Abstract
The hypothalamus is a key regulatory unit of the neuroendocrine system and plays an essential role in energy balance and reproduction. Despite its important role, the molecular mechanisms underlying hypothalamic development are not fully understood. Here, we report molecular analyses of a newly identified murine homeobox gene, Bsx/Bsx1a, that is expressed in the developing and postnatal hypothalamus. We demonstrate that BSX1A is a DNA binding protein and a transcriptional activator. Transcriptional reporter assays identified the C-terminal region of BSX1A as an activation domain. We have isolated an alternative splice form of Bsx1a, designated Bsx1b, which retains the N-terminal region but lacks the homeodomain. Analyses of subcellular localization using transfected cell lines revealed that BSX1A and BSX1B localize in the nuclei and cytoplasm, respectively. Immunohistochemical analyses suggested that both BSX1A and BSX1B are expressed in the neonatal hypothalamus. Taking these data together, we propose that alternative RNA splicing is involved in hypothalamic development/function.
The hypothalamus-pituitary axis is a major component of the neuroendocrine system that controls the homeostasis of energy balance and reproduction. Disturbances of hypothalamic functions cause a variety of human health problems that include growth retardation, obesity, infertility, and hormone-regulated tumor progression. Much has been elucidated in the development of the pituitary in terms of transcriptional regulation; however, relatively little is known about the development of hypothalamic neurons (4, 5, 16, 28, 31, 35).
Although the hypothalamus consists of several cell types located in different regions, only a few transcription factors have been identified that are expressed in the developing hypothalamus. Brn-2 encodes a POU domain transcription factor, and its targeted mutation results in the loss of corticotropin-releasing hormone (CRH), vasopressin (AVP), and oxytocin (OT) in Brn-2-expressing cells. The homozygous mutant mice die in the neonatal period with a decrease in size and weight (18, 26). The basic helix-loop-helix (bHLH)-Per-Arnt-Sim transcription factor Sim1 is expressed in the paraventricular, the supraoptic, and the anterior periventricular nuclei. In Sim1 homozygous mutant mice, OT-, AVP-, thyrotropin-releasing hormone-, CRH-, and somatostatin (SS)-secreting neurons are absent in the Sim1-expressing regions (15). The Sim1 mutant mice die within 24 h after birth. Thus, the phenotype of Sim1 mutant mice overlaps with but is more severe than that of Brn-2 mutant mice. The Sim1 mutant hypothalamus fails to express Brn-2 during development, suggesting that a part of the phenotype of Sim1 mutant mice is due to loss of Brn-2 and that BRN-2 acts downstream of SIM1. Another bHLH-Per-Arnt-Sim transcription factor, Arnt-2, is also involved in hypothalamic development. Arnt-2 mutant mice show a strikingly similar phenotype to the Sim1-deficient mice, consistent with the finding that SIM1 and ARNT-2 form a heterodimer complex in vivo (8, 9, 14). Gsh-1 is a homeobox gene that is expressed in the developing hypothalamus and pituitary gland (32). Gsh-1 homozygous mutant mice exhibit marked dwarfism, infertility, and significant postnatal mortality due to the loss of growth hormone-releasing hormone (GHRH) expression in the arcuate nucleus (ARN) of the hypothalamus and decreased production of pituitary hormones, including growth hormone, thyrotropin, prolactin, adrenocorticotropin, and leutenizing hormone (12). Recently, it has been reported that a bHLH transcription factor, Mash1, is required for the expression of Gsh-1 and that loss of Mash1 results in an absence of GHRH expression (13). Otp encodes a homeodomain-containing transcription factor that is expressed in developing neurons, giving rise to the paraventricular, supraoptic, and anterior periventricular nuclei and ARN. Otp mutant mice die within 2 days after birth and fail to produce AVP, OT, thyrotropin-releasing hormone, CRH, and SS (1, 33).
Recently, a novel hypothalamic homeobox gene, Bsx/Bsx1a, was identified in vertebrates, including human, chicken, zebrafish, and frog (3). The expression of Bsx was detected in a broad region of the developing and postnatal hypothalamus, including the ARN and the dorsomedial nuclei. The Bsx-expressing area includes the neurons that express GHRH and SS, suggesting that BSX/BSX1A is involved in growth control. Thus, Bsx/Bsx1a likely plays a distinct role in hypothalamic development and/or function in comparison with the previously identified transcription factors described above. Since the molecular functions of BSX/BSX1A have not been characterized, here we report biochemical analyses of BSX/BSX1A and its isoform, BSX1B.
MATERIALS AND METHODS
RNA extraction and cDNA cloning.
Total RNA was extracted from embryonic day 12.5 and 14.5 mouse embryos, and cDNA was synthesized as described previously (19, 21). Bsx1a and Bsx1b open reading frames were amplified by PCR using the primers N (5′-GAA TTC ATG AAT CTC AAC TTC ACT TCC-3′), C1 (5′-TCA GAG CAC ATG CGG CCC TG-3′), and C2 (5′-TCA GAG CAC ATG CGG CCC TG-3′). N and C1 were used as first PCR primers and N and C2 as nested PCR primers. The PCR conditions were 94°C for 2 min 30 s, followed by 35 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min, and a final extension of 72°C for 10 min.
Antibody production and immunological analyses.
Anti-BSX1A and anti-BSX1B sera were produced by immunizing rabbits with synthesized peptides, FPHPQ HAELP GKHCR and C-LRPGE KVRNP ALPVD, respectively (Genemed Synthesis). Antibodies were purified from the sera using a SulfoLink kit (PIERCE). pcDNA3-myc (vector), pcDNA3-myc-BSX1A, pcDNA3-myc-BSX1A mutant forms, and pcDNA3-myc-BSX1B were transfected into COS7 or Hs683 (human glioma cell line) cells using FuGENE6 (Roche). After 24 h, the cells were fixed in methanol and incubated with anti-BSX1A, anti-BSX1B, or anti-myc antibody (Sigma) as a primary antibody. The localization of the expressed proteins was visualized using Texas Red-conjugated anti-rabbit immunoglobulin G antibodies (Jackson ImmunoResearch) or fluorescein isothiocyanate-conjugated anti-mouse immunolobulin G antibodies (Sigma). To detect BSX1A and BSX1B in vivo, immunohistochemistry was performed on neonatal mouse brain sections using anti-BSX1A and anti-BSX1B sera (1:50; Zyagen).
Yeast reporter assay.
Yeast media and growth conditions were described previously (20, 22). A yeast expression vector, pAS2.1C, was used to express recombinant GAL4 DNA binding domain (GAL4DBD) fused to BSX1A, BSX1B, or their truncated forms. BSX1ΔC, BSX1ΔN, and BSX1BΔC contained amino acid residues (aa) 1 to 135 and 169 to 232 of BSX1A and 1 to 101 of BSX1B, respectively. The transcriptional activities of the recombinant proteins were evaluated by β-galactosidase (β-Gal) activities and the growth of a Saccharomyces cerevisiae strain (Y190) on His-depleted medium as described previously (22).
Western analysis.
Immunoblotting was performed using anti-GAL4DBD (Clontech) or anti-myc (Sigma) antibodies and Hybond-ECL membrane (Amersham) as described previously (22). 293T (a human embryonic kidney cell line) cells were used to check the mammalian expression plasmids (pcDNA3-myc-BSX1A, pcDNA3-myc-BSX1A mutant forms, and pcDNA3-myc-BSX1B).
Transcriptional reporter assay in mammalian cell lines.
To examine the transcriptional activities of BSX1A in mammalian cell lines, expression and reporter plasmids were transfected using FuGENE6 (Roche), and reporter luciferase activities were measured with a Dual-Luciferase Reporter Assay System (Promega) and a Veritas microplate luminometer (Turner Biosystems). The expression plasmids used were pcDNA3-myc (vector), pcDNA3-myc-BSX1A, pcDNA3-myc-BSX1AΔC, pcDNA3-myc-BSX1AΔAD, and pcDNA3-myc-BSX1AΔNLS. BSX1AΔAD contained aa 1 to 193 of BSX1A, and the arginine residues at positions 109 and 111 were replaced with glycine residues in BSX1AΔNLS. A reporter plasmid was constructed by inserting annealed oligonucleotides (5′-GTACCCCAATTAGCGGATCCCAATTAGCAAGGGTTCCAATTAGCA-3′ and 5′-GATCTGCTAATTGGAACCCTTGCTAATTGGGATCCGCTAATTGGG-3′) into the KpnI/BglII sites of the pGL3-Promoter vector (Promega). Luciferase activities were normalized for transfection efficiency by cotransfecting the pRL-SV40 vector (Promega) and measuring Renilla luciferase activities. Hs683 and Neuro2a (a mouse neuroblastoma cell line) were used for the assay.
EMSA.
To investigate the DNA binding ability of BSX1A, we produced a glutathione S-transferase (GST) fusion protein (GST-BSX1A), its truncated forms (GST-BSX1AΔC and GST-BSX1AΔAD), and GST as described previously (22) and performed electrophoresis mobility shift assays (EMSA) using oligonucleotides that contained a consensus homeodomain binding site (ATTA). A radiolabeled BSX1A binding sequence (BBS) was prepared by annealing oligonucleotides, 5′-GATCCAATTAGC-3′ and 5′-GATCGCTAATTG-3′ and filling in recessed 3′ ends using a Klenow fragment and [α-32P]dCTP. The radiolabeled probe and GST fusion proteins were incubated in the binding buffer [10 mM HEPES, pH 7.7, 50 mM KCl, 1 mM EDTA, 8% glycerol, 0.01 mg/ml poly(dI-dC)] in the presence or absence of BBS and/or mutated BBS (5′-GATCCATGTCTAGATC-3′) as a competitor and separated in a 4% polyacrylamide gel by electrophoresis. The band intensity was measured using ImageQuant (GE Healthcare).
RESULTS
Cloning of Bsx1a and Bsx1b.
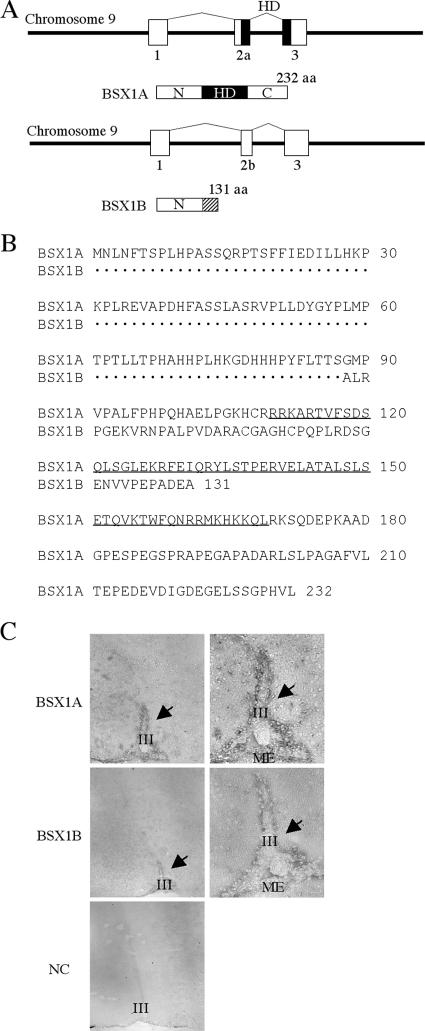
To isolate homeobox genes that are important for nervous system development, we searched the databases (http://www.ncbi.nih.gov and http://www.ensembl.org) and identified a novel homeobox gene sequence. To isolate the new gene, designated Bsx1a, we performed reverse transcription-PCR using mouse embryos as a source of RNA. After we initiated the project, another group isolated the same gene (Bsx) and reported that Bsx/Bsx1a expression was restricted to the nervous system in both embryonic and postnatal stages, including the hypothalamus and pineal gland (3). Comparison of the cDNA sequences of Bsx and Bsx1a identified two silent mutations, probably due to the different mouse strain used. As shown in Fig. 1, BSX1A consists of 232 aa encoded by a gene containing three coding exons on mouse chromosome 9. All exon-intron junctions follow the GT-AG rule. The homeodomain is encoded by the second and third coding exons and is located in the middle of the predicted BSX1A. Hence, BSX1A is divided into three regions: the amino-terminal region, homeodomain region, and carboxy-terminal region. Interestingly, we have found a splice variant of Bsx1a, designated Bsx1b, that utilizes a different splice acceptor site in the second coding exon. As a result, the reading frame is shifted and Bsx1b encodes a 131-aa protein that retains the N region but has no homeodomain. Anti-BSX1A and anti-BSX1B sera detected immunoreactive cells around the third ventricle in the mouse neonatal hypothalamus, suggesting that BSX1A and BSX1B are expressed in vivo (Fig. 1C).
FIG. 1.
Gene structure, schematic representation, deduced amino acid sequences, and expression of BSX1A and BSX1B. (A) Both BSX1A and BSX1B are encoded by three coding exons (squares) on mouse chromosome 9. The horizontal lines indicate noncoding genomic DNA. HD, homeodomain (black squares); N, N-terminal region; C, C-terminal region. The BSX1B-specific region is shaded. (B) Deduced amino acid sequences of BSX1A and BSX1B. The homeodomain sequence is underlined. (C) Immunohistochemistry on neonatal mouse hypothalamic sections using anti-BSX1A and anti-BSX1B sera. On the right are high-magnification images. Immunoreactive cells were detected around the third ventricle (arrows). III, third ventricle; ME, median eminence; NC, negative control.
Subcellular localization of BSX1A and BSX1B.
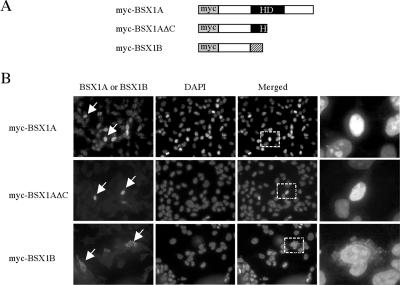
To investigate the subcellular localization of BSX1A, we transfected myc-tagged BSX1A expression vectors (pcDNA3-myc-BSX1A) into COS7 cells and monitored the protein localization using BSX1A antibodies. As shown in Fig. 2, myc-BSX1A was detected in the nuclei of transfected cells, which is consistent with the idea that BSX1A is a transcription factor. Similar results were obtained with myc antibodies, confirming the nuclear localization of myc-BSX1A. When pcDNA3-myc vectors were transfected, no signal was obtained by either antibody (not shown). myc-BSX1AΔC was also located in the nuclei, indicating that the deleted C-terminal region is not necessary for the nuclear localization of BSX1A. However, myc-BSX1B was detected in the cytoplasm of transfected cells by both BSX1B and myc antibodies. This suggests that the N-terminal region of BSX1A does not contain a nuclear localization signal (NLS).
FIG. 2.
Subcellular localization of BSX1A and BSX1B. (A) Schematic representation of myc-tagged proteins expressed in COS7 cells. HD, homeodomain. (B) Immunocytological analyses of myc-tagged protein-transfected COS7 cells. The arrows indicate BSX1A- or BSX1B-immunoreactive cells. The right column contains higher magnifications of parts of the merged images (the dotted squares). Note that BSX1A and BSX1B were detected in the nuclei and cytoplasm, respectively.
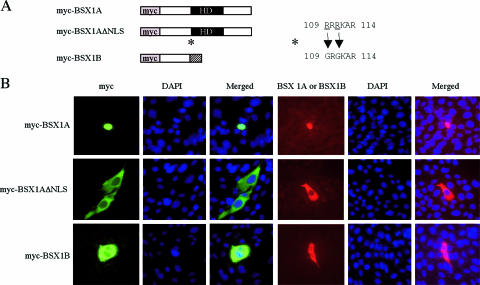
Based on these results, it is likely that BSX1A contains an NLS in its midportion. It is known that basic amino acid (Lys and Arg) stretches located at the beginning of the homeodomain serve as NLSs for several homeoproteins, such as VSX1 and OTX1 (11, 34). Therefore, 109RRRKAR114 of BSX1A is a possible NLS. To confirm the possibility, we mutated 109RRRKAR114 to 109GRGKAR114 (mutations in italics) (ΔNLS) and examined the subcellular localization of BSX1AΔNLS using Hs683 cells (Fig. 3). While the nuclear and cytoplasmic localizations of myc-BSX1A and myc-BSX1B were confirmed in Hs683, myc-BSX1AΔNLS was detected in the cytoplasm, showing that 109RRRKAR114 is involved in the nuclear localization of BSX1A.
FIG. 3.
Subcellular localization of BSX1AΔNLS. (A) Schematic representation of myc-tagged proteins expressed in Hs683 cells. HD, homeodomain. (B) Immunocytological analyses of myc-tagged protein-transfected Hs683cells. The images on the left (green) are myc-immunoreactive signals, and those on the right show BSX1A -or BSX1B-immunoreactive cells (red). Note that BSX1A was detected in the nuclei and BSX1AΔNLS and BSX1B in the cytoplasm.
DNA binding ability of BSX1A.
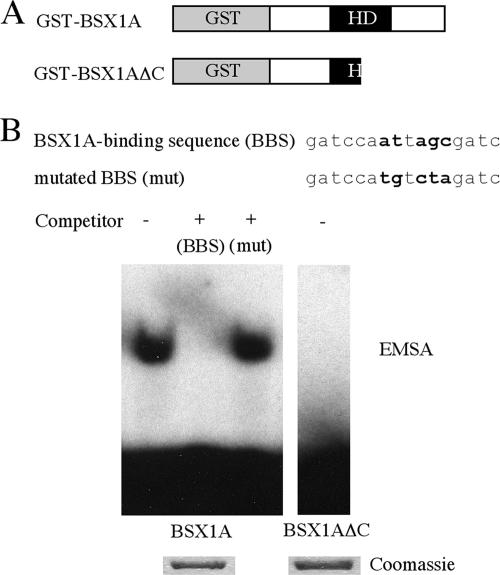
Since BSX1A is a nuclear homeoprotein, we next investigated the DNA binding ability of BSX1A. We produced GST fusion proteins (GST-BSX1A) and performed an EMSA using oligonucleotides that contain a consensus homeodomain binding site, ATTA (7). As shown in Fig. 4, GST-BSX1A bound to a radiolabeled BBS, and a shifted band was detected. The shifted band was still apparent in the presence of the mutated BBS, but not in the wild-type BBS, indicating that the binding of BSX1A to the BBS is a sequence-specific event. When a part of the homeodomain and the C region were deleted (GST-BSX1AΔC), the band shift was not observed, suggesting that the homeodomain is involved in DNA binding.
FIG. 4.
DNA binding ability of BSX1A. (A) Schematic representation of GST fusion proteins used for EMSA. HD, homeodomain. (B) EMSA using BBS. Wild-type and mutated (mut) BBSs were used as competitors. Mutated residues are shown in boldface. Below are Coomassie-stained GST fusion proteins used for EMSA.
Transcriptional activity of BSX1A.
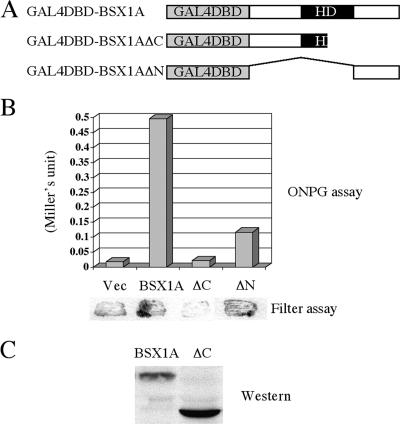
To investigate the transcriptional activity of BSX1A, we utilized a yeast GAL4 reporter system. We constructed yeast expression vectors that expressed full-length or truncated BSX1A fused with GAL4DBD and transformed the plasmids into a yeast strain (Y190). If a GAL4DBD fusion protein has transcriptional activation activity, Y190 can induce β-Gal and HIS3 gene expression, which are monitored by measuring β-Gal activity and by examining growth on His-depleted medium, respectively. As shown in Fig. 5, when full-length BSX1A was expressed in Y190, β-Gal activity was detected, indicating that GAL4DBD-BSX1A has transcriptional activation activity. To identify a transcriptional activation domain, we next expressed the C-region-truncated form of BSX1A (BSX1AΔC) in Y190. GAL4DBD-BSX1AΔC did not show transcriptional activation activity, although a comparable amount of protein was detected by Western analysis. This suggests that the C region is essential for the transcriptional activity of BSX1A and contains a transcriptional activation domain. To confirm this, we fused the C region of BSX1A to GAL4DBD and tested the transcriptional activity. As expected, GAL4DBD-BSX1AΔN showed positive β-Gal activity, suggesting the C-terminal region is involved in the transcriptional activation of BSX1A.
FIG. 5.
Transcriptional activity of BSX1A in yeasts. (A) Schematic representation of full-length or truncated (ΔC or ΔN) BSX1A fused to GAL4DBD. HD, homeodomain. (B) (Top) β-Gal activity measured by a liquid assay using o-nitrophenol-β-d-galactoside (ONPG) as a substrate. The values of the ONPG assay are indicated in Miller units. (Bottom) Results of a β-Gal filter assay using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as a substrate. Vec, vector. (C) Western analysis using anti-GAL4DBD antibody to confirm GAL4DBD fusion protein expression.
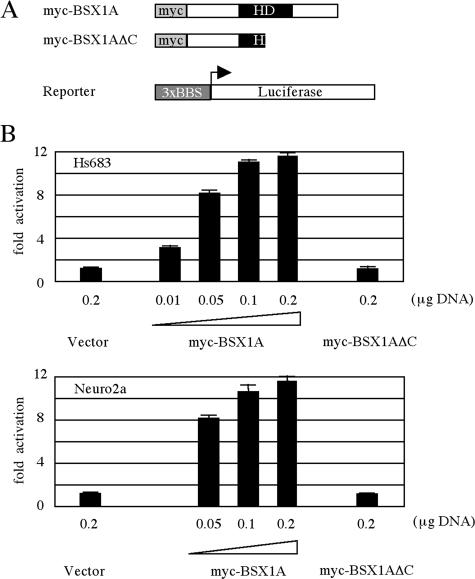
To confirm the findings obtained from the yeast system, we next examined the transcriptional activity in mammalian cell lines (Fig. 6). When pcDNA3-myc-BSX1A was transfected into a human glioma cell line, Hs683, with reporter plasmids, the transcriptional activation was observed to occur in a dose-dependent manner. The transcriptional activation was abolished when the C-terminal region and a part of the homeodomain were truncated. Similar results were obtained using a mouse neuroblast cell line, Neuro2a, indicating that the observed phenomena are not cell type specific. These results are consistent with the idea that BSX1A is a transcriptional activator.
FIG. 6.
Transcriptional activity of BSX1A in mammalian cells. (A) Schematic representation of myc-tagged full-length or truncated (ΔC) BSX1A and a luciferase reporter construct. HD, homeodomain. (B) Luciferase activities of myc-tagged protein-transfected Hs683 (above) and Neuro2a (below) cells; 0.2 μg of reporter plasmid was used for each transfection. All luciferase activities were normalized for transfection efficiency by measuring the Renilla luciferase activity of cotransfected pRL-SV40 vector (0.05 μg). The error bars indicate standard deviations.
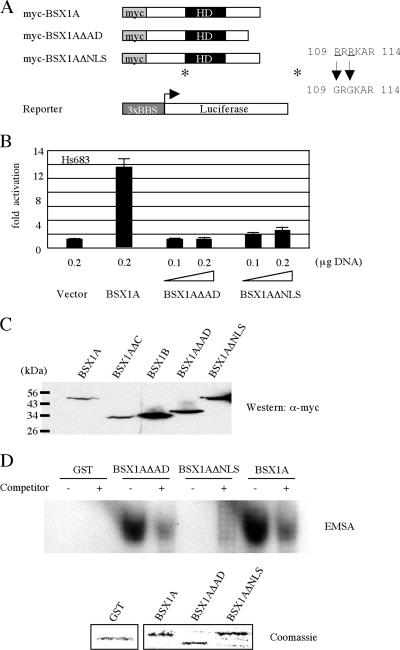
To map a transcriptional activation domain in the C-terminal region of BSX1A, we constructed another truncated form of BSX1A (BSX1AΔAD) that lacked aa 194 to 232 and examined its DNA binding ability and transcriptional activity (Fig. 7). Although GST-BSX1AΔAD retained DNA binding capability, myc-BSX1AΔAD did not activate a reporter gene, indicating that the C-terminal region of BSX1A contains a transcriptional activation domain. myc-BSX1AΔNLS also did not show reporter activation, which is likely due to the loss of DNA binding ability, since GST-BSX1AΔNLS does not bind BBS.
FIG. 7.
Transcriptional activity of mutant BSX1A in mammalian cells. (A) Schematic representation of myc-tagged full-length, truncated (ΔAD), and mutated (ΔNLS) BSX1A and a luciferase reporter construct. A putative NLS (109RRRKAR114) was mutated to 109GRGKAR114 (*). HD, homeodomain. (B) Luciferase activities of myc-tagged protein-transfected Hs683 cells; 0.2 μg of reporter plasmid was used for each transfection. All luciferase activities were normalized for transfection efficiency by measuring the Renilla luciferase activity of cotransfected pRL-SV40 vector (0.05 μg). The error bars indicate standard deviations. (C) Western analysis of myc-tagged wild-type and mutant BSX1A and BSX1B. The expression vectors were transfected 293T cells, and cell lysates were blotted with α-myc antibody (Sigma). (D) EMSA. GST-BSX1AΔAD, but not GST-BSX1AΔNLS, retains the DNA binding ability to BBS. (Below) Coomassie-stained GST fusion proteins used for EMSA.
DISCUSSION
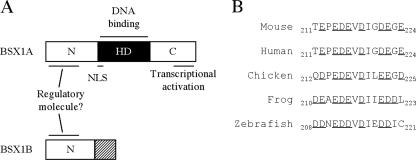
Here, we reported the cloning of a homeobox gene, Bsx1a, and its splice variant, Bsx1b. We verified that BSX1A is a DNA binding protein that functions as a transcriptional activator. BSX1B shares the N-terminal region with BSX1A but does not contain the homeodomain and the C-terminal region responsible for the transcriptional activity of BSX1A (Fig. 8). We demonstrated that BSX1A binds BBS, which includes a consensus homeodomain binding site, ATTA. Although further analyses are required to confirm that BBS is a physiological target sequence of BSX1A, it is noteworthy that GCT/CA/CATTAG/A was identified as a consensus GSH-1 binding site because Gsh-1 is expressed in the ARN and is essential for GHRH expression (12, 17, 32). The common DNA binding sequence and expression site imply that genetic interaction between Bsx1a and Gsh-1 exists in the hypothalamus and that Bsx1a is involved in GHRH production.
FIG. 8.
Functional-domain mapping of BSX1A and BSX1B. (A) The N-terminal regions of BSX1A and BSX1B possibly serve as protein interaction domains. The C-terminal region of BSX1A contains a transcriptional activation domain. The functions of BSX1B-specifc sequences (shaded square) are unknown. HD, homeodomain (black square); N, N-terminal region; C, C-terminal region. (B) An acidic residue cluster in the C-terminal region conserved among vertebrates. Acidic residues are underlined.
We mapped a transcriptional activation domain in the C-terminal region of BSX1A. Glutamine- or proline-rich or acidic regions are known to serve as transcriptional activation domains (30). The C-terminal end of BSX1A contains a number of acidic residues (Glu and Asp), including a cluster, 212EPEDEVDIGDEGE224, that may activate transcription, since the cluster is conserved among other species (Fig. 8).
Our studies of subcellular localization demonstrated that 109RRRKAR114 is an NLS of BSX1A. Arg-rich sequences are known to function as NLSs, since importin β can mediate the nuclear transport of an Arg-rich sequence (23), suggesting that importin β-mediated nuclear localization is a common mechanism to locate homeoproteins in the nuclei. Notably, BSX1AΔNLS lost DNA binding ability, suggesting that 109RRRKAR114 is also important for homeodomain function. Therefore, importin β may play an important role for the functional regulation of homeodomain-containing transcription factors.
Splice variants have been found in several homeobox genes, including Isl1, Lhx9, Pax6, Dmbx1, Prx1, Shox, Pitx2, and Csx1 (2, 6, 10, 21, 24, 25, 27, 29). The size of the transcriptome of homeobox genes seems to be larger than expected, and the regulation of transcription looks more complicated than expected. However, the physiological significance of the isoforms is still largely unknown. Functional domain mapping is one useful approach to investigate the functions of the isoforms. Unique features of BSX1B include its cytoplasmic localization and the lack of a homeodomain. A yeast two-hybrid assay determined that the N-terminal regions of BSX1A and BSX1B serve as protein interaction domains and that BSX1A and BSX1B bind a potential transcriptional regulator (our unpublished results), implying that BSX1B regulates the transcriptional activation of BSX1A by retaining its regulatory molecules in the cytoplasm. Collectively, our findings support the idea that the control of mRNA splicing is involved in hypothalamic development and/or functions.
Acknowledgments
We are grateful to Tara McArthur for Western blotting and critical reading of the manuscript. We thank Christopher Phiel and Leigh Zeidner for technical advice on luciferase assays.
Footnotes
Published ahead of print on 12 March 2007.
REFERENCES
- 1.Acampora, D., S. Mazan, F. Tuorto, V. Avantaggiato, J. L. Tremblay, D. Lazzaro, A. di Carlo, A. Mariano, P. E. Macchia, G. Corte, V. Macchia, J. Drouin, P. Brulet, and A. Simone. 1998. Transient dwarfism and hypogonadism in mice lacking Otx1 reveal prepubescent stage-specific control of pituitary levels of GH, FSH and LH. Development 125:1229-1239. [DOI] [PubMed] [Google Scholar]
- 2.Ando, K., S. Shioda, H. Handa, and K. Kataoka. 2003. Isolation and characterization of an alternatively spliced variant of transcription factor Islet-1. J. Mol. Endocrinol. 31:419-425. [DOI] [PubMed] [Google Scholar]
- 3.Cremona, M., E. Colombo, M. Andreazzoli, G. Cossu, and V. Broccoli. 2004. Bsx, an evolutionary conserved Brain Specific homeoboX gene expressed in the septum, epiphysis, mammillary bodies and arcuate nucleus. Gene Expr. Patterns 4:47-51. [DOI] [PubMed] [Google Scholar]
- 4.Dasen, J. S., and M. G. Rosenfeld. 1999. Combinational codes in signaling and synergy: lessons from pituitary development. Curr. Opin. Gen. Dev. 9:566-574. [DOI] [PubMed] [Google Scholar]
- 5.Dasen, J. S., and M. G. Rosenfeld. 1999. Signaling mechanisms in pituitary morphogenesis and cell fate determination. Curr. Opin. Cell Biol. 11:669-677. [DOI] [PubMed] [Google Scholar]
- 6.Failli, V., M. Rogard, M. Mattei, P. Vernier, and S. Retaux. 2000. Lhx9 and Lhx9a LIM-homeodomain factors: genomic structure, expression patterns, chromosomal localization, and phylogenetic analysis. Genomics 64:307-317. [DOI] [PubMed] [Google Scholar]
- 7.Givens, M. L., N. Rave-Harel, V. D. Goonewardena, R. Kurotani, S. E. Berdy, C. H. Swan, J. L. Rubenstein, B. Robert, and P. L. Mellon. 2005. Developmental regulation of gonadtropin-releasing hormone gene expression by the MSX and DLX homeodomain protein families. J. Biol. Chem. 280:19156-19165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosoya, T., Y. Oda, S. Takahashi, M. Morita, S. Kawauchi, M. Ema, M. Yamamoto, and Y. Fujii-Kuriyama. 2001. Defective development of secretory neurons in the hypothalamus of Arnt2-knockout mice. Genes Cells 6:361-374. [DOI] [PubMed] [Google Scholar]
- 9.Keith, B., D. M. Adelman, and M. C. Simon. 2001. Targeted mutation of the murine arylhydrocarbon receptor nuclear translocator 2 (Arnt2) gene reveals redundancy with Arnt. Proc. Natl. Acad. Sci. USA 98:6693-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, J., and J. D. Lauderdale. 2006. Analysis of Pax6 expression using a BAC transgenic reveals the presence of a paired-less isoform of Pax6 in the eye. Dev. Biol. 292:486-505. [DOI] [PubMed] [Google Scholar]
- 11.Kurtzman, A. L., and N. Schechter. 2001. Ubc9 interacts with a nuclear localization signal and mediates nuclear localization of the paired-like homeobox protein Vsx-1 independent of SUMO-1 modification. Proc. Natl. Acad. Sci. USA 98:5602-5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li, H., P. S. Zeitler, M. T. Valerius, K. Small, and S. S. Potter. 1996. Gsh-1, an orphan Hox gene, is required for normal pituitary development. EMBO J. 15:714-724. [PMC free article] [PubMed] [Google Scholar]
- 13.McNay, D. E. G., M. Pelling, S. Claxton, F. Guillemot, and S.-L. Ang. 2006. Mash1 is required for genetic and subtype differentiation of hypothalamic neuroendocrine cells. Mol. Endocrinol. 20:1623-1632. [DOI] [PubMed] [Google Scholar]
- 14.Michaud, J. L., C. DeRossi, N. R. May, B. C. Holdener, and C. Fan. 2000. Arnt2 acts as the dimerization partner of Sim1 for the development of the hypothalamus. Mech. Dev. 90:253-261. [DOI] [PubMed] [Google Scholar]
- 15.Michaud, J. L., T. Rosenquist, N. R. May, and C. Fan. 1998. Development of neuroendocrine lineages requires the bHLH-PAS transcription factor Sim1. Genes Dev. 12:3264-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullis, P. E. 2000. Transcriptional factors in pituitary gland development and their clinical impact on phenotype. Hormone Res. 54:107-119. [DOI] [PubMed] [Google Scholar]
- 17.Mutsuga, N., Y. Iwasaki, M. Morishita, A. Nomura, E. Yamamori, M. Yoshida, M. Asai, N. Ozaki, F. Kambe, H. Seo, Y. Oiso, and H. Saito. 2001. Homeobox protein Gsh-1-dependent regulation of the rat GHRH gene promoter. Mol. Endocrinol. 15:2149-2156. [DOI] [PubMed] [Google Scholar]
- 18.Nakai, S., H. Kawano, T. Yudate, M. Nishi, J. Kuno, A. Nagata, K. Jishage, H. Hamada, H. Fujii, K. Kawamura, K. Shiba, and T. Noda. 1995. The POU domain transcription factor Brn-2 is required for the determination of specific neuronal lineage in the hypothalamus of the mouse. Genes Dev. 9:3109-3121. [DOI] [PubMed] [Google Scholar]
- 19.Nishijima, I., and A. Ohtoshi. 2006. Characterization of a novel prospero-related homeobox gene, Prox2. Mol. Genet. Genomics 275:471-478. [DOI] [PubMed] [Google Scholar]
- 20.Ohtoshi, A., K. Arai, and H. Masai. 1998. Two recessive modes of growth inhibition by exogenously introduced mutant genes: analysis of mutant CDC28 and CDC7 genes in Sacchromyces cerevisiae. J. Biochem. Mol. Biol. Biophys. 1:253-263. [Google Scholar]
- 21.Ohtoshi, A., I. Nishijima, M. J. Justice, and R. R. Behringer. 2002. Dmbx1, a novel evolutionarily conserved paired-like homeobox gene expressed in the brain of mouse embryos. Mech. Dev. 110:241-244. [DOI] [PubMed] [Google Scholar]
- 22.Ohtoshi, A., and H. Otoshi. 2001. Analysis of β3-endonexin mutants for their ability to interact with cyclin A. Mol. Genet. Genomics 266:664-671. [DOI] [PubMed] [Google Scholar]
- 23.Palmeri, D., and M. H. Malim. 1999. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol. Cell. Biol. 19:1218-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peterson, R. E., S. Hoffman, and M. J. Kern. 2005. Opposing roles of two isoforms of the Prx1 homeobox gene in the chondrogenesis. Dev. Dyn. 233:811-821. [DOI] [PubMed] [Google Scholar]
- 25.Rao, E., R. J. Blaschke, A. Marchini, B. Niesler, M. Burnett, and G. A. Rappold. 2001. The Leri-Weill and Turner syndrome homeobox gene SHOX encodes a cell-type specific transcriptional activator. Hum. Mol. Genet. 10:3083-3091. [DOI] [PubMed] [Google Scholar]
- 26.Schonemann, M. D., A. K. Ryan, R. J. McEvilly, S. M. O'Connell, C. A. Arias, K. A. Kalla, P. Li, P. E. Sawchenko, and M. G. Rosenfeld. 1995. Development and survival of the endocrine hypothalamus and pituitary gland requires the neuronal POU domain factor Brn-2. Genes Dev. 9:3122-3135. [DOI] [PubMed] [Google Scholar]
- 27.Schweickert, A., H. Steinbeisser, and M. Blum. 2001. Differential gene expression of Xenopus Pitx1, Pitx2b and Pitx2c during cement gland, stomodeum and pituitary development. Mech. Dev. 107:191-194. [DOI] [PubMed] [Google Scholar]
- 28.Scully, K. M., and M. G. Rosenfeld. 2002. Pituitary development: regulatory codes in mammalian organogenesis. Science 295:2231-2235. [DOI] [PubMed] [Google Scholar]
- 29.Shiojima, I., I. Komuro, T. Mizuno, R. Aikawa, H. Akazawa, T. Oka, T. Yamazaki, and Y. Yazaki. 1996. Molecular cloning and characterization of human cardiac homeobox gene CSX1. Circ. Res. 79:920-929. [DOI] [PubMed] [Google Scholar]
- 30.Stepchenko, A., and M. Nirenberg. 2004. Mapping activation and repression domains of the vnd/NK-2 homeodomain protein. Proc. Natl. Acad. Sci. USA 101:13180-13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Treier, M., and M. G. Rosenfeld. 1996. The hypothalamic-pituitary axis: co-development of two organs. Curr. Opin. Cell Biol. 8:833-843. [DOI] [PubMed] [Google Scholar]
- 32.Valerius, M. T., H. Li, J. L. Stock, M. Weinstein, S. Kaur, G. Singh, and S. S. Potter. 1995. Gsh-1: a novel murine homeobox gene expressed in the central nervous system. Dev. Dyn. 203:337-351. [DOI] [PubMed] [Google Scholar]
- 33.Wang, W., and T. Lufkin. 2000. The murine Otp homeobox gene plays an essential role in the specification of neuronal cell lineages in the developing hypothalamus. Dev. Biol. 227:432-449. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y. A., A. Okada, C. H. Lew, and S. K. McConnell. 2002. Regulated nuclear trafficking of the homeodomain protein otx1 in cortical neurons. Mol. Cell Neurosci. 19:430-446. [DOI] [PubMed] [Google Scholar]
- 35.Zhu, X., C. R. Lin, G. G. Prefontaine, J. Tollkuhn, and M. G. Rosenfeld. 2005. Genetic control of pituitary development and hypopituitarism. Curr. Opin. Genet. Dev. 15:332-340. [DOI] [PubMed] [Google Scholar]