Abstract
The transcription and DNA repair factor TFIIH is composed of 10 subunits. Mutations in the XPB, XPD, and p8 subunits are genetically linked to human diseases, including cancer. However, no reports of mutations in other TFIIH subunits have been reported in higher eukaryotes. Here, we analyze at genetic, molecular, and biochemical levels the Drosophila melanogaster p52 (DMP52) subunit of TFIIH. We found that DMP52 is encoded by the gene marionette in Drosophila and that a defective DMP52 produces UV light-sensitive flies and specific phenotypes during development: organisms are smaller than their wild-type siblings and present tumors and chromosomal instability. The human homologue of DMP52 partially rescues some of these phenotypes. Some of the defects observed in the fly caused by mutations in DMP52 generate trichothiodystrophy and cancer-like phenotypes. Biochemical analysis of DMP52 point mutations introduced in human p52 at positions homologous to those of defects in DMP52 destabilize the interaction between p52 and XPB, another TFIIH subunit, thus compromising the assembly of the complex. This study significantly extends the role of p52 in regulating XPB ATPase activity and, consequently, both its transcriptional and nucleotide excision repair functions.
Several multiprotein complexes are involved in transcription regulation in eukaryotic cells. TFIIH is one of the basal factors that participate in RNA polymerase II (RNA Pol II) transcription (12, 53). Besides this role, TFIIH also makes a fundamental contribution to the mechanism of nucleotide excision repair (NER) and to the control of the cell cycle (6, 12, 14). TFIIH is composed of 10 subunits that can be divided into subcomplexes. The core subcomplex contains six proteins, p8, p34, p44, p52, p62, and XPB, whereas the Cdk-activating kinase (CAK) complex includes Cdk7, cyclin H, and MAT1. XPD interacts with members of both complexes and seems to act as a bridge between the core and CAK (12). The CAK subcomplex in Saccharomyces pombe and in all higher eukaryotes is involved in cell cycle regulation, acting alone outside the complex (14). TFIIH has several enzymatic activities: XPB and XPD are DNA helicases, Cdk7 is a kinase, and it has been reported that the p44 subunit exhibits a ubiquitin ligase activity (49).
Mutations in either the XPB or XPD gene can lead to three hereditary human disorders: xeroderma pigmentosum (XP), combined symptoms of xeroderma pigmentosum and Cockayne syndrome (XP/CS), and trichothiodystrophy (TTD) (9, 30, 31). Mutations in the p8 protein subunit of TFIIH are also associated with human TTD (19, 39). p8 is important for the stability of TFIIH and has a critical role in DNA repair, where it triggers DNA opening (10).
The TFIIH core subcomplex has a central role in NER: it is required for both global genome repair and transcription-coupled repair (32, 33). In NER, several factors recognize DNA damage caused by UV irradiation. In nontranscribed regions, the DDB and XPC-HR23-β-centrin complexes first bind damaged DNA (48). Then, TFIIH and XPA are recruited; following ATP induction, the helicase activity of TFIIH unwinds the damaged DNA that is ready to be targeted by RPA and XPG/ERCC1-XPF. Following the 5′ and 3′ incisions by XPG and ERCC1-XPF, respectively, the damaged oligonucleotide is removed. The resulting gap is filled by PCNA, RFC, DNA polymerase δ or ɛ, and a DNA ligase (33, 41). In transcribed regions, NER is achieved by the transcription-coupled repair mechanism. In this case, DNA damage is recognized by a stalled RNA Pol II, which with the help of CSB eventually recruits TFIIH and XPG. Then, a pathway similar to global genome repair is followed (15, 26, 27). A third contribution of TFIIH to genome integrity is its role in the cell cycle. The CAK complex phosphorylates Cdks that are fundamental for cell cycle progression and control (29, 37).
During transcription, TFIIH allows the formation of the open complex through its helicase activities. Cdk7 phosphorylates serine 5 in the heptapeptide repeat of the carboxy-terminal domain of the largest RNA Pol II subunit. It has been proposed that this phosphorylation allows RNA Pol II to escape from the promoter and to start transcribing. However, it has recently been documented that in S. pombe a mutation in the Cdk7 homologue gene affects only 5% of all fission yeast transcripts, suggesting that the CTD phosphorylation may be compensated for by other kinases (14, 42).
Little is known about the functions of TFIIH subunits other than XPB, XPD, and the CAK subcomplex. For instance, the p44 subunit is important for the assembly of XPD within the core of TFIIH, and it regulates XPD helicase activity (8). Intriguingly, there are no reports of human diseases related to mutations in TFIIH subunits other than XPB, XPD, and p8. It is possible that mutations in these subunits are extremely deleterious. The lack of knowledge reflects a lack of genetic analysis to study the core subunits p62, p52, p44, and p34 in multicellular organisms. In this work, we present a genetic, molecular, and biochemical analysis of the Drosophila melanogaster homologue (DMP52) of the human p52 (HP52) subunit. We demonstrate that DMP52 is encoded by the gene marionette (mrn) and that mrn is an essential gene. Defective DMP52 produces specific phenotypes in a complex organism during development and chromosomal instability. This study shows that p52 mutations affect the XPB assembly into TFIIH and reduce XPB ATPase activity. Interestingly, some of the defects observed in the fly caused by mutations in DMP52 resemble some of the clinical features found in patients with alterations in TFIIH.
MATERIALS AND METHODS
Drosophila strains.
The Drosophila strains used as controls were OreR, W1118, and red e in all the experiments. The P element insertion lines used were EP3605 (FlyBase identifier [ID] FBti0011698) and EP0572 (FlyBase ID FBti0007856). The mrn alles used were mrn1, mrn3, and mrn5 red e/TM6B (FlyBase ID FBgn0002840) and were kindly provided by M. Fuller (16).
UV irradiation sensitivity assay.
Third-instar red e/TM6B, mrn1, mrn3, and mrn5 red e/TM6B and EP3605/TM6B larvae were irradiated at different UVB light intensities (joules/m2) using a UV stratalinker 2400 (Stratagene). Then, the larvae were allowed to develop into adults, and the emerged population was counted.
Complementation test.
To perform the complementation test, crosses were performed between the different alleles, including mrn1, mrn3, mrn5 red e/TM6B, EP3605, and EP0572/TM6B. Then, the total number of progeny was determined, and the attended class (genotype) was calculated considering the media of the two major classes observed. In these crosses, we were looking for non-tubby progeny. To quantify the larval phenotype, we considered the non-tubby larvae and directly evaluated the percentage of tumor generation.
Tissue dissections and staining.
The dissection of the larvae, the pharates, and the adult sexual gonads was done in phosphate-buffered saline (PBS). The ovaries were mounted directly in 50% Citifluor (Ted Pella, Inc.), while the testes were frozen in liquid N2, fixed in MeOH (−20°C) and acetone (−20°C), washed extensively with 0.1% PBS-Tween 20, and mounted in 50% Citifluor. Both sexual gonads were DAPI (4′,6′-diamidino-2-phenylindole) stained (1:10,000) and washed with 0.1% PBS-Tween 20. All the photographs were taken with conventional optic and fluorescence microscopes.
Transgenic flies.
The complete wild-type (wt) Dmp52 cDNA sequence was obtained by reverse transcription-PCR and inserted in the pCaSper hsp83 vector. The complete wt hp52 cDNA sequence was amplified by PCR and cloned in the pCaSper hsp83 vector. The whole genes were sequenced to confirm their integrity. Transgenic flies were constructed following a standard microinjection protocol (47). One Dmp52 and two hp52 independent lines in the second chromosome and two Dmp52 and two hp52 independent lines in the X chromosome were isolated.
Neuroblast chromosome cytology.
Heteroallelic EP3605/mrn1, mrn3, and mrn5 and heterozygous EP3605/+ organisms were used to obtain mitotic chromosomes from larval neuroblasts following the protocol reported previously (46). Briefly, the brains were dissected in 0.7% sodium chloride and incubated with colchicine for 30 min. Then, the preparation was treated with 0.5% sodium citrate, and the tissue was fixed in 2% formaldehyde-40% acetic acid and squashed. The slides were frozen, dried, and mounted with DAPI. The chromosomes were then visualized in a fluorescence microscope.
Production of recombinant hp52 baculoviruses.
Mutations in the hp52 cDNA were introduced using a site-directed mutagenesis method (36): for mrn5, a change of A 229 to T (K77/stop); for the mrn3 allele, a change of C 604 to T (Q202/stop); and for mrn1, a change of G 928 to A (E310K). New synthetic mutants ware generated by changing G 928 to A plus CG 940 and 941 to GA (E310K/R314D), GC 940 to GA (R314D), and A 929 to G (E310G). The mutant cDNAs were cloned using the appropriate restriction enzymes in pFastBac1 vector (Invitrogen), and the whole genes were sequenced to corroborate the mutations. The corresponding recombinant baculoviruses were produced by the Bacto Bac system (Invitrogen). The recombinant viruses were plaque purified, and viral stocks were prepared by three-step growth amplification.
Purification of recombinant TFIIH complexes.
Typically, 108 cells were infected with combinations of recombinant baculoviruses expressing XPB, XPD, p62, p52, or the different mutant versions, p44, p34, p8, cdk7, cyclin H, and MAT1, as indicated and were collected 48 h after infection. The cells were washed in 1× phosphate-buffered saline-30% glycerol and subjected to Dounce homogenization in 10 ml of buffer A (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 20% glycerol, 0.1% Nonidet P-40, 5 mM β-mercaptoethanol). After centrifugation at 14,000 × g for 30 min at 4°C, the clarified lysates were loaded on a heparin-Ultrogel column (Sepracor) preequilibrated in buffer A. After extensive washing with buffer A containing 300 mM NaCl, the proteins were eluted with buffer A containing 500 mM NaCl. The eluted fractions were dialyzed for 2 h against 50 mM Tris-HCl, pH 7.9, 50 mM KCl, 20% glycerol, 0.1 mM EDTA, and 0.5 mM dithiothreitol and immunopurified using the 1H5 anti-p44 antibody (23).
Transcription and dual-incision NER assays.
Runoff transcription (18, 34) and dual-incision assays (1) were carried out as described previously. Briefly, circular DNA containing a single 1,3-intrastrand d(GpTpG) cisplatin-DNA cross-link (Pt-GTG) was prepared as described previously (45). Repair reactions were carried out in buffer containing 45 mM HEPES, pH 7.8, 70 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 0.3 mM EDTA, 10% glycerol, 2.5 μg of bovine serum albumin, and 2 mM ATP. Each reaction mixture contained 50 ng XPG, 20 ng XPF/ERCC1, 10 ng XPC-hHR23B, 50 ng RPA, 25 ng XPA, and either 1.5 μl of HeLa TFIIH (heparin fraction IV [18]) or the recombinant TFIIH complexes including the p52 wt or mutant subunit. Following preincubation for 10 min at 30°C, 50 ng of Pt-GTG damaged template was added, and the reactions were continued for 90 min at 30°C. The reactions were stopped by rapid freezing. The excised fragments were separated on a 14% urea-polyacrylamide gel after they were annealed with 9 ng of the complementary oligonucleotide and the addition of four α-32P-radiolabeled deoxynucleoside triphosphate (3,000 mCi/mmol) residues by Sequenase V2.1 (USB). Finally, they were visualized by autoradiography as described previously (45).
Interaction assays.
Pairwise protein interactions were characterized by coinfection in Sf9 cells (2.5 × 107) with the corresponding recombinant baculoviruses at a multiplicity of infection of 5. The cells were collected 48 h after infection, washed in 1× phosphate-buffered saline-30% glycerol, and subjected to Dounce homogenization in 2.5 ml of buffer A. The clarified lysates were obtained by centrifugation at 14,000 × g for 30 min at 4°C. Fifty microliters of clarified lysate was adsorbed on 20 μl of protein G-Sepharose beads linked with the appropriate monoclonal antibody (1B3, which recognizes the ATP binding site of XPB, or 1D11, which recognizes the N terminus of p52) in buffer containing 20 mM Tris-HCl, pH 7.5, 50 mM KCl, 10% glycerol, and 0.1 mM EDTA. After 1 h of incubation at 4°C, the beads were washed extensively in buffer C containing 150 mM KCl and resuspended in Laemmli buffer. The proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12.5% acrylamide) and revealed by Western blotting using the appropriate monoclonal antibodies (23).
ATPase assay.
Protein fractions were incubated for 2 h at 30°C in the presence of 1 μCi [γ-32P]ATP (7,000 Ci/mmol; ICN Pharmaceuticals) in a 20-μl reaction volume in 20 mM Tris-HCl, pH 7.9, 4 mM MgCl2, 1 mM dithiothreitol, 50 mg/ml bovine serum albumin, and, when indicated, 120 ng of supercoiled double-stranded DNA (pSK). The reactions were stopped by adding EDTA to 50 mM and sodium dodecyl sulfate to 1% (wt/wt). The reaction mixtures were then diluted fivefold, spotted onto polyethylenimine thin-layer chromatography plates (Merck), run in 0.5 M LiCl-1 M formic acid, and autoradiographed.
Inmunohistochemistry and quantification of fluorescence in nuclear sections of salivary glands.
Salivary glands from third-instar larvae, wt and heteroalelic combinations between the EP3605 and mrn1, mrn3, and mrn5 alleles, were dissected and immunostained as previously described (40). For quantification of fluorescence, we followed the protocol reported previously (40). Briefly, we used confocal microscopy to visualize XPD, XPB, TBP, and histone fluorescence. Representative images of protein fluorescence in nuclear sections from the wt and each genotype of Dmp52 were obtained. The nuclear area (156 pixels for each nucleus) was analyzed for each genotype using a photon-counting program. The protein fluorescence frequencies were obtained as a histogram. The relative fluorescences were shown in a bar chart, with the average frequencies of XPB-TBP, XPB-histones, XPD-TBP, and XPD-histones in the y axis and each genotype of p52 in the x axis. Standard errors were indicated in each plot.
RESULTS
The gene marionette encodes the Drosophila p52 homologue.
In Drosophila, the gene marionette (mrn) was identified in a genetic screen for enhancers of mutants in the β-tubulin 85D gene (β-tub85D) (16). Because mrn alleles fail to complement alleles of β-tub85D and do not map to the β-tub85D locus (16), the mutations define a new locus. mrn maps at position 71C3-E5, which is the same position where the homologue for the Drosophila p52 gene (Dmp52) has been localized, so we hypothesized that mrn and Dmp52 were the same gene.
To confirm that mrn encodes the DMP52 protein, we analyzed two reported EP element insertions that are in or near Dmp52. The transposon EP3605 (FlyBase ID FBti0011698) is inserted 20 nucleotides upstream of the predicted translation initiation codon for Dmp52 (Fig. 1A). The homozygous flies are viable, although adult organisms are sterile and have several morphological defects (see below), suggesting that the EP3605 mutant is hypomorphic for Dmp52. EP0572 (FlyBase ID FBti0007856) is inserted inside the first exon of Dmp52, 5 nucleotides downstream of the translation initiation codon; it is lethal in the homozygous condition. Heteroallelic flies with both EP insertions are also lethal, suggesting that both insertions affect the same locus (Fig. 1B).
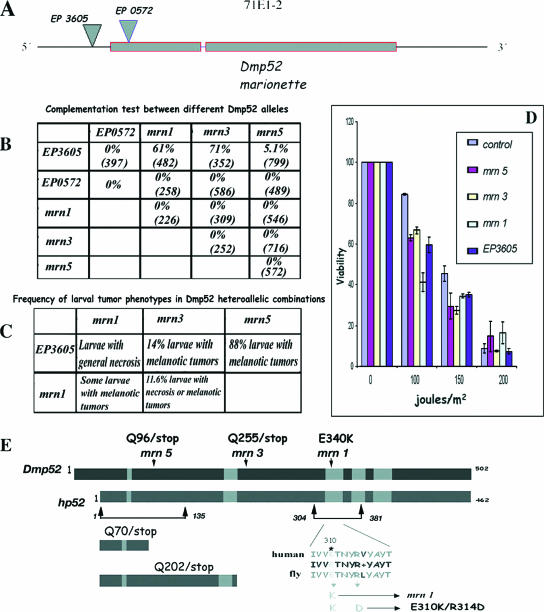
FIG. 1.
Genetic and molecular description of Dmp52 alleles, complementation tests, and UVB irradiation sensitivity assay. (A) Genomic organization of the Dmp52 (mrn) gene and EP insertions. Dmp52 contains two coding exons and a short intron. Dmp52 maps in the cytological position 71E1-2. The positions of the two EP insertions are shown by inverted triangles. EP3605 is inserted at the 5′ end of the gene 20 nucleotides from the start codon. EP0572 is inserted in the first intron of the Dmp52 gene, 5 nucleotides after the start codon, disrupting the DMP52 protein. (B) Complementation test between different mrn point mutations and the EP insertions. The percentage represents the adult flies observed in the expected class. The number in parentheses is the number of expected individuals for each class. Only EP3605 is able to complement, at least partially, the three different mrn point mutation alleles. The other allele combinations are lethal, including EP0572. (C) Frequency of larval tumor phenotypes in the Dmp52 heteroallelic combinations. The percentage represents the larvae observed to have any tumor compared to the healthy class. In the case of the EP3605 allelic combinations, the unaffected populations are presumably those that continue development until adulthood. The larvae of the allelic combinations including at least one copy of Dmp52 (E340K) mrn1 were not quantified. (D) Viability test of the Dmp52 mutants and the parental strain after different UVB irradiation doses. Third-instar larvae heterozygous for Dmp52 mutants were irradiated and then allowed to develop until adulthood. The survival rate is indicated for each strain. The graph represents the results of at least six independent dosage experiments for each genotype. The statistical analysis by analysis of variance indicated a P value of <0.0001 for each mutant compared with the parental strain at 100 and 150 J/m2. The error bars indicate standard errors. (E) Molecular characterization of the Dmp52 point mutants and diagram showing the mutations characterized in the Dmp52 gene introduced into hp52. The gray boxes indicate the regions with highest similarity between the DMP52 and HP52 proteins. The amino acid changes found in the mrn1 (E340K), mrn3 (Q225/stop), and mrn5 (Q96/stop) alleles are indicated by arrows. Truncated peptides, as well as the single and double amino acid changes analyzed in the HP52 protein, are also indicated.
Several ethyl-methane-sulfonate mutant alleles of mrn have been identified (FlyBase ID FBgn0002840) (16). We performed complementation experiments among them and with the EP insertion alleles. In particular, we analyzed the mrn1, mrn3, and mrn5 alleles, which are all lethal when homozygous (Fig. 1B). None of the mrn alleles were able to complement each other, and all failed to complement the EP0572 lethality (Fig. 1B). On the other hand, heteroallelic flies carrying any mrn allele and the EP3605 transposon are semilethal, with the penetrance of the lethality depending on the heteroallelic combination. In mrn5/EP3605 flies, only 5% of the expected class survives, while in mrn3/EP3605 flies, 80% of the expected class of flies develop into adults; in both cases, the adults are sterile (Fig. 1B). In some of the allelelic combinations, a proportion of the individuals develop only up to the third-instar larval (Fig. 1C) or pupal stage. These organisms present melanotic tumors before dying. We hypothesize that the wt mrn product of maternal origin supports variable levels of embryo development.
To confirm that mrn alleles affect the predicted DMP52 protein, we sequenced the Dmp52 mrn1, mrn3, and mrn5 alleles. In mrn5, a change of C 286 to T introduces a stop codon (Q96/stop) (Fig. 1E). In the case of the mrn3 allele, a change of C 673 to T also introduces a stop codon (Q225/stop). Therefore, these two mutants, mrn3 and mrn5, are predicted to result in truncated DMP52 peptides, one of 95 amino acids and the other of 224 (Fig. 1E). In the case of the mrn1 allele, we found a change of G 1018 to A, which changes a glutamic acid to lysine (E340K). The glutamic acid affected in the mrn1 allele is highly conserved in all p52 homologues accessible from GenBank, and it is within a region that is important for the interaction of p52 with XPB in human TFIIH (23). Based on molecular analysis of these three mrn alleles and the complementation experiments with the EP insertions that affect Dmp52, our results prove that mrn is Dmp52.
Heterozygous mutants in DmXPB (haywire) and DmXPD are more sensitive than the wt to UVB irradiation (35, 38, 44); thus, we predicted that the Dmp52 mutants should also be more sensitive than the parental strain. Adult heterozygous Dmp52 mutants (mrn1, mrn3, and mrn5 alleles), as well as a control strain that has the same genetic background and the same balancer chromosome, were irradiated with UVB for various times at a fixed irradiance (see Materials and Methods). The plot presented in Fig. 1D clearly shows that flies heterozygous for Dmp52 lethal alleles are more sensitive to UVB irradiation than the control strain; in particular, the mrn1 mutant is the most sensitive to UVB irradiation among all the tested Dmp52 alleles. The increased UVB irradiation sensitivity is most striking at 100 J/m2, the lowest dosage examined. It is important to stress that one wt Dmp52 allele is present in the balancer chromosome in the analyzed flies, and therefore, all the analyzed mutants were heterozygous.
Developmental phenotypes associated with Dmp52 mutations in the fly.
As we mentioned above, some of the homozygous Dmp52 mutants and heteroallelic combinations die at the stages of third instar larvae and pupae. Interestingly, we noticed that both larvae and pupae homozygous for mrn1, mrn3, and mrn5 alleles and the heteroallelic combinations with the EP3605 mutant are smaller than heterozygous or wt organisms (Fig. 2A and B). This phenotype resembles the minute phenotype, usually caused by protein translation defects. Both larvae and pupae have melanotic tumors in diverse places (Fig. 2F and G). These tumors are aggregates of hematopoietic cells that result from the overproliferation of blood cells and are considered a leukemia-like phenotype (52). In addition, we found that in the specific combination of the mrn3 or mrn5 allele with EP3605, there is a highly reproducible and specific melanization in the larval intersection between the midgut and posterior gut in a region known as the imaginal ring (Fig. 2H and I). The melanization in the imaginal ring could be necrosis, or it could result from abnormal growth of tissue that is subsequently melanized. Organisms that survive the larval stages reach different pupal stages before dying (Fig. 2C). Flies that die at the pharate stage have deformations in the cuticle and exhibit the brittle-bristle phenotype (Fig. 2D and E). These two phenotypes (cuticle deformations and brittle bristles) are identical to those previously reported in haywire (DmXPB) mutants and strongly suggest that these are TFIIH-related defects (35).
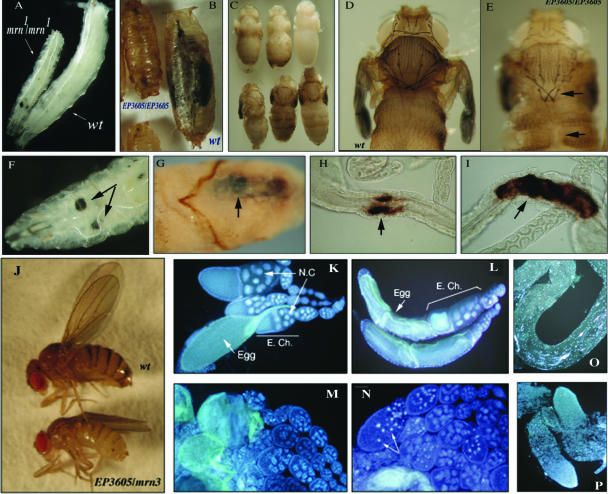
FIG. 2.
Developmental phenotypes associated with mutations in Dmp52. (A) Wt and mrn1/mrn1 mutant third-instar larvae. Note the reduced side of the mutant organism compared to the wt. (B) Wt and EP3605/EP3605 mutant pupae. Note the reduced size of the mutant pupa. (C) Some EP3605/EP3605 and EP3605/mrn1, mrn3, and mrn5 flies can go through metamorphosis but die at different stages before completing development. (D and E) Wt and EP3605/EP3605 pharates, respectively. The mutant organisms have cuticular defects, as well as fragile and deformed bristles (brittle-bristle phenotype). The defects are indicted by arrows. (F and G) mrn1/mrn1 and heteroallelic EP3605/mrn3 larva and pupa, respectively, with melanotic tumors indicated by arrows. (H and I) Presence of melanization in the imaginal ring midgut of heteroallelic EP3605/mrn3 larvae (arrows). This phenotype was always present in the imaginal ring region of the larval gut and extended from this point in both directions. (J) Wt and EP3605/mrn3 adult females. Each genotype is indicated. Note the difference in size between the organisms. (K) Ovarioles from a heterozygous EP3605/+ female. Note that the egg chambers (E. Ch.), nurse cells (N.C.), and oogenesis stages are normal. (L) Ovarioles from an EP3605/EP3605 female. Note that the first oogenesis stages are normal, but the eggs are deformed, apparently resulting from an aberrant chorion (egg shell). (M and N) Ovarioles from heteroalelic EP3605/mrn1 and EP3605/mrn3 females, respectively. Note the aberrant egg chambers, the disrupted nurse cells, and the presence of picnotic nuclei in the eggs (indicated by arrows). (O) Testis from a heterozygous mrn1/+ male. Note that all spermatogenesis stages are present. (P) Testes from an EP3605/mrn1 organism. Note that the testes are aberrant and lack cells progressing through spermatogenesis. The tissue preparation nuclei were visualized by DAPI staining.
The heteroallelic organisms that carry the EP insertion EP3065 and point mutation alleles are semilethal. Interestingly, the organisms that are able to develop to adulthood are smaller than a wt fly, resembling a viable minute phenotype (Fig. 2J). Additionally, these flies have both cuticular-deformation and brittle-bristle phenotypes (data not shown). Although some of the EP3065/mrn (mrn1, mrn3, and mrn5 alleles) heteroallelic flies can develop into adults, they are sterile, as are the homozygous EP3065/EP3065 flies.
Animals were analyzed to determine if sterility was the result of defective gametogenesis. In the case of females, two predominant gametogenesis defects were observed. Figure 2K shows egg chambers with normal oogenesis development in a heterozygous +/mrn3 female. In contrast, in ovarioles from an EP3605/EP3605 female, the first oogenesis stages are normal, but the mature oocytes are deformed (indicated as eggs in Fig. 2L), apparently resulting from an aberrant chorion (egg shell), and therefore the eggs are not viable. In ovarioles from heteroallelic EP3605/mrn1 and EP3605/mrn3 females, aberrant egg chambers were observed and picnotic nuclei containing what looked like apoptotic bodies were present in the nurse cells (indicated in panels M and N), and therefore oogenesis was not completed. In heteroallelic EP3605/mrn1 males, defective testes were present, and spermatogenesis was absent or not completed in most of the organisms (Fig. 2O and P).
Dmp52 is thus similar to haywire (hay), which has a defined deficiency in TFIIH and specific developmental defects. Because Dmp52 fly mutants exhibit specific developmental phenotypes, we infer that some developmental processes are more sensitive than others when TFIIH is subfunctional (21, 35, 53).
The HP52 homologue partially rescues some phenotypes produced by mutations in DMP52.
Although it has been reported that the human XPB homologue is not able to rescue mutations in haywire Drosophila mutants, even if its similarity is 71% (DmXPB) (24), we nevertheless explored whether HP52, which has 50% identity to DMP52, could rescue the mutants that affect Dmp52. To conduct this test, we constructed transgenic flies that express either Dmp52 or the hp52 full-length cDNA under the control of the DmHSP83 promoter using the Casper vector (see Materials and Methods). This promoter is constitutive, and we expected to have moderate expression of the transgenes in all cell types throughout development. We performed the rescue experiments by making crosses between heterozygous point mutation and EP3605 strains with different transgenic lines and then deriving flies homozygous for the Dmp52 mutation and heterozygous for the transgene. First, we looked for rescue of the lethality phenotype. Table 1 shows that, although some homozygous point mutants can develop to adulthood with one copy of the wt Dmp52 gene, the percentage of rescue is low, but as expected, it increases with two transgene copies. The lack of a complete rescue by a single copy of the Dmp52 transgene can be explained if Dmp52 expression is not optimal; components necessary for normal transcript levels could be missing from the promoter or the cDNA or reflect the lack of natural genomic context. In addition, it is known that low levels of transgene transcripts can be influenced by the surrounding chromatin.
TABLE 1.
Rescue of the lethality of homozygous mrn alleles by different transgenic flies
| Mutant genotype | Lethality rescue by transgenic fly (%)a:
|
||||
|---|---|---|---|---|---|
| Dmp52-1 | Dmp52-1/Dmp52-2 | Dmp52-mutb | hp52-2 | hp52-1/hp52-3 | |
| mrn1/mrn1 | 2.4 (540) | 3.4 (256) | 0 (200) | 0 (262) | 2 (111) |
| mrn3/mrn3 | 5.0 (592) | 6.0 (168) | 0 (190) | 1.5 (126) | 3.5 (109) |
| mrn5/mrn5 | 10.4 (281) | 15.0 (267) | NA | NA | 7.8 (153) |
The percentage represents recovery of homozygous flies compared to the expectation for full complementation (number in parentheses). NA, not analyzed.
The Dmp52-mut line is a transgenic line expressing the double point mutant (E340K-R34YE).
In contrast, the sterility phenotype of homozygous EP3605 flies is completely rescued by a single copy of the Dmp52 transgene (Table 2) . As mentioned above, the EP3605 mutant is hypomorphic; we expect that there are reduced Dmp52 transcription levels because of the position of the EP. Therefore, although transgene expression is probably not optimal, it is sufficient for the complete rescue of milder phenotypes.
TABLE 2.
Rescue of the sterility phenotype in heteroallelic organisms by different transgenic flies
| Mutant genotypea | Sterility rescue by transgenic fly (%)b
|
||||
|---|---|---|---|---|---|
| Dmp52-1 | Dmp52-2 | hp52-3 | hp52-4 | Dmp52-mutc | |
| EP3605/EP3605 | 85 | 76 | 95 | 59 | 0 |
| mrn5/EP3605 | 75 | 86 | 100 | 77 | 0 |
| mrn1/EP3605 | 100 | 80 | 0 | 0 | 0 |
| mrn3/EP3605 | 75 | NA | 75 | 87 | NA |
Homozygous EP3605/EP3605 and heteroallelic (EP3605/mrn1, mrn3, and mrn5) adults are 100% sterile.
The percentage represents the individual flies that were fertile and able to generate progeny that developed to larval stage. The total numbers of flies tested were between 10 and 100 for each condition. The numbers of males and females are merged to give a general percentage. There was not bias in sex ratios (data not shown). NA, not analyzed.
The Dmp52-mut line is a transgenic line expressing the double point mutant (E340K-R344E).
In the test of complementation using the hp52 cDNA, a partial rescue of lethality was also observed in homozygous point mutations, but as expected, it was lower than with the Dmp52 transgene (Table 1). Note that two independent transgenic hp52 fly lines rescued, at least partially, the mrn lethal phenotype (Table 1). These lines have a single transgene, and also, the value increases with two copies. Interestingly, with the exception of the mrn1/EP3605 flies, the hp52 transgene was able to fully rescue the fertility defect in homozygous EP3605 flies (Table 2). Note that a Dmp52 double point mutant (E340K/R344D) (Fig. 1E; also see below) transgenic fly was unable to rescue the phenotype in any case. These results demonstrate that the human homologue of Dmp52 is sufficiently conserved to rescue at least some Dmp52 functions during development. The inability of hp52 transgenes to substitute for the mrn1 allele that contains only a single amino acid change is intriguing. Therefore, a more detailed analysis of the protein-protein interactions between p52 and other TFIIH subunits will be required to address this point.
Dmp52 mutations affect nuclear TFIIH levels in vivo.
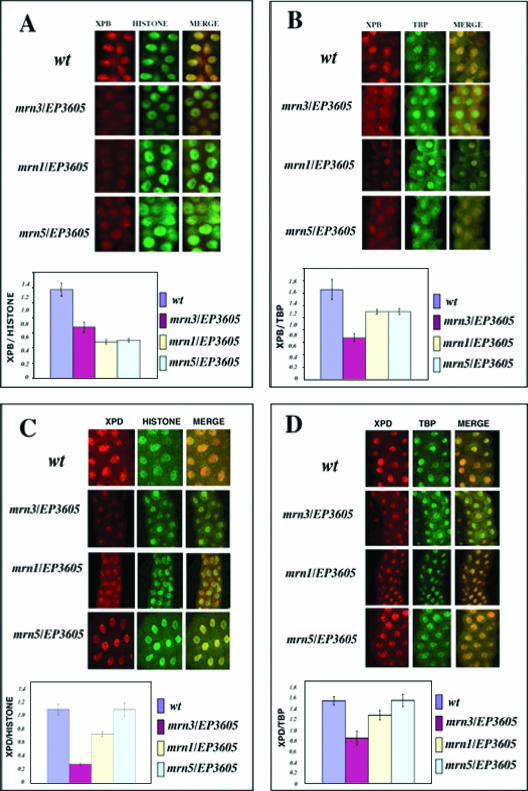
Considering the phenotypes of the fly, we wondered whether these defects could be associated with some particular TTD features. Knowing that reduction in the TFIIH cellular concentration is a typical TTD phenotype (4), we analyzed third-instar salivary gland cells from heteroallelic flies for EP3605, either the mrn1, mrn3, or mrn5 allele, by immunofluorescence of XPB and XPD. Internal controls were made using anti-histone H3 and anti-TBP antibodies. We found that in all the Dmp52 heteroallelic combinations, there was a substantial reduction of the DmXPB protein levels (Fig. 3A and B). With the exception of EP3605/mrn5 organisms, we found that XPD levels were also reduced when Dmp52 was mutated (Fig. 3C and D). This finding parallels the observations reported in XP, XP/CS, and TTD patients, where some of the XPB, XPD, and p8 mutations affect the cellular levels of TFIIH.
FIG. 3.
XPB and XPD protein levels are reduced in the Dmp52 mutants. Wt and heteroallelic mutant third-instar larval salivary glands were costained with either XPB/histone H3, XPB/TBP, or XPD/histone H3 and XPD/PBT antibodies. (A) Costaining and quantification of the XPB/histone ratio. Note that the XPB levels are reduced at least twofold in the mutant nuclei. (B) Costaining and quantification of the XPB/TBP ratio. XPB levels are reduced compared with the control. (C) Costaining and quantification of the XPD/histone ratio. (D) Costaining and quantification of the XPD/TBP ratio. In EP3605/mrn1 and EP3605/mrn3 heteroallelic flies, the XPD levels are reduced. For quantification of fluorescence, we followed the protocol reported previously (31) (see Materials and Methods). At least 10 nuclei for each condition were analyzed. The error bars indicate standard errors. For details, see the text.
Chromosomal instability in Dmp52 mutants.
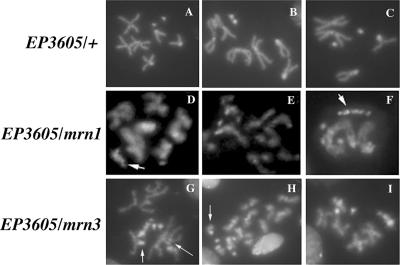
TFIIH is implicated in NER, which is fundamental for genome stability. In humans, mutations in the XPD subunit are linked to cancer. As we have demonstrated in this work, Dmp52 mutants are deficient in DNA repair and as a consequence may have chromosome fragility. Therefore, it was interesting to analyze whether mutations in Dmp52 could generate chromosomal instability. To examine chromosomal integrity, we used neuroblast squashes from heteroalleic Dmp52 mutants and control larvae. In mitotic spreads from Dmp52 mutants (Fig. 4D to I), we observed a high rate of chromosomal aberrations that were evident when comparing wt metaphasic chromosomes (Fig. 4A to C). The most common aberrations were fragmentation of chromosomes and partial chromosome condensation. Acentric chromosome fragments were observed, as well as loss of chromosomal arms (indicated in Fig. 4F, G, and H). In addition, chromosome association and chromosomal rearrangements were found (Fig. 4F and G). These results show for the first time in Drosophila that mutations in one of the TFIIH subunits affect chromosomal integrity.
FIG. 4.
Mutations in Dmp52 cause chromosomal aberrations. (A to C) Mitotic spreads from EP3605/+ neuroblasts, which are identical to the wt. (D to F) Mitotic spreads from EP3605/mrn1 heteroallelic mutants. Note the presence of partially condensed chromosomes and chromosomal fragments, indicated by the arrows. (G to I) Mitotic spreads of EP3605/mrn5 heteroallelic mutants. Note the presence of chromosomal fragments, indicated by arrows, and partially condensed chromatids.
Effect of Dmp52 point mutations on the XPB ATPase activity and the TFIIH transcription and DNA repair activities.
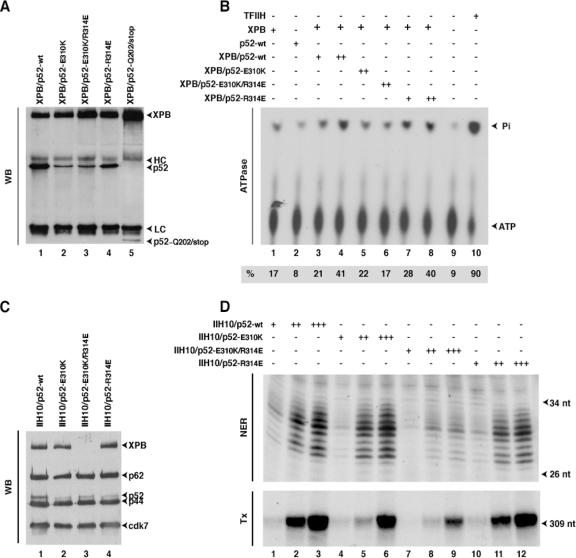
We next asked how the Dmp52 mutations could affect TFIIH functions. The Dmp52 point mutations were introduced by site-directed mutagenesis into the hp52 cDNA at positions Q70/stop (corresponding to mrn5), Q202/stop (corresponding to mrn3), and E310K (corresponding to mrn1), as well as at positions R314D and E310K/R314D, a single and a double mutation (see Discussion) located in a highly conserved region, in order to generate baculoviruses expressing the corresponding proteins. The truncated p52 peptide derived from the Q70/stop mutant was unstable in Sf2 cells (data not shown) and was not analyzed further. Knowing that p52 interacts with XPB (23) and stimulates its ATPase activity to trigger NER (F. Coin, V. Oksenych, and J. M. Egly, submitted for publication), we were wondering what would be the consequences of p52 mutations for XPB ATPase activity. Immunoprecipitations were carried out using insect cell extracts expressing the mutated HP52 proteins together with the wt XPB. p52-E310K and p52-E310K/R314D mutations significantly weakened XPB binding compared to wt p52 or p52-R314D (Fig. 5A). We also observed that the truncated p52-Q202/stop was hardly retained on XPB. There was also a similar reduction of the p52/XPB ratio when we used the p52 antibody to pull down p52 (data not shown). As a consequence, when tested in an ATPase assay, we found that XPB was weakly stimulated by p52-E310K (22%) and p52-E310K/R314D (17%), while wt p52 and p52-R314D were stimulated normally (Fig. 5B). Taken together, our results indicate that the mutations found within a highly conserved region in Drosophila p52 prevent the regulation of XPB ATPase by the p52 regulatory subunit.
FIG. 5.
Mutations in HP52 cause low stimulation of XPB ATPase activity and NER defects. (A) Recombinant XPB and mutant HP52 expressed in insect cells were immunoprecipitated with an anti-XPB antibody and analyzed by Western blotting (WB). The positions of XPB, HP52, the truncated p52-Q202/stop, and the heavy (HC) and light (LC) immunoglobulin chains are indicated. (B) Wt XPB, either alone or in combination with the p52 mutants, was immunoprecipitated with Ab-XPB antibody immobilized on agarose beads. The immobilized protein complexes were washed with 0.4 M KCl and tested in an ATPase assay. TFIIH was used as a control. Quantifications of the inorganic phosphate (Pi) release and ATP were done with a Bio-imaging analyzer, and the ratio presented at the bottom of the figure shows the percentage of the released Pi. (C) Immunopurified recombinant TFIIHs (as indicated at the top of the blot) from insect cell extracts containing all the TFIIH subunits, including either wt HP52 or mutant HP52, were subjected to Western blot analysis using antibodies against XPB, p62, p52, or Cdk7. The TFIIH subunits are indicated by arrowheads. (D) Fifty, 100, and 200 ng of the various IIH10 complexes were tested in a dual-incision assay (NER) containing the recombinant XPC-HR23b, XPA, RPA, XPG, and ERCC1-XPF factors and a closed-circular plasmid containing a single Pt-GTG-DNA cross-link as a template or in a reconstituted transcription assay (Tx) composed of recombinant TFIIB, TFIIF, TBP, TFIIE factors, the purified RNA Pol II, and the adenovirus major late promoter template (11). The sizes of the incision products or transcripts are indicated. nt, nucleotides.
To further analyze the consequences of p52 mutations in transcription and DNA repair, recombinant IIH10 (rIIH10) complexes containing either the wt or modified p52 protein, together with the other nine TFIIH subunits, were produced in H5 insect cells and subjected to a two-step purification process (11). The rIIH10 complexes immunoprecipitated with antibodies directed toward the p44 subunit of TFIIH were next analyzed by Western blotting after being washed with 300 mM KCl (Fig. 5C). The E310K, E310K/R314D, and R314D-p52 proteins were incorporated into the IIH core, as demonstrated by the presence of p62 and cdk7 subunits. However, XPB incorporation was significantly reduced in rIIH10/p52-E310K/R314D compared with rIIH10/p52-wt. It is intriguing that XPB does not play a major role in the stability of rIIH10.
These mutant rIIH10 complexes were tested in both reconstituted transcription and NER assays (11). We observed that the transcription and NER activities of rIIH10/p52-E310K/R314E were significantly reduced compared to rIIH10/p52-wt (Fig. 5D, lanes 7 to 9), underlining how the preservation of the HP52/XPB interacting domains is crucial for the maintenance of both activities. These results also explain why Dmp52 transgenic flies containing this double mutation could not rescue the wt phenotype in Drosophila (Tables 1 and 2). We also noticed that the transcription activity of rIIH10/p52-E310K (which produced the severe phenotype in the fly) was partially (around 25%) inhibited, although NER was rather normal compared to rIIH10/p52-wt (lanes 4 and 5). rIIH10/p52-E314K did not exhibit any deficiency in either transcription or NER activities (lanes 10 to 12).
DISCUSSION
Disturbing TFIIH activities and architecture.
It has previously been demonstrated that HP52 is necessary for the incorporation of XPB into the core of TFIIH (23). Based on this information and because HP52 can rescue some of the DMP52 mutant phenotypes, we introduced mutations into hp52 cDNA to reconstruct the Drosophila mutations and then analyzed their effects in vitro. Here, we have shown that mutations in a highly conserved domain of DMP52 prevent the binding and regulation of XPB ATPase activity, a function that is required for both transcription and DNA repair. With such findings, is intriguing that the p52-E310K mutation that confers UV sensitivity on the fly does not result in an appreciable in vitro biochemical phenotype in humans, although we demonstrated that the mutation does disturb the regulation by p52 of XPB ATPase activity. It thus seems that in humans, the other subunits of TFIIH may circumvent the weakening of the XPB/p52 interaction, allowing TFIIH to be active in both transcription and DNA repair, at least in our in vitro assays. In Drosophila, the other TFIIH subunits are probably unable to compensate for the XPB-p52 interaction defect. However, the importance of residue E310 of p52 for TFIIH transcription and repair functions became clear when a mutation in this residue (E310K) was combined with a mutation in another very conserved residue of p52 (R314E) located 4 amino acids downstream. In this case, p52 was not able to anchor XPB to the core TFIIH, resulting in a TFIIH with low repair and transcription activities. These results show that the region where the HP52/E310R-R314E mutant is located is very important for the interaction with XPB and its subsequent assembly or retention in TFIIH (Fig. 5). The double mutant E310K/R314E does not exist in the fly or human; its construction was based on the high conservation of the residues E310 and R314 in all organisms. Of course, the key element was the mutant mrn1, which has the substitution E310K. As mentioned above, this region binds with XPB, and our results indicate that changes in the amino acid sequence in this domain have a more deleterious effect than a deletion that does not contain this region.
TTD-like phenotypes in Dmp52 mutants.
In the case of the p52 subunit, there are only reports of a conditional mutant in yeast (13), a model in which no developmental effects can be studied. We analyzed two different types of mutations in the Drosophila p52 homologue, EP insertions and three point mutations. With the exception of the EP3605 insertion, all of the analyzed mutants are homozygous lethal. Heteroallelic combinations of EP3605 with the point mutant alleles are semilethal, and adult organisms showing intriguing phenotypes, such as cuticle defects, brittle bristles, and sterility, can be obtained. These two phenotypes are also present in hay (DmXPB) mutants (35). Therefore, we could associate these defects with deficiencies in some TFIIH functions during development. We also observed new phenotypes that we did not find in hay mutants. For instance, homozygous organisms died in the third-instar larval or the pupal stage; these cases were produced by heterozygous females, and the maternally contributed p52 might have sufficed to allow development until these stages. Interestingly, both larvae and pupae homozygous for Dmp52 mutations are significantly smaller than wt organisms. In addition heteroallelic flies with the EP3605 and mrn1, mrn3, or mrn5 mutations can develop to adults but are smaller and sterile. These phenotypes, together with the fragile-bristle phenotype, the reduction of total TFIIH levels, and slow development, resemble TTD in humans and generate a minute-like phenotype in Drosophila. In general, minute phenotypes arise from abnormalities in protein synthesis during development, including a reduction in the ribosomal gene copy number (43); therefore, a reduction in the basal transcription levels from TFIIH mutants may have a similar effect. It is important to note that human patients affected with TTD and CS are short in stature, and therefore, there is an interesting correlation between this human phenotype and the slow-growth phenotype of the Dmp52 gene.
“Cancer-XP-like” phenotypes in Dmp52 mutants.
Some Dmp52 mutant larvae and pupae have melanotic tumors. These tumors are caused mainly by the overgrowth of particular tissues, generally hemocytes, that later melanize (22). Similar tumors can be found in Drosophila mutants affected in other kind of functions, such as transcription modulators and chromatin-remodeling factors, as well in the innate immune system and the JAK/STAT pathway (2, 50, 51), but not in haywire mutants (35). Furthermore, defects in DNA repair in Drosophila, particularly from disruption of the Rad50 gene, which participates in double-strand repair, also generate melanotic tumors. Intriguingly, Rad50 mutants also have cuticle defects (20). Recently, it has been reported that mutations in the damaged DNA binding protein 1 in Drosophila, which participates in NER and in the regulation of the innate immune response, generate chromosomal instability and melanotic tumors (50, 51).
In humans, deficiency in TFIIH activity has been associated with tumor generation, and polymorphisms in XPD are linked to different kinds of cancers (references 7 and 17 and our unpublished results). Current evidence suggests there is no casual relationship between XPD polymorphisms, reduced DNA repair, and increased cancer risk. In addition, it is very well known that patients affected with XP develop skin cancer with high frequency (30). Therefore, it is possible that deficiencies in transcription and DNA repair caused by mutations in Dmp52 are linked to melanotic tumors. In support of this hypothesis, we observed different types and degrees of chromosomal aberrations in mitotic chromosomes in Dmp52 mutants. The most frequent abnormalities were the presence of chromosome breaks and partial chromatid condensation at different mitotic stages. In mammals, chromosomal fragility is common in solid tumors (25), and chromosome instability has been reported in fibroblasts derived from XP individuals, as well as uncontrolled DNA breakage in UV-irradiated XPD from cells of patients (3, 5, 28). However, there are few studies of global genome integrity in patients with deficiencies in TFIIH (17). Therefore, the outcome of combinations of mutations in Dmp52 with mutations in tumor suppressor genes, such as p53 and retinoblastoma in the fly, needs to be further investigated by conducting genetic analysis that is impossible in humans.
Pleiotropy and TFIIH mutants.
Dmp52 mutants are pleiotropic and produce different phenotypes during fly development. We have observed a similar situation in hay mutants (XPB) and have demonstrated that the type of mutation or the subunit affected in TFIIH affects genes differentially (21, 53). Mutations in Dmp52 produce more diverse developmental defects than hay mutations. This phenomenon could be explained by two possibilities that are not mutually exclusive. The first is that mutations in the structural components of TFIIH, such as altered p52, may have a more drastic effect on complex functions, since it is not only important for TFIIH assembly, but also modulates XPB ATPase activity. Alternatively, because TFIIH interacts with multiple factors in transcription and DNA repair, a mutation in p52 may not only affect the assembly of TFIIH and its activities, but can also abolish some of these specific interactions. These two points may also explain the absence of human diseases related to HP52 mutants. That is, such mutations are probably early embryonic lethal in mammals because of the requirements for early embryonic gene transcription. However, we cannot exclude the possibility of the future discovery of a human syndrome caused by mutations in HP52.
When working with multifunctional protein complexes, such as TFIIH, pleiotropic phenotypes are difficult to interpret, because the complexes participate in so many processes. The question to ask is whether pleiotropy can be resolved by identifying the multiple functions a particular protein has within the complex. In our approach to studying TFIIH, we are close to refining the mechanisms, and we can uncover the domains of the protein that are required for each aspect of the biochemical phenotype and thus connect them to the complete organism phenotypes. Then, we can propose a working model to understand TFIIH pleiotropic defects. We think that p52 is a bifunctional protein by virtue of anchoring to the TFIIH complex and, using different domains, by binding to XPB to modulate the activity of the enzyme. Additionally, we could assign specific p52 regions as differentially modulating the NER and transcriptional activities of XPB. Because p52 is bifunctional, we predict that it will be possible to recover mutants that fully distinguish the anchoring and modulation of XPB activities. The existing mutants are a good start, because they identify functional domains, but more detailed structure-function studies are required to understand the mechanisms.
Acknowledgments
We thank V. Walbot for criticism and discussion of the manuscript, V. Barajas for technical advice, and Andres Saralegui for help with the confocal images. We also thank C. Brawn for technical advice. We are grateful to I. Kolb-Cheynel and J. L. Weickert for the production of recombinant baculoviruses.
This study was supported by funds from l'Association de la Recherche contre le Cancer (no. 3113), from l'Institut des Maladies Rares (A03098MS), and from the European Community (no. LSHC-CT-2005-512113.) to J.-M.E. and from PAPIIT/UNAM, CONACyT, and the Howard Hughes Medical Institute to M.Z.
Footnotes
Published ahead of print on 5 March 2007.
REFERENCES
- 1.Araujo, S. J., F. Tirode, F. Coin, H. Pospiech, J. E. Syvaoja, M. Stucki, U. Hubscher, J. M. Egly, and R. D. Wood. 2000. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 14:349-359. [PMC free article] [PubMed] [Google Scholar]
- 2.Bandenhorst, P., M. Voas, I. Rebay, and C. Wu. 2002. Biological functions of ISWI chromatin remodeling complex NURF. Genes Dev. 16:3186-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berneburg, M., L. E. Lowe, T. Nardo, S. Araujo, M. I. Fousteri, M. H. Green, J. Krutmann, R. D. Wood, M. Stefanini, and A. R. Lehmann. 2000. UV damage causes uncontrolled DNA breakage in cells from patients with combined features of XP-D and Cockayne syndrome. EMBO J. 19:1157-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Botta, E., T. Nardo, A. R. Lehmann, J. M. Egly, A. M. Pedrini, and M. Stefanini. 2002. Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum. Mol. Genet. 11:2919-2928. [DOI] [PubMed] [Google Scholar]
- 5.Casati, A., M. Stefanini, R. Giorgi, P. Ghetti, and F. Nuzzo. 1991. Chromosome rearrangements in normal fibroblast from xeroderma pigmentosum homozygotes and heterozygotes. Cancer Genet. Cytogenet. 51:89-101. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., S. Larochelle, X. Li, and B. Suter. 2003. Xpd/Ercc2 regulates CAK activity and mitotic progression. Nature 424:228-232. [DOI] [PubMed] [Google Scholar]
- 7.Clarkson, S. G., and R. D. Wood. 2005. Polymorphisms in the human XPD (ERCC2) gene, DNA repair capacity and cancer susceptibility: an appraisal. DNA Repair 4:1068-1074. [DOI] [PubMed] [Google Scholar]
- 8.Coin, F., J. C. Marinoni, C. Rodolfo, S. Fribourg, A. M. Pedrini, and J. M. Egly. 1998. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat. Genet. 20:184-188. [DOI] [PubMed] [Google Scholar]
- 9.Coin, F., E. Bergmann, A. Tremeau-Bravard, and J. M. Egly. 1999. Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J. 18:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coin, F., L. Proietti De Santis, T. Nardo, O. Zlobinskaya, M. Stefanini, and J. M. Egly. 2006. p8/TTD-A as a repair-specific TFIIH subunit. Mol. Cell 20:215-226. [DOI] [PubMed] [Google Scholar]
- 11.Dubaele, S., L. Proietti De Santis, R. J. Bienstock, A. Keriel, M. Stefanini, B. Van Houten, and J. M. Egly. 2003. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol. Cell 11:1635-1646. [DOI] [PubMed] [Google Scholar]
- 12.Egly, J. M. 2001. TFIIH: from transcription to clinic. FEBS Lett. 498:124-128. [DOI] [PubMed] [Google Scholar]
- 13.Feaver, W. J., N. L. Henry, Z. Wang, X. Wu, J. Q. Svejstrup, D. A. Bushnell, E. C. Friedberg, and R. D. Kornberg. 1997. Genes for Tfb2, Tfb3, and Tfb4 subunits of yeast transcription/repair factor IIH. Homology to human cyclin-dependent kinase activating kinase and IIH subunits. J. Biol. Chem. 272:19319-19327. [DOI] [PubMed] [Google Scholar]
- 14.Fisher, R. P. 2005. Secrets of a double agent: CDK7 in cell cycle control and transcription. J. Cell Sci. 118:5171-5180. [DOI] [PubMed] [Google Scholar]
- 15.Friedberg, E., G. H. Walker, W. Siede, R. D. Wood, R. A. Schultz, and T. Ellenberger. 2006. DNA repair and mutagenesis. ASM Press, Washington, DC.
- 16.Fuller, M. T., C. L. Regan, L. L. Green, B. Robertson, R. Deuring, and T. S. Hays. 1989. Interacting genes identify interacting proteins involved in microtubule function in Drosophila. Cell Motil. Cytoskeleton 14:128-135. [DOI] [PubMed] [Google Scholar]
- 17.Garfinkel, D. J., and A. M. Bailis. 2002. Nucleotide excision repair, genome stability, and human disease: new insight from model systems. J. Biomed. Biotechnol. 2:55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerard, M., L. Fischer, V. Moncollin, J. M. Chipoulet, P. Chambon, and J. M. Egly. 1991. Purification and interaction properties of the human RNA polymerase B(II) general transcription factor BTF2. J. Biol. Chem. 266:20940-20945. [PubMed] [Google Scholar]
- 19.Giglia-Mari, G., F. Coin, J. A. Ranish, D. Hoogstraten, A. Theil, N. Wijgers, N. G. Jaspers, A. Raams, M. Argentini, P. J. van der Spek, E. Botta, M. Stefanini, J. M. Egly, R. Aebersold, J. H. Hoeijmakers, and V. Vermeulen. 2004. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 36:714-719. [DOI] [PubMed] [Google Scholar]
- 20.Gorski, M. M., R. J. Romejin, J. C. J. Eeken, A. W. M. de Jong, B. L. van Veen, K. Szuhai, L. L. Mullunders, W. Ferro, and A. Pasttink. 2004. Disruption of Drosophila Rad50 causes pupal lethality, the accumulation of DNA double-strand breaks, and the induction of apoptosis in the third instar larvae. DNA Repair 3:603-615. [DOI] [PubMed] [Google Scholar]
- 21.Gutierrez, L., C. Merino, M. Vazquez, E. Reynaud, and M. Zurita. 2004. RNA polymerase II 140 wimp mutant and mutations in the TFIIH subunit XPB differentially affect homeotic gene expression in Drosophila. Genesis 40:58-66. [DOI] [PubMed] [Google Scholar]
- 22.Huang. L., S. Ohsako, and S. Tanda. 2005. The lesswright mutation activates Rel-related proteins, leading to overproduction of larval hemocytes in Drosophila melanogaster. Dev. Biol. 280:407-420. [DOI] [PubMed] [Google Scholar]
- 23.Jawhari, A., J.-P. Lainé, S. Dubaele, V. Lamour, A. Poterszman, F. Coin, D. Moras, and J. M. Egly. 2002. p52 mediates XPB function within the transcription/DNA repair factor TFIIH. J. Biol. Chem. 277:31761-31767. [DOI] [PubMed] [Google Scholar]
- 24.Koken, M. H., C. Vreeken, S. A. Bol, N. C. Cheng, I. Jaspers-Dekker, J. H. Hoeijmakers, J. C. Eeken, G. Weeda, and A. Pastink. 1992. Cloning and characterization of the Drosophila homolog of the xeroderma pigmentosum complementation-group B correcting gene, ERCC3. Nucleic Acids Res. 20:5541-5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kops, G. J. P., B. A. A. Weaver, and D. W. Cleeland. 2005. On the road to cancer: aneuploidy and mitotic checkpoint. Nat. Rev. Cancer 5:773-785. [DOI] [PubMed] [Google Scholar]
- 26.Laine, J. P., and J. M. Egly. 2006. Initiation of DNA repair mediated by a stalled RNA polymerase IIO. EMBO J. 25:387-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laine, J. P., and J. M. Egly. 2006. When transcription and repair meet: a complex system. Trends Genet. 22:430-436. [DOI] [PubMed] [Google Scholar]
- 28.Lanza, A., P. Lagomarsini, A. Casati, P. Ghetti, and M. Stefanini. 1997. Chromosomal fragility in the cancer-prone disease xeroderma pigmentosum preferentially involves bands relevant for cutaneous carcinogenesis. Int. J. Cancer 74:654-663. [DOI] [PubMed] [Google Scholar]
- 29.Larochelle, S., J. Pandur, R. P. Fisher, H. Salz, and B. Suter. 1998. Cdk7 is essential for mitosis and for in vivo Cdk-activating kinase activity. Genes Dev. 12:370-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehmann, A. R. 1998. Dual functions of DNA repair genes: molecular, cellular and clinical implications. BioEssays 20:146-155. [DOI] [PubMed] [Google Scholar]
- 31.Lehmann, A. R. 2002. Ageing: repair and transcription keep us from premature ageing. Curr. Biol. 12:550-551. [DOI] [PubMed] [Google Scholar]
- 32.Lehman, A. R. 2003. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and tricothiodystrophy. Biochimie 85:1101-1111. [DOI] [PubMed] [Google Scholar]
- 33.Lindahl, T., P. Karran, and R. D. Wood. 1997. DNA excision repair pathways. Curr. Opin. Genet. Dev. 7:158-169. [DOI] [PubMed] [Google Scholar]
- 34.Marinoni, J. C., M. Rossignol, and J. M. Egly. 1997. Purification of the transcription/repair factor TFIIH and evaluation of its associated activities in vitro. Methods 12:235-253. [DOI] [PubMed] [Google Scholar]
- 35.Merino, C., E. Reynaud, M. Vazquez, and M. Zurita. 2002. DNA repair and transcriptional effects of mutations in TFIIH in Drosophila development. Mol. Biol. Cell 13:3246-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merino, E., J. Osuna, F. Bolivar, and X. Soberon. 1992. A general PCR-based method for single or combinatorial oligonuceotide-directed mutagenesis on pUC/M13 vectors. Biotechnology 12:508-510. [PubMed] [Google Scholar]
- 37.Morgan, D. O. 1995. Principles of CDK regulation. Nature 374:131-134. [DOI] [PubMed] [Google Scholar]
- 38.Mounkes, L. C., R. S. Jones, L. Bee-Choo, W. Gelbart, and M. T. Fuller. 1992. A Drosophila model for xeroderma pigmentosum and Cockayne's syndrome: haywire encodes the fly homologue of ERCC3, a human excision repair gene. Cell 71:925-937. [DOI] [PubMed] [Google Scholar]
- 39.Ranish, J. A., S. Hahn,. Y. Lu, E. C. Yi, X. J. Li, J. Eng, and R. Aebersold. 2004. Identification of TFB5, a new component of general transcription and DNA repair factor IIH. Nat. Genet. 36:707-713. [DOI] [PubMed] [Google Scholar]
- 40.Reynaud, E., H. Lomeli, M. Vazquez, and M. Zurita. 1999. The Drosophila melanogaster homologue of the xeroderma pigmentosum D gene product is located in euchromatic regions and has a dynamic response to UV light-induced lesions in polytene chromosomes. Mol. Biol. Cell 10:1191-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riedl, T., F. Hanaoka, and J. M. Egly. 2003. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 22:5293-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rossignol, M., I. Kolb-Cheynel, and J. M. Egly. 1997. Substrate specificity of the cdk-acticvating kinase (CAK) is altered upon association with TFIIH. EMBO J. 16:1628-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saeboe-Larssen, S., M. Lyamouri, J. Merriam, and M. P. Oksvold. 1998. Ribosomal protein insufficiency and the minute syndrome in Drosophila: a dose-response relationship. Genetics 148:1215-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandoval, M. T., and M. Zurita. 2001. Increased UV light sensitivity in transgenic Drosophila expressing the antisense XPD homologue. Antisense Nucleic Acid Drug Dev. 11:125-128. [DOI] [PubMed] [Google Scholar]
- 45.Shivji, M. K., J. G. Moggs, I. Kuraoka, and R. D. Wood. 1999. Dual-incision assays for nucleotide excision repair using DNA with a lesion at a specific site. Methods Mol. Biol. 113:373-392. [DOI] [PubMed] [Google Scholar]
- 46.Silva, E., S. Tiong, P. Pedersen, E. Homola, A. Royou, B. Fasulo, G. Siriaco, and S. D. Campbell. 2004. ATM is required for telomere maintenance and chromosomal stability during Drosophila development. Curr. Biol. 14:1341-1347. [DOI] [PubMed] [Google Scholar]
- 47.Spradling, A., and G. Rubin. 1982. Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218:341-347. [DOI] [PubMed] [Google Scholar]
- 48.Sugasawa, K., Y. Okuda, M. Saijo, R. Nishi, N. Matsuda, G. Chu, T. Mori, S. Iwai, K. Tanaka, K. Tanaka, and F. Hanaoka. 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121:387-400. [DOI] [PubMed] [Google Scholar]
- 49.Takagi, Y., C. A. Masuda, W.-H. Chang, H. Komori, D. Wang, T. Hunter, C. A. P. Joazeiro, and R. D. Kornberg. 2005. Ubiquitin ligase activity of TFIIH and the transcriptional response to DNA damage. Mol. Cell 18:237-243. [DOI] [PubMed] [Google Scholar]
- 50.Takata, K., H. Yoshida, M. Yamaguchi, and K. Sakaguichi. 2004. Drosophila damaged DNA-binding protein 1 is an essential factor for development. Genetics 168:855-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takata, K., K. Shimanouchi, M. Yamaguchi, M. S. Murakami, G. Ishikawa, R. Takeuchi, Y. Kanai, T. Ruike, R. Nakamura, Y. Abe, and K. Sakaguchi. 2004. Damaged DNA binding protein 1 in Drosophila defense reactions. Biochem. Biophy. Res. Commun. 323:1024-1031. [DOI] [PubMed] [Google Scholar]
- 52.Zynick, D. L., B. G. McGonnigal, and C. R. Dearolf. 1993. Drosophila awdk-pn, a homologue of the metastasis suppressor gene nm23, suppresses the Tum-1 hematopoietic oncogene. Nat. Genet. 4:195-201. [DOI] [PubMed] [Google Scholar]
- 53.Zurita, M., and C. Merino. 2003. The transcriptional complexity of the TFIIH complex. Trends Genet. 19:578-584. [DOI] [PubMed] [Google Scholar]