Abstract
Proper regulation of receptor tyrosine kinase (RTK)-Ras-mitogen-activated protein kinase (MAPK) signaling pathways is critical for normal development and the prevention of cancer. SOS is a dual-function guanine nucleotide exchange factor (GEF) that catalyzes exchange on Ras and Rac. Although the physiologic role of SOS and its CDC25 domain in RTK-mediated Ras activation is well established, the in vivo function of its Dbl Rac GEF domain is less clear. We have identified a novel gain-of-function missense mutation in the Dbl domain of Caenorhabditis elegans SOS-1 that promotes epidermal growth factor receptor (EGFR) signaling in vivo. Our data indicate that a major developmental function of the Dbl domain is to inhibit EGF-dependent MAPK activation. The amount of inhibition conferred by the Dbl domain is equal to that of established trans-acting inhibitors of the EGFR pathway, including c-Cbl and RasGAP, and more than that of MAPK phosphatase. In conjunction with molecular modeling, our data suggest that the C. elegans mutation, as well as an equivalent mutation in human SOS1, activates the MAPK pathway by disrupting an autoinhibitory function of the Dbl domain on Ras activation. Our work suggests that functionally similar point mutations in humans could directly contribute to disease.
Receptor tyrosine kinase (RTK)-Ras-mitogen-activated protein kinase (MAPK) signaling pathways play profound roles in development and, when improperly regulated, can contribute to oncogenesis (8). Although most of the core components of RTK-Ras-MAPK pathways were discovered a decade ago, the diverse mechanisms for positive and negative regulation are still being elucidated. Genetic model organisms, such as Caenorhabditis elegans and Drosophila melanogaster, have been invaluable for the study of the regulation of these pathways. In the C. elegans hermaphrodite, a single epidermal growth factor (EGF)-like ligand and a single EGF receptor (EGFR) family member are required for normal development and behavior (61). A canonical EGFR-Grb2-SOS-Ras-Raf-Mek-MAPK pathway is essential for viability past the first larval stage and for development of the vulva, while an EGF-dependent inositol (1,4,5)-trisphosphate (IP3) pathway that is Ras independent controls ovulation behavior. The vulva develops through the induction of vulval cell fates in three out of six progenitor cells. The initiating events are the production of LIN-3 (EGF) from the anchor cell in the somatic gonad and its stimulation of LET-23 (EGFR) on the underlying P6.p progenitor cell. In the presence of sufficient levels of EGFR signaling and a cooperating signal from a Wnt pathway, Notch receptor ligands are upregulated and stimulate LIN-12 (Notch) on the adjacent P5.p and P7.p progenitors (17). Ultimately, eight progeny are produced from P6.p and seven each from P5.p and P7.p to form a mature 22-cell vulva. The remaining progenitor cells, P3.p, P4.p, and P8.p, fuse with an underlying hypodermal syncytium and do not adopt vulval fates.
Vulval development is well suited for studying EGFR-Ras-MAPK pathway regulation, since small deviations in signaling intensity quantifiably affect vulval development. Too little signaling results in fewer than three progenitor cells adopting vulval fates (vulvaless [Vul]), while excessive signaling results in more than three progenitor cells adopting vulval fates (multivulva [Muv]). Using various genetically defined sensitized backgrounds of either too little or too much signaling, a variety of mechanisms have been identified that regulate output or responsiveness to EGFR-Ras-MAPK signaling (61). These mechanisms include cell-autonomous trans-acting factors, such as SLI-1 (c-Cbl) (45, 96), KSR-1 (54, 87), and GAP-1 (Ras GAP) (37); cell-autonomous intramolecular constraints, such as autoinhibitory determinants within the EGFR (50, 62); and non-cell-autonomous pathways involving heterologous signals from surrounding neurons and muscles (60).
Here, we describe the isolation of a novel gain-of-function mutation in the guanine nucleotide exchange factor (GEF) SOS-1. SOS is a multidomain protein (see Fig. 2B) that in vitro catalyzes exchange on Ras through its CDC25 domain (16, 28) and exchange on Rac through its Dbl domain (65). In Drosophila (9, 75), C. elegans (15), and mouse (69, 92), SOS is essential for development. Whereas it is clear that SOS and its CDC25 domain are crucial for RTK-mediated Ras signaling (9, 15, 69, 75, 92) and that perturbation of this function leads to developmental defects (9, 15, 75), it is less clear to what extent its Dbl domain is also required for normal development. Recent studies have demonstrated that a variety of signals, including phosphatidylinositides (65), protein phosphorylation (82), and protein interactions (80), can regulate SOS Rac GEF activity, suggesting the catalytic activity of the Dbl domain may also be important for development. We found that our gain-of-function mutation in sos-1, which strongly affects an EGFR-Ras-MAPK pathway, lies in the Dbl domain rather than the CDC25 domain. Genetic analysis indicated that the activated mutant SOS-1 predominantly signals through Ras, rather than Rac, suggesting that the Dbl domain has an inhibitory function in Ras signaling that is separate from its catalytic activity on Rac proteins. Molecular modeling further suggested that our mutation may disrupt a recently described in vitro autoinhibitory function of the Dbl domain on CDC25 Ras GEF activity (84). We also found that human SOS1 (hSOS1) can be activated by an equivalent mutation. Together, our data demonstrate a crucial role of the Dbl domain in inhibiting EGFR pathway activity in vivo and suggest that analogous mutations in hSOS1 may contribute to disease.
FIG. 2.
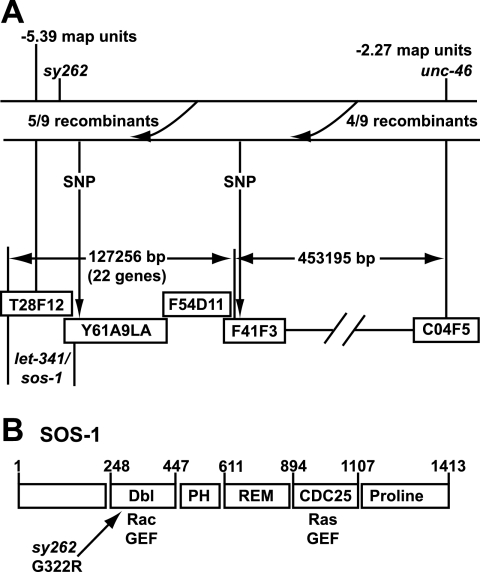
The sy262 mutation maps to the Dbl domain of SOS-1. (A) SNP mapping of sy262. Five out of nine Unc non-sy262 recombinants did not pick up the SNP located at position 9725 of cosmid F41F3, indicating that sy262 is to the left of cosmid F41F3. All the recombinants picked up the SNP at 14897 in the YAC Y61A9LA, which is next to the sos-1 gene. Thus, the sy262 mutation is close to the sos-1 locus. (B) Architecture of the C. elegans SOS-1 protein (GenBank accession number AF251308) and location of the sy262 mutation. The numbers refer to amino acid positions. Domain boundaries were determined by SMART (http://smart.embl-heidelberg.de/), except for the REM domain, whose position was determined by a BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) alignment with hSOS1.
MATERIALS AND METHODS
Strains.
C. elegans was cultured at 20°C (13) unless otherwise indicated. The alleles used were let-23(sy1) and let-23(sy16) (2) and rol-6(e187) (21) on LGII; pha-1(e2123ts) (34) on LGIII; ced-10(n1993) (29), lip-1(zh15) (7), lin-45(sy96) (41), unc-24(e138) (72), lin-3(n378) (30), let-60(n1876) (5), let-60(sy95) (40, 42), let-60(n1531) (5), let-60(sy101) (40, 42), let-60(n1046) (5, 30), dpy-20(e1282) (43), unc-22(s7) (59), and rac-2(ok326) on LGIV; let-341/sos-1(s1031) (44) and unc-46(e177) (13) on LGV; and sli-1(sy143) (45), gap-1(n1691) (37), unc-2(e55) (13), and mig-2(mu28) (98) on LGX.
Isolation, mapping, and molecular identification of the sy262 mutation.
To identify new recessive alleles of the sli-1 locus, an F1 noncomplementation screen was performed. let-23(sy1) males were mutagenized with ethanemethylsulfonate (13) and crossed into marked let-23(sy1); sli-1(sy143) hermaphrodites. The sy262 mutation was discovered as a dominant let-23(sy1) suppressor that was unlinked to the sli-1 locus. Standard genetic approaches assigned linkage to chromosome V. Mapping relative to let-341 (now known to be sos-1) and unc-46 was carried out by crossing let-23(sy1); sy262 males into heterozygous sos-1(s1031) unc-46(e177) hermaphrodites. From let-23(sy1)/+; sos-1 unc-46/sy262 heterozygotes, 12 Unc non-Let recombinants were picked. After homozygosing let-23(sy1) and the recombinant chromosome, it was determined that 12/12 recombinants picked up sy262, indicating sy262 is to the left of unc-46 and close to or to the left of sos-1. Single-nucleotide polymorphism (SNP) mapping (94) was performed by crossing CB4856 Hawaiian C. elegans males into hermaphrodites derived from Bristol, England, carrying let-23(sy1) and the linked sy262 and unc-46 mutations. After homozygosing the let-23 mutation, Unc non-sy262 recombinants were picked. Five out of nine recombinants did not pick up the Hawaiian SNP located at position 9725 of cosmid F41F3, indicating that sy262 is to the left of cosmid F41F3. However, all of the recombinants picked up the SNP in yeast artificial chromosome (YAC) Y61A9LA, which is next to the sos-1 gene. Since we did not find recombinants that crossed over between sy262 and the SNP in Y61A9LA, we speculated that sy262 must be close to the sos-1 locus. Based on these mapping data and our genetic data showing that sy262 suppresses a dominant-negative Ras mutation, but not a Raf reduction-of-function [(rf)] mutation, we speculated that sy262 was a gain-of-function mutation in sos-1. The entire coding regions of the sos-1 cDNAs derived from either wild-type or sy262 mutant worms were sequenced. A single G-to-A transition mutation was found in the sy262 sos-1 cDNA, changing codon 322 from GGA to AGA. The presence of this mutation in sy262 genomic DNA was confirmed by PCR amplifying and sequencing exon 6 from wild-type and sy262 mutant worms.
Vulval induction and gonad ablations.
Vulval development was scored during the L4 stage under Nomarski optics (86). The number of vulval nuclei was used to extrapolate how many of the vulval progenitor cells (VPCs) were induced to adopt vulval fates. A VPC that gave rise to seven or eight great-granddaughters and no hyp7 tissue was scored as 1.0 cell induction. A VPC in which one daughter fused with hyp7 and the other daughter divided to generate three or four great-granddaughter cells was scored as 0.5 cell induction. In wild-type animals, P5.p, P6.p, and P7.p each undergo 1.0 cell induction, whereas the other Pn.p cells do not adopt vulval fates, resulting in a total of 3.0 cell inductions. Animals displaying more than 3.0 cell inductions are Muv, and animals with less than 3.0 cell inductions are Vul. Gonad cells (Z1, Z2, Z3, and Z4) were ablated with a laser microbeam during the L1 stage (4).
RNAi.
Escherichia coli HT115 bacteria containing the respective RNA interference (RNAi) clones were obtained from the Ahringer bacterial feeding library (48). The bacteria were grown overnight in LB with 50 μg/ml ampicillin and then spotted onto NG plates containing 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and 50 μg/ml ampicillin to make RNAi plates. The next day, Po L4 stage worms were seeded onto the plates. After 2 days, the Po worms were transferred to fresh RNAi plates, and progeny were scored 4 to 7 days later.
Plasmid constructions.
A 4,002-bp cDNA encoding isoform 1 of hSOS1 (GenBank accession number L13857) was used in all experiments. A C-terminally FLAG-tagged expression construct was generated by first cloning an N-terminal BamHI/KpnI fragment spanning nucleotides 1 to 3146 into the BglII/KpnI sites of p3XFLAG-CMV-14 (Sigma). A C-terminal fragment spanning nucleotides 3146 to 4000 that replaced the stop codon with an XbaI site was generated by PCR using oligonucleotides hSOS1-1 (5′-AAC TTG AAT CCG ATG GGA AAT AGC-3′) and hSOS1-2 (5′-CTA GCC TAG TCT AGA GGA AGA ATG GGC ATT CTC CAA CAG-3′). This fragment was then ligated to the N-terminal hSOS1 fragment in the FLAG vector via KpnI/XbaI digestion. Site-directed mutagenesis using oligonucleotides hSOS1-3 (5′-C CAT CCA CTA GTA GGA AGC CGC TTT GAA GAC TTA GCA GAG-3′), hSOS1-4 (5′-CTC TGC TAA GTC TTC AAA GCG GCT TCC TAC TAG TGG ATG G-3′), and Accuprime Pfx DNA Polymerase (Invitrogen) was performed on an XhoI hSOS1 subfragment spanning nucleotides 112 to 896 that had been cloned into pBluescript (Stratagene). The mutagenesis changed codon 282 from TGC (Cys) to CGC (Arg). The mutagenized fragment was then used to replace the corresponding region in the wild-type FLAG-tagged construct via XhoI digestion. All constructs were verified by sequencing them.
Transient transfections, growth factor stimulations, and Western blotting.
NIH 3T3 and HEK 293 EBNA cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, penicillin, streptomycin, and glutamine and maintained in a 5% CO2 incubator. The NIH 3T3 cells were plated at 5 × 105 cells/60-mm dish 12 to 24 h prior to transfection. For NIH 3T3 cell transfection, 25 μl of Lipofectamine (Invitrogen) was combined with 300 μl DMEM and 8 μg total DNA and incubated for 30 min at room temperature. Complexes were added to the cells in the presence of 2.4 ml DMEM for 5 hours. The transfected cells were washed once with DMEM or phosphate-buffered saline (PBS) and then starved for an additional 15 to 20 h in serum-free DMEM. HEK 293 EBNA cells were plated at 1.2 × 106 cells/60-mm dish 12 to 24 h prior to transfection. For HEK 293 EBNA cell transfection, 30 μl polyethylenimine (1 mg/ml) was combined with 1.5 ml DMEM and 6.5 μg total DNA and incubated for 15 to 30 min at room temperature. Complexes were added to the cells in a total of 4 ml of DMEM and incubated for 16 h at 37°C. The cells were recovered in complete medium for 8 hours before being starved in serum-free DMEM for 18 to 24 h.
Transfected cells were stimulated with 10 ng/ml human EGF (BD Biosciences) and lysed in NP-40 lysis buffer (1% Nonidet P40, 150 mM NaCl, 50 mM Tris-Cl [pH 8.0]) containing 1 μg/ml antipain, 1 μg/ml aprotinin, 10 μg/ml leupeptin, 1 μg/ml pepstatin A, 20 μg/ml phenylmethylsulfonyl fluoride, 20 mM NaF, 2 mM sodium orthovanadate, and 20 mM beta-glycerophosphate. Protein concentrations were determined by the bicinchoninic acid assay (Pierce).
Following sodium dodecyl sulfate-polyacrylamide gel electrophoresis, proteins were transferred to Immobilon-P membranes (Millipore) in transfer buffer (50 mM Tris-base, 40 mM glycine, 0.04% sodium dodecyl sulfate, 10% methanol) using a semidry transfer apparatus (Owl). The blots were blocked in 5% nonfat dry milk-TBST (50 mM Tris-Cl, 150 mM NaCl, 0.05% Tween-20 [pH 8.0]) and probed with anti-phospho-p44/42 MAPK (Cell Signaling, no. 9101; 1:1,000), anti-FLAG (Sigma, no. F1804; 1:2,000), or anti-p44/42 MAPK (Cell Signaling, no. 9102; 1:2,000). The blots were developed using ECL Plus (Amersham), and the intensities of the bands were determined by densitometric scanning, followed by quantification using ImageJ software (NIH).
SWISS-MODEL.
The three-dimensional structure of the C. elegans SOS-1 Dbl-PH-REM-CDC25 domains was modeled using the crystal structure of the Dbl-PH-REM-CDC25 domains of hSOS1 (crystal structure coordinates 1xd4A.pdb) and the optimize mode of SWISS-MODEL (36, 67, 79), an Internet-based automated comparative protein-modeling server (http://www.expasy.ch/swissmod/SWISS-MODEL.html). In the optimize mode, the SOS-1 protein sequence was aligned with that of hSOS1 using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/).
RESULTS
The sy262 mutation acts at the level of Ras and upstream of Raf.
The sy262 mutation was discovered in a sli-1 (c-Cbl) F1 noncomplementation screen (see Materials and Methods) as a novel mutation that was unlinked to the sli-1 locus but could still strongly suppress a let-23 (EGFR) reduction-of-function mutation (Fig. 1 and Table 1). To determine how the sy262 mutation interacts with the EGFR pathway, we performed a genetic epistasis analysis using standard reduction-of-function mutations in the EGFR-Ras-MAPK pathway (Fig. 1 and Table 1). By itself, the sy262 mutation does not alter vulval development. However, the sy262 mutation strongly suppresses a reduction-of-function mutation in lin-3 (EGF), which likely affects EGF processing, and a mutation in let-23 (EGFR), which affects EGFR localization (47, 56). The sy262 mutation also suppresses a weak dominant-negative S89F mutation in let-60 (Ras) but does not suppress stronger let-60 (Ras) dominant-negative mutations (G10R and G15D) or a relatively weak reduction-of-function mutation in lin-45 (Raf) (40-42). These data suggest that the sy262 locus normally acts upstream of Raf and possibly on Ras.
FIG. 1.
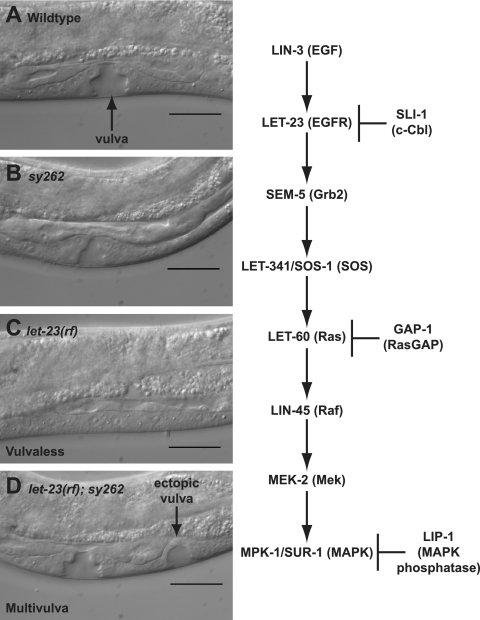
The sy262 mutation suppresses the Vul phenotype of a reduction-of-function mutation in let-23 (EGFR). The animals were photographed during the mid-L4 larval stage using Nomarski optics. (A) Wild type. (B) Homozygous sy262 mutant. (C) Homozygous let-23(sy1) reduction-of-function mutant. (D) Homozygous let-23(sy1); sy262 double mutant. Scale bars = 20 μm. On the right, the names of the C. elegans and human (in parentheses) components of the EGFR pathway are indicated.
TABLE 1.
The sy262 mutation acts at the level of Ras and upstream of Raf
| Genotypea | Vulval inductionb | % Muvc | % Vuld | ne | P valuef |
|---|---|---|---|---|---|
| Wild type | 3.00 | 0 | 0 | 24 | |
| sy262 | 3.00 | 0 | 0 | 20 | |
| lin-3(rf) | 1.40 | 0 | 95 | 21 | |
| lin-3(rf); sy262 | 2.93 | 0 | 10 | 20 | <0.00001 to lin-3(rf) |
| let-23(rf) | 0.23 | 0 | 100 | 20 | |
| let-23(rf); sy262 | 3.80 | 68 | 0 | 22 | <0.00001 to let-23(rf) |
| let-23(rf); sy262/+ | 2.95 | 35 | 20 | 20 | <0.00001 to let-23(rf) |
| let-60(S89Fdn)/+ | 1.55 | 0 | 81 | 21 | |
| let-60(S89Fdn)/+; sy262 | 2.80 | 0 | 18 | 22 | 0.0001 to let-60(S89Fdn)/+ |
| let-60(G10Rdn)/+ | 0.25 | 0 | 100 | 20 | |
| let-60(G10Rdn)/+; sy262 | 0.40 | 0 | 100 | 20 | 0.42 to let-60(G10Rdn)/+ |
| let-60(G15Ddn)/+ | 0.08 | 0 | 100 | 20 | |
| let-60(G15Ddn)/+; sy262 | 0.04 | 0 | 100 | 27 | 0.64 to let-60(G15Ddn)/+ |
| lin-45(rf) | 2.10 | 0 | 70 | 20 | |
| lin-45(rf); sy262 | 2.14 | 0 | 67 | 21 | 0.88 to lin-45(rf) |
The complete genotypes are as follows: (i) sy262 = sy262 him-5(e1490), (ii) lin-3(rf) = lin-3(n378); unc-46(e177), (iii) lin-3(rf); sy262 = lin-3(n378); sy262 unc-46(e177), (iv) let-23(rf) = let-23(sy1), (v) let-23(rf); sy262 = let-23(sy1); sy262 him-5(e1490), (vi) let-23(rf); sy262/+ = let-23(sy1); sy262 unc-46(e177)/+, (vii) let-60(S89Fdn)/+ = unc-24(e138) let-60(sy95)/dpy-20(e1282), (viii) let-60(S89Fdn)/+; sy262 = unc-24(e138) let-60(sy95)/dpy-20(e1282); sy262 unc-46(e177), (ix) let-60(G10Rdn)/+ = let-60(sy101) dpy-20(e1282)/unc-24(e138) unc-22(s7); unc-46(e177), (x) let-60(G10Rdn)/+; sy262 = let-60(sy101) dpy-20(e1282)/unc-24(e138) unc-22(s7); sy262 unc-46(e177), (xi) let-60(G15Ddn)/+ = let-60(n1531) unc-22(s7)/dpy-20(e1282); unc-46(e177), (xii) let-60(G15Ddn)/+; sy262 = let-60(n1531) unc-22(s7)/dpy-20(e1282); sy262 unc-46(e177), (xiii) lin-45(rf) = unc-24(e138) lin-45(sy96), and (xiv) lin-45(rf); sy262 = unc-24(e138) lin-45(sy96); sy262 unc-46(e177).
Average number of vulval progenitor cells adopting vulval fates. Wild type is three. The maximum is six.
The percentage of animals that have more than three vulval progenitor cells adopting vulval fates.
The percentage of animals that have fewer than three vulval progenitor cells adopting vulval fates.
n, number of animals examined.
Statistical significance of the vulval induction value as determined by a two-tailed Student's t test.
The sy262 mutation maps to the Dbl domain of the Ras/Rac GEF SOS-1.
We used three-factor mapping to place sy262 to the left of unc-46 and close to or to the left of let-341 (now known to be sos-1) (see Materials and Methods) on chromosome V. We then used SNPs to determine that sy262 was to the left of cosmid F41F3 and close to the YAC Y61A9LA, which includes the sos-1 locus (Fig. 2A). There are 22 predicted genes between the SNP in F41F3 and sos-1. Given our genetic epistasis results showing that sy262 acts at the level of Ras and upstream of Raf, we hypothesized that the sy262 mutation was a gain-of-function mutation in sos-1. After sequencing the entire coding region of the sos-1 cDNA derived from sy262 mutant animals, we found a single G-to-A mutation in codon 322 that results in a substitution of Arg for Gly in the Dbl domain (Fig. 2B). The presence of the mutation was also confirmed in sy262 genomic DNA.
The sy262 (G322R) mutation predominantly affects Ras signaling.
SOS-1 and its Drosophila and mammalian orthologs are multidomain proteins best characterized for their critical biological roles as Ras GEFs (Fig. 2B) (9, 15, 16, 28, 69, 92). However, in addition to containing a CDC25 Ras GEF domain, SOS-1 also possesses a Dbl domain, which allows the mammalian protein to function as a Rac GEF (65). The location of the sy262 mutation in the Dbl domain suggests at least two models for the way in which the change may create a gain-of-function protein. In one model, the sy262 mutation may relieve one of several previously described forms of inhibition on SOS Rac GEF activity (23, 65, 80-83). For example, the sy262 mutation might disrupt autoinhibition by the PH domain on the Rac GEF activity of the Dbl domain (23, 65, 83). In this model, elevated Rac signaling might bypass some of the requirement for Ras signaling during vulval development. In the second model, the sy262 mutation might relieve inhibition of Ras GEF activity. The second model is particularly appealing, since recent X-ray crystallographic studies of hSOS1 have uncovered an autoinhibitory function of the Dbl domain on CDC25 Ras GEF activity (84).
To distinguish between these models, we examined the sensitivity of sy262 mutant SOS-1 to further mutations in either Ras or Rac (Table 2). For these experiments, we used a background in which the sy262 mutation suppressed a reduction-of-function mutation in let-23 (EGFR). When we additionally lowered Ras levels through a heterozygous strong reduction-of-function mutation in let-60 (Ras), sy262 activity was strongly reduced to only 29% of that seen in the presence of two copies of wild-type Ras. Thus, the activity of SOS-1 G322R is critically dependent on the amount of Ras. This observation is also consistent with the failure of sy262 to suppress strong dominant-negative mutations in Ras (Table 1). C. elegans has three Rac genes: rac-2 (57), ced-10 (71), and mig-2 (98). Although nonnull mutations in let-60 (Ras) by themselves can reduce vulval induction (5, 40) (Table 1), single null mutations in either rac-2 or mig-2 or a single strong reduction-of-function mutation in ced-10 has no effect on vulval development (Table 2). Thus, unlike the requirement for Ras, vulval development is not normally dependent on any single Rac gene. Furthermore, in the sensitized background of sy262 suppression of the let-23 (EGFR) reduction-of-function mutation, rac-2 and ced-10 mutations still have no effect on vulval development. However, a null mutation in mig-2 partially reduced sy262-suppressing activity to 73% of that seen in the presence of MIG-2. The effect of the mig-2 mutation was not enhanced by additional reduction of rac-2 and ced-10 through RNAi. Given that a partial reduction of Ras levels, which by itself is not even sufficient to weaken Ras activity in its known biological pathways (viability, vulval development, and fertility), has a more profound effect on sy262 activity than null mutations and reductions in function of the three Rac genes (1.25 cells induced versus 2.68 cells induced, respectively), it is likely that SOS-1 G322R exerts most of its effect through the Ras pathway (see Discussion).
TABLE 2.
The sy262 mutation predominantly acts through Ras rather than Rac
| Genotypea | Vulval inductionb | % Muvc | % Vuld | ne | P valuef |
|---|---|---|---|---|---|
| Wild type | 3.00 | 0 | 0 | 24 | |
| sy262 | 3.00 | 0 | 0 | 20 | |
| let-23(rf) | 0.23 | 0 | 100 | 20 | |
| let-23(rf); sy262 | 3.80 | 68 | 0 | 22 | |
| let-60(rf)/+ | 3.00 | 0 | 0 | 20 | |
| let-23(rf); let-60(rf)/+; sy262 | 1.25 | 0 | 80 | 22 | <0.00001 to let-23(rf); sy262 |
| rac-2(null) | 3.00 | 0 | 0 | 20 | |
| let-23(rf); rac-2(null); sy262 | 3.65 | 60 | 0 | 20 | 0.50 to let-23(rf); sy262 |
| ced-10(rf) | 3.00 | 0 | 0 | 20 | |
| let-23(rf); ced-10(rf); sy262 | 3.90 | 70 | 5 | 20 | 0.68 to let-23(rf); sy262 |
| mig-2(null) | 3.00 | 0 | 0 | 20 | |
| let-23(rf); sy262; mig-2(null) | 2.85 | 25 | 25 | 20 | 0.002 to let-23(rf); sy262 |
| let-23(rf); rac-2(RNAi); sy262; mig-2(null) | 2.83 | 30 | 30 | 18 | 0.97 to let-23(rf); sy262; mig-2(null) |
| let-23(rf); ced-10(RNAi); sy262; mig-2(null) | 2.55 | 21 | 47 | 19 | 0.45 to let-23(rf); sy262; mig-2(null) |
| let-23(rf); ced10(RNAi); rac-2(RNAi); sy262; mig-2(null) | 2.68 | 30 | 40 | 20 | 0.64 to let-23(rf); sy262; mig-2(null) |
let-23(rf) = let-23(sy1), let-60(rf)/+ = let-60(n1876) unc-22(s7)/dpy-20(e1282), rac-2(null) = rac-2(ok326), ced-10(rf) = ced-10(n1993), and mig-2(null) = mig-2(mu28). In the mig-2(null) strains, sy262 was linked to unc-46(e177). In all other strains, sy262 was linked to him-5(e1490). By themselves, the unc-46 and him-5 mutations do not affect vulval development in let-23(rf) animals.
Average number of vulval progenitor cells adopting vulval fates. Wild type is three. The maximum is six.
The percentage of animals that have more than three vulval progenitor cells adopting vulval fates.
The percentage of animals that have fewer than three vulval progenitor cells adopting vulval fates.
n, number of animals examined.
Statistical significance of the vulval induction value as determined by a two-tailed Student's t test.
The Dbl domain mutation increases the sensitivity of the Ras pathway to upstream signaling.
Although a precise mechanism has not yet been defined, it has been suggested that SOS catalytic activity toward the Ras-MAPK pathway might be positively regulated by growth factor signaling (14, 70). To determine if the G322R change increases SOS-1 basal activity and converts it into a constitutively active GEF, we examined whether the gain-of-function phenotype was still manifested in the complete absence of upstream signaling from EGF and the EGFR. We first tested whether the sy262 mutation could still suppress the Vul phenotype of a let-23 (EGFR) reduction-of-function mutation in the absence of the EGF-producing anchor cell. In wild-type animals, laser ablation of the gonadal primordium, which gives rise to the anchor cell prior to the onset of vulval induction, results in 100% of the animals having absolutely no vulval induction (Table 3). Although a control G13E gain-of-function change in let-60 (Ras) was still able to promote some vulval induction in this background, the SOS-1 G322R could not promote vulval development in the complete absence of EGF (Table 3). Thus, in this assay, SOS-1 G322R does not appear to be constitutively active.
TABLE 3.
The sy262 mutation is strongly dependent on upstream signaling by EGF
| Genotypea | Gonadb | Vulval Inductionc | % Muvd | % Vule | nf | P valueg |
|---|---|---|---|---|---|---|
| Wild type | + | 3.00 | 0 | 0 | 24 | |
| Wild type | − | 0.00 | 0 | 100 | 14 | |
| let-23(rf); sy262 | + | 3.80 | 68 | 0 | 22 | |
| let-23(rf); sy262 | − | 0.00 | 0 | 100 | 19 | |
| let-23(rf); let-60(gf)/+ | + | 4.58 | 100 | 0 | 20 | |
| let-23(rf); let-60(gf) | + | 5.38 | 100 | 0 | 20 | |
| let-23(rf); let-60(gf) | − | 1.17 | 12 | 88 | 16 | 0.01 to wild type gonad ablated |
let-23(rf) = let-23(sy1), let-60(gf) = let-60(n1046gf), and sy262 was linked to him-5(e1490).
The gonadal primordium, which gives rise to the EGF-producing anchor cell, is either present (+) or removed by laser ablation (−).
Average number of vulval progenitor cells adopting vulval fates. Wild type is three. The maximum is six.
The percentage of animals that have more than three vulval progenitor cells adopting vulval fates.
The percentage of animals that have fewer than three vulval progenitor cells adopting vulval fates.
n, number of animals examined.
Statistical significance of the vulval induction value as determined by a two-tailed Student's t test.
In another assay, we examined the ability of the sy262 mutation to suppress the lethality conferred by a homozygous let-23 (EGFR) null mutation. Due to the 100% penetrant lethality of this mutation, we initially constructed a strain in which the sy262 mutation was homozygous but the let-23 (EGFR) null mutation was maintained in a heterozygous state. The let-23 (EGFR) null mutation was linked to a recessive mutation in the rol-6 gene. Thus, in the absence of suppression, 25% of the progeny die as L1 larvae, and adult rolling animals are never seen. If there is 100% suppression of let-23(null) lethality, 25% of the progeny will appear as adult rolling animals, since this is the expected Mendelian frequency of homozygosing the mutant let-23 (EGFR) chromosome from heterozygous animals. In the presence of a homozygous let-60 (Ras) G13E gain-of-function mutation, 26% of the progeny were adult rollers (Table 4). This frequency indicates that the Ras mutation can suppress 100% of the lethality associated with complete loss of the EGFR. These rollers were 100% sterile, since an EGF-dependent IP3 pathway, rather than Ras, is required for hermaphrodite fertility (19). The Ras mutation was also able to drive excessive vulval induction. Almost 100% of the animals had all six progenitor cells adopting vulval fates, rather than the normal three. In contrast to the gain-of-function Ras mutation, the sy262 mutation resulted in only 1.5% of total progeny from heterozygous mothers appearing as adult rollers (Table 4). This translates into a rescue frequency of only 6%, compared to a rescue frequency of 100% for the G13E Ras mutation. The rescued sy262 mutant animals were not rare recombinants, since all were sterile, and on average, only one progenitor cell was induced to adopt a vulval fate. Failure of the sy262 mutation to rescue the sterility defect of let-23 (EGFR) null animals also provides further evidence that the Ras effector arm of the EGFR pathway is specifically affected by the sy262 mutation. Rescue of the EGFR (null)-induced lethality and defective vulval induction was weak but statistically significant. Together, these data indicate that the G322R change only weakly increases the basal activity of SOS-1. However, since SOS-1 G322R has strong rescuing activity in the presence of nonnull mutations in the EGFR pathway (which allows low levels of signaling) (Table 1), we infer that G322R acts by increasing the responsiveness of SOS-1 to upstream signaling.
TABLE 4.
The sy262 mutation displays very weak activity in the absence of functional EGFRs
| Genotypea | % Suppression of let-23(null)-induced lethalityb | P valuec | % Suppression of let-23(null)-induced sterilityd | Vulval inductionf | % Muvh | % Vuli | nj | P valuek |
|---|---|---|---|---|---|---|---|---|
| let-23(null) | 0 (1,091) | NAe | NAg | NAg | NAg | NAg | ||
| let-23(null); sy262 | 6 (1,372) | <0.001 | 0 (16) | 0.98 | 0 | 91 | 11 | 0.013 |
| let-23(null); let-60(gf) | 100 (324) | <0.0001 | 0 (44) | 5.73 | 100 | 0 | 11 | <0.00001 |
The complete genotype of let-23(null) was rol-6(e187) let-23(sy16); let-60(gf) = let-60(n1046gf); sy262 was linked to him-5(e1490).
Suppression of lethality was calculated from strains that were homozygous for the sy262 or let-60 mutation but heterozygous for the rol-6(e187) let-23(sy16) chromosome. In the absence of suppression, rolling adults never appear. In the presence of complete suppression, 25% of total descendants from the rol-6(e187) let-23(sy16) heterozygous parents would be rolling; 100% suppression is normalized to the generation of 25% rollers relative to total progeny. The number in parentheses refers to the total number of progeny examined from rol-6(e187) let-23(sy16) heterozygous parents.
The statistical significance of the observed frequency of viable animals was determined by comparison with let-23(null) animals alone and using a chi square test.
Animals suppressed for let-23(null)-induced lethality were picked as L4 larvae and examined for progeny 4 days later. The number in parentheses refers to the total number of animals examined.
NA, not applicable. Due to the 100% penetrant lethality conferred by the homozygous let-23(null) mutation, these animals could not be examined for sterility; 100% sterility, however, is inferred from reduction-of-function mutations in lin-3 and LET-23 structure-function studies.
Average number of vulval progenitor cells adopting vulval fates. Wild type is three. The maximum is six.
NA, not applicable. Due to the 100% penetrant lethality conferred by the homozygous let-23(null) mutation, these animals could not be examined for vulval induction. However, based on certain combinations of let-23 alleles, it could be inferred that the vulval induction of these animals would be 0.00 and that 0% of the animals would be Muv.
Percentage of animals with more than three vulval progenitor cells adopting vulval fates.
Percentage of animals with fewer than three vulval progenitor cells adopting vulval fates.
n, number of animals examined for vulval induction.
The statistical significance of the vulval induction value was determined by comparison with gonad-ablated wild-type animals and use of a two-tailed Student's t test.
The SOS-1 Dbl domain is a critical developmental inhibitor of Ras signaling.
Our data suggest that during C. elegans vulval development, a major function of the Dbl domain is to prevent excessive Ras activation by the CDC25 Ras GEF domain. To determine the importance of this negative regulatory mechanism relative to that of established trans-acting inhibitors of the EGFR-Ras-MAPK pathway, we compared sy262 suppression of a reduction-of-function mutation in let-23 (EGFR) to that conferred by loss-of-function mutations in sli-1 (c-Cbl) (96), gap-1 (RasGAP) (37), and lip-1 (MAPK phosphatase) (7). We found that the sy262 mutation was a better suppressor of the let-23 (EGFR) mutation than a likely null mutation in lip-1 (MAPK phosphatase) and equivalent in strength to null mutations in either sli-1 (c-Cbl) or gap-1 (RasGAP) (Table 5). To further determine the importance of the Dbl domain for inhibition of the EGFR-Ras-MAPK signaling pathway, we constructed a double mutant harboring the sy262 mutation and a null mutation in gap-1 (RasGAP). Together, these mutations should increase GTP loading on Ras while reducing its GTPase activity. Individually, these single mutations do not disrupt vulval development. However, 25% of double-mutant animals displayed excessive vulval differentiation (Table 5). Thus, the SOS-1 Dbl domain helps provide a critical balance between the opposing GEF and GAP activities that regulate Ras.
TABLE 5.
Magnitude of inhibition of the SOS-1 Dbl domain, c-Cbl, RasGAP, and MAPK phosphatase on the EGFR pathway
| Genotypea | Vulval inductionb | % Muvc | % Vuld | ne | P valuef |
|---|---|---|---|---|---|
| let-23(rf) | 0.23 | 0 | 100 | 20 | |
| sli-1(null) | 3.00 | 0 | 0 | 24 | |
| let-23(rf); sli-1(null) | 3.57 | 60 | 17 | 30 | <0.00001 to let-23(rf) |
| gap-1(null) | 3.00 | 0 | 0 | 36 | |
| let-23(rf); gap-1(null) | 3.74 | 71 | 0 | 21 | <0.00001 to let-23(rf) |
| lip-1(null) | 3.00 | 0 | 0 | 20 | |
| let-23(rf); lip-1(null) | 2.10 | 0 | 52 | 21 | <0.00001 to let-23(rf) |
| sy262 | 3.00 | 0 | 0 | 24 | |
| let-23(rf); sy262 | 3.80 | 68 | 0 | 22 | 0.34 to let-23(rf); sli-1(null), 0.80 to let-23(rf); gap-1(null), and <0.00001 to let-23(rf); lip-1(null) |
| sy262; gap-1(null) | 3.26 | 26 | 0 | 35 | 0.004 to gap-1(null) and sy262 |
let-23(rf) = let-23(sy1), and sli-1(null) = sli-1(sy143). The complete genotype of gap-1(null) is gap-1(n1691) unc-2(e55). lip-1(null) = lip-1(zh15); sy262 was linked to him-5(e1490).
Average number of vulval progenitor cells adopting vulval fates. Wild type is three. The maximum is six.
Percentage of animals with more than three vulval progenitor cells adopting vulval fates.
Percentage of animals with fewer than three vulval progenitor cells adopting vulval fates.
n, number of animals examined.
The statistical significance for the vulval induction value was determine using a two-tailed Student's t test.
A mutation equivalent to sy262 G322R activates hSOS1.
To further study the molecular mechanism of action of the sy262 G322R mutation and to determine whether hSOS1 might be regulated in a similar manner, we sought to introduce the equivalent activating mutation into the hSOS1 cDNA. We used the solved X-ray crystal structure of the Dbl-PH-REM-CDC25 domains of hSOS1 and SWISS-MODEL modeling software (36, 67, 79) to generate a structural model of C. elegans SOS-1 (Fig. 3). In this model, G322 lies in the H3 helix of the Dbl domain. The equivalent residue in hSOS1 appears to be C282, which is not disulfide bonded in the crystal structure. In both the human and C. elegans proteins, these residues appear to be essential to accommodate the bulky side chains from residues in the nearby H2b helix. It is thus conceivable that the G322R change in C. elegans SOS-1 is incompatible with the normal positioning of the H2b helix and that a similar effect might be obtained by a C282R change in hSOS1.
FIG. 3.
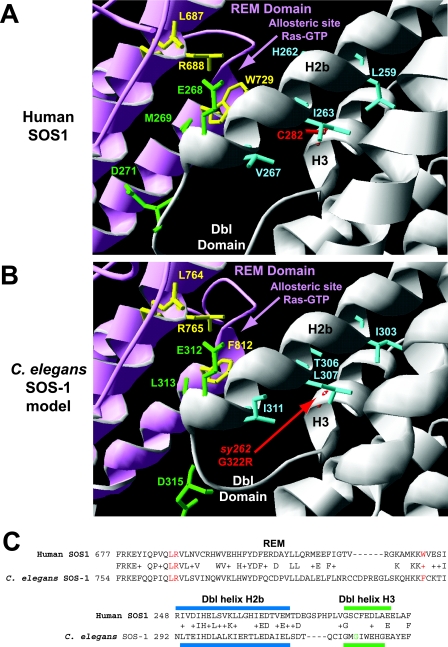
Conservation of the Dbl and REM domains between human and C. elegans SOS proteins. (A) Crystal structure of the Dbl-PH-REM-CDC25 domains from hSOS1 at 3.62 Å (84). The yellow residues (L687, R688, and W729) indicate amino acids in the REM domain that interact with Ras-GTP and are important for Ras-GTP-dependent allosteric stimulation of CDC25 activity. The green residues in the Dbl helix H2b (E268 and M269) and loop (D271) contribute to autoinhibition of the REM domain. Substitution mutations converting all three green amino acids to alanines result in a protein that is hypersensitive to allosteric activation (84). The blue residues (L259, H262, I263, and V267) in helix H2b point toward the surface of helix H3. C282 in helix H3 is in the position analogous to that of the sy262 G322R mutation in the C. elegans Dbl domain. SWISS-MODEL modeling suggested that a C282R change might not be compatible with the normal geometry of the blue residues in helix H2b. (B) SWISS-MODEL of the C. elegans Dbl-PH-REM-CDC25 domains from SOS-1. Note the conservation of the yellow residues (L764, R765, and F812) in the REM domain for allosteric stimulation of CDC25 activity by Ras-GTP, conservation of the green residues (E312, L313, and D315) in the Dbl domain for potential autoinhibition of the REM domain, and conservation of the blue bulky/hydrophobic residues (I303, T306, L307, and I311) that face the surface of Dbl helix H3. SWISS-MODEL modeling suggests that the G322R change in helix H3 may not be compatible with the predicted geometry of the blue residues in helix H2b. (C) BLAST alignments of relevant portions of the REM and Dbl domains between human and C. elegans SOS proteins. The red amino acids in the REM domain are important for binding Ras-GTP. The blue bar indicates the position of Dbl helix H2b (predicted in C. elegans), and the green bar indicates the position of Dbl helix H3 (predicted in C. elegans). The green amino acid is the locations of the sy262 mutation. Plusses indicate conservative amino acid changes between the human and C. elegans proteins.
Since C. elegans SOS-1 G322R predominantly acts by enhancing the signal-dependent output of an EGFR-Ras-MAPK pathway, we examined the ability of transfected hSOS1 C282R to enhance EGF-dependent MAPK activation in serum-starved mammalian cell lines (Fig. 4). In NIH 3T3 cells, compared to transfection of wild-type hSOS1, hSOS1 C282R was able to enhance the amplitude and duration of MAPK activation (Fig. 4A). Although from experiment to experiment there was variability in the extent to which MAPK was activated, we were able to establish statistical significance (P < 0.05) for differences in MAPK activation at 15 and 60 min following EGF stimulation (Fig. 4B). On average, the differences between the activities of the wild-type and mutant proteins were two- to threefold. To further validate this result, we also performed the same experiment in HEK 293 EBNA cells. Once again, we found that compared to wild-type hSOS1, hSOS1 C282R was more potent at promoting EGF-dependent MAPK activation (Fig. 4C). Compared to NIH 3T3 cells, in HEK 293 EBNA cells, the time points at which differences in activity were manifested were more variable. Thus, it was harder to establish statistical significance. Nevertheless, in every experiment we found that at some time point, hSOS1 C282R displayed two- to fourfold more activity than wild-type hSOS1. In conjunction with our genetic data, these data suggest that the G322R change in C. elegans SOS-1, as well as the equivalent C282R change in hSOS1, does indeed enhance EGF-dependent MAPK activation. Thus, it is likely that the Dbl domain normally acts to dampen the amount of EGF-dependent MAPK activation and that the structural mechanism by which this is accomplished is conserved across species.
FIG. 4.
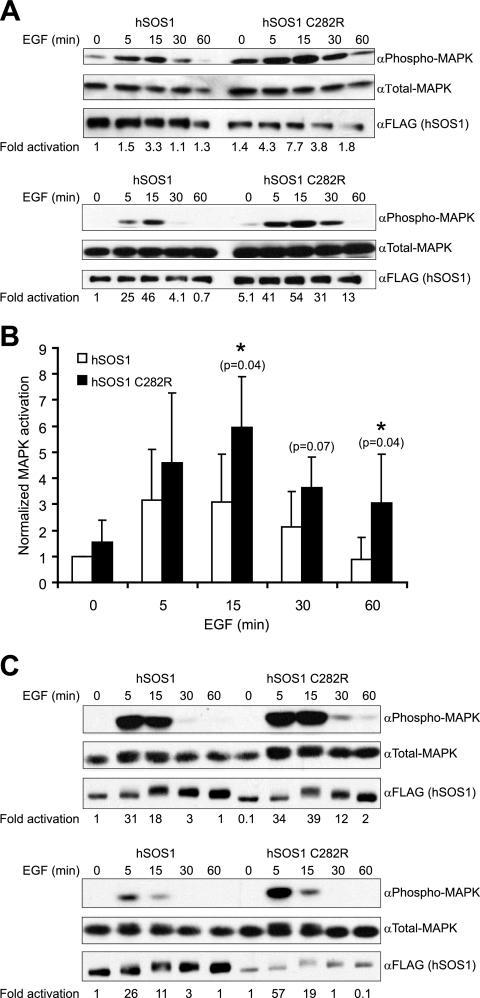
A C282R mutation in hSOS1 is functionally equivalent to the sos-1(sy262) G322R mutation in C. elegans SOS-1. (A) hSOS1 C282R promotes EGF-dependent MAPK activation in NIH 3T3 cells. The cells were transfected with 2 to 4 μg of FLAG-tagged hSOS1 expression vector and 4 μg of HA-ERK1 (MAPK) expression vector. After serum starvation and stimulation with EGF for the indicated times, whole-cell lysates were prepared and analyzed by Western blotting with the indicated antibodies. Phosphorylated and total transfected MAPKs were distinguished from endogenous MAPK by a mobility shift caused by the HA tag on the MAPK in the transfection construct. Representative blots from two independent experiments are shown. Activation (n-fold) was calculated by dividing the α-phospho-MAPK signal by the α-total-MAPK signal and then adjusting the values to that observed with wild-type hSOS1 at the zero time point. (B) Mean results from four independent experiments in NIH 3T3 cells, where the wild-type and mutant hSOS1 constructs were similarly expressed and the control wild-type hSOS1 construct caused similar levels of MAPK activation between experiments. Normalized activation (n-fold) is as described for panel A, except that α-phospho-MAPK/α-total MAPK ratios were also adjusted for slight differences in transfected hSOS1 expression. Statistical significance was assessed using a one-tailed Student's t test. (C) hSOS1 C282R promotes EGF-dependent MAPK activation in HEK 293 EBNA cells. The cells were transfected with 2 to 4 μg of FLAG-tagged hSOS1 expression vector and 2.5 μg of HA-ERK1 (MAPK) expression vector. After serum starvation and stimulation with EGF for the indicated times, whole-cell lysates were prepared and analyzed by Western blotting as described for panel A. Representative blots from two independent experiments are shown.
DISCUSSION
Although SOS is an evolutionarily conserved critical regulator of RTK-Ras-MAPK signaling, the key mechanisms that regulate its biologic activity in vivo are not well-defined. It was initially thought that RTK-dependent translocation of SOS to its substrate Ras at the plasma membrane was the key mechanism that controlled Ras-MAPK activation (3). However, several lines of evidence have suggested that control of SOS activity in the Ras-MAPK pathway might be more complex. First, in Drosophila melanogaster, in the absence of the SEVENLESS RTK or Grb2, SOS still localizes to the plasma membrane, yet Ras signaling is impaired (9, 49). Second, SOS structure-function studies have identified autoinhibitory effects of the N and C termini (20, 51, 93) and have suggested that growth factor signaling might be required to relieve these or other inhibitory mechanisms (14, 70). Third, although their developmental significance is not yet clear, a number of modifications/protein-protein interactions have been described that can alter the activities of either the Rac or Ras GEF domains of SOS. For example, Rac GEF activity of the Dbl domain is autoinhibited through interactions with the PH domain and is relieved by PI3 kinase signaling (23, 65, 83). Rac GEF activity also can be stimulated by protein interactions with Eps8 and Abi-1 (80, 81), as well as tyrosine phosphorylation by Abl (82). Finally, Ras GEF activity is allosterically stimulated through a positive feedback mechanism involving the binding of Ras-GTP to the REM domain (58). A two-pronged mechanism for controlling SOS activity, involving both translocation and signaling-dependent modifications/protein-protein interactions, would be consistent with emerging data regarding regulation of other GEFs. In a basal state, the Rho family GEF Vav is autoinhibited by the insertion of Tyr 174 from the N-terminal acidic region into the Dbl domain active site (1). This autoinhibition can be further compounded by PIP2-dependent interactions between the PH and Dbl domains (39). Signaling leading to phosphorylation of Tyr 174 and the generation of PIP3 disrupts both inhibitory mechanisms and increases Vav catalytic activity (1, 22, 25, 38, 39). Similarly, the catalytic activity of the Ral GEF Ral-GDS is inhibited via its N-terminal REM domain (77). In this case, signaling resulting in PI3 kinase activation promotes the association of another protein, PDK1, with the REM domain and relieves this inhibition (91).
We have discovered a novel gain-of-function mutation in the sos-1 gene that increases EGFR-Ras-MAPK pathway output during C. elegans vulval development. The localization of this mutation to the Dbl domain (G322R) implies that this domain normally confers some type of inhibition on EGFR signaling. SOS-1 G322R restores signaling to animals defective in EGF processing and EGFR localization and overcomes inhibition by one class of dominant-negative Ras protein (Table 1). Given that SOS possesses both Rac and Ras GEF activities and that the sy262 mutation lies within the Rac GEF Dbl domain, it is possible that the mutation acts through one or both of the GTPases. We favor a model in which the predominant effect of the G322R change is on Ras activation. First, SOS-1 G322R does not appear to act through any of the canonical mechanisms implicated in Rac-dependent MAPK activation. Current models involve activation of Pak by Rac and subsequent phosphorylation of either Raf or Mek. Pak can directly phosphorylate Raf on S338 (52), which is an activating modification, or it can phosphorylate Mek and enhance its binding to Raf and MAPK (27, 33, 97). C. elegans Raf has an aspartate residue at the analogous position of S338. Thus, the importance of an acidic charge is conserved, but not the use of protein phosphorylation at this position. Furthermore, our genetic epistasis results indicated that SOS-1 G322R acts upstream of Mek and Raf and possibly Ras activation (Table 1). Second, further genetic analysis indicated that SOS-1 G322R is more sensitive to reductions in Ras than in Rac levels (Table 2). Homozygous null and severe reduction-of-function mutations in two of the three C. elegans Rac genes (rac-2 and ced-10) had no effect on SOS-1 G322R activity, while a null mutation in the third Rac gene, mig-2, had only a partial effect. In fact, even a homozygous null mutation in mig-2 coupled with RNAi-induced reductions in the other Rac genes still allowed SOS-1 G322R to have 69% of its normal activity. In contrast, animals heterozygous for a strong reduction-of-function mutation in Ras retained only 29% of SOS-1 G322R activity. This result is even more striking considering that, by itself, the heterozygous state of the Ras(rf) mutation does not impair any known Ras-dependent pathway, while the homozygous state of the mig-2(null) mutation disrupts Q-cell descendant migration in 85% of animals (98). Finally, by itself, a null mutation in mig-2 (Rac) has no effect on vulval development (Table 2), while reduction-of-function mutations in Ras severely impair vulval development; furthermore, an activated Ras mutant almost fully restores wild-type vulval development to sos-1(null) animals (5, 15, 40). These data suggest that Ras is the key GTPase normally downstream of SOS-1 during vulval development. However, it remains possible that MIG-2 (Rac) partially contributes to the effect of SOS-1 G322R through a novel mechanism or that it functions through an independent pathway parallel to SOS-1 G322R. For example, the Rho family GEF UNC-73 (Trio) acts on MIG-2 (Rac) to regulate vulval cell divisions and cell migrations (53, 85). Furthermore, MIG-2 (Rac) is expressed in cell types other than the VPCs, including neurons (98), which can indirectly regulate vulval development (60).
The Dbl domain could control Ras-MAPK signaling through several mechanisms. Regulatory proteins could constitutively bind to the Dbl domain and sterically interfere with CDC25 Ras GEF activity. Alternatively, the Dbl domain could interfere with CDC25 activity through an autoinhibitory mechanism. trans-acting inhibitors of CDC25 function that bind directly to SOS have not yet been identified. On the other hand, an autoinhibitory function of the Dbl domain on CDC25 activity has been described (84). X-ray crystallographic and biochemical studies indicate that activated Ras (Ras-GTP) can bind to the SOS REM domain and allosterically stimulate the Ras GEF activity of the CDC25 domain (31, 58). However, the SOS crystal structure indicates that accessibility of Ras-GTP to the allosteric site is restricted by the H2b helix of the Dbl domain (84) (Fig. 3A). Specific substitution mutations in the H2b helix at the interface with the REM domain can relieve some of this inhibition (84). A triple mutant of E268A/M269A/D271A has little effect on basal CDC25 activity but increases the sensitivity to Ras-GTP activation by 20-fold. We used a BLAST alignment between the human and C. elegans SOS proteins, along with SWISS-MODEL, to generate a three-dimensional model of C. elegans SOS-1 (Fig. 3B and C). In this model, it appears that the key structural determinants for autoinhibition by the Dbl domain and allosteric stimulation by Ras-GTP are conserved between the two proteins. Our sy262 G322R mutation maps to the H3 helix of the Dbl domain, which lies near the H2b inhibitory helix. In the crystal structure of hSOS1 and our C. elegans SOS-1 model, multiple hydrophobic/bulky side chains in H2b face the surface of H3 (Fig. 3). These side chains are effectively accommodated by G322 and C282 in the worm and human proteins, respectively. The G322R change may prevent H2b from acquiring the correct geometry necessary for blocking the allosteric Ras-GTP site in the REM domain. In fact, consistent with this model, we find that a C282R change in hSOS1 also generates an activated mutant with properties similar to those of the C. elegans SOS-1 G322R. hSOS1 C282R does not display an obvious increase in basal activity but can enhance EGF-dependent MAPK activation (Fig. 4). Thus, if our model is correct, the sy262 mutation would provide strong support for the in vivo existence of the allosteric stimulatory/autoinhibitory functions of the REM and Dbl domains and would demonstrate their critical roles in regulating EGFR-Ras-MAPK signaling during development.
A central question, however, remains regarding the role of EGFR signaling in regulating SOS activity and Ras activation. Although the sy262 G322R mutation weakly increases basal activity (Table 4), mutant SOS-1 is still largely dependent on EGFR signaling for pathway activity (Table 3). This dependence could reflect the requirement for phosphorylated receptors to translocate Grb2-SOS-1 complexes to Ras at the plasma membrane. However, it is also tempting to speculate that SOS-1 itself is modified by activated receptors and that this modification is necessary for its full activity. SOS is phosphorylated immediately following EGFR activation (76), and it has recently been reported that allosteric stimulation of SOS by Ras-GTP requires growth factor signaling, unless SOS is truncated at the N and C termini (12). One interpretation of these results is that receptor signaling is necessary to relieve autoinhibition by the Dbl domain and to allow positive feedback by Ras-GTP. Such feedback may ultimately be necessary to sustain sufficient levels of MAPK activity to drive specific cell fate changes, as seen during vulval development. Thus, the G322R change may bypass the requirement for receptor signaling to allow positive feedback by Ras-GTP but not bypass the requirement for activated receptors to translocate SOS to Ras at the plasma membrane. Alternatively, the G322R change may only partially increase the accessibility of the allosteric site to Ras-GTP. Thus, a subthreshold amount of EGFR signaling might still be required to cooperate with the G322R change to fully allow positive feedback regulation by Ras-GTP.
One unexpected result is that the sos-1(sy262) mutation can suppress one class of dominant-negative Ras mutation, but not another (Table 1). SOS-1 G322R can overcome inhibition conferred by a heterozygous Ras S89F mutation, but not a heterozygous Ras G10R or G15D mutation. SOS-1 G322R also can weakly improve signaling in Ras S89F homozygotes. Ras S89F homozygotes are normally inviable. However, we could recover hermaphrodites homozygous for both the Ras S89F and SOS-1 G322R mutations, and starting with 25 hermaphrodites, we could maintain this population for an additional 2 generations before they died. Thus, consistent with an overexpression analysis of the different classes of dominant-negative Ras mutants (42), we found that the S89F mutant retained some biologic activity and could be regulated by SOS-1.
All nine of the dominant-negative Ras mutations isolated in C. elegans affect residues conserved in human Ras proteins (42). The G10R and G15D changes occur in the phosphate-binding P loop. P-loop mutations, such as those affecting G15, severely impair nucleotide and effector binding while increasing the affinity for GEFs (18, 46, 63, 68). This class of dominant-negative mutant likely acts by sequestering a limiting amount of GEF into a nonproductive signaling complex. S89 is in helix 3 and does not appear to make contact with nucleotides or SOS (10, 66), although an S89F change may affect the positioning of the P loop (Fig. 5). S89F may be a weaker class of dominant-negative mutant due to a weaker perturbation in nucleotide binding and a weaker increased interaction with SOS. As tempting as this model is, it cannot be entirely complete or correct. sur-5 reduction-of-function mutations were isolated as suppressors of a dominant-negative K16N P-loop mutation in Ras (35). sur-5 mutations also suppress other P-loop mutations, but not an S89F mutation. If Ras S89F acted as a dominant negative through the same mechanism as the P-loop mutants, it should also be suppressible by loss of SUR-5, since it is a weaker mutant. These results have led to the proposal that P-loop mutants and the S89F mutant act through different mechanisms (35). One possibility is that there is a second Ras GEF that is selectively inhibited by SUR-5 and, under some conditions, may be able to feed into the Ras pathway (35) (Fig. 6). This notion is supported by evidence that an activated G13E Ras mutant still exhibits EGF-dependent activity in the complete absence of SOS-1 and that the C. elegans genome predicts the existence of at least five other Ras GEFs (15). In one model (Fig. 6, model 1), strong inactivation of SOS-1 by dominant-negative Ras P-loop mutants allows the second Ras GEF to be wired into the Ras pathway. Thus, this class of mutation would be suppressible by a sur-5 mutation, but not by the sos-1(sy262) mutation. In contrast, an S89F Ras mutant may only partially inactivate SOS-1 and not allow the second Ras GEF to wire into the Ras pathway. Thus, this class of mutation would be suppressible by the sos-1(sy262) mutation, but not by a sur-5 mutation. Alternatively (Fig. 6, model 2), the second Ras GEF may be constitutively wired into the Ras pathway but selectively targeted by the S89F Ras mutant. In this case, the S89F mutation would still be suppressible by the sos-1(sy262) mutation, but not by a sur-5 mutation. Selective effects of different dominant-negative Ras mutants on different GEFs may be a general property of Ras proteins. In Saccharomyces cerevisiae, a P-loop dominant-negative RAS2 mutant acts exclusively through a CDC25-dependent mechanism, while a D64Y (D57Y in human H-Ras) mutant can act independently of CDC25 (46).
FIG. 5.
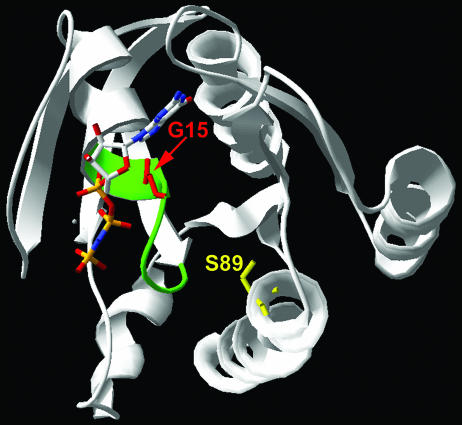
Crystal structure of human H-Ras complexed with the GTP analogue GppNp (66). The phosphate-binding P loop is highlighted in green. The positions of residues that when mutated give rise to two classes of dominant-negative Ras proteins are indicated (G15, red; S89, yellow).
FIG. 6.
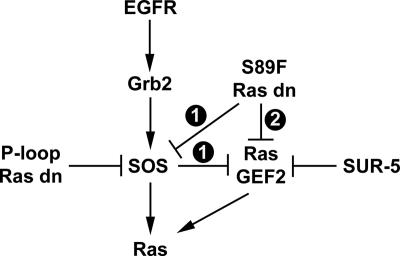
Models for Ras GEF regulation by dominant-negative Ras mutants in C. elegans VPCs. A second Ras GEF (RasGEF2) functions in parallel to SOS. It may be constitutively wired into the Ras pathway, or it may feed in only when SOS is inactivated. RasGEF2 is selectively inhibited by SUR-5. P-loop dominant-negative Ras mutants, such as G15D, strongly inactivate SOS. In model 1, inactivation of SOS by P-loop mutants permits RasGEF2 to function in the Ras pathway. Further inactivation of SUR-5 allows RasGEF2 to promote even more Ras activation. S89F dominant-negative Ras mutants only partially inactivate SOS and fail to allow RasGEF2 to feed into the Ras pathway. Therefore, loss of SUR-5 in an S89F mutant background does not enhance Ras activation. In model 2, RasGEF2 is constitutively wired into the Ras pathway, and S89F dominant-negative Ras mutants selectively inactivate RasGEF2. Thus, in an S89F background, loss of SUR-5 still fails to promote Ras activation.
Although the requirement for the SOS CDC25 domain in regulating RTK-mediated Ras signaling in vivo is well established (9, 15, 69, 75, 92), it is less clear to what extent and how the Dbl domain might regulate development. In fact, the first genetic role for the SOS Dbl domain has just recently been reported. In Drosophila melanogaster, Rac GEF activity from the Dbl domain, but not Ras GEF activity from the CDC25 domain, is necessary for ROBO-mediated axon guidance (32, 95). Our genetic data indicate that the SOS Dbl domain performs an additional critical function during development. It inhibits RTK-Ras-MAPK signaling. This form of inhibition is extremely significant. It is of the same magnitude as established trans-acting inhibitors, such as c-Cbl, RasGAP, and MAPK phosphatase (Table 5). Furthermore, although the worms appeared to be able to cope with the single loss of Dbl-conferred inhibition (through the sy262 mutation), they were on the threshold of having abnormal development. In the further absence of RasGAP, which would also decrease Ras GTPase activity, vulval development was no longer normal, as 25% of the double-mutant animals displayed ectopic vulvae (Table 5). These results highlight the developmental importance of the Dbl domain in properly balancing Ras GEF and GAP activities in order to achieve proper signaling intensity.
The identification of a single missense mutation in the SOS-1 Dbl domain that can significantly alter development in sensitized backgrounds suggests that functionally similar mutations in hSOS1 may contribute to Ras-dependent malignancies. In fact, we found that a C282R mutation in hSOS1, which is analogous to the activating G322R change in C. elegans SOS1, also created a gain-of-function protein (Fig. 4). Moreover, while this work was in revision, it was reported that activating mutations in hSOS1 occur in patients with Noonan syndrome (73, 90). Approximately 50% of Noonan patients carry activating mutations in PTPN11, a positive regulator of Ras signaling (88), and a few other patients carry novel, weakly activating mutations in K-Ras (78). Aside from the developmental abnormalities associated with Noonan syndrome, patients are predisposed to develop juvenile leukemias and myeloproliferative disorders commonly associated with mutations in the Ras pathway (55). Noonan-associated SOS1 mutations occur throughout the gene, affecting the histone fold region, the Dbl domain, the PH domain, the REM domain, the CDC25 domain, and the helical linker between the PH and REM domains (73, 90). Our work agrees with the general conclusions of these studies, namely, that disruption of autoinhibition on the allosteric site promotes Ras activation in vivo. However, not all of the human mutations may act equivalently. Some mutations affecting the Dbl domain, the CDC25 domain, and the helical linker between the PH and REM domains enhance EGF-dependent Ras and MAPK activation, which is consistent with our genetic and biochemical studies with the G322R and C282R mutations (73). Interestingly, one of these mutations, M269R, lies in the H2b helix of the Dbl domain, which directly interacts with the Ras-GTP allosteric site (84). In vitro studies predicted that a mutation at M269 would disrupt this interaction and enhance Ras and MAPK activation in vivo (84). In contrast, a human W729L mutation in the REM domain, which directly affects a residue that mediates binding of Ras-GTP at the allosteric site, appears to act by increasing basal activity toward Ras while having little effect on MAPK activation (90). If the general model from all of these studies is correct, our data provide direct support for the causal link between the human mutations in SOS1 and the associated developmental syndrome. The contributions of activating mutations in the Ras pathway to human cancer have long been appreciated (11). However, the strong activated nature of the classic G12 mutations did not overtly predict the potential contributions of weaker mutations that more subtly alter Ras signaling intensity to cancer and developmental syndromes (6, 24, 26, 64, 74, 78, 89). Our work and the recent discoveries of novel classes of activating mutations in components of the Ras pathway in human disease highlight the complexity with which perturbations in a single signaling pathway can lead to diverse types of human disease.
Acknowledgments
We thank Ben Neel for the hSOS1 cDNA clone and the hemagglutinin (HA)-ERK1 expression vector, Steve Lessnick for HEK 293 EBNA cells and polyethylenimine, and David Virshup for NIH 3T3 cells. We also thank Steve Lessnick, Don Ayer, Scott Kuwada, and members of the Moghal laboratory for critically reading the manuscript and Tommy Wong and Suzanne Elgort for help with figures. We also thank the reviewers for their insightful comments/criticisms about the manuscript. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). We thank the C. elegans Reverse Genetics Core Facility at the University of British Columbia, which is part of the International C. elegans Gene Knockout Consortium, for providing the rac-2(ok326) deletion mutant.
This research was supported by Public Health Services grant R01 GM073184 from the National Institutes of Health to N.M. and the Howard Hughes Medical Institute, for which P.W.S. is an Investigator.
Footnotes
Published ahead of print on 5 March 2007.
REFERENCES
- 1.Aghazadeh, B., W. E. Lowry, X. Y. Huang, and M. K. Rosen. 2000. Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell 102:625-633. [DOI] [PubMed] [Google Scholar]
- 2.Aroian, R., and P. Sternberg. 1991. Multiple functions of let-23, a Caenorhabditis elegans receptor tyrosine kinase gene required for vulval induction. Genetics 128:251-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronheim, A., D. Engelberg, N. Li, N. al-Alawi, J. Schlessinger, and M. Karin. 1994. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell 78:949-961. [DOI] [PubMed] [Google Scholar]
- 4.Bargmann, C., and L. Avery. 1995. Laser killing of cells in Caenorhabditis elegans. Methods Cell Biol. 48:225-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beitel, G., S. Clark, and H. Horvitz. 1990. Caenorhabditis elegans ras gene let-60 acts as a switch in the pathway of vulval induction. Nature 348:503-509. [DOI] [PubMed] [Google Scholar]
- 6.Bentires-Alj, M., J. G. Paez, F. S. David, H. Keilhack, B. Halmos, K. Naoki, J. M. Maris, A. Richardson, A. Bardelli, D. J. Sugarbaker, W. G. Richards, J. Du, L. Girard, J. D. Minna, M. L. Loh, D. E. Fisher, V. E. Velculescu, B. Vogelstein, M. Meyerson, W. R. Sellers, and B. G. Neel. 2004. Activating mutations of the Noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Res. 64:8816-8820. [DOI] [PubMed] [Google Scholar]
- 7.Berset, T., E. F. Hoier, G. Battu, S. Canevascini, and A. Hajnal. 2001. Notch Inhibition of RAS signaling through MAP kinase phosphatase LIP-1 during C. elegans vulval development. Science 291:1055-1058. [DOI] [PubMed] [Google Scholar]
- 8.Blume-Jensen, P., and T. Hunter. 2001. Oncogenic kinase signalling. Nature 411:355-365. [DOI] [PubMed] [Google Scholar]
- 9.Bonfini, L., C. A. Karlovich, C. Dasgupta, and U. Banerjee. 1992. The Son of sevenless gene product: a putative activator of Ras. Science 255:603-606. [DOI] [PubMed] [Google Scholar]
- 10.Boriack-Sjodin, P. A., S. M. Margarit, D. Bar-Sagi, and J. Kuriyan. 1998. The structural basis of the activation of Ras by Sos. Nature 394:337-343. [DOI] [PubMed] [Google Scholar]
- 11.Bos, J. L. 1989. ras oncogenes in human cancer: a review. Cancer Res 49:4682-4689. [PubMed] [Google Scholar]
- 12.Boykevisch, S., C. Zhao, H. Sondermann, P. Philippidou, S. Halegoua, J. Kuriyan, and D. Bar-Sagi. 2006. Regulation of ras signaling dynamics by Sos-mediated positive feedback. Curr. Biol. 16:2173-2179. [DOI] [PubMed] [Google Scholar]
- 13.Brenner, S. 1974. The genetics of Caenorhabditis elegans. Genetics 77:71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrne, J. L., H. F. Paterson, and C. J. Marshall. 1996. p21Ras activation by the guanine nucleotide exchange factor Sos, requires the Sos/Grb2 interaction and a second ligand-dependent signal involving the Sos N-terminus. Oncogene 13:2055-2065. [PubMed] [Google Scholar]
- 15.Chang, C., N. Hopper, and P. Sternberg. 2000. Caenorhabditis elegans SOS-1 is necessary for multiple Ras-mediated developmental signals. EMBO J. 19:3283-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chardin, P., J. H. Camonis, N. W. Gale, L. van Aelst, J. Schlessinger, M. H. Wigler, and D. Bar-Sagi. 1993. Human Sos1: a guanine nucleotide exchange factor for Ras that binds to GRB2. Science 260:1338-1343. [DOI] [PubMed] [Google Scholar]
- 17.Chen, N., and I. Greenwald. 2004. The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev. Cell 6:183-192. [DOI] [PubMed] [Google Scholar]
- 18.Chen, S. Y., S. Y. Huff, C. C. Lai, C. J. Der, and S. Powers. 1994. Ras-15A protein shares highly similar dominant-negative biological properties with Ras-17N and forms a stable, guanine-nucleotide resistant complex with CDC25 exchange factor. Oncogene 9:2691-2698. [PubMed] [Google Scholar]
- 19.Clandinin, T., J. DeModena, and P. Sternberg. 1998. Inositol trisphospate mediates a Ras-independent response to LET-23 receptor tyrosine kinase activation in C. elegans. Cell 92:523-533. [DOI] [PubMed] [Google Scholar]
- 20.Corbalan-Garcia, S., S. M. Margarit, D. Galron, S. S. Yang, and D. Bar-Sagi. 1998. Regulation of Sos activity by intramolecular interactions. Mol. Cell. Biol. 18:880-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox, G., J. Laufer, M. Kusch, and R. Edgar. 1980. Genetic and phenotypic characterization of roller mutants of C. elegans. Genetics 95:317-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crespo, P., K. E. Schuebel, A. A. Ostrom, J. S. Gutkind, and X. R. Bustelo. 1997. Phosphotyrosine-dependent activation of Rac-1 GDP/GTP exchange by the vav proto-oncogene product. Nature 385:169-172. [DOI] [PubMed] [Google Scholar]
- 23.Das, B., X. Shu, G. J. Day, J. Han, U. M. Krishna, J. R. Falck, and D. Broek. 2000. Control of intramolecular interactions between the pleckstrin homology and Dbl homology domains of Vav and Sos1 regulates Rac binding. J. Biol. Chem. 275:15074-15081. [DOI] [PubMed] [Google Scholar]
- 24.Davies, H., G. R. Bignell, C. Cox, P. Stephens, S. Edkins, S. Clegg, J. Teague, H. Woffendin, M. J. Garnett, W. Bottomley, N. Davis, E. Dicks, R. Ewing, Y. Floyd, K. Gray, S. Hall, R. Hawes, J. Hughes, V. Kosmidou, A. Menzies, C. Mould, A. Parker, C. Stevens, S. Watt, S. Hooper, R. Wilson, H. Jayatilake, B. A. Gusterson, C. Cooper, J. Shipley, D. Hargrave, K. Pritchard-Jones, N. Maitland, G. Chenevix-Trench, G. J. Riggins, D. D. Bigner, G. Palmieri, A. Cossu, A. Flanagan, A. Nicholson, J. W. Ho, S. Y. Leung, S. T. Yuen, B. L. Weber, H. F. Seigler, T. L. Darrow, H. Paterson, R. Marais, C. J. Marshall, R. Wooster, M. R. Stratton, and P. A. Futreal. 2002. Mutations of the BRAF gene in human cancer. Nature 417:949-954. [DOI] [PubMed] [Google Scholar]
- 25.Deckert, M., S. Tartare-Deckert, C. Couture, T. Mustelin, and A. Altman. 1996. Functional and physical interactions of Syk family kinases with the Vav proto-oncogene product. Immunity 5:591-604. [DOI] [PubMed] [Google Scholar]
- 26.De Luca, A., I. Bottillo, A. Sarkozy, C. Carta, C. Neri, E. Bellacchio, A. Schirinzi, E. Conti, G. Zampino, A. Battaglia, S. Majore, M. M. Rinaldi, M. Carella, B. Marino, A. Pizzuti, M. C. Digilio, M. Tartaglia, and B. Dallapiccola. 2005. NF1 gene mutations represent the major molecular event underlying neurofibromatosis-Noonan syndrome. Am. J. Hum. Genet. 77:1092-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eblen, S. T., J. K. Slack, M. J. Weber, and A. D. Catling. 2002. Rac-PAK signaling stimulates extracellular signal-regulated kinase (ERK) activation by regulating formation of MEK1-ERK complexes. Mol. Cell. Biol. 22:6023-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egan, S. E., B. W. Giddings, M. W. Brooks, L. Buday, A. M. Sizeland, and R. A. Weinberg. 1993. Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation. Nature 363:45-51. [DOI] [PubMed] [Google Scholar]
- 29.Ellis, R., D. Jacobson, and H. Horvitz. 1991. Genes required for the engulfment of cell corpses during programmed cell death in Caenorhabditis elegans. Genetics 129:79-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferguson, E., and H. Horvitz. 1985. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode C. elegans. Genetics 110:17-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freedman, T. S., H. Sondermann, G. D. Friedland, T. Kortemme, D. Bar-Sagi, S. Marqusee, and J. Kuriyan. 2006. A Ras-induced conformational switch in the Ras activator Son of sevenless. Proc. Natl. Acad. Sci. USA 103:16692-16697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fritz, J. L., and M. F. VanBerkum. 2002. Regulation of rho family GTPases is required to prevent axons from crossing the midline. Dev. Biol. 252:46-58. [DOI] [PubMed] [Google Scholar]
- 33.Frost, J. A., H. Steen, P. Shapiro, T. Lewis, N. Ahn, P. E. Shaw, and M. H. Cobb. 1997. Cross-cascade activation of ERKs and ternary complex factors by Rho family proteins. EMBO J. 16:6426-6438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Granato, M., H. Schnabel, and R. Schnabel. 1994. pha-1, a selectable marker for gene-transfer in C. elegans. Nucleic Acids Res. 22:1762-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gu, T., S. Orita, and M. Han. 1998. Caenorhabditis elegans SUR-5, a novel but conserved protein, negatively regulates LRT-60 ras activity during vulval induction. Mol. Cell. Biol. 18:4556-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 37.Hajnal, A., C. Whitfield, and S. Kim. 1997. Inhibition of Caenorhabditis elegans vulval induction by gap-1 and by let-23 receptor tyrosine kinase. Genes Dev. 11:2715-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han, J., B. Das, W. Wei, L. Van Aelst, R. D. Mosteller, R. Khosravi-Far, J. K. Westwick, C. J. Der, and D. Broek. 1997. Lck regulates Vav activation of members of the Rho family of GTPases. Mol. Cell. Biol. 17:1346-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han, J., K. Luby-Phelps, B. Das, X. Shu, Y. Xia, R. D. Mosteller, U. M. Krishna, J. R. Falck, M. A. White, and D. Broek. 1998. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science 279:558-560. [DOI] [PubMed] [Google Scholar]
- 40.Han, M., R. Aroian, and P. Sternberg. 1990. The let-60 locus controls the switch between vulval and nonvulval cell fates in Caenorhabditis elegans. Genetics 126:899-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Han, M., A. Golden, Y. Han, and P. Sternberg. 1993. C. elegans lin-45 raf gene participates in let-60 RAS-stimulated vulval differentiation. Nature 363:133-140. [DOI] [PubMed] [Google Scholar]
- 42.Han, M., and P. Sternberg. 1991. Analysis of dominant negative mutations of the Caenorhabditis elegans let-60 ras gene. Genes Dev. 5:2188-2198. [DOI] [PubMed] [Google Scholar]
- 43.Hosono, R., K. Hirahara, S. Kuno, and T. Kurihara. 1982. Mutants of C. elegans with Dumpy and Rounded head phenotype. J. Exp. Zool. 224:135-144. [Google Scholar]
- 44.Johnsen, R., and D. Baillie. 1991. Genetic analysis of a major segment [LGV(left)] of the genome of Caenorhabditis elegans. Genetics 129:735-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jongeward, G., T. Clandinin, and P. Sternberg. 1995. sli-1, a negative regulator of let-23-mediated signaling in C. elegans. Genetics 139:1553-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jung, V., W. Wei, R. Ballester, J. Camonis, S. Mi, L. Van Aelst, M. Wigler, and D. Broek. 1994. Two types of RAS mutants that dominantly interfere with activators of RAS. Mol. Cell. Biol. 14:3707-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaech, S., C. Whitfield, and S. Kim. 1998. The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell 94:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin, M. Gotta, A. Kanapin, N. Le Bot, S. Moreno, M. Sohrmann, D. P. Welchman, P. Zipperlen, and J. Ahringer. 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421:231-237. [DOI] [PubMed] [Google Scholar]
- 49.Karlovich, C. A., L. Bonfini, L. McCollam, R. D. Rogge, A. Daga, M. P. Czech, and U. Banerjee. 1995. In vivo functional analysis of the Ras exchange factor son of sevenless. Science 268:576-579. [DOI] [PubMed] [Google Scholar]
- 50.Katz, W., G. Lesa, D. Yannoukakos, T. Clandinin, J. Schlessinger, and P. Sternberg. 1996. A point mutation in the extracellular domain activated LET-23, the Caenorhabditis elegans epidermal growth factor receptor homolog. Mol. Cell. Biol. 16:529-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim, J. H., M. Shirouzu, T. Kataoka, D. Bowtell, and S. Yokoyama. 1998. Activation of Ras and its downstream extracellular signal-regulated protein kinases by the CDC25 homology domain of mouse Son-of-sevenless 1 (mSos1). Oncogene 16:2597-2607. [DOI] [PubMed] [Google Scholar]
- 52.King, A. J., H. Sun, B. Diaz, D. Barnard, W. Miao, S. Bagrodia, and M. S. Marshall. 1998. The protein kinase Pak3 positively regulates Raf-1 activity through phosphorylation of serine 338. Nature 396:180-183. [DOI] [PubMed] [Google Scholar]
- 53.Kishore, R. S., and M. V. Sundaram. 2002. ced-10 Rac and mig-2 function redundantly and act with unc-73 trio to control the orientation of vulval cell divisions and migrations in Caenorhabditis elegans. Dev. Biol. 241:339-348. [DOI] [PubMed] [Google Scholar]
- 54.Kornfeld, K., D. Hom, and H. Horvitz. 1995. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell 83:903-913. [DOI] [PubMed] [Google Scholar]
- 55.Lauchle, J. O., B. S. Braun, M. L. Loh, and K. Shannon. 2006. Inherited predispositions and hyperactive Ras in myeloid leukemogenesis. Pediatr. Blood Cancer 46:579-585. [DOI] [PubMed] [Google Scholar]
- 56.Liu, J., P. Tzou, R. Hill, and P. Sternberg. 1999. Structural requirements for the tissue-specific and tissue-general functions of the Caenorhabditis elegans epidermal growth factor LIN-3. Genetics 153:1257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lundquist, E. A., P. W. Reddien, E. Hartwieg, H. R. Horvitz, and C. I. Bargmann. 2001. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development 128:4475-4488. [DOI] [PubMed] [Google Scholar]
- 58.Margarit, S. M., H. Sondermann, B. E. Hall, B. Nagar, A. Hoelz, M. Pirruccello, D. Bar-Sagi, and J. Kuriyan. 2003. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112:685-695. [DOI] [PubMed] [Google Scholar]
- 59.Moerman, D., and D. Baillie. 1979. Genetic organization in C. elegans: fine-structure analysis of the unc-22 gene. Genetics 91:95-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moghal, N., L. R. Garcia, L. A. Khan, K. Iwasaki, and P. W. Sternberg. 2003. Modulation of EGF receptor-mediated vulva development by the heterotrimeric G-protein Gαq and excitable cells in C. elegans. Development 130:4553-4566. [DOI] [PubMed] [Google Scholar]
- 61.Moghal, N., and P. W. Sternberg. 2003. The epidermal growth factor system in Caenorhabditis elegans. Exp. Cell Res. 284:150-159. [DOI] [PubMed] [Google Scholar]
- 62.Moghal, N., and P. W. Sternberg. 2003. Extracellular domain determinants of LET-23 (EGF) receptor tyrosine kinase activity in Caenorhabditis elegans. Oncogene 22:5471-5480. [DOI] [PubMed] [Google Scholar]
- 63.Munder, T., and P. Furst. 1992. The Saccharomyces cerevisiae CDC25 gene product binds specifically to catalytically inactive Ras proteins in vivo. Mol. Cell. Biol. 12:2091-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niihori, T., Y. Aoki, Y. Narumi, G. Neri, H. Cave, A. Verloes, N. Okamoto, R. C. Hennekam, G. Gillessen-Kaesbach, D. Wieczorek, M. I. Kavamura, K. Kurosawa, H. Ohashi, L. Wilson, D. Heron, D. Bonneau, G. Corona, T. Kaname, K. Naritomi, C. Baumann, N. Matsumoto, K. Kato, S. Kure, and Y. Matsubara. 2006. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat. Genet. 38:294-296. [DOI] [PubMed] [Google Scholar]
- 65.Nimnual, A. S., B. A. Yatsula, and D. Bar-Sagi. 1998. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science 279:560-563. [DOI] [PubMed] [Google Scholar]
- 66.Pai, E. F., U. Krengel, G. A. Petsko, R. S. Goody, W. Kabsch, and A. Wittinghofer. 1990. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J. 9:2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peitsch, M. C. 1996. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24:274-279. [DOI] [PubMed] [Google Scholar]
- 68.Powers, S., K. O'Neill, and M. Wigler. 1989. Dominant yeast and mammalian RAS mutants that interfere with the CDC25-dependent activation of wild-type RAS in Saccharomyces cerevisiae. Mol. Cell. Biol. 9:390-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qian, X., L. Esteban, W. C. Vass, C. Upadhyaya, A. G. Papageorge, K. Yienger, J. M. Ward, D. R. Lowy, and E. Santos. 2000. The Sos1 and Sos2 Ras-specific exchange factors: differences in placental expression and signaling properties. EMBO J. 19:642-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qian, X., W. C. Vass, A. G. Papageorge, P. H. Anborgh, and D. R. Lowy. 1998. N terminus of Sos1 Ras exchange factor: critical roles for the Dbl and pleckstrin homology domains. Mol. Cell. Biol. 18:771-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reddien, P. W., and H. R. Horvitz. 2000. CED-2/CrkII and CED-10/Rac control phagocytosis and cell migration in Caenorhabditis elegans. Nat. Cell Biol. 2:131-136. [DOI] [PubMed] [Google Scholar]
- 72.Riddle, D. 1978. The genetics of development and behavior in I. J. Nematol. 10:1-16. [PMC free article] [PubMed] [Google Scholar]
- 73.Roberts, A. E., T. Araki, K. D. Swanson, K. T. Montgomery, T. A. Schiripo, V. A. Joshi, L. Li, Y. Yassin, A. M. Tamburino, B. G. Neel, and R. S. Kucherlapati. 2007. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat. Genet. 39:70-74. [DOI] [PubMed] [Google Scholar]
- 74.Rodriguez-Viciana, P., O. Tetsu, W. E. Tidyman, A. L. Estep, B. A. Conger, M. S. Cruz, F. McCormick, and K. A. Rauen. 2006. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science 311:1287-1290. [DOI] [PubMed] [Google Scholar]
- 75.Rogge, R. D., C. A. Karlovich, and U. Banerjee. 1991. Genetic dissection of a neurodevelopmental pathway: Son of sevenless functions downstream of the sevenless and EGF receptor tyrosine kinases. Cell 64:39-48. [DOI] [PubMed] [Google Scholar]
- 76.Rozakis-Adcock, M., R. Fernley, J. Wade, T. Pawson, and D. Bowtell. 1993. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature 363:83-85. [DOI] [PubMed] [Google Scholar]
- 77.Rusanescu, G., T. Gotoh, X. Tian, and L. A. Feig. 2001. Regulation of Ras signaling specificity by protein kinase C. Mol. Cell. Biol. 21:2650-2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schubbert, S., M. Zenker, S. L. Rowe, S. Boll, C. Klein, G. Bollag, I. van der Burgt, L. Musante, V. Kalscheuer, L. E. Wehner, H. Nguyen, B. West, K. Y. Zhang, E. Sistermans, A. Rauch, C. M. Niemeyer, K. Shannon, and C. P. Kratz. 2006. Germline KRAS mutations cause Noonan syndrome. Nat. Genet. 38:331-336. [DOI] [PubMed] [Google Scholar]
- 79.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scita, G., J. Nordstrom, R. Carbone, P. Tenca, G. Giardina, S. Gutkind, M. Bjarnegard, C. Betsholtz, and P. P. Di Fiore. 1999. EPS8 and E3B1 transduce signals from Ras to Rac. Nature 401:290-293. [DOI] [PubMed] [Google Scholar]
- 81.Scita, G., P. Tenca, L. B. Areces, A. Tocchetti, E. Frittoli, G. Giardina, I. Ponzanelli, P. Sini, M. Innocenti, and P. P. Di Fiore. 2001. An effector region in Eps8 is responsible for the activation of the Rac-specific GEF activity of Sos-1 and for the proper localization of the Rac-based actin-polymerizing machine. J. Cell Biol. 154:1031-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sini, P., A. Cannas, A. J. Koleske, P. P. Di Fiore, and G. Scita. 2004. Abl-dependent tyrosine phosphorylation of Sos-1 mediates growth-factor-induced Rac activation. Nat. Cell Biol. 6:268-274. [DOI] [PubMed] [Google Scholar]
- 83.Soisson, S. M., A. S. Nimnual, M. Uy, D. Bar-Sagi, and J. Kuriyan. 1998. Crystal structure of the Dbl and pleckstrin homology domains from the human Son of sevenless protein. Cell 95:259-268. [DOI] [PubMed] [Google Scholar]
- 84.Sondermann, H., S. M. Soisson, S. Boykevisch, S. S. Yang, D. Bar-Sagi, and J. Kuriyan. 2004. Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell 119:393-405. [DOI] [PubMed] [Google Scholar]
- 85.Spencer, A. G., S. Orita, C. J. Malone, and M. Han. 2001. A RHO GTPase-mediated pathway is required during P cell migration in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 98:13132-13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sternberg, P., and H. Horvitz. 1986. Pattern formation during vulval development in C. elegans. Cell 44:761-772. [DOI] [PubMed] [Google Scholar]
- 87.Sundaram, M., and M. Han. 1995. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell 83:889-901. [DOI] [PubMed] [Google Scholar]
- 88.Tartaglia, M., E. L. Mehler, R. Goldberg, G. Zampino, H. G. Brunner, H. Kremer, I. van der Burgt, A. H. Crosby, A. Ion, S. Jeffery, K. Kalidas, M. A. Patton, R. S. Kucherlapati, and B. D. Gelb. 2001. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat. Genet. 29:465-468. [DOI] [PubMed] [Google Scholar]
- 89.Tartaglia, M., C. M. Niemeyer, A. Fragale, X. Song, J. Buechner, A. Jung, K. Hahlen, H. Hasle, J. D. Licht, and B. D. Gelb. 2003. Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat. Genet. 34:148-150. [DOI] [PubMed] [Google Scholar]
- 90.Tartaglia, M., L. A. Pennacchio, C. Zhao, K. K. Yadav, V. Fodale, A. Sarkozy, B. Pandit, K. Oishi, S. Martinelli, W. Schackwitz, A. Ustaszewska, J. Martin, J. Bristow, C. Carta, F. Lepri, C. Neri, I. Vasta, K. Gibson, C. J. Curry, J. P. Siguero, M. C. Digilio, G. Zampino, B. Dallapiccola, D. Bar-Sagi, and B. D. Gelb. 2007. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat. Genet. 39:75-79. [DOI] [PubMed] [Google Scholar]
- 91.Tian, X., G. Rusanescu, W. Hou, B. Schaffhausen, and L. A. Feig. 2002. PDK1 mediates growth factor-induced Ral-GEF activation by a kinase-independent mechanism. EMBO J. 21:1327-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang, D. Z., V. E. Hammond, H. E. Abud, I. Bertoncello, J. W. McAvoy, and D. D. Bowtell. 1997. Mutation in Sos1 dominantly enhances a weak allele of the EGFR, demonstrating a requirement for Sos1 in EGFR signaling and development. Genes Dev. 11:309-320. [DOI] [PubMed] [Google Scholar]
- 93.Wang, W., E. M. Fisher, Q. Jia, J. M. Dunn, E. Porfiri, J. Downward, and S. E. Egan. 1995. The Grb2 binding domain of mSos1 is not required for downstream signal transduction. Nat. Genet. 10:294-300. [DOI] [PubMed] [Google Scholar]
- 94.Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston, and R. H. Plasterk. 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28:160-164. [DOI] [PubMed] [Google Scholar]
- 95.Yang, L., and G. J. Bashaw. 2006. Son of sevenless directly links the Robo receptor to rac activation to control axon repulsion at the midline. Neuron 52:595-607. [DOI] [PubMed] [Google Scholar]
- 96.Yoon, C., J. Lee, G. Jongeward, and P. Sternberg. 1995. Similarity of sli-1, a regulator of vulval development in C. elegans, to the mammalian proto-oncogene c-cbl. Science 269:1102-1105. [DOI] [PubMed] [Google Scholar]
- 97.Zang, M., C. Hayne, and Z. Luo. 2002. Interaction between active Pak1 and Raf-1 is necessary for phosphorylation and activation of Raf-1. J. Biol. Chem. 277:4395-4405. [DOI] [PubMed] [Google Scholar]
- 98.Zipkin, I. D., R. M. Kindt, and C. J. Kenyon. 1997. Role of a new Rho family member in cell migration and axon guidance in C. elegans. Cell 90:883-894. [DOI] [PubMed] [Google Scholar]