Abstract
The heterodimeric hypoxia-inducible transcription factors (HIFs) are central regulators of the response to low oxygenation. HIF-α subunits are constitutively expressed but rapidly degraded under normoxic conditions. Oxygen-dependent hydroxylation of two conserved prolyl residues by prolyl-4-hydroxylase domain-containing enzymes (PHDs) targets HIF-α for proteasomal destruction. We identified the peptidyl prolyl cis/trans isomerase FK506-binding protein 38 (FKBP38) as a novel interactor of PHD2. Yeast two-hybrid, glutathione S-transferase pull-down, coimmunoprecipitation, colocalization, and mammalian two-hybrid studies confirmed specific FKBP38 interaction with PHD2, but not with PHD1 or PHD3. PHD2 and FKBP38 associated with their N-terminal regions, which contain no known interaction motifs. Neither FKBP38 mRNA nor protein levels were regulated under hypoxic conditions or after PHD inhibition, suggesting that FKBP38 is not a HIF/PHD target. Stable RNA interference-mediated depletion of FKBP38 resulted in increased PHD hydroxylation activity and decreased HIF protein levels and transcriptional activity. Reconstitution of FKBP38 expression abolished these effects, which were independent of the peptidyl prolyl cis/trans isomerase activity. Downregulation of FKBP38 did not affect PHD2 mRNA levels but prolonged PHD2 protein stability, suggesting that FKBP38 is involved in PHD2 protein regulation.
The response to reduced tissue oxygenation (hypoxia) is characterized by alterations in gene expression, allowing the organism to adapt to hypoxia on the systemic, local, and cellular levels (46). A large number of these target genes are regulated by hypoxia-inducible factor 1 (HIF-1) and HIF-2, the master regulators of oxygen homeostasis in physiological, as well as in pathophysiological, processes (3, 34, 37). HIFs are members of the basic helix-loop-helix/Per-ARNT-Sim (bHLH/PAS) transcription factor family and bind as an α/β heterodimer to cis-regulatory hypoxia response elements (HREs) (47). Oxygen-dependent regulation is mediated by the HIF-α subunits and involves novel enzymatic posttranslational modification pathways that hydroxylate specific amino acids, depending on the oxygen partial pressure (36). A novel family of oxygen-, ferrous iron-, and 2-oxoglutarate-dependent prolyl-4-hydroxylase domain-containing enzymes (PHD1, PHD2, and PHD3) mediate the proteolytic regulation of HIF-α subunits (4, 9, 15). Under normoxic conditions, two conserved prolyl residues within the oxygen-dependent degradation (ODD) domain of HIF-α are hydroxylated and recognized by the von Hippel-Lindau (pVHL) tumor suppressor protein via trans-4-hydroxyprolyl binding (16, 17, 25). pVHL serves as a substrate recognition unit of an E3 ligase complex, targeting HIF-α for degradation by the ubiquitin-proteasome pathway. Because PHDs depend on molecular oxygen as a cosubstrate, hydroxylation is reduced in hypoxia, leading to HIF-α stabilization and accumulation. Thus, PHDs can serve as cellular oxygen sensors and provide a direct link between oxygen availability and HIF-dependent transcriptional regulation. In addition to the regulation of protein stability, hydroxylation of a specific asparaginyl residue within the C-terminal transactivation domain of HIF-α by factor inhibiting HIF (FIH) prevents interaction with the CH1 domains of the p300/CBP transcriptional coactivators, thus reducing the transcriptional activity of HIF-α (13, 21, 22). FIH belongs to the same family as the PHDs, but its Km for oxygen is lower (14, 20), providing an elegant way of fine tuning HIF-α stabilization and target gene induction by the current tissue oxygenation.
All three PHD proteins are able to hydroxylate HIF-α in vitro, but they differ in their expression patterns, as well as in their subcellular localization, at least when overexpressed (1, 26). However, recent analysis of endogenous localization showed expression of all three PHD enzymes predominantly in the cytoplasm (39). PHD2 is ubiquitously expressed and has been shown to be the main hydroxylase responsible for HIF-α modification in normoxia (2). The recent description of a family with erythrocytosis caused by an inherited PHD2 proline point mutation confirmed these data (32). Interestingly, PHD2 and PHD3 are HIF-dependently regulated, constituting a negative feedback mechanism, whereas PHD1 is not regulated by hypoxia (27, 33). In accordance with the complex functions of HIF in various hypoxic adaptation processes, the PHD proteins also seem to be subjected to multilayer regulatory mechanisms, including modification of enzyme activity by posttranslational modifications and protein associations (15, 24). To screen for novel PHD interactors, we applied yeast two-hybrid methodology using PHD2 as bait and identified the peptidyl prolyl cis/trans isomerase FK506-binding protein 38 (FKBP38) as prey.
MATERIALS AND METHODS
Plasmids.
Unless otherwise indicated, cloning work was carried out using Gateway technology (Invitrogen, Basel, Switzerland). Entry vectors were generated by cloning PCR fragments into BamHI/EcoRV-digested (all restriction enzymes were purchased from MBI Fermentas, Labforce, Nunningen, Switzerland) pENTR4 or pENTR/D-TOPO vectors. Full-length PHD1 to -3 (accession numbers NM_053046, NM_022051, and NM_022073, respectively) were amplified by PCR from plasmids pcDNA3.1/PHD1 (29), pcDNA3.1/HA-PHD2 (15), and pcDNA3.1/PHD3 (29). pENTR/PHD2-170-426 was obtained by digesting pENTR/PHD2 with SmaI and NotI, Klenow fill-in, and religation. To obtain pDONR/FKBP38, as well as pDONR/FKBP38Δ98-257, the BP Clonase recombination enzyme mixture (Invitrogen) was used to transfer the corresponding inserts from yeast two-hybrid library vectors into pDONR221. DNA fragments corresponding to amino acids (aa) 3 to 97, aa 99 to 412, and aa 256 to 412 of FKBP38 (accession number AY278607) were amplified by PCR from plasmid pDONR-FKBP38 and cloned into pENTR4. The DNA fragment corresponding to the HIF-2αODD-domain (aa 404 to 569) was amplified by PCR from plasmid pcDNA3/hEPAS and cloned into pENTR/D-TOPO. The inserts of all entry and donor vectors were verified by DNA sequencing (Microsynth, Balgach, Switzerland).
To generate fusion protein expression vectors, entry or donor vectors were recombined in vitro with destination vectors using LR Clonase recombination enzyme mix (Invitrogen). To generate expression plasmids for yeast two-hybrid analysis, the destination vectors pDEST32 (Gal4 DNA-binding domain [Gal4-DBD]) and pDEST22 (Gal4 activation domain [Gal4-AD]) were used. The mammalian Matchmaker vectors pM and pVP16 (Clontech, BD Biosciences, Heidelberg, Germany) were converted to destination vectors by ligation of the Gateway vector conversion cassette reading frame B (Invitrogen) into the EcoRI sites (blunted with Klenow polymerase) of pM and pVP16 to generate pMDEST and pVP16DEST, respectively. Expression vectors for the Gal4-DBD (pM) and VP16AD fusion proteins were obtained after in vitro recombination with the corresponding entry vectors. The mammalian one-hybrid plasmid pM-HIF-1α-370-429-VP16AD was generated by cloning a PCR fragment (using primers 5′-GTCAGAATTCAGAAAATGACTCAGCTATTCACCAA-3′ and 5′-CGATGAATTCGGAATGGTACTTCCTCAAGTTGCT-3′) into the EcoRI site of pM-VP16AD. Plasmids pDEST15 and pDEST17 were used to generate vectors for glutathione S-transferase (GST) and His6 fusion protein expression in bacteria or in rabbit reticulocyte lysates. The plasmid used to generate recombinant GST-HIF-1α-530-826 was described previously (6). pDEST20 was used to generate expression vectors for GST fusion proteins in the baculovirus/Sf9 insect cell system (Invitrogen). pcDNA3.1/nV5-DEST and pcDNA3.1/c-myc-DEST were used to express N-terminal V5- and c-myc-tagged proteins in mammalian cells, respectively. pcDNA3.1/c-myc-DEST was obtained by ligating c-myc oligonucleotides (synthesized by Microsynth) into pcDNA3.1/nV5-DEST digested with EcoRV and HindIII.
Yeast two-hybrid analysis.
Yeast two-hybrid analyses were performed using the ProQuest system according to the manufacturer's instructions (Invitrogen). Full-length PHD2 fused to the Gal4-DBD was expressed from pDEST32-PHD2 in Saccharomyces cerevisiae strain MaV203 (Invitrogen). To test for self-activity, cotransformants of pDEST32/PHD2 and pExpAD502, encoding the Gal4-AD, were examined on selection plates. A Gateway-compatible ProQuest human brain cDNA library was screened on synthetic dropout medium containing 10 mM 3-amino-1,2,4-triazole (Sigma, Buchs, Switzerland). The pExpAD502 plasmids encoding putative PHD2-interacting proteins were isolated, retested for interaction, and tested for self-activity. Inserts of confirmed interactors were sequenced to ensure in-frame coding sequence with the Gal4-AD.
Cell culture and transient transfection.
Human Hep3B hepatoma, HeLa cervical carcinoma, and HEK293 embryonic kidney carcinoma cell lines were cultured in high-glucose Dulbecco's modified Eagle's medium (DMEM) (Sigma) as described previously (6). Transient transfections were performed with the polyethylenimine (Polysciences, Warrington, PA) method (42).
In vitro transcription/translation (IVTT) and GST pull-down.
IVTT reactions were carried out as described by the manufacturer (Promega, Madison, WI) using recombined DEST vectors in the presence of [35S]Met (Hartmann Analytic, Braunschweig, Germany). GST and GST fusion proteins were expressed in Escherichia coli BL21-AI by induction with 0.025% arabinose for 4 h and affinity purified using glutathione-Sepharose columns (GSTrap FF; GE Healthcare, Dübendorf, Switzerland) by liquid chromatography (BioLogic DuoFlow; Bio-Rad, Reinach, Switzerland). Purified GST-tagged proteins (10 μg) were diluted in bead binding buffer (50 mM potassium phosphate, pH 7.5, 150 mM KCl, 1 mM MgCl2, 10% glycerol, 1% TX-100) and incubated with glutathione-Sepharose beads. For pull-down experiments, 20 μl rabbit reticulocyte IVTT reaction mixture was incubated for 2 h at 4°C with bound GST fusion proteins in coimmunoprecipitation (co-IP) buffer (50 mM Tris-HCl, pH 7.6, 2 mM EDTA, 100 mM NaCl, 0.1% TX-100), washed five times with co-IP buffer, boiled in sample buffer (40 mM Tris-HCl, pH 6.8, 1% sodium dodecyl sulfate [SDS], 50 mM β-mercaptoethanol) for 5 min, and separated by SDS-polyacrylamide gel electrophoresis (PAGE). The gels were stained with Coomassie blue and dried, and radioactively labeled proteins were detected by phosphorimaging (Molecular Imager FX; Bio-Rad).
Immunoblotting.
Combined cytoplasmic and nuclear extracts of cultured cells were prepared using 0.4 M NaCl and 0.1% NP-40 in extraction buffer as described previously (23). Nuclear extracts were prepared from isolated nuclei using 0.4 M NaCl. Protein concentrations were determined by the Bradford method using bovine serum albumin as a standard. Immunoblot analyses were performed as previously described (23). The following antibodies were used: mouse monoclonal antibody (MAb) anti-V5 (Invitrogen), MAb anti-c-myc (Roche Diagnostics, Rotkreuz, Switzerland), MAb anti-β-actin (Sigma), MAb anti-HIF-1α (Transduction Laboratories, BD Biosciences), rabbit polyclonal anti-PHD2 antibody (Novus, Abcam, Cambridge, United Kingdom), rabbit polyclonal anti-FKBP38 antibody (7), and secondary polyclonal goat anti-mouse and anti-rabbit antibodies coupled to horseradish peroxidase (Pierce, Perbio, Lausanne, Switzerland). Chemiluminescence detection was performed using Supersignal West Dura (Pierce), and signals were recorded with a charge-coupled device camera (FluorChem8900; AlphaInnotech, Witec, Littau, Switzerland) or by exposure to X-ray film (Fujifilm, Dielsdorf, Switzerland).
Co-IP.
HEK293 cells were transiently cotransfected with pcDNA3.1/V5-FKBP38 and pcDNA3.1/c-myc-PHD2 using the polyethylenimine method. After 24 h, 107 cells were lysed with 1 ml nondenaturing lysis buffer (50 mM Tris-HCl, pH 7.4, 300 mM NaCl, 5 mM EDTA, 1% TX-100, complete protease inhibitors [Roche]). The lysates (500 μl) were precleared with protein A agarose beads (Roche) before incubation with protein A agarose beads bound to monoclonal mouse anti-V5 (2 μg; Invitrogen), anti-c-myc (2 μg; Roche), or nonspecific purified mouse immunoglobulin G (IgG) (2 μg; Sigma) overnight at 4°C. Antibody-protein complexes were washed four times with wash buffer (50 mM Tris-HCl, pH 7.4, 300 mM NaCl, 5 mM EDTA, 0.1% TX-100) and once with phosphate-buffered saline (PBS) and eluted with SDS-PAGE sample buffer.
Immunofluorescence microscopy.
Cells were cultivated on microscope slides, washed twice with ice-cold PBS, and fixed on ice for 30 min with 4% paraformaldehyde. The cells were then permeabilized with 0.1% saponin in PBS. Endogenous FKBP38 was detected with rabbit anti-FKBP38 antibodies, and transfected hemagglutinin (HA)-PHD2 was detected with mouse anti-HA antibodies (Sigma). Immune complexes were visualized with goat anti-rabbit antibody-Alexa 488 or goat anti-mouse antibody-Alexa 568 (Molecular Probes, Invitrogen), respectively. Finally, nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole) (Sigma) for 30 min. After extensive washings with PBS, the slides were mounted and analyzed by confocal laser scanning microscopy (SP1; Leica Microsystems, Switzerland).
Reporter gene and mammalian one- and two-hybrid assays.
Cloning of the HIF-dependent firefly luciferase reporter gene construct pH3SVL was described previously (45). Cells were cotransfected with 500 ng pH3SVL and 20 ng pRL-SV40 Renilla luciferase reporter vector (Promega) to control for differences in transfection efficiency and exposed to normoxic or hypoxic conditions for 16 h. Mammalian one- and two-hybrid analyses were performed using the mammalian Matchmaker system (Clontech, BD Biosciences). HeLa cells were transiently cotransfected with one-hybrid or DBD and AD fusion protein vectors, together with the firefly luciferase reporter vector pGRE5xE1b and pRL-SV40. Luciferase reporter gene activity was determined using the dual-luciferase reporter assay system according to the manufacturer's instructions (Promega).
RNAi.
For silencing FKBP38 by RNA interference (RNAi), a pair of 63-nucleotide oligonucleotides (Microsynth) targeting the sequence 5′-AAGAGUGGCUGGACAUUCUGG-3′ were inserted into pSilencer2.1-U6 hygro digested with BamHI and HindIII (Ambion, Huntingdon, United Kingdom) and transfected into HeLa and Hep3B cells by calcium phosphate coprecipitation. A pool of clones were selected in medium containing 200 μg/ml hygromycin B (Calbiochem, VWR International, Lucerne, Switzerland), and single clones were subsequently isolated by limited dilution. The downregulation of FKBP38 expression by RNAi was analyzed by real-time reverse transcriptase (RT) PCR and immunoblotting and compared to cells containing the control oligonucleotide harboring the vector pSilencer2.1-U6 hygro.
Pulse-chase experiment.
HeLa cells were starved in DMEM (without fetal calf serum [FCS]) lacking Met and Cys for 1 h. The cells were then incubated in the same medium supplemented with 10% FCS (dialyzed against PBS) and [35S]Met/Cys mixture (0.1 mCi/ml; Hartman Analytics, Braunschweig, Germany) for 2 h. After the cells were washed twice with PBS, they were chased with DMEM-10% FCS containing cold Met/Cys (3 mM each). During the chase period, cells were harvested at different time points, and radioactively labeled PHD2 was immunoprecipitated and visualized by phosphorimaging.
In vitro prolyl-4-hydroxylation assay.
HeLa cells were pelleted and Dounce homogenized in cell lysis buffer (100 mM Tris-Cl, pH 7.5, 1.5 mM MgCl2, 8.75% glycerol, 0.01% Tween 20) supplemented with complete protease inhibitors. The lysates were centrifuged for 30 min at 4°C and 20,000 × g, and pVHL-elongin B-elongin C (VBC) binding was determined as previously described (23, 30).
RNA extraction and real-time RT-PCR quantification.
Cells were grown in 15-cm plates under normoxic or hypoxic conditions, and total RNA was extracted as described previously (23). First-strand cDNA synthesis was performed with 3 μg of RNA using Superscript III Moloney murine leukemia virus RT according to the manufacturer's instructions (Invitrogen). Subsequently, mRNA expression levels were quantified with 2 μl of diluted cDNA reaction mixture by real-time PCR using a SybrGreen Q-PCR reagent kit (Sigma) in combination with the MX3000P light cycler (Stratagene, Amsterdam, The Netherlands). Initial template concentrations of each sample were calculated by comparison with serial dilutions of a calibrated standard. To verify RNA integrity and equal input levels, ribosomal protein L28 mRNA was determined, and the data were expressed as ratios relative to L28 levels. Primers were as follows: hL28 forward, 5′-GCAATTCCTTCCGCTACAAC-3′; hL28 reverse, 5′-TGTTCTTGCGGATCATGTGT-3′; hGLUT1 forward, 5′-TCACTGTGCTCCTGGTTCTG-3′; hGLUT1 reverse, 5′-CCTGTGCTCCTGAGAGATCC-3′; hCAIX forward, 5′-GGGTGTCATCTGGACTGTGTT-3′; hCAIX reverse, 5′-CTTCTGTGCTGCCTTCTCACT-3′; hFKBP38 forward, 5′-ACATGACGTTCGAGGAGGAG-3′; hFKBP38 reverse, 5′-GTTGGAAGGTTCCAGCTTCA-3′; hPHD1 forward, 5′-CTGGGCAGCTATGTCATCAA-3′; hPHD1 reverse, 5′-AAATGAGCAACCGGTCAAAG-3′; hPHD2 forward, 5′-GAAAGCCATGGTTGCTTGTT-3′; hPHD2 reverse, 5′-TTGCCTTCTGGAAAAATTCG-3′; hPHD3 forward, 5′-ATCGACAGGCTGGTCCTCTA-3′; and hPHD3 reverse, 5′-CTTGGCATCCCAATTCTTGT-3′.
RESULTS
Identification of FKBP38 as a specific PHD2 interactor.
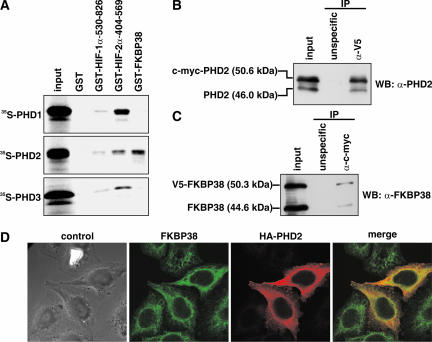
To identify novel PHD2-interacting proteins, human PHD2 expressed as a fusion protein with the Gal4-DBD was used as bait in a yeast two-hybrid screen of a human brain cDNA library. A total of 2 × 106 individual transformants were screened, and several independent cDNA clones encoding FKBP38 were identified based on their abilities to activate the three reporter genes His, Ura, and LacZ (data not shown). In order to confirm these data in a yeast-independent interaction assay, GST pull-down experiments were performed. PHD1, -2, and -3 were subjected to IVTT in the presence of radioactive [35S]Met, incubated with bacterially expressed and purified GST-FKBP38, and precipitated by glutathione-Sepharose. As controls, GST-HIF-1α-530-826 and GST-HIF-2α-404-569 were also expressed and purified as GST fusion proteins. GST alone was used as a noninteraction control. Whereas HIF-α fragments containing the ODD domain interacted with all three PHDs, FKBP38 showed specific association with PHD2 only (Fig. 1A), confirming the previous findings in yeast (data not shown). The stronger binding of PHD2 to HIF-2α-404-569 than to HIF-1α-530-603 might be due to the presence of both hydroxylated proline residues in the recombinant HIF-2α protein (P405 and P531) versus just the C-terminal proline in HIF-1α (P564).
FIG. 1.
FKBP38 interacts specifically with PHD2. (A) IVTT 35S-labeled PHD1, PHD2, and PHD3 were incubated with GST-HIF-1α-530-826, GST-HIF-2α-404-569, GST-FKBP38, or GST alone. Bound proteins were pulled down and visualized by phosphorimaging. (B and C) Total cell extracts from HEK293 cells transiently transfected with V5-FKBP38 and c-myc-PHD2 were incubated with anti-V5 (B) or anti-c-myc (C) antibodies or with isotype-matched control IgG. The antibodies and bound proteins were immunoprecipitated and analyzed by immunoblotting (WB). Coimmunoprecipitated PHD2 and FKBP38 were detected by specific antibodies. IP, immunoprecipitate. (D) Indirect immunofluorescence of FKBP38 and PHD2. HeLa cells were transfected with a plasmid encoding HA-PHD2 and stained with antibodies to FKBP38 (green) and HA (red), followed by fluorescently labeled secondary antibodies.
To confirm the FKBP38:PHD2 interaction in a mammalian system, we transiently transfected HEK293 cells with V5-tagged FKBP38 and c-myc-tagged PHD2. Tag-specific antibodies were then used for co-IPs. V5-FKBP38 interacted with exogenous, as well as with endogenous, PHD2 (Fig. 1B), and c-myc-PHD2 was found in a complex with both overexpressed and cellular FKBP38 (Fig. 1C). Nonspecific isotype-matched mouse IgG served as an immunoprecipitation control.
We next investigated whether FKBP38 colocalizes with PHD2 in HeLa cells. Since the antibodies directed against these two proteins were generated in rabbits, we transiently transfected HA-PHD2 and used mouse anti-HA antibodies to detect PHD2. Immunofluorescence analysis indicated that endogenous FKBP38 colocalized with HA-PHD2 in the cytoplasm of HeLa cells (Fig. 1D).
Mapping of the PHD2 interaction site in FKBP38.
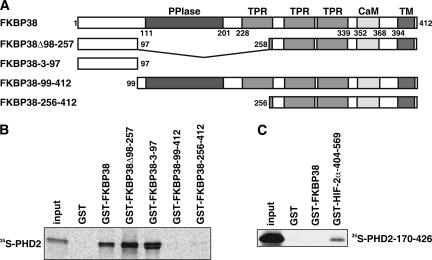
FKBP38 belongs to the enzyme class of peptidyl prolyl cis/trans isomerases (PPIases), and the amino-terminal region of FKBP38 contains a PPIase domain of the FKBP type, harboring PPIase- and FK506-binding activities. In addition, FKBP38 contains three tetratricopeptide repeats, a calmodulin-binding site, and a membrane anchor in its carboxy-terminal half (7, 38). Based on the predicted domain architecture of FKBP38 (Fig. 2A), a series of deletion mutants fused to GST were designed, expressed in bacteria, purified, and analyzed for PHD2 interaction in GST pull-down experiments. Radioactively labeled PHD2 was produced by IVTT, and the interaction of labeled PHD2 with purified GST-FKBP38 deletion constructs, or GST alone, was tested. The N-terminal FKBP38 fragment containing aa 3 to 97 was sufficient to maintain interaction with PHD2 (Fig. 2B). The PPIase domain was not required for PHD2 interaction. On the other hand, an N-terminally truncated PHD2 protein (PHD2 aa 170 to 426) interacted with GST-HIF-2α-404-569 but not with GST-FKBP38 (Fig. 2C). Similar results were obtained in yeast two-hybrid experiments (data not shown). These data suggest that aa 3 to 97 of FKBP38 are required for interaction with aa 1 to 169 of PHD2. The catalytic domain of purified PHD2 is sufficient for HIF-2α ODD domain binding and hydroxylation of HIF-1α-derived peptides (data not shown), suggesting that FKBP38 is not a PHD2 hydroxylation substrate.
FIG. 2.
Mapping of the FKBP38 and PHD2 interaction sites. (A) Schematic representation of the predicted FKBP38 domain architecture and the FKBP38 constructs used. TPR, tetratricopeptide repeats; CaM, calmodulin-binding site; TM, transmembrane domain. (B) IVTT 35S-labeled PHD2 was allowed to interact with GST-FKBP38, GST-FKBP38Δ98-257, GST-FKBP38-3-97, GST-FKBP38-99-412, GST-FKBP38-256-412, or GST alone; the protein complexes were precipitated by glutathione-Sepharose, separated by SDS-PAGE, and visualized by phosphorimaging. (C) IVTT 35S-labeled PHD2-170-426 was tested for interaction with GST-FKBP38, GST-HIF-2α-404-569, or GST alone and analyzed as described above.
FKBP38-PHD2 interaction and FKBP38 gene expression are independent of the oxygen concentration.
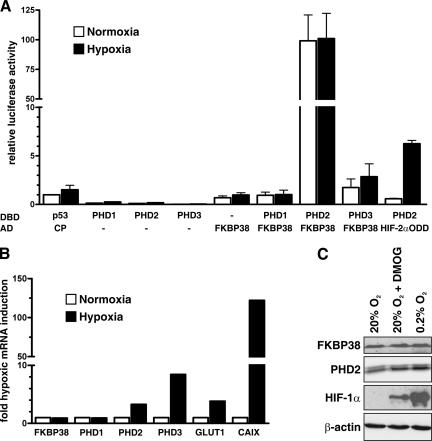
To investigate whether the FKBP38-PHD2 interaction is oxygen-dependent, we expressed PHD1, PHD2, or PHD3 fused to the Gal4-DBD, together with the VP16AD fused to FKBP38, in a mammalian two-hybrid system in HeLa cells. The activity of a cotransfected luciferase reporter gene construct containing five Gal4-DBD sites is greatly enhanced when the AD and DBD fusion proteins interact with each other. As shown in Fig. 3A, luciferase expression was significantly higher when the DBD-PHD2 and AD-FKBP38 fusion constructs were cotransfected than in transfection of either construct alone or AD-FKBP38 in combination with DBD-PHD1 or DBD-PHD3, confirming specific FKBP38-PHD2 interaction in mammalian cells. Cotransfection of DBD-PHD2 with the AD construct fused to FKBP38 deletions confirmed the findings in yeast, as well as the in vitro interaction results mentioned above (data not shown). DBD-p53, together with AD-CP (polyomavirus coat protein), served as a negative control. In contrast to the significantly increased luciferase activity under hypoxic conditions following cotransfection of DBD-PHD2 with AD-HIF-2α-404-569 (HIF-2αODD), luciferase expression remained unchanged following cotransfection of DBD-PHD2 with AD-FKBP38 under hypoxic and normoxic conditions.
FIG. 3.
FKBP38-PHD2 interaction and FKBP38 gene expression are not regulated by oxygen. (A) HeLa cells were transiently transfected with Gal4-DBD and VP16AD fusion protein vectors and Gal4 response element-driven firefly luciferase reporter, as well as a Renilla luciferase control vector. Following transfection, the cells were incubated under normoxic (20% O2) or hypoxic (0.2% O2) conditions, and luciferase reporter gene activities were determined 16 h later. Firefly/Renilla luciferase activity ratios were normalized to the normoxic negative control DBD-p53/AD-CP cotransfection, which was arbitrarily defined as 1. Mean values plus standard errors of the mean are shown for three independent experiments performed in triplicate. (B and C) HeLa cells were cultured under normoxic (20% O2) or hypoxic (0.2% O2) conditions or treated with the PHD inhibitor dimethyloxalylglycine (DMOG) (2 mM). (B) Total RNA was extracted after 8 h of incubation, and mRNA levels of FKBP38, PHD1 to -3, GLUT1, CAIX, and ribosomal protein L28 were quantified by real-time RT-PCR. The transcript levels of these genes were normalized to L28, and hypoxic inductions were calculated (mean, n = 2). (C) Cellular proteins were extracted and separated by SDS-PAGE, and endogenous FKBP38, PHD2, HIF-1α, and β-actin levels were analyzed by immunoblotting.
We next determined mRNA levels in cultured HeLa cells exposed to normoxic or hypoxic conditions by real-time RT-PCR. The known oxygen-regulated HIF target genes PHD2, PHD3, GLUT1, and CAIX, but not FKBP38, PHD1, or L28 control mRNA, were induced by hypoxia (Fig. 3B). FKBP38 protein levels were not regulated by inhibition of PHDs with dimethyloxalylglycine or by hypoxia, whereas HIF-1α, as well as PHD2, protein expression was increased following PHD inhibition or hypoxic exposure (Fig. 3C). In summary, these data demonstrate an oxygen-independent interaction of FKBP38 with PHD2, suggesting that FKBP38 is not a PHD2 substrate but might rather act as a cofactor.
RNAi-mediated FKBP38 silencing increases PHD-dependent HIF-1α hydroxylation.
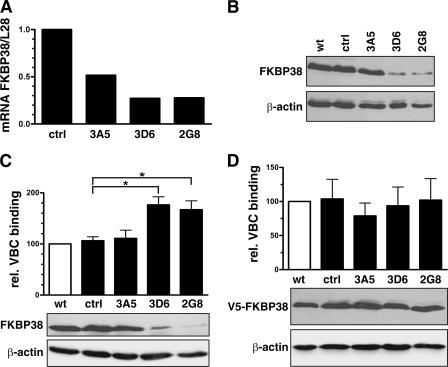
To investigate the functional consequences of the FKBP38-PHD2 interaction with regard to the regulation of PHD activity, FKBP38 mRNA was downregulated by RNAi. HeLa and Hep3B cells were transfected with a FKBP38-silencing construct or with a control plasmid harboring nonspecific oligonucleotides. Pools of clones were generated by hygromycin B selection, and single clones were isolated by limited dilution. As shown in Fig. 4A and B, respectively, FKBP38 RNAi expression resulted in decreased endogenous FKBP38 mRNA, as well as protein levels, in HeLa cells. Similar data were obtained in Hep3B cells (data not shown). The ability of cell lysates derived from FKBP38 RNAi clones to hydroxylate HIF-1α was investigated in an in vitro prolyl-4-hydroxylation assay. PHD hydroxylation activity was measured by binding of the VBC complex to HIF-1α-derived peptides. Decreased FKBP38 resulted in a concomitant significant increase in hydroxylation activity (Fig. 4C). Reconstitution of FKBP38 gene expression using transiently transfected expression vectors normalized prolyl hydroxylation (Fig. 4D), whereas transfection with a lacZ expression vector had no effect (data not shown).
FIG. 4.
Enhanced hydroxylation activity by RNAi-mediated FKBP38 depletion. HeLa cells were transfected with vectors containing FKBP38 RNAi or control oligonucleotides, and stable clones were selected (3A5, 3D6, and 2G8). (A) Total RNA was extracted, and FKBP38 transcript levels were determined by real-time RT-PCR and normalized to L28 mRNA levels (mean, n = 2). (B) Total cell extracts were prepared and analyzed by immunoblotting for FKBP38 and β-actin expression. Total cell extracts of untransfected (C) and FKBP38-reconstituted (D) stable RNAi clones were prepared, and hydroxylation activity was measured using the VBC-binding assay. Shown are mean values of relative VBC binding plus standard errors of the mean of three independent experiments performed in triplicate. P values were obtained by paired t tests (*, P < 0.05). Subsequently, cell extracts were analyzed by immunoblotting for endogenous FKBP38, transfected V5-FKBP38, and β-actin expression. wt, wild type; ctrl, control.
Expression of a functional mutant FKBP38ΔPPIase (FKBP38Δ98-257) resulted in normalized prolyl hydroxylation comparable to transfection with FKBP38 (data not shown).
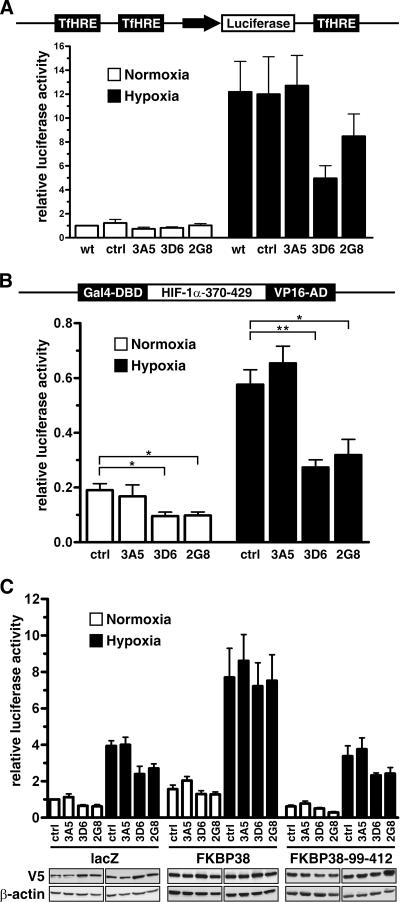
FKBP38 silencing reduces HIF-1α-dependent reporter gene activity.
To further investigate whether increased hydroxylation activity due to FKBP38 depletion affects HIF-dependent reporter gene expression, wild-type HeLa, as well as FKBP38, small interfering RNA (siRNA) cell clones were transiently cotransfected with pH3SVL, containing six HIF-binding sites from the transferrin HRE and a Renilla luciferase control plasmid. Luciferase activity was reduced under hypoxic conditions in FKBP38-depleted cell clones compared with control cells (Fig. 5A). It has recently been reported that PHD2 is able to modulate HIF-1α transcriptional activity (44), and therefore, we established a mammalian one-hybrid assay to investigate HIF-1α protein stability directly. FKBP38 siRNA cell clones were transiently cotransfected with HIF-1α-370-429 fused at the N-terminal end with Gal4-DBD and at the C-terminal end with VP16AD, a firefly luciferase reporter gene construct containing five Gal4-DBDs, and a Renilla luciferase control plasmid. As shown in Fig. 5B, luciferase activity was significantly reduced under normoxic as well as hypoxic conditions in FKBP38-depleted cell clones compared with control cells. Reconstitution of V5-FKBP38 with a FKBP38 expression vector, but not with a lacZ control vector, rescued luciferase activity (Fig. 5C). Overexpression of a functional FKBP38 mutant that does not bind PHD2 (FKBP38-99-412) resulted in luciferase activities comparable to the lacZ control transfections (Fig. 5C). The nonimmunosuppressive small molecule GPI1046 [3-(3-pyridyl)-1-propyl (2S)-1-(3,3-dimethyl-1,2-dioxopentyl)-2-pyrrolidine carboxylate] mimics the FK506 residues interacting with FKBP38. GPI1046 has been shown to preferentially bind FKBP38 and to inhibit the PPIase activity with a Ki of 48 nM (7, 40). Addition of GPI1046, to inhibit FKBP38 PPIase activity, to V5-FKBP38 reconstituted cells had no effect, indicating a PPIase-independent function of FKBP38 in the regulation of HIF-1α hydroxylation (data not shown). Note that relative luciferase activities were generally higher in functional FKBP38 reconstituted clones than in cells transfected with FKBP38, mutant for PHD2 binding, or lacZ, suggesting that overexpression of FKBP38 reduces PHD-dependent degradation of the HIF one-hybrid construct.
FIG. 5.
FKBP38 regulates HIF-dependent reporter gene activity. (A) HeLa wild-type (wt) as well as FKBP38 siRNA cell clones were cotransfected with pH3SVL and pRL-SV40 Renilla luciferase reporter vectors and cultivated for 16 h under normoxic (20% O2) or hypoxic (0.2% O2) conditions before relative luciferase activities were determined. The results are mean values plus standard errors of the mean of five independent experiments performed in triplicate. ctrl, control. (B) Stable FKBP38 RNAi HeLa clones were transiently transfected with Gal4-DBD-HIF-1α-370-429-VP16AD expression vectors (schematically represented) and Gal4 response element-driven firefly luciferase reporter, as well as a Renilla luciferase control vector. (C) HeLa clones were cotransfected with the one-hybrid reporter gene vectors and FKBP38, FKBP38-99-412, or lacZ control expression vectors. Eight hours posttransfection, the cells were cultured under either normoxic or hypoxic conditions for an additional 16 h, and firefly luciferase activities were determined and corrected for Renilla luciferase activity. The results are mean values of relative luciferase activities plus standard errors of the mean of at least three independent experiments performed in triplicate. P values were obtained by paired t tests (**, P < 0.01; *, P < 0.05). Expression of the transfected V5-tagged vectors was verified by immunoblotting against V5 and β-actin.
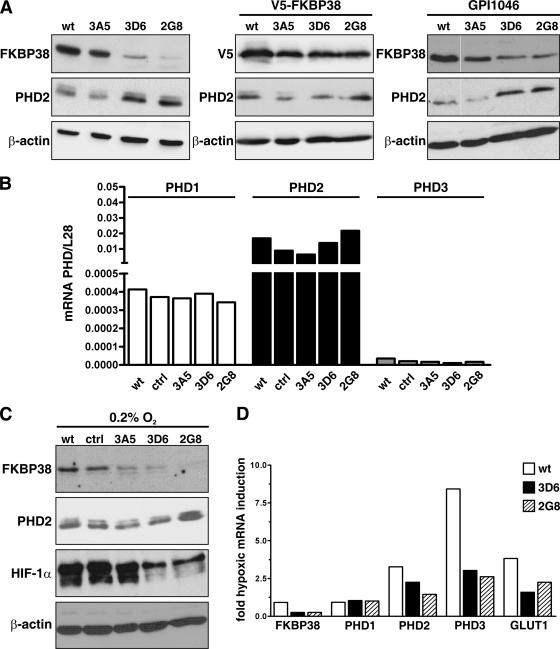
Reduced FKBP38 gene expression leads to increased PHD2 protein levels and attenuates HIF-dependent gene expression.
The enhanced hydroxylation and reduced HIF-1α-dependent one-hybrid reporter gene activity found in FKBP38-depleted cells could be based on modification of the enzymatic activity itself and/or on elevated enzyme abundance. However, incubation of active GST-PHD2 fusion protein purified from baculovirus-infected Sf9 cells with recombinant GST-FKBP38 did not influence PHD2 hydroxylation activity as measured by the VBC capture assay (data not shown). Therefore, we analyzed PHD2 protein levels in FKBP38 RNAi cell clones and observed enhanced PHD2 levels inversely related to FKBP38 (Fig. 6A, left). Transient transfection of these FKBP38 RNAi cells with V5-FKBP38 restored the low constitutive PHD2 protein levels, comparable to those of wild-type HeLa cells (Fig. 6A, middle). Addition of GPI1046 did not influence PHD2 protein levels in wild-type HeLa or FKBP38 RNAi cell clones, again indicating a PPIase-independent effect (Fig. 6A, right). The same results were obtained by treating cells with N-(N′,N′-dimethylcarboxamidomethyl)-cycloheximide, another FKBP38 PPIase inhibitor (reference 8 and data not shown).
FIG. 6.
Increased PHD2 protein stability in FKBP38-depleted cells. (A) Wild-type HeLa cells (wt), stable FKBP38-silenced HeLa clones, V5-FKBP38 reconstituted HeLa cells, or FKBP38-silenced HeLa cells treated with the FKBP38 inhibitor GPI1046 (10 μM) were cultivated under normoxic (20% O2) conditions, and FKBP38, V5-FKBP38, PHD2, or β-actin was detected by immunoblotting. (B) Total RNA was extracted and reverse transcribed into cDNA, and mRNA levels of PHD1, PHD2, and PHD3 were normalized to ribosomal L28 mRNA levels as quantified by real-time RT-PCR (n = 2). ctrl, control. (C) Total hypoxic FKBP38 RNAi cell clone lysates were separated by SDS-PAGE and analyzed for FKBP38, PHD2, HIF-1α, and β-actin protein levels by immunoblotting. (D) Total RNA was extracted after 8 h of incubation in normoxia or hypoxia, and mRNA levels of FKBP38, PHD1 to -3, GLUT1, and ribosomal L28 were quantified by real-time RT-PCR. The transcript levels of these genes were normalized to L28 mRNA, and hypoxic inductions were calculated (n = 2).
PHD1, PHD2, and PHD3 mRNA levels were not affected by downregulation of FKBP38, indicating that PHD2 is posttranscriptionally regulated by FKBP38 (Fig. 6B). To analyze whether FKBP38 is able to attenuate endogenous HIF-1α protein expression by modification of PHD2 abundance, HeLa control cells and FKBP38 RNAi cell clones were cultured under hypoxic conditions and analyzed by immunoblotting. PHD2 protein levels were inversely related to FKBP38 expression even under hypoxic conditions (Fig. 6C). Note that elevated PHD2 protein levels in FKBP38-silenced cells compared to control cells are less pronounced, because hypoxia upregulated PHD2 expression in the control cells. HIF-1α protein was not detectable under normoxic conditions but strongly increased in hypoxic control cells. Strikingly, HIF-1α protein levels markedly decreased in hypoxic FKBP38-depleted cells, suggesting that elevated PHD2 levels lead to increased proteolytic destruction of HIF-1α, even under hypoxic conditions (Fig. 6C).
Reduced HIF-1α protein levels might explain the less pronounced increase in PHD2 levels under hypoxic conditions. Indeed, hypoxia-inducible transcripts of HIF target genes, including PHD2, PHD3, and GLUT-1 but not PHD1, were attenuated two- to threefold in FKBP38 knockdown cell clones (Fig. 6D). These results indicate that increased PHD2 abundance in FKBP38 RNAi cell clones led to elevated HIF-α substrate hydroxylation and subsequent degradation, finally resulting in decreased HIF function in the cell.
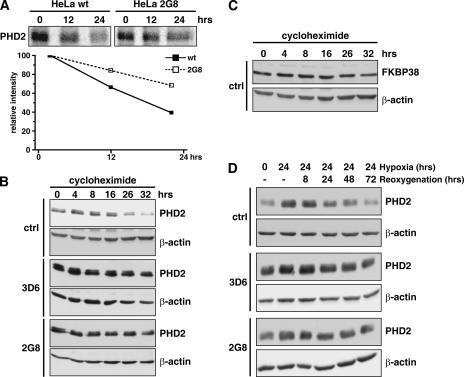
PHD2 protein stability is increased in FKBP38-downregulated cells.
Increased PHD2 protein levels in FKBP38-depleted cells could be due to increased translation or protein stability. To analyze PHD2 protein stability, we performed pulse-chase experiments in wild-type HeLa and FKBP38-silenced HeLa 2G8 cells (Fig. 7A). Whereas the measured half-life of PHD2 was about 20 h in wild-type cells, it was extrapolated to approximately twice as long when FKBP38 was downregulated. In addition, cycloheximide treatment and subsequent analysis of PHD2 protein levels showed a half-life in the same range, as determined by pulse-chase in control HeLa cells, whereas PHD2 protein stability was markedly elevated in FKBP38-downregulated cell clones (Fig. 7B). These experiments suggest a function of FKBP38 in the proteolytic regulation of PHD2. FKBP38 protein levels remained constant for up to 32 h during cycloheximide treatment (Fig. 7C). PHD2 is induced in HeLa cells by incubation under hypoxic conditions for 24 h. After reoxygenation, PHD2 levels remain increased for up to 24 h and afterwards slowly decline. In contrast, in FKBP38-downregulated HeLa cells, PHD2 remained elevated even after 72 h of reoxygenation (Fig. 7D), indicating that FKBP38 is also required for the regulation of PHD2 protein levels under reoxygenation conditions.
FIG. 7.
Increased PHD2 protein stability in FKBP38-silenced cells. (A) The half-lives of PHD2 protein in wild-type HeLa (wt) and FKBP38-depleted (2G8) cells were monitored by a pulse-chase experiment. Cells were metabolically labeled with [35S]Met/Cys for 2 h, followed by chase for the indicated times. PHD2 was immunoprecipitated from the cell lysates with anti-PHD2 antibodies, and signals were detected by autoradiography. Translation was blocked in FKBP38 RNAi HeLa clones by addition of 100 μM cycloheximide and PHD2 (B), as well as FKBP38 (C), and protein levels were determined after the indicated time periods by immunoblotting. Subsequent detection of β-actin served as a loading control. (D) Total lysates of HeLa cells were prepared at the indicated periods of hypoxia or hypoxia followed by reoxygenation, and PHD2 as well as β-actin levels were detected by immunoblotting.
DISCUSSION
Although oxygen-dependent hydroxylation directly links oxygen availability to HIF-α protein stabilization and transcriptional activity, both HIF and PHD hydroxylase activities are likely to be subjected to additional levels of regulation. Indeed, a HIF-dependent regulatory feedback mechanism has been shown to increase PHD2 and PHD3 protein abundance under hypoxic conditions, suggesting a role during reoxygenation (9). We recently found that inducible PHD2 and PHD3 mRNA levels remain upregulated under prolonged hypoxic exposure and that PHDs maintain a functional HIF-α hydroxylation activity even under severe hypoxic conditions (42). Biochemical analysis of a PHD2 protein complex, consisting of approximately 15 proteins with an apparent molecular mass of 320 to 440 kDa, indicated that PHD2 might be part of a large heteromeric complex (15). The formation of stable complexes with other proteins raises the possibility that such proteins might be involved in the regulation of PHD2 function, e.g., through chaperones; modulation of access to the cosubstrates oxygen, ferrous iron, 2-oxoglutarate, and probably ascorbate; and target protein recognition. Because HIF prolyl hydroxylation is a nonreversible process, differences in PHD protein abundance would also be a means of regulating hydroxylation activity.
We identified FKBP38 as a PHD2-associated nonsubstrate protein and showed that silencing of FKBP38 results in increased PHD2 stability, leading to enhanced hydroxylation and attenuated HIF-dependent transcription. Conversely, FKBP38 overexpression resulted in elevated HIF reporter gene activity in transient transfections (data not shown), as well as in one-hybrid assays (Fig. 5C). Whereas PHD1 and PHD3 are targeted for proteasomal degradation by the E3 ubiquitin ligases Siah1a and Siah2 under hypoxic conditions, it is so far unknown how PHD2 is posttranslationally regulated (28). Although the molecular mechanism remains elusive, FKBP38-dependent decreased PHD2 protein stability provides the first data concerning the proteolytic regulation of PHD2 and attributes a novel function to FKBP38.
The enzyme class of PPIases encompasses the immunophilin families of cyclophilins and FKBPs, which were originally discovered as cellular receptors of the immunosuppressive drugs cyclosporine A and FK506, respectively (11, 43). Although structurally unrelated, the two subfamilies share the common enzymatic activity to catalyze the isomerization of the cis and trans conformers of peptide bonds preceding prolines (10). The singularity of peptidyl-prolyl bonds in peptide chains is caused by the formation of an imidic peptide bond at the N terminus of the N-alkylated amino acid proline that is unique among amino acids. The rate of the uncatalyzed interconversion between the prolyl cis/trans isomers is low, pointing to an involvement of PPIases in protein folding. It has been shown for collagen that proline hydroxylation occurs on the nascent polypeptide chain during translation, and subsequent cyclophilin-assisted cis/trans isomerization is the rate-limiting step for collagen helix formation. Procollagen folding is slowed down by the addition of cyclosporine A (41).
Because VHL binds to HIF-α trans-4-hydroxyprolyl residues (16, 17), we speculated that the PPIase FKBP38 might act as a PHD2 cofactor, influencing the conformation of HIF-α prolyl residues and/or the activity of PHD2. However, FKBP38 did not physically interact with HIF-α in yeast or mammalian two-hybrid assays (data not shown), suggesting that FKBP38 does not influence the conformation of the HIF-α ODD domain. On the other hand, the PPIase domain of FKBP38 was not required for PHD2 interaction (Fig. 2), Ca2+/calmodulin-mediated activation of recombinant FKBP38 did not modulate the hydroxylation capacity of PHD2 (data not shown), and the small-molecule FKBP38 PPIase inhibitor GPI1046, as well as N-(N′,N′-dimethylcarboxamidomethyl)-cycloheximide, did not influence PHD2 protein abundance (Fig. 6A and data not shown, respectively). These data provide evidence that the effect of FKBP38 on PHD2 abundance is PPIase independent.
FKBP38 is widely expressed in both adult and embryonic tissues, as well as in different cancer cell lines (5, 18). Recently, knockout mice showed that FKBP38 plays an essential role during development. FKBP38 acts as a negative regulator of the sonic hedgehog (SHH) pathway, and its loss leads to ectopic and ligand-independent activation of SHH signaling, resulting in disturbed cell fate determination and embryonic lethality (5). However, the molecular function of FKBP38 in the SHH pathway remains elusive, and it will be interesting to determine whether PHD2 is involved.
An interaction of FKBP38 with Bcl-2 has also been described (38), but the functional consequences of this association remain controversial. Antiapoptotic (38), as well as proapoptotic (7), effects have been reported, probably depending on the cellular context. The two proteins interacted only upon Ca2+ influx and subsequent FKBP38 activation. However, we did not observe a cofactor-dependent interaction of FKBP38 with PHD2, nor did the addition of Ca2+ to cell lysates alter the hydroxylation activity, and cell viability was not affected by RNAi-mediated downregulation of FKBP38 as measured by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] assays (data not shown). Further potential functions of FKBP38 include antitumor effects by regulation of anti-invasive syndecan 1 and suppression of proinvasive MMP9 and regulation of cell size via the tuberous sclerosis complex (12, 35). Clearly, the increasing amount of data about potential FKBP38 functions suggests that FKBP38 is a multifunctional protein involved in the regulation of many cellular processes, but further studies are required for a better comprehension of the biochemical functions of FKBP38.
In addition to FKBP38, the candidate tumor suppressor ING4 was shown to associate with PHD2, suppressing HIF activity and thereby possibly attenuating HIF target genes involved in tumor growth and angiogenesis (31). It has been proposed that ING4 retains PHD2 in the nucleus in order to degrade HIF upon reoxygenation. However, recent data showed predominant cytoplasmic localization of PHD2 (39). PHD2 has a calculated molecular mass of 46 kDa, enabling it to pass through nuclear pores, and it might be actively excluded from the nucleus by interaction with FKBP38. FKBP38 contains a C-terminal transmembrane domain that anchors the protein in the mitochondrial and endoplasmic reticulum membranes, and our immunofluorescence data also show a patchy cytoplasmic staining for PHD2, indicating an association with subcellular cytoplasmic structures (Fig. 1D). Additionally, it has recently been demonstrated that PHD2 interacts with the HIF-1α C-terminal ODD domain (aa 498 to 603) and thereby reduces the transcriptional activity of this fragment (44). Although we cannot exclude the possibility that attenuation of HIF-1α transcriptional activity by PHD2 is involved in reduced hypoxic HIF-dependent reporter gene (Fig. 5A) as well as HIF target gene (Fig. 6D) induction in FKBP38 siRNA clones, our data suggest that elevated PHD2 protein levels attenuate hypoxic HIF-1α stabilization (Fig. 5B and 6C).
PHDs represent attractive new therapeutic targets to modulate HIF activity, and thus functional understanding of PHD function and regulation is crucial. Recently, it has been reported that an inherited mutation in the active site of PHD2 leads to decreased hydroxylation activity and is associated with familial erythrocytosis (32). These data confirm previous studies with cultured cells implicating PHD2 as the primary PHD isoform responsible for HIF regulation under physiological conditions (2). Interestingly, a screen for genetic mutations in endometrial cancer cells identified PHD2 as being mutated at significantly higher frequencies (19). Expression of wild-type PHD2 in mutated cells induced senescence by negatively regulating HIF-1 expression, suggesting that PHD2 might be a candidate tumor suppressor. Specific upregulation of PHD2 in a FKBP38-dependent manner opens novel possibilities for interfering with one PHD family member specifically, thereby attenuating HIF signaling in cancer cells.
Acknowledgments
We thank D. J. Peet, S. Tan, and W. G. Kaelin, Jr. for gifts of plasmids; I. Flamme and F. Oehme for generous gifts of peptides and helpful advice for the VBC-binding assay; and C. Franke and P. Spielmann for excellent technical assistance.
This work was supported by grants from the Forschungskredit der Universität Zürich (to G.C.), the University Research Priority Program Integrative Human Physiology at the University of Zürich, the Sassella Stiftung (to G.C.), the Hartmann Müller-Stiftung (to G.C.), the BMBF (NBL3 to D.M.K.), DFG (Ka1269/8-1 to D.M.K.), the 6th Framework Programme of the European Commission/SBF grant (EUROXY LSHC-CT-2003-502932/SBF no. 03.0647-2 to R.H.W.), the Olga Mayenfisch Stiftung (to G.C.), and the SNF (3100A0-104219 to R.H.W. and G.C.).
Footnotes
Published ahead of print on 12 March 2007.
REFERENCES
- 1.Appelhoff, R. J., Y. M. Tian, R. R. Raval, H. Turley, A. L. Harris, C. W. Pugh, P. J. Ratcliffe, and J. M. Gleadle. 2004. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 279:38458-38465. [DOI] [PubMed] [Google Scholar]
- 2.Berra, E., E. Benizri, A. Ginouves, V. Volmat, D. Roux, and J. Pouyssegur. 2003. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 22:4082-4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruick, R. K. 2003. Oxygen sensing in the hypoxic response pathway: regulation of the hypoxia-inducible transcription factor. Genes Dev. 17:2614-2623. [DOI] [PubMed] [Google Scholar]
- 4.Bruick, R. K., and S. L. McKnight. 2001. A conserved family of prolyl-4-hydroxylases that modify HIF. Science 294:1337-1340. [DOI] [PubMed] [Google Scholar]
- 5.Bulgakov, O. V., J. T. Eggenschwiler, D. H. Hong, K. V. Anderson, and T. Li. 2004. FKBP8 is a negative regulator of mouse sonic hedgehog signaling in neural tissues. Development 131:2149-2159. [DOI] [PubMed] [Google Scholar]
- 6.Camenisch, G., M. Tini, D. Chilov, I. Kvietikova, V. Srinivas, J. Caro, P. Spielmann, R. H. Wenger, and M. Gassmann. 1999. General applicability of chicken egg yolk antibodies: the performance of IgY immunoglobulins raised against the hypoxia-inducible factor 1α. FASEB J. 13:81-88. [DOI] [PubMed] [Google Scholar]
- 7.Edlich, F., M. Weiwad, F. Erdmann, J. Fanghanel, F. Jarczowski, J. U. Rahfeld, and G. Fischer. 2005. Bcl-2 regulator FKBP38 is activated by Ca2+/calmodulin. EMBO J. 24:2688-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edlich, F., M. Weiwad, D. Wildemann, F. Jarczowski, S. Kilka, M. C. Moutty, G. Jahreis, C. Lucke, W. Schmidt, F. Striggow, and G. Fischer. 2006. The specific FKBP38 inhibitor N-(N′,N′-dimethylcarboxamidomethyl)cycloheximide has potent neuroprotective and neurotrophic properties in brain ischemia. J. Biol. Chem. 281:14961-14970. [DOI] [PubMed] [Google Scholar]
- 9.Epstein, A. C., J. M. Gleadle, L. A. McNeill, K. S. Hewitson, J. O'Rourke, D. R. Mole, M. Mukherji, E. Metzen, M. I. Wilson, A. Dhanda, Y. M. Tian, N. Masson, D. L. Hamilton, P. Jaakkola, R. Barstead, J. Hodgkin, P. H. Maxwell, C. W. Pugh, C. J. Schofield, and P. J. Ratcliffe. 2001. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107:43-54. [DOI] [PubMed] [Google Scholar]
- 10.Fischer, G., and T. Aumüller. 2003. Regulation of peptide bond cis/trans isomerization by enzyme catalysis and its implication in physiological processes. Rev. Physiol. Biochem. Pharmacol. 148:105-150. [DOI] [PubMed] [Google Scholar]
- 11.Fischer, G., B. Wittmann-Liebold, K. Lang, T. Kiefhaber, and F. X. Schmid. 1989. Cyclophilin and peptidyl-prolyl cis-trans isomerase are probably identical proteins. Nature 337:476-478. [DOI] [PubMed] [Google Scholar]
- 12.Fong, S., L. Mounkes, Y. Liu, M. Maibaum, E. Alonzo, P. Y. Desprez, A. D. Thor, M. Kashani-Sabet, and R. J. Debs. 2003. Functional identification of distinct sets of antitumor activities mediated by the FKBP gene family. Proc. Natl. Acad. Sci. USA 100:14253-14258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hewitson, K. S., L. A. McNeill, M. V. Riordan, Y. M. Tian, A. N. Bullock, R. W. Welford, J. M. Elkins, N. J. Oldham, S. Bhattacharya, J. M. Gleadle, P. J. Ratcliffe, C. W. Pugh, and C. J. Schofield. 2002. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 277:26351-26355. [DOI] [PubMed] [Google Scholar]
- 14.Hirsila, M., P. Koivunen, V. Gunzler, K. I. Kivirikko, and J. Myllyharju. 2003. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J. Biol. Chem. 278:30772-30780. [DOI] [PubMed] [Google Scholar]
- 15.Ivan, M., T. Haberberger, D. C. Gervasi, K. S. Michelson, V. Gunzler, K. Kondo, H. Yang, I. Sorokina, R. C. Conaway, J. W. Conaway, and W. G. Kaelin, Jr. 2002. Biochemical purification and pharmacological inhibition of a mammalian prolyl hydroxylase acting on hypoxia-inducible factor. Proc. Natl. Acad. Sci. USA 99:13459-13464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 17.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. von Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 18.Kang, C. B., L. Feng, J. Chia, and H. S. Yoon. 2005. Molecular characterization of FK-506 binding protein 38 and its potential regulatory role on the anti-apoptotic protein Bcl-2. Biochem. Biophys. Res. Commun. 337:30-38. [DOI] [PubMed] [Google Scholar]
- 19.Kato, H., T. Inoue, K. Asanoma, C. Nishimura, T. Matsuda, and N. Wake. 2006. Induction of human endometrial cancer cell senescence through modulation of HIF-1α activity by EGLN1. Int. J. Cancer 118:1144-1153. [DOI] [PubMed] [Google Scholar]
- 20.Koivunen, P., M. Hirsila, V. Gunzler, K. I. Kivirikko, and J. Myllyharju. 2004. Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J. Biol. Chem. 279:9899-9904. [DOI] [PubMed] [Google Scholar]
- 21.Lando, D., D. J. Peet, D. A. Whelan, J. J. Gorman, and M. L. Whitelaw. 2002. Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science 295:858-861. [DOI] [PubMed] [Google Scholar]
- 22.Mahon, P. C., K. Hirota, and G. L. Semenza. 2001. FIH-1: a novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 15:2675-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin, F., T. Linden, D. M. Katschinski, F. Oehme, I. Flamme, C. K. Mukhopadhyay, K. Eckhardt, J. Tröger, S. Barth, G. Camenisch, and R. H. Wenger. 2005. Copper-dependent activation of hypoxia-inducible factor (HIF)-1: implications for ceruloplasmin regulation. Blood 105:4613-4619. [DOI] [PubMed] [Google Scholar]
- 24.Masson, N., R. J. Appelhoff, J. R. Tuckerman, Y. M. Tian, H. Demol, M. Puype, J. Vandekerckhove, P. J. Ratcliffe, and C. W. Pugh. 2004. The HIF prolyl hydroxylase PHD3 is a potential substrate of the TRiC chaperonin. FEBS Lett. 570:166-170. [DOI] [PubMed] [Google Scholar]
- 25.Masson, N., C. Willam, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroxylation. EMBO J. 20:5197-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metzen, E., U. Berchner-Pfannschmidt, P. Stengel, J. H. Marxsen, I. Stolze, M. Klinger, W. Q. Huang, C. Wotzlaw, T. Hellwig-Bürgel, W. Jelkmann, H. Acker, and J. Fandrey. 2003. Intracellular localisation of human HIF-1α hydroxylases: implications for oxygen sensing. J. Cell Sci. 116:1319-1326. [DOI] [PubMed] [Google Scholar]
- 27.Metzen, E., D. P. Stiehl, K. Doege, J. H. Marxsen, T. Hellwig-Bürgel, and W. Jelkmann. 2005. Regulation of the prolyl hydroxylase domain protein 2 (phd2/egln-1) gene: identification of a functional hypoxia-responsive element. Biochem. J. 387:711-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakayama, K., I. J. Frew, M. Hagensen, M. Skals, H. Habelhah, A. Bhoumik, T. Kadoya, H. Erdjument-Bromage, P. Tempst, P. B. Frappell, D. D. Bowtell, and Z. Ronai. 2004. Siah2 regulates stability of prolyl-hydroxylases, controls HIF-1α abundance, and modulates physiological responses to hypoxia. Cell 117:941-952. [DOI] [PubMed] [Google Scholar]
- 29.Oehme, F., P. Ellinghaus, P. Kolkhof, T. J. Smith, S. Ramakrishnan, J. Hutter, M. Schramm, and I. Flamme. 2002. Overexpression of PH-4, a novel putative proline 4-hydroxylase, modulates activity of hypoxia-inducible transcription factors. Biochem. Biophys. Res. Commun. 296:343-349. [DOI] [PubMed] [Google Scholar]
- 30.Oehme, F., W. Jonghaus, L. Narouz-Ott, J. Huetter, and I. Flamme. 2004. A nonradioactive 96-well plate assay for the detection of hypoxia-inducible factor prolyl hydroxylase activity. Anal. Biochem. 330:74-80. [DOI] [PubMed] [Google Scholar]
- 31.Ozer, A., L. C. Wu, and R. K. Bruick. 2005. The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF). Proc. Natl. Acad. Sci. USA 102:7481-7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Percy, M. J., Q. Zhao, A. Flores, C. Harrison, T. R. Lappin, P. H. Maxwell, M. F. McMullin, and F. S. Lee. 2006. A family with erythrocytosis establishes a role for prolyl hydroxylase domain protein 2 in oxygen homeostasis. Proc. Natl. Acad. Sci. USA 103:654-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pescador, N., Y. Cuevas, S. Naranjo, M. Alcaide, D. Villar, M. O. Landazuri, and L. Del Peso. 2005. Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochem. J. 390:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pouyssegur, J., F. Dayan, and N. M. Mazure. 2006. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature 441:437-443. [DOI] [PubMed] [Google Scholar]
- 35.Rosner, M., K. Hofer, M. Kubista, and M. Hengstschlager. 2003. Cell size regulation by the human TSC tumor suppressor proteins depends on PI3K and FKBP38. Oncogene 22:4786-4798. [DOI] [PubMed] [Google Scholar]
- 36.Schofield, C. J., and P. J. Ratcliffe. 2004. Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell. Biol. 5:343-354. [DOI] [PubMed] [Google Scholar]
- 37.Semenza, G. L. 2003. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer 3:721-732. [DOI] [PubMed] [Google Scholar]
- 38.Shirane, M., and K. I. Nakayama. 2003. Inherent calcineurin inhibitor FKBP38 targets Bcl-2 to mitochondria and inhibits apoptosis. Nat. Cell Biol. 5:28-37. [DOI] [PubMed] [Google Scholar]
- 39.Soilleux, E. J., H. Turley, Y. M. Tian, C. W. Pugh, K. C. Gatter, and A. L. Harris. 2005. Use of novel monoclonal antibodies to determine the expression and distribution of the hypoxia regulatory factors PHD-1, PHD-2, PHD-3 and FIH in normal and neoplastic human tissues. Histopathology 47:602-610. [DOI] [PubMed] [Google Scholar]
- 40.Steiner, J. P., M. A. Connolly, H. L. Valentine, G. S. Hamilton, T. M. Dawson, L. Hester, and S. H. Snyder. 1997. Neurotrophic actions of nonimmunosuppressive analogues of immunosuppressive drugs FK506, rapamycin and cyclosporin A. Nat. Med. 3:421-428. [DOI] [PubMed] [Google Scholar]
- 41.Steinmann, B., P. Bruckner, and A. Superti-Furga. 1991. Cyclosporin A slows collagen triple-helix formation in vivo: indirect evidence for a physiologic role of peptidyl-prolyl cis-trans-isomerase. J. Biol. Chem. 266:1299-1303. [PubMed] [Google Scholar]
- 42.Stiehl, D. P., R. Wirthner, J. Koditz, P. Spielmann, G. Camenisch, and R. H. Wenger. 2006. Increased prolyl 4-hydroxylase domain proteins compensate for decreased oxygen levels. Evidence for an autoregulatory oxygen-sensing system. J. Biol. Chem. 281:23482-23491. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, N., T. Hayano, and M. Suzuki. 1989. Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature 337:473-475. [DOI] [PubMed] [Google Scholar]
- 44.To, K. K., and L. E. Huang. 2005. Suppression of hypoxia-inducible factor 1α (HIF-1α) transcriptional activity by the HIF prolyl hydroxylase EGLN1. J. Biol. Chem. 280:38102-38107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wanner, R. M., P. Spielmann, D. M. Stroka, G. Camenisch, I. Camenisch, A. Scheid, D. R. Houck, C. Bauer, M. Gassmann, and R. H. Wenger. 2000. Epolones induce erythropoietin expression via hypoxia-inducible factor-1α activation. Blood 96:1558-1565. [PubMed] [Google Scholar]
- 46.Wenger, R. H. 2002. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 16:1151-1162. [DOI] [PubMed] [Google Scholar]
- 47.Wenger, R. H., D. P. Stiehl, and G. Camenisch. 2005. Integration of oxygen signaling at the consensus HRE. Sci. STKE 306:re12. [DOI] [PubMed]