Abstract
During meiosis, double-strand breaks (DSBs) lead to crossovers, thought to arise from the resolution of double Holliday junctions (HJs) by an HJ resolvase. In Schizosaccharomyces pombe, meiotic crossovers are produced primarily through a mechanism requiring the Mus81-Eme1 endonuclease complex. Less is known about the processes that produces crossovers during the repair of DSBs in mitotic cells. We employed an inducible DSB system to determine the role of Rqh1-Top3 and Mus81-Eme1 in mitotic DSB repair and crossover formation in S. pombe. In agreement with the meiotic data, crossovers are suppressed in cells lacking Mus81-Eme1. And relative to the wild type, rqh1Δ cells show a fourfold increase in crossover frequency. This suppression of crossover formation by Rqh1 is dependent on its helicase activity. We found that the synthetic lethality of cells lacking both Rqh1 and Eme1 is suppressed by loss of swi5+, which allowed us to show that the excess crossovers formed in an rqh1Δ background are independent of Mus81-Eme1. This result suggests that a second process for crossover formation exists in S. pombe and is consistent with our finding that deletion of swi5+ restored meiotic crossovers in eme1Δ cells. Evidence suggesting that Rqh1 also acts downstream of Swi5 in crossover formation was uncovered in these studies. Our results suggest that during Rhp51-dependent repair of DSBs, Rqh1-Top3 suppresses crossovers in the Rhp57-dependent pathway while Mus81-Eme1 and possibly Rqh1 promote crossovers in the Swi5-dependent pathway.
Meiotic cells depend on crossovers for creating genomic rearrangements, a critical step in ensuring genomic vitality. In fact crossing over is essential for spore viability in yeasts, while in mouse cells, blocking crossing over leads to embryonic lethality (6, 17, 35). The classic model for crossing over was first suggested by Robin Holliday, who proposed that crossovers arose by resolution of Holliday junctions (HJs) (27). This model was later refined to include the resolution of a double HJ (dHJ), which remains the major model for crossover formation (48) (Fig. 1).
FIG. 1.
Proposed mechanisms for the generation of crossover and noncrossover products during HR. These mechanisms are derived from previous publications (25, 43, 47).
In meiotic cells, crossing over is initiated at a double-strand break (DSB) induced by Spo11 (35), while in mitotic cells DSBs can arise from exogenous sources, such as exposure to ionizing radiation, or endogenous sources, such as DNA synthesis through a single-strand nick. DSBs not only pose a major problem for cell survival, as a single unrepaired DSB is presumably sufficient to cause cell death (7), but they also are a source of genomic instability (reviewed in references 34, 36, and 50). Genomic instability can lead to loss of information or DNA rearrangements, events associated with cancer in mammalian cells (41).
The cell has two major mechanisms for the repair of DSBs, homologous recombination (HR) and nonhomologous end joining (NHEJ), each used to various degrees in different organisms. HR is characterized as being an error-free process using homologous sequences as the template for repair of the DSB (32, 47) (Fig. 1). In this process, a 3′ single-stranded end is formed that is coated by Rad51, with the aid of mediator proteins Rad52 and the Rad55-Rad57 heterodimer. The resulting nucleoprotein filament finds its homologous sequence by single-end invasion, a process aided by Rad54 (Fig. 1). The 3′ end of the invading strand is extended by a DNA polymerase. At this step, the invading strand can be displaced from the joint molecule, which can now bridge the DSB. Next, DNA synthesis and ligation occur. This process is referred to as synthesis-dependent strand annealing (SDSA) (Fig. 1). In this process crossovers are not predicted to occur, although a model that could lead to crossover has been proposed elsewhere (45). If SDSA does not occur, the joint molecule can go on to form a dHJ. This structure can be resolved by an HJ resolvase, producing either a crossover or noncrossover product (Fig. 1).
An alternative mechanism for crossover formation involves the Mus81-Mms4/Eme1 heterodimer (Fig. 1), a structure-specific endonuclease that is conserved in eukaryotes, including mice and humans (1, 8, 11, 33). The Mus81 complex from budding yeast shows the ability to cleave 3′ flaps and fork structures, as well as a weak activity in cleaving HJs (5). The complex shows a strong preference for substrates that are four-way junctions with an exposed 5′ end at or near the junction crossover point (20, 43, 51). D loops, which are formed during strand invasion, and nicked HJs are examples of substrates that meet these criteria. Mus81 is required in yeasts for repair of meiotic DSBs, as spore viability is greatly reduced in mus81 mutants (8, 33). In fact meiotic crossovers are essentially eliminated in Schizosaccharomyces pombe mus81Δ mutants (43). This suggests that the Mus81 complex is responsible for maturation of crossover products during S. pombe meiosis.
Swi5 is a small protein that acts with Sfr1 during HR in a process that is an alternative to the action of Rhp55/57 (4). In S. pombe, it was shown that while swi5Δ cells are largely insensitive to ionizing radiation and rhp55/rhp57Δ cells show moderate sensitivity, the swi5Δ rhp55/57Δ double mutant is as sensitive to ionizing radiation as an rhp51Δ mutant. More recent studies of Swi5 in S. pombe showed that in meiotic cells, loss of swi5+ significantly decreases the spore viability of rhp57Δ or rhp55Δ cells while dramatically improving the low spore viability of mus81Δ eme1Δ cells, suggesting that Mus81-Eme1 acts downstream of Swi5 (18).
RecQ DNA helicases are found in virtually every organism from bacteria to humans. A common phenotype associated with loss of RecQ helicase activity is increased genomic instability. How RecQ helicases function in maintaining genomic stability is only beginning to be understood, but increasingly, data are demonstrating a role for RecQ helicases in recombination: (i) RecQ mutants show increased rates of HR (3, 23, 26, 42, 54); (ii) loss of HR genes has also been shown to suppress the synthetic interaction between sgs1 and mus81 and between sgs1/rqh1Δ and srs2 (16, 19, 21, 40); (iii) expression of the Escherichia coli HJ resolvase, RusA, partially suppresses the UV and hydroxyurea sensitivities in rqh1Δ cells as well as the synthetic lethality of rqh1Δ mus81Δ (14, 15, 43); and (iv) recently, we reported that the hydroxyurea and UV sensitivities of rqh1Δ cells are suppressed by the loss of a subset of HR genes (28). Together these findings strongly imply that RecQ helicases function through HR to provide genomic stability during both DNA damage and replication arrest.
Two recent studies have provided more evidence for RecQ helicases acting in the late stages of HR. In one study, using HO endonuclease to create an ectopic DSB, Sgs1-Top3 was shown to partially suppress crossover formation during repair by gene conversion (GC) (30). The authors proposed that Sgs1-Top3 acts to resolve dHJs exclusively into noncrossover products. In the other study, Wu and Hickson used in vitro assays to show that the human RecQ helicase (BLM)-topoisomerase IIIα could resolve a synthetic dHJ (53). While RecQ helicases have been implicated in suppressing HR, it is possible that their main role is to suppress crossing over. Human cells lacking the RecQ helicase BLM have very high rates of sister chromosome exchanges (10), which may be representative of increased crossover frequency.
Using a system that generates a unique DSB on a nonessential minichromosome (Ch16) in S. pombe, we reported that Rqh1 acts to block recombination between sister chromatids (29). In the studies reported here we again used the inducible DSB repair system but focused on the roles of Rqh1-Top3 and Mus81-Eme1 in crossing over. Our findings indicate that, as in meiotic cells, crossovers formed in mitotic cells arise primarily through the action of Mus81-Eme1, acting downstream of Swi5. Most GC events, however, are processed through an Rhp57-dependent pathway where Rqh1-Top3 acts to block crossing over. Suppression of crossing over by Rqh1-Top3 requires the helicase activity of Rqh1. In the absence of Rqh1-Top3, Rhp57-dependent but Mus81-Eme1-independent crossovers form. Furthermore, we found that the inviability of cells lacking both Rqh1-Top3 and Mus81-Eme1 activities is suppressed in a swi5Δ background. Together these data provide us with several significant insights into the process of DSB repair by HR in S. pombe.
MATERIALS AND METHODS
Genetic manipulations.
Standard protocols were used for the creation of strains with multiple mutations. The strains used in these studies are listed in Table 1. Cells were propagated on either rich medium, YEA (5% yeast extract, 30% dextrose, and 150-mg/liter adenine), or defined medium containing Edinburgh essential medium (EMM) and 2% dextrose plus appropriate supplements. Throughout these studies, at least two independently derived strains for each genotype described were used. In every case these mutants were found to act identically to each other.
TABLE 1.
Strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| sz1001 | h+ade6-M210 ura4-D18 leu1-32 his3-D Ch16-ade6-M216-MATa−kanMX6 (Th805) | 46 |
| sz1312 | h+ade6-M210 ura4-D18 leu1-32 rhp51::ura4 | 31 |
| sz1260 | h−ade6-M210 ura4-D18 leu1-32 his3-D smt-0 rhp57::ura4 | 49 |
| sz1493 | h−ade6-M210 ura4-D18 leu1-32 his3-D smt-0 swi5::his3 | 4 |
| sz1503 | h−ade6-M210 ura4-D18 leu1-32 his3-D eme1::ura4a | This study |
| sz1245 | h−ade6-M210 ura4-D18 leu1-32 his3-D rqh1::ura4 | 13 |
| sz1335 | h−ade6-M210 ura4-D18 leu1-32 rqh1(rad12)-K547I | 39 |
| sz192 | h−ade6-M210 ura4-D18 leu1-32 top3-15 | This study |
| sz1663 | h−arg1-230 leu1-32 ura4-D18 his3-D | This study |
| sz1058 | h+ade6-M210 leu1-32 ura4-D18 his3-D | Lab strain |
| sz1686 | h+eme1::ura4+arg1-230 leu1-32 ura4-D18 | This study |
| sz1710 | h+swi5::his3+arg1-230 leu1-32 ura4-D18 his3-D | This study |
| sz1493 | h−swi5::his3+smt-0 ura4-D18 leu1-32 his3-D1 ade6-M210 | This study |
| sz1718 | h+swi5::his3 eme1::ura4 ade6-M210 leu1-32 ura4-D18 his3-D | This study |
| sz1720 | h−smt-0 swi5::his3 eme1::ura4 arg1-230 leu1-32 ura4-D18 his3-D | This study |
eme1::ura4+ is a deletion/insertion.
A conditional-lethal top3 mutant was constructed by PCR mutagenesis. The last third of the top3+ gene plus some of the 3′ untranslated sequences were amplified by PCR under conditions designed to generate random mutations in the products. The PCR products were used to transform an S. pombe strain in which ura4+ had been inserted just 3′ to top3+. Transformants were selected for on 5-fluoroorotic acid plates at 25°C. 5-Fluoroorotic acid-resistant colonies were replica plated and incubated at 25°C and 35°C to identify colonies that were not viable at the elevated temperature. The top3 gene from each strain was sequenced to confirm that it contained mutations. One mutant, top3-15, was selected as having near-normal growth at 25°C but was inviable at 35°C.
For meiotic crosses, parental strains were streaked onto fresh YES (yeast extract plus adenine, uracil, histidine, leucine, and arginine) plates for 2 days. Cells were then mixed and spotted onto sporulation plates supplemented with appropriate nutrients. Plates were incubated for 2 days at 25°C and then plated onto YES plates. Colonies were replica plated onto YES plates without adenine and plates with EMM plus adenine, leucine, uracil, and histidine. Plates were incubated for 1 to 2 days and scored.
HO-induced DSB repair assay.
The HO-induced DSB repair assay has been described previously (29, 46). Overnight cultures were inoculated from frozen stocks in EMM plus histidine, uracil, and 8 μM thiamine. The following day half of each culture was washed three times to remove thiamine. From these cells, fresh cultures were started in EMM plus histidine, uracil, and adenine. The thiamine-plus cultures (8 μM thiamine) were diluted into EMM plus histidine, uracil, adenine, and thiamine. Cultures were maintained at the appropriate temperature with constant shaking. At 24-h intervals cultures were diluted into fresh medium to maintain logarithmic growth. At indicated times cells were plated onto yeast extract-thiamine plates and incubated for 3 to 5 days depending on growth rates. Red colonies were scored as chromosome loss (CL) events. White colonies were counted and transferred to 96-well microtiter dishes from which they were stamped onto YEA-thiamine-G418 and yeast extract-thiamine plates. Stamped plates were incubated for 2 to 3 days, and cell growth was noted. Growth on G418 was scored as sister chromatid conversion (SCC) or NHEJ, while no growth on G418 was scored as GC.
Pulsed-field gel electrophoresis (PFGE).
Overnight cultures of individual colonies were propagated in 5-ml cultures in EMM plus histidine, uracil, and leucine. Following overnight growth, cells were counted and approximately 5 × 107 cells were isolated for each sample. Cells were resuspended in 30 μl of 100 mM Na2-EDTA and 1 mM NaN3 and embedded in agarose plugs (Exclu-Sieve; the Nest Group). The plugs were then suspended in 1 ml of spheroplasting solution (1 M sorbitol, 100 mM Na2-EDTA, 40 mM Tris, pH 7.9, 10 mM 2-mercaptoethanol) with 2 mg/ml of lysing enzyme (Sigma) and incubated for 2.5 h at 37°C with shaking. Next 0.25 mg/ml of lyticase (Sigma) was added and cells were incubated at 37°C for an additional 30 min. The plugs were washed for 1 h two times in ETS (250 mM Na2-EDTA, 10 mM Tris, pH 7.9, and 1% sodium dodecyl sulfate) at 55°C. This solution was replaced with 1 ml of SEP buffer (1% N-laurylsarcosine, 0.5 M Na2-EDTA, and 1 mg/ml of proteinase K [ICN]), and the plugs were incubated at 55°C for 48 h. Plugs were then washed three times, 20 min each, in Tris-EDTA before being loaded onto a gel.
PFGE was carried out with a Bio-Rad CHEF II system. Agarose gels (0.6%; Bio-Rad PFGE grade) were run in 1× Tris-acetate-EDTA buffer with circulation at 14°C for 48 h. The gel conditions were 2 V/cm, with an initial switch time of 20 min and a final switch time of 30 min. Chromosomes were visualized by SYBR green (Molecular Probes) staining and UV.
Verification of crossovers.
The compositions of the putative recombinant chromosomes produced by crossing over were analyzed by Southern blotting. Cells from two independent colonies containing putative crossovers were cast in agarose plugs. Following lysis as described above for PFGE, the plugs were washed extensively in restriction buffer and then incubated with AsiSI overnight. The resulting DNA fragments were separated by PFGE in 1× Tris-buffered EDTA at 6 V/cm with switch times of 20 s and 30 s for 24 h. The gel was stained and photographed. Blotting was carried out under neutral conditions. The blot was probed using a random-primer-labeled PCR fragment containing either the ade6+ or rad21+ sequence.
Statistical analysis.
Statistical tests were conducted in order to validate whether crossover frequencies for different mutant strains were similar. For tests performed on mitotic cell crossover frequencies, Fisher's exact test for a 2 × 2 contingency table was utilized due to the small number of crossovers in some tested strains. For tests performed on meiotic cell crossover frequencies, Pearson's χ2 test was appropriate due to the higher cell counts. All tests were planned a priori and run sequentially on Intercooled Stata 9. The analyses employed in this study are based on the assumption of experimental uniformity driven by strict adherence to lab protocols. On this basis, crossover data from multiple experiments were aggregated into a single binomial proportion for each strain.
RESULTS
Analysis of DSB repair using a site-specific endonuclease.
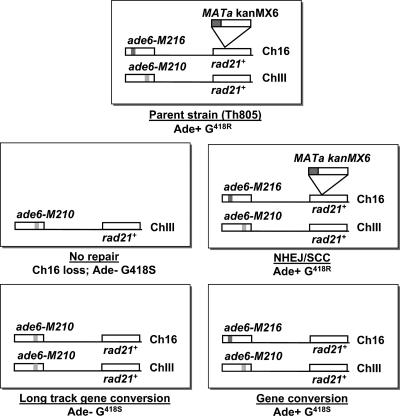
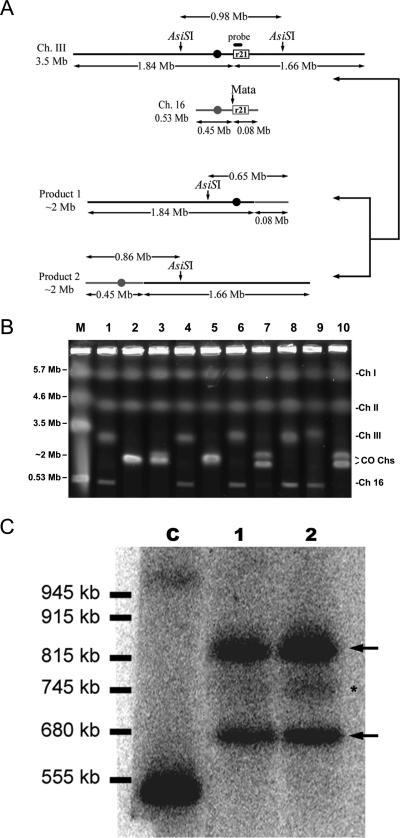
A system for tracking the response of cells to a unique DSB in S. pombe has been described previously and is shown schematically in Fig. 2 (46). The parent strain used in these studies is named Th805 and contains the MATa sequences from Saccharomyces cerevisiae linked to the G418 resistance (G418R) gene contained in the KanMX6 cassette, inserted into rad21+ on Ch16. Ch16 is an 0.535-Mb nonessential minichromosome that consists of the pericentric regions of the 3.5-Mb ChIII. This region of ChIII also contains the ade6+ locus. The ade6-M210 and ade6-M216 alleles are on ChIII and Ch16, respectively, and provide intragenic complementation. Thus, the presence of this minichromosome can be verified by following the ability of the strain to grow in medium lacking adenine. The HO endonuclease, which creates a DSB in Ch16 by cleaving within the MATa sequence, is expressed from a plasmid and is under the control of the thiamine-repressible nmt promoter (44). During DSB repair, GC events between Ch16 and ChIII result in retention of the ade6 markers and lead to loss of the KanMX6 sequence, generating cells that are ade6+ and G418 sensitive (G418S) (Fig. 2). Repair by SCC or NHEJ can also be quantitated in this system (Fig. 2). Nonrepair leads to loss of Ch16 and cells that are both G418S and ade−. Table 2 shows the DSB repair data set obtained for the various genetic backgrounds described in this study. Most importantly, when using this system, crossovers that form during repair of the DSB by GC can be visualized by PFGE (Fig. 3A and B). A crossover event leads to disappearance of both ChIII and Ch16 and the concomitant appearance of two new chromosomes of approximately 2 Mb (Fig. 3A and B). Southern blot analysis was carried out to confirm that these two new chromosomes resulted from a crossover event (Fig. 3A and 3C).
FIG. 2.
Schematic diagram of the DSB repair system. Cells carrying both ChIII and Ch16 are Ade+ and G418R (top center). Failure to repair the DSB leads to loss of Ch16 (scored as CL) and yields a cell that is Ade− and G418S (middle left). Repair of the DSB by GC results in the restoration of the rad21+ sequence on Ch16 and yields a cell that is Ade+ and G418S (bottom right). Long-tract GC events generate cells that are Ade+ and G418S (bottom left). Repair of the DSB by NHEJ or SCC yields cells that are Ade+ and G418R (middle right) and are indistinguishable from the parental strain.
TABLE 2.
DSB repair results at 48 h postinductiona
| Strain | % GC | % SCC/NHEJ | % CL | Total no. of colonies |
|---|---|---|---|---|
| WT (Th805) | 42 ± 3 | 47 ± 2 | 10 ± 2 | 3,615 |
| rhp51 | 2 ± 1 | 47 ± 14 | 51 ± 13 | 3,108 |
| rhp57 | 28 ± 14 | 63 ± 19 | 9 ± 6 | 1,759 |
| swi5 | 34 ± 8 | 35 ± 0 | 32 ± 6 | 4,210 |
| rhp57 swi5 | 1 ± 0 | 42 ± 12 | 57 ± 12 | 2,915 |
| eme1 | 58 ± 20 | 22 ± 14 | 20 ± 6 | 1,167 |
| eme1 rhp57 | 9 ± 6 | 75 ± 17 | 16 ± 11 | 1,817 |
| eme1 swi5 | 13 ± 2 | 57 ± 6 | 30 ± 4 | 3,065 |
| rqh1 | 12 ± 4 | 77 ± 5 | 10 ± 4 | 5,569 |
| rqh1 K547I | 43 ± 7 | 42 ± 5 | 15 ± 4 | 2,425 |
| rqh1 rhp51 | 10 ± 5 | 61 ± 8 | 30 ± 8 | 1,477 |
| rqh1 rhp57 | 22 ± 7 | 65 ± 7 | 14 ± 0 | 1,071 |
| rqh1 swi5 | 12 ± 6 | 75 ± 8 | 13 ± 4 | 2,742 |
| swi5 eme1 rqh1 | 31 ± 8 | 52 ± 8 | 16 ± 3 | 2,420 |
For each genetic background the repair assay was repeated independently at least three times. More than 1,000 colonies were scored per strain. Spontaneous loss of the chromosome was considered when calculating GC and CL frequencies. The average values and standard errors between the independent experiments are shown. We did not observe any measurable loss of viability following HO induction in any strain.
FIG. 3.
Crossovers can be detected by PFGE. (A) (Top) Schematic diagram of the 3.5-Mb ChIII (black) and the 0.53-Mb Ch16 (gray); the relative positions of the centromeres (circles), rad21+ (r21), MATa, and AsiSI sites are indicated. (Middle and bottom) The predicted structures of the crossover products and the estimated sizes of an AsiSI digest are shown. (B) A representative pulsed-field gel showing crossover and noncrossover products from the rqh1Δ background. ChI, ChII, ChIII, and Ch16 are visible in lanes M (marker), 1, 4, 6, 8, and 9, representing noncrossovers. Lanes 2, 3, 5, 7, and 10 contain ChI, ChII, and the two crossover chromosomes (CO Chs) of approximately 2 Mb. In some lanes (3, 7, and 10) the crossover chromosomes separate into distinct bands, while they migrate as a single band in others (lanes 2 and 5). We assume that these differences are due to variations in the size of ribosomal DNA sequences found on ChIII. (C) Southern blot of AsiSI-digested chromosomal DNA from a noncrossover control (lane C) and two individual crossover colonies (lanes 1 and 2). All three samples were recovered from the WT background. Bands were visualized with a radiolabeled probe generated from rad21+ sequences. The indicated sizes are estimated based on a commercial S. cerevisiae chromosome marker. An AsiSI digest of chromosomes from a noncrossover colony yields a 0.98-Mb fragment from ChIII and an intact Ch16 (lane C). A digest of chromosomes from a crossover colony yields predicted fragments of 0.72 Mb and 0.78 Mb as indicated in the middle and bottom subpanels of panel A. The actual sizes of the AsiSI fragments (arrows) differ slightly from the predicted sizes due to uncertainty about the telomere lengths of Ch16. *, band likely due to a partial AsiSI digest based on the fact that two separate probes (ade6+ and rad21+) detected the same product.
GC and crossovers depend on Rhp51.
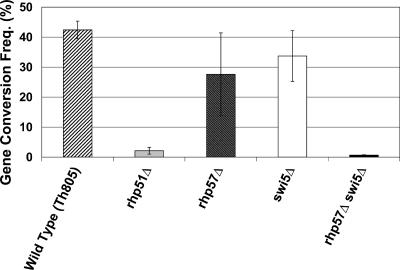
Rhp51 is central to the HR process during DSB repair, so predictably the loss of rhp51+ severely limited the formation of GC products and crossover intermediates in this system. In wild-type (WT) cells, at 48 h post-HO induction, 42% ± 3% of DSBs are repaired by GC (Fig. 4; Table 2) while 10% ± 2% are not repaired, seen as CLs (Table 2). Not surprisingly, as shown in Table 2, only 2% ± 1% of DSBs in a Th805 rhp51Δ strain are repaired by GC by 48 h (Fig. 4; Table 2), with the majority of DSBs failing to be repaired (51% ± 13% CL [Table 2]). This reduction in GC frequency in the rhp51Δ background is consistent with a previous report using the HO-induced DSB repair system (46). Since GCs were rare in the absence of Rhp51, only 38 GCs were isolated and analyzed. None contained crossovers (Table 3; Fig. 5A). Thus, GC and crossover formation during DSB repair are dependent on Rhp51.
FIG. 4.
Comparison of GC frequencies in the WT, rhp51Δ, rhp57Δ, swi5Δ, and rhp57Δ swi5Δ backgrounds.
TABLE 3.
Crossover formation in the different genetic backgroundsa
| Strain | Total no. of GCs | Crossovers
|
SE | |
|---|---|---|---|---|
| No. | % | |||
| WT (Th805) | 104 | 13 | 12.5 | 3.2 |
| rhp51Δ | 38 | 0 | 0.0 | 0.0 |
| rhp57Δ | 51 | 15 | 29.4 | 6.4 |
| swi5Δ | 158 | 6 | 3.8 | 1.5 |
| rhp57Δ swi5Δ | 32 | 0 | 0.0 | 0.0 |
| eme1Δ | 70 | 1 | 1.4 | 1.4 |
| eme1Δ rhp57Δ | 44 | 2 | 4.5 | 3.1 |
| eme1Δ swi5Δ | 88 | 3 | 3.4 | 1.9 |
| rqh1Δ | 79 | 32 | 40.5 | 5.5 |
| rqh1Δ rhp51Δ | 16 | 0 | 0.0 | 0.0 |
| rqh1Δ rhp57Δ | 30 | 0 | 0.0 | 0.0 |
| rqh1Δ swi5Δ | 113 | 37 | 32.7 | 4.4 |
| swi5Δ eme1Δ rqh1Δ | 92 | 33 | 35.9 | 5.0 |
| rqh1 K547I | 68 | 25 | 36.8 | 5.8 |
| rqh1 K547I rhp57Δ | 114 | 6 | 5.3 | 2.1 |
| top3-15 | 71 | 17 | 23.9 | 5.1 |
| eme1Δ top3-15 | 110 | 17 | 15.5 | 3.4 |
PFGE was used to determine whether colonies that had repaired the DSB by GC had experienced a crossover. These data represent the results of a minimum of three independent experiments. The standard error for each strain was calculated assuming crossover percentage as a binomial variable. For comparisons discussed in the text a Pearson χ2 test was performed.
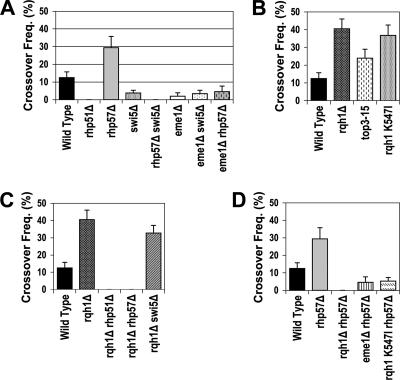
FIG. 5.
Comparisons of crossover frequencies in selected genetic backgrounds. (A) WT (Th805), rhp51Δ, rhp57Δ, swi5Δ, rhp57Δ swi5Δ, eme1Δ, eme1Δ swi5Δ, and eme1Δ rhp57Δ. (B) WT (Th805), rqh1Δ, top3-15, and rqh1 K547I. (C) WT (Th805), rqh1Δ, rqh1Δ rhp51Δ, rqh1Δ rhp57Δ, and rqh1Δ swi5Δ. (D) WT (Th805), rhp57Δ, rqh1Δ rhp57Δ, eme1Δ rhp57Δ, and rqh1 K547I rhp57Δ.
Swi5 and Rhp57 represent two branches of the Rhp51 GC pathway.
Previous studies have shown that Rhp55-Rhp57 and Swi5-Sfr1 represent alternative processes in HR in S. pombe (4, 18, 24, 28). We wished to determine if they represented the sole pathways for Rhp51-dependent DSB repair. In our DSB repair assay, loss of swi5+ slightly reduced the frequency of repair by GC to 34% ± 8% (Table 2; Fig. 4), as did deletion of rhp57+ (28% ± 14% [Table 2; Fig. 4]); both reductions were much less than that seen with the loss of rhp51+. We next created a Th805 rhp57Δ swi5Δ strain for analysis. When a DSB was induced and the resulting colonies were analyzed, the majority of colonies that formed had lost Ch16 (57% ± 12% [Table 2]), comparable to the results observed in rhp51Δ cells (51% ± 13% [Table 2]), and GC frequencies were also essentially the same as in rhp51Δ (1% ± 0% compared to 2% ± 1% [Table 2; Fig. 4]). As with Th805 rhp51Δ, the low number of GC events limited the number of colonies available for crossover analysis. Of 32 Th805 rhp57Δ swi5Δ colonies analyzed, none contained crossovers (Table 3; Fig. 5A). These data provide further evidence that Swi5-Sfr1 and Rhp55-Rhp57 represent parallel pathways for Rhp51-dependent GC.
Crossovers in WT cells depend on the action of Mus81-Eme1.
We next turned our attention to the process of crossover formation. We first determined the crossover frequency in WT cells by analyzing the chromosomes of colonies that arose from cells that had repaired their DSB by GC. Out of 104 colonies examined, 13 (12.5% ± 3.2%) contained crossovers (Fig. 5A; Table 3). We next investigated the mechanism by which these crossovers arose. Recent studies of S. pombe have suggested that during meiosis, crossovers arise largely through the action of Mus81-Eme1 (8, 18, 20, 43). To ask if the crossovers formed during mitotic DSB repair in WT cells depend on Mus81-Eme1, we created a Th805 eme1Δ strain. PFGE analysis of chromosomes from 70 eme1Δ colonies that repaired their DSBs by GC revealed a single crossover (Fig. 5A; Table 3). This result shows that the crossovers formed during GC of DSBs in WT cells depend on the action of Mus81-Eme1. These data strengthen an earlier genetic study which suggested that Mus81-dependent and -independent crossovers form in vegetative cells (43). As far as we are aware, this is the first direct observation demonstrating a role for Mus81-Eme1 in crossover formation in mitotic cells during the repair of a DSB. These data contrast with studies of S. cerevisiae using a similar DSB repair system, which concluded that Mus81/Mms4 played no role in crossover formation in mitotic cells (30).
Eme1 acts downstream of Swi5 in crossover formation.
Previous studies have shown that the extremely low spore viability of mus81Δ/eme1Δ cells is largely suppressed by the additional loss of Swi5 and suggest that Eme1 acts downstream of Swi5 in meiosis (18). Having shown that mitotic crossovers depend on Eme1, we next wanted to know if Eme1 also functions downstream of Swi5 in mitotic cells. We checked the frequency of crossovers in colonies that had repaired their DSBs by GC in the swi5Δ background and found that of 158 colonies analyzed only six contained crossovers, a rate of 3.8% ± 1.5% (Table 3; Fig. 5A). This is a 3.3-fold reduction from the WT value and indicates that Swi5 plays a significant role in the generation of crossovers in WT cells. We next created a Th805 eme1Δ swi5Δ strain to ask if the remaining crossovers formed in the absence of Swi5 depend on Eme1. Analysis of chromosomes from 88 colonies identified three that had crossovers, a rate of 3.4% ± 1.9%, not significantly different from the 3.8% ± 1.5% observed in a swi5Δ background (Table 3; Fig. 5A; P = 1). Thus, the crossovers formed in the absence of Swi5 do not depend on Eme1 and it appears that the Eme1-dependent crossovers observed in WT cells also require Swi5.
When we blocked repair by the Rhp57-dependent pathway using Th805 rhp57Δ, crossovers increased to 29.4% ± 6.4% (Table 3; Fig. 5A). In this background, only Swi5-dependent GCs are produced and crossovers should be largely dependent on Mus81-Eme1. As expected, when we tested this possibility in an rhp57Δ eme1Δ background we found that only 4.6% ± 3.1% of GCs formed crossovers (Table 3; Fig. 5A). Together these data suggest that Mus81-Eme1 acts downstream of Swi5 to produce crossovers, while Rhp57-mediated GCs largely result in noncrossover products (only 3.8% ± 1.5% of GCs in a swi5Δ background resulted in crossovers [Table 3; Fig. 5A]).
While Ellermeier et al. (18) had shown that loss of swi5+ largely suppressed the spore inviability of a mus81 mutant, the frequency of crossover formation in a swi5Δ mus81Δ background was not shown. If Mus81/Eme1 acts downstream of Swi5 in crossover formation, then the meiotic crossover frequency of a swi5 mus81 double mutant should be similar to that of a swi5 mutant. To test this possibility, we created WT, swi5Δ, eme1Δ, and swi5Δ eme1Δ strains and determined their intergenic meiotic crossover frequencies using ade6 and arg1 on ChIII as markers. The results in Table 4 show that the meiotic crossover frequencies of these markers in swi5Δ cells compared to those in swi5Δ eme1Δ cells were not significantly different (13.9% and 12.0%, respectively; P = 0.36). These data further support a model of Mus81/Eme1 acting downstream of Swi5 in promoting crossovers.
TABLE 4.
Eme1 acts downstream of Swi5 in meiotic crossover formation
| Straina | R1b | R2b | Totalc | % ± SEd | cMe | Fold reductionf |
|---|---|---|---|---|---|---|
| WT | 190 | 209 | 1,112 | 35.9 ± 1.4 | 63.3 | |
| swi5Δ | 43 | 42 | 700 | 12.1 ± 1.2g | 13.9 | 4.6 |
| eme1Δ | 12 | 2 | 899 | 1.6 ± 0.4 | 1.6 | 39.6 |
| swi5Δ eme1Δ | 57 | 39 | 900 | 10.7 ± 1.0g | 12.0 | 5.3 |
Data were obtained for crosses between ade6 and arg1 single mutants of the indicated backgrounds: WT (sz1663 × sz1058), swi5Δ (sz1710 × sz1493), eme1Δ (sz1503 × sz1686), and swi5Δ eme1Δ (sz1718 × sz1720). Table 1 shows the genotypes.
R1 is the total number of prototrophs and R2 is the total number of double auxotrophs found in each background.
Three independent crosses of at least two different isolates were carried out for each strain.
Percentage of total recombinants and standard error for proportions.
Calculated from the formula of Haldane (1919), x = −1/2ln(1 − 2R), where x is the genetic distance in morgans and R is the recombinant fraction.
The ratio of average centimorgans in the WT to average centimorgans in each mutant strain.
The difference between these two values is not statistically significant. Analysis was done by χ2, P = 0.36.
Rqh1 blocks crossovers from forming during DSB repair.
RecQ mutants in several organisms show elevated levels of crossing over. Both Sgs1 and BLM have been proposed to suppress crossovers by resolving dHJs into noncrossover products. We previously constructed a Th805 rqh1Δ strain and observed that the rqh1Δ mutant showed a reduced frequency of DSB repair by GC (29). We measured the level of crossing over during GC in the rqh1Δ background and found that the crossover frequency increased more than threefold over the WT value, to 40.5% ± 5.5% (Table 3; Fig. 5B). These data indicate that Rqh1 functions to efficiently suppress crossover formation.
Many RecQ helicases act together with topoisomerase III (Top3) in various processes. In S. cerevisiae, a top3 mutant was shown to share a common phenotype with an sgs1 mutant in a DSB repair assay (30). In S. pombe, top3 mutants are inviable (22, 39). We constructed a conditional-lethal mutant, the top3-15 strain, and analyzed how inactivation of Top3 affected crossover frequencies. At semipermissive temperatures (32°C), 23.9% ± 5.1% of GCs contained crossovers in this background (Table 3; Fig. 5B), a nearly twofold increase over the WT value. The finding that crossovers were not elevated to the same level as seen in an rqh1Δ background is not surprising, considering that the assay was carried out at semipermissive temperatures.
The helicase activity of Rqh1 is essential for its role in preventing crossovers.
Helicase-dead Rqh1 mutants have intermediate phenotypes for DNA damage sensitivity and suppression of top3Δ lethality compared to the deletion mutant (2, 39). We recently reported that while rqh1Δ mutants show a decreased frequency of DSB repair by GC, a helicase-dead mutant (rqh1:K547I [39]) showed near-WT frequencies of GC (29). These data indicate that Rqh1 has both helicase-dependent and -independent functions. We asked whether the helicase function of Rqh1 plays a role in crossover suppression during GC. PFGE analysis of chromosomes from Th805 rqh1 K547I cells that had repaired the DSB by GC revealed that 25 of 68 (36.8% ± 5.9% [Table 3; Fig. 5B]) had chromosome patterns consistent with repair involving a crossover. This demonstrates the requirement of the helicase activity of Rqh1 in suppressing crossing over during GC.
Rqh1 suppresses crossing over downstream of Rhp57.
Various phenotypes of RecQ mutants are suppressed by loss of HR genes such as RAD51, RAD55, and RAD57 (21, 28, 37, 40). This has led to proposals that RecQ helicases function during the later stages of HR. We asked whether the increased frequency of crossovers in the rqh1Δ mutant depends on the function of proteins that act in the earlier stages of HR such as Rhp51, Rhp57, and Swi5. We analyzed 16 GCs recovered from the Th 805 rqh1Δ rhp51Δ mutant and saw that none of these GCs resulted in crossovers (Table 3; Fig. 5C). This result shows that crossovers in the rqh1Δ mutant depend on the activity of Rhp51 and that Rqh1 is not required to suppress crossovers in Rhp51-independent GCs.
Next, we measured the frequency of crossovers in the Th805 rqh1Δ rhp57Δ background. We found that the crossover frequency of this mutant was 0% (0 of 30 colonies analyzed [Table 3; Fig. 5C]) versus the rqh1Δ mutant background of 40.5% ± 5.5% (Table 3; Fig. 5C). This shows that the bulk of the crossovers in the rqh1Δ mutant arise through an Rhp57-dependent process.
In the absence of Swi5, GCs are processed through the Rhp57-dependent pathway. When we examined GCs in an rqh1Δ swi5Δ background, we found that 32.7% ± 4.4% contained crossovers (Table 3; Fig. 5C). Consistent with the above results, most crossovers that form in the absence of Rqh1 do not depend on Swi5, and Rqh1 acts primarily in the Rhp57-dependent pathway to block crossing over.
Rqh1 is required for Swi5-dependent crossover formation.
The total absence of crossovers that we observed in the rqh1Δ rhp57Δ double mutant presented us with a conundrum. Our data indicate that the majority of GCs are processed by the Rhp57-dependent pathway where they encounter Rqh1-Top3 and crossing over is blocked. In the absence of Rhp57, Swi5 remains as the only pathway for Rhp51-dependent GCs, as demonstrated above. We observed elevated levels of Mus81-Eme1-dependent crossovers in the rhp57Δ background (rhp57Δ, 29.4% ± 6.4% of GCs; rhp57Δ eme1Δ, 4.6% ± 3.1% of GCs [Table 3; Fig. 5D]). Yet, no crossovers form in the rqh1Δ rhp57Δ background (Table 3; Fig. 5D). All of the GCs in the rhp57Δ background are Swi5 dependent, and many should result in crossovers through the action of Mus81-Eme1. The fact that the rqh1Δ rhp57Δ double mutant does not behave like an rhp57Δ single mutant implies that Rqh1 has another Rhp57-independent function. Since our data suggest that in the absence of Rhp57 crossovers depend on Swi5 and Eme1, this Rqh1 function likely influences the Swi5-dependent pathway. Our data do not allow us to conclude whether Rqh1 acts upstream or downstream of Swi5 or whether it acts with Mus81-Eme1 directly or in some independent event. Using the rqh1 helicase-dead mutant (rqh1 K547I), we asked if Rqh1's helicase activity was necessary for promoting crossovers in the Swi5-dependent pathway. In the rhp57Δ rqh1 K547I background crossover frequencies were also reduced compared to those in the rhp57Δ background (rhp57Δ rqh1 K547I, 5.3% ± 2.1% of GCs, versus rhp57Δ, 29.4% ± 6.4% of GCs [Table 3; Fig. 5D]). This indicates that the helicase activity is required for this Rhp57-independent function of Rqh1.
Evidence that crossovers arise by two different mechanisms during DSB repair.
One remaining question was whether crossovers formed in the absence of Rqh1 depend on Mus81-Eme1. This question could not be answered directly, as the rqh1Δ mus81/eme1Δ mutant is inviable (9, 14). In S. cerevisiae the synthetic lethality between mus81-eme1 and sgs1 is rescued by the loss of genes of the RAD52 epistasis group (19). A similar suppression is not observed in S. pombe (16; M. Whitby, personal communications). Swi5 represents a parallel pathway for HR in S. pombe, and it is known that deletion of swi5+ can suppress the meiotic defect of mus81Δ (18). We speculated that the loss of Swi5 activity might suppress rqh1Δ eme1Δ synthetic lethality. Thus, we made the appropriate crosses and found that a swi5Δ rqh1Δ eme1Δ triple mutant was viable. Analysis of this suppression will be described elsewhere. We created a Th805 swi5Δ rqh1Δ eme1Δ strain and tested it in the DSB repair assay. First, the frequencies of CLs, GCs, and SCC/NHEJ were measured (Table 2). As with several other strains lacking Rqh1, compared to WT cells, GC frequencies are reduced while SCC numbers are elevated. More importantly, we found that in this triple mutant, 33 of 92 colonies (35.9% ± 5.0% [Table 3]) that repaired their DSBs by GC contained crossovers. Consistent with this finding, the excess crossovers found in a top3-15 mutant were also not dependent on Eme1 (15.5% ± 3.5% of GCs in a top3-15 eme1Δ double mutant produced crossovers [Table 3]). These data indicate that the crossovers observed in the rqh1Δ background do not require Eme1 and reveal an efficient Mus81-Eme1-independent mechanism for generating crossover products in S. pombe, as has been seen in budding yeast (30).
DISCUSSION
There are several significant findings in this paper that provide new insights into the process of GC during DSB repair in S. pombe. First, similarly to meiotic cells, the crossovers that form during DSB repair in mitotic cells appear to arise primarily through the action of Mus81-Eme1. Second, Mus81-Eme1 appears to function downstream of Swi5 in crossover formation in mitotic and meiotic cells alike. Third, we found that Rqh1-Top3 acts downstream of Rhp57 to block crossover formation during DSB repair. Fourth, in cells lacking Rqh1-Top3 activity, crossovers form that do not depend on Mus81-Eme1, suggesting that a second mechanism for crossover formation exists in S. pombe. Finally, our studies indicate that Rqh1 has an Rhp57-independent function that could be involved in the production of Mus81-Eme1-dependent crossovers.
Crossover formation in mitotic cells.
Similarly to results reported for S. cerevisiae (30), we found that crossovers formed only in a small fraction of WT cells that repaired an HO-induced DSB by GC. However, in contrast to S. cerevisiae, where no evidence was found to implicate Mus81-Mms4/Eme1 in mitotic crossovers (30), we found that in the absence of Eme1, crossovers rarely formed (1 out of 70). This suggests that most crossover formation in S. pombe WT cells requires Mus81-Eme1, a finding that is consistent with other studies of S. pombe, where Mus81-Eme1 has been shown to be involved in the primary mechanism for crossover formation in meiotic cells (8, 18, 43). Direct comparisons between the S. pombe and S. cerevisiae site-specific DSB systems should take into consideration that there is a region of heterology at the DSB site in the S. pombe system that does not exist in the S. cerevisiae system. However, there is no reason to believe that Mus81-Eme1 would be involved in processing this heterology, as evidenced by similar levels of GC and CL in WT and eme1 mutant backgrounds (Table 2).
We found that, in mitosis as well as in meiosis, Mus81-Eme1 appears to act downstream of Swi5 to produce crossovers. This conclusion is based on the following. In an swi5Δ background, a low number of crossovers formed, 3.8%, compared to 12.5% in WT cells. These Swi5-independent crossover events are not dependent on Mus81-Eme1, as seen in our analysis of swi5Δ eme1Δ cells, where crossovers occurred at a similar frequency (3.4% versus 3.8%, P = 1). These results indicate that loss of Swi5 prevents Mus81-Eme1-dependent crossovers from forming and are consistent with Mus81-Eme1 acting downstream of Swi5 in this process. Interestingly in meiotic cells loss of Swi5 also blocks Eme1-dependent crossovers. As shown in Table 4, meiotic crossover frequencies in swi5Δ and swi5Δ eme1Δ backgrounds are not significantly different, suggesting that the mechanisms for crossover formation are similar in mitotic and meiotic cells.
Mus81-Eme1 appears not to be the only mechanism for crossover formation in S. pombe. We found that in various genetic backgrounds, Mus81-Eme1-independent crossovers formed during GC of DSBs. The most striking example was seen in cells lacking Rqh1-Top3 activity, where crossovers were detected in 40.5% of rqh1Δ colonies that had repaired the DSB by GC. We went on to show that these crossovers arose through a Mus81-Eme1-independent process. Because of the synthetic lethality between rqh1− and mus81−/eme1− we could not create the double mutant strain. Our finding that the inviability of the rqh1Δ eme1Δ mutant could be overcome by loss of swi5+ provided an approach to test whether the crossovers formed in an rqh1Δ background depend on Mus81-Eme1. In the rqh1Δ swi5Δ eme1Δ strain, crossovers formed in 35.9% of colonies that repaired the HO-induced DSB by GC. This is similar to the number of crossovers found in a single rqh1Δ mutant (40.5% versus 35.9%, P = 0.63) and indicates that the majority of crossovers formed in the absence of Rqh1-Top3 arise by a process that is independent of Mus81-Eme1. How these crossovers form cannot be predicted from the current data.
In parallel experiments we took advantage of a conditional-lethal mutant of the top3 strain, the top3-15 strain, previously constructed in our laboratory. At semipermissive temperatures, 23.9% of GC events in a Th805 top3-15 background resulted in crossovers, an almost twofold increase over the value for WT cells. When we analyzed Th805 top3-15 eme1Δ colonies that had repaired their DSBs by GC, 15.5% contained crossovers. These results are consistent with our finding in the rqh1Δ swi5Δ eme1Δ background and provide further evidence that a Mus81-Eme1-independent mechanism for forming crossovers exists in S. pombe. Only two mechanisms for crossover formation have been proposed in eukaryotes: Mus81-Eme1/Mms4 cleavage of a nicked dHJ and the resolution of a dHJ by a resolvase. While the existence of an as-yet-undescribed process for crossover formation cannot be ruled out, based on our current understanding we suggest that these Mus81-Eme1-independent crossovers arise by resolution of a dHJ, presumably through an HJ resolvase.
Crossover suppression by Rqh1.
Our results demonstrate that Rqh1 plays a major role in crossover suppression in S. pombe. This is based on our findings that in the absence of Rqh1, crossovers increase 28% (from 12.5% of GC events in WT cells to 40.5% in rqh1Δ cells). The suppression of crossovers by Rqh1 is dependent on its helicase activity, demonstrated by the similarity in results between our helicase-dead and deletion mutants. However, the mechanism of this suppression is not clear from our studies. The Rqh1 helicase could prevent crossover formation by promoting SDSA, either by displacing the invading strand to limit conversion tract length or by blocking second-strand end capture to prevent dHJ formation. Another possibility is that Rqh1 blocks crossovers by facilitating dissolution, where RecQ compresses the dHJ, which is then deconcatenated by Top3 (53). This process was proposed by Wu and Hickson and is based on in vitro studies using purified BLM, human topoisomerase IIIα, and BLAP75 to resolve a synthetic dHJ (52, 53). This mechanism has also been proposed for suppression of crossovers by Sgs1-Top3 in S. cerevisiae, although no direct evidence has been provided yet (30).
From our data, we cannot explain the mechanism by which Rqh1 suppresses crossovers. We have evidence that suggests that Rqh1-Top3 may play a larger role in suppressing crossovers in fission yeast while Sgs1-Top3 in S. cerevisiae appears to play a less extensive role in crossover suppression; loss of Sgs1-Top3 led to a 2.4-fold increase in crossover frequencies, from 4.8% ± 1% in WT cells to 11.7% ± 2.4% in sgs1 cells (30). In addition we found that in S. pombe crossover suppression by Rqh1-Top3 requires Rqh1's helicase activity. This result contrasts with conclusions for S. cerevisiae, where it has been reported that suppression of crossover formation by Sgs1-Top3 is independent of its helicase activity (38). However, the extent to which dissimilar experimental systems may contribute to this difference cannot be determined. While the high level of suppression is more in keeping with a role for Rqh1 in promoting SDSA, further studies will need to be carried out to determine the mechanism of crossover suppression by Rqh1.
Are Swi5-dependent and Rhp57-dependent intermediates different?
Swi5-Sfr1 and Rhp55-Rhp57 represent the only Rhp51-dependent pathways for GCs during DSB repair. This is supported by our findings that only 1% of DSBs were repaired by GC in a rhp57Δ swi5Δ double mutant, a number similar to that found in an rhp51Δ background. In WT cells, the majority of GC events are processed by the Rhp57-dependent pathway, while most crossovers form via the Swi5-dependent pathway. Interestingly, in the absence of Swi5, a small number of crossovers form downstream of Rhp57 (3.8% of GCs result in crossovers in swi5Δ cells) and therefore arise through the same intermediate that Rqh1 normally recognizes to produce noncrossovers. These Rhp57-dependent crossovers do not require Mus81-Eme1 (3.4% of GCs result in crossovers in a swi5Δ eme1Δ background), suggesting that this intermediate is not a substrate for Mus81-Eme1 activity. Likewise, in the absence of Rqh1, crossovers increase dramatically (from 12.5% in the WT to 40.5% in rqh1Δ cells), and yet these crossovers form independently of Mus81-Eme1 activity (40.6% of GCs form crossovers in an rqh1Δ eme1Δ swi5Δ strain). While our studies do not directly address the structural characteristics of the intermediate that forms downstream of Rhp57, they suggest that this intermediate is different from the one that forms downstream of Swi5. Furthermore we found that Rqh1 cannot block crossover formation downstream of Swi5 (in the rhp57Δ strain 29.4% of GCs formed Mus81-Eme1-dependent crossovers). This increase in crossover formation occurs in the presence of the Rqh1 protein, indicating that Rqh1 cannot prevent the Swi5-dependent intermediates from forming crossovers. Together, these data support a model where the two pathways of Rhp51-dependent DSB repair form distinct intermediates.
Rqh1 appears to participate in Mus81-Eme1 crossover formation.
Results from these studies suggest that Rqh1 contributes to the Mus81-Eme1 process for crossover formation in mitotic cells. In an rhp57Δ rqh1+ background, crossovers were detected in 29.4% of colonies that repaired their DSBs by GC. This result would seem to be explained by more GC events being diverted down the Swi5-dependent pathway that are converted into crossovers by Mus81-Eme1, a result consistent with our finding that crossovers were reduced to 4.6% in an rhp57Δ eme1Δ double mutant. However in an rqh1Δ rhp57Δ background, no crossovers were detected, which suggested that Rqh1-Top3 might be required in crossover formation by Mus81-Eme1. There are human data that seem to support this idea. Zhang et al. recently reported that human Mus81 and BLM protein colocalize at sites of replication arrest (55). Furthermore, they showed that BLM stimulated the endonuclease activity of Mus81. Together these data provide an intriguing possibility: that Rqh1 acts downstream of Swi5 in a process that is important for Mus81-Eme1 function in forming crossovers.
Model.
Based on the findings reported here, combined with previous results, we propose the following model for the process of DSB repair by GC in S. pombe (Fig. 6). Essentially all GC events occur through an Rhp51-dependent process (GCs were 2% in rhp51Δ cells), which has two subpathways, one that is Rhp57 dependent and one that is Swi5 dependent (GCs were 1% in rhp57Δ swi5Δ cells). Most GC events are processed down the Rhp57-dependent pathway, where Rqh1-Top3 efficiently blocks crossover formation. Our results do not allow us to conclude whether Rqh1 promotes noncrossovers through SDSA or dissolution of dHJs. In the absence of Rqh1, crossovers likely form through the random resolution of a dHJ. A smaller fraction of GC events are processed by the Swi5-dependent pathway, where intermediates can be acted on by Mus81-Eme1. Both crossovers and noncrossovers form in this pathway (29.4% of GCs formed crossovers in an rhp57Δ background), and the crossovers are largely dependent on Mus81-Eme1 (4.6% of GCs formed crossovers in an rhp57Δ eme1Δ background). We cannot determine from these data whether intermediates processed by Mus81-Eme1 all result in crossovers, as one proposed mechanism for Mus81-Eme1 suggests (43). It also seems clear that a Mus81-Eme1-independent process exists downstream of Swi5 that leads to noncrossovers. The proteins involved in this process remain to be identified.
FIG. 6.
Model for the process of crossover formation in S. pombe following DSB repair. Two pathways can process DSBs into Rhp51-dependent GCs, Swi5-Sfr1 and Rhp55-Rhp57. In the Swi5-Sfr1 pathway an intermediate forms that is processed by Mus81-Eme1 into crossovers. A separate process produces noncrossovers. In the Rhp55-Rhp57 pathway, Rqh1-Top3 largely blocks crossover formation, although a low number of crossovers can form in a swi5 mutant. In cells lacking Rqh1-Top3 activity, crossovers readily form. CO, crossover; NCO, noncrossover.
ADDENDUM
While the manuscript was being revised, a paper by Cromie et al. reported on data for S. pombe suggesting that only single HJs form during meiosis (12). This could also be the case in mitotic cells; however, this result does not change our interpretation of the data presented in this paper.
Acknowledgments
We appreciate the expert technical assistance of Gloria Osorio and Rachel M. Lytle. We also want to thank William Friedewald and Shing Lee for guidance on the statistical analysis of the data.
This research was funded by NIH grant CA072647. J.C.H. is a Ruth L. Kirschstein Fellow, GM20376.
Footnotes
Published ahead of print on 12 March 2007.
REFERENCES
- 1.Abraham, J., B. Lemmers, M. P. Hande, M. E. Moynahan, C. Chahwan, A. Ciccia, J. Essers, K. Hanada, R. Chahwan, A. K. Khaw, P. McPherson, A. Shehabeldin, R. Laister, C. Arrowsmith, R. Kanaar, S. C. West, M. Jasin, and R. Hakem. 2003. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 22:6137-6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad, F., C. D. Kaplan, and E. Stewart. 2002. Helicase activity is only partially required for Schizosaccharomyces pombe Rqh1p function. Yeast 19:1381-1398. [DOI] [PubMed] [Google Scholar]
- 3.Ajima, J., K. Umezu, and H. Maki. 2002. Elevated incidence of loss of heterozygosity (LOH) in an sgs1 mutant of Saccharomyces cerevisiae: roles of yeast RecQ helicase in suppression of aneuploidy, interchromosomal rearrangement, and the simultaneous incidence of both during mitotic growth. Mutat. Res. 504:157-172. [DOI] [PubMed] [Google Scholar]
- 4.Akamatsu, Y., D. Dziadkowiec, M. Ikeguchi, H. Shinagawa, and H. Iwasaki. 2003. Two different Swi5-containing protein complexes are involved in mating-type switching and recombination repair in fission yeast. Proc. Natl. Acad. Sci. USA 100:15770-15775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastin-Shanower, S. A., W. M. Fricke, J. R. Mullen, and S. J. Brill. 2003. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1-Rad10. Mol. Cell. Biol. 23:3487-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baudat, F., K. Manova, J. P. Yuen, M. Jasin, and S. Keeney. 2000. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell 6:989-998. [DOI] [PubMed] [Google Scholar]
- 7.Bennett, C. B., A. L. Lewis, K. K. Baldwin, and M. A. Resnick. 1993. Lethality induced by a single site-specific double-strand break in a dispensable yeast plasmid. Proc. Natl. Acad. Sci. USA 90:5613-5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates III, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107:537-548. [DOI] [PubMed] [Google Scholar]
- 9.Boddy, M. N., A. Lopez-Girona, P. Shanahan, H. Interthal, W. D. Heyer, and P. Russell. 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20:8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaganti, R. S., S. Schonberg, and J. German. 1974. A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc. Natl. Acad. Sci. USA 71:4508-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciccia, A., A. Constantinou, and S. C. West. 2003. Identification and characterization of the human mus81-eme1 endonuclease. J. Biol. Chem. 278:25172-25178. [DOI] [PubMed] [Google Scholar]
- 12.Cromie, G. A., R. W. Hyppa, A. F. Taylor, K. Zakharyevich, N. Hunter, and G. R. Smith. 2006. Single Holliday junctions are intermediates of meiotic recombination. Cell 127:1167-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davey, S., C. S. Han, S. A. Ramer, J. C. Klassen, A. Jacobson, A. Eisenberger, K. M. Hopkins, H. B. Lieberman, and G. A. Freyer. 1998. Fission yeast rad12+ regulates cell cycle checkpoint control and is homologous to the Bloom's syndrome disease gene. Mol. Cell. Biol. 18:2721-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doe, C. L., J. S. Ahn, J. Dixon, and M. C. Whitby. 2002. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277:32753-32759. [DOI] [PubMed] [Google Scholar]
- 15.Doe, C. L., J. Dixon, F. Osman, and M. C. Whitby. 2000. Partial suppression of the fission yeast rqh1(-) phenotype by expression of a bacterial Holliday junction resolvase. EMBO J. 19:2751-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doe, C. L., and M. C. Whitby. 2004. The involvement of Srs2 in post-replication repair and homologous recombination in fission yeast. Nucleic Acids Res. 32:1480-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edelmann, W., P. E. Cohen, B. Kneitz, N. Winand, M. Lia, J. Heyer, R. Kolodner, J. W. Pollard, and R. Kucherlapati. 1999. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat. Genet. 21:123-127. [DOI] [PubMed] [Google Scholar]
- 18.Ellermeier, C., H. Schmidt, and G. R. Smith. 2004. Swi5 acts in meiotic DNA joint molecule formation in Schizosaccharomyces pombe. Genetics 168:1891-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fabre, F., A. Chan, W. D. Heyer, and S. Gangloff. 2002. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA 99:16887-16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaillard, P. H., E. Noguchi, P. Shanahan, and P. Russell. 2003. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol. Cell 12:747-759. [DOI] [PubMed] [Google Scholar]
- 21.Gangloff, S., C. Soustelle, and F. Fabre. 2000. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25:192-194. [DOI] [PubMed] [Google Scholar]
- 22.Goodwin, A., S. W. Wang, T. Toda, C. Norbury, and I. D. Hickson. 1999. Topoisomerase III is essential for accurate nuclear division in Schizosaccharomyces pombe. Nucleic Acids Res. 27:4050-4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrigan, J. A., and V. A. Bohr. 2003. Human diseases deficient in RecQ helicases. Biochimie 85:1185-1193. [DOI] [PubMed] [Google Scholar]
- 24.Haruta, N., Y. Kurokawa, Y. Murayama, Y. Akamatsu, S. Unzai, Y. Tsutsui, and H. Iwasaki. 2006. The Swi5-Sfr1 complex stimulates Rhp51/Rad51- and Dmc1-mediated DNA strand exchange in vitro. Nat. Struct. Mol. Biol. 13:823-830. [DOI] [PubMed] [Google Scholar]
- 25.Heyer, W. D., K. T. Ehmsen, and J. A. Solinger. 2003. Holliday junctions in the eukaryotic nucleus: resolution in sight? Trends Biochem. Sci. 28:548-557. [DOI] [PubMed] [Google Scholar]
- 26.Hickson, I. D. 2003. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer 3:169-178. [DOI] [PubMed] [Google Scholar]
- 27.Holliday, R. 1964. A mechanism for gene conversion in fungi. Genet. Res. 5:282-304. [DOI] [PubMed] [Google Scholar]
- 28.Hope, J. C., M. Maftahi, and G. A. Freyer. 2005. A postsynaptic role for Rhp55/57 that is responsible for cell death in Deltarqh1 mutants following replication arrest in Schizosaccharomyces pombe. Genetics 170:519-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hope, J. C., S. M. Mense, M. Jalakas, J. Mitsumoto, and G. A. Freyer. 2006. Rqh1 blocks recombination between sister chromatids during double strand break repair, independent of its helicase activity. Proc. Natl. Acad. Sci. USA 103:5875-5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ira, G., A. Malkova, G. Liberi, M. Foiani, and J. E. Haber. 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115:401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang, Y. K., Y. H. Jin, Y. S. Shim, M. J. Kim, E. J. Yoo, R. H. Seong, S. H. Hong, and S. D. Park. 1995. Evidences for possible involvement of Rhp51 protein in mitotic events including chromosome segregation. Biochem. Mol. Biol. Int. 37:329-337. [PubMed] [Google Scholar]
- 32.Johnson, R. D., and M. Jasin. 2001. Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans. 29:196-201. [DOI] [PubMed] [Google Scholar]
- 33.Kaliraman, V., J. R. Mullen, W. M. Fricke, S. A. Bastin-Shanower, and S. J. Brill. 2001. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. J15:2730-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kanaar, R., J. H. Hoeijmakers, and D. C. van Gent. 1998. Molecular mechanisms of DNA double strand break repair. Trends Cell Biol. 8:483-489. [DOI] [PubMed] [Google Scholar]
- 35.Keeney, S. 2001. Mechanism and control of meiotic recombination initiation. Curr. Top. Dev. Biol. 52:1-53. [DOI] [PubMed] [Google Scholar]
- 36.Khanna, K. K., and S. P. Jackson. 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27:247-254. [DOI] [PubMed] [Google Scholar]
- 37.Klein, H. 2001. Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Delta with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157:557-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lo, Y. C., K. S. Paffett, O. Amit, J. A. Clikeman, R. Sterk, M. A. Brenneman, and J. A. Nickoloff. 2006. Sgs1 regulates gene conversion tract lengths and crossovers independently of its helicase activity. Mol. Cell. Biol. 26:4086-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maftahi, M., C. S. Han, L. D. Langston, J. C. Hope, N. Zigouras, and G. A. Freyer. 1999. The top3(+) gene is essential in Schizosaccharomyces pombe and the lethality associated with its loss is caused by Rad12 helicase activity. Nucleic Acids Res. 27:4715-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maftahi, M., J. C. Hope, L. Delgado-Cruzata, C. S. Han, and G. A. Freyer. 2002. The severe slow growth of Deltasrs2 Deltarqh1 in Schizosaccharomyces pombe is suppressed by loss of recombination and checkpoint genes. Nucleic Acids Res. 30:4781-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mills, K. D., D. O. Ferguson, and F. W. Alt. 2003. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 194:77-95. [DOI] [PubMed] [Google Scholar]
- 42.Myung, K., A. Datta, C. Chen, and R. D. Kolodner. 2001. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27:113-116. [DOI] [PubMed] [Google Scholar]
- 43.Osman, F., J. Dixon, C. L. Doe, and M. C. Whitby. 2003. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12:761-774. [DOI] [PubMed] [Google Scholar]
- 44.Osman, F., E. A. Fortunato, and S. Subramani. 1996. Double-strand break-induced mitotic intrachromosomal recombination in the fission yeast Schizosaccharomyces pombe. Genetics 142:341-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paques, F., and J. E. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prudden, J., J. S. Evans, S. P. Hussey, B. Deans, P. O'Neill, J. Thacker, and T. Humphrey. 2003. Pathway utilization in response to a site-specific DNA double-strand break in fission yeast. EMBO J. 22:1419-1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Symington, L. S. 2002. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 66:630-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szostak, J. W., T. L. Orr-Weaver, R. J. Rothstein, and F. W. Stahl. 1983. The double-strand-break repair model for recombination. Cell 33:25-35. [DOI] [PubMed] [Google Scholar]
- 49.Tsutsui, Y., T. Morishita, H. Iwasaki, H. Toh, and H. Shinagawa. 2000. A recombination repair gene of Schizosaccharomyces pombe, rhp57, is a functional homolog of the Saccharomyces cerevisiae RAD57 gene and is phylogenetically related to the human XRCC3 gene. Genetics 154:1451-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Gent, D. C., J. H. Hoeijmakers, and R. Kanaar. 2001. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2:196-206. [DOI] [PubMed] [Google Scholar]
- 51.Whitby, M. C., F. Osman, and J. Dixon. 2003. Cleavage of model replication forks by fission yeast Mus81-Eme1 and budding yeast Mus81-Mms4. J. Biol. Chem. 278:6928-6935. [DOI] [PubMed] [Google Scholar]
- 52.Wu, L., C. Z. Bachrati, J. Ou, C. Xu, J. Yin, M. Chang, W. Wang, L. Li, G. W. Brown, and I. D. Hickson. 2006. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc. Natl. Acad. Sci. USA 103:4068-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, L., and I. D. Hickson. 2003. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426:870-874. [DOI] [PubMed] [Google Scholar]
- 54.Yamagata, K., J. Kato, A. Shimamoto, M. Goto, Y. Furuichi, and H. Ikeda. 1998. Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases. Proc. Natl. Acad. Sci. USA 95:8733-8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, R., S. Sengupta, Q. Yang, S. P. Linke, N. Yanaihara, J. Bradsher, V. Blais, C. H. McGowan, and C. C. Harris. 2005. BLM helicase facilitates Mus81 endonuclease activity in human cells. Cancer Res. 65:2526-2531. [DOI] [PubMed] [Google Scholar]