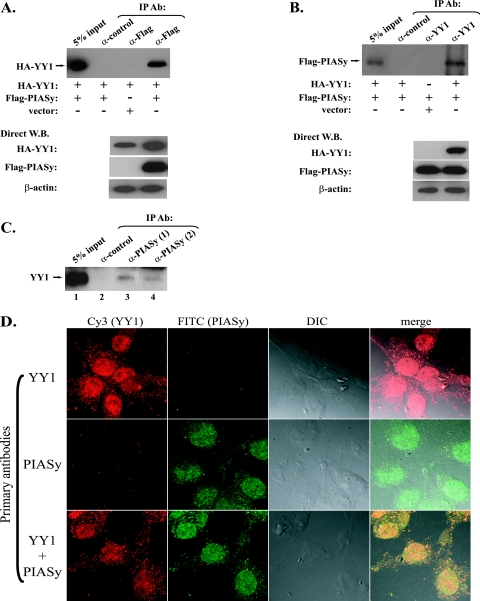
SUMO E3 ligases do not possess intrinsic enzymatic activity but act as adapters to bring the SUMO E2 (Ubc9) and substrate proteins together (15). Therefore, we asked whether YY1 physically associates with PIASy. In the coimmunoprecipitation experiments using the cell lysates from Cos-7 cells cotransfected with HA-YY1 and Flag-PIASy, we observed that a Flag antibody (D-8; Santa Cruz) could pull down HA-YY1 (Fig. 4A), while in the reciprocal immunoprecipitation a YY1 antibody (H-10; Santa Cruz) could also pull down the Flag-PIASy (Fig. 4B), suggesting the in vivo association of YY1 and PIASy. We further studied the association between the endogenous YY1 and PIASy. As a result, two different PIASy antibodies (P-18 and I-19; Santa Cruz), but not the control antibody, could individually pull down YY1 from the Cos-7 cell lysates in the immunoprecipitation study (Fig. 4C, compare lanes 3 and 4 with lane 2). We failed to detect PIASy protein in the samples immunoprecipitated by YY1 antibodies, which could be due to the weak detection in Western blotting by the available PIASy antibodies.
FIG. 4.
YY1 and PIASy interact and colocalize in vivo. (A) In Cos-7 cells, HA-YY1 plasmid was cotransfected with either Flag-PIASy plasmid or an empty vector as indicated below the top panel. Total cell lysates were immunoprecipitated (IP) with a control antibody (Ab [α-control]) or anti-Flag antibody (D-8 [α-Flag]) as indicated at the top, followed by Western blotting (W.B.) with an anti-HA antibody (F-7). The direct Western blots of HA-YY1, Flag-PIASy, and β-actin are shown in the lower panel. (B) Cos-7 cells were cotransfected by the plasmids indicated below the top panel. Total cell lysates were immunoprecipitated with a control antibody or the anti-YY1 antibody (H-10 [α-YY1]) as indicated at the top. The samples were immunoblotted with the anti-Flag antibody (D-8). The direct Western blots of HA-YY1, Flag-PIASy, and β-actin are shown in the lower panel. (C) The interaction of endogenous YY1 and PIASy. Cos-7 cell lysates (1.5 mg) were individually incubated with 2 mg of control, PIASy (I-19), and PIASy (P-18) antibodies (α-PIASy) overnight at 4°C. After extensive washing, the samples were analyzed by Western blotting using YY1 (H-10) antibody. (D) Colocalization of YY1 and PIASy. Cos-7 cells cultured on coverslips were treated with mouse anti-YY1 (H-10) antibody (top panel), goat anti-PIASy (F-20) antibody (middle panel), or both of the antibodies (bottom panel). All of the coverslips were then incubated with Cy3-conjugated rabbit anti-mouse and FITC-conjugated donkey anti-goat secondary antibodies. Images were captured with a Zeiss confocal microscope (Oberkochen, Germany). Columns show (from left to right) red fluorescence emitted by Cy3, green fluorescence emitted by FITC, a differential interference contrast (DIC) image, and merged images.
The association of YY1 and PIASy led to our query of whether there was any correlation between their subcellular localizations. We first observed good colocalization of the fusion proteins of YY1-EGFP and PIASy-RFP when they were cotransfected in Cos-7 cells (see Fig. S2 in the supplemental material). Furthermore, we attempted to investigate the localizations of the endogenous YY1 and PIASy. YY1 (H-10) and PIASy (F-20) antibodies were used to detect the endogenous YY1 and PIASy, respectively, followed by the incubation of the corresponding fluorescence-conjugated secondary antibodies. The slides were visualized using a Zeiss confocal microscope. As shown in Fig. 4D, endogenous YY1 and PIASy are both predominantly localized inside the nuclei (top and middle panels). A small portion of YY1 is present in cytoplasm, which is consistent with previously reported results (44). When both the primary antibodies for YY1 and PIASy were used, the merged fluorescence of the two proteins showed a well-overlapped image (Fig. 4D, bottom panel), indicating that YY1 and PIASy colocalize in the nucleus.
Mapping of the domains required for YY1-PIASy interaction.
To further determine whether the interaction between YY1 and PIASy is direct, we carried out the in vitro protein binding experiments. As shown in Fig. 5, GST-YY1 can pull down His-PIASy (Fig. 5A, lane 3) and GST-PIASy can also pull down His-YY1 (Fig. 5B, lane 3). In both pull-down experiments, the negative control is GST alone, while the positive control is GST-p53, which is known to interact with both PIASy and YY1 (42, 58). The in vitro binding of YY1 and PIASy indicated that YY1 and PIASy directly bind to each other. We further conducted experiments to delimit the regions responsible for the interaction between YY1 and PIASy. In the experiments to identify the PIASy binding site on YY1, we determined that aa 331 to 414 of YY1 are both necessary and sufficient for YY1 to interact with PIASy (Fig. 5A, lanes 6 to 8). Likewise, the binding domain of YY1 was mapped to the N-terminal portion, most likely among aa 100 to 202, of PIASy (Fig. 5B).
FIG. 5.
YY1 and PIASy directly interact in vitro. (A) Identification of the YY1 domain that interacts with PIASy. GST-YY1 fusion proteins are diagrammed in the top panel. The purified GST-YY1 proteins (3 μg each) were incubated with purified His-PIASy (2 μg) at 4°C for 4 h. After extensive washing, the samples were analyzed by Western blotting using the PIASy antibody (D-8). The inputs of the GST and GST fusion proteins are shown in the bottom panel. The results of the protein interaction are summarized on the right. (B) Identification of the PIASy domain that interacts with YY1. GST-PIASy fusion proteins are diagrammed in the top panel. The purified GST-PIASy proteins (3 μg each) were incubated with purified His-YY1 (1.5 μg) at 4°C for 4 h. After extensive washing, the samples were analyzed by Western blotting using the YY1 antibody (H-10). The inputs of GST and GST fusion proteins are shown in the bottom panel. The results of their binding affinity to YY1 and their capability of enhancing YY1 sumoylation (see Fig. 6A) are summarized on the right. ΔRF, RING finger deletion.
Determining the functional domain of PIASy in stimulating YY1 sumoylation.
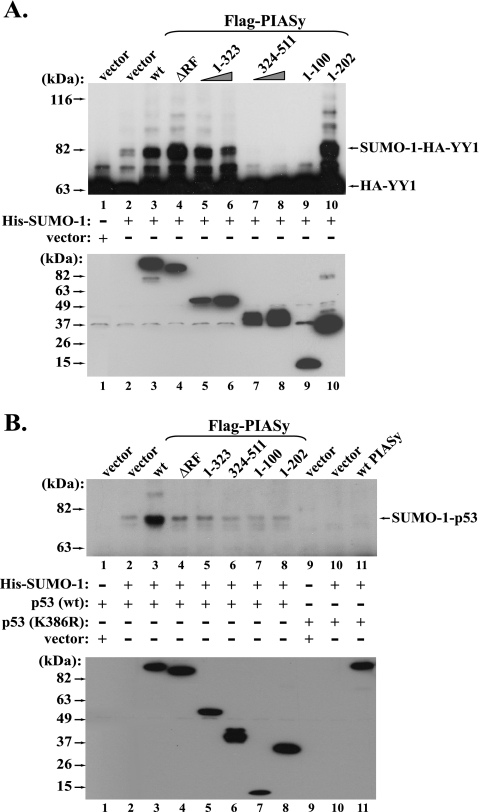
To investigate the structural basis of PIASy in enhancing YY1 sumoylation, we sought to further identify the functional domains of PIASy in stimulating YY1 sumoylation. It has been indicated in multiple reports that the RING fingers of PIAS proteins are the crucial domains in enhancing protein sumoylation. Previous reports have shown that deletion or amino acid point mutations in the RING finger could always decrease or mostly abolish their SUMO E3 ligase activity (2, 7, 18, 22, 50). We therefore asked if the removal of the RING finger of PIASy could also affect its SUMO E3 activity to YY1 sumoylation. Surprisingly, the generated PIASy ΔRING finger (aa 324 to 368) mutant still retained a strong capability of enhancing YY1 sumoylation (Fig. 6A, lane 4), suggesting that the RING finger of PIASy is dispensable in its SUMO E3 activity to mediate SUMO-1 conjugation to YY1. We further examined effects of other PIASy mutants. The N-terminal portion (aa 1 to 323) of PIASy that lacks both the RING finger and the rest of the C-terminal region still kept strong SUMO E3 activity (Fig. 6A, lanes 5 and 6). However, the PIASy C-terminal region (aa 324 to 511) that contains the RING finger domain and the SUMO-interacting motif (39, 50) not only lost the SUMO E3 activity, but also significantly decreased the YY1 sumoylation, suggesting a dominant-negative role (Fig. 6A, compare lanes 7 and 8 with lanes 1 and 2). To further analyze the functional domain of the N-terminal region of PIASy, we tested the two short N-terminal fragments of PIASy. Strikingly, the PIASy aa 1 to 202 mutant could greatly stimulate YY1 sumoylation (Fig. 6A, lane 10), while the aa 1 to 100 fragment of PIASy acted as a dominant-negative mutant (Fig. 6A, compare lane 9 with lanes 1 and 2). It is noteworthy that these studied PIASy proteins did not exert their effects on YY1 sumoylation in a casual fashion. Instead, whether they enhance or inhibit YY1 sumoylation is well correlated to their interaction with YY1 (Fig. 5B, right panel).
FIG. 6.
Functional domains of PIASy in enhancing the in vivo sumoylation of YY1 and p53. (A) Functional domains of PIASy in stimulating YY1 sumoylation. In Cos-7 cells, plasmids expressing HA-YY1 and His-SUMO-1 were cotransfected with an empty vector, Flag-PIASy wt, or its different mutants as labeled at the top of lanes 2 to 10. The control sample contains only HA-YY1 without His-SUMO-1 (lane 1). His-SUMO-1-conjugated HA-YY1 proteins were analyzed as described above and immunoblotted with the anti-HA antibody (F-7). The lower panel shows the direct Western blot of the Flag-PIASy proteins using the anti-Flag antibody (D-8). ΔRF, RING finger deletion. (B) Functional domains of PIASy in stimulating p53 sumoylation. In Cos-7 cells, plasmids expressing wt p53 (lanes 1 to 8) or the p53 K386R mutant (lanes 9 to 11) were respectively cotransfected with either an empty vector or His-SUMO-1 (as indicated in the middle panel), as well as the plasmids expressing Flag-PIASy proteins labeled at the top. His-SUMO-1-conjugated p53 was analyzed as described above and immunoblotted with an anti-p53 antibody (DO-1; Santa Cruz). The levels of the Flag-PIASy proteins were detected by Western blotting using the anti-Flag antibody (D-8) and are shown in the lower panel.
We next asked whether the effects of PIASy mutants on YY1 sumoylation could be extended to the situation in vitro. The PIASy wt and mutants used in Fig. 6A were subcloned into a His tag vector. The proteins were expressed and purified from bacteria, followed by the experiments to determine their effects on the in vitro sumoylation of YY1. As shown in Fig. S3 in the supplemental material, only wt PIASy exhibited stimulation of YY1 in vitro sumoylation, while none of the ΔRING finger, aa 1 to 323 and aa 1 to 202 mutants retained the function in promoting SUMO conjugation to YY1. In addition, we observed that the presence of PIASy N-terminal region (aa 1 to 323) significantly decreased the basal level of YY1 sumoylation (compare lanes 7 and 8 with lane 2 in Fig. S3 in the supplemental material). Actually, a similar effect was also present in the in vivo YY1 sumoylation (lanes 5 and 6 in Fig. 6A). Currently, the mechanism of this effect is still unclear, but we predict that it may be due to stoichiometric inhibition. In sum, the effect of the wt and mutated PIASy proteins on the in vitro YY1 sumoylation did not resemble their effects in vivo, suggesting the essential distinction between in vivo and in vitro sumoylation of YY1 mediated by PIASy.
The observation that some of the PIASy mutants retained SUMO E3 activity toward the in vivo sumoylation of YY1 was indeed unsuspected. We therefore asked if these PIASy mutants could also show the same effects on the in vivo sumoylation of p53, which is another substrate of PIASy (7). We therefore tested the effects of these PIASy proteins on p53 sumoylation. As shown in Fig. 6B, besides the enhancement of wt PIASy on p53 sumoylation, the ΔRING mutant showed a mild effect (Fig. 6B, lane 4). However, none of the other mutants could demonstrate either a positive or dominant-negative effect on in vivo p53 sumoylation (compare lanes 5 to 8 with lanes 1 and 2), as they did with YY1, suggesting the PIASy-mediated in vivo YY1 sumoylation is exceptional.
The PIASy-YY1 association increases the stability of YY1.
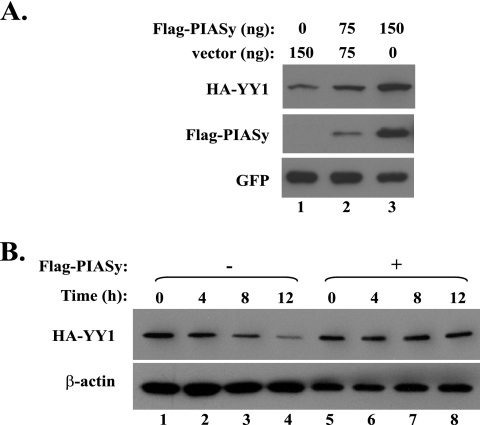
We next asked how PIASy could affect the homeostasis of YY1. When the plasmids expressing HA-YY1 were cotransfected with Flag-PIASy into Cos-7 cells, we observed a significant increase in HA-YY1 steady-state level compared with the cotransfection of an empty vector (Fig. 7A, compare lanes 2 and 3 with lane 1). Therefore, we used cycloheximide to block the protein synthesis and examined the stability of YY1 with the cotransfection of either an empty vector or the plasmid expressing PIASy. As shown in Fig. 7B, YY1 stability was significantly increased in the presence of PIASy (Fig. 7B, compare lanes 5 to 8 with lanes 1 to 4). We further asked whether the effect of PIASy on YY1 stability depends on YY1 sumoylation. We tested the stability of YY1 wt and K288R mutant in Cos-7 cells with cotransfected His-SUMO-1 and/or Flag-PIASy. As shown in Fig. S4A and S4B in the supplemental material, neither wt YY1 nor the K288R mutant exhibited a response to the His-SUMO-1 overexpression, but they could be stabilized by transfected Flag-PIASy, suggesting that PIASy stabilizes YY1 in a sumoylation-independent manner.
FIG. 7.
Overexpression of PIASy-stabilized YY1 in vivo. (A) Coexpression of PIASy increases the steady-state level of YY1 protein. In Cos-7 cells, plasmid expressing HA-YY1 was cotransfected with an empty vector or 75 and 150 ng of Flag-PIASy plasmid. The steady-state protein levels of HA-YY1, PIASy, and the cotransfected GFP proteins were immunoblotted with anti-HA (F-7), anti-Flag (D-8), and anti-GFP (B-2; Santa Cruz) antibodies, respectively. (B) Overexpressed PIASy increases the stability of YY1. In Cos-7 cells, plasmid expressing HA-YY1 was cotransfected with an empty vector or Flag-PIASy plasmid. Two days posttransfection, the cells were treated with 10 μg/ml cycloheximide to block protein synthesis and the samples were collected at the time points labeled at the top. After the starting HA-YY1 levels in the two groups were normalized, the HA-YY1 protein at each time point was analyzed by Western blotting using the anti-HA antibody F-7. β-Actin immunoblotted by an anti-β-actin antibody (MAB1510; Chemicon) was used as the loading control.
PIASy-mediated sumoylation represses the transcriptional activity of YY1.
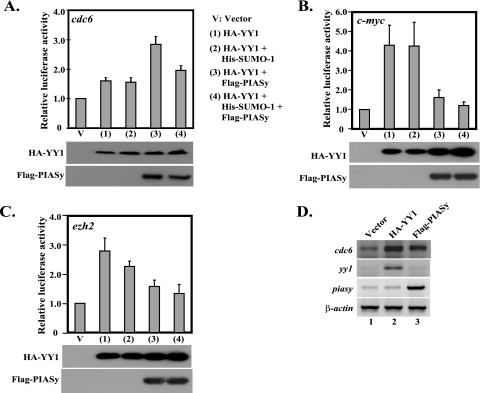
We next asked whether the sumoylation of YY1 could affect its biological function. As a transcription factor, YY1 has been reported to regulate the expression of many genes, such as cdc6 and c-myc (48, 52). In addition, the consensus binding site of YY1 is present in the promoters of many other genes, including ezh2 (data not shown). We therefore study the effect of PIASy-mediated YY1 sumoylation using the reporter constructs for the cdc6, c-myc, and ezh2 promoters. The luciferase reporter plasmids of these promoters were individually cotransfected with an empty vector or HA-YY1 in the presence of His-SUMO-1 and/or Flag-PIASy, followed by the assay to determine the luciferase activity in the cell lysates. YY1 could stimulate the expression of these three promoters (compare lanes 1 and V in Fig. 8A to C). The addition of His-SUMO-1 did not greatly alter the expression but only slightly decreased the transcription of the ezh2 promoter (compare lanes 2 and 1 in Fig. 8A to C). However, the introduction of PIASy exhibited differential effects on YY1-mediated transcription on these promoters. While PIASy enhanced the YY1-mediated cdc6 transcription, it significantly inhibited the YY1-mediated transcription of c-myc and ezh2 (compare lanes 3 and 1 in Fig. 8A to C). With the addition of SUMO-1 in the presence of PIASy, we observed a slightly decreased expression of all three promoters compared with PIASy alone (compare lanes 4 and 3 in Fig. 8A to C). Overall, the results from the luciferase reporter assay indicate that YY1 sumoylation, especially in the presence of PIASy, exerts an inhibitory effect on the transcriptional activity of YY1.
FIG. 8.
Transcriptional activity of YY1 is affected by PIASy and/or PIASy-mediated sumoylation. (A to C) In Cos-7 cells, plasmids (as indicated on the right side of panel A) were cotransfected with luciferase reporter vectors for cdc6 (A), c-myc (B) and ezh2 (C) and plasmid expressing the SEAP driven by a β-actin promoter. The relative luciferase activity of each sample is shown based on the normalization by the levels of the coexpressed SEAP. The error bars represent the standard variations within triplicate transfections. The Western blots of the transfected HA-YY1 and Flag-PIASy are shown below the graphs. (D) PIASy enhances the expression of an endogenous YY1-dependent gene. Cos-7 cells were transfected by an empty vector or plasmids expressing HA-YY1 and Flag-PIASy, respectively. Two days posttransfection, the total RNAs from the transfected cells were extracted and analyzed by semiquantitative RT-PCR for the expression of the cdc6, yy1, piasy, and β-actin genes.
We further inquired whether PIASy introduction could alter the expression of any endogenous YY1-dependent gene. Therefore, we carried out semiquantitative RT-PCR to examine endogenous cdc6 gene expression upon the exogenous expression of PIASy. As a target gene of YY1 (52), the introduction of exogenous HA-YY1 led to significant activation of the cdc6 gene after being normalized by β-actin expression (compare lane 2 with lane 1 in Fig. 8D). Meanwhile, increased PIASy expression also resulted in elevated expression of the endogenous cdc6 gene (compare lane 3 with lane 1 in Fig. 8D), which is consistent with the result obtained from the cdc6-luciferase reporter experiment.
DISCUSSION
As a multifunctional protein, YY1 regulates both gene expression and protein modifications. Actually, these two types of regulation are closely related, because YY1-mediated protein modifications play a regulatory role in gene expression as well. For example, YY1 has been shown to regulate histone acetylation, deacetylation, and methylation that consequently affect chromatin structure and mediate gene expression. Meanwhile, YY1 also possesses functions that are independent of its transcriptional activity, represented by mediating the ubiquitination and acetylation of p53 (17, 58).
Multiple reports have suggested that the posttranslational modifications of transcription factors regulate their stability, subcellular transport, protein interaction, etc. Since YY1 is involved in both gene transcription and protein modifications, it is reasonable to predict that the modifications of YY1 itself may alter the function of YY1 and exert a differential impact on various biological processes. Indeed, the high content of lysine residues and the association with various protein modifiers imply YY1 has great potential to possess different modifications. Nevertheless, the posttranslational modifications of YY1 still remain understudied and only its phosphorylation and acetylation have been reported (5, 67).
In the last few years, protein sumoylation has emerged as an important modification for many proteins that play regulatory roles in cell cycle control, DNA repair, protein relocalization, and cell signaling(19). In this study, we have provided strong evidence to demonstrate that YY1 can be sumoylated both in vivo and in vitro. We have also identified lysine 288 as a major sumoylation site of YY1. YY1 directly interacts with Ubc9, consistent with our observation that YY1 could be sumoylated in vitro in the absence of any SUMO E3 ligase. Furthermore, we discovered PIASy as a SUMO E3 ligase in facilitating YY1 sumoylation both in vivo and in vitro.
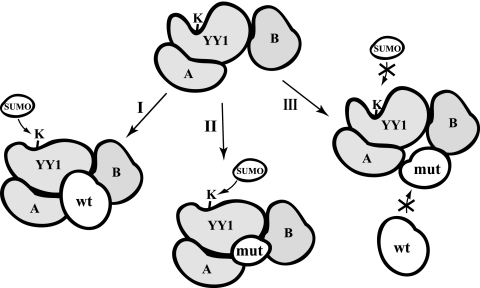
The SUMO E3 ligases in PIAS family have two domains that are crucial in facilitating protein sumoylation: the RING finger domain that binds to Ubc9 (20, 23) and the SUMO-interacting motif (39) that binds to SUMOs. To our best knowledge, the RING finger domains of the PIAS proteins are indispensable to their SUMO E3 ligase activity with all of the identified substrates (2, 7, 18, 22, 36, 50). Either deletion or single-amino-acid mutations could dramatically decrease or, in most cases, abolish their SUMO E3 activity. However, in our study, the PIASy-mediated in vivo YY1 sumoylation does not follow this canonical pattern depending on the integrity of the PIASy protein. Neither the RING finger domain nor the SUMO-interacting motif (50) of PIASy was required in mediation of YY1 sumoylation. Instead, all of the examined truncated PIASy mutants that were capable of binding to YY1 could also accelerate in vivo YY1 sumoylation in the same manner as wt PIASy (Fig. 6A). On the contrary, the PIASy mutants that have lost interaction with YY1 could prevent in vivo YY1 sumoylation (Fig. 6A). Importantly, these unexpected effects of the PIASy mutants on YY1 sumoylation did not apply to that of p53, which still followed a pattern dependent on the integrity of the PIASy protein (Fig. 6B). These observations indicate that PIASy-mediated YY1 sumoylation follows a unique pattern. Based on our observation in this study, we hypothesize that the in vivo binding of wt PIASy or its YY1-interacting mutants leads to the conformational change of YY1 to make Lys288 more accessible to the SUMOs (Fig. 9, paths I and II). Therefore, they are able to enhance the SUMO conjugation to YY1. On the contrary, the PIASy mutants that lost their interaction with YY1 may be incorporated into the sumoylation machinery but are unable to reach the YY1 molecule to generate the conformation susceptible to sumoylation (path III in Fig. 9). Instead, their presence in the sumoylation complex blocks the entry of the endogenous PIASy and therefore inhibits the basal sumoylation of YY1. It is worthy of notice that this model is only appropriate to the PIASy-mediated in vivo YY1 sumoylation (Fig. 6A) but not the in vitro scenario (see Fig. S3 in the supplemental material). We reasoned that this was because YY1 sumoylation stimulated by these PIASy mutants may require other components that can be provided in vivo but are absent in vitro. Actually, our proposed model is consistent with the result shown in Fig. 3D that the siRNA-mediated depletion of endogenous PIASy severely decreased the in vivo sumoylation of YY1. Although Ubc9 may directly recognize the substrate proteins and mediate in vitro SUMO conjugation, our data and previous reports (24, 60) indicated that the depletion of certain SUMO E3 ligases could nearly eliminate the in vivo sumoylation of special targets.
FIG. 9.
Schematic model of YY1 sumoylation mediated by PIASy proteins. The sumoylation of YY1 may be regulated by a sumoylation complex consisting of a number of components, simplified as A and B proteins in the model. The possibility of SUMO conjugation to YY1 in the presence of wt PIASy (I) and PIASy mutants that kept (II) or lost (III) YY1 interaction are illustrated. In the scenario of wt PIASy or YY1-interacting PIASY mutants (paths I and II), the binding of PIASy leads to a conformational change of YY1 that makes SUMO more accessible. However, the PIASy mutants deficient in YY1 interaction cannot reach YY1 to cause this conformational change (path III). They will block the access of the endogenous PIASy and consequently play a dominant-negative role.
The colocalization of the endogenous YY1 and PIASy in cell nuclei (Fig. 4D) suggests that the two proteins may work as functional partners in certain biological processes. We observed that PIASy-mediated YY1 sumoylation repressed its transcriptional activity (Fig. 8A to C). We postulate that the PIASy-mediated YY1 sumoylation may either prevent its interaction with transcriptional coactivators or lead to recruitment of corepressors. Without coexpressed PIASy, changes in the transcriptional activity with exogenously provided SUMO-1 were very marginal. This could be explained by fact that the sumoylation of the transfected YY1 requires the presence of sufficient PIASy. When only SUMO-1 was overexpressed, the machinery for YY1 sumoylation could have been saturated and therefore the limited sumoylation of YY1 was not enough to cause obvious changes in its transcriptional regulation.
The effect of YY1 sumoylation in the absence of transfected PIASy is very marginal, and therefore we did not convincingly detect a significant difference in the transcriptional activities between the YY1 wt and K288R mutant with the promoters tested (data not shown). This may imply that the YY1 sumoylation does not play a major role in mediating its transcriptional function under a normal physiological condition. However, it is still possible that YY1 sumoylation may be important for regulating its activity very transiently or under certain physiological conditions.
The functional interplay of PIASy and YY1 should not be limited to the PIASy-mediated YY1 sumoylation. As shown in Fig. 7, PIASy stabilizes YY1 without the overexpression of SUMO-1. This is consistent with the observation that both the YY1 wt and the K288R mutant exhibited the same response to the exogenously provided SUMO-1 and/or PIASy (see Fig. S4 in the supplemental material), implying that the effect of PIASy on YY1 stability is independent of YY1 sumoylation. Although we could not convincingly demonstrate any significant change in the endogenous YY1 expression in Cos-7 cells upon PIASy depletion or overexpression (data not shown), we predict that the YY1-PIASy interplay may be important in certain scenarios, such as exposure to genomic stresses, or in some other cell types. Especially, PIASy has been demonstrated to regulate cellular senescence and apoptosis by mediating the function of p53 and pRb (7, 42), both of which directly interact with YY1 (45, 58). Therefore, YY1 and PIASy may be partners in a macromolecular complex that can change its components and alter its function in response to different physiological alterations.
Knowledge about protein sumoylation has enormously increased in the past few years. However, the mechanisms of regulation by SUMO conjugation are far from being understood. Meanwhile, since protein sumoylation is a dynamic and transient process, its study has also been restricted by many technical limitations. Although our finding in the PIASy-YY1 physical and functional interplay revealed another type of YY1 modification that potentially modulates its function in certain cellular processes, we are still in a very primitive stage in pursuing the biological roles of YY1 sumoylation. Future research will be needed to further characterize the functional roles of YY1 sumoylation in YY1-mediated gene expression and protein modifications. Due to the pivotal role of YY1 in tumorigenesis, these studies will provide new insights into tumor development and progression.
Supplementary Material
Acknowledgments
We thank Scott Cramer, Nathan Wall, and Johnathan Whetstine for critical reading of the manuscript. We thank Suzy Torti, Grace Gill, and Constantinos Koumenis for helpful discussion and advice. Frauke Melchior generously provided the reagents and protocol for in vitro translation. Charles J. Sherr kindly sent us the His-tagged SUMO constructs. Yang Shi and El Bachir Affar generously provided some other reagents. Mark Willingham and Ken Grant helped us with the cell immunostaining and confocal microscopy.
This work was supported by the startup fund from the Comprehensive Cancer Center of Wake Forest University to G.S.
Footnotes
Published ahead of print on 12 March 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Atchison, L., A. Ghias, F. Wilkinson, N. Bonini, and M. L. Atchison. 2003. Transcription factor YY1 functions as a PcG protein in vivo. EMBO J. 22:1347-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azuma, Y., A. Arnaoutov, T. Anan, and M. Dasso. 2005. PIASy mediates SUMO-2 conjugation of topoisomerase-II on mitotic chromosomes. EMBO J. 24:2172-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baek, S. H. 2006. A novel link between SUMO modification and cancer metastasis. Cell Cycle 5:1492-1495. [Epub ahead of print 17 July 2006.] [DOI] [PubMed] [Google Scholar]
- 4.Bayer, P., A. Arndt, S. Metzger, R. Mahajan, F. Melchior, R. Jaenicke, and J. Becker. 1998. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280:275-286. [DOI] [PubMed] [Google Scholar]
- 5.Becker, K. G., P. Jedlicka, N. S. Templeton, L. Liotta, and K. Ozato. 1994. Characterization of hUCRBP (YY1, NF-E1, delta): a transcription factor that binds the regulatory regions of many viral and cellular genes. Gene 150:259-266. [DOI] [PubMed] [Google Scholar]
- 6.Begon, D. Y., L. Delacroix, D. Vernimmen, P. Jackers, and R. Winkler. 2005. Yin Yang 1 cooperates with activator protein 2 to stimulate ERBB2 gene expression in mammary cancer cells. J. Biol. Chem. 280:24428-24434. [DOI] [PubMed] [Google Scholar]
- 7.Bischof, O., K. Schwamborn, N. Martin, A. Werner, C. Sustmann, R. Grosschedl, and A. Dejean. 2006. The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol. Cell 22:783-794. [DOI] [PubMed] [Google Scholar]
- 8.Bode, A. M., and Z. Dong. 2004. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 4:793-805. [DOI] [PubMed] [Google Scholar]
- 9.Brown, J. L., D. Mucci, M. Whiteley, M. L. Dirksen, and J. A. Kassis. 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1:1057-1064. [DOI] [PubMed] [Google Scholar]
- 10.Caretti, G., M. Di Padova, B. Micales, G. E. Lyons, and V. Sartorelli. 2004. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 18:2627-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, L., and J. Chen. 2003. MDM2-ARF complex regulates p53 sumoylation. Oncogene 22:5348-5357. [DOI] [PubMed] [Google Scholar]
- 12.Furlong, E. E. M., T. Rein, and F. Martin. 1996. YY1 and NF1 both activate the human p53 promoter by alternatively binding to a composite element, and YY1 and E1A cooperate to amplify p53 promoter activity. Mol. Cell. Biol. 16:5933-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia, E., C. Marcos-Gutierrez, M. del Mar Lorente, J. C. Moreno, and M. Vidal. 1999. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 18:3404-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill, G. 2005. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15:536-541. [DOI] [PubMed] [Google Scholar]
- 15.Gill, G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18:2046-2059. [DOI] [PubMed] [Google Scholar]
- 16.Gordon, S., G. Akopyan, H. Garban, and B. Bonavida. 2006. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 25:1125-1142. [DOI] [PubMed] [Google Scholar]
- 17.Gronroos, E., A. A. Terentiev, T. Punga, and J. Ericsson. 2004. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc. Natl. Acad. Sci. USA 101:12165-12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross, M., R. Yang, I. Top, C. Gasper, and K. Shuai. 2004. PIASy-mediated repression of the androgen receptor is independent of sumoylation. Oncogene 23:3059-3066. [DOI] [PubMed] [Google Scholar]
- 19.Hay, R. T. 2005. SUMO: a history of modification. Mol. Cell 18:1-12. [DOI] [PubMed] [Google Scholar]
- 20.Hochstrasser, M. 2001. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 107:5-8. [DOI] [PubMed] [Google Scholar]
- 21.Inouye, C. J., and E. Seto. 1994. Relief of YY1-induced transcriptional repression by protein-protein interaction with the nucleolar phosphoprotein B23. J. Biol. Chem. 269:6506-6510. [PubMed] [Google Scholar]
- 22.Ji, Z., C. Degerny, N. Vintonenko, J. Deheuninck, B. Foveau, C. Leroy, J. Coll, D. Tulasne, J. L. Baert, and V. Fafeur. 2007. Regulation of the Ets-1 transcription factor by sumoylation and ubiquitinylation. Oncogene 26:395-406. (First published 24 July 2006; doi: 10.1038/sj.onc.1209789.) [DOI] [PubMed] [Google Scholar]
- 23.Johnson, E. S. 2004. Protein modification by SUMO. Annu. Rev. Biochem. 73:355-382. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, E. S., and A. A. Gupta. 2001. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106:735-744. [DOI] [PubMed] [Google Scholar]
- 25.Kagey, M. H., T. A. Melhuish, S. E. Powers, and D. Wotton. 2005. Multiple activities contribute to Pc2 E3 function. EMBO J. 24:108-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kagey, M. H., T. A. Melhuish, and D. Wotton. 2003. The polycomb protein Pc2 is a SUMO E3. Cell 113:127-137. [DOI] [PubMed] [Google Scholar]
- 27.Kahyo, T., T. Nishida, and H. Yasuda. 2001. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8:713-718. [DOI] [PubMed] [Google Scholar]
- 28.Kotaja, N., U. Karvonen, O. A. Jänne, and J. J. Palvimo. 2002. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 22:5222-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kracklauer, M. P., and C. Schmidt. 2003. At the crossroads of SUMO and NF-kappaB. Mol. Cancer 2:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwek, S. S., J. Derry, A. L. Tyner, Z. Shen, and A. V. Gudkov. 2001. Functional analysis and intracellular localization of p53 modified by SUMO-1. Oncogene 20:2587-2599. [DOI] [PubMed] [Google Scholar]
- 31.Last, T. J., A. J. van Wijnen, M. J. Birnbaum, G. S. Stein, and J. L. Stein. 1999. Multiple interactions of the transcription factor YY1 with human histone H4 gene regulatory elements. J. Cell Biochem. 72:507-516. [PubMed] [Google Scholar]
- 32.Lee, J. S., K. M. Galvin, R. H. See, R. Eckner, D. Livingston, E. Moran, and Y. Shi. 1995. Relief of YY1 transcriptional repression by adenovirus E1A is mediated by E1A-associated protein p300. Genes Dev. 9:1188-1198. [DOI] [PubMed] [Google Scholar]
- 33.Lee, T. C., Y. Zhang, and R. J. Schwartz. 1994. Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene 9:1047-1052. [PubMed] [Google Scholar]
- 34.Long, J., D. Zuo, and M. Park. 2005. Pc2-mediated sumoylation of Smad-interacting protein 1 attenuates transcriptional repression of E-cadherin. J. Biol. Chem. 280:35477-35489. [DOI] [PubMed] [Google Scholar]
- 35.Lund, A. H., and M. van Lohuizen. 2004. Epigenetics and cancer. Genes Dev. 18:2315-2335. [DOI] [PubMed] [Google Scholar]
- 36.Mabb, A. M., S. M. Wuerzberger-Davis, and S. Miyamoto. 2006. PIASy mediates NEMO sumoylation and NF-kappaB activation in response to genotoxic stress. Nat. Cell Biol. 8:986-993. [DOI] [PubMed] [Google Scholar]
- 37.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 38.Melchior, F., M. Schergaut, and A. Pichler. 2003. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 28:612-618. [DOI] [PubMed] [Google Scholar]
- 39.Minty, A., X. Dumont, M. Kaghad, and D. Caput. 2000. Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J. Biol. Chem. 275:36316-36323. [DOI] [PubMed] [Google Scholar]
- 40.Miyauchi, Y., S. Yogosawa, R. Honda, T. Nishida, and H. Yasuda. 2002. Sumoylation of Mdm2 by protein inhibitor of activated STAT (PIAS) and RanBP2 enzymes. J. Biol. Chem. 277:50131-50136. [DOI] [PubMed] [Google Scholar]
- 41.Muller, S., C. Hoege, G. Pyrowolakis, and S. Jentsch. 2001. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell. Biol. 2:202-210. [DOI] [PubMed] [Google Scholar]
- 42.Nelson, V., G. E. Davis, and S. A. Maxwell. 2001. A putative protein inhibitor of activated STAT (PIASy) interacts with p53 and inhibits p53-mediated transactivation but not apoptosis. Apoptosis 6:221-234. [DOI] [PubMed] [Google Scholar]
- 43.O'Carroll, D., S. Erhardt, M. Pagani, S. C. Barton, M. A. Surani, and T. Jenuwein. 2001. The Polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21:4330-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palko, L., H. W. Bass, M. J. Beyrouthy, and M. M. Hurt. 2004. The Yin Yang-1 (YY1) protein undergoes a DNA-replication-associated switch in localization from the cytoplasm to the nucleus at the onset of S phase. J. Cell Sci. 117:465-476. [DOI] [PubMed] [Google Scholar]
- 45.Petkova, V., M. J. Romanowski, I. Sulijoadikusumo, D. Rohne, P. Kang, T. Shenk, and A. Usheva. 2001. Interaction between YY1 and the retinoblastoma protein. Regulation of cell cycle progression in differentiated cells. J. Biol. Chem. 276:7932-7936. [DOI] [PubMed] [Google Scholar]
- 46.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 47.Rezai-Zadeh, N., X. Zhang, F. Namour, G. Fejer, Y. D. Wen, Y. L. Yao, I. Gyory, K. Wright, and E. Seto. 2003. Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1. Genes Dev. 17:1019-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riggs, K. J., S. Saleque, K. K. Wong, K. T. Merrell, J. S. Lee, Y. Shi, and K. Calame. 1993. Yin-yang 1 activates the c-myc promoter. Mol. Cell. Biol. 13:7487-7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubinson, D. A., C. P. Dillon, A. V. Kwiatkowski, C. Sievers, L. Yang, J. Kopinja, D. L. Rooney, M. M. Ihrig, M. T. McManus, F. B. Gertler, M. L. Scott, and L. Van Parijs. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 33:401-406. [DOI] [PubMed] [Google Scholar]
- 50.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sacher, M., B. Pfander, C. Hoege, and S. Jentsch. 2006. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nat. Cell Biol. 8:1284-1290. [DOI] [PubMed] [Google Scholar]
- 52.Schlisio, S., T. Halperin, M. Vidal, and J. R. Nevins. 2002. Interaction of YY1 with E2Fs, mediated by RYBP, provides a mechanism for specificity of E2F function. EMBO J. 21:5775-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seto, E., B. Lewis, and T. Shenk. 1993. Interaction between transcription factors Sp1 and YY1. Nature 365:462-464. [DOI] [PubMed] [Google Scholar]
- 54.Shi, Y., J. S. Lee, and K. M. Galvin. 1997. Everything you have ever wanted to know about Yin Yang 1 … . Biochim. Biophys. Acta 1332:F49-F66. [DOI] [PubMed] [Google Scholar]
- 55.Shi, Y., E. Seto, L. S. Chang, and T. Shenk. 1991. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67:377-388. [DOI] [PubMed] [Google Scholar]
- 56.Shrivastava, A., S. Saleque, G. V. Kalpana, S. Artandi, S. P. Goff, and K. Calame. 1993. Inhibition of transcriptional regulator Yin-Yang-1 by association with c-Myc. Science 262:1889-1892. [DOI] [PubMed] [Google Scholar]
- 57.Srinivasan, L., and M. L. Atchison. 2004. YY1 DNA binding and PcG recruitment requires CtBP. Genes Dev. 18:2596-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sui, G., E. B. Affar, Y. Shi, C. Brignone, N. R. Wall, P. Yin, M. Donohoe, M. P. Luke, D. Calvo, S. R. Grossman, and Y. Shi. 2004. Yin Yang 1 is a negative regulator of p53. Cell 117:859-872. [DOI] [PubMed] [Google Scholar]
- 59.Sui, G., and Y. Shi. 2005. Gene silencing by a DNA vector-based RNAi technology. Methods Mol. Biol. 309:205-218. [DOI] [PubMed] [Google Scholar]
- 60.Takahashi, Y., T. Kahyo, E. A. Toh, H. Yasuda, and Y. Kikuchi. 2001. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 276:48973-48977. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi, Y., A. Toh-e, and Y. Kikuchi. 2001. A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene 275:223-231. [DOI] [PubMed] [Google Scholar]
- 62.Thomas, M. J., and E. Seto. 1999. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 236:197-208. [DOI] [PubMed] [Google Scholar]
- 63.Usheva, A., and T. Shenk. 1996. YY1 transcriptional initiator: protein interactions and association with a DNA site containing unpaired strands. Proc. Natl. Acad. Sci. USA 93:13571-13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, L., J. L. Brown, R. Cao, Y. Zhang, J. A. Kassis, and R. S. Jones. 2004. Hierarchical recruitment of polycomb group silencing complexes. Mol. Cell 14:637-646. [DOI] [PubMed] [Google Scholar]
- 65.Wu, F., and A. S. Lee. 2001. YY1 as a regulator of replication-dependent hamster histone H3.2 promoter and an interactive partner of AP-2. J. Biol. Chem. 276:28-34. [DOI] [PubMed] [Google Scholar]
- 66.Yang, W. M., Y. L. Yao, J. M. Sun, J. R. Davie, and E. Seto. 1997. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J. Biol. Chem. 272:28001-28007. [DOI] [PubMed] [Google Scholar]
- 67.Yao, Y.-L., W.-M. Yang, and E. Seto. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21:5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou, Q., and D. A. Engel. 1995. Adenovirus E1A243 disrupts the ATF/CREB-YY1 complex at the mouse c-fos promoter. J. Virol. 69:7402-7409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou, Q., R. W. Gedrich, and D. A. Engel. 1995. Transcriptional repression of the c-fos gene by YY1 is mediated by a direct interaction with ATF/CREB. J. Virol. 69:4323-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.