Abstract
The PeBoW complex is essential for cell proliferation and maturation of the large ribosomal subunit in mammalian cells. Here we examined the role of PeBoW-specific proteins Pes1, Bop1, and WDR12 in complex assembly and stability, nucleolar transport, and preribosome association. Recombinant expression of the three subunits is sufficient for complex formation. The stability of all three subunits strongly increases upon incorporation into the complex. Only overexpression of Bop1 inhibits cell proliferation and rRNA processing, and its negative effects could be rescued by coexpression of WDR12, but not Pes1. Elevated levels of Bop1 induce Bop1/WDR12 and Bop1/Pes1 subcomplexes. Knockdown of Bop1 abolishes the copurification of Pes1 with WDR12, demonstrating Bop1 as the integral component of the complex. Overexpressed Bop1 substitutes for endogenous Bop1 in PeBoW complex assembly, leading to the instability of endogenous Bop1. Finally, indirect immunofluorescence, cell fractionation, and sucrose gradient centrifugation experiments indicate that transport of Bop1 from the cytoplasm to the nucleolus is Pes1 dependent, while Pes1 can migrate to the nucleolus and bind to preribosomal particles independently of Bop1. We conclude that the assembly and integrity of the PeBoW complex are highly sensitive to changes in Bop1 protein levels.
The nucleolus is the site of the highly regulated, evolutionarily conserved processes of rRNA transcription, pre-rRNA processing, and ribosome subunit assembly (30). The mammalian 18S, 5.8S, and 28S rRNAs are derived from a single 47S precursor (pre-rRNA), which is processed to the mature species through a series of endonucleolytic, exonucleolytic, and modification steps (5, 31). Mature rRNAs are assembled into 40S and 60S ribosomal subunits (8). The biogenesis of ribosomes is a major expenditure of cellular resources that needs to be tightly coordinated with cell cycle progression. Doubling of the translational machinery is essential for continuous cell proliferation to sustain the equilibrium between cell growth and cell division (16, 24). The recent years have revealed interesting links between the nucleolus and cell cycle regulation. Ribosome biogenesis is highly sensitive to cellular stresses such as chemotherapeutic agents like actinomycin D. Ribosomal proteins like L11 are no longer incorporated into nascent ribosomes. They accumulate as free proteins and bind and inactivate the E3 ubiquitin ligase Hdm2 that targets the tumor suppressor p53 for degradation (21, 33). Thus, p53 accumulates and elicits cell cycle arrest and apoptosis. Alternatively, it was shown that a subset of ribosomes contained cytoplasmic p53 covalently linked to 5.8S rRNA (7). Therefore, the export of intact ribosomal subunits to the cytoplasm may be important for p53 degradation as well (27). Despite these remarkable connections between ribosome synthesis and other cellular processes, little is known about the mammalian ribosome biogenesis machinery compared to yeast.
We have recently characterized a nucleolar complex of endogenous Pes1, Bop1, and WDR12 in mammalian cells, termed the PeBoW complex (14). Interestingly, expression of N-terminal or C-terminal truncations of Pes1, Bop1, or WDR12 in mammalian cells blocked processing of the 32S pre-rRNA into mature 28S rRNA and triggered p53-dependent cell cycle arrest (10, 14, 20, 23). Apparently, the PeBoW complex is a good target for the generation of dominant-negative mutants and plays a crucial role in rRNA processing and maturation of the large ribosomal subunit.
Coimmunoprecipitation assays showed that dominant-negative mutant forms of Pes1 were indeed incorporated into the PeBoW complex, suggesting that they block its function by building up dead-end complexes that prevent further essential interactions (10, 20). Thus, the amount of PeBoW components needs to be tightly controlled and adjusted to the rate of ribosome synthesis and proliferation. Quiescent or serum-starved cells exhibit low levels of Pes1, Bop1, and WDR12 that are induced by the proto-oncogene c-Myc upon cell cycle entry (14). c-Myc is overexpressed in a variety of human malignancies, suggesting a coordinated upregulation of the PeBoW complex in tumor cells. But interestingly, amplification of the gene for Bop1, but not that for Pes1, was frequently found in colorectal cancers, associated with an increase in Bop1 mRNA (18). The PeBoW complex may play an additional role in mitosis, as transient overexpression of Bop1 increased the percentage of multipolar spindles. Depletion of Pes1 or Bop1 also caused an increase in abnormal mitotic figures (17).
These observations underline the importance of a functional PeBoW complex playing a role in the cross talk between ribosome biogenesis and the cell division cycle. However, it is unknown how the integrity of the PeBoW complex is controlled in mammalian cells.
In this study, we investigated how changes in the abundance of individual PeBoW components affect its functionality. We show that overexpression of Bop1 disturbs cell proliferation and ribosome biogenesis by titrating endogenous WDR12 into a Bop1/WDR12 subcomplex and that its negative effects could be rescued by coexpression of WDR12, but not Pes1. Further, Bop1 was found to be essential for the copurification of Pes1 and WDR12, thus arguing for an indirect interaction mediated by Bop1. Finally, depletion of individual PeBoW components by RNA interference revealed a strong interdependence of their protein levels. Thus, the integrity of the PeBoW complex is tightly controlled by protein-protein interactions and highly sensitive to elevated levels of Bop1.
MATERIALS AND METHODS
Tissue culture.
TGR-1 rat fibroblasts, U2OS osteosarcoma cells, and H1299 lung carcinoma cells (non-small-cell lung carcinoma) were cultured in Dulbecco modified Eagle medium with 10% fetal bovine serum at 8% CO2. For generation of polyclonal cell lines, 6 × 105 cells were transfected with the respective pRTS-1 plasmids by using Polyfect (QIAGEN) and stably selected in the presence of 200 μg/ml hygromycin B and/or 1 μg/ml puromycin for 10 to 14 days. Conditional gene expression was induced with 1 μg/ml doxycycline. Pes1 and WDR12 carrying a C-terminal hemagglutinin (HA) tag are of human origin, whereas Bop1 carrying an N-terminal HA tag is mouse specific. Therefore, we analyzed different cell types and species.
RNA analysis and 32P in vivo labeling.
Total RNA was isolated with Trifast (PeqLab). Two micrograms of total RNA for detection of ITS-1 and ITS-2 or 10 μg of total RNA for analyzing the endogenous mRNA levels was separated on a 1% agarose-formaldehyde gel and blotted onto Hybond N+ membranes (GE Healthcare). The following 32P-end-labeled DNA oligonucleotides were used to visualize rRNA precursors: ITS-1 (human specific), 5′-CCTCCGCGCCGGAACGCGCTAGGTACCTGGACGGCGGGGGGGCGGACG-3′; ITS-2 (human specific), 5′-GCGGCGGCAAGAGGAGGGCGGACGCCGCCGGGTCTGCGCTTAGGGGGA-3′; ITS-1 (rat specific), 5′-GGACCAGACCCGACACCCTGCCACCGCACACCTGTCCCGAAACCCCCT-3′; ITS-2 (rat specific), 5′-GCCCCGGGGAGCGGGCCCTGCGAGCAGACTCCCAGCCGCGCGACGCGA-3′; 18S rRNA (human and rat specific), 5′-CACCCGTGGTCACCATGGTAGGCACGGCGACTACCATCGAAAGTTGATAG-3′.
Metabolic labeling of rRNA has been described elsewhere (14).
Production of antibodies.
Monoclonal antibodies (MAbs) against human nucleostemin and human Nog1 were generated as previously described (14). For immunization, we used a glutathione S-transferase (GST)-nucleostemin fusion protein and a Nog1-specific peptide coupled to ovalbumin (Peptide Specialty Laboratories GmbH, Heidelberg, Germany). The Nog1 peptide sequence is ESKEKNTQGPRMPRTAKKVQRTVLEKC. The nucleostemin (7H3) MAb belongs to the immunoglobulin G2b subclass, and the Nog1 (1D8) MAb belongs to the immunoglobulin G2a subclass. Polyclonal antibodies against mouse Bop1 were raised by immunization of guinea pigs with a mixture of the peptides GKPHMSPASLPGKRRLEPDQELQIQ and SQEHTQVLLHQVSRRRSQSPFRRSHG.
Immunoblotting, immunofluorescence, and immunoprecipitation.
For immunoblotting, cells were directly lysed with 2× sodium dodecyl sulfate (SDS) loading buffer (100 mM Tris/HCl, 200 mM dithioerythritol, 4% SDS, 10 mM EDTA, 0.2% bromophenol blue, 20% glycerol). Whole-cell lysates were separated by SDS-polyacrylamide gel electrophoresis and blotted onto nitrocellulose membranes (GE Healthcare). Immunodetection was performed with anti-HA (3F10; Roche), anti-Pes1 (8E9), anti-Bop1 (6H11), anti-WDR12 (1B8), antinucleostemin (7H3), anti-Nog1 (1D8), anti-NPM1 (clone FC82291; Sigma Aldrich), antitubulin (Sigma Aldrich), anti-p53 (PAb240; Dianova), and anti-c-Myc (N-262; Santa Cruz Biotechnology, Inc.). Recombinant mouse Bop1 protein was detected with a 1:10,000 dilution of polyclonal mouse Bop1-specific guinea pig antibodies in methanol-acetone-fixed cells. Immunofluorescence and immunoprecipitation have been described elsewhere (14).
Native gel electrophoresis.
Cells (3 × 106) were lysed in 100 μl lysis buffer (50 mM Tris-HCl [pH 8.0], 1% NP-40, 150 mM NaCl, phosphatase inhibitors, protease inhibitors) at 4°C for 20 min. A 7.5-μl volume of 2× sample buffer (125 mM Tris-HCl [pH 6.8], 30% glycerol, 0.02% bromophenol blue) was added to 7.5 μl of total lysate and separated by polyacrylamide gel electrophoresis (6.5%) in the absence of SDS at 4°C. Blotting was performed in the absence of methanol. Immunoblotting was performed as described above.
siRNA transfection.
The day before transfection, ∼5 × 104 to 105 cells were seeded in six-well plates. Five microliters of 20 μM control, Pes1-, Bop1-, or WDR12-specific small interfering RNA (siRNA) was diluted in 150 μl of OptiMEM (Invitrogen). One hundred fifty microliters of OptiMEM containing 5 μl of Oligofectamine (Invitrogen) was added, and the mixture was incubated for 15 min. Finally, 600 μl of OptiMEM was added and the mixture was applied to cells after aspiration of the culture medium. Cells were incubated for 5 to 6 h. The following sequences (sense) were used: Pes1 UTR, CCAGAGGACCUAAGUGUGAdTdT; Pes1 ORF, AGGUCUUCCUGUCCAUCAAdTDT; Bop1 UTR, UCGUGCUGAAGUCAACAGAdTdT; Bop1 ORF, AUGGCAUGGUGUACAAUGAdTdT; WDR12 UTR-1, CGUACGUUUCCGUGGGCAAdTdT; WDR12 UTR-2, CGCUUACCUGUGCAGUCUAdTdT; Control (nonspecific siRNA), UUCUCCGAACGUGUCACGUdTdT.
Knockdown-knock-in assay.
Exogenous gene expression (Pes1, Bop1, or WDR12) was activated for 3 days by treatment with 1 μg/ml doxycycline and then maintained throughout the subsequent course of two siRNA transfections with siRNAs directed against the 3′ untranslated region (UTR) of the Pes1, Bop1, or WDR12 mRNA.
Cell fractionation and sucrose gradients.
Cells were harvested by trypsinization and washed three times with cold phosphate-buffered saline. Cells (3 × 106) were lysed in 100 μl lysis buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 0.5% NP-40, protease inhibitors) at 4°C for 20 min. Cytoplasmic fractions were isolated after centrifugation. The pelleted nuclei were washed three times with cold lysis buffer A and then lysed in 100 μl lysis buffer B (50 mM Tris-HCl [pH 8.0], 1% NP-40, 150 mM NaCl, protease inhibitors) at 4°C for 20 min. Sucrose gradients have been described elsewhere (10).
RESULTS
Overexpression of Bop1 negatively affects proliferation and processing of pre-rRNA.
We aimed to investigate the control of PeBoW complex integrity by increasing or decreasing the amount of its individual members. First, we determined the proliferation rates of TGR-1 and H1299 cells overexpressing Pes1, Bop1, or WDR12. Cells were stably transfected with pRTS constructs (1) conditionally expressing the respective HA-tagged wild-type forms or luciferase in a doxycycline-dependent manner. Equal numbers of cells were seeded in the presence of doxycycline, and cell numbers were determined after 6 days. Overexpression of Bop1-HA reduced the cell count up to 41% in TGR-1 and to 20% in H1299 cells (Fig. 1A, lanes 3 and 7) compared to control cells expressing luciferase (lanes 1 and 5). Overexpression of neither WDR12-HA (lanes 2 and 6) nor Pes1-HA (lanes 4 and 8) significantly affected cell proliferation. Expression of the HA-tagged forms of Pes1, Bop1, and WDR12 was verified by Western blot analysis (Fig. 1B).
FIG. 1.
Overexpression of Bop1 inhibits cell proliferation and pre-rRNA processing. (A) Equal numbers of H1299 and TGR-1 cells stably transfected with the indicated constructs were seeded in the presence of 1 μg/ml doxycycline. After 6 days, cells were trypsinized and counted by trypan blue exclusion. The histogram depicts the cell counts relative to that of the mock-treated cell line expressing luciferase. Error bars indicate standard deviations. (B) Expression levels of HA-tagged proteins were determined with the anti-HA antibody 3F10. Equal loading was verified by immunodetection of α-tubulin. (C) The proliferation of stable polyclonal H1299 cell lines expressing the indicated proteins was analyzed as described for panel A. (D) Expression levels of HA-tagged proteins were determined as described for panel B. (E) Diagram of the primary 47S rRNA transcript and the major rRNA intermediates. Positions of the hybridization probes are depicted. ETS, external transcribed spacer. (F) Northern blot analysis of rRNA precursors. Total RNA was extracted from subconfluent H1299 cells expressing the indicated HA-tagged proteins for 24 h. Equal amounts of total RNA were separated by agarose-formaldehyde gel electrophoresis and hybridized with probes specific for ITS-1 and ITS-2 of the rRNA intermediates. As a loading control, blots were incubated with a probe specific for 18S rRNA. (G) Ratios of 47S-45S/18S and 32S/18S rRNAs.
Next, we tested whether the antiproliferative effect of Bop1 overexpression resulted from an altered stoichiometry of the PeBoW complex and could be alleviated or rescued by coexpression of either Pes1 or WDR12. H1299 cells were stably transfected with two individual pRTS constructs harboring a puromycin or a hygromycin resistance gene. The enhanced green fluorescent protein of the last one was replaced with monomeric red fluorescent protein to better monitor coexpression (data not shown). Cells expressing Pes1, Bop1, or WDR12 or combinations thereof were seeded in equal cell numbers. The number of cells was determined in multiples and compared with the mean cell count of a mock-treated cell line after 6 days (Fig. 1C). The negative effect of Bop1 overexpression on cell proliferation could be rescued by coexpression of WDR12, but not Pes1 (Fig. 1C, lanes 4 and 6). Expressing WDR12 together with Pes1 did not alter the proliferation rate (Fig. 1C, lane 7). Equal expression levels were determined by Western blot analysis (Fig. 1D).
The PeBoW complex is involved in pre-rRNA processing. Therefore, we studied the maturation of rRNA in cells overexpressing individual PeBoW components. A scheme of mammalian RNA-processing pathways is shown in Fig. 1E. Pes1, Bop1, WDR12, and combinations thereof were stably expressed in H1299 and TGR-1 (data not shown) cells for 24 h. Total RNA was isolated and analyzed by Northern blot analysis with probes specific for internal transcribed spacer 1 (ITS-1) and ITS-2 of the ribosomal pre-rRNA (Fig. 1F and G). Bop1-overexpressing cells accumulated the 47/45S and 32S pre-rRNAs by twofold (Fig. 1F and G, lanes 2) whereas overexpression of WDR12 or Pes1 did not interfere with pre-rRNA processing (Fig. 1F and G, lanes 3 and 5). The Bop1-mediated aberrant accumulation of pre-rRNAs could be completely reversed through coexpression of WDR12, but not Pes1 (Fig. 1F and G, lanes 4 and 6). These results are in line with the proliferation experiments showing that coexpression of WDR12 but not Pes1 alleviates the negative effects of Bop1 overexpression.
Overexpression of Bop1 titrates endogenous WDR12 into a Bop1/WDR12 subcomplex.
These results prompted us to investigate the impact of Bop1 overexpression on the PeBoW complex in more detail. We examined whether ectopically expressed Pes1, Bop1, or WDR12 affected the abundance of the endogenous PeBoW proteins. Endogenous Pes1 and WDR12 can be discriminated from the human HA-tagged forms by their lower molecular weights. Recombinant rodent HA-Bop1 is not recognized by the human-specific Bop1 MAb. Overexpression of HA-Bop1, but not WDR12-HA, Pes1-HA, or luciferase, strongly reduced the steady-state levels of endogenous Bop1 in H1299 cells after 6 days (Fig. 2A). However, such a decrease in the endogenous Bop1 protein level was not detected in cells overexpressing Bop1 for only 1 day (data not shown). We did not observe a decrease in the protein levels of endogenous proteins WDR12 and Pes1 (Fig. 2A, lanes 2 and 4) after expression of Pes1-HA and WDR12-HA, respectively.
FIG. 2.
(A) Overexpression of Bop1 reduces the endogenous Bop1 protein level. H1299 cells were stably transfected with the indicated constructs. After 6 days, endogenous Pes1, Bop1, and WDR12 protein levels were analyzed by Western blotting. Human-specific anti-Bop1 MAb 6H11 does not recognize recombinant mouse Bop1. Expression levels of HA-tagged proteins were determined with anti-HA antibody 3F10. Equal loading was verified by immunodetection of α-tubulin. (B) Overexpression of Bop1 titrates endogenous WDR12 into a Bop1/WDR12 subcomplex. Total cell lysates of U2OS cells expressing the indicated proteins were separated by native gel electrophoresis after 1 day of expression. Pes1, Bop1, and WDR12 were visualized by immunoblotting. Nonincorporated proteins or complexes containing the respective factor are indicated. (C) Overexpression of Bop1-HA replaces endogenous Bop1 in PeBoW complex formation. Total lysates of U2OS cells expressing luciferase or Bop1-HA were separated by native gel electrophoresis after 6 days of expression. Pes1, Bop1, WDR12, and Bop1-HA were visualized by immunoblotting. An asterisk indicates monomeric Bop1-HA. Double asterisks indicate the Bop1/WDR12 subcomplex. (D) Total lysates of U2OS cells expressing the indicated proteins were separated by native gel electrophoresis. WDR12 were visualized by immunoblotting. Nonincorporated proteins or complexes containing the respective factor are indicated. Expression levels of HA-tagged proteins were determined by Western blot analysis with anti-HA antibody 3F10. Equal loading was verified by immunodetection of α-tubulin. (E) Cytoplasmic and nuclear fractions of H1299 cells expressing the indicated genes were analyzed by native gel electrophoresis as described for panel B. Complexes were visualized by immunostaining with HA-specific antibodies.
We have previously shown that the intact PeBoW complex can be visualized by native gel electrophoresis. Therefore, we established stable U2OS cell lines that conditionally overexpressed HA-tagged Pes1, Bop1, WDR12, or luciferase for 1 day and analyzed the PeBoW complex by native gel electrophoresis (14). Immunoblot analysis of all three proteins showed a single band representing the PeBoW complex (Fig. 2B). Overexpression of WDR12 (Fig. 2B, lane 3) and Pes1 (lane 2) resulted in faster-migrating bands indicating free nonincorporated protein. In contrast, overexpression of Bop1 led to the formation of an additional complex consisting of Bop1 and WDR12 but lacking Pes1 (Fig. 2B, lane 6). The immunoblot analysis of WDR12 further indicated that the overexpression of Bop1 titrated free endogenous WDR12 into this subcomplex (Fig. 2B, lane 6). Bop1-overexpressing cells lack monomeric WDR12, in contrast to Pes1-overexpressing cells. The overexpression of another nucleolar protein, nucleostemin or Nog1, did not influence the PeBoW complex (Fig. 2B, lanes 4 and 5).
To verify that recombinant Bop1 is incorporated into the PeBoW complex, we performed native gel electrophoresis after 6 days of Bop1 overexpression (Fig. 2C). Recombinant Bop1, which is only recognized by the HA-specific but not the human-specific Bop1 MAb, replaced the endogenous Bop1 in PeBoW complex assembly (Fig. 2C, lanes 2 and 4).
As overexpression of Bop1 titrates endogenous WDR12 into a Bop1/WDR12 subcomplex without Pes1, we tested whether additional coexpression of Pes1 restores PeBoW complex formation. U2OS cells were stably transfected with two or three individual pRTS constructs. We performed native gel electrophoresis after 6 days of coexpression. The immunoblot of WDR12 shows that monomeric WDR12 (Fig. 2D, lane 1) builds a subcomplex with coexpressed Bop1 (lane 2) but not with coexpressed Pes1 (lane 3). The recombinant expression of all three PeBoW subunits is sufficient for the establishment of the PeBoW complex (Fig. 2D, lane 4). Equal expression levels were determined by Western blot analysis (Fig. 2D).
Overexpression of Bop1 and Pes1 induces a Bop1/Pes1 subcomplex.
Overexpression of Bop1 did not reveal a detectable subcomplex with the endogenous Pes1, possibly because of its low stability. To test whether such a complex can generally be formed, we overexpressed Bop1 and Pes1 together. Cell fractionation experiments revealed a Bop1/Pes1 subcomplex in the nuclear fraction but not in the cytoplasmic fraction (Fig. 2E). In contrast, the Bop1/WDR12 subcomplex appeared only in the cytoplasmic fraction and was absent from the nuclear fraction.
Knockdown of Pes1 induces a Bop1/WDR12 subcomplex.
Overexpression of Bop1 resulted in the formation of an incomplete PeBoW complex containing Bop1 and WDR12 but not Pes1. Therefore, we aimed to investigate whether this Bop1/WDR12 subcomplex would also appear in cells depleted of Pes1. We performed endogenous Pes1, Bop1, and WDR12 siRNA knockdown experiments. U2OS cells were transfected two times with siRNAs directed against the UTR or open reading frame of WDR12, Bop1, or Pes1 mRNA. All siRNAs induced a strong reduction of the respective proteins within 2 days after the last transfection (Fig. 3A). We also tested whether knockdown of Pes1, Bop1, or WDR12 interfered with rRNA processing. Metabolic labeling of nascent rRNA revealed that cells depleted of a single PeBoW component failed to produce mature 28S rRNA (Fig. 3B).
FIG. 3.
Bop1 is the core factor of the PeBoW complex. (A) U2OS cells were transfected twice with the indicated siRNAs. Endogenous protein levels were analyzed by Western blotting 2 days after the last transfection. α-Tubulin is shown as a loading control. (B) Cells were treated as described for panel A and metabolically labeled with [32P]orthophosphate for 60 min 1 day after the last transfection. Subsequently, cells were incubated for 2 h in regular culture medium. Labeled rRNAs are indicated. Ethidium bromide staining is shown as a loading control. (C) Total cell lysates of U2OS cells transfected twice with the indicated siRNAs were separated by native gel electrophoresis 1 day after the last transfection. Endogenous Pes1, Bop1, and WDR12 were visualized by immunoblotting. Nonincorporated proteins or complexes containing the respective factor are indicated. (D) U2OS cells were transfected at days 0 and 1 with either control or Bop1-specific siRNA. Total cell lysates were subjected to immunoprecipitation with antibodies either against Pes1 (8E9), Bop1 (6H11), WDR12 (1B8), or the isotype control coupled to protein G-Sepharose beads 2 days after the last transfection. Equivalent amounts of total lysates used for immunoprecipitation (IP) were loaded in each lane or 20% thereof for the input. Immunodetection was performed with the anti-Pes1 (8E9) or anti-WDR12 (1B8) antibody, respectively. An asterisk indicates cross-reactivity to immunoglobulin molecules.
To study the integrity of the PeBoW complex in cells depleted of its individual components, we performed native gel electrophoresis 1 day after the last siRNA transfection (Fig. 3C). Immunoblot analysis of Pes1, Bop1, and WDR12 showed that knockdown of either protein led to the disappearance of an intact PeBoW complex (Fig. 3C, lanes 3 to 5, 8 to 10, and 13 to 15). Bop1- or Pes1-depleted cells accumulated free nonincorporated WDR12 (Fig. 3C, lanes 3 and 4), whereas knockdown of WDR12 and Bop1 led to an accumulation of monomeric Pes1 (lanes 13 and 14). We were not able to detect nonincorporated Bop1 in this assay (Fig. 3B, lanes 6 to 10), possibly because of the low stability of free Bop1 (see below). Interestingly, Pes1-depleted cells exhibited a subcomplex of WDR12 and Bop1 (Fig. 3C, lanes 5, 10, and 15), as observed in Bop1-overexpressing cells (Fig. 2B).
Bop1 mediates the interaction of Pes1 and WDR12.
WDR12 and Bop1, as well as Pes1 and Bop1, can form a subcomplexes, but we failed to detect a Pes1/WDR12 subcomplex. Therefore, we studied the interaction of Pes1, Bop1, and WDR12 in more detail. U2OS cells were transfected two times with either control or Bop1-specific siRNA, and coimmunoprecipitation experiments were performed (Fig. 3D). In control cells, WDR12 and Pes1 were specifically immunoprecipitated and also coimmunoprecipitated (Fig. 3D, lanes 3 to 5). In contrast, WDR12 and Pes1 could not be coimmunoprecipitated in Bop1-depleted cells (Fig. 3D, lanes 8 and 9). Therefore, we suppose that the interaction of Pes1 and WDR12 is indirect and mediated by the core factor Bop1.
Interdependent stability of PeBoW components.
Native gel electrophoresis revealed that Pes1-, Bop1-, or WDR12-depleted cells lack intact PeBoW complexes. Therefore, we analyzed whether depletion of single PeBoW components affected the abundance of the other components. H1299 cells were transfected twice with siRNA against the 3′ UTR of the Pes1, Bop1, or WDR12 mRNA, and cells were harvested daily. The strongest reduction of the respective proteins was observed 3 days after the last siRNA transfection (Fig. 4A). Knockdown of WDR12 slightly reduced the abundance of Bop1 but not of Pes1. In contrast, depletion of Pes1 resulted in a concomitant loss of Bop1 and WDR12. Furthermore, the knockdown of Bop1 strongly reduced the protein levels of Pes1 and WDR12. The abundances of three other nucleolar proteins, Nog1, nucleostemin, and nucleophosmin (NPM1), were not affected. In addition to their nucleolar localization, Nog1 and NPM1 were found in preribosomal complexes and shown to function in the processing of the 28S rRNA (15, 25, 29). These results indicate that the interdependence of the steady-state levels of individual PeBoW components is specific for and restricted to this subcomplex (Fig. 4B).
FIG. 4.
Specific interdependency of the PeBoW components. (A) H1299 cells were transfected two times with the indicated siRNAs. Effects on the Pes1, Bop1, and WDR12 protein levels were monitored at day 3 after the last transfection by Western blot analysis. (B) A schematic overview of the interdependency of the PeBoW components Pes1, Bop1, and WDR12 is shown. The number of arrows indicates the extent of downregulation.
Knockdown-knock-in experiments reconstitute steady-state levels of PeBoW proteins.
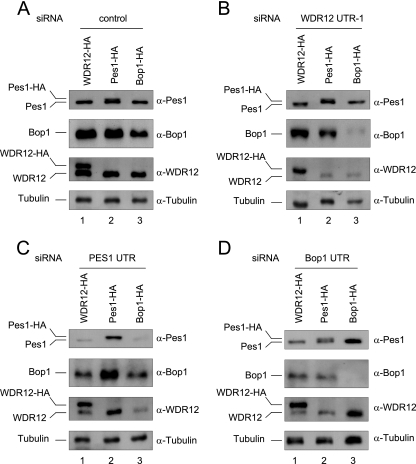
To verify that the mutual reduction of PeBoW proteins after knockdown of a single component is not an off-target effect, we aimed to rescue the depletion phenotype specifically by coexpression of the respective wild-type protein by a knockdown-knock-in approach. H1299 cells were stably transfected with pRTS constructs conditionally expressing the respective HA-tagged form of Pes1, Bop1, or WDR12 in a doxycycline-dependent manner. Exogenous gene expression was activated for 3 days and then maintained throughout the subsequent course of two siRNA transfections. We used siRNAs directed against the 3′ UTRs of the Pes1, Bop1, and WDR12 mRNAs. These siRNAs were used to specifically deplete the endogenous, but not the HA-tagged, recombinant proteins.
The endogenous Pes1, Bop1, and WDR12 protein levels were studied in cells depleted of the endogenous protein and additionally expressing either of the HA-tagged PeBoW components (Fig. 5). The endogenous, as well as the exogenous HA-tagged, Pes1 and WDR12 proteins were detected by the MAbs but could be discriminated by their increased molecular weights and slower migration due to the C-terminal HA tag. The MAb specific for human Bop1 did not recognize recombinant mouse Bop1-HA. First of all, overexpression of Bop1-HA down-regulated the endogenous Bop1, as described above (Fig. 5A to D, lanes 3), consistent with our previous results (Fig. 2A). Knockdown of WDR12 slightly reduced endogenous Bop1 protein levels but not endogenous Pes1 protein levels (Fig. 5B). This reduction of Bop1 was rescued upon coexpression of WDR12-HA (Fig. 5B, lane 1), but not Pes1-HA or Bop1-HA (lanes 2 and 3). Depletion of endogenous Pes1 caused a concomitant loss of Bop1 and WDR12 (Fig. 5C) that was alleviated by expression of Pes1-HA (Fig. 5C, lane 2), but not WDR12-HA or Bop1-HA (lanes 1 and 3).
FIG. 5.
Knockdown-knock-in assay of stable polyclonal H1299 cell lines expressing Pes1-HA, Bop1-HA, or WDR12-HA. Cells were treated with (A) control or (B) WDR12 (3′ UTR)-, (C) Pes1 (3′ UTR)-, or (D) Bop1 (3′ UTR)-specific siRNA. Endogenous Pes1, Bop1, and WDR12 protein levels were analyzed 2 days after the last transfection by Western blotting. Human-specific anti-Bop1 MAb 6H11 does not recognize recombinant mouse Bop1. α-Tubulin is shown as a loading control.
In addition, the knockdown of Bop1 reduced the Pes1 and WDR12 protein levels (Fig. 5D). The expression of Bop1-HA (Fig. 5D, lane 3) but not WDR12-HA or Pes1-HA (lanes 1 and 2) fully prevented this reduction. Thus, these reconstitution experiments verify that the Pes1, Bop1, and WDR12 protein levels reciprocally depend on each other after experimentally induced knockdown of any one of them.
Interdependence of PeBoW protein levels is transcription independent.
In parallel with the Western blot analysis, mRNA levels of Pes1-, Bop1-, or WDR12-depleted cells were investigated by Northern blot analysis (Fig. 6A and B). The knockdown of Pes1, Bop1, or WDR12 strongly reduced the mRNA levels of the respective genes but did not reduce the mRNA levels of other PeBoW components. We even observed a slight increase in mRNA levels. Therefore, the interdependency of the PeBoW proteins is not caused by regulation of the abundance of their mRNA but suggests a translational or posttranslational mechanism.
FIG. 6.
Knockdown of Pes1, Bop1, or WDR12 does not influence the steady-state mRNA levels of the PeBoW components. (A) U2OS cells were transfected twice with the indicated siRNAs. Levels of the mRNAs of Pes1, Bop1, and WDR12 were analyzed 2 days after the last transfection by Northern blot analysis. As a loading control, blots were incubated with a probe specific for 18S rRNA. (B) The histogram depicts quantification of the mRNA levels of Pes1, Bop1, and WDR12 relative to that of a cell line transfected with control siRNA. Error bars indicate standard deviations.
Interdependence of PeBoW proteins is caused by protein destabilization.
These results prompted us to study whether the stability of the PeBoW proteins is reduced in cells depleted of the other PeBoW components. First we analyzed the stability of endogenous PeBoW proteins in untreated cells. Therefore, U2OS cells were incubated with cycloheximide for different time intervals to block de novo protein synthesis (Fig. 7A). The levels of the Pes1, Bop1, and WDR12 proteins were barely affected over 24 h. In contrast, short-lived proteins such as p53 and c-Myc rapidly decreased within a few hours. Hence, endogenous PeBoW proteins are quite stable.
FIG. 7.
Knockdown of Pes1, Bop1, or WDR12 influences the stability of the PeBoW components. (A) U2OS cells were treated with 50 μg/ml cycloheximide (CHX) for the indicated times. The stability of the indicated endogenous proteins was monitored by Western blot analysis. Immunodetection was performed with antibodies against Pes1, Bop1, WDR12, p53 (PAb240), and c-Myc (N-262). Equal loading was verified by immunodetection of α-tubulin. (B to D) H1299 cells stably transfected with the indicated constructs were treated two times with control or WDR12 (3′ UTR)-, Pes1 (3′ UTR)-, or Bop1 (3′ UTR)-specific siRNA. Expression of HA-tagged proteins was induced 2 days after the last siRNA transfection for 6 h by addition of 0.1 μg/ml doxycycline and stopped by its removal. The stability of Pes1-HA, Bop1-HA, and WDR12-HA was analyzed by Western blotting with anti-HA antibody 3F10. α-Tubulin is shown as a loading control.
Next, we aimed to assess whether the turnover of PeBoW proteins is increased in Pes1, Bop1, or WDR12 knockdown cells. Therefore, we monitored the abundance of HA-tagged PeBoW proteins at different time points after a pulse expression in cells depleted of the endogenous PeBoW members. H1299 cells conditionally expressing the respective HA-tagged forms were treated with WDR12 (3′ UTR)-, Pes1 (3′ UTR)-, or Bop1 (3′ UTR)-specific siRNA. Two days after the last siRNA transfection, pulse expression of HA-tagged proteins was performed for 6 h by addition and subsequent removal of doxycycline. The stability of Pes1-HA, Bop1-HA, and WDR12-HA was analyzed by Western blotting with the anti-HA antibody. In control siRNA-treated cells, WDR12-HA and Pes1-HA exhibited only a little turnover within 24 h, consistent with the cycloheximide time course experiments (Fig. 7B and C, lanes 1 to 4). The decline in Pes1 was more pronounced between 24 and 48 h after the pulse expression. The levels of Bop1-HA already decreased substantially within 24 h (Fig. 7D, lanes 1 to 4). This apparent discrepancy between the pulse-chase and the cycloheximide time course experiment suggests that Bop1 is only stable in the context of the PeBoW complex but unstable as a free protein. These results support the observations that the amounts of Bop1 are tightly controlled in mammalian cells (Fig. 2A).
The stability of Bop1-HA was further diminished in Pes1-depleted cells (Fig. 7D, lanes 9 to 12) but not in WDR12-depleted cells (lanes 5 to 8). WDR12-HA protein levels were significantly reduced within 24 h in cells lacking endogenous Bop1 (Fig. 7B, lanes 5 to 8) or Pes1 (lanes 9 to 12), and the stability of Pes1 was dependent on the presence of Bop1 (Fig. 7C, lanes 9 to 12) but not WDR12 (lanes 5 to 8). All of these results are fully in line with the mutual dependency of endogenous PeBoW protein levels observed after siRNA knockdown (Fig. 4). In conclusion, the abundance and stability of Pes1, Bop1, and WDR12 are interdependent.
Nucleolar localization of Bop1 requires Pes1.
Bop1 overexpression induces subcomplexes of Bop1/WDR12 and Bop1/Pes1. To study the subcellular distribution of these complexes, we preformed indirect immunofluorescence assays with Bop1-specific antibodies. Recombinantly expressed Pes1 and WDR12 proteins show predominant nucleolar staining (10, 14). In contrast, recombinant Bop1 localized exclusively to the cytoplasm in H1299 and TGR-1 cells (Fig. 8A and B). Only cells expressing minor levels of Bop1 also revealed nucleolar staining (data not shown). The cytoplasmic localization of Bop1 was not affected by coexpression of WDR12. However, coexpression of Pes1 fostered the nucleolar localization of Bop1. These results are in line with the cell fractionation experiments in Fig. 2E and further suggest that the titration of WDR12 into the Bop1/WDR12 subcomplex retains WDR12 in the cytoplasm.
FIG. 8.
Pes1 is required for nucleolar localization of Bop1. Bop1-HA expressed in (A) H1299 cells and (B) TGR-1 cells was detected by indirect immunofluorescence assay. Coexpression of luciferase (control), WDR12, and Pes1 is indicated. Recombinant proteins were induced by doxycycline for 30 h, and cells were fixed with methanol-acetone. Bop1 was stained by guinea pig polyclonal antibodies specific for mouse Bop1 (poly-α-Bop1). Nuclei were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI).
Do subcomplexes of PeBoW bind to preribosomal particles?
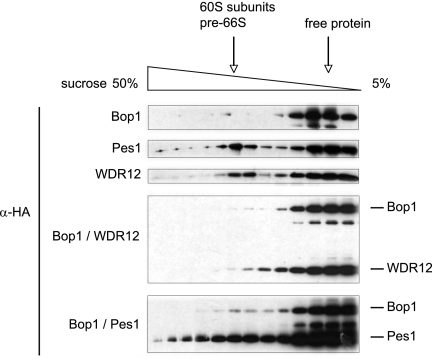
The association of Bop1/WDR12 and Bop1/Pes1 with preribosomal complexes was studied by sucrose gradient centrifugation with TGR-1 total cell lysates (Fig. 9). All recombinant proteins accumulated in low-molecular-weight fractions, most likely because of an access of overexpressed free protein in total lysates. However, recombinant Pes1 and WDR12, but not Bop1, exhibited specific enrichment in high-molecular-weight fractions that cofractionated with ribosomal particles. Importantly, coexpression of Bop1 abolished the association of WDR12 with preribosomal particles, while coexpression of Pes1 led to a significant albeit restricted increase in preribosome-associated Bop1. The results are compatible with the model in which overexpression of Bop1 leads to the formation of two subcomplexes, Bop1/WDR12 and Bop1/Pes1. The former is retained in the cytoplasm and inhibits the translocation of WDR12 to the nucleolus, the later enables the transport of Bop1 to the nucleolus, but a functional PeBoW complex cannot assemble in the absence of WDR12.
FIG. 9.
Association of recombinant PeBoW components with large preribosomal particles after overexpression. Shown is a Western blot analysis of sucrose gradient fractions of TGR-1 cells expressing Bop1-HA, Pes1-HA, and WDR12-HA or coexpressing Bop1-HA/WDR12-HA and Bop1-HA/Pes1-HA. Fractions containing the 60S/pre-66S subunits (28S rRNA) are indicated.
DISCUSSION
Ribosome biogenesis is an essential and evolutionarily highly conserved process. Mature 60S and 40S subunits contain different species of rRNA and ribosomal proteins (8). Numerous trans-acting factors are required along the maturation of ribosomes for processing and modification of rRNA and preribosome assembly (6). So far, it remains elusive how the timely and sterically correct assembly of so many different factors and rRNAs into macromolecular preribosomal complexes is achieved. It is thought that several factors preassemble into subcomplexes that orchestrate the formation and maturation of preribosomes (3). The yeast small-subunit (SSU) processome, the macromolecular correlate of the terminal knob structure of nascent primary rRNA transcripts, was previously shown to consist of distinct subcomplexes (4, 11, 19, 21). Depletion of individual SSU processome components affected ribosome synthesis differently, depending on the complex that they associated with (9). Another subcomplex identified in yeast, containing the factors Nop7p, Erb1p, and Ytm1p, is involved in the maturation of the large ribosomal subunit (11, 19, 21). We have previously characterized the homologous mammalian complex (PeBoW complex), consisting of Pes1, Bop1, and WDR12 (14). The PeBoW complex is critical for mammalian ribosome biogenesis, as it proved to be highly susceptible for the generation of dominant-negative mutants that specifically abrogated the synthesis of the 28S rRNA (10, 14, 20, 28). However, it is unknown how the integrity of the PeBoW complex is preserved. First of all, the amount of each component must be adjusted coordinately to the rate of ribosome biogenesis. In mammalian cells that transit from a quiescent into a proliferating state, this concerted action may be accomplished by the oncogenic transcription factor c-Myc, which upregulates the mRNAs of Pes1, Bop1, and WDR12 in conjunction with many other genes involved in ribosome biogenesis (26). However, this global transcriptional regulation might be insufficient for fine-tuned control of PeBoW protein levels, as the respective mRNAs are relatively long-lived (unpublished results). Therefore, we aimed to investigate how altered abundances of individual PeBoW proteins affect complex integrity. In proliferating cells, Pes1, Bop1, and WDR12 are relatively stable factors, as revealed by cycloheximide treatment blocking de novo protein synthesis. However, their stability is dramatically reduced in cells depleted of a single PeBoW protein. We identified a specific pattern of their destabilization. For example, pulse-chase expression of tagged wild-type WDR12 or Pes1 in Bop1-depleted cells demonstrated that the half-life of both was strongly reduced compared with that of control siRNA-treated cells, but the half-life of Pes1 was not affected in WDR12-depleted cells. Thus, individual PeBoW factors are only stable in the context of an intact PeBoW complex (Fig. 10). These results also explain the observation that the amounts of Pes1, Bop1, and WDR12 are interdependent in a specific manner. Continuously disturbing the stoichiometry of their expression levels within the cell by RNA interference triggers a cascade of reduced stabilization and thus increased degradation. But it is noteworthy that the extent of the mutual destabilization mediated by knockdown through RNA interference significantly differed, depending on the depleted PeBoW component. Loss of WDR12 had no detectable effect on Pes1 expression levels, whereas loss of Bop1 destabilized Pes1 and caused its concomitant reduction. This observation might be explained by our finding that the interaction of Pes1 and WDR12 is indirect and mediated by Bop1, as revealed by coimmunoprecipitation experiments. Similar results were reported in a recent study that investigated the physical interaction of the corresponding yeast homologues by GST pull-down assays. Nop7p, the homologue of Pes1, interacted with Erb1p but not Ytm1p, the homologues of Bop1 and WDR12, respectively (22). The fact that knockdown of Pes1 very strongly codepletes Bop1 suggests that the Pes1-dependent codepletion of WDR12 is mediated by the reduction of Bop1. As loss of WDR12 only slightly affected the abundance of Bop1, the amount of Bop1 is still sufficient to prevent significant destabilization of Pes1. These results indicated a crucial role for Bop1 in PeBoW complex assembly and integrity. This notion was further supported by our studies of cells overexpressing Bop1. Excess amounts of Bop1 impaired cell proliferation and rRNA processing, and this could be rescued by coexpression of WDR12, but not Pes1. It was previously shown that truncation mutant forms of Bop1, but not wild-type Bop1, impaired cell proliferation and rRNA maturation (23, 28). Indeed, we can fully confirm that dominant-negative Bop1Δ potently blocks rRNA processing and cell cycle progression. Also, in our experiments, Bop1Δ elicited a profound cell cycle arrest, in contrast to wild-type Bop1, as revealed by a bromodeoxyuridine-light assay investigating the efficiency of reversible cell cycle arrests (unpublished results). However, the comparison of mock-treated cells with wild-type Bop1- and Bop1Δ-overexpressing cells in long-term proliferation assays revealed an inhibitory effect of Bop1 (1). Similarly, overexpressed Bop1 disturbs rRNA processing, although less severely than Bop1Δ, but significantly compared to that in control cells.
FIG. 10.
Model for transport and assembly of Pes1, Bop1, and WDR12 in the nucleolus. Cytoplasmic Pes1, Bop1, and WDR12 are unstable. Bop1 requires Pes1 for nucleolar transport, while nucleolar transport of WDR12 is blocked by high Bop1 levels. PeBoW complex formation occurs in the nucleolus and stabilizes all three proteins.
We also found that the levels of Bop1 within a cell are tightly controlled. Enforced expression of HA-tagged Bop1 was partially compensated for by a decrease in endogenous Bop1. Pulse-chase-expressed, tagged Bop1 was also less stable than Pes1 or WDR12. Thus, any excess of Bop1 is counteracted by its rapid degradation. Therefore, recombinant Bop1 that largely replaces endogenous Bop1, as the tagged form is also incorporated into the PeBoW complex, provokes the degradation of nonincorporated endogenous Bop1. Further, Bop1 contains a PEST motif that is generally thought to mediate destabilization. Our data suggest that the PEST sequence of Bop1 might render the protein unstable in its free form, but incorporation into the PeBoW complex blocks the function of the PEST motif and protects it from rapid degradation.
In addition, depletion of Pes1 triggered the formation of a Bop1/WDR12 subcomplex. Inadequately high levels of Bop1 likewise sequestered endogenous or recombinant WDR12 into this incomplete PeBoW complex. Apparently, the Bop1/WDR12 subcomplex resulted from a relative deficiency of Pes1, as additionally providing sufficient amounts of Pes1 restored PeBoW formation. Alternatively, overexpression of Bop1 and Pes1 induced the formation of a Bop1/Pes1 subcomplex. Thus, overexpression of Bop1 leads to sequestration of Pes1 and WDR12 in two subcomplexes, each containing Bop1 (Fig. 10). In addition, cell fractionation, indirect immunofluorescence, and sucrose gradient centrifugation experiments demonstrated that the subcomplexes behave differently. While the Bop1/WDR12 subcomplex is retained in the cytoplasm, the Bop1/Pes1 subcomplex is located in the nucleolus (Fig. 10). It is further evident that Bop1 requires the help of Pes1 for translocation into the nucleolus. We could recently show that two domains are essential for the nucleolar transport of Pes1, the Bop1 interaction domain and the BRCT domain (10, 13).
Studies of preribosomal complexes in yeast suggest that the Bop1 and Pes1 homologues Erb1p and Nop7p assemble at the preribosome prior to the WDR12 homologue Ytm1p (2, 22). Overexpressed WDR12 is largely located in the nucleolus (14), suggesting that in mammals nucleolar transport of WDR12 also occurs independently of Pes1 and Bop1 and that the PeBoW complex assembles in the nucleolus. Interdependency of factors has recently also been reported for the activity of the SSU processome in yeast (32).
In the present study, we have identified a crucial role for Bop1 in PeBoW homeostasis. As the gene for Bop1 is frequently amplified in colorectal cancers (18), its deregulation might be involved in carcinogenesis. Overexpression of Bop1, as well as expression of a dominant negative C-terminally truncated Bop protein, affects ribosome biogenesis (28). Whether Bop1 overexpression increases the risk of cancer by impairment of the PeBoW complex or by interaction with other cellular factors like the Cdc14 phosphatase (12), which is important for exit of mitosis, remains unclear and deserves further investigation. We also cannot rule out the possibility that overexpression of Bop1 induces p53 and thereby selects for mutants inactivating p53.
We have conclusively shown that the PeBoW complex is essential for ribosome biogenesis and that its integrity is controlled by its interdependent subunits Pes1, Bop1, and WDR12. The PeBoW complex demonstrates that the stability of free and complex-associated proteins can differ substantially. This complex-specific mutual dependency of protein stability might serve as a control mechanism to accurately adjust expression levels and to ensure correct subcomplex assembly, subsequently required for the proper maturation of a large macromolecule such as the ribosome.
Acknowledgments
We thank Dimitri Pestov for providing HA-Bop1 cDNA.
This work was supported by the Deutsche Forschungsgemeinschaft (EI 216/8-1, SFB684, SFB-Transregio5).
We have no conflict of interest.
Footnotes
Published ahead of print on 12 March 2007.
REFERENCES
- 1.Bornkamm, G. W., C. Berens, C. Kuklik-Roos, J. M. Bechet, G. Laux, J. Bachl, M. Korndoerfer, M. Schlee, M. Hölzel, A. Malamoussi, R. D. Chapman, F. Nimmerjahn, J. Mautner, W. Hillen, H. Bujard, and J. Feuillard. 2005. Stringent doxycycline-dependent control of gene activities using an episomal one-vector system. Nucleic Acids Res. 33:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dez, C., and D. Tollervey. 2004. Ribosome synthesis meets the cell cycle. Curr. Opin. Microbiol. 7:631-637. [DOI] [PubMed] [Google Scholar]
- 3.Dosil, M., and X. R. Bustelo. 2004. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90S pre-ribosomal particle. J. Biol. Chem. 279:37385-37397. [DOI] [PubMed] [Google Scholar]
- 4.Dragon, F., J. E. Gallagher, P. A. Compagnone-Post, B. M. Mitchell, K. A. Porwancher, K. A. Wehner, S. Wormsley, R. E. Settlage, J. Shabanowitz, Y. Osheim, A. L. Beyer, D. F. Hunt, and S. J. Baserga. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichler, D. C., and N. Craig. 1994. Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 49:197-239. [DOI] [PubMed] [Google Scholar]
- 6.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 7.Fontoura, B. M., C. A. Atienza, E. A. Sorokina, T. Morimoto, and R. B. Carroll. 1997. Cytoplasmic p53 polypeptide is associated with ribosomes. Mol. Cell. Biol. 17:3146-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 9.Gallagher, J. E., D. A. Dunbar, S. Granneman, B. M. Mitchell, Y. Osheim, A. L. Beyer, and S. J. Baserga. 2004. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes Dev. 18:2506-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimm, T., M. Hölzel, M. Rohrmoser, T. Harasim, A. Malamoussi, A. Gruber-Eber, E. Kremmer, and D. Eick. 2006. Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res. 34:3030-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harnpicharnchai, P., J. Jakovljevic, E. Horsey, T. Miles, J. Roman, M. Rout, D. Meagher, B. Imai, Y. Guo, C. J. Brame, J. Shabanowitz, D. F. Hunt, and J. L. Woolford, Jr. 2001. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell 8:505-515. [DOI] [PubMed] [Google Scholar]
- 12.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 13.Hölzel, M., T. Grimm, M. Rohrmoser, A. Malamoussi, T. Harasim, A. Gruber-Eber, E. Kremmer, and D. Eick. 2007. The BRCT domain of mammalian Pes1 is crucial for nucleolar localization and rRNA processing. Nucleic Acids Res. 35:789-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hölzel, M., M. Rohrmoser, M. Schlee, T. Grimm, T. Harasim, A. Malamoussi, A. Gruber-Eber, E. Kremmer, W. Hiddemann, G. W. Bornkamm, and D. Eick. 2005. Mammalian WDR12 is a novel member of the Pes1-Bop1 complex and is required for ribosome biogenesis and cell proliferation. J. Cell Biol. 170:367-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen, B. C., Q. Wang, C. T. Kifer, and M. Parsons. 2003. The NOG1 GTP-binding protein is required for biogenesis of the 60S ribosomal subunit. J. Biol. Chem. 278:32204-32211. [DOI] [PubMed] [Google Scholar]
- 16.Jorgensen, P., and M. Tyers. 2004. How cells coordinate growth and division. Curr. Biol. 14:R1014-R1027. [DOI] [PubMed] [Google Scholar]
- 17.Killian, A., N. Le Meur, R. Sesboue, J. Bourguignon, G. Bougeard, J. Gautherot, C. Bastard, T. Frebourg, and J. M. Flaman. 2004. Inactivation of the RRB1-Pescadillo pathway involved in ribosome biogenesis induces chromosomal instability. Oncogene 23:8597-8602. [DOI] [PubMed] [Google Scholar]
- 18.Killian, A., N. Sarafan-Vasseur, R. Sesboue, F. Le Pessot, F. Blanchard, A. Lamy, M. Laurent, J. M. Flaman, and T. Frebourg. 2006. Contribution of the BOP1 gene, located on 8q24, to colorectal tumorigenesis. Genes Chromosomes Cancer 45:874-881. [DOI] [PubMed] [Google Scholar]
- 19.Krogan, N. J., W. T. Peng, G. Cagney, M. D. Robinson, R. Haw, G. Zhong, X. Guo, X. Zhang, V. Canadien, D. P. Richards, B. K. Beattie, A. Lalev, W. Zhang, A. P. Davierwala, S. Mnaimneh, A. Starostine, A. P. Tikuisis, J. Grigull, N. Datta, J. E. Bray, T. R. Hughes, A. Emili, and J. F. Greenblatt. 2004. High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13:225-239. [DOI] [PubMed] [Google Scholar]
- 20.Lapik, Y. R., C. J. Fernandes, L. F. Lau, and D. G. Pestov. 2004. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol. Cell 15:17-29. [DOI] [PubMed] [Google Scholar]
- 21.Lohrum, M. A., R. L. Ludwig, M. H. Kubbutat, M. Hanlon, and K. H. Vousden. 2003. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3:577-587. [DOI] [PubMed] [Google Scholar]
- 22.Miles, T. D., J. Jakovljevic, E. W. Horsey, P. Harnpicharnchai, L. Tang, and J. L. Woolford, Jr. 2005. Ytm1, Nop7, and Erb1 form a complex necessary for maturation of yeast 66S preribosomes. Mol. Cell. Biol. 25:10419-10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pestov, D. G., Z. Strezoska, and L. F. Lau. 2001. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G1/S transition. Mol. Cell. Biol. 21:4246-4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudra, D., and J. R. Warner. 2004. What better measure than ribosome synthesis? Genes Dev. 18:2431-2436. [DOI] [PubMed] [Google Scholar]
- 25.Savkur, R. S., and M. O. Olson. 1998. Preferential cleavage in pre-ribosomal RNA byprotein B23 endoribonuclease. Nucleic Acids Res. 26:4508-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlosser, I., M. Hölzel, M. Murnseer, H. Burtscher, U. H. Weidle, and D. Eick. 2003. A role for c-Myc in the regulation of ribosomal RNA processing. Nucleic Acids Res. 31:6148-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherr, C. J., and J. D. Weber. 2000. The ARF/p53 pathway. Curr. Opin. Genet. Dev. 10:94-99. [DOI] [PubMed] [Google Scholar]
- 28.Strezoska, Z., D. G. Pestov, and L. F. Lau. 2002. Functional inactivation of the mouse nucleolar protein Bop1 inhibits multiple steps in pre-rRNA processing and blocks cell cycle progression. J. Biol. Chem. 277:29617-29625. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi, N., M. Yanagida, S. Fujiyama, T. Hayano, and T. Isobe. 2003. Proteomic snapshot analyses of preribosomal ribonucleoprotein complexes formed at various stages of ribosome biogenesis in yeast and mammalian cells. Mass Spectrom. Rev. 22:287-317. [DOI] [PubMed] [Google Scholar]
- 30.Thiry, M., and D. L. Lafontaine. 2005. Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 15:194-199. [DOI] [PubMed] [Google Scholar]
- 31.Warner, J. R. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 24:437-440. [DOI] [PubMed] [Google Scholar]
- 32.Wehner, K. A., J. E. Gallagher, and S. J. Baserga. 2002. Components of an interdependent unit within the SSU processome regulate and mediate its activity. Mol. Cell. Biol. 22:7258-7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, Y., G. W. Wolf, K. Bhat, A. Jin, T. Allio, W. A. Burkhart, and Y. Xiong. 2003. Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 23:8902-8912. [DOI] [PMC free article] [PubMed] [Google Scholar]