Abstract
Transcription of the Xist gene triggers X chromosome inactivation in cis and is therefore silenced on the X chromosome that remains active. DNA methylation contributes to this silencing, but the mechanism is unknown. As methylated DNA binding proteins (MBPs) are potential mediators of gene silencing by DNA methylation, we asked whether MBP-deficient cell lines could maintain Xist repression. The absence of Mbd2 caused significant low-level reactivation of Xist, but silencing was restored by exogenous Mbd2. In contrast, deficiencies of Mbd1, MeCP2, and Kaiso had no detectable effect, indicating that MBPs are not functionally redundant at this locus. Xist repression in Mbd2-null cells was hypersensitive to the histone deacetylase inhibitor trichostatin A and to depletion of the DNA methyltransferase Dnmt1. These synergies implicate Mbd2 as a mediator of the DNA methylation signal at this locus. The presence of redundant mechanisms to enforce repression at Xist and other loci is compatible with the hypothesis that “stacking” of imperfect repressive tendencies may be an evolutionary strategy to ensure leakproof gene silencing.
The Xist gene plays a central role in X chromosome dosage compensation in eutherian mammals (7, 45). The product of the gene is a nontranslated RNA that localizes to the inactive X chromosome from which it is transcribed (4, 5). Female embryonic stem cells and cells of the inner cell mass express the Xist gene from both parentally derived X chromosomes, but later expression from one X chromosome is enhanced, while expression from the other allele is extinguished (35, 41, 42). Elevated expression of Xist is the earliest known marker of X inactivation and leads to formation of a compartment that lacks transcription machinery (8) and a sequence of alterations to chromatin and DNA that secure the inactive chromosomal state.
This study concerns the active X chromosome, whose Xist allele is silenced embryonically and remains inert for the duration of that somatic lifetime. Stable Xist silencing on the active X chromosome in females and on the solitary X chromosome of males is presumably of paramount importance if inactivation of all X chromosomes is to be avoided. Early studies showed that the promoter of the silenced Xist gene is heavily methylated at CpG sites, whereas the active Xist promoter is methylation free (1), and this has been confirmed in detail by bisulfite sequencing (31). The hypothesis that silencing is mediated by DNA methylation was verified experimentally through studies of DNA methylation-deficient mice, which showed demethylation of the Xist promoter and frequent reactivation of the gene (1, 36). More recently, it has been shown that de novo DNA methylation is not required for the initial establishment of Xist silencing on the active X chromosome but is essential for the stability of the silent state (40).
There is ample evidence that DNA methylation can maintain repression of local gene expression, but the mechanism of silencing is less well characterized. In model systems, two kinds of mechanisms are shown to be potentially involved: (i) direct blocking of access by transcription factors to DNA due to the presence of methyl-CpG within the factor binding site (44) and (ii) attraction of proteins with an affinity for methyl-CpG that recruit corepressors to effect transcriptional shutdown (2, 3). Seven mammalian methylated DNA binding proteins (MBPs) are transcriptional repressors with an affinity for methylated DNA: four of these (MeCP2, MBD2, MBD1, and MBD4) are members of the MBD family (18, 24, 27), and three are members of the Kaiso family (13, 37). Several of these MBPs are shown to associate with corepressor complexes that have the potential to modify chromatin structure (12, 21, 32, 33, 46).
The biological importance of MBPs can be assessed using mice that carry null mutations in the respective genes. In this way, endogenous target genes that bind to a methyl-CpG binding protein (MBP) in vivo and are developmentally deregulated in the absence of that protein have begun to emerge. Ectopic expression of interleukin-4 (IL-4) and gamma interferon (IFN-γ) in T helper cells of the immune system was reported to occur in Mbd2−/− mice (20), and increased basal expression of the brain-derived neurotrophic factor gene was observed in neurons of Mecp2-null mice (9, 29). In transformed cell lines, cases of aberrant gene silencing by DNA methylation have also been shown to involve Mbd2 (28) and Kaiso (46). Here we have asked whether the DNA methylation-mediated silencing of the Xist gene on the active X chromosome depends upon methyl-CpG binding proteins. We find that Mbd2 is involved but that the other tested MBPs are not implicated. Mbd2 mediates robust histone deacetylation at the Xist chromatin, and its activity depends on the “maintenance DNA methyltransferase” Dnmt1.
MATERIALS AND METHODS
Western blotting.
For Western blotting, proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted onto a Protran membrane (Schleicher & Schuell). Blots were blocked in 5% milk-Tris-buffered saline, incubated with primary antibody (anti-Dnmt1 PATH52 [1:10,000; a gift from Tim Bestor] or anti-glyceraldehyde-3-phosphate dehydrogenase [anti-GAPDH; 1:5000; Abcam]) for 120 min. After being washed and incubated with secondary antibody (horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibody; 1:10,000; Amersham), proteins were visualized using enhanced chemiluminescence.
Retroviral infection.
The Flag-tagged version of the mouse Mbd2 cDNA was subcloned into the pBMN-ZIN retroviral vector, which permits expression of a Flag-tagged protein of interest and of the neomycin resistance gene from a bicistronic mRNA. Virus was produced in Phoenix cells after transfecting either pBMN-ZIN Flag-Mbd2 or empty pBMN-ZIN (mock transfection), and then mutant or wild-type (wt) mouse fibroblasts were infected with the virus according to the Nolan Lab protocols (http://www.stanford.edu/group/nolan/protocols/pro_helper_dep.html). Infected cell lines were maintained under G418 selection to ensure expression in the vast majority of the cells. The efficiency of infection was verified by indirect immunofluorescence using the anti-Flag M2 monoclonal antibody (Sigma).
PCR.
For reverse transcription, TRI reagent (Sigma) was used to purify total RNA from male mouse tail fibroblasts. RNA (0.5 μg) was DNase treated (RQ DNase; Promega) and reverse transcribed in a 50-μl reaction mixture containing 1 mM deoxynucleoside triphosphate mix, 5 μM of random hexamer primers, 1 U of RNasin (Promega), and 1 U of Moloney murine leukemia virus reverse transcriptase (Promega). Control samples without the reverse transcriptase served to check for contaminating DNA and were consistently negative. For real-time (RT)-PCR, a mastermix of 1.2 μl of cDNA sample was mixed with 10 μl of iQ SYBR green supermix (Bio-Rad) and 7.2 μl distilled water followed by 1.6 μl of the appropriate primers (3.2 pmol/μl). Reactions were performed in an iCycler iQ multicolor RT-PCR detection system (Bio-Rad) using the following steps: 95°C for 1 min 30 s for predenaturation, followed by 95°C for 30 s, 60°C for 30 s, and 72°C for 15 s, for a total of 45 cycles. Signals were normalized to the GAPDH signal. Primers used for RT-PCR were Dnmt1-3F (AAGTGCCCCGAGTGTG), Dnmt1-3R (AGGTGGAGTCGTAGATGG), Gapdh p3 (TACCCCCAATGTGTCCGTCG), Gapdh p4 (CCTGCTTCACCACCTTCTTG), XIST-4R (CACATTGCTTGATCACGCTG), XIST-10F (ACTGCTACCCTACTCATGCC), XIST-7F (AGCAAGCCCACAATTCTGG), and XIST-11R (GGACTGCCAGCAGCCTATAC).
For PCR genotyping of the different cell lines, 28 cycles were performed as described above, followed by an additional extension step of 2 min at 72°C. The primers were Kaiso-F (TCCTCTTGCAGAATACCGCACC), Kaiso-R (AGT CCCCAGAAGGATCTTGAC), M2b-RT-F-3′ (GATGAAGACATTAGGAAACAGG), M2b-RT-R-3′ (GTCCATGTCAATGTCTACTTCC), MBD1_F (AGACGGCAGATGCTCCAGAGG), MBD1-Ex15-R (CCCAGGCTGAAAATCTCCGTG), MeCP2-F (TCGCTCTGCTGGAAAGTATGATGTA), and MeCP2 (CTCCCACCTTTTAAAATATGCAATCA).
Chromatin immunoprecipitation (ChIP).
Cells were cross-linked for 10 min by adding formaldehyde to the culture medium to a final concentration of 1%. Cells were washed twice with ice-cold phosphate-buffered saline (supplemented with 0.5 mM phenylmethylsulfonyl fluoride), harvested, and resuspended in 100 μl ice-cold lysis buffer (1% sodium dodecyl sulfate, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], 0.5 mM phenylmethylsulfonyl fluoride, and Complete protease inhibitor cocktail [Roche Diagnostics]). After incubation on ice for 10 min, 500 μl of the cell suspension was sonicated on ice for 3 min at a 30% duty cycle for 5-s pulses with 10-s gaps (Branson 250 Sonifier). The cross-links were reversed by adding 1.2 μl 5 M NaCl per 20 μl chromatin at 65°C for 4 to 5 h. Washed Sepharose A (Amersham Biosciences) was used for rabbit and mouse antibodies and Sepharose G for the sheep Mbd2 antibody. Prewashed beads were incubated with 1 ml of chromatin sample for 2 h at 4°C and centrifuged at 1,000 rpm. The antibodies (10 μg control polyclonal or 5 μl Mbd2 antiserum) were added to the supernatant and rotated at 4°C overnight. S923 is an anti-Mbd2 antibody that was raised in this laboratory (33), anti-acetylated H4 antibodies were from Upstate, and the anti-Flag antibody was from Sigma. The irrelevant control antibody was rabbit polyclonal anti-alpha amylase antibody (Sigma). DNA fragments were amplified using primers XIST-7F and XIST-11R (see above) and the following primers: F2 (GTGTCCAAGACGCGGAGATAC), R2 (CAAACATGGCTGGAGCAAGCC), F3 (CGTGATACGGCTATTCTCGAG), and R3 (CCATTGCTACACACCAGAACA). The sequences for primers XIST-7F/XIST-11R are given above.
siRNA transfection.
Short interfering RNA (siRNA) against Dnmt1 was designed according to the siRNA user guide from the Tuschl laboratory, Rockefeller University. The siRNA D1-4 targeting sequence 5′-AAGTCGGACAGTGACACCCTT-3′, siRNA D1-22 targeting sequence 5′-AAGTGCAAGGCGTGCAAAGAT-3′, and siRNA D1-33 targeting sequence 5′-AACTTCGTGTCCTACAGACGC-3′ were synthesized by QIAGEN custom siRNA service. Male murine tail fibroblasts (wt and Mbd2 knockout) were seeded at 20% confluence and grown for 16 to 24 h in Dulbecco modified Eagle's medium (Cambrex) supplemented with 4 mM l-glutamine (Cambrex), 10% donor bovine serum (Gibco), and 10 units/ml penicillin-streptomycin solution (Cambrex). The next day, 40 μM siRNA was transfected into the cells using Oligofectamine reagent (Invitrogen) by following the manufacturer's instructions. Control cells were treated with the same amount of scrambled negative-control siRNA (no. 1 from Ambion). Cells were split 48 h later and transfected twice more before being harvested for analysis.
Bisulfite sequencing.
Bisulfite sequencing was conducted essentially as described previously (34). DNA was purified using the PCR purification kit (QIAGEN) and eluted in 30 μl distilled water. The CpG-rich region of the Xist promoter was amplified by two rounds of PCR amplification using nested primers. For the first round of amplification, primers BS3 (TAAAGGTTTAATAAGATGTTAGAA) and BS4 (AAATAAATCCACCATACACA) were used for a total of 45 cycles with an annealing temperature of 54°C; for the second round of amplification, primers BS5 (TGTAATTTTTGTGGTTATTTTTTTT) and BS6 (ATATTCCCCCAAAACTCCTTAAATA) were used and annealed at 58°C for a total of 35 cycles. Both PCR amplification reactions were carried out in a 50-μl reaction mixture containing 3.2 pmol of each primer, 1 mM dCTP and dGTP, 2 mM dATP and dTTP, 1.25 U of Taq DNA polymerase (Roche), and 1× reaction buffer. PCR products were separated on a 2.5% agarose gel and purified using a gel extraction kit (QIAGEN). PCR cycles were carried out as follows: 94°C for 45 s, 54°C or 58°C for 45 s, and 72°C for 45 s. Purified PCR products were cloned using TOPO TA cloning kits (Invitrogen) before sequencing.
In situ hybridization.
Mouse Xist RNA was detected by in situ hybridization using probe GPT16, a 6-kb DNA probe spanning most of exon 1 which was a gift from T. Nesterova. Hybridization was performed as described previously (11). RNase treatment abolished the hybridization signal (data not shown). Between 500 and 3,000 nuclei of cells of each genotype/drug treatment were scored for localized Xist expression. The aggregate number of Xist-positive nuclei was used to derive an overall percentage. Cells were treated with 5-azacytidine for 5 days and with trichostatin A (TSA) for 24 h. When both drugs were used together, TSA was added at the end of the TSA treatment regime.
RESULTS
Xist silencing is compromised by the absence of Mbd2.
We chose to study Xist repression in male cells, since the analysis is uncomplicated by the additional presence of an inactive X chromosome. Simian virus 40-transformed fibroblast cell lines were derived from the tails of mice that were either wt or mutated in one or more of the methyl-CpG binding domain protein genes, Mbd2, Mecp2, or Kaiso (19, 38). In addition, male Mbd1-null mouse embryonic fibroblasts were examined (48). Reverse transcriptase PCR of RNA from independent tail fibroblast cell lines (Fig. 1A) and embryonic fibroblasts (Fig. 1B) used in this study established that wt cells bear all of these genes, whereas single, double, and triple mutant cells lack only the appropriate mRNAs. Dnmt1 was expressed in each cell line.
FIG. 1.
Expression of Xist in mouse fibroblasts lacking Mbd2. (A) Reverse transcriptase PCR analysis of expression of Mbd2, Kaiso, MeCP2, Mbd1, and Dnmt1 in a selection of tail fibroblast cell lines that lack one or more of the corresponding genes due to targeted mutation. Genotypes (shown above panel) match the expression of each gene. (B) Same as for panel A, but using Mbd1−/− and control mouse embryonic fibroblasts. (C) Diagram of the Xist gene showing exons (black bars) and the CpG island region (black lollipops) that becomes methylated on the active X chromosome. PCR primers XIST-7F/XIST-11R and XIST-4R/XIST-10F distinguish the processed RNA from genomic DNA. Primers F2/R2 and F3/R3 were used for analysis of immunoprecipitated chromatin. The gray box denotes the region tested by bisulfite sequencing. Chr X, chromosome X; FR2, primer pair F2/R2; FR3, primer pair F3/R3; 7/11, primer pair XIST-7F/XIST-11R; 4/10, primer pairs XIST-4R/XIST-10F. (D) RT-PCR analysis of Xist expression in cell lines lacking one or more MBPs as shown in panel A. The primers were XIST-7F/XIST-11R (darkly shaded bars) and XIST-4R/XIST-10F (lightly shaded bars). Expression levels relative to Gapdh mRNA as an internal control were normalized by setting the level in wt cells to 1. (E) Bisulfite analysis of the Xist gene promoter region (see panel B) confirms persistence of heavy methylation in Mbd2−/− fibroblasts.
The Xist gene is highly repressed in wt male cells, but low-level expression was detected by quantitative RT-PCR using two primer pairs (XIST-4R/XIST-10F and XIST-7F/XIST-11R) that amplified different regions of the Xist sequence and are specific for the spliced RNA product (Fig. 1C). We asked whether the absence of any of the MBPs caused increased expression of the Xist gene. Comparison of mutants with mutations in the Mbd2, Mbd1, Mecp2, and Kaiso genes with wt cells showed a consistent ∼3-fold increase in Xist expression in Mbd2-deficient cells but no significant change in the other single mutant cell lines (Fig. 1D). In particular, Mbd2−/− Mecp2−/Y double mutants and Mbd2−/− Mecp2−/Y Kaiso−/Y triple mutant cells showed levels of Xist induction that were similar to those of Mbd2−/− single mutant cells (Fig. 1D). The slightly more elevated expression seen in Mecp2−/y Mbd2−/− double mutant than in Mbd2−/− single mutant cells was consistently seen in this pair of cell lines but not in independent lines with the same genotypes (data not shown). We were therefore unable to conclude that MeCP2 contributes to the silencing of Xist. The experiments implicate Mbd2 in Xist repression but argue against the involvement of Mbd1, MeCP2, or Kaiso in this process. To test whether the absence of Mbd2 affected the degree of methylation in the Xist CpG island, we carried out bisulfite sequencing of the region in Mbd2−/− cells. Complete methylation was observed at all tested CpGs (Fig. 1E), suggesting that the absence of the Mbd2 repressor, rather than the alteration of Xist gene methylation in the mutant cells, is responsible for the effect.
Mbd2 binds the Xist gene and restores defective silencing.
The Xist promoter is silenced, in part at least, by dense methylation of its CpG-rich promoter (36). As Mbd2 is a protein that can recruit the NuRD corepressor to methylated DNA, we considered the hypothesis that Mbd2 binds directly to the Xist gene and contributes to the formation of transcriptionally inert chromatin. To test this, we used ChIP to determine whether Mbd2 protein is associated with the silent Xist allele. Precipitated DNA was amplified by primer pairs corresponding to the CpG island (F3/R3) and downstream regions of the transcription unit (XIST-7F/XIST-11R; Fig. 1B). The data showed that both regions of Xist were precipitated with an Mbd2-specific antibody (Fig. 2A). That this antibody was genuinely specific for Mbd2 was demonstrated by parallel immunoprecipitation of Mbd2-null cells, which reproducibly detected no Xist sequences (Fig. 2A).
FIG. 2.
Mbd2 associates with the Xist gene and restores Xist repression in Mbd2-null cells. (A) Immunoprecipitation (IP) of cross-linked chromatin with an anti-Mbd2 antibody reveals that endogenous Mbd2 is associated with the CpG island (primers F3/R3) and downstream regions (primers XIST-7F/XIST-11R) of the methylated Xist gene. in, input; -ve, negative. (B) Exogenous Mbd2 is expressed from retroviral vector pBMN-ZIN Flag-Mbd2 (pBM) or empty vector pBMN-ZIN (pB) in Mbd2-null cells as detected on a Western blot using antibodies against Mbd2 or the Flag epitope tag. (C) Elevated Xist expression in Mbd2-null cells is abolished by exogenous Mbd2 expression. Levels of Xist RNA were measured by quantitative PCR analysis as described in the legend for Fig. 1B and C using primers XIST-7F/XIST-11R and XIST-4R/XIST-10F. Rel., relative. (D) Exogenous Mbd2 associates with the Xist gene in Mbd2-null cells. ChIP of cells transformed with the Mbd2-expressing vector (pBMN-ZIN Flag-Mbd2) or empty vector (pB) was done as described for panel A.
To be sure that Mbd2 was the key factor behind Xist reactivation in Mbd2-null cells, we asked whether exogenous Mbd2 expressed from a retroviral vector (pBMN-ZIN Flag-Mbd2) was able to reverse the derepression of Xist seen in Mbd2−/− fibroblasts. Western blots confirmed that the retrovirus directed the expression of Mbd2 protein (Fig. 2B). Quantitative RT-PCR established that the full repression level characteristic of wt cells was restored in Mbd2-deficient cells by the expression of retrovirally encoded Mbd2 (Fig. 2C). These results establish that the absence of Mbd2 is responsible for the increased Xist expression in these cells and effectively rule out the possibility that secondary changes in the cell lines, unrelated to the absence of Mbd2, are responsible. To find out whether the virus-expressed Mbd2 localized to the Xist gene, we carried out ChIP with the anti-Mbd2 antibody. Cells transfected with empty virus were negative for immunoprecipitated Xist gene fragments, but cells expressing retrovirus-encoded Mbd2 gave a positive signal when amplified with Xist gene primers (Fig. 2D). The data show that endogenous Mbd2 is normally bound to the methylated Xist gene and, when absent, can be replaced by exogenous Mbd2, which both restores repression and binds to the methylated Xist gene.
Mbd2 reinforces histone deacetylation at the Xist gene.
As Mbd2 can associate with a corepressor complex that contains histone deacetylases (HDACs) (12, 33), we asked whether Xist expression in the absence of Mbd2 was more sensitive to the HDAC inhibitor TSA. Using a quantitative PCR assay (Fig. 3A), we observed that treatment of the wt cells with TSA induced Xist expression approximately 2.5-fold. As shown above, the absence of Mbd2 also induced Xist two- to threefold, but when these mutant cells were additionally treated with TSA, a 17-fold induction of Xist was observed (see Fig. 3A). The result was verified using independently isolated tail fibroblast lines from different mice with the same genotype (data not shown). The synergy between the removal of Mbd2 and treatment with TSA indicates that Mbd2 deficiency renders Xist repression more sensitive to inhibition of HDACs.
FIG. 3.
Synergy between histone deacetylation and Mbd2 in silencing the Xist gene. (A) TSA collaborates with Mbd2 deficiency to cause enhanced activation of Xist transcription as measured by both RT-PCR analysis of Xist RNA using primers XIST-7F/XIST-11R (upper panel) and counting of Xist-positive (+ve) cells in the microscope (lower panel). Xist RNA was visualized in treated and untreated cells by in situ hybridization as described in Materials and Methods. (B) Xist-positive foci (red) within TSA-treated Mbd2-null cell nuclei stained with DAPI (4′,6′-diamidino-2-phenylindole). (C) Enhanced histone acetylation in Mbd2-null cells treated with TSA as measured by ChIP with an antibody against pan-acetyl H4 and quantitative RT-PCR of the Xist gene (primers XIST-7F/XIST-11R; see Fig. 1C). Values are expressed as the percentage of input precipitated. As a control, samples were precipitated with an irrelevant antibody and processed the same way (right-hand bar in each pair).
In order to directly visualize Xist expression, we performed fluorescence in situ hybridization using a probe against Xist RNA. We observed a discrete focus of Xist accumulation in the nuclei in about 1% of Mbd2−/− cells (Fig. 3B), but no Xist foci were seen in wt cells (not shown). TSA treatment caused ∼0.3 to 0.5% of wt cells to become Xist RNA positive, whereas TSA-treated Mbd2-null cells gave 10 times more Xist RNA-positive cells (3 to 5%) (Fig. 3A, lower panel). The close similarity between the RNA measurements and the fraction of Xist-positive nuclei (Fig. 3A, compare upper and lower panels) indicates that both the absence of Mbd2 and the exposure to TSA cause activation of Xist by increasing the probability that a particular nucleus will activate Xist, leading to the coating of the X chromosome with the RNA.
Is the deacetylated status of Xist gene chromatin on the active X chromosome (23, 30) altered by the absence of Mbd2? An antibody against the acetylated N-terminal tail of histone H4, a marker of transcriptionally active chromatin, precipitated equivalent low levels of the methylated Xist CpG island from wt and Mbd2−/− cells in a ChIP experiment (Fig. 3C). We conclude that H4 deacetylation at Xist is maintained despite the absence of Mbd2. Mbd2−/− cells were, however, significantly more sensitive to TSA treatment. H4 acetylation at the Xist gene was ∼6-fold higher in wt but ∼80-fold higher in Mbd2−/− cells after exposure to TSA (Fig. 3C). The TSA sensitivities of Xist chromatin acetylation and Xist gene expression therefore match, with Mbd2-null cells being approximately 1 order of magnitude more sensitive than wt cells.
The role of Dnmt1 in silencing of Xist.
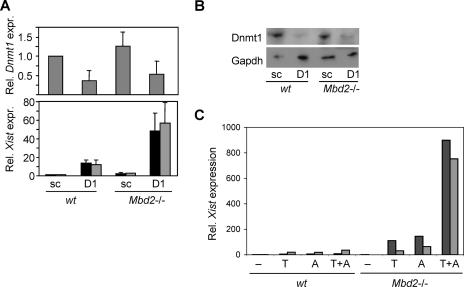
Previous work has shown that depletion of the “maintenance” DNA methyltransferase Dnmt1 compromises Xist repression (17, 36). We verified this in our fibroblasts using siRNA directed against the Dnmt1 transcript. When Dnmt1 expression was reduced in wt cells by about 50% using siRNA, as measured by quantitative RT-PCR and Western blotting (Fig. 4A and B), a 10- to 15-fold increase in Xist expression was observed (average of four independent experiments). This level of Xist induction following moderate depletion of Dnmt1 is greater than that caused by the absence of Mbd2 (two- to fourfold), indicating that Dnmt1 causes transcriptional repression of Xist via pathways that do not involve Mbd2. Depletion of Dnmt1 in Mbd2−/− cells, however, consistently caused a synergistic induction of Xist expression 50 to 80 times that of the wt level (Fig. 4A). This synthetic effect of double deficiency for Mbd2 and Dnmt1 points to cooperation between these proteins in bringing about effective Xist silencing.
FIG. 4.
Xist expression is dependent on Mbd2 and Dnmt1. (A) Quantitative PCR analysis of the effects of siRNA against Dnmt1 (D1) on transcription of Dnmt1 (upper panel) and Xist (lower panel) in wt (+/+) and Mbd2-null (−/−) cells. Scrambled siRNA (sc) was a negative control. Rel., relative; expr., expression. (B) Depletion of Dnmt1 in wt and Mbd2−/− cells as determined by Western blotting in comparison with GAPDH. (C) Induction of Xist expression by treatment of wt and Mbd2-null cells with TSA (T), 5-azacytidine (A), or both drugs (T+A). Xist expression levels were normalized against the level of expression of Gapdh RNA.
We next asked whether Dnmt1 deficiency, as induced by the drug 5-azacytidine, would have a synergistic effect on Xist gene derepression when combined with TSA treatment (6). When these treatments were combined using Mbd2-deficient cells, Xist RNA was induced ∼800-fold (Fig. 4C). In contrast, wt cells responded much more weakly to the double treatment with TSA and 5-azacytidine, showing a ∼20-fold induction of Xist. These results strongly reinforce the notion that Mbd2 shares a repressive pathway with both Dnmt1 and HDACs.
DISCUSSION
DNA methylation at the Xist locus is read by Mbd2.
We demonstrate that the methyl-CpG binding protein Mbd2 is required for the leakproof silencing of the Xist gene in mouse fibroblasts. In wt male fibroblasts, Xist-bearing cells are undetectable, but about 1% of Mbd2-null cells exhibit a coating of the X chromosome by Xist RNA. Thus, the absence of Mbd2 increases the probability that Xist gene reactivation will occur. Previous studies reported that artificial demethylation reactivated Xist in about 1% of hypomethylated human fibroblasts (17), while in Dnmt1-deficient differentiated mouse embryonic stem cells, about 15% of cells carried Xist (36). Therefore, deficiency of either DNA methylation or Mbd2 increases the probability of Xist reactivation but does not cause Xist reexpression in every cell. This weakening of Xist repression recalls the effects of Mbd2 deficiency on T helper cells, where it was found that only a proportion of Mbd2-null T helper cells reactivate IL-4 and IFN-γ inappropriately (20). As in the case of Xist, a lack of Mbd2 increased the probability of inappropriate activation of IL-4 and IFN-γ.
Expression of Xist is synergistically increased when both Mbd2 and Dnmt1 are deficient. The simplest interpretation of the relationship between these two proteins is that Mbd2 reads the DNA methylation signal that has been added to the locus by Dnmt1. The data also make clear, however, that Mbd2 is not the only mediator of silencing through Dnmt1, as the induction of Xist expression by Dnmt1 deficiency is consistently greater than that caused by the absence of Mbd2 (∼15-fold versus ∼2.5-fold). The additional contribution of Dnmt1 could be caused by direct interference of DNA methylation with transcription factor access or by the involvement of MBP repressors other than those tested here. An alternative possibility is that Dnmt1 itself contributes to Xist repression through its association with HDACs (14, 39). Under this scenario, Mbd2 may read the methylation signal while Dnmt1 directly participates in the repression process. The mechanism by which Dnmt1 might target repression is unclear, as it is not known to specifically complex with DNA except during catalysis following DNA replication. A potential interaction between Dnmt1 and Mbd2 could in theory contribute to specificity (43).
Xist was induced ∼20-fold by TSA treatment of Mbd2-null cells, whereas either Mbd2 deficiency or TSA treatment of wt cells induced Xist only two- to threefold each. In agreement with these results, TSA treatment resulted in ∼8-fold higher Xist chromatin acetylation levels in Mbd2-null cells than in wt cells. Biochemical data have consistently shown Mbd2 to be associated with the NuRD corepressor complex, which includes HDAC1 and HDAC2 (12, 25, 26, 33, 47). Therefore, Mbd2-null cells are likely to recruit less HDAC1 and HDAC2 to methylated sites at the Xist gene, and this may explain the heightened sensitivity of the Xist gene to TSA treatment. An alternative hypothesis is that there is increased turnover of acetyl groups on histone H4 in Mbd2−/− cells, with the steady-state acetylation levels remaining much the same (see Fig. 3C). Inhibition with TSA might then enhance H4 acetylation by perturbing the steady-state level of HDAC activity, resulting in Xist transcription. The mutual interdependence of Mbd2, histone deacetylation, Dnmt1, and Xist gene expression is well illustrated by the massive induction of Xist (2 to 3 orders of magnitude above that of wt cells) when Mbd2-null cells are simultaneously depleted in Dnmt1 and exposed to TSA. The data are compatible with a scenario in which DNA methylation recruits NuRD, which in turn causes local deacetylation of chromatin, leading to leakproof transcriptional silencing of Xist.
Do MBPs work solo or as a team?
Our findings shed light on the specificity of methyl-CpG binding proteins. Model experiments have shown previously that Kaiso, Mbd1, Mbd2, and MeCP2 can repress transcription, but do they compete to repress a particular methylated gene? Given their common DNA binding sequence specificities, it might be expected that the absence of one MBP would be compensated for by the continued presence of the others. An alternative scenario proposes that a particular MBP is dedicated to a specific subset of genes that is different from the subsets that are targeted by other MBPs. Recent analysis of target sites for Mbd2 and MeCP2 showed clearly that these proteins occupy distinct sites in vivo, strongly favoring the second hypothesis (22). Our data are also compatible with distinct, nonoverlapping functions for MBPs, as Xist repression depends on Mbd2 but is unaffected by the removal of Mbd1, MeCP2, or Kaiso. Even weak collaborations between these proteins should have been detected by combining the absence of Mbd2 with deficiencies of other Mbd proteins. No such synthetic effects were seen, leading to the conclusion that, in these cells at least, only Mbd2 participates in Xist repression. The earlier observation that the neurological phenotype seen in Mecp2-null mice is not enhanced in Mecp2 Mbd2 double null mice (16) also fits with independent MBP functioning.
Gene silencing by “stacking” repressive tendencies.
Although Mbd2 does not appear to collaborate with other MBPs at the Xist locus, it does collaborate with other gene-silencing mechanisms. Deacetylation of histone H4 was superficially normal in the chromatin of both wt and Mbd2-null cells (see Fig. 3C). It follows that HDACs are active at the Xist locus even when Mbd2 is not present and must be recruited by other components of the machinery that has evolved to repress Xist. The Mbd2 pathway is evidently just one of several epigenetic mechanisms that collaborate to efficiently silence Xist.
Why should repression mechanisms be redundant in this way? One possibility is that each mechanism is moderately efficient but not efficient enough to guarantee the continuous silence of Xist in all cells. By enlisting several such mechanisms at a single locus, the probability that all will fail simultaneously is reduced. A paradigm for redundant, repressive mechanisms working in tandem is X chromosome inactivation. Here, Xist RNA, polycomb group proteins, histone deacetylation, histone (H3K9) methylation, histone H2aX substitution, late replication, and DNA methylation are all involved in silencing. The synergy of several of these mechanisms has been demonstrated experimentally (10) and resembles the redundancy of Xist repression mechanisms reported here. Conceivably, it is more parsimonious from an evolutionary perspective to “stack” imperfect silencers in this way than to create a single “perfect” silencing mechanism. A prerequisite of the “stacked tendency” model is that each imperfect mechanism in the stack is independent of the others (i.e., does not share limiting components). If so, their likelihoods of failure can be multiplied together, resulting in efficient shutdown of the gene concerned. For example, three independent, 90% efficient mechanisms directed at the same target gene would combine to give 99.9% efficient silencing. A prediction of this model is that the loss of any one member of the stack (e.g., by mutation) would leave repression largely intact but would be manifest as an increase in transcriptional leakiness. This may help to explain the relatively mild phenotype resulting from gene mutations of some MBPs and of certain other proteins that target corepressors to DNA, for example, Ikaros (15).
Acknowledgments
We thank Aimée Deaton and Heather Owen for commenting on the manuscript and Fred Gage (Salk Institute, California) for Mbd1-null fibroblasts.
This work was supported by the Wellcome Trust, Cancer Research UK, and fellowship grant He 45162-1 from the Deutsche Forschungsgesellschaft (DFG) to A.H.
Footnotes
Published ahead of print on 12 March 2007.
REFERENCES
- 1.Beard, C., E. Li, and R. Jaenisch. 1995. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 9:2325-2334. [DOI] [PubMed] [Google Scholar]
- 2.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6-21. [DOI] [PubMed] [Google Scholar]
- 3.Boyes, J., and A. Bird. 1991. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64:1123-1134. [DOI] [PubMed] [Google Scholar]
- 4.Brockdorff, N., A. Ashworth, G. F. Kay, V. M. McCabe, D. P. Norris, P. J. Cooper, S. Swift, and S. Rastan. 1992. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71:515-526. [DOI] [PubMed] [Google Scholar]
- 5.Brown, C. J., B. D. Hendrich, J. L. Rupert, R. G. Lafreniere, Y. Xing, J. Lawrence, and H. F. Willard. 1992. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71:527-542. [DOI] [PubMed] [Google Scholar]
- 6.Cameron, E. E., K. E. Bachman, S. Myohanen, J. G. Herman, and S. B. Baylin. 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 21:103-107. [DOI] [PubMed] [Google Scholar]
- 7.Chadwick, B. P., and H. F. Willard. 2003. Barring gene expression after XIST: maintaining facultative heterochromatin on the inactive X. Semin. Cell Dev. Biol. 14:359-367. [DOI] [PubMed] [Google Scholar]
- 8.Chaumeil, J., P. Le Baccon, A. Wutz, and E. Heard. 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 20:2223-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, W. G., Q. Chang, Y. Lin, A. Meissner, A. E. West, E. C. Griffith, R. Jaenisch, and M. E. Greenberg. 2003. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302:885-889. [DOI] [PubMed] [Google Scholar]
- 10.Csankovszki, G., A. Nagy, and R. Jaenisch. 2001. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol. 153:773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duthie, S. M., T. B. Nesterova, E. J. Formstone, A. M. Keohane, B. M. Turner, S. M. Zakian, and N. Brockdorff. 1999. Xist RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum. Mol. Genet. 8:195-204. [DOI] [PubMed] [Google Scholar]
- 12.Feng, Q., and Y. Zhang. 2001. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 15:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filion, G. J. P., S. Zhenilo, S. Salozhin, D. Yamada, E. Prokhortchouk, and P.-A. Defossez. 2006. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol. Cell. Biol. 26:169-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuks, F., W. A. Burgers, A. Brehm, L. Hughes-Davies, and T. Kouzarides. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 24:88-91. [DOI] [PubMed] [Google Scholar]
- 15.Georgopoulos, K., M. Bigby, J. H. Wang, A. Molnar, P. Wu, S. Winandy, and A. Sharpe. 1994. The Ikaros gene is required for the development of all lymphoid lineages. Cell 79:143-156. [DOI] [PubMed] [Google Scholar]
- 16.Guy, J., B. Hendrich, M. Holmes, J. E. Martin, and A. Bird. 2001. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 27:322-326. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, R. S., T. K. Canfield, A. M. Stanek, E. A. Keitges, and S. M. Gartler. 1998. Reactivation of XIST in normal fibroblasts and a somatic cell hybrid: abnormal localization of XIST RNA in hybrid cells. Proc. Natl. Acad. Sci. USA 95:5133-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrich, B., and A. Bird. 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18:6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hendrich, B., J. Guy, B. Ramsahoye, V. A. Wilson, and A. Bird. 2001. Closely related proteins Mbd2 and Mbd3 play distinctive but interacting roles in mouse development. Genes Dev. 15:710-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutchins, A. S., A. C. Mullen, H. W. Lee, K. J. Sykes, F. A. High, B. D. Hendrich, A. P. Bird, and S. L. Reiner. 2002. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol. Cell 10:81-91. [DOI] [PubMed] [Google Scholar]
- 21.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19:187-191. [DOI] [PubMed] [Google Scholar]
- 22.Klose, R. J., S. A. Sarraf, L. Schmiedeberg, S. M. McDermott, I. Stancheva, and A. P. Bird. 2005. DNA binding specificity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol. Cell 19:667-678. [DOI] [PubMed] [Google Scholar]
- 23.Komura, J., S. A. Sheardown, N. Brockdorff, J. Singer-Sam, and A. D. Riggs. 1997. In vivo ultraviolet and dimethyl sulfate footprinting of the 5′ region of the expressed and silent Xist alleles. J. Biol. Chem. 272:10975-10980. [DOI] [PubMed] [Google Scholar]
- 24.Kondo, E., Z. Gu, A. Horii, and S. Fukushige. 2005. The thymine DNA glycosylase MBD4 represses transcription and is associated with methylated p16INK4a and hMLH1 genes. Mol. Cell. Biol. 25:4388-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kransdorf, E. P., S. Z. Wang, S. Z. Zhu, T. B. Langston, J. W. Rupon, and G. D. Ginder. 2006. MBD2 is a critical component of a methyl cytosine-binding protein complex isolated from primary erythroid cells. Blood 108:2836-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Guezennec, X., M. Vermeulen, A. B. Brinkman, W. A. M. Hoeijmakers, A. Cohen, E. Lasonder, and H. G. Stunnenberg. 2006. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 26:843-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis, J. D., R. R. Meehan, W. J. Henzel, I. Maurer-Fogy, P. Jeppesen, F. Klein, and A. Bird. 1992. Purification, sequence and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69:905-914. [DOI] [PubMed] [Google Scholar]
- 28.Lin, X., and W. G. Nelson. 2003. Methyl-CpG-binding domain protein-2 mediates transcriptional repression associated with hypermethylated GSTP1 CpG islands in MCF-7 breast cancer cells. Cancer Res. 63:498-504. [PubMed] [Google Scholar]
- 29.Martinowich, K., D. Hattori, H. Wu, S. Fouse, F. He, Y. Hu, G. Fan, and Y. E. Sun. 2003. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302:890-893. [DOI] [PubMed] [Google Scholar]
- 30.McCabe, V., E. J. Formstone, L. P. O'Neill, B. M. Turner, and N. Brockdorff. 1999. Chromatin structure analysis of the mouse Xist locus. Proc. Natl. Acad. Sci. USA 96:7155-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonald, L. E., C. A. Paterson, and G. F. Kay. 1998. Bisulfite genomic sequencing-derived methylation profile of the xist gene throughout early mouse development. Genomics 54:379-386. [DOI] [PubMed] [Google Scholar]
- 32.Nan, X., H.-H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393:386-389. [DOI] [PubMed] [Google Scholar]
- 33.Ng, H.-H., Y. Zhang, B. Hendrich, C. A. Johnson, B. M. Burner, H. Erdjument-Bromage, P. Tempst, D. Reinberg, and A. Bird. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23:58-61. [DOI] [PubMed] [Google Scholar]
- 34.Nuber, U. A., S. Kriaucionis, T. C. Roloff, J. Guy, J. Selfridge, C. Steinhoff, R. Schulz, B. Lipkowitz, H. H. Ropers, M. C. Holmes, and A. Bird. 2005. Up-regulation of glucocorticoid-regulated genes in a mouse model of Rett syndrome. Hum. Mol. Genet. 14:2247-2256. [DOI] [PubMed] [Google Scholar]
- 35.Panning, B., J. Dausman, and R. Jaenisch. 1997. X chromosome inactivation is mediated by Xist RNA stabilization. Cell 90:907-916. [DOI] [PubMed] [Google Scholar]
- 36.Panning, B., and R. Jaenisch. 1996. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 10:1991-2002. [DOI] [PubMed] [Google Scholar]
- 37.Prokhortchouk, A., B. Hendrich, H. Jorgensen, A. Ruzov, M. Wilm, G. Georgiev, A. Bird, and E. Prokhortchouk. 2001. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 15:1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prokhortchouk, A., O. Sansom, J. Selfridge, I. M. Caballero, S. Salozhin, D. Aithozhina, L. Cerchietti, F. G. Meng, L. H. Augenlicht, J. M. Mariadason, B. Hendrich, A. Melnick, E. Prokhortchouk, A. Clarke, and A. Bird. 2006. Kaiso-deficient mice show resistance to intestinal cancer. Mol. Cell. Biol. 26:199-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 40.Sado, T., M. Okano, E. Li, and H. Sasaki. 2004. De novo DNA methylation is dispensable for the initiation and propagation of X chromosome inactivation. Development 131:975-982. [DOI] [PubMed] [Google Scholar]
- 41.Sheardown, S. A., S. M. Duthie, C. M. Johnston, A. E. Newall, E. J. Formstone, R. M. Arkell, T. B. Nesterova, G. C. Alghisi, S. Rastan, and N. Brockdorff. 1997. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell 91:99-107. [DOI] [PubMed] [Google Scholar]
- 42.Sun, B. K., A. M. Deaton, and J. T. Lee. 2006. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol. Cell 21:617-628. [DOI] [PubMed] [Google Scholar]
- 43.Tatematsu, K. I., T. Yamazaki, and F. Ishikawa. 2000. MBD2-MBD3 complex binds to hemi-methylated DNA and forms a complex containing DNMT1 at the replication foci in late S phase. Genes Cells 5:677-688. [DOI] [PubMed] [Google Scholar]
- 44.Watt, F., and P. L. Molloy. 1988. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 2:1136-1143. [DOI] [PubMed] [Google Scholar]
- 45.Wutz, A. 2003. RNAs templating chromatin structure for dosage compensation in animals. Bioessays 25:434-442. [DOI] [PubMed] [Google Scholar]
- 46.Yoon, H. G., D. W. Chan, A. B. Reynolds, J. Qin, and J. Wong. 2003. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol. Cell 12:723-734. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, Y., H.-H. Ng, H. Erdjument-Bromage, P. Tempst, A. Bird, and D. Reinberg. 1999. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 13:1924-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao, X., T. Ueba, B. R. Christie, B. Barkho, M. J. McConnell, K. Nakashima, E. S. Lein, B. D. Eadie, A. R. Willhoite, A. R. Muotri, R. G. Summers, J. Chun, K. F. Lee, and F. H. Gage. 2003. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc. Natl. Acad. Sci. USA 100:6777-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]