Abstract
The assembly of nucleosomes into chromatin is essential for the compaction of DNA and inactivation of the DNA template to modulate and repress gene expression. The nucleosome assembly protein 1, NAP1, assembles nucleosomes independent of DNA synthesis and was shown to enhance coactivator-mediated gene expression, suggesting a role for NAP1 in transcriptional regulation. Here, we show that Alien, known to harbor characteristics of a corepressor of nuclear hormone receptors such as of the vitamin D receptor (VDR), binds in vivo and in vitro to NAP1 and modulates its activity by enhancing NAP1-mediated nucleosome assembly on DNA. Furthermore, Alien reduces the accessibility of the histones H3 and H4 for NAP1-promoted assembly reaction. This indicates that Alien sustains and reinforces the formation of nucleosomes. Employing deletion mutants of Alien suggests that different regions of Alien are involved in enhancement of NAP1-mediated nucleosome assembly and in inhibiting the accessibility of the histones H3 and H4. In addition, we provide evidence that Alien is associated with chromatin and with micrococcus nuclease-prepared nucleosome fractions and interacts with the histones H3 and H4. Furthermore, chromatin immunoprecipitation and reimmunoprecipitation experiments suggest that NAP1 and Alien localize to the endogenous CYP24 promoter in vivo, a VDR target gene. Based on these findings, we present here a novel pathway linking corepressor function with nucleosome assembly activity.
The nucleosome is the basal structural unit of chromatin consisting of two histone H2A-H2B dimers, a histone H3-H4 tetramer, and DNA wrapped around histone octamers. Chromatin assembly occurs by depositing first the histone H3-H4 tetramer prior to assembling two heterodimers of the histones H2A and H2B to form nucleosomes (18, 22). Transcriptional regulation is associated with rearrangements of chromatin structure that include histone modifications and changes in nucleosome structure (2, 4, 20, 33, 36). Chromatin compaction and nucleosome arrangement itself represent a barrier and a repressive state that has to be overcome for transcriptional activation. The amino-terminal histone tails protruding from the nucleosome were shown not to significantly participate in nucleosome formation but, rather, present variable docking sites for other proteins and protein complexes to regulate chromatin compaction (20). Activation or repression of gene expression on the chromatin level can also be regulated by enzymatic activities that reorganize and rearrange the chromatin template (5, 36). ATP-dependent chromatin remodeling complexes along with histone chaperones mobilize nucleosomes. These activities can lead to an increase of accessibility for transcription factors and also facilitate histone exchange reactions within the nucleosome (5, 17, 23). Histone exchange reactions are important since they allow histone variants to be incorporated into nucleosomal array independent of DNA synthesis. The deposition of histone variants, such as H2A.Z, has severe consequences for gene regulation as they are known to localize to specific transcriptional domains and to heterochromatin (14, 21).
Recent experiments showed that nucleosomal structures are disrupted at promoters upon transcription in yeast, suggesting that there is an inverse correlation between gene activation and nucleosome assembly (15). While sliding mechanisms have been proposed, other studies support the notion that nucleosome displacement can occur via disassembly of the nucleosome in part by histone removal (2, 17, 36). Analysis of the inducible interleukin-2 gene suggests the loss of histones also from this mammalian promoter upon activation (8). Histone chaperones play an essential role in the context of histone depletion, as was shown for the yeast PHO5 and PHO8 promoters (1). Here, the nucleosome disassembly is carried out by the histone chaperone ASF1 and is an essential prerequisite for transcriptional activation. Interestingly, for gene repression an ASF1-independent reassembly of the nucleosome was observed (1). Thus, the current understanding about regulatory processes in transcription includes nucleosome disassembly for gene activation and histone deposition and nucleosome assembly to establish the basal or repressed state.
The highly conserved protein Alien was characterized as a corepressor for a variety of nuclear hormone receptors including the vitamin D receptor (VDR) (10, 24, 28). Alien enhances nuclear hormone receptor-mediated transcriptional silencing and possesses an intrinsic transcriptional silencing function. One mechanism of Alien-mediated silencing was shown to be histone deacetylase dependent.
Here, we provide evidence that Alien is a chromatin-associated protein, is present in micrococcal nuclease (MNase)-prepared nucleosome fractions, and binds specifically to the histones H3 and H4. Furthermore, we identified the histone chaperone nucleosome assembly protein 1 (NAP1) as a new interaction partner for Alien. NAP1 is known to possess chromatin assembly and histone binding capabilities (11). Recently, it was also reported that NAP1 autonomously exchanges histones and incorporates histone variants into intact nucleosomes (21, 26). This NAP1 activity functions independent of nucleosome remodeling complexes. Different studies implicate NAP1 in transcriptional regulation processes, such as deletion of NAP1 in yeast, which leads to down- and up-regulation of about 10% of all open reading frames (25). In in vitro studies NAP1 facilitates transcription factor binding to nucleosomal sites, which requires the nucleosome disassembly (40). Moreover, it was shown that NAP1 associates in mammalian cells with the coactivator p300/CBP, leading to an enhancement of p300-mediated transcriptional activation (3, 32).
Our investigations suggest that NAP1 possesses an intrinsic repression function when tethered to DNA. Furthermore, using NAP1-mediated nucleosome assembly experiments, we show that Alien increases the efficiency of nucleosome assembly. Alien also reduces histone accessibility for NAP1 through interaction with the histones H3 and H4. We hypothesize that Alien is able to repress transcription by reinforcing and sustaining a compact nucleosomal structure. Chromatin immunoprecipitation (ChIP) and re-ChIP experiments indicate that Alien, NAP1, and VDR are corecruited at hormone regulatory sites in vivo. Taken together, these data point toward a novel link between corepressors and nucleosome assembly activity.
MATERIALS AND METHODS
The yeast two-hybrid screen.
The yeast two-hybrid screen (Matchmaker Two-Hybrid System 3; Clontech) was performed as described in Eckey et al. (10a) with three selections for growth and lacZ gene expression. For the yeast two-hybrid interaction analysis, Saccharomyces cerevisiae strain EGY48 was used, and transformation as well as a liquid culture assay monitoring lacZ gene activity was described previously (10).
GST pull-down experiments.
Glutathione S-transferase (GST) pull-down experiments were performed essentially as described elsewhere (24). Bacterial expression of GST, GST-human NAP1 (hNAP1), or GST-Alien was induced with 0.2 mM isopropyl-β-d-thiogalactopyranoside for 3 h at 37°C in Escherichia coli strain HB101. Purification of the GST fusion proteins and interaction studies with in vitro translated [35S]methionine-labeled constructs (TNT kit; Promega) were employed according to the manufacturer's protocol. The sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels were stained with Coomassie brilliant blue to ensure equal loading of GST fusion proteins. The bound and labeled proteins were visualized by fluorography.
Plasmids.
The VP16-NAP1 fusion was generated by inserting the cDNA from the yeast screen library into pEG202 encoding the B42-DNA binding domain. pJG-Alien was described elsewhere (10). The full-length hNAP1 protein was cloned by reverse transcription-PCR from HeLa cells using the primers 5′-TCTTAGCGCTTGGCAGACATTGACAACAAAG (FW) and 5′-GCCAGGATCCTCCTGCTTCACTGCTGCTTG (BW) into pcDNA3-ATG, a pcDNA3 vector (Invitrogen) derivative with a Kozak sequence. NAP1 cDNA was inserted into pCS2-MT for eukaryotic expression as a myc-tagged protein.
Full-length Drosophila NAP1 (dNAP1) cDNA was obtained from J. T. Kadonaga and cloned into pcDNA3-ATG. In vitro translation of Alien, pGST-linker, and pGST-Alien was described earlier (10). GST-Alien deletions were generated using pGEX-2T (Amersham) or pGST-linker by in-frame insertions of Alien cDNA into the polylinker. Human and Drosophila GST-NAP1 fusions were generated by in-frame insertions into pGST-linker.
Vectors for mammalian expression systems were pUAS4x-TATA-Luc and pCMV-LacZ (where CMV is cytomegalovirus) as reporter plasmids and pABgal94 (10) for eukaryotic expression of GAL4 DNA binding domain fusions (amino acids [aa] 1 to 94 of GAL4). Partial cDNA from mNAP1 obtained from a mouse cDNA library was expressed as a GAL fusion for the mammalian two-hybrid assay. hNAP1 full-length protein was expressed as a GAL fusion for transcriptional regulation analysis. Alien was expressed as a VP16 fusion for mammalian two-hybrid experiments using in-frame insertion into the pCMV-VP16.
Reporter gene studies.
Cell culture and cell transfections were performed as described earlier (24). HeLa, HEK293, and CV-1 cells were cotransfected with the amounts of reporter plasmid indicated in the figure legends, the expression plasmids, and 0.2 μg of the pCMV-Lac-Z reporter plasmid for internal normalization.
Plasmid supercoiling assays.
Double CsCl purified, supercoiled pBR322 DNA (100 ng) was relaxed by incubation with 5 U of topoisomerase I (Promega) at 37°C for 1 h in assembly buffer (10 mM Tris, pH 7.5, 100 mM NaCl, 2 mM MgCl2, 0.5 mM dithiothreitol, 0.1 mg/ml bovine serum albumin [BSA]) in a volume of 15 μl. In a separate reaction, 400 ng of purified core histones (Roche) was added to GST or GST-dNAP and the purified protein indicated in the figure legends prior to incubation at 37°C for 15 min in assembly buffer. The topoisomerase and histone incubations were then combined (total volume of 30 μl plus 15 μl for each additional protein incubation) and incubated at 37°C for 1 h for histone deposition. The protein amounts were kept constant at every stage by the addition of BSA. The reactions were terminated and deproteinized by the addition of the stop buffer (0.2 μg/μl proteinase K, 1% SDS, 20 mM EDTA). After phenol-chloroform extraction and ethanol precipitation, the DNAs were separated in an agarose gel and stained with ethidium bromide.
Coimmunoprecipitation.
HEK293 cells were transfected with a vector expressing human myc-tagged NAP1. After lysis of the cells (20 mM Tris, pH 7.5, 200 mM NaCl, 0.5% NP-40, protease inhibitor cocktail complete [Roche)]) the, extract was incubated together with a mixture (1:1) of protein A-Sepharose- and protein G-agarose-coupled Alien-specific polyclonal or myc-specific antibody (9E10; Santa Cruz) followed by several washing steps with washing buffer (20 mM Tris, pH 7.5, 100 mM NaCl, 0.25% NP-40). The retained proteins were run on an SDS-PAGE gel and blotted, followed by immunodetection with either anti-Alien or myc antibody (9E10; Santa Cruz) or anti-histone H3 pan-antibody (07-690; Upstate). For detection of the histone H3-Alien interaction, cells were treated with formaldehyde as for ChIP (31). After the beads were washed with low and high salt, LiCl washing buffer, and Tris-EDTA buffer, the SDS loading buffer was added, and immunoblotting was carried out using the anti-histone H3 pan-antibody for detection.
Fractionations and chromatin preparations.
Nucleosome fractions (chromatin fractions) were prepared from HEK293 cells as previously described (19). The presence of nucleosomes in the chromatin fraction was confirmed by DNA agarose gel electrophoresis and SDS-PAGE followed by Coomassie staining. All collected fractions were then subjected to immunoblot analysis. The membranes with the wet-blot-transferred proteins were incubated overnight in blocking solution (5% nonfat milk in phosphate-buffered saline with 0.1% Tween). After antibody incubation the membrane was subjected to fluorography analysis using an ECL kit (Amersham). Actin (H-196) and nucleophosmin (H-106) antibodies were purchased from Santa Cruz.
Histone binding assays.
Bacterially expressed GST, GST-Alien, and deletions were bound to glutathione-Sepharose beads and blocked with BSA before incubation with 10 μg of core histones (Roche) in the presence of BSA for 1 h. Beads were washed extensively with a buffer containing 20 mM Tris-HCl, pH 7.5, 270 mM KCl, 5 mM MgCl, 1% NP-40, 1 mM dithiothreitol, 10 μg/ml BSA, and protease inhibitor cocktail complete (Roche).
ChIP assays.
ChIP and re-ChIP were performed essentially as described earlier (31). MCF-7 cells growing in Dulbecco's modified Eagle medium supplemented with 10% charcoal-stripped fetal calf serum were transfected with 5 μg of pCS+MT-NAP1 (myc-tagged NAP) using DOTAP [N(1-(2,3-dioleoyloxy)propyl)-N,N,N-trimethylammonium methyl-sulfate; Roche]. Prior to transfection (2 h) the medium was replaced, and 6 h after transfection cells were washed with phosphate-buffered saline twice and maintained in Dulbecco's modified Eagle medium containing serum. At 48 h posttransfection, the cells were treated with 10−7 M 1,25-dihydroxyvitamin D3 [1,25(OH)2D3; active vitamin D3] (Sigma-Aldrich) for 60 min prior to cross-linking with formaldehyde. For PCR the following primers were used: CYP24P1, 5′-GTCCAGGCTGGGGGTATCTG; CYP24P2, 5′-CCAATGAGCACGCAGAGGAGG; CYP24controlP1, 5′-CAGTCATTAGCCCCTCCAGAC; and CYP24controlP2, 5′-ACTACCACCAGCCAGGGAGG. Antibodies used were anti-VDR (H-81; Santa Cruz); anti-c-myc (9E10; Santa Cruz), and anti-Alien (10).
RESULTS
NAP1 as a novel interacting partner for Alien.
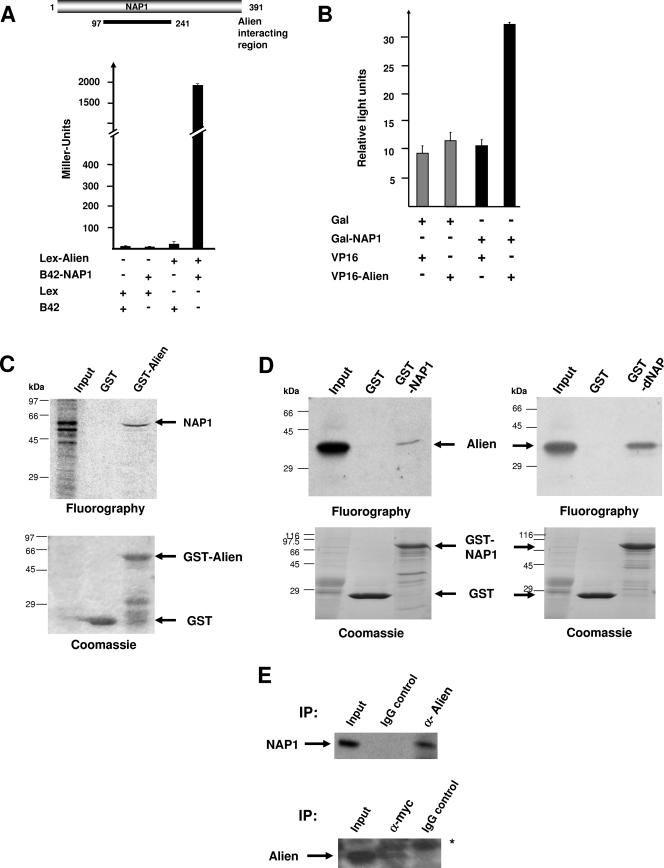
To identify novel interacting factors for Alien, we performed a stringent yeast two-hybrid screen with a cDNA library from mouse embryonic day 9.5 (E9.5) to E10.5 using the Matchmaker 3 system (Clontech) with multiple internal stringent selection steps in S. cerevisiae strain AH109. All 122 isolated cDNAs were individually retransformed in the other yeast strain, Y187, to identify false positives by using other selection criteria. Analysis of the remaining cDNAs surprisingly showed that 28% of these emerged as cDNAs of NAP1 (Table 1). In three other yeast two-hybrid screens using the same cDNA library but unrelated proteins as bait, no NAP1 cDNA was obtained. Although the various isolated NAP1 cDNAs from the yeast two-hybrid screen differed in length, all contained an overlapping region of NAP1, which suggests that a minimal region encompassing aa 140 to 241 of NAP1 is necessary for interaction with Alien (data not shown). The strongest measurable interaction with Alien was observed using the cDNA spanning aa 97 to 241, which was confirmed in a different yeast strain, EGY48 (Fig. 1A). As negative controls, Alien as prey together with the empty bait vector or a reverse setup was used. Only the combination of Alien with NAP yielded high β-galactosidase units, confirming the interaction of Alien with NAP in yeast cells. Notably, this region is highly conserved between NAP1 derived from human and mouse with only one conserved amino acid being exchanged (not shown). Also, this region of hNAP1 shares high homologies, 67% on the amino acid level, with the Drosophila nucleosome assembly protein dNAP.
TABLE 1.
Results of yeast two-hybrid screens
| Bait | No. of colonies after stringent selection | No. of colonies with NAP cDNA |
|---|---|---|
| Alien | 122 | 34 |
| Unrelated proteins | ||
| p52 | 101 | 0 |
| p33 | 16 | 0 |
| p66 | 198 | 0 |
FIG. 1.
Identification of NAP1 as an interaction partner for Alien in vitro and in vivo. (A) NAP1 cDNAs were isolated from a yeast two-hybrid screen performed in strain AH109, and interactions were verified the Y187 yeast strain using a mouse embryonic VP16-cDNA library from E9.5 to 10.5 and Gal-Alien as bait. Schematic view of NAP1 with the interaction region of NAP1 with Alien is depicted. The NAP1 cDNA and the Alien cDNA were subsequently cloned into the yeast two-hybrid vectors with the B42 activation domain as the prey and the LexA DNA binding domain as the bait, respectively. The interaction was further tested in the yeast strain EGY48 in liquid yeast cultures for expression of β-galactosidase. As controls, the empty LexA and the empty B42 vectors were transformed. Interaction is monitored by increased β-galactosidase activity and normalized to the number of yeast cells (Miller units). (B) Mammalian two-hybrid experiments were performed using CV1 cells transiently transfected with mammalian vectors expressing Gal-NAP1, VP16-Alien, and a luciferase reporter harboring four Gal4 binding sites. Luciferase values were normalized to β-galactosidase units obtained with the internal cotransfected control vector pCMV-LacZ. (C) Bacterially expressed GST or GST-Alien was incubated with in vitro translated and [35S]methionine-labeled NAP1 in GST pull-down assays. The bound material was separated by SDS-PAGE. Radiolabeled proteins were detected by fluorography. Coomassie staining visualized the proteins bound to glutathione beads. The input lane corresponds to 10% of the amount used. (D) Bacterially expressed GST or GST-NAP1 was incubated with in vitro translated and [35S]methionine-labeled Alien in the GST pull-down assay. SDS-PAGE separated proteins were visualized by Coomassie staining followed by fluorography to detect radiolabeled Alien. (E) Coimmunoprecipitation of endogenous Alien with transfected myc-tagged NAP1 was performed with HEK293 cells. Whole-cell extracts were prepared and incubated with the IgG control, with anti-myc, or with anti-Alien antibody for immunoprecipitation. Immunoprecipitates were separated by SDS-PAGE, and Western analysis was performed with anti-myc antibody to detect myc-tagged NAP1 (upper panel) or with anti-Alien antibody (lower panel) in the immunoprecipitation. The asterisk represents an unspecific band. IP, immunoprecipitation.
These findings in yeast were confirmed using the mammalian two-hybrid system with the isolated partial NAP1 cDNA as a Gal4 DNA binding domain fusion and full-length human Alien as a VP16-activator fusion. A specific interaction between Alien and NAP1 was observed, as reflected by the increase of the UAS-driven reporter activity (Fig. 1B). As a control, the empty Gal or VP16 vectors in combination with VP16-Alien or Gal-NAP1, respectively, exhibited no significant activation of the reporter.
To test for direct binding of NAP1 to Alien, GST pull-down experiments were used. In vitro translated full-length NAP1 cDNA was incubated with bacterially expressed and purified GST or GST-Alien (Fig. 1C). In vitro translated NAP1 gave rise to two prominent bands, and Alien preferentially binds the slower-migrating band, which might represent incomplete translation, proteolytic products, or a posttranslational modified form of NAP1 (30). GST alone as a negative control did not retain NAP1. Using the reverse experimental setup with a bacterial GST-NAP1 fusion and in vitro translated Alien also resulted in specific binding. However, in relation to the input lane the binding of the in vitro translated material was weaker (Fig. 1, compare panel D to panel C). Also, a specific interaction of Alien with dNAP was observed (Fig. 1D).
To assess whether Alien and NAP1 are complexed in vivo, coimmunoprecipitation experiments were carried out. Endogenous Alien was immunoprecipitated from human embryonal kidney cells (HEK293) transfected with a myc-tagged full-length NAP1 vector. The presence of NAP1 was analyzed by immunoblotting using an anti-myc antibody. The anti-Alien antibody, but not preimmune serum or immunoglobulin G (IgG) as controls, led to a specific coimmunoprecipitation of NAP1 (Fig. 1E). Similar results were obtained with the reciprocal experimental setup (Fig. 1E), suggesting that Alien and NAP1 are complexed in vivo. Taken together, these data suggest that Alien is an interaction partner for NAP1.
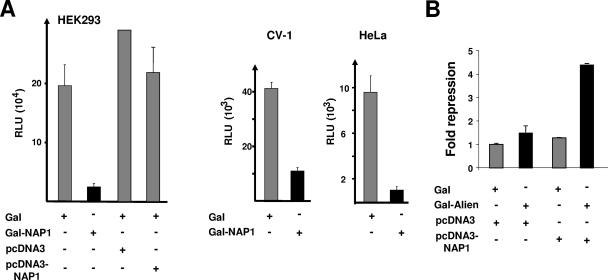
NAP1 was previously shown to be involved in transcriptional regulation mediated by CBP/p300 (3, 32). Since we identified NAP1 as an interaction partner for Alien having characteristics as a transcriptional repressor, we wondered whether NAP1 itself harbors any intrinsic transcriptional function. For that purpose we used full-length NAP1 as a Gal-NAP1 fusion containing the minimal GAL4 DNA binding domain (aa 1 to 94) (10) and examined its influence on the expression of a reporter gene possessing four Gal4 binding sites. Cotransfection experiments were performed in two different cell lines: HEK293 and CV1 with either Gal-NAP1 or Gal alone as control (Fig. 2A). NAP1 exhibited a silencing function in all cell lines tested, reducing promoter activity by about four- to eightfold, depending on the cell line. Since transfection of the unfused NAP1 expression vector did not significantly change the reporter activity (Fig. 2A), the results indicate that the NAP1-mediated silencing function observed is dependent on the DNA binding of NAP1. In line with this, under conditions of weak Alien-mediated repression, the overexpression of NAP1 enhanced the silencing function of Alien (Fig. 2B). Thus, the data support the notion that NAP1 harbors an intrinsic and transferable silencing function.
FIG. 2.
NAP1 harbors an intrinsic and transferable transcriptional silencing function. (A) To test for intrinsic transcriptional properties of full-length NAP1, an expression vector coding for Gal-NAP1 was generated and tested in the mammalian cell lines HEK 293, CV1, and HeLa. As a reporter, plasmid pUAS4x-TATA-Luc (1 μg) was cotransfected and tested for luciferase activity. Expression vectors for unfused NAP1 (pcDNA3-NAP1) and the corresponding empty vector as well as a plasmid expressing Gal alone were used as controls. RLU, relative light units. (B) Overexpression of NAP1 enhances Gal-Alien-mediated repression. The Gal-Alien expression vector (0.5 μg) was cotransfected with the NAP1 expression vector in a 1:6 molar ration. The empty expression vector served as a negative control. The values obtained with Gal alone were set arbitrarily as 1, and the data represent the relative increase in repression.
Alien is associated with chromatin.
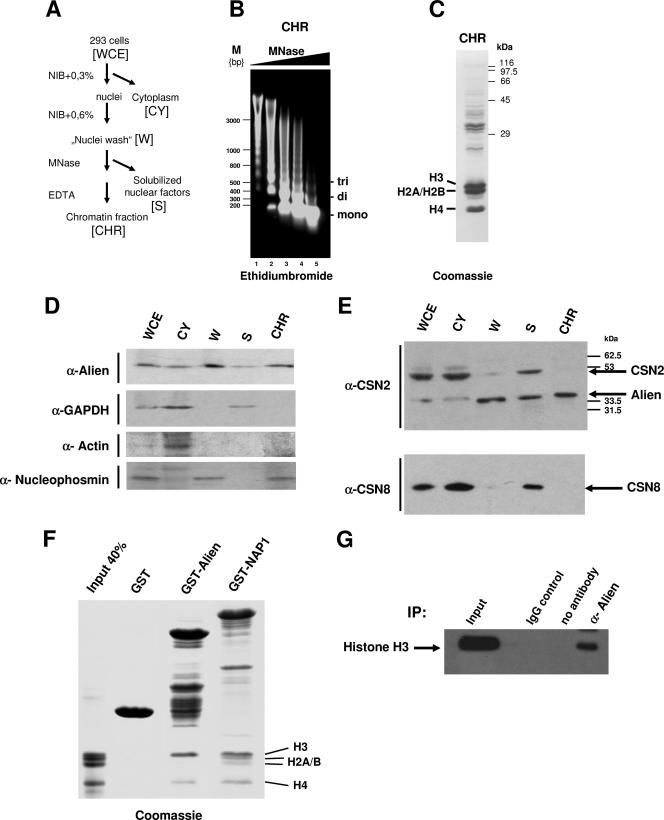
Based on the association of NAP1 with Alien, we addressed the question of whether Alien is associated with nucleosomes and chromatin in vivo. Therefore, cell extracts (HEK293) were prepared (Fig. 3A) including MNase digestion of chromatin, and fractions were analyzed for the presence of Alien. Extraction of the DNA from the chromatin fraction and its detection on ethidium bromide-stained agarose gels revealed that the majority consists of DNA fragments corresponding to mono- and oligonucleosomes (Fig. 3B). Coomassie staining of the chromatin preparation (Fig. 3B, lane 4) showed that the majority of the isolated proteins are the core histones (Fig. 3C) and additional bands, which presumably represent chromatin-associated factors. Western analysis of the different fractions determined that Alien is present in all fractions including the nuclear fraction (Fig. 3D, W), as well as the supernatant of the nuclei eluate (S), and is apparent in the chromatin preparation (CHR). As a control, the localization of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was determined, which is mainly present in the cytoplasmic fraction and partially in the supernatant after MNase digestion (Fig. 3D). Accordingly, a nuclear localization of GAPDH has been described earlier (34). Also, detection of the predominantly nuclear nucleophosmin and the predominantly cytoplasmic actin were used as markers for fractionation (Fig. 3D).
FIG. 3.
Alien is associated with chromatin in vivo. (A) Schematic view of the chromatin fractionation including MNase digestion of human embryonal kidney cell line HEK293. (B) Ethidium bromide staining of the isolated genomic DNA from the chromatin fractions after incubation with increasing amount of MNase. Depicted are the migrations of DNA fragments corresponding to DNA wrapped around mono-, di-, and trinucleosomes. (C) Coomassie staining of the chromatin fraction confirms the presence of the core histones as major components. (D) Western analysis of the fractions performed with antibodies specific for Alien, GAPDH, actin, and nucleophosmin revealed the specific presence of each protein. (E) Identification of the signalosome subunits CSN2 and CSN8 in the fractions. The anti-CSN2 antibody recognized both Alien and CSN2. (F) GST or the indicated GST fusions were incubated with the histones H3, H2A, H2B, and H4. The input lane corresponds to 40% of the histones used. After several washing steps, bound proteins were separated by SDS-PAGE and visualized by Coomassie staining. NAP1 interacts with all four core histones whereas Alien interacts specifically with histones H3 and H4. (G) Immunoprecipitation of Alien with histone H3 from HEK293 cells. IgG and the beads alone (no antibody) were used as internal controls for immunoprecipitation (IP); Western analyses were performed with histone H3 antibody. WCE, whole-cell extracts; CY, cytoplasmic fraction; W, supernatant of nuclei washing steps; S, solubilized nuclear factors after MNase digestion; CHR, chromatin fraction; α, anti; M, molecular size marker.
A longer form of Alien (CSN2) was shown to be part of the COP9 signalosome (CSN) complex (6, 9). To differentiate whether Alien is cofractionating with components of the signalosome complex CSN2 and CSN8, specific antibodies were employed. The anti-CSN8 antibody directed against the subunit 8 of CSN was used for Western analysis. Interestingly, CSN8 was detected specifically in the cytoplasm and in the supernatant after MNase treatment but not in the chromatin fraction (Fig. 3E). Similarly, a differential distribution of Alien and CSN2 was confirmed by using a commercial antibody recognizing both isoforms (Fig. 3E). These findings indicate that Alien has distinct affinities compared to some signalosome components and is associated with chromatin.
Since the major component of the chromatin fraction is represented by the core histones, we investigated whether Alien might directly bind to histones. Therefore, GST pull-down experiments with purified histones were performed. GST as a negative control and GST-NAP1 as a positive control were compared with GST-Alien for their ability to bind to histones (Fig. 3F). All four core histones (Fig. 3F, Input) were incubated with the GST or GST fusion proteins, and after stringent washes the bound proteins were analyzed by SDS-PAGE and Coomassie staining. The slower-migrating bands represent the expressed and stained GST fusions. The migration of the histones is indicated. As expected, GST-NAP1 retained all four core histones, H3, H2A, H2B, and H4, which is in line with previous findings (26). Remarkably, Alien associates specifically with high preference with the histone H3 and the histone H4 (Fig. 3F). To confirm an interaction of Alien with histones, a an immunoprecipitation assay of endogenous Alien from cross-linked HEK293 cell extracts and subsequent Western analysis revealed coprecipitated histone H3 (Fig. 3G). In cells histones are complexed within nucleosomes. Therefore, from these coimmunoprecipitation experiments a specificity of Alien-histone interaction cannot be concluded, but the data confirm that Alien is associated with histones in vivo. In summary the data reveal that Alien is chromatin associated and preferentially binds to the core histones H3 and H4.
Alien enhances NAP1-mediated nucleosome assembly activity.
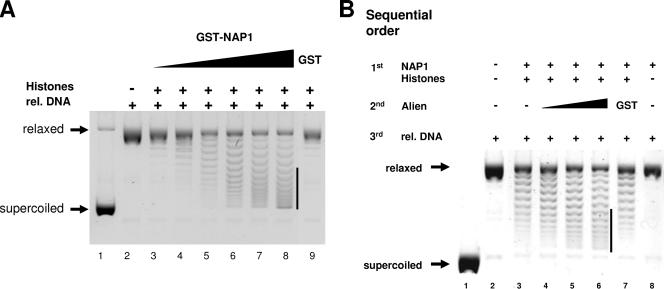
Next, we addressed the functional role of Alien in NAP1-mediated histone deposition. NAP1 is known to be involved in nucleosome assembly and efficiently loads histones onto DNA in classical supercoiling assays used for detection of nucleosome assembly activity (11, 13). The core particle formation in this assay is reflected by a faster electrophoretic migration of the initially relaxed plasmid DNA. To test for a functional role of Alien, we performed supercoiling assays using bacterially purified GST-NAP1, histones, topoisomerase I for relaxation, and a supercoiled pBR322 plasmid (Fig. 4). First, the supercoiling activity was tested with different amounts of GST-NAP1. As shown by the appearance of supercoiled DNA, the combined incubation of histones with increasing amounts of NAP1 led to a dose-dependent histone deposition onto plasmid DNA (Fig. 4A, lanes 3 to 8), which was not achieved by the addition of histones and GST alone (Fig. 4A, lane 9). Under conditions in which NAP1 less efficiently promotes core particle formation by employing limited amounts of NAP1, Alien was added to the supercoiling assay in the sequential order indicated in Fig. 4B. Addition of bacterially purified GST served as a control. Alien alone did not exhibit supercoiling activity (data not shown). Analysis of histone assembly revealed an enhancement of NAP1 activity by Alien (Fig. 4B, compare lanes 6 and lane 7), which was dose dependent (Fig. 4B, lanes 4 to 6). Thus, the data suggest that Alien enhances the histone deposition mediated by NAP1.
FIG. 4.
Alien enhances nucleosome assembly. (A) Supercoiling assays were performed by adding topoisomerase I relaxed plasmid DNA, all four core histones, and increasing amounts of bacterially expressed and purified GST-NAP1 (13.3 ng/μl to 66.6 ng/μl). The bar highlights the changes in supercoiling. The total protein amounts were kept identical by complementing with BSA. Purified GST alone served as a negative control. The migration of the relaxed and supercoiled DNA is indicated. (B) Alien increased the nucleosome assembly reaction in supercoiling assays when Alien was added after NAP1 and histone preincubation. Please note that limiting amounts of NAP1 (33 ng/μl) were used. The total protein amounts were kept constant by the addition of BSA. Addition of purified GST served as an additional control. rel DNA, relaxed DNA.
Alien inhibits histone H3 and H4 accessibility for NAP1.
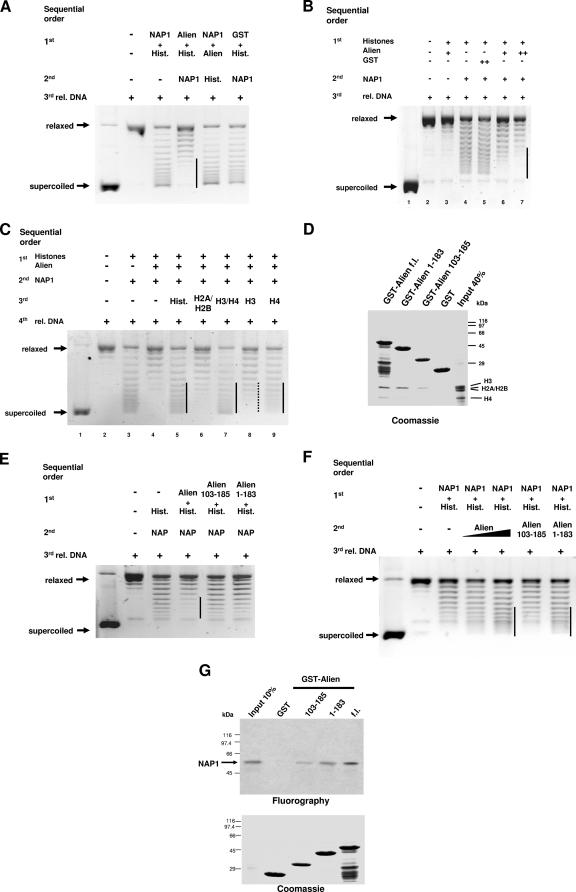
Based on the interaction of Alien with the histones H3 and H4, we were interested whether Alien might influence histone accessibility for their deposition. For that purpose we changed the order of incubation in the supercoiling assay and preincubated histones first with Alien prior to the addition of NAP1 to perform the nucleosome assembly (Fig. 5A). Indeed, NAP1-mediated nucleosome deposition was strongly reduced when Alien was preincubated with histones but was unaffected when GST as an unrelated protein was employed. Increasing amounts of NAP1 reversed, in part, the Alien-mediated inhibition (data not shown), suggesting a steady-state reaction. Preincubation of Alien with NAP1 prior to the addition of histones did not inhibit nucleosome deposition. This suggests that Alien inhibits nucleosome assembly if it is preincubated with histones. To test whether the inhibition of the histone accessibility is dependent on the amount of Alien, increasing doses of Alien were tested in the same sequential order. The total protein amounts were kept constant. The decline in generation of supercoiled DNA reflects a dose-dependent inhibition of nucleosome core particle deposition by Alien (Fig. 5B). These data indicate that the repression of NAP1 activity by Alien is exerted presumably by inhibition of histone accessibility for NAP1.
FIG. 5.
Alien inhibits the accessibility of the histones H3 and H4. (A) Preincubation of Alien with histones (see the sequential order of incubation) was analyzed in supercoiling experiments. As controls, Alien was preincubated with GST-NAP1 or GST was preincubated with histones. The protein amounts were kept constant by adding BSA. (B) To test for dose dependency, supercoiling assays were performed with preincubation of increasing amounts of purified Alien with histones. Alien and GST protein concentrations range from 16.4 ng/μl (+) to 38.6 ng/μl (++). (C) Addition of the histones H3 and H4 abrogate the Alien-mediated block of nucleosome assembly activity. Alien was preincubated with histones followed by incubation with NAP1. A total of 0.4 μg of the indicated histones was added after preincubation of Alien with histones and NAP1 in supercoiling assays. (D) The Alien mutants with deletions of aa 1 to 183 and aa 103 to 185 lack significant binding to histone H4. GST or the indicated GST fusions were incubated with the core histones H3, H2A, H2B, and H4 in GST pull-down assays. After several stringent washing steps, proteins were separated by SDS-PAGE and visualized by Coomassie staining. (E) Supercoiling assays were performed with preincubation of identical protein amounts of full-length Alien or the indicated Alien deletions (aa 1 to 183 and aa 103 to 185), as described in panel A. The total protein amount was equalized with BSA. (F) The Alien mutant encompassing aa 1 to 183, but not the mutant aa 103 to 185, is able to enhance nucleosome assembly activity. Purified full-length Alien, Alien harboring aa 1 to 183, or Alien harboring aa 103 to 185 were tested for their ability to enhance nucleosome assembly activity in supercoiling assays. (G) The interaction of in vitro translated and 35S-labeled NAP1 with full-length (fl) GST-Alien and the mutants encompassing aa 1 to 183 or aa 103 to 185 were analyzed with GST pull-down experiments. The GST proteins used were visualized by Coomassie staining. Fluorography detected the amount of the retained [35S]methionine-labeled NAP1. The input lane corresponds to 10% of the amount used. rel DNA, relaxed DNA.
If, indeed, the underlying mechanism is the lack of histone accessibility, further addition of histones should rescue the inhibition by Alien. Therefore, additional histones, either as a mix of all core histones or as mixtures of H2A-H2B or H3-H4, were employed (Fig. 5C). In fact, histones H3-H4 were able to abrogate Alien-mediated inhibition of NAP1-mediated nucleosome deposition, whereas additional amounts of histones H2A-H2B had only very marginal effects (Fig. 5C, compare lane 4 with 6 and 7). This is in line with the preferential binding of Alien to the histones H3 and H4 (Fig. 2F). Testing histone H3 or histone H4 individually, the Alien-mediated inhibition was less efficiently rescued compared to the combined addition of histones H3-H4. Interestingly, histone H4 alone, and to a minor extent also histone H3, significantly reversed the blocking compared to histones H2A-H2B (Fig. 5C, compare lane 6 with 8 and 9).
In conclusion, the findings suggest that the accessibility of histone H4 for histone deposition is preferentially inhibited by Alien, and histones H3 and H4 are likely to be combined targeted. Thus, we speculate that the inhibitory effect is due to the interaction between Alien and histones H3 and H4.
Thus, the data suggest that Alien interacts with the histones H3 and H4. If Alien is allowed to interact prior to the NAP1-histone interaction, it can sequester H3 and H4 from NAP1 and decrease nucleosome assembly. This is reversed if the reaction is supplemented with additional histones H3 and H4. However, if NAP1 is allowed to interact with histones prior to Alien, Alien appears to enhance nucleosome assembly.
To get more insight into specificity, different Alien deletion mutants were generated and tested for histone binding (Fig. 5D). Using GST pull-down experiments the Alien deletion mutant encompassing aa 1 to 183 revealed a strongly reduced binding to histone H4, whereas the interaction with histone H3 was similar to wild-type Alien. The Alien deletion mutant encompassing aa 103 to 185 did not bind to histone H4 and showed a similar binding to histone H3 as the deletion mutant lacking aa 1 to 183, considering the protein amounts used (Fig. 5D).
Furthermore, these deletion mutants were subjected to supercoiling assays to analyze the specificity and their potential to inhibit histone deposition mediated by NAP1 (Fig. 5E). Both mutants (aa 1 to 183 and aa 103 to 185) were unable to block the histone assembly reaction effectively. This further indicates the specificity of Alien to inhibit histone deposition and is in accordance with the strongly reduced binding of these deletion mutants to histone H4. Moreover, these two Alien deletions were subjected to supercoiling assays to assess their ability to enhance the NAP1-mediated nucleosome assembly. The Alien mutant aa 1 to 183 specifically enhanced nucleosome assembly mediated by NAP1 to a comparable extent as wild-type Alien, whereas the mutant aa 103 to 185 failed to enhance this activity (Fig. 5F). Tests of these deletions for interaction with NAP1 in GST pull-down assays showed that both mutants retained binding ability, although reduced, to NAP1 (Fig. 5G). Thus, specific regions of Alien enhance nucleosome assembly and inhibit histone accessibility.
NAP1, Alien, and VDR are corecruited to hormone regulatory chromatin sites in vivo.
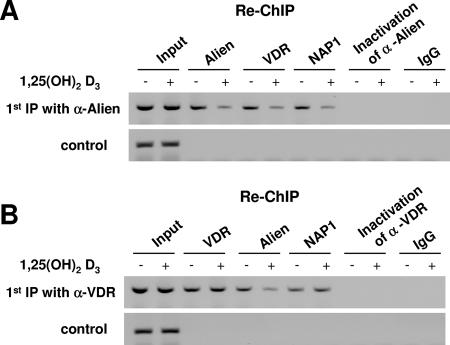
Alien was shown to be a corepressor of VDR recruited at the endogenous CYP24 promoter at VDR target sites in vivo (24, 28). We wondered whether Alien, VDR, and NAP1 are corecruited in vivo to the VDR regulatory sites and whether the recruitment may be regulated by 1,25(OH)2D3 hormone. For that purpose we applied ChIP with a first antibody and re-ChIP with the antibodies indicated in Fig. 6. Due to lack of an appropriate NAP1 antibody, we transfected MCF-7 cells with an expression vector encoding myc-tagged NAP1. The anti-Alien antibody was used for the first immunoprecipitation, and after a second immunoprecipitation with the same antibody, genomic PCR of the CYP24 promoter was performed. The results suggest a recruitment of Alien in the absence of ligand and a reduced recruitment in the presence of hormone (Fig. 6A). This confirms a ligand-sensitive recruitment of Alien at the VDR chromatic sites of the endogenous CYP24 promoter (24). Reprecipitation of the Alien first immunoprecipitate with either VDR antibody in the second immunoprecipitation or anti-myc antibody for myc-NAP1 revealed a PCR product of the same genomic locus, suggesting that Alien, VDR, and NAP1 are corecruited in vivo. Consistently, hormone treatment reduces the amount of the immunoprecipitated genomic DNA. As negative controls inactivation of anti-Alien antibody or the IgG control did not lead to precipitation of the genomic CYP24 locus. In addition, an upstream chromosomal location of the CYP24 promoter lacking vitamin D responsive elements (VDREs) did not show significant localization of Alien.
FIG. 6.
NAP1, Alien, and VDR are corecruited in vivo to the vitamin D3 responsive chromatin site. ChIP and re-ChIP experiments were performed on the CYP24 promoter known to be repressed by VDR in the absence of ligand and activated in the presence of 1,25(OH)2D3. MCF-7 cells were transfected with the expression plasmid encoding myc-NAP1 and were untreated or treated with hormone for 60 min prior to ChIP experiments. The upper panel shows the recruitment to the VDREs with a direct repeat spaced by 3 nucleotides whereas the lower control panel shows PCR results from the same material amplifying an upstream chromosomal location of the CYP24 promoter lacking VDREs. (A) ChIP and re-ChIP experiments were performed with the anti-Alien antibody for the first immunoprecipitation. These immunoprecipitates were subjected to a second immunoprecipitation with the antibodies against the indicated factors. Anti-myc antibody was used to precipitate myc-tagged NAP1. As a specificity control, inactivated anti-Alien antibody or IgG was used. (B) The same experimental setup as in panel A was used except that the anti-VDR antibody was employed in the first immunoprecipitation. α, anti.
By performing re-ChIP experiments using anti-VDR antibody for the first immunoprecipitation and anti-VDR antibody for the subsequent second immunoprecipitation, the recruitment of VDR was detected (Fig. 6B). In agreement with our previous findings, VDR is recruited to the hormone regulatory chromatin sites of the CYP24 gene independent of the addition of hormone (24). Immunoprecipitation with anti-Alien antibody for the second immunoprecipitation yielded fewer PCR products in the presence of the hormone, confirming a hormone-sensitive localization of Alien at these chromatin sites in vivo. Re-ChIP was used to analyze for the presence of NAP1 in the VDR immunoprecipitate (Fig. 6B). Interestingly, the NAP1 immunoprecipitation experiments suggest that NAP1 is recruited to these chromatic sites in vivo simultaneously with VDR. Hormone treatment did not significantly affect the amount of the immunoprecipitated genomic DNA, indicating that NAP1 is recruited ligand independently. Since an association of NAP1 with p300/CBP was reported and p300/CBP is a known coactivator for nuclear hormone receptors, it is possible that in the presence of hormone NAP1 is associated with VDR via p300/CBP. These data suggest an in vivo corecruitment of VDR, Alien, and NAP1 to the same chromatin sites.
DISCUSSION
It is known that the chromatin structure is altered toward a higher accessibility for transcription factors to potentiate gene activation (33). It has been suggested that a more open chromatin state caused by gene activation, apart from histone modifications and alterations in the nucleosomal array, is also a result of changes in nucleosome integrity due to histone displacement (4). This is supported by the notion that for histone-variant incorporation dependent on transcriptional activation, the nucleosomes themselves have to be disrupted and reassembled (14). Remarkably, it was shown that histone chaperones play an important role in these processes (14, 29, 37). Equally important to chromatin opening is the establishment of a repressed state of chromatin structure both before and after gene activation.
The complex purification of the histone H2A.Z, for example, revealed an association of this histone variant with the SWR1 remodeling complex and NAP1 (23). Interestingly, NAP1 has an intrinsic histone exchange ability in vitro, which is not dependent on the presence of other proteins such as remodeling complex subunits (26). The reported association of NAP1 with remodeling complexes may nevertheless imply that NAP1 could operate in concert with such factors to regulate gene expression. Remodeling factors that were originally thought to be involved only in gene activation were also shown to participate in transcriptional silencing (12, 41). Accordingly, nucleosome deposition and arrangement can represent a barrier for transcriptional activation. Thus, it is tempting to speculate that the histone chaperones are also of importance for the compaction of chromatin. Here, we provide first evidence that a transcriptional corepressor influences nucleosome deposition and assembly through the regulation of a histone chaperone activity.
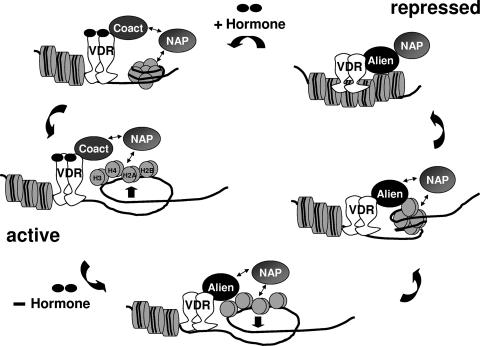
Liganded VDR activates target genes, such as the CYP24 gene, whereas unliganded VDR is known to silence genes and to recruit corepressors, such as Alien (28, 39). The CYP24 gene is highly inducible by 1,25(OH)2D3 mediated by two VDREs with direct repeats spaced by 3 nucleotides and is repressed in the absence of hormone (24, 28, 38). Using ChIP experiments, we observed recruitment of Alien specifically in a hormone-sensitive manner to the chromatin sites harboring the VDREs. Re-ChIP experiments suggest the presence of VDR or NAP1 in the absence of ligand on the VDRE containing the genomic locus of CYP24, whereas in the presence of 1,25(OH)2D3, only reduced amounts were immunoprecipitated. This is line with a hormone-sensitive recruitment of Alien, whereas ligand-independent occupancy of VDR was detected. Re-ChIP of the VDR ChIP experiments revealed that NAP1 binds to the same genomic locus in a ligand-independent manner, whereas, as expected, Alien is recruited hormone sensitively. One explanation for NAP1 recruitment could be that NAP1 is chromatin associated to augment activation in the presence of hormone potentially through conjunction with p300/CBP, whereas it enhances repression in the absence of hormone (hypothetical model depicted in Fig. 7). We hypothesize that the mode of repression by NAP1 is mediated through the depositing of nucleosomes on DNA in a replication-independent manner, leading to compaction and to an inaccessible chromatin template. Our data suggest that Alien reinforces that process by supporting nucleosome assembly. This may be achieved by binding of Alien to NAP1 to enhance its activity. On the other hand, we find that Alien binds specifically to the histones H3 and H4 and inhibits NAP-1 activity, and the functional tests show that additional amounts of histones H3-H4 rescue nucleosome assembly activity. NAP1 was originally reported to bind to histones H2A-H2B in cytosolic or crude whole-cell extracts in vivo (7, 13), which, however, may not exclude the binding of NAP1 to the histones H3 and H4 in chromatin. Consistently with other publications, NAP1 not only binds to the histones H2A-H2B but also interacts with the core histones H3 and H4 (Fig. 4E) (16, 21, 22). Notably, Alien interacts with NAP1 in a region that is suggested to be involved in histone interaction (27), which could imply that the interaction between histone and NAP is compromised in the presence of Alien. Therefore, one likely mechanism is that Alien blocks the nucleosome assembly function by binding to the histones H3 and H4 to prevent their accessibility for NAP1. Interestingly, it was also shown that in addition to nucleosome assembly another NAP1-regulated activity is the release of histones H3-H4, which is, however, dependent on the presence of the histones H2A-H2B (16). It is therefore hypothetically possible that Alien, on one hand, promotes nucleosome assembly leading to a more compacted chromatin structure and, on the other hand, shifts the steady state toward prevention of histone displacement mediated by NAP1. Thereby, Alien also drives the equilibrium toward a more repressed chromatin state.
FIG. 7.
Hypothetical model of Alien-mediated modulation of nucleosome assembly. According to previously published data, in the presence of hormone, VDR associates with coactivators, such as p300/CBP. Since it was previously shown that NAP1 enhances p300/CBP-mediated gene activation, we hypothesize that NAP1 recruitment also occurs in the presence of ligand. This enhancement could be achieved by histone displacement. Removal of hormone leads to dissociation of coactivators from VDR. The unliganded VDR associates with Alien, which then enhances the activity of NAP1-mediated nucleosome assembly. After deposition Alien inhibits the accessibility of the histones H3 and H4 for NAP1, thus preventing histone displacement and driving the equilibrium toward a more repressed chromatin state.
These results indicate that Alien increases nucleosome formation by a poorly understood mechanism that may include the following activities, in part or in combination: (i) augmenting NAP1-mediated histone deposition through direct interactions with NAP1 and histones H3 and H4 during assembly, (ii) stabilizing nucleosomes postassembly through direct interactions with NAP1 and histones H3 and H4, or (iii) stabilizing nucleosomes postassembly by occluding NAP1 directly or indirectly.
Nucleosome deposition and chromatin compaction are known to lead to a repressed state (4, 33, 36). Notably, an intrinsic transcriptional repression function was observed when NAP1 was tethered to DNA. In line with this observation, very recently findings revealed that NAP1 is associated with the heterochromatic protein 2, HP2, in Drosophila, suggesting that NAP1 is in part associated with heterochromatin (35). Whether this is a direct cause through nucleosome deposition or is mediated by other interacting proteins has to be investigated.
Another level of complexity of NAP1 function arises from reports that NAP1 can be posttranslationally modified, which was suggested to regulate its assembly activity (30). Hence, one could envisage that these modifications might regulate the association of specific proteins such as Alien.
NAP1 is a ubiquitous and abundant nuclear protein and may be spread throughout chromatin in association with nucleosomes. It is conceivable that the NAP1 function must be controlled in an ordered manner at specific chromatin sites to regulate expression of individual genes in a specific manner. It could be envisaged that the association of NAP1 with coregulators—either coactivators or corepressors—might dictate its specific function. Here, we provide such a link between nuclear hormone receptor corepressors and nucleosome assembly activity.
Acknowledgments
We are grateful to M. Beato for providing baculovirus-expressed NAP1, C. Englert for the mouse cDNA library, and J. T. Kadonaga for dNAP1 cDNA. We thank M. Lutz and J. Boeke for critically reading the manuscript.
This work was supported by the DFG grant BA 1457/3.
Footnotes
Published ahead of print on 5 March 2007.
REFERENCES
- 1.Adkins, M. W., S. R. Howar, and J. K. Tyler. 2004. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14:657-666. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad, K., and S. Henikoff. 2002. Epigenetic consequences of nucleosome dynamics. Cell 111:281-284. [DOI] [PubMed] [Google Scholar]
- 3.Asahara, H., S. Tartare-Deckert, T. Nakagawa, T. Ikehara, F. Hirose, T. Hunter, T. Ito, and M. Montminy. 2002. Dual roles of p300 in chromatin assembly and transcriptional activation in cooperation with nucleosome assembly protein 1 in vitro. Mol. Cell. Biol. 22:2974-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeger, H., D. A. Bushnell, R. Davis, J. Griesenbeck, Y. Lorch, J. S. Strattan, K. D. Westover, and R. D. Kornberg. 2005. Structural basis of eukaryotic gene transcription. FEBS Lett. 579:899-903. [DOI] [PubMed] [Google Scholar]
- 5.Cairns, B. R. 2005. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 15:185-190. [DOI] [PubMed] [Google Scholar]
- 6.Chamovitz, D. A., and D. Segal. 2001. JAB1/CSN5 and the COP9 signalosome. A complex situation. EMBO Rep. 2:96-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, L., S. S. Loranger, C. Mizzen, S. G. Ernst, C. D. Allis, and A. T. Annunziato. 1997. Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry 36:469-480. [DOI] [PubMed] [Google Scholar]
- 8.Chen, X., J. Wang, D. Woltring, S. Gerondakis, and M. F. Shannon. 2005. Histone dynamics on the interleukin-2 gene in response to T-cell activation. Mol. Cell. Biol. 25:3209-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cope, G. A., and R. J. Deshaies. 2003. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell 114:663-671. [DOI] [PubMed] [Google Scholar]
- 10.Dressel, U., D. Thormeyer, B. Altincicek, A. Paululat, M. Eggert, S. Schneider, S. P. Tenbaum, R. Renkawitz, and A. Baniahmad. 1999. Alien, a highly conserved protein with characteristics of a corepressor for members of the nuclear hormone receptor superfamily. Mol. Cell. Biol. 19:3383-3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Eckey, M., S. P. Tenbaum, and A. Baniahmad. 2003. Mixed lineage kinase 2 enhances trans-repression of Alien and nuclear receptors. Mol. Cell. Endocrinol. 213:71-78. [DOI] [PubMed] [Google Scholar]
- 11.Fujii-Nakata, T., Y. Ishimi, A. Okuda, and A. Kikuchi. 1992. Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. J. Biol. Chem. 267:20980-20986. [PubMed] [Google Scholar]
- 12.Fyodorov, D. V., M. D. Blower, G. H. Karpen, and J. T. Kadonaga. 2004. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 18:170-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito, T., M. Bulger, R. Kobayashi, and J. T. Kadonaga. 1996. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 16:3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamakaka, R. T., and S. Biggins. 2005. Histone variants: deviants? Genes Dev. 19:295-310. [DOI] [PubMed] [Google Scholar]
- 15.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36:900-905. [DOI] [PubMed] [Google Scholar]
- 16.Levchenko, V., and V. Jackson. 2004. Histone release during transcription: NAP1 forms a complex with H2A and H2B and facilitates a topologically dependent release of H3 and H4 from the nucleosome. Biochemistry 43:2359-2372. [DOI] [PubMed] [Google Scholar]
- 17.Lorch, Y., B. Maier-Davis, and R. D. Kornberg. 2006. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl. Acad. Sci. USA 103:3090-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 19.Macfarlan, T., S. Kutney, B. Altman, R. Montross, J. Yu, and D. Chakravarti. 2005. Human THAP7 is a chromatin-associated, histone tail-binding protein that represses transcription via recruitment of HDAC3 and nuclear hormone receptor corepressor. J. Biol. Chem. 280:7346-7358. [DOI] [PubMed] [Google Scholar]
- 20.Margueron, R., P. Trojer, and D. Reinberg. 2005. The key to development: interpreting the histone code? Curr. Opin. Genet. Dev. 15:163-176. [DOI] [PubMed] [Google Scholar]
- 21.Mazurkiewicz, J., J. F. Kepert, and K. Rippe. 2006. On the mechanism of nucleosome assembly by histone chaperone NAP1. J. Biol. Chem. 281:16462-16472. [DOI] [PubMed] [Google Scholar]
- 22.McBryant, S. J., Y. J. Park, S. M. Abernathy, P. J. Laybourn, J. K. Nyborg, and K. Luger. 2003. Preferential binding of the histone (H3-H4)2 tetramer by NAP1 is mediated by the amino-terminal histone tails. J. Biol. Chem. 278:44574-44583. [DOI] [PubMed] [Google Scholar]
- 23.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 24.Moehren, U., U. Dressel, C. A. Reeb, S. Vaisanen, T. W. Dunlop, C. Carlberg, and A. Baniahmad. 2004. The highly conserved region of the co-repressor Sin3A functionally interacts with the co-repressor Alien. Nucleic Acids Res. 32:2995-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohkuni, K., K. Shirahige, and A. Kikuchi. 2003. Genome-wide expression analysis of NAP1 in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 306:5-9. [DOI] [PubMed] [Google Scholar]
- 26.Park, Y. J., J. V. Chodaparambil, Y. Bao, S. J. McBryant, and K. Luger. 2005. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J. Biol. Chem. 280:1817-1825. [DOI] [PubMed] [Google Scholar]
- 27.Park, Y. J., and K. Luger. 2006. The structure of nucleosome assembly protein 1. Proc. Natl. Acad. Sci. USA 103:1248-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polly, P., M. Herdick, U. Moehren, A. Baniahmad, T. Heinzel, and C. Carlberg. 2000. VDR-Alien: a novel, DNA-selective vitamin D(3) receptor-corepressor partnership. FASEB J. 14:1455-1463. [DOI] [PubMed] [Google Scholar]
- 29.Ray-Gallet, D., J. P. Quivy, C. Scamps, E. M. Martini, M. Lipinski, and G. Almouzni. 2002. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 9:1091-1100. [DOI] [PubMed] [Google Scholar]
- 30.Regnard, C., E. Desbruyeres, J. C. Huet, C. Beauvallet, J. C. Pernollet, and B. Edde. 2000. Polyglutamylation of nucleosome assembly proteins. J. Biol. Chem. 275:15969-15976. [DOI] [PubMed] [Google Scholar]
- 31.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 32.Shikama, N., H. M. Chan, M. Krstic-Demonacos, L. Smith, C. W. Lee, W. Cairns, and N. B. La Thangue. 2000. Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell. Biol. 20:8933-8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sims, R. J., III, S. S. Mandal, and D. Reinberg. 2004. Recent highlights of RNA-polymerase-II-mediated transcription. Curr. Opin. Cell Biol. 16:263-271. [DOI] [PubMed] [Google Scholar]
- 34.Sirover, M. A. 2005. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J. Cell Biochem. 95:45-52. [DOI] [PubMed] [Google Scholar]
- 35.Stephens, G. E., H. Xiao, D. H. Lankenau, C. Wu, and S. C. Elgin. 2006. Heterochromatin protein 2 interacts with Nap-1 and NURF: a link between heterochromatin-induced gene silencing and the chromatin remodeling machinery in Drosophila. Biochemistry 45:14990-14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svejstrup, J. Q. 2003. Transcription. Histones face the FACT. Science 301:1053-1055. [DOI] [PubMed] [Google Scholar]
- 37.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 38.Väisanen, S., T. W. Dunlop, C. Frank, and C. Carlberg. 2004. Using chromatin immunoprecipitation to monitor 1α,25-dihydroxyvitamin D3-dependent chromatin activity on the human CYP24 promoter. J. Steroid Biochem. Mol. Biol. 89-90:277-279. [DOI] [PubMed] [Google Scholar]
- 39.Väisanen, S., T. W. Dunlop, L. Sinkkonen, C. Frank, and C. Carlberg. 2005. Spatio-temporal activation of chromatin on the human CYP24 gene promoter in the presence of 1α,25-dihydroxyvitamin D3. J. Mol. Biol. 350:65-77. [DOI] [PubMed] [Google Scholar]
- 40.Walter, P. P., T. A. Owen-Hughes, J. Cote, and J. L. Workman. 1995. Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol. Cell. Biol. 15:6178-6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue, Y., J. Wong, G. T. Moreno, M. K. Young, J. Cote, and W. Wang. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2:851-861. [DOI] [PubMed] [Google Scholar]