Abstract
Two distinct pathways for the degradation of mammalian cyclin E have previously been described. One pathway is induced by cyclin E phosphorylation and is dependent on the Cul1/Fbw7-based E3 ligase. The other pathway is dependent on the Cul3-based E3 ligase, but the mechanistic details of this pathway have yet to be elucidated. To establish the role of Cul3 in the degradation of cyclin E in vivo, we created a conditional knockout of the Cul3 gene in mice. Interestingly, the biallelic loss of Cul3 in primary fibroblasts derived from these mice results in increased cyclin E expression and reduced cell viability, paralleling the loss of Cul3 protein expression. Cell cycle analysis of viable, Cul3 hypomorphic cells shows that decreasing the levels of Cul3 increases both cyclin E protein levels and the number of cells in S phase. In order to examine the role of Cul3 in an in vivo setting, we determined the effect of deletion of the Cul3 gene in liver. This gene deletion resulted in a dramatic increase in cyclin E levels as well as an increase in cell size and ploidy. The results we report here show that the constitutive degradation pathway for cyclin E that is regulated by the Cul3-based E3 ligase is essential to maintain quiescence in mammalian cells.
Cyclin E is a positive regulator of proliferation in mammalian fibroblasts, and, thus, its levels are tightly regulated in cells (26). Cyclin E functions by associating primarily with the cyclin-dependent kinase, Cdk2, and activating its kinase activity. This activation results in the phosphorylation of key substrates by Cdk2, thereby driving cells into S phase (45). Cyclin E is synthesized just prior to the onset of S phase in response to the E2F family of transcription factors (2, 14, 34) and is then rapidly degraded by the ubiquitin-dependent proteolytic pathway (5, 57). Both mammalian Cullin 1 (Cul1) and mammalian Cullin 3 (Cul3) have been implicated in the degradation of cyclin E. Cul1 mouse knockouts arrest early in development, with a subset of cells containing high levels of cyclin E (8, 53). Cul3 knockouts also arrest early in development but different subsets of cells, distinct from those seen in Cul1 knockouts, have elevated levels of cyclin E. In vitro studies have shown that Cul1 can ubiquitinate cyclin E (24), and Cul3 has been shown to bind and ubiquitinate cyclin E in a transient transfection system (48). Previous work in the identification of Cul3 showed that it was responsible for a constitutive pathway of cyclin E degradation. This pathway did not require cyclin E to have been phosphorylated on threonine 380, whereas Cul1 complexes have been shown to be involved in induced degradation of cyclin E following phosphorylation at this site (51, 60).
Amplification of the cyclin E gene and over expression of cyclin E protein is seen frequently in human malignancies (including breast and ovarian cancer) (10). One study showed that 10% of female transgenic mice engineered to express human cyclin E in mammary glands developed mammary carcinomas (1). Furthermore, a majority of the transgenic mouse mammary epithelial cells that overexpress cyclin E exhibited characteristics that are required for tumor development (1). Cyclin E-deficient mice acquire increased resistance to oncogenic transformation by c-Myc, H-Ras, and dominant-negative p53, expressed in combination or alone (15). Lastly, cell lines derived from human tumors over expressing cyclin E rarely show an increase in cyclin E mRNA, implying that the error is posttranscriptional (40). Thus, errors in cyclin E proteolysis are likely a major contributor to tumor progression.
Because of a significant role in cellular biology and, by extension, human health, cellular proteolysis in general has been widely studied. It has been shown that the majority of cellular proteolysis is regulated by the ubiquitin-dependent proteolytic pathway. The hallmark of this pathway involves the attachment of ubiquitin molecules to lysine residues on target substrates, thereby creating a signal for degradation by the 26S proteasome. The ubiquitin-dependent proteolytic pathway is responsible for the degradation of specific proteins that are involved in maintaining a variety of events such as the cell stress response, DNA repair, transcription, cell cycle regulation, and cellular transformation. This complex pathway involves a multistep process facilitated by three major enzymatic activities: an E1 or ubiquitin-activating enzyme, an E2 or ubiquitin-conjugating enzyme, and an E3 or ubiquitin ligase (21). The E1 enzyme attaches ubiquitin to itself via a thiol-ester bond in a reaction that requires ATP, thus activating the ubiquitin molecule. Subsequently, the E1 transfers the activated ubiquitin molecule to one of many E2 ubiquitin-conjugating enzymes (37). The E2 enzyme is then capable of transferring ubiquitin to one of many lysine side chains on target proteins in concert with an E3 ubiquitin ligase, which confers specific substrate recognition to the system (11, 19). After multiple rounds of ubiquitination, often resulting in ubiquitin chains, the ubiquitin-conjugated substrate is recognized by the 26S proteasome, resulting in degradation of the substrate and recycling of the ubiquitin molecules (21).
One major class of E3 ligases includes those that contain proteins called cullins. The cullin-based E3 ligases are large multisubunit complexes in which the cullin subunit serves as a scaffold for the assembly of a substrate recognition subunit as well as an E2 ubiquitin-conjugating enzyme (9). Members of the cullin family are all highly homologous in their C-terminal regions, the portion necessary for binding an essential RING-finger protein called Roc1/Rbx1 (62), which is thought to be the E2 docking site. The Nedd8 (neural precursor cell expressed, developmentally down-regulated 8) conjugation site also resides in the C-terminal region of all cullins (20). Nedd8 is required for activation of most cullin-dependent E3 ligases, although it is still unclear how this modification is necessary for all cullin-dependent activity. In contrast, cullins have very little sequence similarity at their N termini, which have been shown to contain a region required for binding the proteins involved in substrate recognition (62). For example, the Cul1-dependent E3 ligase, also known as the SCF complex (Skp1-Cul1-F box), binds an adapter molecule called Skp1, which in turn binds one of many substrate recognition proteins that contain a motif called an F box. These substrate recognition proteins bind Skp1 via their F-box domain and in addition contain a protein-protein interaction domain involved in binding specific substrates (49). The hallmark of substrate recognition of the SCF complex is that substrate phosphorylation is a requirement.
Cul3 has been shown to form a similar yet distinct ligase complex. Unlike Cul1, the N terminus of Cul3 is required for binding one of many BTB domain-containing proteins, a family of proteins that have been shown to contain both the adapter and substrate recognition moiety in a single polypeptide (13, 16, 39, 58). Additionally, there are no data describing any requirement for phosphorylation to enhance recognition of the substrate by the Cul3-based E3 ligase. Cul3 is required for a variety of important biological processes. For example, In Drosophila, eye disc formation requires degradation of the transcription factor Cubitus interruptus (Ci) via both Cul3- and Cul1-dependent E3 ligases in a cell-type-specific manner. Cul1 is responsible for Ci proteolysis in cells that are anterior to the morphogenic furrow of the developing eye disc while Cul3 is responsible for degradation of Ci in the cells that are posterior to the morphogenic furrow (36). Furthermore, Cul1-mediated Ci proteolysis is dependent on Nedd8 conjugation in contrast to Cul3-mediated degradation of Ci, which requires the Nedd8 modification of Cul3 only in cells directly adjacent to the anterior/posterior boundary (morphogenic furrow); the remaining posterior cells do not require Nedd8 modification of Cul3. Additionally, Cul3 has been shown to be required for neuronal arborization and proper elaboration of dendrites in an Nedd8-dependent manner in certain neurons in Drosophila (63). In Caenorhabditis elegans Cul3 is necessary for synaptic signaling and plasticity by targeting GLR-1, a subunit of the AMPA (α-amino-3-hydroxy-5-methyl-isoxazole-4-propionic acid)-type glutamate receptors, for degradation (47). Cul3 was shown to bind a BTB-containing protein called Kel-8 (kelch-repeat containing protein 8), which in turn recognizes GLR-1, ultimately targeting it for degradation. Cul3 also associates with another neuronally expressed BTB/Kelch-containing protein, actinfilin, which in turn regulates the levels of the GluR6 receptor (46). Another process in C. elegans that requires Cul3 is the meiosis-mitosis transition. This regulation is mediated by Cul3 binding to the BTB domain-containing protein MEL-26 and degrading MEI-1 (27, 38, 39). Mammalian Cul3 binds to Keap-1, another BTB domain-containing protein, and negatively regulates the Nrf2 transcription factor that promotes cell survival during oxidative stress (7, 23). Cul3 also binds RhoBTB2, a potential tumor suppressor commonly deleted in breast and lung cancer cell lines (55). The binding of Cul3 to RhoBTB2 may represent a unique interaction in which the BTB domain-containing protein (RhoBTB2) may itself be the substrate for degradation. Cul3 also negatively regulates DNA topoisomerase 1 (TOP1) which is important for releasing torsional stress in DNA during replication, transcription, and DNA repair (61). In a similar theme, Cul3 has also been shown to be required for X chromosome inactivation by regulating the monoubiquitination of Bmi-1 and MACROH2A1 via the BTB domain-containing protein SPOP (speckle-type POZ protein) (18). In this case, it is not clear how this type of ubiquitination reaction is accomplished or how it regulates the activity of those molecules.
The mechanisms involved in the recognition and subsequent degradation of cyclin E are poorly understood. To this end, we sought to identify the primary proteins involved in the degradation of cyclin E. In previous work, we identified an E3 ligase called Cul3 in a yeast two-hybrid screen searching for proteins that interact with cyclin E not bound to Cdk2. We were able to show that Cul3 binds and ubiquitinates cyclin E in a transient transfection system (48). To further characterize Cul3 function in vivo, we created a Cul3 knockout mouse and discovered that deletion of the Cul3 gene resulted in early embryonic lethality. Additionally, we observed an increase in cyclin E in a subset of cells in the extraembryonic tissue, including the giant trophoblast cells, which had lost their ability to undergo endoreduplication because of the excess cyclin E (48). From those studies, we proposed a new pathway for cyclin E degradation that is Cul3 mediated.
To date, all the data associated with the function of Cul3 in vivo is specific to certain systems and is for the most part associated with development. Additionally, the role of Cul3 in cyclin E degradation has not been clearly established. Previous work supported a causal link between cyclin E steady-state levels and Cul3 function but provided no mechanistic link. To definitively show that Cul3 plays a physiologically relevant role in degrading cyclin E in vivo, we have constructed a Cul3 conditional knockout mouse. Here, we show that Cul3 is a critical regulator of cyclin E in primary fibroblasts and hepatocytes. The observed change in the regulation of cyclin E in cells with reduced levels of Cul3 is due to a defect in normal cyclin E degradation. This Cul3-dependent regulatory pathway plays an essential role in animals, since Cul3-mediated degradation of cyclin E is required for liver cells to maintain a quiescent state. Thus, loss of Cul3 in hepatocytes results in up-regulation of cyclin E, causing an increase in DNA synthesis and the formation of micronuclei. These results closely mimicked results obtained from experiments in which cyclin E was overexpressed, implying that cyclin E is the major substrate of the Cul3-based E3 ligase.
MATERIALS AND METHODS
Cell culture.
Mouse embryonic fibroblasts were cultured from embryonic day 12.5 embryos. The embryos were minced separately in 100-mm tissue culture plates containing 2 ml of trypsin using a surgical blade and were further broken down by pipetting vigorously. The embryonic tissue in trypsin was placed at 37°C for 10 min. After incubation, fresh medium was added to the minced embryo, and 5 h later (once fibroblasts had begun to adhere to the tissue culture plate) the culture was washed vigorously with phosphate-buffered saline (PBS) to remove remaining tissue and other nonfibroblast cells. Fresh medium was added to the culture and changed daily.
Construction of the Cul3 floxed allele.
The insert from a Cul3 lambda genomic clone containing 15.4 kb spanning exons 4 to 7 was cloned into pBSII (Strategene) at the NotI site. A loxP site was cut out of pBS246 (Life Technologies) with BamHI and XmnI and blunted; it was then inserted into a unique SnaBI site just downstream of exon 7 and screened for orientation by sequencing (pJS1108). pJS1008 was cut with NotI, blunted, and cut with BamHI. A 1-kb promoter-proximal fragment from that digest was then cloned into the targeting plasmid pPNT (52) that was digested with EcoRI (blunted) and BamHI (pJS1109). pJS1108 was then cut with BamHI and NotI, and the rest of the clone (14.4 kb) was inserted into BamH1-Not1-digested pJS1109. This insertion removed the neomycin (Neo) resistance gene in pPNT and resulted in the insertion of a 15.4-kb genomic piece of DNA containing exons 4 to 7 of the Cul3 gene in which a loxP site was inserted near the 3′ end (pJS1110). A cassette was then constructed using oligonucleotides (forward, 5′-GGATCCCTATAACTTCGTATAATGTATGCTATACGAAGTTATCGCTTTGAAGTTCCTATTCCGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCAGAGCGCTTGATCTATAGATCATGAGTGTA-3′; reverse, 5′-GGATCCCAAGCGCTCTGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCGGAATAGGAACTTCAAAGCGAATTCTACCGGGTAGGGGAGGCG-3′; bold indicates loxP, underlining indicates frt, and bold with underlining indicates the Neo site) and PCR that contained BamHI sites at either end, and a Neo gene flanked by FRT (Flp recombination target) sites. This construct also contained a loxP site at the 3′ end of the Neo gene in the same orientation as the loxP site in the Cul3 genomic construct. This cassette was inserted into the unique BamHI site in pJS1110 and screened for orientation such that the Neo gene was inserted backwards compared to the Cul3 gene and the loxP site was Cul3 promoter proximal. Thus, the addition of the Cre recombinase would delete both the Neo gene as well as exons 4 to 7 of the Cul3 gene in the normal Cul3 genomic position (pJS1112). This plasmid was then assayed for Cre-mediated recombination by cotransfection of pJS1112 and pBS185 (a Cre recombinase-expressing vector; Life Technologies) to ensure that the loxP sites remained functional. The plasmid was then linearized at the unique Not1 site and used for electroporation into mouse embryonic stem cells as described previously (48). Selected clones were then screened for the presence of both loxP sites, and correct clones were used to make animals.
Retroviral transfection and infection.
Retroviral ecotropic packaging cells (ATCC) were transfected with a vector containing a puromycin resistance gene and the Cre recombinase gene. The viral supernatant was harvested at 4-h intervals starting at 24 h after transfection and stored at 4°C on ice. Mouse embryonic fibroblasts (MEFs) were cultured to 60% confluence for viral infection. The viral supernatant was supplemented with 10% fetal bovine serum (FBS), and a 4 μg/ml final concentration of hexadimethrine bromide (polybrene; Sigma) was added to the MEFs at 4-h intervals over a 15- to 24-h period. For puromycin selection, 5 μg/ml puromycin (Sigma) was added to the virally infected MEFs 48 h after initial viral infection.
siCyclinE knockdown.
MEFs were seeded at 20% confluence onto 60-mm plates 24 h prior to transfection. For each sample, a small interfering RNA (siRNA) directed against cyclin E1 (siCyclinE; 200 nmol) in 150 μl of Opti-MEM I (Invitrogen) was added to 6 μl of Lipofectamine 2000 (Invitrogen) dissolved in 150 μl of Opti-MEM I. The mixture was added to each plate with 3 ml of Dulbecco's modified Eagle medium and 10% FBS containing no antibiotics. Twenty-four hours later each sample was placed in fresh medium and then analyzed the following day. The RNA oligonucleotide sequence used for cyclin E1 was AGGUGUGCGAAGUCUAUAAUU.
Recombination and genotyping.
In order to analyze MEFs for recombination at the loxP sites located in the Cul3 floxed allele, two primer sequences flanking the 5′ and 3′ loxP sites were used: the forward primer 5′-GGAAACCTAAAGTTTTTATGCATG-3′ and the reverse primer 5′-TTTGTCTGGACCAAATATGGCAGCCCAAACC-3′. The amplified PCR product is 1.3 kb for recombination between the two loxP sites, and no product is detected when recombination does not occur.
Genotyping to detect the presence of the Cul3flox and/or Cul3+ alleles used the following primer sequences flanking the 3′ loxP cloning site: the forward primer 5′-CAGGTTGTATTTTAACTGCTTAAATGTCAAAACCT-3′ and the reverse primer 5′-TTTGTCTGGACCAAATATGGCAGCCCAAACC-3′.
Detection of apoptosis, flow cytometry, and bromodeoxyuridine (BrdU) staining.
For apoptosis, MEFs were seeded onto coverslips prior to experimental conditions. Cells were stained for apoptosis using terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) according to the manufacturer's instructions (Roche). Cells were also stained with 4′,6′-diamidino-2-phenylindole (DAPI) to allow identification of the nucleus. The images were acquired and analyzed using a fluorescent microscope and Metamorph software.
Flow cytometry was used to analyze cells from a 100-mm tissue culture plate at 75% confluence. Cells were harvested and fixed in ethanol. They were then stained with propidium iodide (0.01 mg/ml propidium iodide) followed by data collection on a FACSCaliber flow cytometer (Becton Dickinson) using CellQuest software to analyze the data.
To measure BrdU incorporation, MEFs were supplemented with 1 μg/ml BrdU (Roche) and 1 mg/ml uridine followed by incubation at 37°C for 30 min. Ascorbic acid was added to the MEFs to stop the BrdU incorporation reaction (final concentration, 67 mM). Anti-BrdU (Roche) was added to the cells for 30 min at 37°C. An ABC Elite Vectastain kit (Vector) and the Vector NovaRed substrate kit (for peroxidase) were used for application and detection of the secondary antibody.
Immunoblotting.
The following primary antibodies were used: 1:200 dilution anti-Cul3 antibody (48), 1:2,000 affinity purified cyclin E (48), and 1:2,500 anti-myc antibody (Santa Cruz). Membranes were placed in primary antibodies overnight at room temperature and then transferred to horseradish peroxidase-conjugated secondary antibodies, diluted 1:10,000, for 30 min at room temperature. Bands were visualized using enhanced chemiluminescence (Amersham) and exposed to X-ray film (Kodak) and/or digital acquisition using an AlphaInnotech FluorChem SP system.
RNA isolation and quantitative reverse transcription-PCR (RT-PCR).
MEFs were cultured to 75% confluence and harvested for RNA. The cells were resuspended in 1 ml of Trizol (Invitrogen) and 0.2 ml of chloroform. The aqueous phase was transferred to 1 ml of isopropanol, inverted, and incubated at −20°C for 30 min. Then the RNA was pelleted by centrifugation for 15 min at 14,000 rpm at 4°C. The RNA pellet was washed with 1 ml of 75% ethanol in diethylpyrocarbonate-treated water and then finally resuspended in 20 μl of diethylpyrocarbonate-treated water. One microgram of RNA was used to synthesize cDNA according to the manufacturer's instructions (Bio-Rad). For the quantitative real-time PCR, iTaq SYBR Green super mix with ROX was used following the manufacturer's instructions. Ten nanograms of cDNA was used for each reaction. To amplify Cul3 (173 bp) the following primers were used: forward primer, 5′-AATGAGGAAATAGAGCGGGTGA-3′; and reverse primer, 5′-ATGGCAAGCAAGGTCTTCTGTC-3′. To amplify cyclin E1 (161 bp) the following primers were used: forward primer, 5′-AATTGGGGCAATAGAGAAGAGGT-3′; and reverse primer, 5′-TGGAGCTTATAGACTTCGCACA-3′. To amplify cyclin E2 (131 bp) the following primers were used: forward primer, 5′-AGGAATCAGCCCTTGCATTATC-3′; and reverse primer, 5′-CCCAGCTTAAATCTGGCAGAG-3′. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control for each sample using the following primers (227 bp): forward primer, 5′-GTGAAGGTCGGTGTGAACGG-3′; and reverse primer, 5′-CTCCTGGAAGATGGTGATGG-3′. Each reaction was analyzed using the Applied Biosystems 7300/7500 PCR machine and SDS software.
Half-life determination.
For endogenous Cul3 half-life determination, MEFs were seeded to 30 to 40% confluence onto 60-mm dishes 24 h prior to the experiment. Cells were then washed and 3 ml of fresh medium without l-methionine and l-cysteine, supplemented with 10% dialyzed FBS (Gibco), was added to each plate for 2 h. The medium was then removed and replaced with 1 ml of methionine- and cysteine-free medium supplemented with 200 μCi of Expre35S35S protein labeling mix (Perkin Elmer), followed by incubation for 45 min at 37°C. At time zero fresh medium was added to the plates, and cells were harvested at the indicated time points (see Fig. 2D). The cells were subsequently sonicated in the presence of radioimmunoprecipitation assay (RIPA) buffer (1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 150 mM NaCl, 0.01 M sodium phosphate, pH 7.2, and 2 mM EDTA) containing protease inhibitors and then immunoprecipitated using anti-Cul3 antibody affinity purified against a peptide from the amino-terminal portion of the Cul3 coding region. Endogenous cyclin E half-lives were determined as described above except that the labeling time was 1 h and the antibody used for immunoprecipitation was polyclonal anti-cyclin E (M20; Santa Cruz Biotechnology). For exogenous cyclin E half-life determination, each 100-mm tissue culture plate of MEFs was infected separately with pBabe-myc-tagged cyclin E. After infection, cells were seeded onto 60-mm tissue cultures plates and incubated for 24 h. The cells were treated with 2.4 μM cycloheximide (Sigma) and then harvested at the indicated time points (see Fig. 6C).
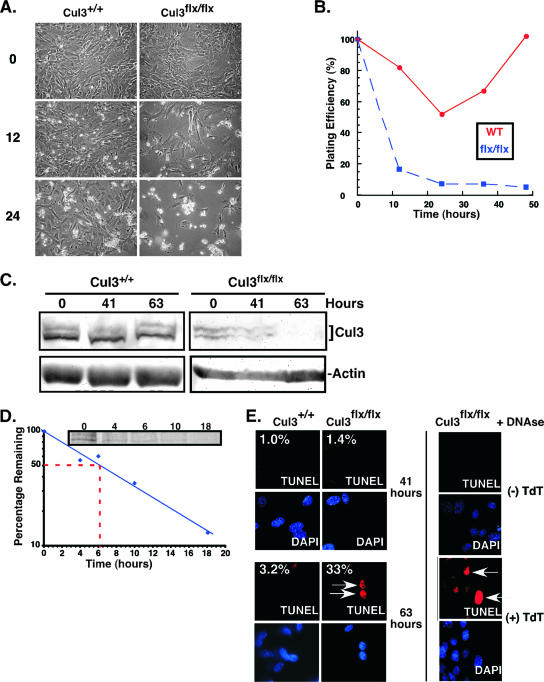
FIG. 2.
Loss of Cul3 results in decreased cell viability. Cul3+/+ and Cul3flox/flox MEFs were infected with a Cre recombinase-expressing retrovirus that also expressed the puromycin resistance gene. (A) Photographs of cells were taken at the indicated time points (hours) after addition of puromycin. The left panels show wild-type MEFs, and the right panels show Cul3flox/flox MEFs. (B) A plot of the percentage of cells remaining versus time after addition of puromycin. Blue dashed line represents Cul3flox/flox MEFs, and the solid red line represents Cul3+/+ MEFs. (C) Immunoblot analysis of relative Cul3 expression levels in Cul3flox/flox and Cul3+/+ Cre recombinase-infected MEFs. Time indicates hours after introduction of the Cre recombinase. (D) Cul3 half-life was measured in wild-type MEFs using 35S pulse labeling for 1 h and harvesting at different time points after the label was removed. Data were plotted as the percentages of the time zero signal remaining. The red dashed line indicates the time when 50% remained, at approximately 6.3 h. Inset shows a gel of the half-life. (E) Determination of apoptosis using TUNEL on Cul3flox/flox and Cul3+/+ MEFs at 41 and 63 h postinfection with Cre-recombinase (left). Tetramethylrhodamine-dUTP was used to visualize TUNEL-positive cells (in red), and DAPI was used to stain the nuclei (in blue). For controls (right) noninfected Cul3flox/flox MEFs were treated with DNase in the presence (+) or absence (−) of terminal deoxytransferase (TdT). Arrows indicate TUNEL-positive cells.
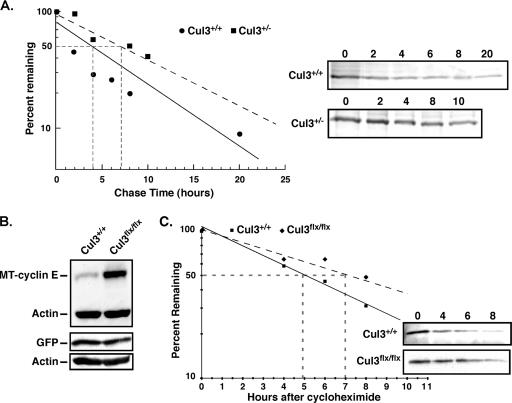
FIG. 6.
Cul3 regulates steady-state levels of cyclin E. (A) Plot of endogenous cyclin E half-life measured in Cul3+/+ and Cul3+/− MEFs using 35S pulse-labeling for 1 h and chase times as indicated. These data were collected using a Typhoon (GE) phosphorimager. Panels on the right show labeled cyclin E at the indicated time points. (B) Cul3+/+ and Cul3flox/flox MEFs infected with viruses expressing MT-cyclin E or GFP. Relative levels of protein were determined with immunoblots using anti-myc antibody or anti-GFP antibody. Immunoblot for actin is used as control to demonstrate even loading. (C) Half-life of MT-cyclin E at different time points after addition of cycloheximide in Cul3+/+ or Cul3flox/flox MEFs. (Insets) Immunoblots of immunoprecipitated MT-cyclin E at different time points after addition of cycloheximide in Cul3+/+ and Cul3flox/flox MEFs. Bands were quantified using digital imaging.
Kinase assays.
Fibroblasts were harvested from 10-cm plates in log phase, and the cells were lysed in RIPA buffer as described. Extracts were incubated in anti-cyclin E antibody (M20; Santa Cruz) with gentle mixing for 6 h, followed by the addition of protein A-conjugated Sepharose beads. After an additional incubation, the beads were pelleted by incubation on ice, and the supernatant was discarded. The beads were washed twice with RIPA buffer and then twice with histone wash buffer (25 mM Tris-HCl, pH 7.5, 70 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol). Kinase reactions were performed with 0.1 μg/μl histone H1, 10 μM cold ATP, and 0.2 μCi/μl of [γ-32P]ATP. A total of 30 μl of histone wash buffer containing histone and ATP was added to the washed beads, followed by incubation at 37°C for 30 min. The reaction was terminated by the addition of sodium dodecyl sulfate sample buffer, and the boiled samples were applied to 12% polyacrylamide gels. After electrophoresis, the gels were blotted to membranes, and the membranes were placed on film or imaged using a phosphorimager (Typhoon; GE Healthcare).
Adenoviral introduction and analysis of liver tissue.
Mice were injected with 1 × 108 infectious particles/g of weight with either Cre-adenovirus, cyclin E-expressing adenovirus, or control virus. Livers were subsequently removed and shock-frozen in liquid nitrogen or used for fixation in 4% Formol to process for paraffin sections.
For immunoblotting, livers were crushed in liquid nitrogen, and the tissue powder was dissolved in RIPA buffer. The lysates were sonicated and centrifuged for 20 min. Protein concentration was determined with a Bio-Rad DC protein assay. For Western blotting 100 μg per lane was loaded, and blots were probed with the following antibodies: actin (clone 4; ICN) and Cullin 3 (H-293; Santa Cruz).
β-Catenin staining was performed on 3-μm paraffin sections, deparaffinized, and subjected to a decreasing ethanol series. Sections were boiled in Tris-EDTA antigen unmasking solution, pH 9.0, and blocked in 1% bovine serum albumin in PBS for 1 h. Antibody was incubated for 2 h at room temperature at a dilution of 1:50 in 0.05% bovine serum albumin-PBS (catalog no. 610155; BD Biosciences). Sections were washed in PBS and counterstained with DAPI. They were subsequently mounted in Vectashield mounting medium (Vector). Pictures were taken on a Leica DM5000B fluorescence microscope.
Feulgen quantification.
To analyze DNA content 3-μm sections were deparaffinized, subjected to a decreasing ethanol series, and incubated in 4% buffered formol. After a washing step in distilled water, sections were incubated in 5 M HCl for exactly 1 h, washed in PBS, and incubated for 20 min in Schiffs reagent. To stop the reaction sections were washed in water. This was followed by a 20-min incubation in sulfite water. Sections were dehydrated and mounted in Corbit mounting medium. Analyses were done with an Ahrens ICM Cytometry System.
RESULTS
Construction and characterization of a Cul3 conditional knockout allele.
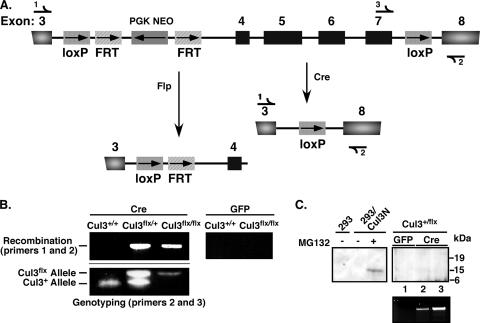
We have previously shown that a deletion of the Cul3 gene in mice results in early embryonic lethality (48). In this study we created a conditional or “floxed” Cul3 allele (Cul3flox) to accomplish two goals: (i) to clarify the role of Cul3 in cyclin E degradation, given the fact that Cul1 can also degrade cyclin E, and (ii) to test the contribution of cyclin E accumulation to the phenotype of the Cul3 knockout since other substrates have been identified. Using the Cre/lox system, in which Cul3 exons 4 to 7 were flanked with loxP sites, site-specific recombination catalyzed by action of the Cre recombinase (Fig. 1A) resulted in the creation of a Cul3 null allele. This deletion removed the same region of the Cul3 gene described in the original knockout (48). In addition to the loxP sites, there is a Neo resistance gene inserted in the third intron that was used to select for homologous recombinants. The Neo resistance gene is flanked with FRT sites, which recombine in the presence of the Flp recombinase and result in the removal of the Neo resistance gene (3) (Fig. 1A). Cul3+/+, Cul3flox/+, and Cul3flox/flox MEFs were tested for loxP recombination after retroviral infection with Cre recombinase for 48 h. Cre-mediated recombination results in the ability of the primers to amplify a 1.3-kb band (Fig. 1B); the same primers are unable to amplify a product in the absence of recombination (Fig. 1B, left panel). Recombination occurred only in the Cul3flox/flox and Cul3flox/+ MEFs (Fig. 1B left panel, lanes 2 and 3). As a negative control, cells were infected with a green fluorescent protein (GFP)-expressing vector and tested for loxP recombination. Those cells had no detectable recombination (Fig. 1B, right panel). Since we were concerned that instead of a null allele we had constructed a potentially dominant-negative truncation mutant, we transfected a construct that was predicted to make the same size protein as would be encoded by the first three exons. The antibodies we used to detect Cul3 were able to detect the transfected truncation only in the presence of the proteasome inhibitor MG132 (Fig. 1C, left panel, lane 3). No similar band was seen in cells that had the Cul3 gene deleted using the Cre recombinase (Fig. 1C, right panel, lanes 2 and 3) or in any of the genotypes used in this study (not shown). Thus, a small N-terminally truncated form of Cul3 that might still be expressed following the deletion of Cul3 exons 4 to 7 was not detected, and the extremely high rates of turnover of such a possible peptide cannot contribute to the observed phenotypes.
FIG. 1.
Construction of a Cul3 conditional knockout allele (floxed, Cul3flox). (A) Diagram illustrating recombination of loxP sites flanking Cul3 exons 4 to 7 via Cre recombinase and recombination of FRT sites flanking the phosphoglycerate kinase (PGK)-Neo cassette via the Flp recombinase. Half-arrows represent primers used to detect loxP recombination using PCR (see below). (B) PCR was used to detect Cre-induced loxP recombination in MEFs infected with Cre recombinase (left upper panel) or GFP (right upper panel). The lower left panel shows genotyping of the promoter distal loxP site for the Cul3flox (upper band) allele or the Cul3+ alleles (lower band). (C) A Cul3 N-terminal truncated protein (Cul3N) was created by placing a stop codon at amino acid 141, mimicking a stop after exon 3 and making a 14,853-Da protein. HEK293 cells were transfected with a plasmid that expresses this Cul3 truncated protein and then treated without MG132 (left panel) or left untreated. The anti-Cul3 antibodies used in this study were able to detect the truncated Cul3 (left panel). Immunoblot analysis of Cul3+/flox MEF lysates from cells infected with EGFP or Cre recombinase for 48 h (in duplicate) and then selected for infection with puromycin are shown in the right panel in which no truncated protein was detected. Verification that the correct region was deleted was determined by PCR analysis for loxP recombination in Cul3+/flox MEFs infected with enhanced GFP or Cre recombinase for 48 h in the presence (lanes 1 and 3) or absence (lane 2) of puromycin (lower panel).
From crosses with mice that were heterozygous for a wild-type and a Cul3 floxed allele (Cul3+/flox), we generated MEFs that are homozygous for the wild-type allele (Cul3+/+) and homozygous for the Cul3 floxed allele (Cul3flox/flox). In addition, we generated MEFs heterozygous for the wild-type allele and the Cul3 knockout null allele (Cul3+/−) from crosses of animals with the same genotype. Cells homozygous for the Cul3 floxed allele with the Neo resistance gene removed (Cul3floxΔN/floxΔN) were obtained from animals that were previously crossed to Flp-recombinase-expressing mice (Jackson Laboratory) carrying the Cul3flox allele. We also attempted to make Cul3flox/− and/or Cul3floxΔN/− animals and MEFs because they could potentially recombine more efficiently, since only one allele contained loxP sites. However, we were unable to obtain any animals (0 obtained out of 46 pups examined) with these genotypes. Six independent crosses of Cul3flox/flox and Cul3+/− mice resulted in a total of 26 pups, all of which were Cul3flox/+ (expect 50%) and none of which were Cul3flox/− (expect 50%). Four independent crosses of Cul3floxΔN/floxΔN and Cul3+/− mice resulted in a total of 20 pups, all of which were Cul3floxΔN/+ (expect 50%) and none of which were Cul3floxΔN/− (expect 50%). Therefore, we concluded that the floxed allele was haploinsufficient for Cul3 expression.
Deletion of the Cul3 gene results in decreased cell viability in MEFs.
Previously, using a complete Cul3 knockout, we observed that most of the cells appeared normal in the developing embryos lacking Cul3 (48). Thus, we concluded that not all cell types need Cul3 to survive. MEFs provide a model system to study cell cycle phenotypes in primary cells. In order to determine if MEFs require Cul3 to proliferate, Cul3+/+ and Cul3flox/flox MEFs were infected with a retrovirus containing the Cre recombinase gene and a puromycin resistance gene (pBabe-Cre), which is maximally expressed within 24 to 36 h after initial infection. At 48 h after infection, the cells were subjected to puromycin selection. Loss of Cul3 resulted in a decrease in the number of adherent cells (Fig. 2A) after 12 h in puromycin-supplemented medium (60 h after infection). After 24 h of puromycin selection (72 h after infection), Cul3flox/flox Cre-infected MEFs continued to decrease in cell number, whereas Cul3+/+ MEFs continued to proliferate until they reached 100% confluence (Fig. 2B). In non-drug-selected cells, we also observed a decrease in Cul3 protein levels 41 h after infection with Cre recombinase and a drop in Cul3 protein to nearly undetectable levels after 63 h in the Cul3flox/flox MEFs (Fig. 2C, right panel). This is in contrast to Cul3+/+ MEFS, in which no reduction of Cul3 protein levels was seen (Fig. 2C, left panel). This rapid loss of Cul3 protein and associated reduction in cell viability was surprising because it implied that Cul3 had a short half-life such that the level of protein drops to a critical point within 12 h after loss of the gene (Fig. 2A). To determine if this was the case, we measured the half-life of endogenous Cul3 in Cul3+/+ MEFs and found it to be 6.3 h (Fig. 2D). This short half-life is consistent with the observation that very shortly after deletion of the Cul3 gene (less than two half-lives), cells reach a threshold level of Cul3 and thus are unable to progress.
The loss of viable cells as a result of deletion of the Cul3 gene led us to hypothesize that either the cells were dying by entering a cell death pathway or that they were losing their ability to adhere to the surface of the tissue culture dish. Since we had seen an increase in the number of apoptotic cells in developing embryos that lacked Cul3 (our unpublished observations), we decided to look at apoptosis in these MEFs. We performed TUNEL measurements on Cul3+/+ and Cul3flox/flox MEFs infected with pBabe-Cre at 41 and 63 h after infection to determine if apoptosis was occurring due to loss of Cul3. We observed that after 41 h, there was no significant difference in the percent of TUNEL-positive cells between the Cul3flox/flox and Cul3+/+ MEFs (1.4% and 1%, respectively). However, at 63 h, Cul3flox/flox MEFs had 33% TUNEL-positive cells compared to 3.2% found in Cul3+/+ (Fig. 2E). This increase in the number of TUNEL-positive cells is consistent with the cause being a decrease in Cul3 expression levels, especially at 63 h where Cul3 levels are nearly undetectable (Fig. 2C, right panel). The loss of plating efficiency and the increase in TUNEL-positive cells are similar to what is seen during anoikis (12). Cells infected with GFP-expressing virus showed no difference in the percentage of apoptotic cells at any of the time points (not shown).
Cul3 floxed alleles have decreased Cul3 expression.
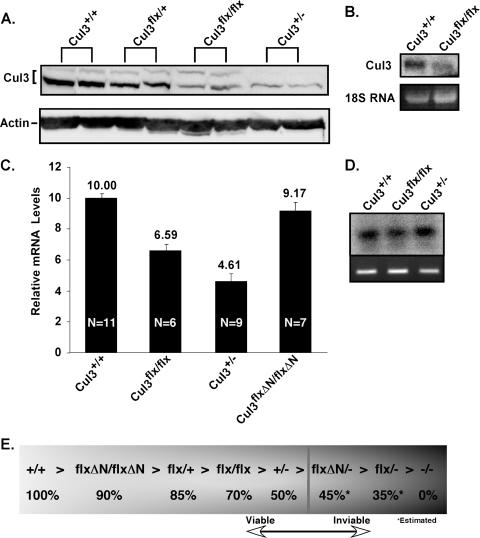
We were unable to obtain animals or cells containing both a knockout allele and a floxed allele; thus, we speculated that the floxed allele might have reduced levels of Cul3 expression and that these reduced levels are insufficient for cell survival. Therefore, we decided to examine the Cul3 expression levels resulting from the floxed allele by using viable cells that contain at least one floxed allele. Immunoblot analysis of the Cul3+/+, Cul3flox/+, Cul3flox/flox, and Cul3+/− MEFs revealed that they have different Cul3 expression levels. By defining Cul3+/+ MEFs as 100% expression, we observed that none of the genotypes expressed normal levels of Cul3 (Fig. 3A). As we had observed previously, Cul3+/− MEFs express 50% of normal levels (Fig. 3A, lanes 1 and 2 versus lanes 7 and 8). The Cul3flox/flox MEFs expressed significantly less Cul3 than wild type with levels comparable to that seen in Cul3+/− MEFs (Fig. 3A, lanes 5 and 6). To determine if this reduced expression is due to changes in the amount of mRNA, we measured the relative amounts of Cul3 message using Northern blot analysis in either Cul3+/+ cells (Fig. 3B, lane 1) or Cul3flox/flox cells (Fig. 3B, lane 2). We saw significantly less Cul3 message in Cul3flox/flox cells compared to Cul3+/+. In order to precisely measure the amount of Cul3 transcript in cells, we used quantitative real-time PCR (Fig. 3C). Wild-type levels were defined as 100% (Fig. 3C, first bar), and the others were plotted as a percentage of wild type. We observed that Cul3flox/flox cells contain 66% (Fig. 3C, second bar) of wild-type levels and that Cul3+/− cells contained 46% (Fig. 3C, third bar). To verify that the amplified product was Cul3, we performed Southern blot analysis on the amplified material and probed for Cul3 using a probe that did not overlap the amplifying primers (Fig. 3D). We also measured relative protein levels using fluorescent secondary antibodies and observed that quantitative measurements of Cul3 protein expression levels from immunoblotting closely matched the results from the quantitative real-time PCR (data not shown).
FIG. 3.
Relative Cul3 expression levels. (A) Cul3+/+, Cul3flox/+, Cul3flox/flox, and Cul3+/− MEFs were analyzed for Cul3 expression using a Cul3 immunoblot. (B) RNA was isolated from Cul3+/+ and Cul3flox/flox MEFs and analyzed by Northern blotting (upper panel; left lane is wild type, and right lane is homozygous floxed). 18S rRNA staining is shown as a loading control (bottom panel). (C) cDNA was isolated from Cul3+/+, Cul3flox/flox, Cul3+/−, or Cul3floxΔN/floxΔN MEFs and analyzed by quantitative real-time PCR for Cul3 expression. The relative expression levels of Cul3 transcript are given arbitrary units after normalization to GAPDH. N, the number of independent samples used for each experiment. (D) The amplified Cul3 cDNA product was run on an agarose gel and detected by ethidium bromide (bottom panel) and then analyzed by Southern blotting using a Cul3 probe (upper panel). (E) Diagram illustrating relative levels of Cul3 expression represented for each genotype as a percentage of wild-type expression. The asterisks indicate estimated levels of Cul3 expression levels in the Cul3floxΔN/− and the Cul3flox/− cells since they were nonviable. The vertical bar indicates the estimated Cul3 expression level that is required for cell viability. Allele abbreviations: + and Cul3+, wild-type allele; Cul3floxΔN, the floxed allele with the Neo gene removed; Cul3flox, the floxed allele with the Neo gene present; − and Cul3−, the Cul3 knockout allele.
It has been observed that introduction of a selectable marker in a mouse gene can interfere with expression of the gene of interest (33). To determine if the Neo resistance gene was responsible for the observed changes in Cul3 transcription, we generated MEFs that were homozygous for the floxed allele but with the Neo resistance gene removed by the activity of the Flp recombinase. We designated this genotype Cul3floxΔN/floxΔN. Quantitative real-time PCR revealed that removal of the Neo resistance gene restored Cul3 transcript levels close to that of Cul3+/+ (Fig. 3C, bar 4).
By combining the data obtained from quantitative real-time PCR and quantitative immunoblot analysis of material obtained from MEFs with different genotypes, we were able to determine the relative Cul3 expression levels in our allelic series (Fig. 3E): Cul3+/+, 100%; Cul3floxΔN/floxΔN, 90%; Cul3flox/+, 85%; Cul3flox/flox, 70%; Cul3+/−, 50%. From these data we estimated the Cul3 expression level of genotypes we were not able to recover: Cul3floxΔN/−, 45%; Cul3flox/−, 35%; Cul3−/−, 0% (Cul3− has previously been determined to be a null allele [48]). We can use this information to make a prediction of the minimal amount of Cul3 expression needed for cell viability to be greater than 45% of wild type. This means that the Cul3+/− MEFs are expressing the least amount of Cul3 necessary to remain viable.
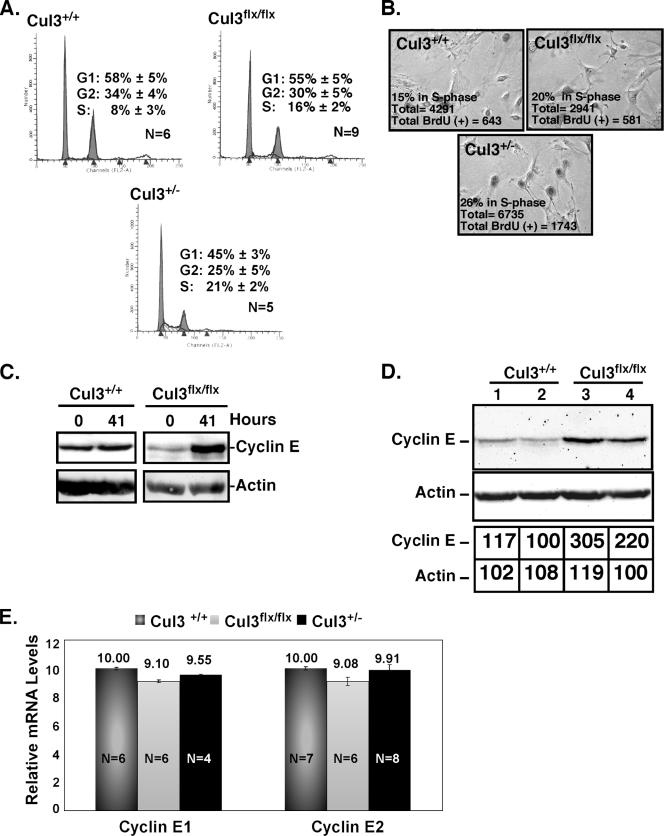
Reducing levels of Cul3 results in changes in cell cycle profiles and an increase in the levels of cyclin E.
Since we identified an allelic series of fibroblasts in which the expression of Cul3 varies from sufficient to almost lethal, we reasoned that these cells might also vary in their ability to degrade key Cul3 substrates. Thus, these cells may display phenotypes due to reduced Cul3 expression that would provide clues to Cul3 function in cells. Since we initially identified Cul3 as a crucial regulator of the cell cycle promoter cyclin E, we decided to examine our allelic series for changes in cell cycle profiles using flow cytometry. We found by comparison of the Cul3 hypomorphs to wild-type MEFs that they showed an increase in the percentage of cells in S phase ranging from 8% in wild type (Fig. 4A, upper left panel) to 21% in the lowest Cul3-expressing cells, Cul3+/− (Fig. 4A, lower left panel). Partial restoration of Cul3 expression levels by deletion of the Neo resistance gene resulted in a restoration of normal cell cycle profiles (data not shown).
FIG. 4.
Low Cul3 expression changes normal cell cycle profiles. (A) The cell cycle profiles of Cul3+/+, Cul3flox/flox, and Cul3+/− MEFs were determined using flow cytometry, and data were plotted as number of events versus DNA content. N, the number of independent measurements used for each experiment. (B) BrdU staining was used to determine the percentage of cells in S phase in Cul3+/+, Cul3flox/flox, and Cul3+/− MEFs. (C) Immunoblotting of endogenous cyclin E 41 h after infection with Cre recombinase in Cul3+/+ and Cul3flox/flox MEFs. A loading control immunoblot for actin is shown (below). (D) Immunoblotting of endogenous cyclin E from Cul3+/+ and Cul3flox/flox MEFs. The data were obtained on a digital camera, and the quantification of the signal for each lane is shown as a percentage of lowest value (lower table). (E) Quantitative real-time PCR of cyclin E1 and cyclin E2 message levels in Cul3+/+, Cul3flox/flox, and Cul3+/− MEFs. Cyclin E transcript levels were given arbitrary units after normalization to GAPDH mRNA. Error bars and values are represented above each bar. N, the number of independent measurements.
We then directly measured the percentage of cells in S phase by examining BrdU incorporation. We observed that, similar to the flow cytometry estimates for the number of cells in S phase, MEFs expressing low levels of Cul3 showed an increase in the number of cells in S phase compared to wild type (Fig. 4B). The Cul3flox/flox cells had 20% of the cells in S phase (Fig. 4B, panel 2) compared to 15% in the wild type (Fig. 4B, panel 1), and the Cul3+/− MEFs had 26% of the cells in S phase (Fig. 4B, panel 3).
In prior work, we determined that Cul3 regulated cyclin E in some cells in the developing embryo (48). We speculated that cyclin E might also be a Cul3 substrate in fibroblasts. To address this question we examined how loss of Cul3 protein affected the steady-state levels of cyclin E. Extracts from Cul3flox/flox and Cul3+/+ MEFs, with and without infection by a Cre recombinase-expressing virus, were prepared for immunoblot analysis to determine the levels of cyclin E protein in cells with a deletion of Cul3 (Fig. 4C). In Cul3+/+ MEFs, there was no obvious change in the steady-state levels of cyclin E after 41 h of infection compared to cells that were not infected (Fig. 4C, left panel). However, we observed that the levels of cyclin E were dramatically elevated in Cul3flox/flox MEFs after 41 h of infection, a point at which there is significant loss of Cul3 protein compared to Cul3+/+ (Fig. 4C, right panel, and 2C).
Since the Cul3 hypomorphic cells had an increase in the percentage of cells in S phase, which could be caused by changes in cyclin E expression, we decided to determine if cyclin E levels were abnormal in cells containing Cul3 hypomorphic alleles. Relative cyclin E protein levels were determined among Cul3 hypomorphs using immunoblot analysis (Fig. 4D). We found that Cul3flox/flox MEFs have an increase in steady-state cyclin E levels (Fig. 4D, lanes 3 and 4 versus lanes 1 and 2). Digital quantification of the bands showed an approximately twofold increase in cyclin E steady-state levels in Cul3flox/flox MEFs versus wild type (Fig. 4D). In order to determine if this increase was due to an increase in cyclin E transcription or to a posttranscriptional mechanism, we determined the relative cyclin E transcript levels in these hypomorphic cells compared to wild type using quantitative real-time PCR. We observed that the levels of both cyclin E1 and E2 transcripts are indistinguishable among the Cul3 hypomorphs (Fig. 4E). To verify that the measured amplicon was either cyclin E1 or cyclin E2, the products were hybridized to probes that do not overlap the amplifying primers (data not shown).
Reduction of cyclin E levels in vivo reverses the hypomorphic Cul3 phenotype.
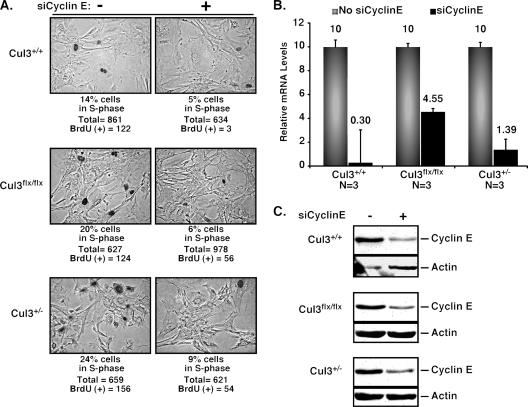
It is possible that Cul3 regulates cell cycle distribution through a mechanism involving modulation of cyclin E levels. We have shown that low Cul3 expression causes an increase in the number of cells in S phase. We have also shown that these same cells have elevated levels of cyclin E. Others have shown that ectopic expression of cyclin E in fibroblasts causes an increase in the number of cells in S phase (25, 35, 43). To directly test the hypothesis that cyclin E contributes to the observed changes in cell cycle profiles, we reduced the expression of cyclin E using siRNA and measured BrdU incorporation. Quantitative RT-PCR showed knockdown of cyclin E transcript levels to 45% of wild type, or less, among all the genotypes (Fig. 5B). Furthermore, cyclin E protein levels were significantly reduced (Fig. 5C, right lanes) compared to cells without the addition of siCyclinE (Fig. 5C, left lanes). BrdU incorporation showed that Cul3flox/flox and Cul3+/− MEFs with siCyclinE had a 60 to 70% decrease in the percentage of cells in S phase (Fig. 5A, right panels) compared to cells without siCyclinE (Fig. 5A, left panels). Control double-stranded RNA against an unrelated BTB domain-containing protein had no effect on the levels of cyclin E protein, mRNA, or on the cell cycle profiles (not shown). Thus, the observed increase in the steady-state levels of cyclin E directly contributes to the changes in cell cycle profiles in Cul3 hypomorphic cells.
FIG. 5.
Elevated cyclin E is responsible for the increase in cells in S phase. (A) BrdU staining was used to determine the percentage of cells in S phase in Cul3+/+, Cul3flox/flox, and Cul3+/− MEFs with and without transfection of siCyclinE. (B) cDNA was isolated from Cul3+/+, Cul3flox/flox, and Cul3+/− MEFs with and without transfection of siCyclinE and analyzed using quantitative RT-PCR for expression of cyclin E. The relative expression levels of cyclin E transcript are given arbitrary units after normalization to a GAPDH control. N, the number of independent measurements. (C) Immunoblotting of endogenous cyclin E from Cul3+/+, Cul3flox/flox, and Cul3+/− MEFs with and without transfection of siCyclinE.
The half-life of cyclin E is longer in cells expressing low levels of Cul3.
Because cyclin E levels are elevated and there was no observed change in cyclin E transcription, we speculated that it was likely that these cells were deficient in their ability to degrade cyclin E. We tested this hypothesis by measuring the half-life of cyclin E in Cul3 hypomorphic cells (Fig. 6). Using the method of pulse-chase labeling, we observed that cells with low levels of Cul3 had a significant increase in the half-life of endogenous cyclin E compared to wild-type cells (Fig. 6A). This difference was close to 3 h. To eliminate any potential contribution of differential transcriptional regulation of cyclin E in the cell types tested, we also measured the half-life of exogenously introduced cyclin E that was tagged with a myc epitope (MT-cyclin E). We observed that the steady-state levels of exogenously expressed cyclin E were elevated in Cul3flox/flox MEFs compared to wild-type MEFS (Fig. 6B, upper panel). This is in contrast to a GFP control in which GFP levels were identical in both genotypes (Fig. 6B, lower panel). This observation is similar to the increased steady-state levels of endogenous cyclin E we observed in Cul3flox/flox MEFs compared to wild-type MEFS (Fig. 4A). In order to determine if this increase in steady-state levels was caused by a change in cyclin E degradation, we measured the half-life of exogenous cyclin E in wild-type and Cul3flox/flox cells. Consistent with a general reduction in ability to degrade cyclin E as was seen with endogenous cyclin E, the half-life of exogenous cyclin E was significantly longer in the Cul3flox/flox MEFs (Fig. 6C). We observed an almost 2-h difference in the half-lives of the two genotypes. Thus, we conclude that a reduction of Cul3 levels in cells directly increases the half-life of cyclin E.
Loss of Cul3 in hepatocytes induces endoreduplication and the formation of multiple micronuclei.
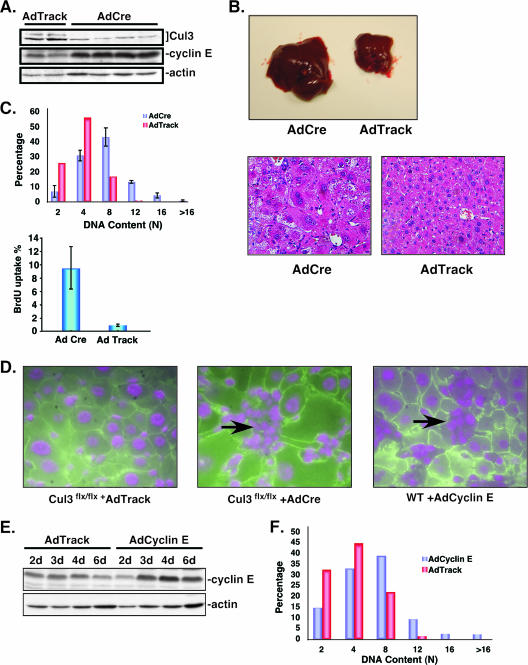
After showing that loss of Cul3 expression in primary fibroblasts resulted in up-regulation of cyclin E levels and an increase in the percentage of cells in S phase, a major question remains: What are the consequences of reducing Cul3 levels in cells that are not continuously proliferating? To determine whether expression of Cul3 in nonproliferating cells is required to maintain quiescence, we chose to target hepatocytes because only 5% of all liver cells are actively proliferating under physiological conditions. To induce deletion of Cul3 we infected Cul3flox/flox mice by tail vein injection with an adenovirus expressing Cre recombinase. This procedure is known to result in a very high number of adenovirus-infected hepatocytes leading to deletion of the floxed sequence in almost 100% of all liver cells (6). Figure 7A shows Cul3 levels in several mice infected with a Cre-expressing adenovirus or an AdTrack control for 3 weeks. Although levels of Cul3 were significantly reduced in Cre-infected Cul3flox/flox mice, residual Cul3 expression was still detectable. This is most likely due to infiltrating blood cells or stromal components in the liver that are not targeted by the Cre adenovirus. Importantly, and in accordance with our experiments in MEFs, reduction of Cul3 expression led to a massive increase in cyclin E levels compared to AdTrack control mice, indicating that Cul3 contributes to the control of cyclin E expression in quiescent cells. As shown in Fig. 7B, loss of Cul3 in the liver had a significant impact on liver morphology, with a striking increase in liver size. The livers of Cul3 knockout mice were on average three times heavier than control livers. This remarkable increase in organ mass was caused by a massive increase in cell and nuclear size, as is shown in Fig. 7B. To test whether the observed increase in nuclear size was caused by an increase in DNA content, we stained liver sections with the Feulgen (29) reagent and performed cytometric studies. Figure 7C shows quantification of these results (upper panel). While AdTrack-infected livers showed a normal composition of liver cell nuclei with DNA contents between 2N and 4N, nuclei from Cul3flox/flox cells infected with a Cre-expressing adenovirus showed a significant increase in ploidy with DNA contents reaching up to 32N (>16N on plot). Additionally, similar to the results seen in Cul3 hypomorphic MEFs, we observed an increase in the number of BrdU-positive hepatocytes in Cul3-deleted tissue compared to controls (Fig. 7C, lower panel). In addition to polyploidization, we also observed widespread signs of genetic instability in livers of Cul3flox/flox mice 3 weeks after infection with Cre adenovirus. Figure 7D shows liver sections stained with a β-catenin antibody to visualize cell borders and counterstained with DAPI. Mice with reduced expression of Cul3 showed a high proportion of cells with multiple micronuclei that are generated because of chromosome breakage or a dysfunction of the mitotic apparatus. Recently, the formation of micronuclei has been used as a marker for defects in the degradation of cyclin E (42). Given that loss of Cul3 led to a massive induction of cyclin E expression, we wondered if, indeed, the dysregulation of this cyclin might be the cause for the observed liver phenotype, namely, the induction of polyploidization and the formation of micronuclei. To test this prediction, we infected wild-type mouse livers with an adenovirus-expressing cyclin E. As shown in Fig. 7E, 3 days after infection cyclin E levels were greatly elevated in liver tissue. The increased levels of cyclin E, however, did not differ from what we found in livers of Cul3 knockout mice (Fig. 7A). In agreement with our prediction that the dysregulation of cyclin E was indeed responsible for the induction of polyploidization and the generation of micronuclei, we found that cyclin E-overexpressing livers had DNA contents of up to 32N (Fig. 7F) and showed many cells with multiple micronuclei (Fig. 7D). Together, these experiments indicate that the activity of the Cul3-dependent degradation complex is essential to maintain quiescence in hepatocytes by keeping the levels of cyclin E low.
FIG. 7.
Loss of Cul3 in hepatocytes leads to cell growth, polyploidization, and multiple micronuclei. (A) Immunoblot of liver lysates showing decreased levels of Cul3 in adenovirus Cre-treated mice compared to control animals; bracket indicates Cul3. The Cul3 knockout also leads to accumulation of cyclin E in hepatocytes. (B) Livers of conditional Cul3 knockout and control mice, 3 weeks after the knockout of Cul3 by intravenous injection of adenovirus-Cre and control adenovirus. The pictures show a huge increase in liver mass caused by an increase in cell and nuclear size. Below are sections from livers stained with hematoxylin and eosin. The figure shows the grossly enlarged cells in Cul3flox/flox livers infected with a Cre-expressing virus compared to cells in livers from animals infected with control virus (right). (C) Paraffin sections of livers were Feulgen stained and analyzed for DNA content (top). The conditional Cul3 knockout in hepatocytes leads to an increase in DNA content (blue bars) in contrast to control mice that were injected with a control adenovirus (red bars). The diagram shows the percentage of cells with DNA contents of 2N to 32N. Liver sections from BrdU-injected Cul3flox/flox mice infected for 3 weeks with adenovirus expressing the Cre recombinase (AdCre) or an empty vector control virus (AdTrack) were stained for BrdU incorporation (as previously described) (bottom). (D) Cul3 knockout hepatocytes and hepatocytes expressing cyclin E by adenoviral expression develop multiple micronuclei (arrows). No micronuclei were seen after infection with a control virus. (E) Immunoblot of cyclin E in liver lysates of mice injected by intravenous injection with cyclin E-expressing adenovirus or control virus. Expression of cyclin E in hepatocytes can be detected 3 to 6 days after injection and does not exceed the observed endogenous cyclin E levels seen in Cul3 knockout mice. (F) Cyclin E-overexpressing hepatocytes (blue bars) show an increase in ploidy compared to controls (red bars) as measured by Feulgen staining.
DISCUSSION
In an earlier study, we identified Cul3 as a potential E3 ligase for the degradation of cyclin E in mammalian cells. We showed that Cul3 could bind cyclin E and increase the incidence of addition of ubiquitin chains to cyclin E. However, those observations are not direct evidence for Cul3-mediated degradation of cyclin E. We decided that a more sophisticated tool for the in vivo analysis of the role of this cyclin E degradation pathway was necessary. To this end, we created a Cul3 knockout mouse. Surprisingly, the knockout resulted in early embryonic lethality. Further examination of the knockout embryos revealed that even though the entire embryo lacked Cul3, only a subset of cells were TUNEL positive, and, therefore, we concluded that Cul3 was not essential for all cellular functions. Additionally, several groups have used siRNA to knock down Cul3 in cell lines and have not found Cul3 to be essential for viability (7, 23, 61). Thus, we concluded that the essential nature of Cul3 was cell type specific.
In order to determine the role of Cul3 in cyclin E degradation in light of the observation that Cul1 can also degrade cyclin E and to establish a physiological function for Cul3, we created a conditional or floxed Cul3 allele (Cul3flox). This system circumvents the early lethality of Cul3 knockouts, yet allows us to knock down Cul3 expression in a controlled manner to observe its essential functions. Here, we show that loss of Cul3 results in a dramatic increase in the levels of cyclin E. These data are consistent with the idea that Cul3 regulates cyclin E in cells by acting as a cyclin E-specific E3 ligase. We also show that removal of the Cul3 gene results in a decrease in cell density on plates. We determined that the mechanism for this loss of plating efficiency is because the cells are entering an apoptotic pathway upon loss of Cul3. Although a role for Cul3 in apoptosis has not been described, we suggest that one potential mechanism that may induce apoptosis in the absence of Cul3 is the loss of the oscillating balance of cell cycle regulators. Deregulation of cell cycle components or an increase in G1 cyclins is one of many apoptotic signals (50). Cyclin E has also been proposed to participate in promotion of both proliferation and apoptosis (31). There are other examples of a switch between proliferation and apoptosis. In the case of the myc oncogene, overexpression of myc can have two outcomes. If antiproliferative signals are present such as high levels of p53, cells that are overexpressing myc enter an apoptotic pathway. However, under proliferative permissive conditions such as when the p53/ARF pathway is down-regulated, high levels of myc contribute to unscheduled proliferation (41).
Our finding that deletions of the Cul3 gene lead to apoptosis limited the extent of our analysis. We took advantage of the fact that the floxed allele is haploinsufficient. By combining different alleles, we found that Cul3 expression varies dramatically among this series of Cul3 hypormorphs. We were able to use these hypomorphs to examine the effect of reduced levels of Cul3 in primary fibroblasts since the allelic series spans a large range of Cul3 expression levels. We observed profound changes in cyclin E levels that correlated well with reduced levels of Cul3 as well as with changes in cell cycle profiles. In addition, we were able to reverse the loss of the Cul3 phenotype on the cell cycle by reducing the levels of cyclin E. Lastly, we showed that reducing levels of Cul3 in cells directly reduced the ability of the cell to degrade cyclin E. Taken together, these data demonstrate that Cul3 is a major regulator of cyclin E in primary fibroblasts. In addition, since siRNA against cyclin E was able to reverse the cell-cycle phenotype seen in Cul3 hypomorphs and overexpression of cyclin E mimicked the loss of Cul3 in liver, it seems likely that cyclin E is an essential target of the Cul3-based E3 ligase.
Cul1 has also been shown to be involved in mammalian cyclin E degradation. Are these two redundant degradation pathways? Alternatively, is it possible that Cul1 is not a major player in fibroblasts? It is also formally possible that loss of Cul3 results in loss of Cul1 by some unknown mechanism. In order to test that hypothesis, we measured the relative levels of both Cul1 and Fbw7 in cells that express low levels of Cul3 (see Fig. S1 in the supplemental material). Neither Cul1 nor Fbw7 changes in abundance in cells that express low levels of Cul3, showing that these observations are not the result of changes in the levels of the Cul1-based E3 ligase. Previous work with Cul3 showed it recognized a distinct pool of cyclin E: the monomeric pool that was not bound to Cdk2 (48). Cul1 has been shown to recognize phosphorylated cyclin E through its F box containing substrate recognition subunit Fbw7 (24, 32, 51, 59) and through another F box containing protein Skp2 (60). The phosphorylation of cyclin E at several sites has been implicated in the formation of the degron that is recognized by the Cul1-based E3 ligase (54, 59). In addition, threonine 380 has been implicated as being crucial for cyclin E turnover (5, 57). A mouse knock-in model, in which the equivalent threonine (T393) was changed to an alanine, resulted in an increase in the half-life of cyclin E (30). We have observed that Cul3 does not require cyclin E to be phosphorylated on threonine 380 to recognize it as a substrate (48). We also proposed that Cul3-dependent degradation of cyclin E represented a unique pathway that was distinct from the Cul1-mediated pathway. Thus, it remains likely that a different region of cyclin E comprises the Cul3 recognition motif. There are many examples in which rapidly degraded substrates have multiple, distinct, degrons that are independently recognized by different E3 ligases (4, 22, 28, 44). In addition, there is evidence that degradation substrates are recognized differently in different cell types (17). In Drosophila Cul1 and Cul3 have also been shown to recognize and degrade the same substrate, the transcription factor Ci, during different times in the development of the eye (36).
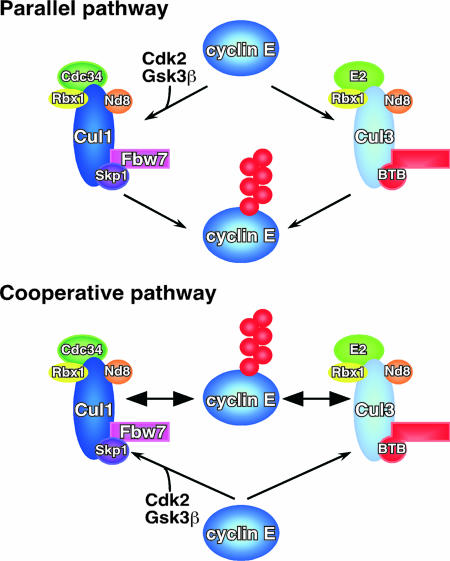
We suggest that there are two possible modes in which both E3 ligase complexes can be responsible for degradation of the same substrate (Fig. 8). In one mode, the “parallel” model, we propose that both the Cul1/Fbw7 ligase and the Cul3 ligase degrade cyclin E and recognize different regions, or degrons. These are independent from each other; therefore, loss of function of either results in a modest, yet measurable, increase in the levels of cyclin E. The other mode implies “cooperation” between the two ligases in which one may add mono-ubiquitin that is then converted to chains by the other. The model suggests that a reduction in function of either ligase results in accumulation of cyclin E that is capable of binding and activating Cdk2. We have determined that the excess cyclin E we observe results in an increase in cyclin E/Cdk2 activity (see Fig. S2A in the supplemental material) and this excess cyclin E/Cdk2 activity is responsible for the abnormal number of cells in S-phase (see Fig. S2B in the supplemental material). This is consistent with the idea that cyclin E that is normally degraded by Cul3 is still capable of binding to Cdk2. A determination of the Cul3 recognition sequence on cyclin E will help clarify this issue.
FIG. 8.
A model for cyclin E degradation regulated by two E3 ligases. Both Cul1 and Cul3 are involved the regulation of cyclin E levels in mammalian cells. There are two possible mechanisms for these two ligase complexes to share a substrate. One, the parallel pathway model shown in the upper panel, would propose that both complexes work on the ubiquitination of cyclin E independently through different degrons and signaling pathways. In this model, both ligases are required for efficient degradation; however, loss of either one results in a modest reduction in cyclin E levels. The other model, the cooperative pathway shown in the lower panel, suggests that both complexes are needed, either because one passes the substrate to the other or each one marks cyclin E differently, and both sets of signals are required for efficient degradation. Ubiquitin molecules are represented by the multiple red spheres attached to cyclin E.
Here, we show that the Cul3-dependent pathway is essential for proper turnover of cyclin E and for normal cell viability in primary fibroblasts. We also show that Cul3 is essential for maintenance of a quiescent state in liver; loss of Cul3 in liver cells results in an increase in DNA content caused by abnormal entrance into S phase. It is not clear how loss of Cul3 leads to quiescent cells entering the cell cycle. However, this observation is not without precedent. Deletion of a component of another cullin-based E3 ligase, Apc2, also results in hepatocytes reentering the cell cycle (56). In that case, the mechanism is also unknown; however, the investigators suggest that cell cycle entry is possibly due to the accumulation of intracellular proteins capable of promoting proliferative growth. In this study, we know that at least one such protein, cyclin E, does accumulate when Cul3 is deleted.
It is interesting that decreasing the level of Cul3 gradually (as we see in the allelic series) shows graded degrees of cyclin E dysregulation. This observation implies that Cul3 might act as a haploinsufficient tumor suppressor protein since slight alterations in the activity of the complex (for example by posttranslational modifications) could already have a major impact on proliferation and genetic instability. The Cul3-dependent cyclin E degradation pathway appears to be distinct from the pathway that requires the Cul1-based E3 ligase since we see no changes in levels of the Cul1 ligase and we do not see a requirement for threonine 380 phosphorylation. Thus, both cyclin E degradation pathways, the Cul1-mediated phosphorylation-triggered pathway and the Cul3-mediated pathway, represent essential modes for cyclin E degradation in cells. It is possible that these two E3 ligases have different recognition requirements for cyclin E that represent different cellular requirements for the removal of cyclin E. It is also possible that there is cell type specificity for the two ligases such that each one is responsible for cyclin E degradation in distinct cell types. The work presented here establishes that the Cul3-mediated pathway for cyclin E degradation in primary fibroblasts is a major mechanism for the regulation of steady-state levels of cyclin E. In addition, this pathway functions in hepatocytes and represents a constitutive pathway of cyclin E degradation that does not require mitogenic signaling and, thus, is an essential component of the machinery required to maintain quiescence.
Supplementary Material
Acknowledgments
This work was supported by the Emerald Foundation (J.D.S.), The Leukemia and Lymphoma Society (J.D.S.), the Rhode Island Biomedical Research Infrastructure Network (BRIN, National Center for Research Resources, NIH, to J.D.S.) and the Center of Biological Research Excellence (COBRE, National Center for Research Resources, NIH, to J.D.S.; grant number P20 RR015578).
We thank K. Mowry, T. Serio, and R. Sheaff for critical reading of the manuscript.
Footnotes
Published ahead of print on 5 March 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bortner, D. M., and M. P. Rosenberg. 1997. Induction of mammary gland hyperplasia and carcinomasin transgenic mice expressing human cyclin E. Mol. Cell. Biol. 17:453-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Botz, J., K. Zerfass-Thome, D. Spitkovsky, H. Delius, B. Vogt, M. Eilers, A. Hatzigeorgiou, and P. Jansen-Durr. 1996. Cell cycle regulation of the murine cyclin E gene depends on an E2F binding site in the promoter. Mol. Cell. Biol. 16:3401-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchholz, F., P. O. Angrand, and A. F. Stewart. 1996. A simple assay to determine the functionality of Cre or FLP recombination targets in genomic manipulation constructs. Nucleic Acids Res. 24:3118-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, P., P. Johnson, T. Sommer, S. Jentsch, and M. Hochstrasser. 1993. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast Mat α2. Cell 74:357-369. [DOI] [PubMed] [Google Scholar]
- 5.Clurman, B. E., R. J. Sheaff, K. Thress, M. Groudine, and J. M. Roberts. 1996. Turnover of cyclin E by the ubiquitin-proteasome pathway is regulated by cdk2 binding and cyclin phosphorylation. Genes Dev. 10:1979-1990. [DOI] [PubMed] [Google Scholar]
- 6.Colnot, S., T. Decaens, M. Niwa-Kawakita, C. Godard, G. Hamard, A. Kahn, M. Giovannini, and C. Perret. 2004. Liver-targeted disruption of Apc in mice activates beta-catenin signaling and leads to hepatocellular carcinomas. Proc. Natl. Acad. Sci. USA 101:17216-17221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullinan, S. B., J. D. Gordan, J. Jin, J. W. Harper, and J. A. Diehl. 2004. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 24:8477-8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dealy, M. J., K. V. Nguyen, J. Lo, M. Gstaiger, W. Krek, D. Elson, J. Arbeit, E. T. Kipreos, and R. S. Johnson. 1999. Loss of Cul1 results in early embryonic lethality and dysregulation of cyclin E. Nat. Genet. 23:245-248. [DOI] [PubMed] [Google Scholar]
- 9.Deshaies, R. J. 1999. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15:435-467. [DOI] [PubMed] [Google Scholar]
- 10.Donnellan, R., and R. Chetty. 1999. Cyclin E in human cancers. FASEB J. 13:773-780. [DOI] [PubMed] [Google Scholar]
- 11.Finley, D. 1992. The yeast ubiquitin system, p. 539-582. In E. W. Jones, J. R. Pringle, and J. R. Broach (ed.), The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Frisch, S. M., and H. Francis. 1994. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 124:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furukawa, M., Y. J. He, C. Borchers, and Y. Xiong. 2003. Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat. Cell Biol. 5:950-951. [DOI] [PubMed] [Google Scholar]
- 14.Geng, Y., E. N. Eaton, M. Picon, J. M. Roberts, A. S. Lundberg, A. Gifford, C. Sardet, and R. A. Weinberg. 1996. Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene 12:1173-1180. [PubMed] [Google Scholar]
- 15.Geng, Y., Q. Yu, E. Sicinska, M. Das, J. E. Schneider, S. Bhattacharya, W. M. Rideout, R. T. Bronson, H. Gardner, and P. Sicinski. 2003. Cyclin E ablation in the mouse. Cell 114:431-443. [DOI] [PubMed] [Google Scholar]
- 16.Geyer, R., S. Wee, S. Anderson, J. Yates, and D. A. Wolf. 2003. BTB/POZ domain proteins are putative substrate adaptors for Cullin 3 ubiquitin ligases. Mol. Cell 12:783-790. [DOI] [PubMed] [Google Scholar]
- 17.Herbst, A., S. E. Salghetti, S. Y. Kim, and W. P. Tansey. 2004. Multiple cell-type-specific elements regulate Myc protein stability. Oncogene 23:3863-3871. [DOI] [PubMed] [Google Scholar]
- 18.Hernandez-Munoz, I., A. H. Lund, P. van der Stoop, E. Boutsma, I. Muijrers, E. Verhoeven, D. A. Nusinow, B. Panning, Y. Marahrens, and M. van Lohuizen. 2005. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl. Acad. Sci. USA 102:7635-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hershko, A. 1991. The ubiquitin pathway of protein degradation and proteolysis of ubiquitin-protein conjugates. Biochem. Soc. Trans. 19:726-729. [DOI] [PubMed] [Google Scholar]
- 20.Hori, T., F. Osaka, T. Chiba, C. Miyamoto, K. Okabayashi, N. Shimbara, S. Kato, and K. Tanaka. 1999. Covalent modification of all members of human cullin family proteins by NEDD8. Oncogene 18:6829-6834. [DOI] [PubMed] [Google Scholar]
- 21.Jentsch, S. 1992. The ubiquitin-conjugation system. Annu. Rev. Genet. 26:179-207. [DOI] [PubMed] [Google Scholar]
- 22.Johnson, P. R., R. Swanson, L. Rakhilina, and M. Hochstrasser. 1998. Degradation signal masking by heterodimerization of MATalpha2 and MATa1 blocks their mutual destruction by the ubiquitin-proteasome pathway. Cell 94:217-227. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi, A., M. I. Kang, H. Okawa, M. Ohtsuji, Y. Zenke, T. Chiba, K. Igarashi, and M. Yamamoto. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24:7130-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koepp, D. M., L. K. Schaefer, X. Ye, K. Keyomarsi, C. Chu, J. W. Harper, and S. J. Elledge. 2001. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294:173-177. [DOI] [PubMed] [Google Scholar]
- 25.Koff, A., F. Cross, J. Schumacher, K. Leguellec, M. Philippe, and J. M. Roberts. 1991. Human cyclin E, a new cyclin that interacts with two members of the CDC2 gene family. Cell 66:1217-1228. [DOI] [PubMed] [Google Scholar]
- 26.Koff, A., A. Giordano, D. Desai, K. Yamashita, J. W. Harper, S. Elledge, T. Nishimoto, D. O. Morgan, B. R. Franza, and J. M. Roberts. 1992. Formation and activation of a cyclin E-cdk2 complex during the G1 phase of the human cell cycle. Science 257:1689-1694. [DOI] [PubMed] [Google Scholar]
- 27.Kurz, T., L. Pintard, J. H. Willis, D. R. Hamill, P. Gonczy, M. Peter, and B. Bowerman. 2002. Cytoskeletal regulation by the Nedd8 ubiquitin-like protein modification pathway. Science 295:1294-1298. [DOI] [PubMed] [Google Scholar]
- 28.Laney, J. D., and M. Hochstrasser. 1999. Substrate targeting in the ubiquitin system. Cell 97:427-430. [DOI] [PubMed] [Google Scholar]
- 29.Levinson, J. W., S. Retzel, and J. J. McCormick. 1977. An improved acriflavine-Feulgen method. J. Histochem. Cytochem. 25:355-358. [DOI] [PubMed] [Google Scholar]
- 30.Loeb, K. R., H. Kostner, E. Firpo, T. Norwood, K. Tsuchiya, B. E. Clurman, and J. M. Roberts. 2005. A mouse model for cyclin E-dependent genetic instability and tumorigenesis. Cancer Cell 8:35-47. [DOI] [PubMed] [Google Scholar]
- 31.Mazumder, S., E. L. DuPree, and A. Almasan. 2004. A dual role of cyclin E in cell proliferation and apoptosis may provide a target for cancer therapy. Curr. Cancer Drug Targets 4:65-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moberg, K. H., D. W. Bell, D. C. Wahrer, D. A. Haber, and I. K. Hariharan. 2001. Archipelago regulates cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 413:311-316. [DOI] [PubMed] [Google Scholar]
- 33.Nagy, A. 2000. Cre recombinase: the universal reagent for genome tailoring. Genesis 26:99-109. [PubMed] [Google Scholar]
- 34.Ohtani, K., J. DeGregori, and J. R. Nevins. 1995. Regulation of the cyclin E gene by transcription factor E2F1. Proc. Natl. Acad. Sci. USA 92:12146-12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohtsubo, M., A. M. Theodoras, J. Schumacher, J. M. Roberts, and M. Pagano. 1995. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol. 15:2612-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ou, C.-Y., Y.-F. Lin, Y.-J. Chen, and C.-T. Chien. 2002. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosphila eye development. Genes Dev. 16:2403-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pickart, C. M., and I. A. Rose. 1985. Functional heterogeneity of ubiquitin carrier proteins. J. Biol. Chem. 260:1573-1581. [PubMed] [Google Scholar]
- 38.Pintard, L., T. Kurz, S. Glaser, J. H. Willis, M. Peter, and B. Bowerman. 2003. Neddylation and deneddylation of Cul-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr. Biol. 13:911-921. [DOI] [PubMed] [Google Scholar]
- 39.Pintard, L., J. H. Willis, A. Willems, J.-L. F. Johnson, M. Srayko, T. Kurz, S. Glaser, P. E. Mains, M. Tyers, B. Bowerman, and M. Peter. 2003. The BTB protein MEL-26 is a substrate-specific adaptor of the CUL-3 ubiquitin-ligase. Nature 425:311-316. [DOI] [PubMed] [Google Scholar]
- 40.Porter, P. L., K. E. Malone, P. Heagerty, L. Gatti, G. Alexander, E. Firpo, J. R. Daling, and J. M. Roberts. 1997. Expression of cell cycle regulators p27kip1 and cyclin E, alone and in combination, correlate with survival in young breast cancer patients. Nat. Med. 3:222-225. [DOI] [PubMed] [Google Scholar]
- 41.Puisieux, A., S. Valsesia-Wittmann, and S. Ansieau. 2006. A twist for survival and cancer progression. Br. J. Cancer 94:13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajagopalan, H., P. V. Jallepalli, C. Rago, V. E. Velculescu, K. W. Kinzler, B. Vogelstein, and C. Lengauer. 2004. Inactivation of hCDC4 can cause chromosomal instability. Nature 428:77-81. [DOI] [PubMed] [Google Scholar]
- 43.Resnitzky, D., M. Gossen, H. Bujard, and S. I. Reed. 1994. Acceleration of the G1/S phase transition by expression of cyclin D1 and cyclin E with and inducible system. Mol. Cell. Biol. 14:1669-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richter Ruoff, B. W. D. H. H. M. 1994. Degradation of the yeast MAT alpha 2 transcriptional regulator is mediated by the proteasome. FEBS Lett. 354:50-52. [DOI] [PubMed] [Google Scholar]
- 45.Roberts, J. M., and C. J. Sherr. 2003. Bared essentials of CDK2 and cyclin E. Nat. Genet. 35:9-10. [DOI] [PubMed] [Google Scholar]
- 46.Salinas, G. D., L. A. Blair, L. A. Needleman, J. D. Gonzales, Y. Chen, M. Li, J. D. Singer, and J. Marshall. 2006. Actinfilin is a CUL3 substrate adaptor, linking GluR6 kainate receptor subunits to the ubiquitin-proteasome pathway. J. Biol. Chem. 281:40164-40173. [DOI] [PubMed] [Google Scholar]
- 47.Schaefer, H., and C. Rongo. 2006. KEL-8 is a substrate receptor for CUL3-dependent ubiquitin ligase that regulates synaptic glutamate receptor turnover. Mol. Biol. Cell 17:1250-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singer, J. D., M. Gurian-West, B. Clurman, and J. M. Roberts. 1999. Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev. 13:2375-2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skowyra, D., K. L. Craig, M. Tyers, S. J. Elledge, and J. W. Harper. 1997. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell 91:209-219. [DOI] [PubMed] [Google Scholar]
- 50.Slingerland, J., and M. Pagano. 1998. Regulation of the cell cycle by the ubiquitin pathway. Results Probl. Cell Differ. 22:133-147. [DOI] [PubMed] [Google Scholar]
- 51.Strohmaier, H., C. H. Spruck, P. Kaiser, K. A. Won, O. Sangfelt, and S. I. Reed. 2001. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in breast cancer cell lines. Nature 413:316-322. [DOI] [PubMed] [Google Scholar]
- 52.Tybulewicz, V. L., C. E. Crawford, P. K. Jackson, R. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 53.Wang, Y., S. Penfold, X. Tang, N. Hattori, P. Riley, J. W. Harper, J. C. Cross, and M. Tyers. 1999. Deletion of the Cul1 gene in mice causes arrest in early embryogenesis and accumulation of cyclin E. Curr. Biol. 9:1191-1194. [DOI] [PubMed] [Google Scholar]
- 54.Welcker, M., J. Singer, K. R. Loeb, J. Grim, A. Bloecher, M. Gurien-West, B. E. Clurman, and J. M. Roberts. 2003. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol. Cell 12:381-392. [DOI] [PubMed] [Google Scholar]
- 55.Wilkins, A., Q. Ping, and C. L. Carpenter. 2004. RhoBTB2 is a substrate of the mammalian Cul3 ubiquitin ligase complex. Genes Dev. 18:856-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wirth, K. G., R. Ricci, J. F. Gimenez-Abian, S. Taghybeeglu, N. R. Kudo, W. Jochum, M. Vasseur-Cognet, and K. Nasmyth. 2004. Loss of the anaphase-promoting complex in quiescent cells causes unscheduled hepatocyte proliferation. Genes Dev. 18:88-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Won, K. A., and S. I. Reed. 1996. Activation of cyclin E/CDK2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. EMBO J. 15:4182-4193. [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, L., Y. Yue, J. Reboul, P. Vaglio, T.-H. Shin, M. Vidal, S. J. Elledge, and J. W. Harper. 2003. BTB proteins are substrate-specific adaptors in an SCF-like modular ubiquitin ligase containing CUL-3. Nature 425:316-321. [DOI] [PubMed] [Google Scholar]
- 59.Ye, X., G. Nalepa, M. Welcker, B. M. Kessler, E. Spooner, J. Qin, S. J. Elledge, B. E. Clurman, and J. W. Harper. 2004. Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J. Biol. Chem. 279:50110-50119. [DOI] [PubMed] [Google Scholar]
- 60.Yeh, K. H., T. Kondo, J. Zheng, L. M. Tsvetkov, J. Blair, and H. Zhang. 2001. The F-box protein SKP2 binds to the phosphorylated threonine 380 in cyclin E and regulates ubiquitin-dependent degradation of cyclin E. Biochem. Biophys. Res. Commun. 281:884-890. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, H. F., A. Tomida, R. Koshimizu, Y. Ogiso, S. Lei, and T. Tsuruo. 2004. Cullin 3 promotes proteasomal degradation of the topoisomerase I-DNA covalent complex. Cancer Res. 64:1114-1121. [DOI] [PubMed] [Google Scholar]
- 62.Zheng, N., B. Schulman, L. Song, J. Miller, P. Jeffrey, P. Wang, C. Chu, D. Koepp, S. Elledge, M. Pagano, R. Conaway, J. Conaway, J. Harper, and N. Pavletich. 2002. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 416:703-709. [DOI] [PubMed] [Google Scholar]
- 63.Zhu, S., R. Perez, M. Pan, and T. Lee. 2005. Requirement of Cul3 for axonal arborization and dendritic elaboration in Drosophila mushroom body neurons. J. Neurosci. 25:4189-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.