Abstract
Nuclear poly(A) polymerase (PAP) polyadenylates nascent mRNAs, promoting their nuclear export, stability, and translation, while the related cytoplasmic polymerase GLD-2 activates translation of deadenylated mRNAs. Here we characterize the biochemical activity of fission yeast Schizosaccharomyces pombe Cid1, a putative cytoplasmic PAP implicated in cell cycle checkpoint controls. Surprisingly, Cid1 has robust poly(U) polymerase activity in vitro, especially when isolated in native multiprotein complexes. Furthermore, we found that upon S-phase arrest, the 3′ ends of actin mRNAs were posttranscriptionally uridylated in a Cid1-dependent manner. Finally, Hs2 (ZCCHC6), a human ortholog of Cid1, shows similar activity. These data suggest that uridylation of mRNA forms the basis of an evolutionarily conserved mechanism of gene regulation.
Pre-mRNA undergoes various co- and posttranscriptional modifications, such as the addition of the 5′ cap structure, splicing, cleavage at the poly(A) site, and polyadenylation (36). These modifications are thought to stabilize the RNA, enable export from the nucleus, and stimulate translation (13, 24). Stabilization of mRNA by the poly(A) tail is dependent on poly(A) binding protein (Pab1) (6), which requires a tail of at least 11 adenylyl residues to bind RNA in vitro (9, 20). In addition, increasing poly(A) tail length enhances the translational rate (35, 42, 43). The loss of the poly(A) tail is often the initial step in the degradation of mRNA, usually occurring before both 5′→3′ degradation and exosome-mediated 3′→5′ degradation (8, 45). Thus, control of poly(A) tail length represents a key step in the regulation of gene expression.
In the cytoplasm, poly(A) tail length is controlled by the opposing functions of deadenylation and poly(A) addition. In contrast to the canonical nuclear poly(A) polymerase (PAP), cytoplasmic PAPs are thought to extend existing poly(A) tails of mature messages, thereby stabilizing these transcripts and/or increasing their translation. We previously identified Cid1 as a cytoplasmic PAP in Schizosaccharomyces pombe (38). The cid1 gene was identified in an overexpression screen as conferring resistance to the combination of hydroxyurea, an inhibitor of ribonucleotide reductase and, thus, DNA replication, and caffeine, which overrides the corresponding S-M checkpoint (51). Cid1 is one of a family of six related proteins in S. pombe; these proteins are involved in a wide variety of processes, such as the RNA interference pathway and the turnover of aberrant nuclear RNAs (29, 55). Cid13 is another cytoplasmic PAP in fission yeast that has been shown to target mRNA encoding the small subunit of ribonucleotide reductase (41). cid13Δ mutant cells have reduced deoxynucleoside triphosphate pools and are therefore sensitive to hydroxyurea. Cytoplasmic PAPs have also been described in metazoans. For instance, in maturing Xenopus laevis oocytes, xGld-2 extends the poly(A) tails of its target mRNAs (2). Recently, it has been shown that PARN, a poly(A)-specific RNase, and xGld-2 reside in the same ribonucleoprotein complex (19). Though xGld-2 acts constitutively on its target RNAs, the opposing action of PARN maintains short poly(A) tails on these messages. Once the oocytes mature, PARN is expelled and the message is polyadenylated by xGld-2 by default. Other homologs, identified in Caenorhabditis elegans, mice, and humans, are thought to be involved in such processes as germ cell development and neuronal synaptic plasticity (2, 30, 40, 50). However, not all of these novel polymerases are involved in stabilizing transcripts. The only Saccharomyces cerevisiae members of the Cid1 family, Trf4 and Trf5, have been shown to function in a nuclear RNA surveillance pathway as part of the TRAMP complex, which activates RNA for exosome-mediated degradation by the addition of an oligo(A) tail (18, 21, 48, 56). This oligoadenylation pathway may share a common evolutionary origin with an analogous pathway in bacteria. In S. pombe, Cid14 has been identified as the functional homolog of Trf4/Trf5 (55), and a similar process has also been implicated in the degradation of pre-mRNA species in human cells (54). The ability of an oligo(A) tail to destabilize RNA, while a poly(A) tail stabilizes it, may well be linked to the inability of an oligo(A) tail to bind Pab1.
While the presence and length of a poly(A) tail are key determinants of RNA stability, several studies have highlighted the importance of other oligonucleotide tails. For instance, the addition of uridylyl residues has been implicated in many RNA metabolic pathways, such as RNA editing in trypanosomes (1), microRNA (miRNA)-directed mRNA cleavage (22, 44), miRNA stability (22), and U6 snRNA 3′ end formation and maintenance (5, 47). Investigations of miRNA-directed cleavage in Arabidopsis thaliana revealed that the 5′ product of the cleavage event was often oligouridylated; this modification may activate the RNA decay machinery (44). The addition of such oligonucleotide tracts after miRNA-directed cleavage is likely to be conserved, since a similar phenomenon occurs in the generation of mature Epstein-Barr virus DNA polymerase mRNA (12, 34). Posttranscriptional oligouridylation of other noncoding RNAs, such as U6 snRNA, has also been observed (5). While most U6 molecules are blocked with a cyclic 2′,3′-phosphate 3′ end group (25), it is now thought that the oligouridylation of U6 acts to regenerate molecules that do not yet contain the terminal cyclic phosphate group and are therefore subject to trimming by various exonucleases (5). The enzyme responsible for posttranscriptional U6 terminal uridylyl-transferase (TUTase) activity in human cells has recently been identified and is a member of the Cid1 family (46). Since uridylation of polyadenylated mRNAs has not been observed, it has been suggested that this modification is restricted to small, noncoding RNAs or cleaved transcripts, which are targeted for decay.
In our initial investigations of Cid1 (38), we noticed that in addition to having PAP activity, Cid1 possessed substantial poly(U) polymerase (PUP) activity, though it was a weak poly(C) and poly(G) polymerase (PCP and PGP, respectively). Here we show that the PUP activity of recombinant Cid1 produced in Escherichia coli out-competes its PAP activity under physiologically relevant conditions. In addition, Cid1-containing complexes purified from S. pombe also possess strong PUP activity but weak PAP activity. Critically, we demonstrate that polyadenylated actin mRNA is subject to Cid1-dependent uridylation during S-phase arrest; these data underscore a role for Cid1 PUP activity in vivo. The Cid1 ortholog Hs2 shows strikingly similar nucleotide preferences when it is isolated in a complex from human cells. These data are the first to demonstrate nonadenylyl tailing of polyadenylated mRNAs and suggest that uridylation may represent a conserved pathway of mRNA regulation.
MATERIALS AND METHODS
Primers.
All oligonucleotide sequences are shown in Table S4 in the supplemental material.
Fission yeast strains and methods.
The conditions for growth and maintenance were as described previously (28). S. pombe strains are described in the figure legends. Strains were grown at 30°C, except where stated, in yeast extract with supplements (YE5S) or appropriately supplemented Edinburgh minimal medium (EMM2). When necessary, gene expression from plasmids containing the nmt1 promoter (26) was repressed by the addition of 30 μM thiamine to the growth medium. RNA was isolated as described in reference 28.
Plasmids and strains.
pGEX6P-1 truncated Cid1 (tCid1) was generated with a 1.2-kb BamHI-XhoI product by an Expand PCR system (Roche) with primers 3′ XhoI and OR1. pREPNTAP-Cid1 was kindly provided by A. Stevenson. Primers 5′ NTAPcid1 and 3′ NTAPcid1 were used to generate a SalI-NotI PCR product encoding the Cid1 DADA mutation, which was then cloned into pREPNTAP to create pREPNTAP-Cid1 DADA.
To create pcDNA-Hs2-TAP, Hs2 was amplified from the HeLa cell cDNA and from clone KIAA1711 (Kazusa DNA Research Institute, Japan) with primers Hs2 5′ and Hs2 3′. Once generated, the Hs2 fragment was digested with KpnI and NotI and then cloned into pcDNA3.1(+) (Invitrogen) to generate pcDNA-Hs2. The TAP sequence was subcloned from pcDNA-TAP (37) with NotI and XbaI into pcDNA-HS2 to make the plasmid pcDNA-Hs2-TAP. To create pcDNA-Hs2/DADA-TAP, the 5′ half of the Hs2 DADA mutant was amplified with the primers DADA-R and Hs2 5′; the 3′ half of Hs2 DADA was amplified with the primers DADA-F and Hs2 3′. Hs2 DADA was then formed by stitch PCR. Once generated, Hs2-DADA was digested and cloned as described above.
Preparation of recombinant protein.
E. coli BL21(DE3) pLysS cells, transformed with pGEX6P-1 tCid1, were grown to an A600 of 0.5, equilibrated to the induction temperature of 24°C, and induced with 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG). After 20 h, cells were harvested and pellets were frozen at −20°C. These pellets were then thawed on ice, and the cells were lysed with BugBuster (Novagen) supplemented with benzonase (Novagen). Lysate was then clarified by centrifugation at 16,000 × g for 25 min at 4°C. The supernatant was incubated with 40 μl of glutathione-Sepharose (Amersham), rocking at 4°C for 1 h. The beads were washed four times with HEPES-buffered saline (HBS) (40 mM HEPES [pH 7], 200 mM NaCl) and incubated overnight with 1 μg 3C protease at 4°C (kindly provided by J. Endicott). The supernatant was removed and again incubated with glutathione-Sepharose for 1 h. Finally, the supernatant was collected after 30 s of centrifugation at 16,000 × g and the protein concentration was determined by the Bradford assay. tCid1 (2 to 4 pmol) was then used for polymerase assays as described below.
Tandem affinity purification of Cid1 complexes.
Expression and immunoglobulin G (IgG) purification of TAP-Cid1 were similar to those described by Motamedi et al. (29), with the following modifications. After incubation of the cellular extract with IgG FastFlow Sepharose slurry (Amersham), the Sepharose beads were washed twice with cold lysis buffer (6 mM Na2HPO4, 4 mM NaH2PO4, 1% NP-40, 2 mM EGTA, 50 mM NaF, 0.1 mM Na3VO4) and then three times with cold HBS. The IgG-Sepharose beads (2 to 4 μl) with immobilized Cid1 complexes were then used for polymerase assays as described below.
Tandem affinity purification of Hs2 complexes.
The tandem affinity purification method (37) was used to purify Hs2 protein. Briefly, 293T cells in an 80-mm dish were transfected with 10 μg of pcDNA-Hs2-TAP using 20 μl of Lipofectamine 2000 transfection reagent (Invitrogen). Cells were harvested at 30 h posttransfection, and lysates were prepared by resuspending cells in Tris lysis buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.5% Igepal CA-630 [Sigma], 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 1 Complete Mini, EDTA-free protease inhibitor cocktail tablet [Roche]/10 ml of solution) on ice. Hs2-TAP was purified by incubating 350 μl of cell lysate with IgG FastFlow Sepharose slurry (Amersham) at 4°C for 2 h. The Sepharose beads were washed four times with wash buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.1% Igepal CA-630, 1 mM PMSF) and three times with cold HBS. IgG-Sepharose beads (4.5 μl) with immobilized Hs2 complexes were then used for the polymerase assays described below.
In vitro assays.
Template-independent polymerization assays were carried out as described previously (38, 49), using, when appropriate, 0.5 mM ATP, CTP, GTP, or UTP (Promega). Under physiologically relevant conditions, CTP, GTP, and UTP were each used at a final concentration of 0.05 mM. When appropriate, 120 units of yeast PAP (Amersham) was used for polymerization reaction mixtures. Polymerization reaction mixtures were incubated for 30 to 45 min at 37°C. When appropriate, Cid1-tailed RNA was digested with 1 μg RNase A (Sigma) and/or 10 units RNase T1 (Roche) for 10 to 30 min. Reaction products were separated by 7 M urea-12% polyacrylamide gel electrophoresis. Analysis was preformed with AIDA software.
To determine nucleotide incorporation, reactions were performed as described above with the addition of either [α-32P]ATP (Amersham) or [α-32P]UTP (Amersham). In noncompetitive reactions, cold nucleotide was also included (0.5 mM ATP or 0.05 mM UTP). In competitive reactions, cold nucleotides were used at the following concentrations: 0.5 mM ATP, 0.05 mM CTP, 0.05 mM GTP, and 0.05 mM UTP. After incubation, reactions were stopped with the addition of STOP buffer (20 mM Tris-Cl [pH 7.5], 0.1 M NaCl, 10 mM EDTA). Measurement of incorporation was then carried out as described previously (7). Briefly, after reaction mixtures were absorbed on GF/C discs (Whatman), the discs were placed in 5% trichloroacetic acid (Sigma). The discs were washed five times in 5% trichloroacetic acid, once in ethanol (Sigma), and finally once in acetone (BDH Chemicals, United Kingdom). After drying, incorporation was measured by scintillation counting.
To perform ATPase assays, reactions were performed as described above with Cid1 or Cid1 DADA complexes; however, these reaction mixtures lacked (A)15 RNA and contained [γ-32P]ATP (Amerhsam) and 0.5 mM ATP. As a negative control, reaction mixtures without protein were included; as a positive control, 120 units of yeast PAP were incubated with unlabeled (A)15 in addition to [γ32P]ATP and 0.5 mM ATP. At the appropriate time points, samples were added to STOP buffer. To determine free Pi or PPi, samples were treated as described previously (53). Briefly, after the addition of 5% activated charcoal (Sigma) in 50 mM NaH2PO4, samples were incubated on ice for 15 min. Samples were clarified by centrifugation at 16,000 × g, and the extraction was repeated. 32P release was then measured by scintillation counting. To calculate the percentage of hydrolyzed ATP, the background degradation was subtracted from each condition, and then, using a counting efficiency of 80%, counts per minute were converted to μCi.
HSC-RACE analysis.
Hybrid-selection circularized rapid amplification of cDNA ends (HSC-RACE) analysis of actin mRNA was performed as described previously (54). As described elsewhere (10), T7 transcription of a template made by PCR amplification using primers 5′ T7 actin and 3′ T7 actin was used to generate the biotinylated probe. To release selected RNA from streptavidin-coated paramagnetic beads (Promega), 50 pmol of the specific DNA primer RNase H actin and, when appropriate, 50 pmol of oligo(dT) were added. RNA, ligated by T4 RNA ligase (New England Biolabs), was reverse transcribed using the primer RT Actin with SuperScript II (Invitrogen) according to the manufacturer's guidelines. One-twentieth of the cDNA was PCR amplified with the divergent primers RT Actin and Seq Actin, using Taq polymerase (Cancer Research UK). When appropriate, [α-32P]dCTP (Amersham) was included in PCRs. PCR products were cloned into pCR2.1 (Invitrogen) following the manufacturer's guidelines and then sequenced.
Northern blot analysis.
Ten micrograms of RNA was separated by 7 M urea-12% polyacrylamide gel electrophoresis. After being transferred to Hybond-N+ membranes (Amersham), RNA was cross-linked using a UV cross-linker (Hoefer) according to the manufacturer's guidelines. Membranes were then prehybridized in ExpressHyb hybridization solution (BD Biosciences). Since U6 contains an intron, to generate a probe for the mature message, U6 RNA was reverse transcribed by SuperScript II with the primer 3′ U6. cDNA was then amplified with 3′ U6 and 5′ U6. PCR products were labeled with [α32P]dCTP (Amerhsam) and the Rediprime II labeling system (Amersham) following the manufacturer's instructions. After overnight hybridization at 60°C, membranes were washed twice with 2× SSC (0.3 NaCl, 0.03 M sodium citrate [pH 7]) with 0.05% sodium dodecyl sulfate (SDS) for 20 min at 40°C. Next, membranes were washed twice with 0.1× SSC (0.015 NaCl, 1.5 mM sodium citrate [pH 7]) with 0.1% SDS for 20 min at 40°C. Finally, membranes were exposed to PhosphorImager screens.
RESULTS
Recombinant Cid1 shows a preference for UTP over ATP.
In order to better characterize the biochemical properties of Cid1, we first used recombinant protein produced in E. coli. Because full-length Cid1 was unstable during purification (data not shown), tCid1, which lacked the first 31 residues and had increased stability, was used for further experiments. The preparation was determined to be 95% pure as judged by Coomassie staining (Fig. 1A).
FIG. 1.
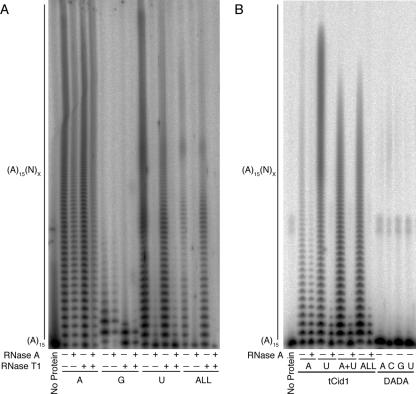
Recombinant tCid1 is both a strong PAP and PUP. (A) After induction with IPTG, glutathione S-transferase-tagged tCid1 was isolated from E. coli BL21(DE3) pLysS cells with glutathione-Sepharose. tCid1 was released from the glutathione-Sepharose by cleavage with 3C protease. (B) tCid1 was incubated with a 5′-labeled (A)15 primer and 0.5 mM of either ATP, CTP, GTP, or UTP for 30 min at 37°C (A, C, G, or U, respectively). Reaction products were then separated by 12% urea-polyacrylamide gel electrophoresis. As a negative control, (A)15 was incubated with ATP but without tCid1 (No Protein). (C to F) Densitometry plots of the ATP, CTP, GTP, and UTP lanes in panel B.
When tCid1 was incubated with a 5′ 32P-labeled (A)15 primer and either ATP, CTP, GTP, or UTP (Fig. 1B), it displayed activity similar to that described previously (38): tCid1 had more PUP than PAP activity, with relatively weak PCP and PGP activity. This activity was specific to Cid1 since the catalytically inactive proteins with mutation D101A, D103A (DADA), or D160A showed no activity (Fig. 2B and data not shown). The PUP activity was stronger than the PAP activity of tCid1 as judged by two different criteria. First, with ATP, while 44% of tCid1-generated products were extended by more than 30 nucleotides, when UTP was present, 59% of products were extended by this amount. This accumulation of high-molecular-weight species is clearly visible on the densitometry plot of the polyacrylamide gel (Fig. 1C to F; also see Fig. S1 in the supplemental material). The stronger PUP activity of tCid1 was also reflected in the average number of nucleotides added to each primer. With CTP and GTP, tCid1 had weak activity and added 1.5 nucleotides; with ATP, tCid1 added 34.5 nucleotides. However, with UTP, the average tail extension was 51 nucleotides. It should also be noted that when UTP was the nucleotide donor, reaction products showed a periodicity, as seen by the relative accumulation of species at approximately 15 nucleotide intervals (Fig. 1B and F). In comparison, when ATP was used, the tCid1-generated products did not show such a distribution (Fig. 1C). This may be due to base pairing between the Cid1-generated poly(U) tail and the (A)15 primer. We consider it unlikely that the observed PUP activity is an in vitro artifact given the markedly different structures of adenine and uracil. In contrast, despite the structural similarities between ATP and GTP, the PGP activity of Cid1 was negligible in comparison to PAP and PUP activity.
FIG. 2.
Recombinant tCid1 prefers to incorporate UMP over AMP. (A) tCid1 was incubated with a 5′-labeled (A)15 primer and a 0.5 mM concentration of various nucleotides: ATP, GTP, and UTP or ATP, CTP, GTP, and UTP (A, G, U, or ALL, respectively). These reaction products were then incubated with RNase A, RNase T1, or both, as indicated, and separated by gel electrophoresis. (B) tCid1 was incubated with 0.5 mM ATP (A), 0.5 mM UTP (U), 0.5 mM ATP and 0.05 mM UTP (AU), or 0.5 mM ATP, 0.05 mM CTP, 0.05 GTP, and 0.05 mM UTP (ALL). The DADA mutant was incubated with either 0.5 mM ATP, CTP, GTP, or UTP (A, C, G, or U, respectively).
In order to determine the relative strengths of these activities, we repeated the assay, this time incubating tCid1 with several nucleotides simultaneously. Products were then subjected to an RNase A-RNase T1 double digestion (Fig. 2A). RNase A cleaves after pyrimidine residues, while RNase T1 cleaves after guanosines. By employing both RNases, a heteropolymeric RNA is digested to oligo(A) tracts with a terminal cytidylyl, guanylyl, or uridylyl residue. Therefore, with a 5′-labeled (A)15 primer, the RNase double digestion allows estimation of the position of the first-incorporated nonadenylyl residue (although because both RNases leave a 3′ phosphate, degradation products migrate slightly faster on gel electrophoresis than might be expected).
Surprisingly, when all four nucleotides were added to the same reaction mixture in equimolar ratios, the Cid1-generated tails were completely cleaved by RNase A, though not significantly by RNase T1 (Fig. 2A). Thus, tCid1 initially incorporated either a cytidylyl or uridylyl residue. Because of the weak PCP activity, as shown in Fig. 1B, we consider it unlikely that a cytidylyl residue was the first to be incorporated. The resistance of tCid1-generated products to RNase T1 treatment shows that the polymerase did not appreciably incorporate GMP when presented with all four nucleotides. The products generated in this situation also displayed the periodic product accumulation mentioned above, suggesting not only that uridine was the first nucleotide incorporated but also that tCid1-generated tails contain oligo(U) tracts. The slight degradation of homopolymeric poly(A) tails upon RNase A treatment may reflect nonspecific activity by the endoribonuclease. However, in comparison, RNase A-mediated degradation was nearly complete when UTP or all four nucleotides were used. Therefore, the robust PUP activity of tCid1 is able to out-compete its PAP activity. In contrast to tCid1, the products generated by yeast PAP in the presence of all four nucleotides were refractory to digestion by both RNases A and T1 (see Fig. S2 in the supplemental material). These data strongly suggest that the ability of tCid1 to incorporate uridylyl residues instead of adenylyl residues is specific to this polymerase.
We next determined whether UTP could still out-compete ATP at physiologically relevant ratios, given that ATP is present in a 2- to 10-fold molar excess over UTP in vivo (3, 31). tCid1 was incubated with (A)15 and ATP in a 10-fold molar excess over UTP or with the combination of UTP, CTP, and GTP (Fig. 2B). Given the lack of GMP incorporation at equimolar ratios, only RNase A was used to digest the tCid1-generated products. In the presence of excess ATP, tCid1 again preferred initially to incorporate UMP as seen by the ability of RNase A to cleave tCid1 tails made in the presence of either ATP and UTP or in the presence of all four nucleotides (Fig. 2B). From these experiments, it is clear not only that tCid1 possesses strong PUP activity but also that this activity is able to out-compete its PAP activity.
Cid1 complexes from S. pombe possess enhanced PUP activity.
Unlike canonical, nuclear PAPs, Cid1 lacks an RNA recognition motif and, thus, is likely to require interacting proteins to direct its activity to specific RNAs; presumably, such associating proteins could affect the nucleotide usage of Cid1. Therefore, we isolated tandem affinity purified Cid1 complexes from S. pombe and tested the PAP, PCP, PGP, and PUP activities of these complexes (Fig. 3A). As determined by mass spectrometry, these complexes contained poly(A) binding protein and several other putative RNA-binding proteins (A. Stevenson, A. Akoulitchev, and C. J. Norbury, unpublished data). Surprisingly, tandem affinity purified Cid1 showed much weaker PAP activity than the PUP activity seen with recombinant tCid1. PCP and PGP activities were again weak in comparison to PUP activity. Tandem affinity purified Cid1 DADA complexes showed no activity, though similar amounts of Cid1 were present in the purified material, as determined by Western blotting (Fig. 3A and data not shown). The reduced PAP activity is not due to any contaminating ATPase that might reduce the effective concentration of ATP sufficiently to mask the PAP activity of Cid1 complexes (see Fig. S3 in the supplemental material). Thus, we conclude that these data are consistent with an in vivo role for Cid1 PUP activity.
FIG. 3.
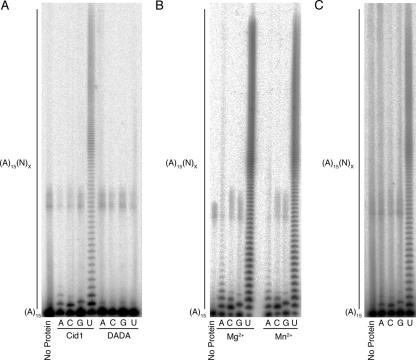
Cid1 complexes purified from S. pombe show strong PUP but weak PAP activity. (A) cid1Δ cells (h− cid1::ura4+ leu1-32 ura4-D18), transformed with pREPNTAP Cid1 or pREPNTAP DADA, were grown to saturation in minimal medium lacking thiamine. TAP-Cid1 complexes were purified with IgG-Sepharose. Polymerase assays were performed as described in the legend to Fig. 1. (B) Cid1 complexes were incubated with a 5′-labeled (A)15 primer, either 1 mM Mg2+ or 1 mM Mn2+ (shown with Mg2+ and Mn2+), and either ATP, CTP, GTP, or UTP (A, C, G, or U, respectively). (C) cdc27 cells (h− cdc27-P11 leu1-32) transformed with pREPNTAP Cid1 were grown to mid-exponential log phase in minimal medium lacking thiamine at 30°C. These cells were then shifted to 36°C for 4 h before being harvested. Purified Cid1 complexes were incubated with a 5′-labeled (A)15 primer and either ATP, CTP, GTP, or UTP (A, C, G, or U, respectively).
Nevertheless, in principle, the activity of Cid1 might be affected by the cation used in vitro. Members of the polymerase β (Pol β) superfamily, which includes Cid1, coordinate a divalent cation through the three catalytic aspartate residues; in most cases, the cation is Mg2+ (32). However, coordination of Mn2+ can occur and can affect the specificity of the polymerase due to increased reactivity (33). For instance, the in vitro PAP activities of both Trf4 and Cid13 are affected by the divalent cation used (14, 41). As Cid1 complexes still showed strong PUP activity and weak PAP, PCP, and PGP activities in the presence of either Mg2+ or Mn2+ (Fig. 3B), the activities of the polymerase appear to be unaffected by the change in cation. Given this finding, we used Mg2+ in all subsequent experiments.
We have previously speculated that Cid1 is normally inactive and that upon S-phase arrest, it acts on cytoplasmic RNAs to activate the S-M checkpoint and promote cell survival (39). Cells lacking Cid1 and containing temperature-sensitive Pol δ alleles, such as cdc27-P11, fail to arrest properly in their cell cycle after the shift to the restrictive temperature (52). It is possible that this activation reflects a change in Cid1 binding partners, which may influence nucleotide usage. To test this possibility, Cid1 complexes were purified from cdc27-P11 cells that, during exponential growth, had been shifted to the restrictive temperature. These complexes, like those isolated from exponentially growing cells, showed very strong PUP activity with weak PAP, PCP, and PGP activities (Fig. 3C). Indeed, we have isolated Cid1 complexes from cells grown under a variety of conditions but have never found them to possess processive PAP activity or lack strong PUP activity (Fig. 3 and data not shown). Thus, we conclude that the binding partners of Cid1 or some as-yet-unidentified modifications, not carried out in E. coli, either stimulate its PUP activity or inhibit its PAP activity, suggesting that the former is more relevant in vivo.
The PUP activity of Cid1 complexes is able to effectively out-compete its PAP activity.
We next determined whether Cid1 complexes, like recombinant tCid1, preferred to incorporate UMP in the presence of an excess of ATP. To do this, we first repeated the RNase digestion assay (Fig. 4A). It was clear that when Cid1 complexes from S. pombe were presented with all four nucleotides at physiological ratios, the polymerase initially incorporated UMP. To further investigate the nucleotide usage of Cid1 complexes, we performed incorporation experiments with [α-32P]ATP or [α-32P]UTP (Fig. 4B and C). Complexes were incubated with [α-32P]ATP and unlabeled ATP (for noncompetitive conditions) or with [α-32P]ATP and all four nucleotides at physiologically relevant concentrations (for competitive conditions). Cid1 did incorporate AMP in the presence of CTP, GTP, and UTP; however, this was reduced fivefold in comparison with the incorporation during noncompetitive reactions. Confirming the result obtained from the RNase digestion experiment, this experiment demonstrates that UTP is able to out-compete ATP.
FIG. 4.
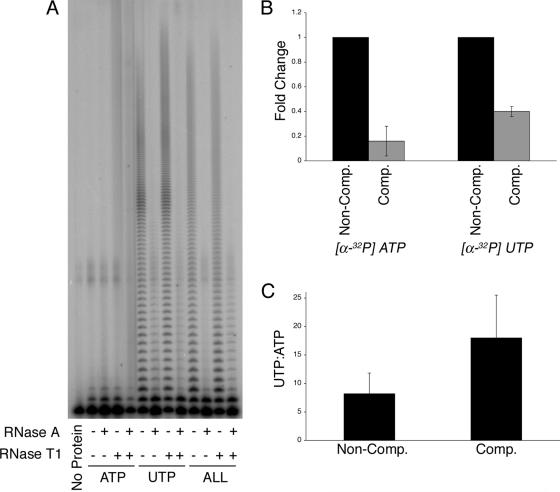
Cid1 complexes prefer to incorporate UMP over AMP. (A) Cid1 complexes were incubated with a 5′-labeled (A)15 primer and various nucleotides, as follows: 0.5 mM ATP, 0.5 mM UTP, or 0.5 mM ATP with 0.05 mM CTP, 0.05 mM GTP, and 0.05 mM UTP (ATP, UTP, or ALL, respectively). These reaction products were then incubated with RNase A, RNase T1, or both, as indicated, and then separated by gel electrophoresis. (B and C) Under noncompetitive conditions (Non-Comp.), Cid1 complexes were incubated with [α-32P]ATP with 0.5 mM ATP or with [α-32P]UTP with 0.05 mM UTP. Under competitive conditions (Comp.), Cid1 complexes were incubated with either [α-32P]ATP or [α-32P]UTP as well as 0.5 mM ATP, 0.05 mM CTP, 0.05 mM GTP, and 0.05 mM UTP. Three independent experiments were performed. Incorporation was measured by scintillation counting. (B) To determine the effect of nucleotide competition, incorporation under competitive conditions was normalized to noncompetitive conditions. (C) After normalization of the 10-fold difference in ATP and UTP levels, the ratio of incorporated [α-32P]UMP to [α-32P]AMP was obtained for both noncompetitive and competitive conditions.
When the corresponding experiment was performed with [α-32P]UTP, we observed a twofold reduction in 32P incorporation upon the addition of all four nucleotides. This reduction is unlikely to be explained by the out-competition of UTP by ATP. More likely, the reduced incorporation of [α-32P]UMP reflects the possibility that the average length of products generated in the presence of all four nucleotides was decreased relative to that observed when Cid1 was incubated with UTP alone (for example, see Fig. 2A and 4A). When incorporation levels of [α-32P]AMP and [α-32P]UMP under noncompetitive conditions were compared, it was evident that Cid1 complexes had eightfold stronger PUP activity than PAP activity. Furthermore, under competitive conditions, Cid1 incorporated on average 15 uridylyl residues for every adenylyl residue. These findings strongly suggest that Cid1 possesses PUP activity within the cell.
Nucleotide usage by Cid1 complexes depends on the RNA substrate.
Given the strong PUP activity of Cid1 when presented with an (A)15 primer, it is conceivable that in subsequent rounds of polymerization, Cid1 could be presented with 3′ ends other than a poly(A) tail, such as an oligo(U) tail. We therefore investigated the nucleotide usage by Cid1 complexes with (U)15 primers (Fig. 5A to F). As expected, Cid1 displayed strong PUP activity when incubated with (U)15. Surprisingly, when presented with (U)15, Cid1 possessed stronger PAP activity than was seen with an (A)15 primer.
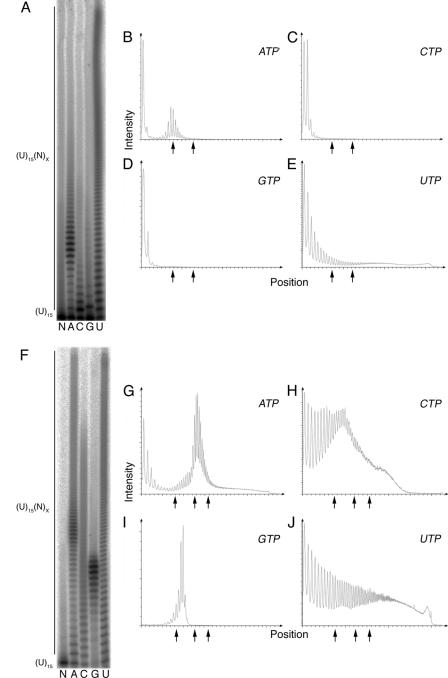
FIG. 5.
Cid1 activity and processivity vary with the nature of the RNA primer. (A) Cid1 complexes were incubated with 5′-labeled (U)15 and either 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, or 0.5 mM UTP (A, C, G, or U, respectively). As a negative control, (U)15 was incubated without Cid1 (lane N). (B to E) Densitometry plots of the ATP, CTP, GTP, and UTP lanes in panel A. Arrows mark positions of 10-, 20-, and 30-nucleotide extensions. (F) tCid1 was incubated with 5′-labeled (U)15 and either 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, or 0.5 mM UTP (A, C, G, or U, respectively). As before, (U)15 was incubated without protein (lane N). (G to J) Densitometry plots of the ATP, CTP, GTP, and UTP lanes in panel F.
These same experiments were repeated with tCid1, giving broadly similar results (Fig. 5F to J). As with the (A)15 primer (Fig. 1B), tCid1 displayed strong PAP and PUP activity with (U)15 (Fig. 5F). Surprisingly, with (U)15, tCid1 possessed enhanced PCP and PGP activities, extending primers with as many as 40 and 8 nucleotides, respectively. Nevertheless, as with those differences seen with the (A)15 primer, the observed differences between the activities of tCid1 and Cid1 complexes with (U)15 likely reflect the absence of regulatory subunits in recombinant preparations. With both (A)15 and (U)15, Cid1 complexes displayed stronger PUP activity than PAP, PCP, and PGP activities, relative to those seen with recombinant tCid1. We conclude that the PUP activity of Cid1 is likely to be dominant over its PAP activity in vivo.
Actin transcripts are uridylated upon S-phase arrest.
We next sought in vivo evidence for Cid1 PUP activity. Given the recent identification of the U6 TUTase as a member of the Cid1 family (46), we wished to determine whether Cid1 was the fission yeast ortholog of the human U6 TUTase. However, Northern blot analysis did not provide any evidence of posttranscriptional uridylation of U6 in S. pombe, nor did Cid1 affect steady-state levels of U6 transcripts (see Fig. S4 in the supplemental material). Concluding that U6 snRNA is unlikely to be a target of Cid1, we therefore began to explore the possibility that Cid1 might uridylate mRNA.
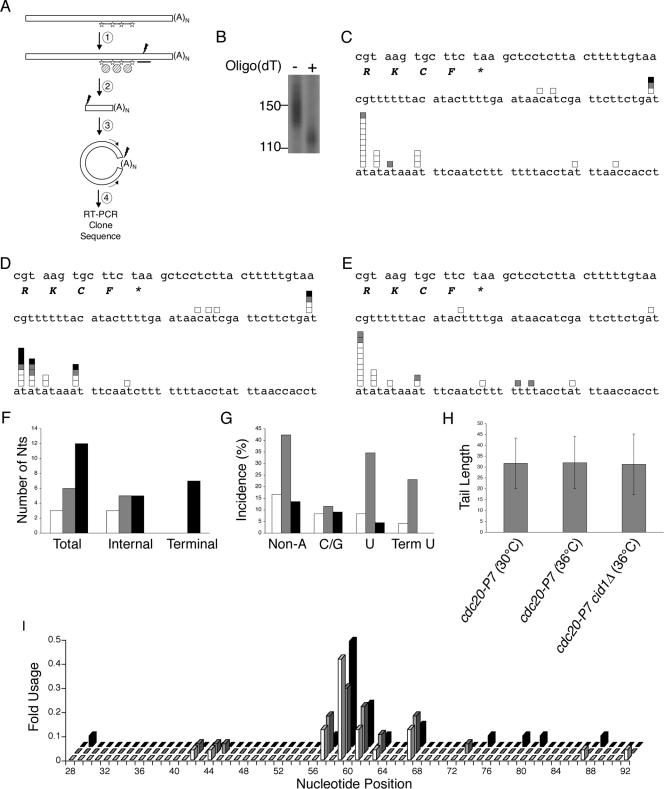
By employing an oligo(dT)-primed reverse transcription, earlier studies of the S. pombe transcriptome would have been unable to identify terminal nonadenylyl residues. Accordingly, we decided to employ a transcript-specific, tail-independent approach, 3′ HSC-RACE analysis, to investigate the presence of such residues on polyadenylated mRNAs. In this approach (54), a biotinylated probe is used to select transcripts on magnetic streptavidin beads (Fig. 6A, step 1); RNAs are then released from the beads by RNase H-directed cleavage (step 2). Cleaved RNAs are next intramolecularly ligated to capture the 3′ tail (step 3). Reverse transcription and PCR with divergent primers across the ligated junction allow subsequent sequencing of the transcript tail and poly(A) site (step 4).
FIG. 6.
Polyadenylated actin mRNAs are uridylated in a Cid1-dependent manner during S-phase arrest. (A) In HSC-RACE analysis, transcripts are first selected with a biotinylated probe (marked with stars) on streptavidin magnetic beads (striped circles) (step 1). DNA oligonucleotide-directed RNase H cleavage releases the selected transcript (step 2). T4 RNA ligase circularizes the released RNA (step 3); reverse transcription and PCR with divergent primers (arrows) allow visualization, cloning, and sequencing of products (step 4). (B) mRNA from exponentially grown h− leu1-32 (wild-type) cells was subjected to actin HSC-RACE analysis (marked with —). HSC-RACE analysis was also performed where oligo(dT) was added during the RNase H cleavage step (denoted by +). (C) HSC-RACE products of actin transcripts isolated from h− cdc20-P7 (cdc20-P7) cells growing exponentially at the permissive temperature of 30°C. The final four codons and stop codon (marked by *) are shown, as well as the 3′ UTR. White boxes indicate pure poly(A) tails; gray boxes indicate poly(A) tails with internal cytidylyl, guanylyl, or uridylyl residues; black boxes indicate poly(A) tails with a terminal uridylyl residue. (D and E) HSC-RACE products of actin transcripts isolated from h− cdc20-P7 cells (D) or from h− cdc20-P7 cid1::ura4+ ura4-D18(cdc20-P7 cid1Δ) cells (E) that, during exponential growth, had been shifted to 36°C for 4 h. (F) The numbers of total, internal, and terminal nonadenylyl residues (Nts) are shown (white, cytidylyl residues; gray, guanylyl residues; black, uridylyl residues). (G) The percentage of tails with nonadenylyl residues (total nonadenylyl [Non-A] residues, total cytidylyl and guanylyl [C/G] residues, total uridylyl [U] residues, and terminal uridylyl [Term U] residues) is shown. Tails from cdc20-P7 cells grown at the permissive temperature are plotted in white; tails from cdc20-P7 cells shifted to 36°C for 4 h are shown in gray; tails from cdc20-P7 cid1Δ cells shifted to 36°C for 4 h are shown in black. (H) The average poly(A) tail length of actin transcripts for the three conditions tested. (I) The n-fold usage of sites of poly(A) cleavage is shown (white, gray, and black coding is as shown in panel G).
In the absence of an identified target of Cid1, we decided to look at the abundant mRNA products of the actin (act1) gene. First, we analyzed mRNAs from wild-type cells to determine the average poly(A) tail length (Fig. 6B). To do this, we compared HSC-RACE products with those where oligo(dT) had been included during the RNase H step of the protocol (Fig. 6A, step 2); this inclusion results in cleavage of the poly(A) tail before intramolecular ligation and, thus, allows estimation of its length. PCR was performed in the presence of [α-32P]dCTP, and products were separated by gel electrophoresis. HSC-RACE products generated with oligo(dT) were, on average, 30 nucleotides smaller than those made without oligo(dT). We conclude that the poly(A) tails of actin transcripts are approximately 30 nucleotides long during exponential growth.
Next, we investigated the nature of actin mRNA tails isolated from conditions where Cid1 is known to be necessary for the S-M checkpoint. Cells lacking Cid1 and containing temperature-sensitive Pol ɛ alleles, such as cdc20-P7, fail to arrest properly in their cell cycle after the shift to the restrictive temperature of 36°C (52). RNA was isolated from cdc20-P7 cells that had been shifted to 36°C for 4 h during exponential growth, and HSC-RACE analysis was performed. Strikingly, a significant proportion of actin transcripts (23%) were terminally uridylated under these conditions (Fig. 6D and G; also see Table S2 in the supplemental material).
Given previous work (38, 51, 52) and our biochemical analysis, we hypothesized that this uridylation might depend on S-phase arrest and be catalyzed by Cid1. When actin mRNA tails from cdc20-P7 cells grown at the permissive temperature were examined, only 1 of 24 tails (4%) contained a terminal uridylyl residue (Fig. 6C and G; also see Table S1 in the supplemental material). This result supports a role for S-phase arrest in the uridylation of actin mRNA. Moreover, transcripts from cdc20-P7 cid1Δ cells that had been shifted to the restrictive temperature completely lacked terminal uridylation (Fig. 6E and G; also see Table S3 in the supplemental material). There were no significant differences in the incidence of internal nonadenylyl residues (Student's t test). Being condition- and Cid1-independent, these residues are likely to be derived from PCR-induced mutations and/or occasional nonadenylyl incorporation by nuclear PAP (Fig. 6F). Thus, these data suggest that this 3′ modification is dependent upon S-phase arrest and are consistent with a role for Cid1 PUP activity in vivo.
Consistent with the in vitro data showing little PAP activity in Cid1 complexes, analysis of the poly(A) tail length showed no significant difference between all three conditions tested (Fig. 6H). Like those observed in wild-type cells (Fig. 6B), poly(A) tails were, on average, 30 nucleotides in length. In addition, it is unlikely that tail length is an important factor for Cid1-dependent uridylation as there was no noticeable correlation between poly(A) tail length and terminal uridylation. Finally, while the sites of poly(A) cleavage (Fig. 6I) appear more variable than those in other organisms, there were no significant differences in these cleavage sites under the three conditions analyzed. These sites, as determined by RNase protection analysis, were also the same as those observed during exponential growth (data not shown). In conclusion, the terminal uridylation of actin mRNAs represents the only significant change observed upon S-phase arrest. Thus, we have described for the first time posttranscriptional uridylation of polyadenylated mRNAs; this modification is Cid1 dependent and is likely to occur cytoplasmically.
Cid1 PUP activity is conserved in human cells.
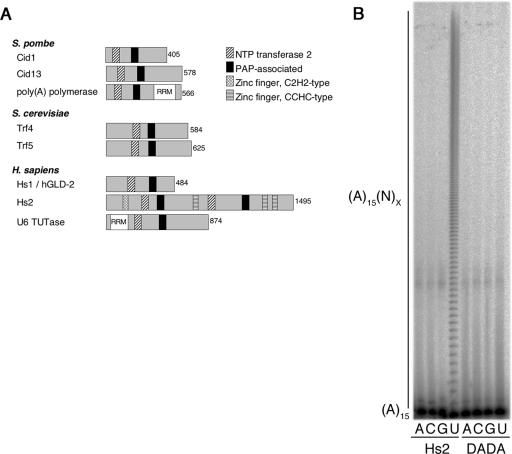
The human genome encodes seven Cid1 family polymerases, as follows: PAPD4, ZCCHC6, ZCCHC11, PAPD1, U6 TUTase (RBM21), POL5, and PAPD5. While the roles of the majority of these enzymes have yet to be determined, it is likely that they have functions similar to those of their fission yeast orthologs. In addition, while characterizing the U6 TUTase, Trippe et al. described two biochemically distinct activities capable of transferring uridylyl residues: one specific for the U6 snRNA and another that uridylated many RNAs (46). Given our findings with fission yeast Cid1, we suspected that another Cid1 family protein might be responsible for the second of these two activities.
Hs2 (ZCCHC6) is a larger protein than Cid1, containing two PAP-associated domains and two nucleotidyl transferase motifs (Fig. 7A). However, only one of these sites is likely to be catalytically active since the other lacks the aspartate triad necessary for cation coordination. In addition, Hs2 possesses several zinc-knuckle motifs, which may allow it, as well as its binding partners, to bind RNA. We purified TAP-tagged Hs2 following its expression in 293T cells. Native tandem affinity purified Hs2 complexes were then incubated with 5′-labeled (A)15 and either ATP, CTP, GTP, or UTP. Like Cid1 complexes isolated from S. pombe, Hs2 complexes had very little PAP, PCP, or PGP activity (Fig. 7B). Remarkably, Hs2 also displayed efficient PUP activity. We conclude that the PUP activity seen with Cid1 is conserved in Hs2. Moreover, these data suggest that the mRNA uridylation we have described in fission yeast is also likely to occur in human cells.
FIG. 7.
Cid1 uridylation activity is evolutionarily conserved. (A) The domain structure of various GLD-2/Cid-related polymerases. PAP and U6 TUTase each contain an RNA recognition motif (RRM). Note that only two of the six S. pombe Cid1-like proteins and three of the seven human proteins are shown. (B) Hs2 complexes were isolated from 293T cells transfected with either pcDNA-Hs2-TAP or pcDNA-Hs2/DADA-TAP (marked with Hs2 or DADA, respectively) on IgG-Sepharose and then incubated with 5′-labeled (A)15 and either 0.5 mM ATP, 0.5 mM CTP, 0.5 mM GTP, or 0.5 mM UTP (A, C, G, or U, respectively).
DISCUSSION
Here we have presented evidence that the PUP activity of Cid1 is stronger than its PAP activity, that actin mRNA is uridylated in a Cid1-dependent manner during S-phase arrest, and that this PUP activity is evolutionarily conserved. Use of an RNase A-RNase T1 double digestion demonstrated that recombinant tCid1 initially preferred to add uridylyl residues to an (A)15 primer. Further biochemical analysis with tandem affinity purified Cid1 complexes showed an even stronger bias toward PUP activity. Using HSC-RACE to analyze tails of actin transcripts, we have shown for the first time 3′ posttranscriptional uridylation of polyadenylated mRNA. This modification was observed only during S-phase arrest and was dependent on Cid1. Finally, Hs2, a human member of the Cid family, also displayed striking PUP activity but very weak PAP, PCP, and PGP activity. Taken together, these data suggest conservation of PUP activity in the noncanonical polymerases Cid1 and Hs2 and that this activity may form the basis of a conserved gene regulation pathway.
There have been numerous recent investigations into the polymerase activities of other Cid1 family proteins. The initial investigation of Cid13 (41) demonstrated that this polymerase utilized ATP effectively and CTP weakly. No transferase activity was seen with UTP or GTP. Studies of the S. cerevisiae Cid family proteins Trf4 and Trf5 have also described PAP activity (15, 21, 48, 56). While LaCava et al. did observe weak PGP activity in Trf4 preparations after protracted incubation (21), others have not reported any activity other than PAP for both Trf4 and Trf5 (14, 15, 48). Those studies of S. cerevisiae Cid1-related proteins used both bacterially expressed protein and tandem affinity purified complexes. Given the lack of PUP activity in other members of the Cid1 family, with the exception of the human U6 TUTase, it is likely then that the PUP activity seen with Cid1 and Hs2 is specific to these polymerases.
Our sequence analysis of actin mRNAs provides further evidence for the relevance of Cid1 PUP activity. We observed terminal uridylation during S-phase arrest in 23% of these mRNAs. In cells lacking Cid1, no such uridylation was observed. However, it was surprising, given the rampant PUP activity of tCid1 and Cid1 complexes in vitro, that only one terminal uridine was found in these cases. We suspect that the lack of longer oligo(U) tails may be because even a short oligo(U) tail could form a hairpin with the poly(A) tail; this structure would be then refractory to ligation and subsequent reverse transcription-PCR. Alternatively, it is possible that in vivo Cid1 adds only a single uridine, which is sufficient to alter the fate of the mRNA in some way. Finally, it is also possible that the addition of U-rich tails results in the rapid degradation of targeted mRNA (see below). Work is ongoing to distinguish between these possibilities and to determine whether longer oligo(U) tails are generated in vivo.
In addition, the HSC-RACE analysis of actin transcripts has allowed the sites of poly(A) cleavage to be determined. The sites of cleavage appear to be more variable than in other organisms and, unlike the situation with higher eukaryotes (36), very few transcripts (3%) were cleaved after a CA dinucleotide. This variability may reflect divergence from the AAUAAA consensus that has been suggested to be common in fission yeast genes (17); for instance, in the actin 3′ untranslated region (UTR), the poly(A) site sequence appears to be AAUAA. Previous work on actin mRNA in S. pombe (27) demonstrated the presence of two additional poly(A) sites, one 400 nucleotides downstream of the site mapped in this study and another 600 nucleotides downstream. It will be interesting to investigate whether transcripts derived from the downstream poly(A) sites are also subject to uridylation, as we suspect that motifs in the 3′ UTR could direct Cid1 activity.
The existence of a heterogeneous tail that contains uridylyl residues presents many questions. In particular, does such a tract stabilize or destabilize RNA? There is indirect evidence for both possibilities. Oligo(U) tailing of human U6 transcripts implies a role in RNA stabilization, as posttranscriptional uridylation is thought to regenerate a functional U6 end after the 3′ oligo(U) tract has been trimmed by exonucleases (5). Moreover, it has also been observed in vitro that oligo(U) tracts are able to protect RNA from exosome-mediated degradation (11). This protection is dependent on a cellular protein that has yet to be identified. However, several studies also suggest that a terminal oligo(U) tract may destabilize RNA. In A. thaliana, oligo(U) tails are added both to the upstream mRNA product of miRNA-directed cleavage and to the miRNAs themselves (22, 44). In both cases, such uridylation either leads to or is coincident with degradation, though a causal relationship between these tails and decay is far from established. Investigations into the degradation machinery in bacteria have suggested that oligo(U) tracts might preferentially target an RNA for degradation and that these tracts can be shortened by exoribonucleases (16, 23). Nevertheless, it is conceivable that, like poly(A) tails, the effect of oligo(U) tails may be dependent on length and cellular environment. Short oligo(A) tails created by the TRAMP complex lead to degradation of various RNAs, while long poly(A) tails, competent to bind Pab1, stabilize their RNA. Contextual conditions might similarly affect the ability of a poly(U) binding protein to bind and stabilize the RNA.
In one attractive model, the Cid1-generated oligo(U) tract on an mRNA would be able to base pair with the poly(A) tail created by the canonical, nuclear PAP. A hairpin of this sort may itself be stable enough to prevent exosome-mediated degradation, or alternatively, it may be bound by stabilizing proteins. Another possibility is that the creation of AU-rich tails might generate de novo AU-rich elements (AREs), similar to those described in the 3′UTRs of many tightly regulated mRNAs. Canonical AREs often destabilize these mRNA molecules by binding ARE-binding proteins, which then recruit the exosome and lead to rapid degradation (4). Cid1-generated tails may be fed into a specialized version of ARE-mediated decay. It is also conceivable that the addition of a Cid1 tail allows a form of translational reprogramming necessary for cell survival following S-phase arrest. In any case, the interesting problem remains of how Cid1 target RNAs are regulated as the cell recovers from arrest.
The PUP activities of Cid1 and Hs2 raise many questions, not only about the roles of these polymerases in the cell cycle but also about RNA biology itself. Indeed, the use of nucleotides other than adenine in RNA tails may represent an unanticipated mechanism of gene regulation.
Supplementary Material
Acknowledgments
We thank other members of the laboratory, Steve West and Nick Proudfoot for helpful discussions and critical reading of the manuscript, Kieran Finan for assistance and advice with incorporation experiments, and Jane Endicott and Aleksandra Watson for help in the early stages of tCid1 characterization.
This work was supported by Cancer Research UK, the Medical Research Council, and the Association for International Cancer Research. O.S.R. was supported by a scholarship from the Rhodes Trust.
Footnotes
Published ahead of print on 12 March 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aphasizhev, R. 2005. RNA uridylyltransferases. Cell. Mol. Life Sci. 62:2194-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnard, D. C., K. Ryan, J. L. Manley, and J. D. Richter. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119:641-651. [DOI] [PubMed] [Google Scholar]
- 3.Cao, D., J. J. Leffert, J. McCabe, B. Kim, and G. Pizzorno. 2005. Abnormalities in uridine homeostatic regulation and pyrimidine nucleotide metabolism as a consequence of the deletion of the uridine phosphorylase gene. J. Biol. Chem. 280:21169-21175. [DOI] [PubMed] [Google Scholar]
- 4.Chen, C. Y., R. Gherzi, S. E. Ong, E. L. Chan, R. Raijmakers, G. J. Pruijn, G. Stoecklin, C. Moroni, M. Mann, and M. Karin. 2001. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107:451-464. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Y., K. Sinha, K. Perumal, and R. Reddy. 2000. Effect of 3′ terminal adenylic acid residue on the uridylation of human small RNAs in vitro and in frog oocytes. RNA 6:1277-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coller, J. M., N. K. Gray, and M. P. Wickens. 1998. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 12:3226-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook, P. R., and F. Gove. 1992. Transcription by an immobilized RNA polymerase from bacteriophage T7 and the topology of transcription. Nucleic Acids Res. 20:3591-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Decker, C. J., and R. Parker. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7:1632-1643. [DOI] [PubMed] [Google Scholar]
- 9.Deo, R. C., J. B. Bonanno, N. Sonenberg, and S. K. Burley. 1999. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell 98:835-845. [DOI] [PubMed] [Google Scholar]
- 10.Dye, M. J., and N. J. Proudfoot. 1999. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol. Cell 3:371-378. [DOI] [PubMed] [Google Scholar]
- 11.Ford, L. P., and J. Wilusz. 1999. 3′-Terminal RNA structures and poly(U) tracts inhibit initiation by a 3′->5′ exonuclease in vitro. Nucleic Acids Res. 27:1159-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furnari, F. B., M. D. Adams, and J. S. Pagano. 1993. Unconventional processing of the 3′ termini of the Epstein-Barr virus DNA polymerase mRNA. Proc. Natl. Acad. Sci. USA 90:378-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu, M., and C. D. Lima. 2005. Processing the message: structural insights into capping and decapping mRNA. Curr. Opin. Struct. Biol. 15:99-106. [DOI] [PubMed] [Google Scholar]
- 14.Haracska, L., R. E. Johnson, L. Prakash, and S. Prakash. 2005. Trf4 and Trf5 proteins of Saccharomyces cerevisiae exhibit poly(A) RNA polymerase activity but no DNA polymerase activity. Mol. Cell. Biol. 25:10183-10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houseley, J., and D. Tollervey. 2006. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 7:205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang, H., J. Liao, and S. N. Cohen. 1998. Poly(A)- and poly(U)-specific RNA 3′ tail shortening by E. coli ribonuclease E. Nature 391:99-102. [DOI] [PubMed] [Google Scholar]
- 17.Humphrey, T., P. Sadhale, T. Platt, and N. Proudfoot. 1991. Homologous mRNA 3′ end formation in fission and budding yeast. EMBO J. 10:3503-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadaba, S., A. Krueger, T. Trice, A. M. Krecic, A. G. Hinnebusch, and J. Anderson. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18:1227-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, J. H., and J. D. Richter. 2006. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell 24:173-183. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn, U., and E. Wahle. 2004. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta 1678:67-84. [DOI] [PubMed] [Google Scholar]
- 21.LaCava, J., J. Houseley, C. Saveanu, E. Petfalski, E. Thompson, A. Jacquier, and D. Tollervey. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121:713-724. [DOI] [PubMed] [Google Scholar]
- 22.Li, J., Z. Yang, B. Yu, J. Liu, and X. Chen. 2005. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 15:1501-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lisitsky, I., and G. Schuster. 1999. Preferential degradation of polyadenylated and polyuridinylated RNAs by the bacterial exoribonuclease polynucleotide phosphorylase. Eur. J. Biochem. 261:468-474.10215858 [Google Scholar]
- 24.Luna, R., S. Jimeno, M. Marin, P. Huertas, M. Garcia-Rubio, and A. Aguilera. 2005. Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol. Cell 18:711-722. [DOI] [PubMed] [Google Scholar]
- 25.Lund, E., and J. E. Dahlberg. 1992. Cyclic 2′,3′-phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNA's. Science 255:327-330. [DOI] [PubMed] [Google Scholar]
- 26.Maundrell, K. 1990. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265:10857-10864. [PubMed] [Google Scholar]
- 27.Mertins, P., and D. Gallwitz. 1987. A single intronless action gene in the fission yeast Schizosaccharomyces pombe: nucleotide sequence and transcripts formed in homologous and heterologous yeast. Nucleic Acids Res. 15:7369-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 29.Motamedi, M. R., A. Verdel, S. U. Colmenares, S. A. Gerber, S. P. Gygi, and D. Moazed. 2004. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell 119:789-802. [DOI] [PubMed] [Google Scholar]
- 30.Nakanishi, T., H. Kubota, N. Ishibashi, S. Kumagai, H. Watanabe, M. Yamashita, S. Kashiwabara, K. Miyado, and T. Baba. 2006. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev. Biol. 289:115-126. [DOI] [PubMed] [Google Scholar]
- 31.Osorio, H., E. Carvalho, M. del Valle, M. A. Gunther Sillero, P. Moradas-Ferreira, and A. Sillero. 2003. H2O2, but not menadione, provokes a decrease in the ATP and an increase in the inosine levels in Saccharomyces cerevisiae. An experimental and theoretical approach. Eur. J. Biochem. 270:1578-1589. [DOI] [PubMed] [Google Scholar]
- 32.Pelletier, H., and M. R. Sawaya. 1996. Characterization of the metal ion binding helix-hairpin-helix motifs in human DNA polymerase beta by X-ray structural analysis. Biochemistry 35:12778-12787. [DOI] [PubMed] [Google Scholar]
- 33.Pelletier, H., M. R. Sawaya, W. Wolfle, S. H. Wilson, and J. Kraut. 1996. A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase beta. Biochemistry 35:12762-12777. [DOI] [PubMed] [Google Scholar]
- 34.Pfeffer, S., M. Zavolan, F. A. Grasser, M. Chien, J. J. Russo, J. Ju, B. John, A. J. Enright, D. Marks, C. Sander, and T. Tuschl. 2004. Identification of virus-encoded microRNAs. Science 304:734-736. [DOI] [PubMed] [Google Scholar]
- 35.Preiss, T., M. Muckenthaler, and M. W. Hentze. 1998. Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA 4:1321-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 37.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 38.Read, R. L., R. G. Martinho, S. W. Wang, A. M. Carr, and C. J. Norbury. 2002. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. USA 99:12079-12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Read, R. L., and C. J. Norbury. 2002. Roles for cytoplasmic polyadenylation in cell cycle regulation. J. Cell. Biochem. 87:258-265. [DOI] [PubMed] [Google Scholar]
- 40.Rouhana, L., L. Wang, N. Buter, J. E. Kwak, C. A. Schiltz, T. Gonzalez, A. E. Kelley, C. F. Landry, and M. Wickens. 2005. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA 11:1117-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saitoh, S., A. Chabes, W. H. McDonald, L. Thelander, J. R. Yates, and P. Russell. 2002. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109:563-573. [DOI] [PubMed] [Google Scholar]
- 42.Sheets, M. D., C. A. Fox, T. Hunt, G. Vande Woude, and M. Wickens. 1994. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 8:926-938. [DOI] [PubMed] [Google Scholar]
- 43.Sheets, M. D., M. Wu, and M. Wickens. 1995. Polyadenylation of c-mos mRNA as a control point in Xenopus meiotic maturation. Nature 374:511-516. [DOI] [PubMed] [Google Scholar]
- 44.Shen, B., and H. M. Goodman. 2004. Uridine addition after microRNA-directed cleavage. Science 306:997. [DOI] [PubMed] [Google Scholar]
- 45.Shyu, A. B., J. G. Belasco, and M. E. Greenberg. 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 5:221-231. [DOI] [PubMed] [Google Scholar]
- 46.Trippe, R., E. Guschina, M. Hossbach, H. Urlaub, R. Luhrmann, and B. J. Benecke. 2006. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12:1494-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trippe, R., B. Sandrock, and B. J. Benecke. 1998. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 26:3119-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanacova, S., J. Wolf, G. Martin, D. Blank, S. Dettwiler, A. Friedlein, H. Langen, G. Keith, and W. Keller. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3:e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wahle, E., G. Martin, E. Schiltz, and W. Keller. 1991. Isolation and expression of cDNA clones encoding mammalian poly(A) polymerase. EMBO J. 10:4251-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, L., C. R. Eckmann, L. C. Kadyk, M. Wickens, and J. Kimble. 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419:312-316. [DOI] [PubMed] [Google Scholar]
- 51.Wang, S. W., C. Norbury, A. L. Harris, and T. Toda. 1999. Caffeine can override the S-M checkpoint in fission yeast. J. Cell Sci. 112:927-937. [DOI] [PubMed] [Google Scholar]
- 52.Wang, S.-W., T. Toda, R. MacCallum, A. L. Harris, and C. Norbury. 2000. Cid1, a fission yeast protein required for S-M checkpoint control when DNA polymerase δ or ɛ is inactivated. Mol. Cell. Biol. 20:3234-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, W., D. Takezawa, S. B. Narasimhulu, A. S. Reddy, and B. W. Poovaiah. 1996. A novel kinesin-like protein with a calmodulin-binding domain. Plant Mol. Biol. 31:87-100. [DOI] [PubMed] [Google Scholar]
- 54.West, S., N. Gromak, C. J. Norbury, and N. J. Proudfoot. 2006. Adenylation and exosome-mediated degradation of cotranscriptionally cleaved pre-messenger RNA in human cells. Mol. Cell 21:437-443. [DOI] [PubMed] [Google Scholar]
- 55.Win, T. Z., S. Draper, R. L. Read, J. Pearce, C. J. Norbury, and S. W. Wang. 2006. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol. Cell. Biol. 26:1710-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyers, F., M. Rougemaille, G. Badis, J. C. Rousselle, M. E. Dufour, J. Boulay, B. Regnault, F. Devaux, A. Namane, B. Seraphin, D. Libri, and A. Jacquier. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121:725-737. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.