Abstract
The cooperation of stem cell factor (SCF) and erythropoietin (Epo) is required to induce renewal divisions in erythroid progenitors, whereas differentiation to mature erythrocytes requires the presence of Epo only. Epo and SCF activate common signaling pathways such as the activation of protein kinase B (PKB) and the subsequent phosphorylation and inactivation of Foxo3a. In contrast, only Epo activates Stat5. Both Foxo3a and Stat5 promote erythroid differentiation. To understand the interplay of SCF and Epo in maintaining the balance between renewal and differentiation during erythroid development, we investigated differential Foxo3a target regulation by Epo and SCF. Expression profiling revealed that a subset of Foxo3a targets was not inhibited but was activated by Epo. One of these genes was Cited2. Transcriptional control of Epo/Foxo3a-induced Cited2 was studied and compared with that of the Epo-repressed Foxo3a target Btg1. We show that in response to Epo, the allegedly growth-inhibitory factor Foxo3a associates with the allegedly growth-stimulatory factor Stat5 in the nucleus, which is required for Epo-induced Cited2 expression. In contrast, Btg1 expression is controlled by the cooperation of Foxo3a with cyclic AMP- and Jun kinase-dependent Creb family members. Thus, Foxo3a not only is an effector of PKB but also integrates distinct signals to regulate gene expression in erythropoiesis.
Forkhead transcription factors regulate a multitude of developmental processes (40, 45). Subclass O (Foxo) of Forkhead transcription factors can be phosphorylated by protein kinase B (PKB), which results in transcriptional inactivation through nuclear export and cytosolic retention by 14-3-3 proteins (8, 13, 14, 39, 43, 68). Initial studies on the function of Foxo proteins in hematopoiesis pointed to a role in apoptosis and cell cycle regulation (9, 14, 17, 24). On the other hand, Foxo1 induces survival and maturation in thymocytes (46), and the activation of Foxo3a in erythroblasts induces differentiation, indicating that the role of Foxo proteins in hematopoiesis is diverse and probably cell type specific (4).
Erythroblasts can be expanded in vitro using serum-free medium supplemented with erythropoietin (Epo), stem cell factor (SCF), and glucocorticoids, which reflects the in vivo expansion of erythroblasts under stress conditions (7, 12, 25, 70). Immortal cultures of erythroblasts can reproducibly be established from p53−/− mice (60, 70). These cultures remain dependent on Epo, SCF, and glucocorticoids for their expansion and retain the ability to undergo complete differentiation into erythrocytes in the presence of Epo. The expansion of these cultures is dependent on Epo-induced activation of the tyrosine kinase receptor Ron/Stk (60), whereas differentiation relies on Epo-induced Stat5 phosphorylation (26). Both Epo and SCF activate the phosphatidylinositol 3-kinase (PI3K)-PKB pathway, although SCF induces phosphorylation of PKB more strongly (70). The inhibition of PI3K abrogates Epo/SCF-induced expansion of in vitro cultures, inducing differentiation instead, suggesting that pathways downstream of PI3K-PKB control the proliferation of erythroblasts (70). This was corroborated by in vivo experiments. Mice lacking the PI3K subunit p85 displayed transient fetal anemia with reduced numbers of burst-forming units-erythroid and CFU-erythroid (37). The lack of p85 did not increase apoptosis of erythroblasts and mast cells but decreased proliferation (29, 37, 48). Foxo3a−/− mice displayed compensated anemia with reticulocytosis, suggesting normal expansion but defects in erythrocyte maturation (18).
Both SCF and Epo were able to inhibit the expression of Foxo3a target genes Cdkn1b (p27KIP) and Btg1. Nevertheless, SCF delays erythroid differentiation, while Epo enables erythroid differentiation. By consequence, Foxo3a targets may be differentially regulated by Epo and SCF. Two lines of evidence support this. First, Foxo proteins integrate a variety of signaling pathways, and examples show cooperation with transforming growth factor β signaling, IκB kinase, Wnt signaling, and the Jak-Stat pathway (27, 36, 44, 61). Furthermore, Foxo transcription factors can mediate gene expression independent of their DNA binding ability, underlining the importance of transcriptional coregulators (57, 68). Second, although Epo and SCF have overlapping functions (i.e., activation of the PI3K and Ras-mitogen-activated protein kinase pathways), there are also differences. Epo specifically activates the Jak-Stat pathway and the tyrosine kinase receptor Ron, which recruits the adaptor Gab1 (21, 67). Other targets are induced by both Epo and SCF but may have stimulus-specific effects. Epo and SCF activate Btk, but only SCF-induced Btk protects from Trail-induced apoptosis (60). Along similar lines, Epo and SCF may induce the differential regulation of Foxo3a target genes.
Finally, it is not known to what extent Foxo3a is involved in Epo/SCF-mediated repression of gene expression following factor deprivation (42), nor do we know to what extent all Foxo3a target genes are regulated by Epo/SCF-induced activation of the PI3K-PKB pathway. To analyze the relation between Epo- and SCF-controlled signal transduction and Foxo3a activity, we investigated Foxo3a-, Epo-, and SCF-induced gene expression on an expressed-sequence-tag (EST) microarray containing 17,000 cDNAs (17K EST cDNA array). We found that Foxo3a target genes are differentially affected by growth factor stimulation, and we analyzed two clusters of Foxo3a-upregulated genes in more detail. One cluster encompasses target genes that are repressed by Epo/SCF and upregulated in differentiation (Cdkn1b/p27Kip, Btg1, Ccng2/Cyclin G2, and Ulk1), while the other cluster contains genes that are less obviously inhibited by Epo/SCF and that are not upregulated during differentiation (Dcn, Sesn1, and Cited2). Interestingly, Cited2 appeared to be a Foxo3a target gene that was induced instead of repressed by Epo. Data presented demonstrate that the alleged growth-stimulatory transcription factor Stat5 cooperates with the alleged growth-inhibitory transcription factor Foxo3a to control the expression of Cited2. In contrast, the upregulation of Btg1 during differentiation appeared to be reinforced by the cooperation of Foxo3a with the cyclic AMP (cAMP)-responsive transcription factor CREB/ATF1. Our data imply that Foxo3a functions to integrate and transmit multiple signals that cooperate to regulate the gene expression program of erythroblasts.
MATERIALS AND METHODS
Cells and reagents.
BA/F3 cells were cultured in RPMI 1640 (Invitrogen) supplemented with 10% fetal calf serum (HyClone; PerBio) and 10 ng/ml murine interleukin-3 (IL-3) (supernatant). 293T cells were cultured in Dulbecco's modified Eagle's medium-10% fetal calf serum and transfected by calcium phosphate as described previously (4). Erythroid progenitors derived from E14 fetal livers and the erythroid cell line I/11 were cultured in Stempro medium (Invitrogen) supplemented with 0.5 U/ml Epo (a kind gift of Ortho-Biotech, Tilburg, The Netherlands), 100 ng/ml SCF (supernatant), and 1 μM dexamethasone (Sigma-Aldrich) (70). To induce the differentiation of erythroblasts, the cells were cultured in Stempro medium supplemented with 5 U/ml Epo and 0.5 mg/ml iron-loaded transferrin (Scipac). Stable Foxo3a(A3):ER-expressing I/11 clones were generated using the retroviral expression vector pBabe as described previously (4). To activate Foxo3a(A3):ER, 50 nM 4-hydroxytamoxifen (4OHT; Sigma-Aldrich) was added to expansion conditions. Stat5−/− fetal livers were obtained from mice harboring a complete deletion of the Stat5a/b gene locus (20). For stimulation, cells were incubated for 4 h in plain IMDM (Invitrogen) and stimulated at 37°C with 200 ng/ml SCF or 5 U/ml Epo. Reactions were stopped by the addition of ice-cold phosphate-buffered saline. LY294002 was obtained from Alexis (Switzerland). cAMP was measured by enzyme immunoassay (EIA; Pharmacia) according to the manufacturer's instructions. Antibodies used in this study were anti-phospho-Foxo3a (catalog no. 06-951; Upstate Biotechnology), anti-Foxo3a, anti-Myc (9E10), anti-Stat3 (C-20), anti-Stat5 (N-20), anti-phospho-CREB-1 (Ser133), and anti-CREB-1 (24H4B) (Santa Cruz). Concentrated aliquots of the CREB/ATF1, Stat3, and Stat5 antibodies (Santa Cruz) were used for electrophoretic mobility shift assay (EMSA).
cDNA array hybridizations and analysis.
Total RNA was generated from cells treated with or without 4OHT. Dual labeling was used to hybridize the cDNAs pairwise to a custom-made 17K EST cDNA microarray. Profiles for factor-deprived and Epo- and SCF-restimulated cells were obtained in the same experiment but were reported previously (42). For a description of the procedures compliant with minimum information about a microarray experiment, see the supplemental material. Hierarchical clustering of data was performed and visualized with the Spotfire application using Euclidean distance and average linkage to assess distance.
Real-time Q-PCR.
cDNA synthesis and quantitative reverse transcription-PCR were performed using TaqMan technology (PE Applied Biosystems model 7700 or 7900 sequence detector) and SYBR green detection (Applied Biosystems) of double-stranded DNA as described previously (42). For primers, see Table 1. The threshold cycle values of the RNase inhibitor were used for normalization. Melting curves were performed to determine the specificity of the quantitative PCR (Q-PCR) product.
TABLE 1.
Oligonucleotides used
| Assay | Identity | Sequence (5′→3′) | F/Ra |
|---|---|---|---|
| Q-PCR | Btg1 | GCAGGAGCTGCTGGCAG | F |
| Q-PCR | Btg1 | TGCTACCTCCTGCTGGTGA | R |
| Q-PCR | Ccng2 | TGAAACCGAAACACCTGTCC | F |
| Q-PCR | Ccng2 | TCGAGTTTATCGAGGCTGAGA | R |
| Q-PCR | Ulk-1 | TACCAGAATGTTCTCAGTGG | F |
| Q-PCR | Ulk-1 | TGCTCCATGAGGGTCTCC | R |
| Q-PCR | Sesn1 | TCTGATGTGACAAGGTGACA | F |
| Q-PCR | Sesn1 | TGTTACCGCCAACACGGTC | R |
| Q-PCR | Dcn | TGGGCGGCAACCCACTG | F |
| Q-PCR | Dcn | TCAGGCTGGGTGCATCAAC | R |
| Q-PCR | Cited2 | TGAACCACGGGCGCTTCC | F |
| Q-PCR | Cited2 | TGGCGTGCCTGATGCCGC | R |
| Q-PCR | RNase inhibitor | TCCAGTGTGAGCAGCTGAG | F |
| Q-PCR | RNase inhibitor | TGCAGGCACTGAAGCACCA | R |
| ChIP | Cited2-DBE | GCAAGTAACTGTGTATGTGCA | F |
| ChIP | Cited2-DBE | GTACGTGTGCTTCTGCTAAG | R |
| ChIP | Cited2-SRE | TTTATTTGCAAGTCAATGAGCC | F |
| ChIP | Cited2-SRE | TGGTAATCGCTTTGTAAATAA | R |
| ChIP | Cited2 nonconserved DBE | GGAGCTTTCACACGCGCCTCC | F |
| ChIP | Cited2 nonconserved DBE | CTGCGGAAAGGAAGTGGCGTA | R |
| ChIP | Cis-SRE | TCCCTGCACTTCAATAGGTCG | F |
| ChIP | Cis-SRE | CCAGGCGCCTCCTAATCT | R |
| ChIP | Btg1-DBE | GGAGCTTTCACACGCGCCTCC | F |
| ChIP | Btg1-DBE | CTGCGGAAAGGAAGTGGCGTA | R |
| EMSA | btg-1 CRE | AGCTGAGCAGATTACGTCAGCTCCTA | F |
| EMSA | btg-1 CRE | AGCTGAGGAGCTGACGTAATCTGCTA | R |
| EMSA | btg-1 CRE mutated | AGCTGAGCAGATTTGGTCAGCTCCTC | F |
| EMSA | btg-1 CRE mutated | AGCTGAGGAGCTGACCAAATCTGCTC | R |
| EMSA | c-Jun | AGCTGAGCAGATTACCTCAGCTCCTC | F |
| EMSA | c-Jun | AGCTGAGGAGCTGAGGTAATCTGCTC | R |
| EMSA | creb | AGCTGAGCAGATGACATCAGCTCCTC | F |
| EMSA | creb | AGCTGAGGAGCTGATGTCATCTGCTC | R |
| EMSA | β-Casein | AGCTAGATTTCTAGGAATTCAATCC | F |
| EMSA | β-Casein | AGCTGGATTGAATTCCTAGAAATCT | R |
F/R, forward (F) or reverse (R) orientation.
Western blotting and antibodies.
Cell lysis, preparation of nuclear and cytoplasmic extracts, immunoprecipitation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and Western blotting were performed as described previously (3, 69). Lysate of 20 × 106 cells was used for one immunoprecipitation.
Constructs.
To construct the expression plasmids of Foxo3a deletion mutants, portions of Foxo3a were amplified by PCR using the following primers: 5′-AATGGATCCGGAAAAGCCCCCCGGCGGC-3′ (forward) and 5′-AATCTCGAGTTCAGCCTGGCACCCAGCTCT-3′ (reverse) for ΔN (G244 to the end), 5′-AATGGATCCGCAGAGGCACCGGCTTCCCC-3′ (forward) and 5′-AATCTCGAGTTCACTTGCTTACTGAAGGTGACAGG-3′ (reverse) for the ΔC mutant (the first Met to K360), and 5′-AATGGATCCGCGGCTGGGGGCTCCGGGCA-3′ (forward) and 5′-AATCTCGAGTTCACTTGCTTACTGAAGGTGACAGG-3′ (reverse) for the ΔNC mutant. PCR products were digested with BamHI and XhoI (both sites are included in the forward and the reverse primers described above, respectively) and subcloned into the corresponding sites of the Myc-tagged fusion protein expression vector pcDNA3/Myc (53). The resultant plasmids were verified by sequencing for the amplified regions and the junctions with the vector. The dominant negative Stat5 construct was described previously (72).
The mouse Cited2 promoter was cloned into pGL3-basic (Promega) using primers 5′-CCTATTGCTCCACTGAACAAT-3′ (forward) and 5′-CTCACCTTCCGTCTTTGCGATTTC-3′ (reverse) and the Expand High Fidelity PCR system (Roche). For promoter alignment between human (GenBank accession number AF129290) and murine (accession number NT039491) Cited2, we used the DNAMAN program, version 5.2.9. The transcriptional start site has been adapted from the published human Cited2 promoter, and the positions of Foxo and Stat5 binding sites were numbered accordingly (47). Mutations in the Cited2 promoter were made using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. The Daf16 binding element (DBE) was mutated using forward primer 5′-GATCGCTGAGTTTAAATACAGAGCAGGGAC-3′; the Stat5 site in the Cited2 promoter was mutated using forward primer 5′-TGCTCCACTGAACAATTCAAGCTTCAAGGAAGACTAGTAGC-3′. The cAMP-responsive element (CRE) was mutated using forward primer 5′-AGCAGATTTGGTCAGCTCCTC-3′. The respective opposite strand was used as the reverse primer.
Luciferase reporter assays.
Ba/F3 cells (10 × 106 cells) were electroporated (0.28 kV; capacitance, 960 μFD) with a maximum of 20 μg of DNA. After recovery for several hours in normal medium, cells were washed and grown overnight in the presence of SCF and stimulated for 7 h with IL-3 and/or LY294002 the next day. Luciferase activity was measured using the Steady-Glo system (Promega). Transfection efficiency was determined by cotransfecting lacZ and analyzing β-galactosidase activity.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed by using an acetyl-histone H3 immunoprecipitation kit according to the manufacturer's instructions (catalog no. 17-245; Upstate). Approximately 75 × 106 erythroblasts (I/11) were used for a specific condition. DNA was purified by phenol-chloroform extraction and dissolved in 200 μl water. Two microliters was used for PCR. Primers used are listed in Table 1. Antibodies to precipitate complexes harboring Foxo3a (sc-11251), Stat5 (sc835), and c-myc (9E10) were obtained from Santa Cruz.
EMSA.
Nuclear extracts were prepared and EMSA was performed as described previously by using the oligonucleotide probes shown in Table 1 (65).
RESULTS
Identification of Foxo3a targets.
To study the effect of Epo- and SCF-induced signaling on Foxo3-regulated gene expression, we used I/11 erythroblast clones stably expressing an inducible, activated Foxo3a mutant [Foxo3a(A3):ER] in which the three inhibitory PKB phosphorylation sites were mutated to alanine residues and which was fused to the ligand binding domain of the estrogen receptor to induce transcriptional activity with 4OHT (4, 24). cDNAs derived from Foxo3a(A3):ER-expressing erythroblasts and control cells induced by 4OHT under expansion conditions were hybridized to 17K EST cDNA arrays (42). Potential Foxo3a target genes were selected using an arbitrary threshold of a >1.75-fold change (positive or negative) for the ratio of plus/minus 4OHT in Foxo3a(A3):ER clones and <1.3-fold for the same genes in the vector control clone to exclude nonspecific effects of 4OHT. This threshold was previously used for SCF-regulated gene expression profiling on the same arrays and allowed us to analyze the 1% most regulated genes (42). This analysis yielded 299 potential target genes. Expression of the potential Foxo3a target genes was clustered with data on the regulation of these genes by Epo, SCF, and dexamethasone, detected on the same arrays (Fig. 1; see the supplemental material) (42). This analysis divided Foxo3a upregulated genes into three clusters. Cluster A (22 genes) contains genes exhibiting the expected pattern: upregulated by Foxo3a activation and downregulated by Epo- and SCF-induced signaling. Cluster A includes all genes known to be Foxo targets, such as Cdkn1b (p27Kip), Btg1, and Ccng2 (Cyclin G2) (Table 2) (4, 49). In contrast, clusters D (42 genes; Table 3) and E (5 genes) contain Foxo3a-upregulated genes that are either less prominently downregulated by Epo/SCF signaling or even upregulated by Epo/SCF signaling. Genes downregulated upon Foxo3a activation are also divided into a cluster that is repressed by Epo and SCF (cluster C) and a cluster that is less prominently regulated by Epo and SCF (cluster B), but these clusters have not been analyzed in this study.
FIG. 1.
Dendrogram of Foxo3a target genes and their regulation by Epo, SCF, and Dex. I/11 cells expressing Foxo3a(A3):ER or a control vector were treated with or without 4OHT (50 nM) for 6 h under expansion conditions (Epo/SCF/Dex). cDNAs were hybridized to 17K EST arrays in pairs using dual labeling with fluorochromes. Genes were selected when 4OHT induced or repressed expression >1.75-fold in Foxo3a(A3):ER-expressing cells (ratio of plus 4OHT/minus 4OHT, “Foxo3a′”) and <1.3-fold in control cells (ratio of plus 4OHT/minus 4OHT, “EV”). These genes were clustered with data on the regulation of these genes by Epo (E), SCF (S), Epo/SCF (ES), Epo/SCF/Dex (ESD), Epo/SCF/2k112.993 (ES2k), and Dex (D) by Spotfire software using Euclidean distance and average linkage to assess distance. Five major clusters of target genes (A to E) are indicated on the left side. Black ovals indicate the branches of the distinct clusters. Red indicates upregulation, and green indicates repression by 4OHT or growth factors. The original data as well as genes and values corresponding to this graph are presented in the supplemental material.
TABLE 2.
Foxo3a-upregulated, Epo/SCF-downregulated genes (cluster A)
| GenBank accession no. | Genea | Gene description | Expression (fold)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| F14b | Ec | Sc | ESc | ESDc | ESZkc | Dc | EVb | |||
| AI451894 | Cyclin G2 | Ccng2 | 1.30 | −1.20 | −1.04 | −1.95 | −1.31 | −2.04 | 0.08 | −0.05 |
| AI846040 | LIM motif-containing protein kinase 2 | Limk2 | 0.96 | −0.84 | −1.35 | −2.02 | −0.51 | −1.44 | 0.51 | −0.12 |
| AI846647 | Carnitine palmitoyltransferase 1 | Cpt1 | 1.05 | −0.98 | −0.89 | −1.45 | −1.24 | −0.71 | −0.11 | −0.09 |
| AI843786 | Cyclin-dependent kinase inhibitor 1B/p27Kip | Cdkn1b | 1.33 | −0.45 | −1.00 | −1.48 | −1.28 | −1.37 | 0.11 | −0.17 |
| AI853707 | Cell cycle progression 1 | Ccpg1 | 1.25 | −0.08 | −0.69 | −1.35 | −0.62 | −0.95 | 0.01 | 0.09 |
| AI451891 | RIKEN cDNA 4833420G17 gene | EST | 0.83 | −0.49 | −0.78 | −1.31 | −1.00 | −0.86 | 0.02 | 0.02 |
| AI596353 | NFKB inhibitor-interacting Ras-like protein 2 | Nkiras2 | 0.94 | −0.50 | −0.82 | −0.97 | −0.98 | −0.84 | −0.13 | 0.14 |
| AI850194 | Unc-51-like kinase 1 | Ulk1 | 0.83 | −0.48 | −0.67 | −0.74 | −1.05 | −0.92 | −0.16 | 0.03 |
| AI429475 | Exportin 7 | Xpo7 | 1.13 | −0.61 | −0.68 | −0.74 | −0.89 | −0.66 | 0.19 | 0.02 |
| AI843236 | Retinoblastoma-like 2/p130 | Rbl2 | 0.89 | −0.36 | −0.61 | −0.83 | −0.77 | −0.98 | 0.00 | −0.28 |
| AI845268 | t-complex 11-like 2 | Tcp11l2 | 0.92 | −0.41 | −0.46 | −0.74 | −0.73 | −0.82 | 0.05 | −0.18 |
| AI450702 | ATPase, Na+/K+ transporting, beta 3 polypeptide | Atp1b3 | 0.82 | −0.33 | −0.73 | −0.56 | −0.53 | −0.93 | 0.05 | −0.10 |
| AI449499 | Glucocorticoid-induced transcript 1 | Glcci1 | 0.81 | −0.66 | −0.56 | −0.71 | −0.49 | −0.83 | 0.05 | 0.18 |
| AI450119 | RIKEN cDNA 1110038D17 gene | EST | 0.99 | −0.48 | −0.44 | −0.75 | −0.62 | −0.64 | 0.04 | 0.14 |
| AI847059 | PTEN-induced putative kinase 1 | Pink1 | 0.92 | −0.38 | −0.61 | −0.93 | −0.39 | −0.53 | 0.15 | 0.22 |
| AI854419 | Solute carrier family 12, member 6 | Slc12a6 | 0.90 | −0.41 | −0.66 | −0.68 | −0.44 | −0.43 | 0.04 | −0.09 |
| AI845739 | Phosphoglucomutase 2-like 1 | Pgm2l1 | 0.86 | −0.32 | −0.80 | −0.50 | −0.57 | −0.44 | 0.23 | −0.01 |
| AI851307 | RIKEN cDNA C630043F03 | EST | 0.95 | −0.34 | −0.50 | −0.31 | −0.66 | −0.69 | 0.13 | 0.10 |
| AI835817 | Thymus-expressed acidic protein | Trp53inp1 | 1.28 | −0.93 | −0.39 | −0.64 | −0.15 | −0.80 | −0.12 | 0.08 |
| AI450899 | AMP deaminase 2 | Ampd2 | 1.90 | −0.85 | −0.76 | −0.86 | −0.58 | −0.86 | 0.30 | 0.29 |
| AI848411 | B-cell translocation gene 1 | Btg1 | 1.77 | −0.49 | −1.06 | −1.14 | 0.09 | −1.30 | 0.75 | −0.37 |
| AI448121 | IMAGE:558102 | EST | 0.81 | −0.18 | −1.00 | −0.73 | 0.09 | −0.80 | 0.53 | 0.31 |
Abbreviations are according to the Unigene database (NCBI). Genes in boldface type have been validated.
F14 is a Foxo3a(A3):ER-expressing clone; EV, vector control. The 2-log expression ratio is ± 4OHT.
The 2-log expression ratio of plus/minus ligand. E, Epo; S, SCF; D, dexamethasone. Zk, Zk112.993, an antagonist of the glucocorticoid receptor.
TABLE 3.
Foxo3a-upregulated genes not regulated by Epo/SCF (cluster D)
| GenBank accession no. | Genea | Gene description | Expression (fold)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| F14b | Ec | Sc | ESc | ESDc | ESZkc | Dc | EVb | |||
| AI449437 | ATPase, Cu2+ transporting, alpha polypeptide | Atp7a | 0.99 | −0.48 | −0.09 | −0.27 | 0.09 | −0.99 | −0.15 | −0.26 |
| AI450792 | RIKEN cDNA B230106I24 gene | EST | 0.82 | −0.21 | 0.05 | −0.19 | −0.08 | −1.00 | 0.19 | 0.04 |
| AI662267 | Homeodomain-interacting protein kinase 1 | Hipk1 | 0.88 | −0.21 | 0.01 | −0.48 | 0.01 | −0.67 | −0.07 | −0.01 |
| AI451316 | Transcription factor 12 | Tcf12 | 0.81 | −0.23 | −0.02 | −0.39 | 0.28 | −0.33 | −0.17 | 0.16 |
| AI843965 | Sestrin | Sesn1 | 0.87 | −0.10 | −0.12 | −0.31 | −0.70 | −0.79 | −0.14 | −0.15 |
| AI325508 | Expressed sequence AW049829 | EST | 1.04 | −0.28 | −0.32 | −0.21 | −0.53 | −0.44 | −0.23 | 0.22 |
| AI415470 | Neuropathy target esterase | Nte | 0.92 | −0.22 | −0.18 | −0.23 | −0.41 | −0.47 | −0.22 | 0.09 |
| AI449513 | Glutamate receptor, ionotropic, AMPA3 | Gria3 | 0.91 | −0.37 | −0.06 | −0.48 | −0.23 | −0.18 | −0.45 | 0.17 |
| AI447150 | Insulin-like growth factor I receptor | Igf1-R | 1.03 | −0.10 | −0.05 | −0.16 | −0.18 | −0.21 | −0.32 | 0.31 |
| AI852445 | DnaJ homolog, subfamily C, member 12 | Dnajc12 | 0.84 | 0.15 | −0.20 | −0.20 | −0.65 | −0.26 | 0.06 | 0.09 |
| AI845479 | Oxysterol binding protein-like 9 | Osbpl9 | 0.88 | −0.19 | −0.23 | −0.13 | −0.19 | −0.53 | 0.07 | 0.07 |
| AI451237 | Protein geranylgeranyltransferase type I, beta | Pggt1b | 0.87 | −0.14 | −0.17 | −0.08 | −0.24 | −0.47 | 0.07 | 0.17 |
| NM_008138 | Guanine nucleotide binding protein, alpha-inhibiting 2 | Gnai2 | 0.84 | −0.11 | −0.22 | −0.11 | −0.29 | −0.17 | 0.26 | 0.22 |
| AI842614 | NAD(P)H:menadione oxidoreductase 1 | Nmor1 | 1.00 | −0.02 | −0.01 | −0.18 | −0.40 | −0.40 | 0.13 | −0.21 |
| AI448727 | Core promoter element binding protein | Copeb | 1.28 | −0.22 | −0.19 | −0.42 | −0.23 | −0.49 | 0.28 | 0.03 |
| NM_007799 | Cathepsin E | Ctse | 1.17 | −0.09 | −0.18 | −0.24 | −0.36 | −0.22 | 0.24 | 0.06 |
| AI414015 | RIKEN cDNA D030011O10 gene | EST | 1.05 | 0.05 | −0.32 | −0.93 | 0.35 | 0.04 | −0.13 | 0.26 |
| AI845199 | Selenoprotein P, plasma, 1 | Sepp1 | 0.92 | 0.14 | −0.10 | −0.69 | −0.11 | 0.02 | 0.36 | −0.14 |
| AI449375 | MAD homolog 4-interacting transcription coactivator 1 | Mitc1 | 1.17 | −0.33 | −0.54 | 0.03 | −0.26 | −0.02 | −0.23 | 0.01 |
| NM_007482 | Arginase 1 | Arg1 | 0.81 | −0.07 | −0.70 | 0.26 | −0.18 | −0.06 | 0.34 | −0.05 |
| AI464367 | Unc4.1 homeobox | Uncx4.1 | 0.81 | −0.18 | −0.14 | 0.28 | 0.38 | −0.18 | −0.14 | −0.17 |
| AI661009 | RIKEN cDNA C330018D20 | EST | 0.95 | −0.45 | −0.12 | −0.26 | −0.12 | −0.09 | 0.18 | −0.05 |
| AI449354 | Zinc finger protein 60 | Zfp60 | 0.89 | −0.02 | −0.23 | −0.05 | 0.01 | −0.12 | 0.06 | −0.03 |
| AI452320 | Prolyl endopeptidase-like | Prepl | 0.83 | 0.06 | −0.22 | 0.13 | 0.16 | −0.09 | 0.13 | −0.06 |
| AI465319 | Thioredoxin domain-containing 1 | Txndc1 | 0.91 | −0.01 | −0.04 | 0.01 | 0.02 | 0.02 | 0.13 | 0.09 |
| D44443 | Endogenous mouse mammary tumor virus | Mtv1 | 0.86 | 0.10 | −0.08 | 0.06 | 0.13 | 0.04 | 0.44 | 0.19 |
| D44443 | Endogenous mouse mammary tumor virus | Mtv1 | 0.86 | 0.10 | −0.08 | 0.06 | 0.13 | 0.04 | 0.44 | 0.19 |
| NM_021099 | Kit oncogene | CKit | 1.30 | 0.04 | 0 | −0.30 | −0.03 | −0.08 | 0.14 | 0.12 |
| AI846778 | Decorin | Dcn | 1.38 | 0.28 | 0.04 | −0.32 | 0.09 | −0.14 | 0.11 | 0.07 |
| NM_007781 | Colony-stimulating factor 2 receptor, beta 2 | Csf2rb | 1.56 | 0.18 | −0.08 | 0.11 | −0.16 | −0.08 | 0.31 | 0.08 |
| AI430768 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | Cited2 | 0.9 | 0.23 | −0.34 | −0.06 | −0.32 | 0 | −0.61 | 0.28 |
| AI838934 | SFT2 domain-containing 2 | Sft2d2 | 0.88 | 0.56 | −0.51 | 0.02 | 0.19 | 0.27 | −0.38 | −0.06 |
| AI429552 | Expressed sequence AW146242 | EST | 0.91 | 0.21 | 0.07 | 0.38 | −0.06 | 0.36 | −0.22 | 0.01 |
| AI323564 | Neurofibromatosis 2 | Nf2 | 0.87 | 0.03 | 0.08 | 0.30 | 0.06 | 0.14 | 0.09 | 0.36 |
| AA123949 | Complement receptor 2 | Cr2 | 0.81 | 0.03 | 0.28 | 0.45 | 0.16 | 0.33 | 0.1 | −0.02 |
| AI385712 | Vinculin | Vcl | 0.92 | −0.16 | 0.33 | 0.27 | 0.31 | 0.15 | 0.12 | 0.17 |
| AI413346 | N-Acylsphingosine amidohydrolase 1 | Asah1 | 1.04 | 0.04 | 0.17 | 0.49 | 0.27 | 0.19 | 0.05 | 0.18 |
| AI452330 | Cysteine-rich hydrophobic domain 1 | Chic1 | 1.09 | 0.13 | 0.15 | 0.35 | 0.33 | 0.13 | 0.34 | 0.18 |
| AI844042 | Protein tyrosine phosphatase, non-receptor-type substrate 1 | Ptpns1 | 0.85 | 0.33 | 0.3 | 0.56 | 0.26 | 0.59 | 0.08 | 0.07 |
| NM_007781 | Colony-stimulating factor 2 receptor, beta 2 | Csf2rb | 1.45 | 0.10 | −0.07 | 0.44 | 0.01 | 0.52 | 0 | 0.10 |
| AI426361 | Cytoplasmic polyadenylation element binding protein 4 | Cpeb4 | 1.00 | 0.40 | −0.41 | 0.70 | 0.43 | 0.53 | 0 | 0.08 |
Abbreviations are according to the Unigene database (NCBI). Genes in boldface type have been validated.
F14 is a Foxo3a(A3):ER-expressing clone; EV, vector control. The 2-log expression ratio is ± 4OHT.
The 2-log expression ratio of plus/minus ligand. E, Epo; S, SCF; D, dexamethasone; Zk, Zk112.993.
Differential regulation of Foxo3a target genes by PI3K and during erythroid differentiation.
To further investigate the differential regulation of Foxo3a target genes by Epo/SCF signaling with respect to regulation by PI3K and during differentiation, we examined the regulation of selected Foxo3a-upregulated genes with different physiological roles in cell cycle regulation, survival, and signal transduction to gene expression from clusters A and D: Btg1, Ulk1 (Unc-51-like kinase), and Ccng2 (cyclin G2) to represent cluster A and Dcn (decorin), Sesn1 (sestrin), and Cited2 (CBP/p300-interacting transactivator with a Glu/Asp-rich C-terminal domain) to represent cluster D.
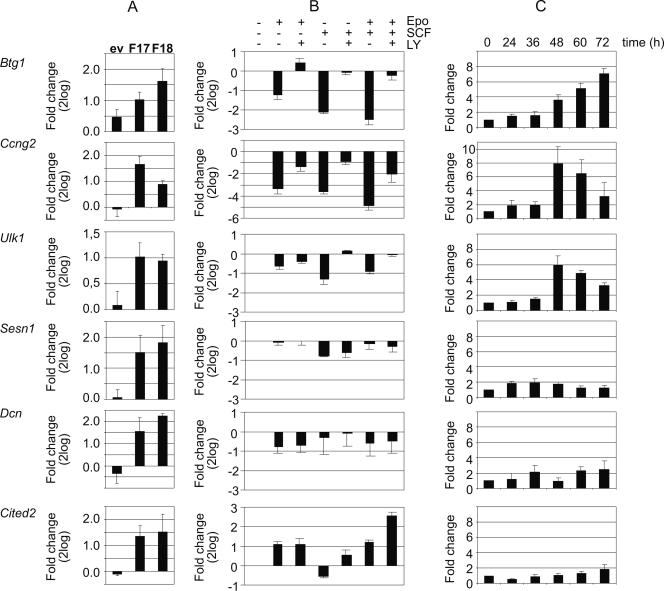
All targets were induced two- to fourfold within 2 h in two independent Foxo3a(A3):ER clones (F17 and F18) but not in control cells (Fig. 2A). Quantitative expression analysis following factor deprivation and subsequent restimulation with Epo, SCF, or Epo/SCF of I/11 cells confirmed that the expression of cluster A targets Btg1, Ccng2, and Ulk1 was subject to PI3K-dependent repression by Epo, SCF, and Epo/SCF (Fig. 2B). In contrast, cluster D targets Dcn and Sesn1 were less-than-twofold downregulated by Epo/SCF, and inhibition of PI3K had no effect on their expression (Fig. 2B). Notably, the expression of Cited2 was repressed by SCF but upregulated by Epo and by Epo plus SCF. Inhibition of PI3K did not affect Epo-induced upregulation of Cited2 but further increased Epo/SCF-induced expression. Thus, not all Foxo3a-targeted genes showed the predictable response to PI3K activity during factor deprivation and restimulation. We next examined whether PI3K-dependent expression correlates with expression during differentiation. We have demonstrated increased Foxo3a expression during erythroid differentiation, with a concurrent decrease in PKB activity and Foxo3a phosphorylation (4). This decrease in PKB phosphorylation during differentiation correlates with EpoR activity. Epo-induced signaling occurs in early phases of differentiation but does not affect late differentiation (71). Consequently, the expression of Btg1 and Cdkn1b sharply increases from the moment unphosphorylated Foxo3a accumulates in differentiating erythroblasts (4). The expression of the cluster A genes Ccng2 and Ulk1 similarly increased 48 h after differentiation induction. However, none of the cluster D genes was significantly upregulated during differentiation (Fig. 2C).
FIG. 2.
Expression of Foxo3a target genes in response to growth factors and during differentiation of erythroblasts. (A) Independent Foxo3a(A3):ER-overexpressing clones and control clones were treated with 50 nM 4OHT for 2 h. The expression ratio (2-log values) of plus 4OHT/minus 4OHT is shown for selected target genes, indicated at the left side of the panels, in the control clone (ev) and Foxo3a(A3):ER clones F17 and F18. (B) I/11 cells were factor deprived and subsequently stimulated for 2 h with Epo, SCF, or Epo plus SCF in the presence or absence of the PI3K inhibitor LY294002 (LY) (15 μM). The expression ratio (2-log values) of various conditions over factor-deprived cells is shown for the selected target genes. (C) I/11 cells were differentiated, and samples were taken every 12 h until the end of erythroid differentiation (72 h). Transcript levels from the selected genes were compared between the start of the differentiation experiment (arbitrarily set at 1) and ensuing time points. In all experiments, transcript levels were determined using poly(A) mRNA and real-time PCR. Bars indicate the averages of at least three experiments, and error bars indicate standard deviations.
In conclusion, Btg1, Ccng2, and Ulk1 (and Cdkn1b [4]) are repressed by Epo and SCF via a PI3K-dependent pathway and induced late in differentiation. In contrast, Dcn and Sesn1 are hardly affected by the PI3K-PKB pathway in response to factor deprivation and restimulation by Epo/SCF and are also not upregulated during differentiation. The observed induction of Cited2 by Epo and Foxo3a as well as its repression by SCF demonstrates the differential regulation of Foxo3a target genes by Epo and SCF.
Epo induces a nuclear Foxo3a/Stat5 complex.
To examine the differential regulation of Foxo3a targets in the presence of Epo, we investigated the expression of Cited2. Cited2 is a transcriptional cofactor that is able to bind CBP/p300, which results in the inhibition of Hif1. A Cited2 deficiency results in multiple developmental failures resulting in early embryonal lethality (5, 73, 74). Cited2 is a cytokine- and growth factor-inducible gene transcriptionally controlled by the Jak/Stat pathway (62). Because Epo robustly activates Stat5 in erythroblasts, we examined a possible association between endogenous Foxo3a and Stat5. Notably, Epo causes partial and SCF causes almost complete phosphorylation of Foxo3a, whereas Stat5 is phosphorylated exclusively by Epo (4, 70). Following factor deprivation and restimulation with Epo and/or SCF, Foxo3a appeared to coimmunoprecipitate with Stat5 from total cell lysate under all conditions (Fig. 3A). However, Foxo3a-Stat5 complexes in the nucleus were detected specifically in lysates of Epo-stimulated cells (Fig. 3A). With respect to gene regulation, the association of Foxo3a and Stat5 in the nucleus is important. Therefore, we examined the role of Epo and SCF in the cellular distribution of Foxo3a, Stat5, and the Foxo3a-Stat5 complex. Both Epo and SCF induced transient translocation of Foxo3a from the nucleus to the cytoplasm, but Epo induced partial phosphorylation of Foxo3a and consequently only partial exclusion of Foxo3a from the nucleus. In the presence of Epo plus SCF, phosphorylation and nuclear exclusion of Foxo3a were nearly complete (Fig. 3B). Foxo3a always coimmunoprecipitated with Stat5 in cytoplasmic fractions (exposure of cytoplasmic and nuclear fraction is not equal; phosphorylated and nonphosphorylated Foxo3a show as one band in Fig. 3C). However, the nuclear localization of Stat5 and nuclear Foxo3a/Stat5 association were induced by Epo (Fig. 3C). Nuclear extracts stained with the Foxo3a antibody show a slower-migrating, aspecific background band in serum-starved cells (t = 0 min), as shown before (Fig. 3C) (4).
FIG. 3.
Epo induces Foxo3a-Stat5 complex formation. (A) I/11 erythroblasts were factor deprived (4 h) and restimulated with Epo (E) (20 min with 5 U/ml), SCF (S) (20 min with 200 ng/ml), or both (ES) as indicated below the blots. Stat5 was immunoprecipitated from whole-cell and nuclear extracts and stained for Foxo3a (upper panels) and Stat5 (lower panels). (B and C) I/11 erythroblasts were factor deprived and restimulated with Epo (E) or with Epo plus SCF (ES) for the indicated times (minutes). (B) Western blots (WB) containing total cytoplasmic (C) and nuclear (N) extracts were stained for Foxo3a. The low-mobility phosphorylated form (P-Foxo3a) and faster-mobility unphosphorylated form (Foxo3a) of Foxo3a are indicated. (C) Stat5 immunoprecipitates from cytoplasmic (C) or nuclear (N) fractions of Epo-stimulated cells were stained for Foxo3a and Stat5. (D) Schematic representation of the constructed Foxo3a deletion mutants. wt Foxo3a comprises amino acids 1 to 673. The DNA binding domain (Forkhead box) is depicted in gray. The N-terminal mutant lacks the first 243 amino acids, and the C-terminal deletion mutant lacks amino acids 361 to 673. (E) Stat5, wt Foxo3a, and the N-terminal and C-terminal deletion mutants were expressed in 293T cells as indicated. Expression in total cell lysates was checked by staining membranes using a Stat5 and Myc-tagged antibody as indicated. (F) Stat5 immunoprecipitations (IP) from total lysates stained with Stat5 (lower panel). Nonspecific binding was verified by treating total lysate with beads only (lane 2). Stat5 immunoprecipitation blots were stained with Myc-tagged antibody to detect coprecipitating Foxo3a (upper panel, full-length Foxo3a; middle panel, ΔN and ΔC Foxo3a deletion mutants). (G) Anti-Myc-tagged immunoprecipitations from total lysates backstained with Myc antibody (lower panel) or stained with Stat5 antibody (upper panel). Nonspecific binding was verified by treating total lysate with beads only (lane 2).
To verify which Foxo3a domain is required for the Stat5 interaction, we constructed different Foxo3a mutants: an N-terminal deletion mutant Foxo3a lacking amino acids 1 to 243 (ΔN), a C-terminal deletion mutant lacking amino acids 361 to 673 (ΔC), and a mutant containing mainly the DNA-binding domain (ΔNC) (Fig. 3D). The Foxo3a constructs were Myc tagged and expressed together with Stat5 in 293T cells (Fig. 3E to H). Expression of the ΔN Foxo3a mutant was low compared to that of other constructs, probably due to a less stable protein product, because other ΔN mutants tested were also expressed at lower levels (data not shown). Stat5 was efficiently immunoprecipitated from total lysates (Fig. 3F, lower panel), and no Foxo3a or Stat5 was precipitated in a nonspecific manner (Fig. 3F and G, lane 2). Full-length Foxo3a and the ΔC and the ΔNC Foxo3a mutants precipitated with Stat5 (Fig. 3F). In reverse, Stat5 immunoprecipitated with all Foxo3a constructs (Fig. 3G). This indicates that Stat5 associates with Foxo3a through a domain located within the overlapping C-terminal domain of amino acids 244 to 360. The fact that the ΔN Foxo3a mutant could not be detected in a Stat5 immunoprecipitation (Fig. 3F), as well as the low levels of Stat5 coprecipitated by the ΔN Foxo3a mutant (Fig. 3G), can be explained by the low expression levels of the ΔN Foxo3a mutant (Fig. 3E).
In conclusion, the allegedly proliferation-inhibiting factor Foxo3a and proliferation-promoting factor Stat5 associate, which may result in the regulation of common target genes such as Cited2. The nuclear localization of this complex is maximal in the presence of Epo, which seems to be due to the fact that only Epo induces the concurrent nuclear localization of Stat5 and Foxo3a.
Foxo3a- and Stat5-responsive elements in the Cited2 promoter.
Next, we investigated the potential transcriptional control of the Cited2 promoter by the Foxo3a/Stat5 complex. Alignment of a mouse genomic DNA fragment from chromosome 10 (GenBank accession number NW_001030408) to a previously described fragment at positions −3300 to +18 encompassing the human CITED2 promoter (47) revealed one conserved, palindromic Stat5 response element (SRE) (at bp −1207 bp) and one conserved Foxo3a binding element (DBE at bp −872). Both sites are located in a highly conserved promoter region and are separated by 335 bp (Fig. 4A). We employed ChIP assays to test the binding of the Epo-induced Foxo3a/Stat5 complex to these sites (Fig. 4B and C). I/11 erythroblasts were factor deprived (4 h), which was followed by 20 min of Epo stimulation. PCR analysis revealed that immunoprecipitated Stat5-DNA complexes isolated from Epo-stimulated cells contain the SRE of Cis, an established Stat5 target gene, but not the DBE of Btg1 (Fig. 4B). In reverse, Foxo3a-DNA complexes isolated from factor-deprived cells contain the DBE of Btg1 but not the SRE of Cis (Fig. 4C). In addition, the binding of Foxo3a to the Btg1 promoter was reversed upon Epo stimulation (Fig. 4C). Importantly, both the DBE and the SRE of Cited2 were detected in Stat5-DNA complexes (Fig. 4B) and in Foxo3a-DNA complexes only in response to Epo stimulation (Fig. 4C), which is in accordance with the nuclear Foxo3a-Stat5 interaction following Epo stimulation. These data corroborate that the regulation of the two Foxo3a target genes Cited2 and Btg1 follows opposite kinetics, with Cited2 being upregulated and Btg1 being downregulated in response to Epo stimulation. Apparently, the Foxo3a-Stat5 complex is able to escape the repression by the PI3K-PKB pathway induced by Epo.
FIG. 4.
Cited2 promoter analysis and ChIP. (A) Alignment of the mouse (upper line) and human (lower line) Cited2 promoters containing the DBE and the SRE (both in boldface type). Positions of these elements are based on promoter A identified in the human CITED2 promoter (47). Primers (F, forward; R, reverse) indicated by arrows were used for ChIP in B and C. (B and C) ChIPs using a Stat5 (B) or Foxo3a (C) antibody were performed in I/11 cells factor deprived for 4 h and subsequently stimulated with Epo for 20 min. An anti-Myc (αmyc) ChIP was used as a negative control, and input DNA represents samples before immunoprecipitation. The DNA ladder shows bands from 100 to 500 bp with 100-bp increments. PCR was used to detect the SRE or DBE in Cited2, Btg1, and Cis as indicated on the right-hand side.
Transcriptional activation of Cited2 by the Foxo3a/Stat5 complex.
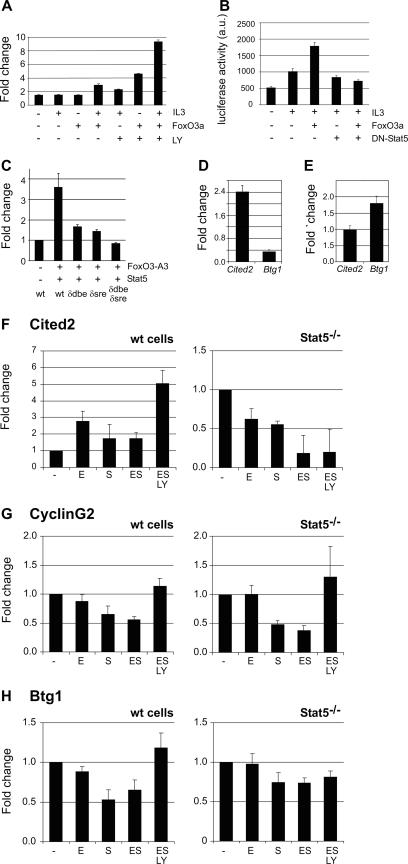
Because sonication results in DNA fragments of approximately 500 bp, whereas the DBE and SRE in the Cited2 promoter are separated by 335 bp, the data are not conclusive regarding the question whether the Epo-induced Foxo3a/Stat5 complex binds the SRE, the DBE, or both sites. Therefore, a Cited2 promoter fragment (positions −1228 to +269) was cloned in front of a luciferase reporter (40-fold increase in luciferase in Ba/F3 and 293HEK cells) (data not shown). Because I/11 or other factor-dependent erythroblast cultures cannot be transfected efficiently, we tested factor-dependent promoter activity in Ba/F3 lymphoblastic cells. IL-3 induces predominantly Stat5 phosphorylation, whereas SCF is required for the adequate activation of PI3K and full phosphorylation of Foxo3a in these cells (data not shown), which mirrors the cooperation of Epo and SCF in erythroblasts. Ba/F3 cells were transfected with the Cited2 promoter in the presence or absence of wild-type (wt) Foxo3a. Cells were IL-3 deprived in the presence of SCF, which prevents the activation of wt Foxo3a. Subsequently, cells were treated with combinations of IL-3 and 15 μM LY294002 to activate endogenous Stat5 and Foxo3a, respectively (Fig. 5A). In the absence of active Foxo3a (no LY294002), IL-3 stimulation only modestly increased promoter activity. In contrast, active Foxo3a (plus LY294002) induced Cited2 promoter activity fourfold, which was enhanced to ninefold upon stimulation with IL-3 (Fig. 5A). Cotransfection of dominant-negative Stat5 (mSTAT5AΔ750) (72), however, inhibited the activation of the Cited2 promoter (Fig. 5B). Mutation of either the SRE or the DBE was sufficient to abolish Foxo3a/Stat5-stimulated promoter activity (Fig. 5C), indicating that both sites are required for the Foxo3a-Stat5 complex to induce Cited2 expression. The fact that Foxo3a cannot activate the Cited2 promoter alone, but only in cooperation with Stat5, provides a mechanism for the differential regulation of Cited2 and Btg1 by Foxo3a. This was confirmed by comparing Cited2 and Btg1 promoter activities. Whereas IL-3 stimulation of Ba/F3 cells activated the Cited2 promoter, it repressed Btg1 promoter activity more than twofold (Fig. 5D). In addition, growth factor depletion induced the Btg1 promoter but did not affect the activity of the Cited2 promoter (Fig. 5E).
FIG. 5.
Foxo3a and Stat5 cooperate to induce Cited2 promoter activity. For promoter studies, Ba/F3 cells were electroporated with Cited2-luciferase constructs (positions −1128 to +269 of the Cited2 promoter) and a β-galactosidase construct to correct for transfection efficiency. Luciferase activity is expressed as arbitrary units (a.u.). (A) Ba/F3 cells were transfected with the Cited2-reporter construct together with wt Foxo3a, where indicated, and grown overnight in the presence of SCF. Subsequently, cells were treated (8 h) with combinations of IL-3 and the PI3K inhibitor LY (15 μM). Luciferase activity is presented as induction (n-fold) compared to Cited2 promoter activity in the presence of SCF only (first lane). (B) Ba/F3 cells were transfected with combinations of the Cited2 promoter, Foxo3a, and dominant-negative Stat5 (mStat5AΔ750). Cells were incubated overnight in the presence or absence of IL-3. (C) Activities of different Cited2 promoter mutants with a mutated DBE (δDBE), a mutated SRE (δSRE), or a double-mutated construct (δDBE/δSRE) relative to that of the wt promoter. Ba/F3 cells were transfected with the different promoters together with Stat5 and Foxo3a-A3 expression constructs grown overnight in the presence of IL-3, after which promoter activity was analyzed. (D) wt Cited2 and Btg1 promoter constructs were transfected into Ba/F3 cells, grown overnight in the presence of SCF alone, and stimulated the next day with IL-3 for 8 h. Luciferase activity is presented as the ratio with IL-3 to that without. (E) As in D, cells were treated the next day with LY294002 (15 μM). (F to H) Primary erythroid progenitors were grown for 6 days from E14 fetal livers derived from Stat5−/− embryos and wt littermates. Cells were factor deprived (−) and restimulated with Epo (E), SCF (S), Epo plus SCF (ES), or Epo plus SCF plus LY294002 (ESLY), as described in the legend of Fig. 2. The expression of Cited2 (F), Cyclin G2 (G), and Btg1 (H) was measured by Q-PCR, and changes (n-fold) were normalized to RNase inhibitor. Values represent means and standard deviations of three experiments.
To further evaluate the role of Stat5 in the Foxo3a-dependent expression regulation of Cited2 and other identified target genes, we cultured erythroid progenitors from fetal livers of Stat5-deficient embryos and wt littermates. After 6 days of culture, the progenitors were factor deprived and restimulated with Epo and/or SCF. RNA was isolated, and expression of Cited2, Cyclin G2, and Btg1 was assessed by Q-PCR (Fig. 5F to H). In wt primary erythroid progenitors, the regulation of these Foxo3a target genes was similar to what was observed in I/11 cells (Fig. 2). The effects of SCF are less pronounced because the primary culture contained partly differentiated cells that had lost their SCF response. In Stat5−/− erythroid progenitors, however, Cited2 expression was not upregulated in the presence of Epo or Epo/SCF plus LY (Fig. 5F). The expression of Cited2, Cyclin G2, and Btg1 in the absence of factor was not significantly different in Stat5−/− cells compared to that in wt cells. The observation that Cyclin G2 and Btg1 are regulated similarly in Stat5−/− and wt cells indicates that the observed association of Foxo3a and Stat5 in the cytoplasm does not affect Foxo3a function in general.
CREB family members contribute to Btg1 expression.
The Foxo3a-binding DBE sites in the Cited2 and Btg1 promoters are identical consensus sites. The presence of the SRE in the Cited2 promoter and the nuclear association between Foxo3a and Stat5 may explain why Cited2 and Btg1 are differentially regulated by Epo stimulation. Indeed, reanalysis of the Btg1 promoter did not reveal any potential SRE. Instead, we found a CRE at position −204, i.e., 15 bp downstream of the DBE in the Btg1 promoter (Fig. 6A) (4). Promoter activity was assayed in NIH 3T3 cells using luciferase activity as a reporter. Mutation of the CRE or the DBE repressed basal promoter activity to a similar extent, and repression was not enhanced when both elements were deleted (Fig. 6B). In addition, costimulation by Foxo3a and cAMP significantly enhanced Btg1 promoter activity (Fig. 6C).
FIG. 6.
Expression of Btg1 is controlled by Foxo3a binding to its specific site (DBE) and by factors binding to a CRE located close to the DBE. (A) Alignment of the mouse and human Btg1 promoters. The putative transcription start site is indicated as +1, and transcribed sequence is in boldface and italic type. The TATA box, the DBE, and the CRE are boxed. (B) Btg1 promoter fragments (wt or containing a mutated DBE, a mutated CRE, or both) were cloned into a luciferase reporter construct and transfected in NIH 3T3 cells, and luciferase activity was measured 24 h after transfection. Values were normalized for transfection efficiency using β-galactosidase activity encoded by a cotransfected expression plasmid. Data are presented as a ratio to the activity of the wt promoter. (C) The wt promoter reporter construct was cotransfected with or without the Foxo3a-A3 expression construct, and cells were treated with db-cAMP (10 μM) overnight. Reporter activity was measured 16 h after transfection. Data are presented as a ratio to the activity of the wt promoter in the absence of Foxo3a or db-cAMP. Values represent means and standard deviations of three experiments.
To examine whether the cAMP pathway regulates Btg1 expression, we examined which stimulus effectively induced the phosphorylation of Creb in erythroblasts. I/11 cells were factor deprived and subsequently treated with dibutyryl-cAMP (db-cAMP) (10 μM) and ligands of prostaglandin E2 (PGE2) (10 μM) and adrenergic receptor (norepinephrine) (100 μM) (56). All three compounds induced the phosphorylation of nuclear Creb and Atf1 (Fig. 7A). Since the induction of Btg1 by Foxo3a is most prominent during differentiation, we determined the activity of the cAMP pathway in differentiation. Levels of cAMP showed a marked increase during differentiation, reaching a maximum between 40 and 60 h after induction of differentiation (Fig. 7B), concomitant with strongly increased Btg1 levels (4). The phosphorylation of Creb and Atf1 followed the levels of cAMP during differentiation, while the expression of Creb and Atf1 was constant for 48 h and dropped only during the final stages of differentiation, when total proteins levels decrease (Fig. 7C). To investigate the binding of Creb/Atf1 to the CRE site in the Btg1 promoter, nuclear extracts were incubated with an oligonucleotide probe encompassing the Btg1 CRE site and tested in an EMSA. Two protein complexes bound to the oligonucleotide probe in both expanding, nondifferentiating erythroblasts and 48-h-differentiated cells following differentiation induction in the presence of Epo alone. Competition with wt, but not with a CRE mutated probe, inhibited the binding of both complexes, indicating that they bind specifically to the CRE (Fig. 7D and E). The CRE element in the Btg1 promoter (TTACGTCA) is not completely similar to the common palindromic CRE sequence (TGANNTCA) (51, 59). Instead, it resembles the cJun/Atf2 binding site (TTACCTCA), through which cJun/Atf2 regulates, for instance, cJun expression (34, 65, 66). To discriminate between Creb/Atf1 and cJun/Atf2 binding to the CRE element in the Btg1 promoter, probes with an optimal Creb (TGACATCA) or an optimal cJun/Atf2 (TTACCTCA) binding site were used as competitors for the Btg1 CRE probe. A Creb/Atf1 probe competed efficiently with both complexes, while the cJun/Atf2 probe inhibited binding of only the slower-migrating complex (Fig. 7D and E). Consistent with these findings, the faster-migrating complex but not the slower-migrating complex could be supershifted upon the addition of the Creb/Atf1 antibody (Fig. 7D and E).
FIG. 7.
The CRE in the Btg1 promoter binds CREB/ATF1 as well as c-Jun/ATF2. (A) I/11 cells were factor deprived (4 h) and stimulated with db-cAMP (db) (10 μM), prostaglandin E2 (E2) (10 μM), or norepinephrine (NE) (100 μM) for 5, 10, or 20 min. Cytoplasmic and nuclear extracts were prepared and stained with an antibody recognizing phosphorylated serines 133 and 63 in CREB and ATF1, respectively. (B) Intracellular cAMP levels were determined using a cAMP enzyme immunoassay. (C) I/11 cells were induced to differentiate, and cytoplasmic and nuclear extracts were generated at 24-h intervals and stained with an antibody recognizing phosphorylated serines 133 and 63 in CREB and ATF1, respectively. As a loading control, the same blot was stained for total CREB/ATF1. (D and E) Nuclear extracts from expanding (D) and 48-h-differentiated (E) I/11 cells were incubated with a 32P-labeled oligonucleotide probe derived from the Btg1 promoter encompassing the CRE. Binding of protein complexes was assessed by EMSA. To verify specific binding, we added 10- and 100-fold excesses of oligonucleotide probe: wt (wt CRE) (lanes 2 and 3) or a mutated CRE (mut-CRE) (lanes 4 and 5). To identify the protein complexes, an excess of a CREB/ATF1-specific (CREB oligo) (lanes 6 and 7) or a c-Jun/ATF2-specific (ATF2 oligo) (lanes 8 and 9) oligonucleotide probe was added. In supershift experiments, anti-CREB/ATF1 (lane 11) or anti-Stat5 (lane 12) was added. Arrows at the right-hand side indicate the mobility of c-Jun/ATF2 and CREB/ATF1 complexes.
In conclusion, Creb/Atf1 and cJun/Atf2 complexes are able to bind the CRE in the Btg1 promoter. The binding of these complexes did not differ between 0 and 48 h. This is not unexpected, because the phosphorylation of Creb/Atf1 and cJun/Atf2 controls their transcriptional activity but not DNA binding. Activation of the cAMP pathway at the final stage of differentiation may contribute to the upregulation of Btg1.
DISCUSSION
From the Foxo3a target genes identified in this study, only a minority was prominently downregulated in response to Epo or SCF. Most Foxo3a target genes showed either no apparent regulation by growth factors, a more complex regulation, or even an opposite regulation. This suggests that these targets require the cooperation of Foxo3a with additional transcription factors regulated by different stimuli. Stat5 appeared to be one such transcription factor, binding to Foxo3a to control the expression of Cited2. Epo enhances Cited2 expression as it potently induces the nuclear translocation of Stat5 and only partially phosphorylates Foxo3a, allowing Foxo3a to remain in the nucleus. Also, the expression pattern of Btg1 appeared to be reinforced by cooperation with factor-dependent transcription factor activity, i.e., with Creb/Atf1 and cJun/Atf2, which are activated late in differentiation and following stress, respectively. Our data show how Foxo3a takes part in the integration of distinct signaling pathways to modulate the balance between proliferation and differentiation of erythroblasts in response to its environment.
Differential regulation of Foxo3a target genes.
Foxo target genes are induced in response to various stress situations including oxidative stress, DNA damage, and growth factor depletion (17, 68). These stress conditions inhibit cell growth but regulate distinct target genes; Cdkn1b/p27Kip and Bim are upregulated in response to factor deprivation, while Gadd45 is induced in response to DNA damage and oxidative stress (15, 24, 30, 43, 50, 64). Oxidative stress induces the acetylation and phosphorylation of Foxo concomitant with nuclear localization and the association of Foxo proteins with the acetylase p300/CBP-associated factor and subsequently with the deacetylase Sirt1, which decreases the interaction of Foxo proteins with p300 (15, 68). The interaction of Foxo with Sirt-1 repressed the Foxo-induced expression of Bim, Cdkn1b, Peck, and IGFBP1 but induced the expression of Gadd45 (15, 52). As a result, cell cycle arrest and replicative senescence were enhanced, and apoptosis was suppressed (15). In addition to differential target gene regulation in response to stress (32), this study deals with differential target gene regulation in response to signaling.
Because we performed single hybridizations, the list of putative target genes may contain errors. However, expression data for Epo, SCF, Epo/SCF, Epo/SCF/Dex, and Epo/SCF/ZK profiles are highly consistent (42). Moreover, we validated selected target genes in distinct Foxo3a(A3):ER-expressing clones, which confirmed that Foxo3a target genes in cluster D (e.g., Sesn1, Dcn, and Cited2) are not involved in a cellular response to factor deprivation. Strikingly, genes in clusters A and D represent different cellular processes. For example, the cluster D representative Sestrin1 (Sesn1) regenerates the antioxidant potential of thioredoxin, decorin (Dcn) modulates growth factor signaling, and Cited2 modulates the interaction of p300/CBP with transcription factors (1, 11, 16, 38). In contrast, cluster A representative Cyclin G2 (Ccng2) as well as p27Kip/Cdkn1b and Rbl2 (p130Rb2) inhibit cell cycle progression (6), whereas other genes in cluster A have a functional role in mature erythrocytes. Our study showed that this diverse set of Foxo3a target genes enables us to understand how Foxo3a cooperates with various environmental signals to control the expression of the appropriate set of target genes in response to specific conditions.
Foxo3a integrates various signaling pathways.
We showed that an Epo-induced Foxo3a/Stat5 complex regulates Cited2 expression. Epo-induced Cited2 expression was not affected by the PI3K inhibitor LY294002, which is not surprising, since Epo strongly induces Stat5 activation, while it causes only weak Foxo3a phosphorylation. Nevertheless, it is surprising that a ChIP with anti-Foxo3a antibodies precipitates the DBE of Cited2 only in presence of Epo, while the DBE of Btg1 is present predominantly in the absence of Epo. The two DBE sequences are both full consensus sites. This suggests that the affinity of Foxo3a for its recognition site is not very strong and may require stabilization through the interaction with other transcription factors. It is striking that Foxo-associated factors are controlled by signaling pathways that have such diverse functions. Whereas Stat5 signaling is associated with proliferation, the cAMP pathway is associated with differentiation and cell cycle arrest (33). Recently, Foxo was shown to interact with β-catenin, the central effector of the Wnt-signaling pathway assumed to impose “stem-cellness” to progenitor cells (27, 58). Thus, Foxo3a functions to integrate various signaling pathways, and this may explain its versatile function. Complex formation with other transcription factors also explains how a mutant Foxo1, incapable of DNA binding, retains the ability to regulate the expression of part of its target genes (57). This mutant lost the ability to induce apoptosis but was still able to cause a G1 arrest, underscoring that different mechanisms of target gene activation are associated with distinct cellular responses.
Regulation of Cited2 expression by Foxo3a and Stat5.
Cited2 is a modulator of transcription that has been implicated in developmental processes and cancer (5, 73). Its regulation of important regulatory processes such as Hif1 activity and the transforming growth factor β pathway (74) suggests that Cited2 expression requires tight control. Foxo3a and Stat5 do not control expression per se, but they are important to adapt Cited2 expression to environmental conditions. This seems to be true for most of the identified target genes. For instance, Btg1, Cyclin G2, and Sestrin are known as p53 target genes, but they are expressed in p53-deficient I/11 cells in the absence of growth factor (i.e., activation of Foxo3a). Thus, in response to distinct stress factors, p53 and Foxo3a can activate expression independent of each other.
The phosphorylation of Stat5 does not seem to affect the association of Foxo3a with Stat5, because coimmunoprecipitation from cytoplasmic lysates is equally efficient in the presence or absence of Epo, and no phosphorylated Stat5 can be detected in the absence of Epo. Instead, it is colocalization in the nucleus that induces the transcriptional activity of a Stat5/Foxo3a complex in the presence of Epo.
The interaction of Foxo3a and Stat5 involves a domain immediately C terminal of the DNA binding domain. The Foxo3a domain at positions 244 to 360 is an important site for Foxo3a regulation. It contains the PKB phosphorylation sites S253, localized in the Forkhead domain, and S314, localized in the phosphorylation patch (68). Importantly, S253 phosphorylation of Foxo by PKB is regarded as being the triggering event leading to Foxo phosphorylation, resulting in nuclear export. Stat5 binding to the region at positions 244 to 360 may thus explain how Foxo3a escapes negative control by the PI3K pathway and is able to stimulate the transcription of Cited in response to Epo stimulation.
We did not observe a marked regulation of Cited2 during differentiation. This is not surprising because Cited2 expression in response to Epo (differentiation) or Epo plus SCF (renewal) is similar (Fig. 2). Notably, Stat5 is expressed early but not late in differentiation, while Foxo3a expression increases from the start of differentiation induction to reach maximal levels 48 h after differentiation induction, when Stat5 expression becomes undetectable (4, 26). The reduced expression of Stat5 most likely precludes the upregulation of Cited2 when Foxo3a expression in the nucleus increases.
Role of cAMP signaling in erythropoiesis.
cAMP levels rise late in differentiation. Erythroid cells express Gαs-coupled receptors, among which are the adrenergic receptor and receptors for thrombin and PGE2 (56). Notably, the PGE2 receptor is upregulated late in differentiation (W. J. Bakker and M. von Lindern, unpublished data; 25), whereas prostaglandin dehydrogenase (Pgdh1) is induced by dexamethasone in expanding erythroblasts and rapidly downregulated during differentiation (42; Bakker and von Lindern, unpublished). As a consequence, PGE2 activity is restricted to the final phase of erythroid differentiation when it controls hemoglobin expression, particularly Ala-S, the rate-limiting enzyme in heme synthesis, and NF-E2, a transcription factor binding the β-globin enhancer (2, 22, 31, 54, 55, 63).
The CRE site in the Btg1 promoter also recruits cJun and/or Atf-2. The cJun/Atf2 complex is not regulated by the cAMP-PKA pathway but is regulated through Jun kinase (JNK). JNK is a stress-activated kinase, and cJun/Atf2 transcriptional activity is strongly enhanced in response to genotoxic stress (23, 41, 66). Interestingly, Btg1 expression is induced in response to DNA damage (19). The binding of Creb/Atf1 to the CRE site in the Btg1 promoter accelerates Foxo3a-mediated expression late in differentiation, while the binding of cJun/Atf2 following the activation of JNK may enhance Foxo-mediated expression in response to oxidative or genotoxic stress.
In conclusion, we identified putative Foxo3a target genes that were assigned to different clusters based on their regulation by Epo and SCF signal transduction. Examples from each cluster show that Foxo3a integrates PI3K-PKB signals with other signaling pathways. On the Btg1 promoter, PI3K-PKB signaling is integrated with cAMP signaling. Surprisingly, we found that the cooperation of Foxo3a and Stat5, i.e., the cooperation of pathways generally regarded to inhibit and promote proliferation, respectively, induced the expression of Cited2. The data strengthen the notion that Foxo proteins function at the heart of signal-dependent gene regulation and in complexes that integrate multiple signals to decide between cell death or survival, senescence, and functional maturation.
Supplementary Material
Acknowledgments
We thank H. van Dam (LUMC, Leiden, The Netherlands) and I. P. Touw for many critical discussions regarding the work presented, Kim Birkenkamp (UMC, Utrecht, The Netherlands) for detailed information on Ba/F3 cell electroporation, Herbert Auer (IMP, Vienna, Austria) for his patient help with the array analysis, Mark Kerenyi (IMP, Vienna, Austria) for Stat5−/− fetal livers, and Victor de Jager (Erasmus MC) for his help in bioinformatics.
This work was supported by grants from the Dutch Cancer Society (EUR 2000-2230), The Netherlands Organization for Scientific Research (050-10-051), the European Union (HPRN-CT-2000-00083), an Erasmus fellowship to T.B.V.D., and a fellowship of the Dutch Academy of Arts and Sciences to M.V.L.
Footnotes
Published ahead of print on 12 March 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abdel-Wahab, N., S. J. Wicks, R. M. Mason, and A. Chantry. 2002. Decorin suppresses transforming growth factor-beta-induced expression of plasminogen activator inhibitor-1 in human mesangial cells through a mechanism that involves Ca2+-dependent phosphorylation of Smad2 at serine-240. Biochem. J. 362:643-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, N. C., H. Erdjument-Bromage, M. B. Davidson, P. Tempst, and S. H. Orkin. 1993. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature 362:722-728. [DOI] [PubMed] [Google Scholar]
- 3.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakker, W. J., M. Blazquez-Domingo, A. Kolbus, J. Besooyen, P. Steinlein, H. Beug, P. J. Coffer, B. Lowenberg, M. Von Lindern, and T. B. Van Dijk. 2004. FoxO3a regulates erythroid differentiation and induces BTG1, an activator of protein arginine methyl transferase 1. J. Cell Biol. 164:175-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bamforth, S. D., J. Braganca, C. R. Farthing, J. E. Schneider, C. Broadbent, A. C. Michell, K. Clarke, S. Neubauer, D. Norris, N. A. Brown, R. H. Anderson, and S. Bhattacharya. 2004. Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat. Genet. 36:1189-1196. [DOI] [PubMed] [Google Scholar]
- 6.Bates, S., S. Rowan, and K. H. Vousden. 1996. Characterisation of human cyclin G1 and G2: DNA damage inducible genes. Oncogene 13:1103-1109. [PubMed] [Google Scholar]
- 7.Bauer, A., F. Tronche, O. Wessely, C. Kellendonk, H. M. Reichardt, P. Steinlein, G. Schutz, and H. Beug. 1999. The glucocorticoid receptor is required for stress erythropoiesis. Genes Dev. 13:2996-3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biggs, W. H., III, W. K. Cavenee, and K. C. Arden. 2001. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm. Genome 12:416-425. [DOI] [PubMed] [Google Scholar]
- 9.Birkenkamp, K. U., and P. J. Coffer. 2003. Regulation of cell survival and proliferation by the FOXO (Forkhead box, class O) subfamily of Forkhead transcription factors. Biochem. Soc. Trans. 31:292-297. [DOI] [PubMed] [Google Scholar]
- 10.Reference deleted.
- 11.Braganca, J., J. J. Eloranta, S. D. Bamforth, J. C. Ibbitt, H. C. Hurst, and S. Bhattacharya. 2003. Physical and functional interactions among AP-2 transcription factors, p300/CREB-binding protein, and CITED2. J. Biol. Chem. 278:16021-16029. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 12.Broudy, V. C., N. L. Lin, G. V. Priestley, K. Nocka, and N. S. Wolf. 1996. Interaction of stem cell factor and its receptor c-kit mediates lodgment and acute expansion of hematopoietic cells in the murine spleen. Blood 88:75-81. [PubMed] [Google Scholar]
- 13.Brownawell, A. M., G. J. Kops, I. G. Macara, and B. M. Burgering. 2001. Inhibition of nuclear import by protein kinase B (Akt) regulates the subcellular distribution and activity of the Forkhead transcription factor AFX. Mol. Cell. Biol. 21:3534-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunet, A., A. Bonni, M. J. Zigmond, M. Z. Lin, P. Juo, L. S. Hu, M. J. Anderson, K. C. Arden, J. Blenis, and M. E. Greenberg. 1999. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96:857-868. [DOI] [PubMed] [Google Scholar]
- 15.Brunet, A., L. B. Sweeney, J. F. Sturgill, K. F. Chua, P. L. Greer, Y. Lin, H. Tran, S. E. Ross, R. Mostoslavsky, H. Y. Cohen, L. S. Hu, H. L. Cheng, M. P. Jedrychowski, S. P. Gygi, D. A. Sinclair, F. W. Alt, and M. E. Greenberg. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303:2011-2015. [DOI] [PubMed] [Google Scholar]
- 16.Budanov, A. V., A. A. Sablina, E. Feinstein, E. V. Koonin, and P. M. Chumakov. 2004. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science 304:596-600. [DOI] [PubMed] [Google Scholar]
- 17.Burgering, B. M., and G. J. Kops. 2002. Cell cycle and death control: long live Forkheads. Trends Biochem. Sci. 27:352-360. [DOI] [PubMed] [Google Scholar]
- 18.Castrillon, D. H., L. Miao, R. Kollipara, J. W. Horner, and R. A. DePinho. 2003. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science 301:215-218. [DOI] [PubMed] [Google Scholar]
- 19.Cortes, U., C. Moyret-Lalle, N. Falette, C. Duriez, F. E. Ghissassi, C. Barnas, A. P. Morel, P. Hainaut, J. P. Magaud, and A. Puisieux. 2000. BTG gene expression in the p53-dependent and -independent cellular response to DNA damage. Mol. Carcinog. 27:57-64. [PubMed] [Google Scholar]
- 20.Cui, Y., G. Riedlinger, K. Miyoshi, W. Tang, C. Li, C.-X. Deng, G. W. Robinson, and L. Hennighausen. 2004. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 24:8037-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Damen, J. E., H. Wakao, A. Miyajima, J. Krosl, R. K. Humphries, R. L. Cutler, and G. Krystal. 1995. Tyrosine 343 in the erythropoietin receptor positively regulates erythropoietin-induced cell proliferation and Stat5 activation. EMBO J. 14:5557-5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datta, M. C. 1985. Prostaglandin E2 mediated effects on the synthesis of fetal and adult hemoglobin in blood erythroid bursts. Prostaglandins 29:561-577. [DOI] [PubMed] [Google Scholar]
- 23.Devary, Y., R. A. Gottlieb, L. F. Lau, and M. Karin. 1991. Rapid and preferential activation of the c-jun gene during the mammalian UV response. Mol. Cell. Biol. 11:2804-2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dijkers, P. F., R. H. Medema, J. W. Lammers, L. Koenderman, and P. J. Coffer. 2000. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 10:1201-1204. [DOI] [PubMed] [Google Scholar]
- 25.Dolznig, H., F. Boulme, K. Stangl, E. M. Deiner, W. Mikulits, H. Beug, and E. W. Mullner. 2001. Establishment of normal, terminally differentiating mouse erythroid progenitors: molecular characterization by cDNA arrays. FASEB J. 15:1442-1444. [DOI] [PubMed] [Google Scholar]
- 26.Dolznig, H., B. Habermann, K. Stangl, E. M. Deiner, R. Moriggl, H. Beug, and E. W. Mullner. 2002. Apoptosis protection by the epo target bcl-x(l) allows factor-independent differentiation of primary erythroblasts. Curr. Biol. 12:1076-1085. [DOI] [PubMed] [Google Scholar]
- 27.Essers, M. A., L. M. de Vries-Smits, N. Barker, P. E. Polderman, B. M. Burgering, and H. C. Korswagen. 2005. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science 308:1181-1184. [DOI] [PubMed] [Google Scholar]
- 28.Reference deleted.
- 29.Fukao, T., T. Yamada, M. Tanabe, Y. Terauchi, T. Ota, T. Takayama, T. Asano, T. Takeuchi, T. Kadowaki, J. H. Ji, and S. Koyasu. 2002. Selective loss of gastrointestinal mast cells and impaired immunity in PI3K-deficient mice. Nat. Immunol. 3:295-304. [DOI] [PubMed] [Google Scholar]
- 30.Furukawa-Hibi, Y., K. Yoshida-Araki, T. Ohta, K. Ikeda, and N. Motoyama. 2002. FOXO forkhead transcription factors induce G(2)-M checkpoint in response to oxidative stress. J. Biol. Chem. 277:26729-26732. [DOI] [PubMed] [Google Scholar]
- 31.Garingo, A. D., M. Suhasini, N. C. Andrews, and R. B. Pilz. 1995. cAMP-dependent protein kinase is necessary for increased NF-E2.DNA complex formation during erythroleukemia cell differentiation. J. Biol. Chem. 270:9169-9177. [DOI] [PubMed] [Google Scholar]
- 32.Greer, E. L., and A. Brunet. 2005. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 24:7410-7425. [DOI] [PubMed] [Google Scholar]
- 33.Guillemin, M. C., E. Raffoux, D. Vitoux, S. Kogan, H. Soilihi, V. Lallemand-Breitenbach, J. Zhu, A. Janin, M. T. Daniel, B. Gourmel, L. Degos, H. Dombret, M. Lanotte, and H. De The. 2002. In vivo activation of cAMP signaling induces growth arrest and differentiation in acute promyelocytic leukemia. J. Exp. Med. 196:1373-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herr, I., H. van Dam, and P. Angel. 1994. Binding of promoter-associated AP-1 is not altered during induction and subsequent repression of the c-jun promoter by TPA and UV irradiation. Carcinogenesis 15:1105-1113. [DOI] [PubMed] [Google Scholar]
- 35.Reference deleted.
- 36.Hu, M. C., D. F. Lee, W. Xia, L. S. Golfman, F. Ou-Yang, J. Y. Yang, Y. Zou, S. Bao, N. Hanada, H. Saso, R. Kobayashi, and M. C. Hung. 2004. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell 117:225-237. [DOI] [PubMed] [Google Scholar]
- 37.Huddleston, H., B. Tan, F. C. Yang, H. White, M. J. Wenning, A. Orazi, M. C. Yoder, R. Kapur, and D. A. Ingram. 2003. Functional p85alpha gene is required for normal murine fetal erythropoiesis. Blood 102:142-145. [DOI] [PubMed] [Google Scholar]
- 38.Iozzo, R. V., D. K. Moscatello, D. J. McQuillan, and I. Eichstetter. 1999. Decorin is a biological ligand for the epidermal growth factor receptor. J. Biol. Chem. 274:4489-4492. [DOI] [PubMed] [Google Scholar]
- 39.Jacobs, F. M., L. P. van der Heide, P. J. Wijchers, J. P. Burbach, M. F. Hoekman, and M. P. Smidt. 2003. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J. Biol. Chem. 278:35959-35967. [DOI] [PubMed] [Google Scholar]
- 40.Kaestner, K. H., W. Knochel, and D. E. Martinez. 2000. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 14:142-146. [PubMed] [Google Scholar]
- 41.Kawasaki, H., L. Schiltz, R. Chiu, K. Itakura, K. Taira, Y. Nakatani, and K. K. Yokoyama. 2000. ATF-2 has intrinsic histone acetyltransferase activity which is modulated by phosphorylation. Nature 405:195-200. [DOI] [PubMed] [Google Scholar]
- 42.Kolbus, A., M. Blazquez-Domingo, S. Carotta, W. Bakker, S. Luedemann, M. von Lindern, P. Steinlein, and H. Beug. 2003. Cooperative signaling between cytokine receptors and the glucocorticoid receptor in the expansion of erythroid progenitors: molecular analysis by expression profiling. Blood 102:3136-3146. [DOI] [PubMed] [Google Scholar]
- 43.Kops, G. J., and B. M. Burgering. 1999. Forkhead transcription factors: new insights into protein kinase B (c-akt) signaling. J. Mol. Med. 77:656-665. [DOI] [PubMed] [Google Scholar]
- 44.Kortylewski, M., F. Feld, K. D. Kruger, G. Bahrenberg, R. A. Roth, H. G. Joost, P. C. Heinrich, I. Behrmann, and A. Barthel. 2003. Akt modulates STAT3-mediated gene expression through a FKHR (FOXO1a)-dependent mechanism. J. Biol. Chem. 278:5242-5249. [DOI] [PubMed] [Google Scholar]
- 45.Lai, E., K. L. Clark, S. K. Burley, and J. E. Darnell, Jr. 1993. Hepatocyte nuclear factor 3/fork head or “winged helix” proteins: a family of transcription factors of diverse biologic function. Proc. Natl. Acad. Sci. USA 90:10421-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leenders, H., S. Whiffield, C. Benoist, and D. Mathis. 2000. Role of the forkhead transcription family member, FKHR, in thymocyte differentiation. Eur. J. Immunol. 30:2980-2990. [DOI] [PubMed] [Google Scholar]
- 47.Leung, M. K., T. Jones, C. L. Michels, D. M. Livingston, and S. Bhattacharya. 1999. Molecular cloning and chromosomal localization of the human CITED2 gene encoding p35srj/Mrg1. Genomics 61:307-313. [DOI] [PubMed] [Google Scholar]
- 48.Lu-Kuo, J. M., D. A. Fruman, D. M. Joyal, L. C. Cantley, and H. R. Katz. 2000. Impaired kit- but not FcepsilonRI-initiated mast cell activation in the absence of phosphoinositide 3-kinase p85alpha gene products. J. Biol. Chem. 275:6022-6029. [DOI] [PubMed] [Google Scholar]
- 49.Martínez-Gac, L., M. Marqués, Z. García, M. R. Campanero, and A. C. Carrera. 2004. Control of cyclin G2 mRNA expression by forkhead transcription factors: novel mechanism for cell cycle control by phosphoinositide 3-kinase and forkhead. Mol. Cell. Biol. 24:2181-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Medema, R. H., G. J. Kops, J. L. Bos, and B. M. Burgering. 2000. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature 404:782-787. [DOI] [PubMed] [Google Scholar]
- 51.Montminy, M. 1997. Transcriptional regulation by cyclic AMP. Annu. Rev. Biochem. 66:807-822. [DOI] [PubMed] [Google Scholar]
- 52.Motta, M. C., N. Divecha, M. Lemieux, C. Kamel, D. Chen, W. Gu, Y. Bultsma, M. McBurney, and L. Guarente. 2004. Mammalian SIRT1 represses forkhead transcription factors. Cell 116:551-563. [DOI] [PubMed] [Google Scholar]
- 53.Obata, Y., K. Yamamoto, M. Miyazaki, K. Shimotohno, S. Kohno, and T. Matsuyama. 2005. Role of cyclophilin B in activation of interferon regulatory factor-3. J. Biol. Chem. 280:18355-18360. [DOI] [PubMed] [Google Scholar]
- 54.Pilz, R. B. 1993. Impaired erythroid-specific gene expression in cAMP-dependent protein kinase-deficient murine erythroleukemia cells. J. Biol. Chem. 268:20252-20258. [PubMed] [Google Scholar]
- 55.Ponka, P. 1999. Cell biology of heme. Am. J. Med. Sci. 318:241-256. [DOI] [PubMed] [Google Scholar]
- 56.Porzig, H., R. Gutknecht, G. Kostova, and K. Thalmeier. 1995. G-protein-coupled receptors in normal human erythroid progenitor cells. Naunyn Schmiedebergs Arch. Pharmacol. 353:11-20. [DOI] [PubMed] [Google Scholar]
- 57.Ramaswamy, S., N. Nakamura, I. Sansal, L. Bergeron, and W. R. Sellers. 2002. A novel mechanism of gene regulation and tumor suppression by the transcription factor FKHR. Cancer Cell 2:81-91. [DOI] [PubMed] [Google Scholar]
- 58.Rattis, F. M., C. Voermans, and T. Reya. 2004. Wnt signaling in the stem cell niche. Curr. Opin. Hematol. 11:88-94. [DOI] [PubMed] [Google Scholar]
- 59.Sassone-Corsi, P. 1995. Transcription factors responsive to cAMP. Annu. Rev. Cell Dev. Biol. 11:355-377. [DOI] [PubMed] [Google Scholar]
- 60.Schmidt, U., E. Van Den Akker, M. Parren-Van Amelsvoort, G. Litos, M. De Bruijn, L. Gutierrez, R. W. Hendriks, W. Ellmeier, B. Lowenberg, H. Beug, and M. Von Lindern. 2004. Btk Is required for an efficient response to erythropoietin and for SCF-controlled protection against TRAIL in erythroid progenitors. J. Exp. Med. 199:785-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seoane, J., H. V. Le, L. Shen, S. A. Anderson, and J. Massague. 2004. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell 117:211-223. [DOI] [PubMed] [Google Scholar]
- 62.Sun, H. B., Y. X. Zhu, T. Yin, G. Sledge, and Y. C. Yang. 1998. MRG1, the product of a melanocyte-specific gene related gene, is a cytokine-inducible transcription factor with transformation activity. Proc. Natl. Acad. Sci. USA 95:13555-13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Surinya, K. H., T. C. Cox, and B. K. May. 1997. Transcriptional regulation of the human erythroid 5-aminolevulinate synthase gene. Identification of promoter elements and role of regulatory proteins. J. Biol. Chem. 272:26585-26594. [DOI] [PubMed] [Google Scholar]
- 64.Tran, H., A. Brunet, J. M. Grenier, S. R. Datta, A. J. Fornace, Jr., P. S. DiStefano, L. W. Chiang, and M. E. Greenberg. 2002. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science 296:530-534. [DOI] [PubMed] [Google Scholar]
- 65.van Dam, H., M. Duyndam, R. Rottier, A. Bosch, L. de Vries-Smits, P. Herrlich, A. Zantema, P. Angel, and A. J. van der Eb. 1993. Heterodimer formation of cJun and ATF-2 is responsible for induction of c-jun by the 243 amino acid adenovirus E1A protein. EMBO J. 12:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Dam, H., D. Wilhelm, I. Herr, A. Steffen, P. Herrlich, and P. Angel. 1995. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 14:1798-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van den Akker, E., T. van Dijk, M. Parren-van Amelsvoort, K. S. Grossmann, U. Schaeper, K. Toney-Earley, S. E. Waltz, B. Lowenberg, and M. von Lindern. 2004. Tyrosine kinase receptor RON functions downstream of the erythropoietin receptor to induce expansion of erythroid progenitors. Blood 103:4457-4465. [DOI] [PubMed] [Google Scholar]
- 68.Van Der Heide, L. P., M. F. Hoekman, and M. P. Smidt. 2004. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem. J. 380:297-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Dijk, T. B., E. van Den Akker, M. P. Amelsvoort, H. Mano, B. Lowenberg, and M. von Lindern. 2000. Stem cell factor induces phosphatidylinositol 3′-kinase-dependent Lyn/Tec/Dok-1 complex formation in hematopoietic cells. Blood 96:3406-3413. [PubMed] [Google Scholar]
- 70.von Lindern, M., E. M. Deiner, H. Dolznig, M. Parren-Van Amelsvoort, M. J. Hayman, E. W. Mullner, and H. Beug. 2001. Leukemic transformation of normal murine erythroid progenitors: v- and c-ErbB act through signaling pathways activated by the EpoR and c-Kit in stress erythropoiesis. Oncogene 20:3651-3664. [DOI] [PubMed] [Google Scholar]
- 71.von Lindern, M., W. Zauner, G. Mellitzer, P. Steinlein, G. Fritsch, K. Huber, B. Lowenberg, and H. Beug. 1999. The glucocorticoid receptor cooperates with the erythropoietin receptor and c-Kit to enhance and sustain proliferation of erythroid progenitors in vitro. Blood 94:550-559. [PubMed] [Google Scholar]
- 72.Wang, D., D. Stravopodis, S. Teglund, J. Kitazawa, and J. Ihle. 1996. Naturally occurring dominant negative variants of Stat5. Mol. Cell. Biol. 16:6141-6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Withington, S. L., A. N. Scott, D. N. Saunders, K. Lopes Floro, J. I. Preis, J. Michalicek, K. Maclean, D. B. Sparrow, J. P. Barbera, and S. L. Dunwoodie. 2006. Loss of Cited2 affects trophoblast formation and vascularization of the mouse placenta. Dev. Biol. 294:67-82. [DOI] [PubMed] [Google Scholar]
- 74.Yin, Z., J. Haynie, X. Yang, B. Han, S. Kiatchoosakun, J. Restivo, S. Yuan, N. R. Prabhakar, K. Herrup, R. A. Conlon, B. D. Hoit, M. Watanabe, and Y. C. Yang. 2002. The essential role of Cited2, a negative regulator for HIF-1alpha, in heart development and neurulation. Proc. Natl. Acad. Sci. USA 99:10488-10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.