Abstract
Plants can defend themselves from herbivorous insects by emitting volatile chemical signals that attract natural enemies of the herbivore. For example, maize seedlings attacked by beet armyworm larvae (Spodoptera exigua) produce a mixture of terpenoid and indole volatiles that serve to attract parasitic wasps. A key step in terpenoid biosynthesis is the conversion of acyclic prenyl diphosphates to terpenoid compounds by specific terpenoid synthases (cyclases). We have cloned a maize sesquiterpene cyclase gene, stc1, by transposon tagging and have identified two deletion mutations of the gene. The stc1 gene is located on chromosome 9S and does not seem to have a closely related ortholog in the maize genome. It is induced 15- to 30-fold in maize leaves by beet armyworm larvae feeding or by application of purified volicitin, the insect-derived elicitor, at a mechanically wounded site. stc1 induction is systemic, because undamaged leaves of the same plant show a similar increase in stc1 transcription. Analysis of volatiles from volicitin-treated seedlings revealed that a major naphthalene-based sesquiterpene was present in wild-type seedlings but absent in the Ac-insertion and x-ray-deletion mutants. Therefore, we have identified a maize gene that responds to caterpillar herbivory by producing a chemical defense signal that most likely serves to attract natural enemies of the herbivore.
Recent studies have demonstrated that plant volatiles released in response to insect damage serve as chemical cues that attract natural enemies of the herbivore. This plant-mediated parasitoid–host interaction has been termed the tritrophic interaction. When maize is attacked by beet armyworm (BAW) larvae, both the damaged and undamaged leaves of the same plants emit a bouquet of volatiles that selectively attract parasitoids of the caterpillar. Female parasitic wasps deposit their eggs in the caterpillars, which are eventually devoured by the emerging wasp larvae (1–3). A similar exploitation of volatile chemicals has been reported in other plant species (4–8). Interestingly, terpenoids are regularly found among the herbivore-induced plant volatiles of different plant species (8, 9).
Plant leaves also release volatile terpenoids when they are mechanically wounded and treated with oral secretions of the herbivore (1, 5), indicating that an elicitor of plant volatiles is present in the insect's saliva. Different types of elicitors have been found. For example, a β-glucosidase from the regurgitant of Pieris brassicae (cabbage-white butterfly) larvae cleaves terpenoids stored as β-glucosides in the cabbage plant (10). In contrast, a small compound called volicitin [N-(17 hydroxylinolenoyl)-l-glutamine] from the regurgitant of Spodoptera exigua (BAW) larvae causes systemic release of a blend of volatile terpenoids when applied to mechanically damaged corn seedlings (11).
How the insect-derived elicitor triggers plant defense genes to produce volatile terpenoids is unclear. Studies in cotton and maize have uncovered important aspects of the type of gene expression that may be involved in this terpenoid production. In cotton, insect injury results in de novo synthesis of terpenoids (12), suggesting that genes for terpenoid biosynthesis are induced. In maize seedlings, the release of terpenoids occurs transiently between 6–18 h after insect damage (13), suggesting that the genes for terpenoid biosynthesis may be transcriptionally regulated. Insect damage leads to the production of a systemic signal, because undamaged leaves of maize seedlings release the same blend of monoterpenes and sesquiterpenes as insect-damaged leaves (2). These observations suggest that terpenoid biosynthesis is a major target of regulation.
Biochemical analysis has defined hydroxymethyl glutaryl (HMG)-CoA reductase and terpenoid synthase as two key regulatory enzymes of terpenoid biosynthesis. HMG-CoA reductase catalyzes the committed reaction of the general terpenoid synthesis pathway. Terpenoid synthases catalyze the divergent reactions that produce the vast variety of terpenoids seen in plants. In several plant species, terpenoid synthase genes are induced by fungal elicitors (14, 15), leading to the generation of antimicrobial sesquiterpene phytoalexins. Insect damage also results in the up-regulation of genes for these enzymes. For example, potato leaves accumulate HMG-CoA reductase mRNA more rapidly after application of an insect regurgitant to a mechanically wounded site than after mechanical wounding alone (16). In grand fir, both wounding and insect damage induce terpenoid biosynthesis (17), most likely through induction of terpenoid synthase (18).
Plant defense terpenoids are secondary metabolites, and the corresponding synthases are likely to be conserved enzymes encoded by families of genes (19). To date, more than 30 cDNA clones for terpenoid synthase genes have been isolated (19), including some that may function in the formation of volatile defense signal chemicals (20). Because mutations in these genes result in altered chemotypes that are normally difficult to detect, no knockout mutants for these candidate genes have been identified. As a result, genetic studies of plant defense against insects have been limited to analyses of quantitative trait loci among natural plant populations (21). Therefore, elucidating the roles of individual terpenoid signal chemicals in the tritrophic interaction remains a challenge.
We report here the isolation of a maize sesquiterpene cyclase gene, stc1, and the characterization of wild-type and mutant alleles. We present evidence that stc1 can be induced by insect injury, insect oral secretion, or purified volicitin and have characterized these patterns of induction. Through the analysis of gene knockout mutations in an isogenic background, we have identified the volatile terpenoid product of the gene.
Materials and Methods
Plant Genetic Stocks.
All of the genetic stocks used in this study were in a W22 isogenic background. The Stc1-McC wild-type allele was introgressed into W22 together with bronze Bz-McC, an allele of the bz locus with which it is closely linked. It is the normal progenitor allele of the Ac insertion mutations used in this study. stc1-m6087 is an insertion mutation generated by Ac transposition from the nearby bz donor site. It is linked to the Dissociation reporter allele bz-m2(DI), which monitors Ac activity, and was referred to earlier as simply Ac6087 (22). sh-bz-X2 and sh-bz-X3 are x-ray-induced deletions of a large chromosome segment that includes the sh and bz loci (23). As shown here, they are also deleted for the stc1 locus, which is located between sh and bz in 9S. Both deletions show reduced pollen transmission but are homozygous viable.
DNA and RNA Extraction, Probe Hybridization, and DNA Sequencing.
Leaf DNA was extracted by a urea-extraction procedure (24). Total RNA was extracted by using TRIzol reagent according to the manufacturer's protocol (Life Technologies, Rockville, MD). DNA (10 μg) digested with restriction enzymes and total RNA (20 μg) were resolved on a 0.8% and a 1.2% agarose gel, respectively, and then transferred to Zeta-probe membrane (25). 32P-labeled probes were generated with Ready-To-Go DNA labeling beads (Amersham Pharmacia). The probes used in this study were STC-323, corresponding to the distalmost 2.5-kb KpnI–BamHI fragment from a Bz-McC λ genomic clone (26, 27) and a 1.5-kb PstI internal fragment from an stc1 cDNA clone. The hybridization and membrane wash followed the procedure of Church and Gilbert (28). DNA sequencing was carried out as described (29).
cDNA Cloning, Reverse Transcription–PCR (RT-PCR), Primer Extension, and Rapid Amplification of cDNA Ends (RACE).
A cDNA library was constructed from poly(A) RNA extracted from wild-type juvenile leaves (6–8 weeks), following essentially the instructions from Stratagene. RT-PCR was performed by using the Titan One Tube kit from Hoffmann–La Roche. PCR primers used in this study were as follows: 5′ primers, TCCATCTGGGGCGATTTCTTCCTC and GGAATGGTCTAGAGCTTCAGTCAAG; 3′ primers, ACACTCGGTAAATAACTCGCACC and CCTGTATCTTCACGCGAGCCACTTC. RT-PCR products were cloned into the pGEMT vector (Promega). Primer extension was carried out as described (30) by using a primer with the sequence CATGACAACTGGTGGTGGCGG. RACE analysis followed the manufacturer's instructions (CLONTECH). Total RNA from wild-type juvenile leaves was used as a template for RT-PCR, primer extension, and RACE.
Volatile Collection and GC-MS Analysis.
The collection apparatus for volatile compounds, which used a push/pull technique (13), was located in a growth chamber (33°C; 50% humidity) supplemented with artificial light. In the time course experiment, three 10-day-old seedlings were cut above the root at 0600, immediately immersed in a volicitin solution (5 μl of volicitin in 500 μl of 50 mM phosphate buffer, pH 8.3; ref. 11) and kept in complete darkness. Filtered air was passed from the top down over the plants and out of the glass sleeve through a column with SuperQ absorbent (Alltech Associates) traps. Volatile compounds were collected every 3 h beginning at 1200 and ending at 2400. In a second set of experiments, three 10-day-old seedlings each of wild type, stc1-m6087, sh-bz-X2, and sh-bz-X3 were treated with volicitin at 2200 (day 1), as described above. Volatile compounds were collected at 1000 (day 2) for 3 h. Volatile collection and analysis were also conducted from seedlings that had 5 μl of volicitin or regurgitant applied to razor blade wounds on their second leaf.
The samples were injected into a Hewlett–Packard model 5890 GC by using a 5:1 split ratio at 250°C. The OV 101 methyl silicon column (Quadrex, New Haven, CT) was held at 40°C for 5 min and then programmed to 240°C in 103 min. The structure of the unique volatile component in the wild-type seedlings was determined by MS analysis. All experiments were conducted in duplicate or triplicate.
Results
Identification of a Maize Sesquiterpene Cyclase Gene.
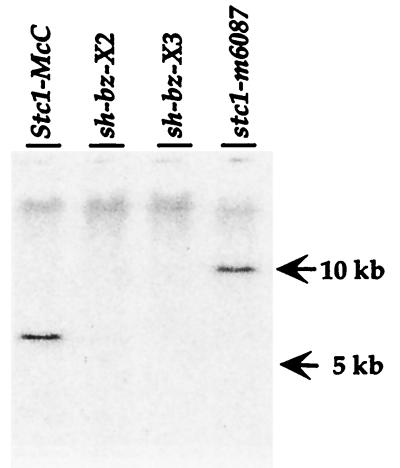
The transposable element Ac tends to transpose to nearby chromosome locations (22, 31). Ac6087 is a transposed Ac from the mutable allele bz-m2(Ac) that reinserted closely distal to the bz locus in 9S (22). Hybridization of a 2.5-kb fragment from the centromere-distal end of a 16-kb Bz-McC lambda clone (26, 27) to DNA from Ac6087 digested with several enzymes revealed a band that was 4.6 kb larger than that of wild type (Fig. 1), confirming that Ac had transposed to a nearby location. The band was absent in the deletion mutations sh-bz-X2 and sh-bz-X3, corroborating that the probe detects a single copy sequence that lies adjacent to the bz locus in the maize genome. By using a combination of restriction enzymes (EcoRI and XhoI) that cut within Ac, we determined by Southern blot analysis that Ac6087 was inserted with its 5′ end toward the telomere (data not shown).
Figure 1.
Southern blot analysis of wild type and mutants. Genomic DNA (10 μg) from wild type (Stc1-McC), the deletion mutations sh-bz-X2 and sh-bz-X3, and the Ac insertion mutation stc1-m6087 was digested with SstI and hybridized with an stc1 probe. The arrows on the right indicate the position of DNA size markers in the same gel.
The portion of the lambda genomic clone flanking the Ac6087 insertion site was subcloned into the pBluescript KS(+) vector and sequenced. A search with this sequence against the GenBank database revealed extensive amino acid sequence homology to sesquiterpenoid cyclase genes from other plants. Therefore, we designated the gene stc1, for the first sesquiterpenoid cyclase gene in maize. An almost full-length cDNA for stc1 was assembled from a leaf cDNA clone and various RT-PCR products. The genomic DNA sequence was used as a template to design oligonucleotide RT-PCR primers corresponding to the conserved domains of the terpenoid cyclases. The transcription start site (+1) was determined by primer extension and verified by RT-PCR with an upstream oligonucleotide primer based on the genomic sequence spanning the start site. The 3′ end of the gene was determined by 3′ RACE analysis and confirmed by sequencing three independent cDNA clones. Introns and exons were defined by comparing the nucleotide sequences of the Stc1-McC genomic DNA and cDNA clones. The putative translation start site was predicted assuming that the first in-frame ATG in an appropriate sequence context is used in vivo. The Stc1-McC mRNA has a 198-bp 5′ untranslated leader and a 52-bp 3′ untranslated tail. It is produced by the splicing of six introns, all of which have the conserved GT and AG dinucleotides at their respective 5′ and 3′ ends. The direction of transcription of stc1 in 9S is opposite to that of bz, the distance between the nearest poly(A) addition sites in Stc1-McC and Bz-McC (26) being just 1.3 kb.
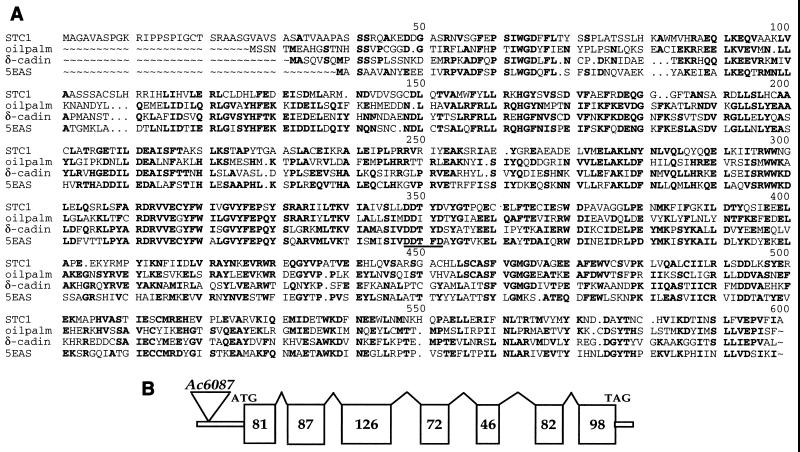
A comparison of the translated STC1 peptide sequence with other terpenoid cyclases from the database (including cyclases for monoterpenes, sesquiterpenes, and diterpenes) revealed similarities of 30–40%, the highest being to sesquiterpene cyclases. In addition, all of the key conserved amino acids common among the terpenoid cyclases are conserved in STC1, including the absolutely conserved aspartate-rich motif DDXXD (Fig. 2A). Another feature of the stc1 gene is that the sizes of its exons, except the first, are conserved relative to those of other terpenoid cyclase genes (Fig. 2B; ref. 32).
Figure 2.
(A) Amino acid sequence alignment between the maize STC1 protein and sesquiterpene cyclases from oil palm (GenBank accession no. AAC31570), Gossypium arboreum [δ-cadinene synthase (δ-cadin), Swissprot Q39760], and Nicotiana tabaccum [5-epiaristolochene synthase (5EAS), Swissprot Q40577]. The sequences were aligned with the GCG program PILEUP. A residue that is highly conserved appears in bold. Gaps are represented by dots. Amino acids corresponding to the conserved DDXXD motif are underlined. (B) stc1 gene structure. Exons are represented as open boxes, the numbers referring to the approximate number of amino acids in each exon. The 5′ and 3′ untranslated regions (UTRs) are shown as double lines, and the Ac6087 insertion is shown as an inverted triangle in the 5′ UTR. The stc1 genomic sequence has been deposited in GenBank (accession no. AF296123).
The junction sequence of the Ac6087 insertion site was resolved by sequencing a PCR product from Ac6087 genomic DNA amplified with Ac- and stc1- based primers. Ac6087 created an 8-bp direct repeat of bases 102–109 in the 5′ UTR of the stc1 gene and was inserted in the same direction of transcription as the stc1 gene (Fig. 2B). We have designated the new insertion allele stc1-m6087.
stc1 Is Developmentally Regulated in Maize Leaves.
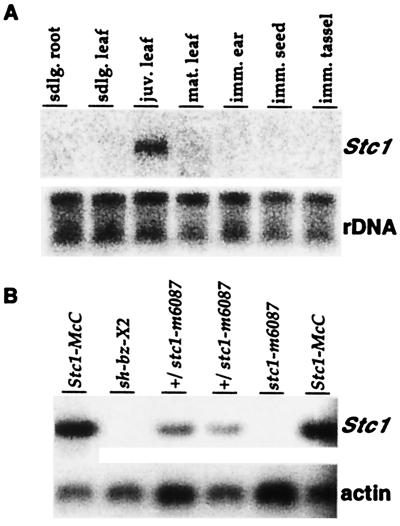
The expression pattern of stc1 was examined by Northern blot analysis. Wild-type maize was grown in a growth chamber, and total RNA was prepared from 10-day-old seedling roots, leaves, and sheaths; juvenile plant leaves (6 weeks); mature plant leaves (10 weeks); immature ears (3–6 cm); immature tassels (2–3 days preemergence); and immature seed (14 days after pollination). As can be seen in Fig. 3A, stc1 is expressed exclusively in juvenile leaves. Similar results were obtained in two other experiments. Field-grown maize plants also express stc1 in juvenile leaves, but at a higher level (data not shown).
Figure 3.
Northern blot analysis of the stc1 transcript. (A) Expression of the stc1 gene in different wild-type tissues: 20 μg of total RNA from each tissue were resolved in a 1.2% agarose gel and hybridized sequentially with an stc1 probe and with a genomic rDNA probe from tobacco. (B) Expression of stc1 in wild-type and mutant leaves: 1 μg of juvenile leaf mRNA from several different genotypes was resolved in a 1.2% agarose gel and hybridized with an stc1 probe and with a maize actin cDNA probe. The following genotypes were compared: wild type (Stc1-McC), the deletion mutation sh-bz-X2, and the Ac insertion mutation stc1-m6087 in homozygous and heterozygous condition.
stc1 transcript levels in juvenile leaves of the stc1-m6087 insertion mutant (Fig. 3B) and the sh-bz-X2 and sh-bz-X3 deletion mutants (data not shown) were analyzed by Northern blotting. No transcript was detected in the deletion mutations, as expected, or in the Ac insertion mutation, which indicates that the Ac insertion in the 5′ UTR of the stc1 gene terminates stc1 transcription. Because of the deletion of a large chromosome fragment that measures at least 2 centimorgans and includes the region between sh and bz, sh-bz-X2 and sh-bz-X3 differ from wild type in seed morphology, plant stature, and pollen transmission (23). However, stc1-m6087 is phenotypically indistinguishable from wild type.
Wounding and Insect Injury Induces the Expression of the stc1 Gene in Maize.
Several lines of evidence point to the possibility that the stc1 gene might be involved in plant defense. First, plants are known to use sesquiterpenes as defense chemicals (32). Second, insect-damaged corn seedlings release volatile monoterpenes and sesquiterpenes that attract parasitoids that attack the herbivorous insects (1). Third, field-grown plants express stc1 at a higher level than chamber-grown plants (data not shown), suggesting that stc1 might be responsive to environmental stresses.
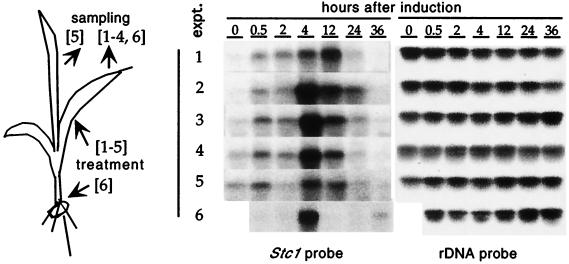
Therefore, we tested whether stc1 transcript levels in corn seedlings changed in response to herbivore damage. As can be seen in Fig. 4, mechanical wounding alone induced stc1 gene expression in seedlings; this induction reached a 30-fold maximum 12 h after treatment. However, induction of stc1 transcription by herbivore damage or various treatments resembling it was much stronger (5-fold higher) and earlier (peaking at 4 h vs. 12 h) than by mechanical damage alone. Induction was consistent among all treatments, which included placing starved insects on the leaf, applying either insect oral secretion or purified volicitin to artificially damaged sites on the leaf, and immersing the base of a seedling cut immediately above the root into purified volicitin. In all treatments, systemic induction of stc1 was evident in the upper and lower undamaged leaves of the damaged plant, although it peaked later (12 h) than in the damaged leaves. Finally, stc1 transcript was almost undetectable in untreated seedlings or in treated seedlings 24 h after induction. The above experiment was replicated three times, with similar results.
Figure 4.
Induction of the stc1 transcript. For treatments 1–5, the second leaf of 10-day-old wild-type seedlings was wounded artificially (treatment 1), fed upon by BAW larvae (treatment 2), wounded and treated with 5 μl of BAW oral regurgitant (treatment 3), or wounded and treated with 5 μl of purified volicitin (treatments 4 and 5). For treatment 6, the seedling was cut just above the root, and the stem was immersed immediately in a volicitin solution (treatment 6). The treated leaf (second leaf for treatments 1–5) or untreated leaf (third leaf for treatment 5; second leaf for treatment 6) was harvested after induction for 0, 0.5, 2, 4, 12, 24, and 36 h in the dark. One seedling was used per treatment and time point except for damage by BAW (three repeats with the same result). All samples were frozen in liquid N2 and stored at −80°C. Total RNA (20 μg) was resolved on a 1.2% agarose gel and hybridized first to an stc1 probe and then to a maize 23S rDNA probe.
In summary, our results show that (i) insect damage or an insect elicitor causes an elevated and rapid induction of the stc1 transcript in maize; (ii) the pattern of induction is independent of the type of damage but dependent on the presence of the elicitor; and (iii) the stc1 transcript is induced transiently and systemically.
The stc1 Mutants Lack a Sesquiterpene Derived from Naphthalene.
Terpenoids are synthesized from the intermediates of the central isoprenoid biosynthesis pathway by a series of chemical reactions that include cyclization and other modifications. Cyclization is the first committed reaction and is catalyzed by a terpenoid cyclase. Cyclases are responsible for the formation of an extremely diverse group of isoprenoid compounds and may be the target for regulating the production of these compounds.
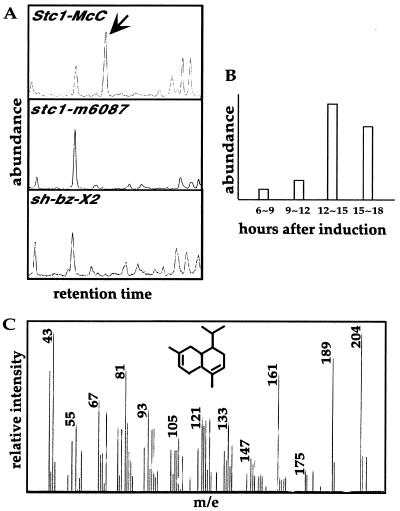
Because the stc1 null mutations (stc1-m6087, sh-bz-X2, and sh-bz-X3) accumulate no stc1 gene transcripts, we anticipated that one or more terpenoid compounds would be missing in the mutants relative to wild type. After volicitin treatment, volatile compounds were collected from wild type and the nearly isogenic null mutants and separated by GC. The GC profiles thus generated were compared to determine whether any peaks were missing in the mutants. Fig. 5A shows the only region of the GC spectrum that differed between the mutants and wild type (sh-bz-X3 gave the same results as sh-bz-X2; data not shown). As can be seen, a peak corresponding to a major volatile compound in wild type is missing in all of the null mutants. Production of the stc1-specific compound in wild-type seedlings was monitored for 16 h after volicitin treatment. The compound was detectable 6 h after treatment (Fig. 5B). It peaked after 12 h, i.e. about 8 h later than the stc1 transcript.
Figure 5.
GC profiles of volicitin-induced volatiles in various genotypes. (A) At 10-days-old, seedlings were cut above the root, immediately immersed in a volicitin solution, and kept in complete darkness for 12 h. Volatiles were collected from treated seedlings in a special chamber provided with artificial light and purified air. GC profiles are for volatiles from wild type (Stc1-McC), the Ac insertion mutation stc1-m6087, and the deletion mutation sh-bz-X2. The unique peak in wild type is indicated by an arrow. This experiment was repeated three times with the same result. (B) Time course production of the Stc1-McC-specific compound from wild-type seedlings. The induction treatment was the same as above. (C) MS identification of the Stc1-McC-specific compound. The unique peak in the Stc1-McC GC was analyzed by MS. The MS spectrum of the Stc1-McC-specific compound matched that of the sesquiterpenoid naphthalene, 1,2,4a,5,8,8a–hexahydro-4,7-dimethyl-1-(1-methylethyl)-,(1α,4aβ,8aα) from the chemical database (Inset).
The structure of this compound was resolved by MS. The MS profile of the stc1-specific compound, which is shown in Fig. 5C, matched that of the sesquiterpene naphthalene, 1,2,4a,5,8,8a-hexahydro-4,7-dimethyl-1-(1-methylethyl)-(1α,4aβ,8aα).
Discussion
Characterization of Wild-Type and Mutant stc1 Alleles.
Ac6087 is one of several transpositions of Ac from the mutable allele bz-m2(Ac) that map very close to the bz donor locus (22). As part of a project designed to characterize the nature of Ac insertion sites in the bz genomic region, we isolated the sequence flanking Ac in transposed Ac6087 (tac6087) and compared it to the GenBank sequence database. The tac6087 sequence had significant similarity to terpenoid synthase genes from other organisms, the highest being to the sesquiterpene cyclase gene from oil palm (GenBank accession no. AAC31570), the only monocot sesquiterpene cyclase in the database at the time. Other features of the tac6087 sequence indicate that it is a sesquiterpene cyclase gene. Its transcript is the spliced product of seven exons that are conserved in number and size relative to other sesquiterpene cyclase genes. Its translated peptide sequence has the key amino acid motif DDXXD, characteristic of all sesquiterpene cyclases (33). Because this gene is the first sesquiterpene cyclase gene reported in maize, we have designated it stc1.
The STC1 conceptual protein has an N-terminal signal peptide that most likely targets the protein to the chloroplast. This peptide is about 40 amino acids long, is rich in serine and other hydrophobic amino acids, and has the conserved cleavage site motif of other chloroplast transit peptides (34). Sesquiterpenoids were believed to be synthesized only in the cytoplasm, because sesquiterpene synthases lacked N-terminal signal peptides. However, it was discovered recently that higher plants are capable of generating isopentenyl diphosphate, the precursor of all terpenoids, by alternative pathways in the cytosolic and plastidial compartments (35). Several sesquiterpenes are reportedly synthesized in the cytoplasm from deoxyxylose 5-phosphate, a chloroplast-made precursor (36). The fact that STC1 possesses a signal peptide suggests that the chloroplast may be capable of synthesizing sesquiterpenes as well.
We have characterized four mutant alleles of stc1. These include the Ac insertion mutant alleles stc1-m6087 and stc1-m6067 and the deletion mutant alleles sh-bz-X2 and sh-bz-X3 (23). In stc1-m6087, Ac is inserted in the 5′ UTR of the gene in the same direction of transcription as stc1. This orientation of Ac should cause RNA transcripts from stc1-m6087 to terminate prematurely within Ac (37). As expected, no stc1 transcript was detected in this mutant or in the sh-bz-X2 and sh-bz-X3 mutants, which are deleted for the stc1 gene. The deletions of the entire stc1 gene enabled us to confirm that the band hybridizing to the stc1 probe in DNA and RNA blots is stc1-specific, that stc1 is a single copy gene in maize, and that all three mutants are stc1 null mutants. In addition to stc1-m6087, we have identified a second null allele produced by transposition of Ac into the stc1 gene. In this allele, stc1-m6067, Ac is inserted in the first exon in opposite transcriptional orientation relative to stc1. Although these two Ac insertion lines are indistinguishable phenotypically from wild type, they are both stc1 knockouts. As in stc1-m6087, no stc1 transcript is detected in either the juvenile leaves or the volicitin-treated seedling leaves of stc1-m6067 plants. An examination of the maize expressed sequence tag database (http://gremlin3.zool.iastate.edu/cgi-bin/nph-blast/ZMDB) reveals that there are other genes in maize related to stc1, but none show more than 67% identity with stc1 at the nucleotide level.
Although stc1-m6087 and stc1-m6067 are null mutations, homozygous plants did not show an obvious mutant phenotype. This result is not surprising, because stc1 is not normally expressed in maize seedlings and its sesquiterpene product is a nonessential secondary metabolite. Volatile sesquiterpenes produced by insect-damaged maize seedlings presumably serve as chemical signals that attract natural enemies of the herbivores (1). If stc1 is a defense gene involved in the production of these chemiosignals, it should be induced by insect damage and insect elicitor and its product should be a volatile sesquiterpene. As discussed below, our data confirmed both expectations.
Several features of the observed induction of stc1 suggest a role for the gene in defense against herbivorous insect larvae. First, the elicitor of stc1 induction is volicitin, a compound produced in the larvae's salivary glands. The same pattern of stc1 transcript induction was observed when 10-day-old wild-type maize seedlings were damaged by BAW larvae, treated with insect regurgitant, or treated with purified volicitin. Second, the induction of stc1 is transient. Transcript levels became detectable 1 h after treatment, peaked after 4 h, and were no longer detectable at 24 h. Third, the induction of stc1 is systemic. The stc1 transcript can be detected in the upper and lower undamaged leaves of the plants that have been damaged elsewhere. Fourth, stc1 transcription can be induced by mechanical wounding alone, albeit at a much weaker level and a later time than when wounding is accompanied by volicitin treatment.
To determine the nature of the stc1-specific compound in maize, the volatiles emitted from volicitin-treated wild-type and mutant seedlings were analyzed by GC and MS (Fig. 5; data not shown for stc1-m6067). A comparison of the GC profiles of wild-type and mutant seedlings allowed the identification of a major peak that was present in the former but absent in the latter. Therefore, this peak corresponds to the stc1-specific compound. Release of this compound peaked around 12 h after induction. The MS profile of the stc1-specific compound shows that it is most likely the sesquiterpene naphthalene, 1,2,4a,5,8,8a–hexahydro-4,7-dimethyl-1-(1-methylethyl)-(1α,4aβ,8aα). The time course of release of the stc1 sesquiterpenoid was similar to that of maize terpenoids previously reported to be produced under herbivore attack (38). This observation lends further support to our premise that stc1 is a defense gene involved in the production of a terpenoid chemiosignal. Insect attraction by plant-emitted chemicals is a quantitative trait that relies on a statistical demonstration of differences in insect behavior. The volatile mix produced by maize plants under herbivore attack is complex and under the control of several genes, of which stc1 is just one. The results presented here argue that it should be possible to identify other maize quantitative trait loci involved in the tritrophic interaction by a simple molecular assay, namely their induction by volicitin.
Regulation of stc1 Transcription.
The biosynthesis of terpenoids is energetically very costly (39) and, therefore, should be tightly controlled. Inducible production of terpenoids would be beneficial over constitutive production (40), especially for seedlings that have limited energy resources. In addition to being energetically economic, a pulse of volatiles is also a suitable signal. Normally in corn seedlings, the stc1 transcript is barely detectable, and the major classes of terpenoids are not produced (data not shown). The level of stc1 transcript rises in mature leaves, suggesting that the stc1 product may serve different functions in mature plants and in seedlings.
Plant terpenoids are involved in functions as diverse as growth regulation, UV/heat protection, and defense against pathogens and pests. Plants respond to developmental and environmental cues by either producing different terpenoids selectively or by using the same terpenoids for multiple roles. The stc1 gene in corn is induced by wounding and by insect oral secretion in a pattern similar to that of the gene for HMG-CoA reductase in the potato (16). Because the product of HMG-CoA reductase is used as a substrate in terpenoid synthesis, it is possible that genes for both the HMG-CoA reductase and terpenoid synthase are coordinately regulated.
The pattern of stc1 transcription also resembles that of proteinase-inhibitor genes and of the indole biosynthetic gene Igl in that all three are induced by insects and regulated at the transcriptional level (ref. 41, 42). Proteinase inhibitors serve as direct defense chemicals against herbivores, although insects may develop some level of resistance to them (43). The STC1-catalyzed sesquiterpenoid and the IGL-catalyzed indole, on the other hand, probably serve as indirect defense chemicals. Chemicals that serve in indirect defense are thought to have evolved later, possibly from direct defense chemicals (1).
Genetic Control of Terpenoid Production in Maize.
We have noticed that naphthalene, 1,2,4a,5,8,8a–hexahydro-4,7-dimethyl-1-(1-methylethyl)-(1α,4aβ,8aα), is not a major component of terpenoid mixtures previously reported in maize, suggesting considerable line-to-line variation in terpenoid production. Such genotypic variation has been discussed by Turlings et al. (44) and has been observed in our preliminary analysis of five different maize inbreds (B.S. and H.K.D., unpublished data). One possible explanation for this observation is that different maize lines carry alleles of the various terpenoid biosynthetic genes that are expressed at different levels. Consistent with this explanation, we have observed line-to-line variation in stc1 transcript levels and in the promoter sequence of the stc1 alleles carried by these lines (data not shown). A common feature of these promoters, however, should be a volicitin-responsive element. We have now begun a characterization of the Stc1-McC promoter in transgenic maize with the objective of defining the sequence corresponding to the volicitin-responsive element in the stc1 gene.
The Value of an Ac Insertion Library for Functional Genomics.
The transposable element Ac tends to insert preferentially into hypomethylated DNA sequences (45–47), the component of the maize genome where most genes reside (48, 49). In the maize Ac insertion library that we have generated, almost all of the sequences flanking Ac insertions that have been isolated are either single copy or low copy DNA sequences (X. Yan and H.K.D., unpublished data). This observation indicates that the transposable element Ac is indeed an effective gene-searching engine in maize. Yet, most Ac insertions in our maize Ac insertion library have no obvious mutant phenotype. This finding is not surprising because many traits in plants are controlled by genes with minor effects (quantitative trait loci) that would be missed in a conventional mutagenesis screen designed to identify gross changes in phenotype. Because it is feasible to determine by molecular analysis whether a gene knockout has been obtained, Ac tagging could be a particularly useful tool for the functional characterization of genes that have subtle or no obvious phenotypes when mutated. The results of the present study support the validity of that strategy.
Acknowledgments
We thank Dr. Chunlin Wang for assistance at the GC/MS facility and Matthew Cowperthwaite, Huihua Fu, and Xianghe Yan for comments on the manuscript. B.S. is a recipient of a Busch Predoctoral Fellowship from Rutgers University. This research was supported in part by National Science Foundation Plant Genome Grant DBI 98–13364.
Abbreviations
- BAW
beet armyworm
- HMG
hydroxymethyl glutaryl
- RACE
rapid amplification of cDNA ends
- RT-PCR
reverse transcription–PCR
- STC
sesquiterpene cyclase
- UTR
untranslated region
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF296123).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.240284097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.240284097
References
- 1.Turlings T C J, Tumlinson J H, Lewis W J. Science. 1990;250:1251–1253. doi: 10.1126/science.250.4985.1251. [DOI] [PubMed] [Google Scholar]
- 2.Turlings T C J, Tumlinson J H. Proc Natl Acad Sci USA. 1992;89:8399–8402. doi: 10.1073/pnas.89.17.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dicke M, Bruin J, Sabelis M W. Curr Top Plant Physiol. 1993;11:182–196. [Google Scholar]
- 4.Hopke J, Donath J, Blerchert S, Boland W. FEBS Lett. 1994;352:146–150. doi: 10.1016/0014-5793(94)00948-1. [DOI] [PubMed] [Google Scholar]
- 5.Dicke M, van Baarlen P, Wessels R, Dijkman H. J Chem Ecol. 1993;19:581–599. doi: 10.1007/BF00994327. [DOI] [PubMed] [Google Scholar]
- 6.Scutareanu P, Drukker B, Bruin J, Posthumus M A, Sabelis M W. J Chem Ecol. 1997;23:2241–2260. [Google Scholar]
- 7.Shimoda T, Takayabashi J, Ashihara W, Takafuji A. J Chem Ecol. 1997;23:2033–2048. [Google Scholar]
- 8.De Moraes C M, Lewis W J, Pare P W, Alborn H T, Tumlinson J H. Nature (London) 1998;393:570–572. [Google Scholar]
- 9.Takabayashi J, Dicke M. Trends Plant Sci. 1996;1:109–113. [Google Scholar]
- 10.Mattiacci L, Dicke M, Posthumus M A. Proc Natl Acad Sci USA. 1995;92:2036–2040. doi: 10.1073/pnas.92.6.2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alborn H T, Turlings T C J, Jones T H, Stenhagen G, Loughrin J H, Tumilson J H. Science. 1997;276:945–949. [Google Scholar]
- 12.Pare P W, Tumlinson J H. Plant Physiol. 1997;114:1161–1167. doi: 10.1104/pp.114.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loughrin J H, Manukian A, Heath R R, Turlings T C J, Tumlinson J H. Proc Natl Acad Sci USA. 1994;91:11836–11840. doi: 10.1073/pnas.91.25.11836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi D, Ward B L, Bostock R M. Plant Cell. 1992;4:1333–1344. doi: 10.1105/tpc.4.10.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Facchini P J, Chappell J. Proc Natl Acad Sci USA. 1992;89:11088–11092. doi: 10.1073/pnas.89.22.11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korth K L, Dixon R A. Plant Physiol. 1997;115:1299–1305. doi: 10.1104/pp.115.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steele C L, Lewinsohn E, Croteau R. Proc Natl Acad Sci USA. 1995;92:4164–4168. doi: 10.1073/pnas.92.10.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funk C, Lewinsohn E, Vogel B S, Steele C L, Croteau R. Plant Physiol. 1994;106:999–1005. doi: 10.1104/pp.106.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohlmann J, Meyer-Gauen G, Croteau R. Proc Natl Acad Sci USA. 1998;95:4126–4133. doi: 10.1073/pnas.95.8.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crock J, Wildung M, Croteau R. Proc Natl Acad Sci USA. 1997;94:12833–12838. doi: 10.1073/pnas.94.24.12833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMullen M D, Byrne P F, Snook M E, Wiseman B R, Lee E A, Widstrom N W, Coe E H. Proc Natl Acad Sci USA. 1996;93:1996–2000. doi: 10.1073/pnas.93.17.8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dooner H K, Belachew A. Genetics. 1989;122:447–457. doi: 10.1093/genetics/122.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mottinger J P. Genetics. 1970;64:259–271. doi: 10.1093/genetics/64.2.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greene B, Walko R, Hake S. Genetics. 1994;138:1275–1285. doi: 10.1093/genetics/138.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Q, Shen B Z, Dai X K, Hei M H, Saghai Maroof M A, Li Z B. Proc Natl Acad Sci USA. 1994;91:8675–8679. doi: 10.1073/pnas.91.18.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ralston E J, English J J, Dooner H K. Genetics. 1988;119:185–197. doi: 10.1093/genetics/119.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu H, Dooner H K. Genome Res. 2000;10:866–873. doi: 10.1101/gr.10.6.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan X, Martinez-Ferez I M, Kavchok S, Dooner H K. Genetics. 1999;152:1733–1740. doi: 10.1093/genetics/152.4.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim K N, Fisher D K, Gao M, Guiltinan M J. Plant Mol Biol. 1998;38:945–956. doi: 10.1023/a:1006057609995. [DOI] [PubMed] [Google Scholar]
- 31.Greenblatt I M. Genetics. 1984;108:471–485. doi: 10.1093/genetics/108.2.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chappell J. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:521–547. [Google Scholar]
- 33.Chappell J. Plant Physiol. 1995;107:1–6. doi: 10.1104/pp.107.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gavel Y, Heijne G V. FEBS Lett. 1990;261:455–458. doi: 10.1016/0014-5793(90)80614-o. [DOI] [PubMed] [Google Scholar]
- 35.Lange B M, Croteau R. Arch Biochem Biophys. 1999;365:170–174. doi: 10.1006/abbi.1999.1168. [DOI] [PubMed] [Google Scholar]
- 36.McCaskill D, Croteau R. Planta. 1995;197:49–56. [Google Scholar]
- 37.Wessler S. Science. 1988;242:399–405. doi: 10.1126/science.2845581. [DOI] [PubMed] [Google Scholar]
- 38.Turlings T C J, Loughrin J H, McCall P J, Rose U S R, Lewis W J, Tumlinson J H. Proc Natl Acad Sci USA. 1995;92:4169–4174. doi: 10.1073/pnas.92.10.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gershenzon J. J Chem Ecol. 1994;20:1281–1328. doi: 10.1007/BF02059810. [DOI] [PubMed] [Google Scholar]
- 40.Baldwin I T. Proc Natl Acad Sci USA. 1998;95:8113–8118. doi: 10.1073/pnas.95.14.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koiwa H, Bressan R, Hasegawa P. Trends Plant Sci. 1996;2:379–384. [Google Scholar]
- 42.Frey M, Stettner C, Paré P W, Schmelz E A, Tumlinson J H, Gierl A. Proc. Natl. Acad. Sci. USA. 2000. http://www.pnas.org/cgi/doi/10.1073/pnas.260499897 10.1073/pnas.260499897. http://www.pnas.org/cgi/doi/10.1073/pnas.260499897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jongsma M A, Bakker P L, Peters J, Bosch D, Stiekema W J. Proc Natl Acad Sci USA. 1995;92:8041–8045. doi: 10.1073/pnas.92.17.8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turlings T C J, Scheepmaker J W A, Vet L E M, Tumlinson J H, Lewis W J. J Chem Ecol. 1990;16:1577–1589. doi: 10.1007/BF01014091. [DOI] [PubMed] [Google Scholar]
- 45.Chen J, Greenblatt I M, Dellaporta S L. Genetics. 1987;117:109–116. doi: 10.1093/genetics/117.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Greenblatt I M, Dellaporta S L. Genetics. 1992;130:665–676. doi: 10.1093/genetics/130.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bennetzen J L, Schrick K, Springer P S, Brown W E, SanMiguel P. Genome. 1994;37:565–576. doi: 10.1139/g94-081. [DOI] [PubMed] [Google Scholar]
- 48.Antequera F, Bird A P. EMBO J. 1988;7:2295–2299. doi: 10.1002/j.1460-2075.1988.tb03072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carels N, Barakat A, Bernardi G. Proc Natl Acad Sci USA. 1995;92:11057–11060. doi: 10.1073/pnas.92.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]