Abstract
The SWR1 complex (SWR1-C)-dependent deposition of the histone variant Htz1 on promoter nucleosomes is typical of Saccharomyces cerevisiae genes whose expression is frequently reprogrammed. Although this epigenetic marking is of significant physiological importance, the determinants of Htz1 deposition, the conditions that set off SWR1-C occupancy, and the implications of Htz1 in transcriptional initiation are issues that remain unresolved. In this report, we addressed these questions by investigating the GAL1 promoter. We show that Htz1 is required for efficient Mediator recruitment and transcription only when the GAL1 promoter is under the influence of the Tup1 corepressor. In fact, we show that it is Tup1 that specifies Htz1 deposition for the promoter nucleosome covering the transcription start site. This deposition occurs rapidly following transcriptional repression, and it correlates with a Tup1-independent transient recruitment of the SWR1 complex. We propose that Tup1 cooperates with SWR1-C and specifies Htz1 deposition at GAL1, thereby marking the promoter for rapid neutralization from its repressive effects.
Promoter chromatin organization is a fundamental determinant of the dynamics of transcriptional states of eukaryotic genes. In the course of transcriptional activation or the establishment of transcriptional repression, promoter-resident nucleosomal histones serve as substrates for a variety of modifying and/or remodeling activities that have their reorganization as a result. This process is often facilitated by the presence of specific histone variants in these nucleosomes (for a review, see reference 2). A major class of such histone variants includes isoforms of histone H2A that differ from the canonical histone in both the length and sequence of the C terminus (38). Among those, H2A.Z is present in all eukaryotic species, and its functional impact ranges from the maintenance of heterochromatin and high-order chromatin structure in mammals (9, 33) to an involvement in both heterochromatin and euchromatin in lower eukaryotes (1, 25). In Saccharomyces cerevisiae, H2A.Z is encoded by the nonessential HTZ1 gene, and initial studies focusing on its genome-wide location revealed that it imposes a barrier in the spreading of the Sir complex-dependent heterochromatin (25). It was recently established that Htz1, the yeast H2A.Z homologue, is preferentially present in promoter regions of euchromatic genes (13, 21, 32, 44) and that this presence is inversely proportional to transcription rate and occupancy of RNA polymerase II (PolII) (21). In fact, studies on specific promoters, such as those of GAL1 and PHO5, have shown that Htz1 is present only when they are silent, being evicted upon transcriptional activation (1, 35), a fact most likely reflecting the partial nucleosome loss from transcriptionally active promoters (4, 34, 36). Finally, it has been shown for both these model promoters that the presence of Htz1 is actually required for proper transcriptional activation (13, 21, 44).
The fact that Htz1 contributes to the transcriptional activation of previously repressed genes has led to the hypothesis that this epigenetic marking serves genes whose expression is constantly reprogrammed (22). The obvious question then concerns the identification of the signal(s) that determines the recruitment of the elicitors of Htz1 deposition to such promoters. The only identified elicitor, SWR1-C, a multisubunit protein complex, has been shown to replace Htz1-H2B with H2A-H2B dimers in vitro, while a deletion of SWR1, the gene encoding the catalytic Swr1 ATPase subunit of SWR1-C, abolishes Htz1 deposition in vivo (18, 19, 27). Recent studies have implicated two promoter features as being signals that target SWR1-C recruitment and Htz1 depositions to euchromatic loci. One feature is acetylation of nucleosomal histones, a modification that targets SWR1-C recruitment through the mediation of its Bdf1 subunit, which contains two tandem bromodomains shown to bind acetylated histone H3 and H4 tails (20, 24). In concert with this notion, inactivation of the Gcn5 HAT subunit of SAGA resulted in decreased levels of Htz1 deposition at certain loci, and a similar defect was observed when Bdf1 was inactivated (32, 44). The second feature is based on a common nucleosomal architecture, according to which the Htz1-containing nucleosomes are located upstream and downstream of a nucleosome-free region that encompasses the transcription start site (TSS) (32). Strikingly, a single 22-bp DNA promoter sequence was found to be sufficient for both the formation of a nucleosome-free region and Htz1 deposition in the two flanking nucleosomes. This sequence element consists of a binding site for the Myb-related DNA binding protein Reb1 and an adjacent dT:dA tract, and both motifs are required for Htz1 deposition (32).
Although these motifs and the corresponding chromatin architecture are frequently found in yeast promoters, they by no means cover the entire repertoire of Htz1-containing promoters. One such case is the well-studied GAL1 promoter. This Htz1-containing promoter lacks a sequence element that contains Reb1 binding sites adjacent to dT:dA tracks (28). Moreover, GAL1 lacks a nucleosome-free region encompassing the TSS; instead, it bears a well-positioned nucleosome (nucleosome −1) covering the TSS (5, 41). In addition, it is hard to envision how histone acetylation can offer targeting for SWR1-C since the GAL1 promoter is subjected to transcriptional repression by the Tup1 corepressor complex known to be involved in the recruitment of histone deacetylases (6, 43). It follows that GAL1 is a notable exception from the general rules established from whole-genome studies, and its well characterized repression-activation mechanisms offer a unique opportunity to expand the rules that determine Htz1 deposition to this category of drastically reprogrammable genes.
In this report, we have investigated the regulatory factors that determine Htz1 deposition at specific GAL1 promoter nucleosomes as well as the conditions that trigger SWR1-C recruitment. We show that the Tup1 corepressor specifies Htz1 deposition at a single GAL1 promoter nucleosome upon glucose repression and that this deposition marks the promoter for efficient Mediator recruitment and rapid transcriptional activation. This mechanism is not restricted to the GAL1 promoter since we also show that Tup1 is required for Htz1 deposition at the SUC2 promoter.
MATERIALS AND METHODS
Yeast strains and media.
Standard synthetic yeast media were used: yeast extract-peptone-glucose (2% glucose) and yeast extract-peptone-galactose (2% galactose). Cells were shifted from glucose to galactose following a 15-min wash in sterile water at 30°C. Cells were shifted from galactose to glucose by removing yeast extract-peptone-galactose and adding yeast extract-peptone-glucose with no water wash interval. FT5, a Gal+ derivative of S288c (42), was used to epitope tag Htz1 and Swr1 with 3-hemagglutinin (3-HA) and 9-Myc epitopes, respectively, according to methods described previously (17). These tagged strains exhibited normal growth with 2% formamide as previously described (15). 9-Myc Srb4 and 3-HA Gcn5 were similarly constructed in strain FT5, while the 3-HA TATA-binding protein (TBP)-expressing strain was a gift from K. Struhl. Total deletions of HTZ1, HDA1, and SWR1 were generated using a PCR-based strategy as described previously (17). The FT5 derivatives swi2Δ, tup1Δ, and cyc8Δ were described previously (30, 39).
Gene expression analysis and nucleosome remodeling assays.
Total yeast RNA was isolated from yeast cells grown to an optical density at 600 nm of ∼0.60 to 0.8 by the acid hot-phenol method as described previously (41). Micrococcal nuclease assays were performed using nystatin-permeabilized spheroplasts (41). Following MNase digestions, the nucleosomal structure of the GAL1 promoter was analyzed by the indirect end-labeling method, cutting by PvuII, and using the GAL1 BsaI-PvuII fragment as a probe, as described previously (41).
Chromatin IPs.
All chromatin immunoprecipitations (IPs) were performed as described previously (37), with the following modifications. The cross-linking of HA-Htz1-bearing strains was done for 20 min, while the cross-linking of 9-Myc Swr1 was prolonged to 1 h. An excess amount of antibody was used in all cases in order to deplete antigen. The following commercially available antibodies were used: polyclonal antibody specific for the HA and Myc epitopes (Y-11 and A-14, respectively; Santa Cruz Biotechnologies), polyclonal antibody against the DNA binding domain of Gal4 activator protein and PolII (Sc-577 and C-21, respectively; Santa Cruz Biotechnologies), and, finally, polyclonal serum against H3 (ab1791; Abcam). Immunoprecipitated as well as input DNAs were amplified by a 26-cycle PCR in the presence of [α-32P]dATP and tested for linearity by using template serial dilutions, and products were analyzed in a 7% polyacrylamide gel. The DNA fragments were visualized through autoradiography and quantified with a PhosphorImager (Molecular Dynamics). PCR products were quantified, and numbers indicating the ratio of IP over input PCR products were calculated. The oligonucleotide primers used were as follows (coordinates are given relative to the ATG [+1]): GAL1 (upstream activation sequence [UAS]/upstream repression sequence) primers from positions −370 to −169, GAL1 (core promoter) from positions −179 to + 43, GAL1 open reading frame (ORF) primers from positions +1342 to +1558, the PHO5 promoter from positions −367 to +40, and TEL6 primers that amplify an intergenic region proximal to the right arm of telomere VI (VIR) (positions 269322 to 269339 and 269679 to 269698, respectively) (40). For experiments involving mononucleosomes, fragmentation of cross-linked protein-DNA complexes was accomplished by extensive MNase digestion, as described previously (37). Primers amplifying the nucleosome-protected DNA fragments were designed in accordance with methods described previously (13). Mononucleosome prevalence was verified by PCR using a 200-bp amplicon flanking the above-described nucleosome regions.
RESULTS
Htz1 is required for efficient Mediator recruitment at the GAL1 promoter.
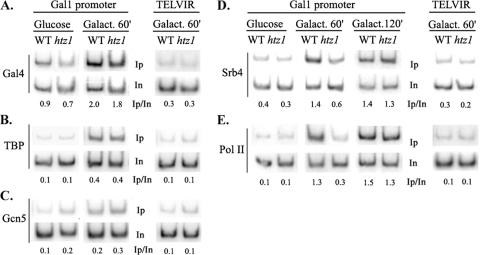
Clues for the identification of factors determining Htz1 deposition at the GAL1 promoter nucleosomes can be offered by gaining an understanding of the functional significance of such a deposition in transcriptional dynamics. It was reported previously that Htz1 is required for the efficient transcriptional activation of GAL1 without being involved in the efficiency of the binding of the Gal4 activator (1). In order to identify the step at which Htz1 exerted its function, we examined the recruitment of coactivators known to be required for GAL1 transcriptional activation in yeast strains lacking the HTZ1 gene (htz1Δ strains). As shown in Fig. 1A and B, chromatin IP analysis revealed that the extent of Gal4 and TBP recruitment was comparable between wild-type and htz1Δ strains grown under either repressive (glucose) or inducing (60 min in galactose) conditions. As expected, neither Gal4 nor TBP displayed detectable binding on an irrelevant intergenic region located proximal to the right arm of chromosome VI (Fig. 1A and B, right). In accordance with the fact that functions mediated by the SAGA coactivator complex dictate TBP recruitment on this promoter (3, 8), the weak increase in the occupancy of the Gcn5 histone HAT subunit of SAGA observed early in the course of transcriptional induction was also not affected by the lack of the Htz1 histone variant (Fig. 1C). By contrast, monitoring of Srb4, a core Mediator subunit, revealed that the recruitment of Mediator was severely compromised in the htz1Δ strain grown in galactose medium (60 min), and as a consequence, RNA PolII (monitored by the large Rpb1 subunit) failed to be recruited on the GAL1 promoter under the same conditions (Fig. 1D and E). No recruitment was again observed on the telomere VIR intergenic region (Fig. 1D and E, right). These results suggested a positive function for the Htz1-containing nucleosomes in the recruitment of the Mediator complex on the GAL1 promoter during the process of transcriptional activation. This positive role of Htz1 was prominent at early stages of galactose induction; at later time points (galactose for 120 min), the levels of Mediator and PolII occupancy of GAL1 at the htz1 mutant strain were comparable to those observed in the wild-type strain (Fig. 1D and E), consistent with the similar levels of GAL1 expression at this time point (see below).
FIG. 1.
Htz1 is required for Mediator recruitment at the GAL1 promoter. The recruitment of various components of the preinitiation complex on GAL1 (left) and on the telomere VIR (TELVIR) (right) was monitored by chromatin IP followed by PCR amplification. Yeast cultures were grown exponentially in yeast extract-peptone-glucose and shifted in galactose (Galact.)-containing medium either for 60 min (A, B, and C) or for 60 min and 120 min (D and E). (A) Recruitment of the Gal4 activator protein. (B) Recruitment of HA-tagged TBP. (C) Recruitment of HA-tagged Gcn5 HAT subunit of SAGA. (D) Recruitment of the Myc-tagged Mediator subunit Srb4. (E) Recruitment of the Rpb1 subunit of PolII. Primers used in panels A, B, and C amplify the GAL1 UAS region, while primers used in panels D and E amplify the GAL1 core promoter (see also Fig. 6 for a physical map). Inputs (In) and immunoprecipitated (Ip) DNAs were PCR amplified in the presence of [32P]dATP, and numbers indicate immunoprecipitated/input ratios as quantified by phosphorimaging. WT, wild type.
The Htz1 requirement for GAL1 activation is compromised in the absence of the Tup1 corepressor.
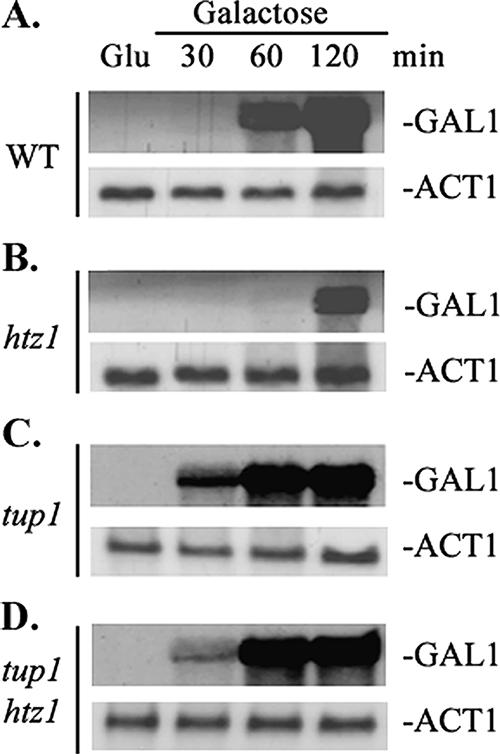
The process of transcriptional activation of the GAL1 gene requires the alleviation of the negative function of the Tup1 corepressor. Besides recruiting histone deacetylases (see above), the Tup1 transcriptional repression function is also mediated by physical interactions between the Tup1 repression domain and specific Mediator subunits (14, 29, 44). Given the above-described results, one could postulate that Htz1 antagonizes this Tup1-mediated repression. We tested this hypothesis genetically. The GAL1 transcriptional defect of the htz1Δ strain was manifested as a significant delay (60 min versus 120 min) in GAL1 activation kinetics upon shifting glucose-grown yeast cultures to galactose medium (Fig. 2A and B). In a tup1Δ mutant strain, GAL1 transcription was induced even more rapidly than in the wild-type strain (Fig. 2C), and similar rapid kinetics were observed in the tup1Δ htz1Δ double mutant strain (Fig. 2D). Thus, alleviation of Tup1-mediated repression compromises the Htz1 requirement for transcriptional activation of GAL1 transcription.
FIG. 2.
Deletion of TUP1 overcomes the Htz1 requirement for GAL1 activation. Northern blot analysis of GAL1 mRNA isolated from (A) wild-type (WT), (B) htz1Δ, (C) tup1Δ, and (D) double htz1Δtup1Δ yeast mutant strains is shown. Cells were grown in yeast extract-peptone-glucose and shifted to yeast extract-peptone-galactose medium. At the indicated time points, total RNA was extracted, fractionated by formaldehyde agarose gel, and hybridized with 32P-labeled GAL1 probe and ACT1 probe (serving as a loading control).
Htz1 deposition at GAL1 depends on the Tup1 corepressor.
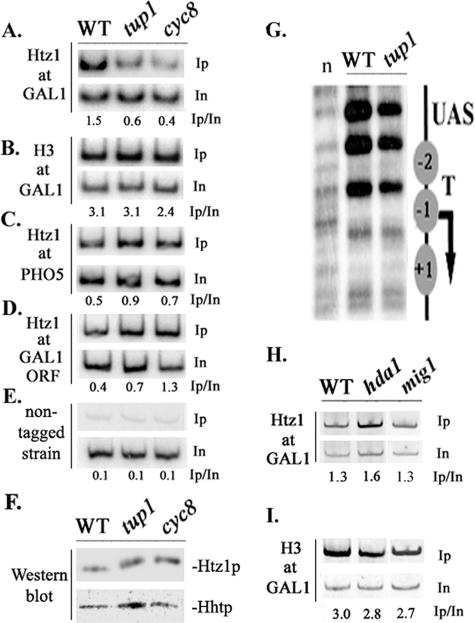
Given the above-described finding, it is possible that Htz1-containing nucleosomes do not even exist in the absence of this corepressor complex. This possibility was tested by examining the levels of Htz1 present on the GAL1 promoter in tup1Δ and cyc8Δ strains lacking either one of the corepressor's subunits. Using epitope-tagged Htz1-expressing strains grown under repressive conditions (glucose), we observed that Htz1 levels deposited at the GAL1 promoter were significantly lower in the tup1Δ strain than those observed in the wild-type strain (Fig. 3A). In the cyc8Δ yeast mutant strain, we observed an even more severe defect in Htz1 deposition (Fig. 3A). In contrast to the Htz1 variant, the levels of histone H3 were comparable in wild-type, tup1Δ, and cyc8Δ strains, a fact suggesting that the underlying nucleosomal density was not affected by these mutations (Fig. 3B). This was substantiated through micrococcal nuclease analysis, which revealed that Tup1 was not required for the positioning of the promoter nucleosomes (Fig. 3G). On the other hand, Htz1 deposition at the Tup1-independent PHO5 promoter as well as at the coding sequence of the GAL1 gene was actually enriched in tup1Δ or the cyc8Δ strains (Fig. 3C and D), although both mutant strains express the Htz1 protein (relative to histone H3) at levels comparable to those observed for the wild-type strain (Fig. 3F). Finally, as shown in Fig. 3H, Htz1 was normally deposited at the GAL1 promoter in a yeast mutant strain lacking HDA1, the histone deacetylase known to function with Tup1 in the same pathway (43), and similar results were observed with histone H3 (Fig. 3I). Htz1 and H3 were also normally deposited at GAL1 in a yeast strain lacking MIG1 (Fig. 3H and I), the DNA-binding repressor protein shown to be important for full transcriptional repression function yet dispensable for Cyc8-Tup1 recruitment to the GAL1 promoter (30). In addition, Htz1 was normally deposited at the GAL1 promoter in strains lacking either Rpd3 or Cti6 subunits of the Sin3-Rpd3 histone deacetylase complex (16) (data not shown). The results presented above clearly indicate that Htz1 deposition at the GAL1 promoter depends on the Tup1-Cyc8 corepressor complex and that this effect does not involve localized histone deacetylation through Hda1 or the function of additional repressor proteins such as Mig1.
FIG. 3.
Htz1 deposition at the GAL1 promoter depends on the Tup1 corepressor. Chromatin IPs were performed in glucose-grown wild-type (WT), tup1Δ, and cyc8Δ mutant strains all expressing an HA epitope-tagged Htz1 histone variant. Htz1 or H3 deposition in these strains was monitored for the GAL1 (A and B), the PHO5 (C), and the GAL1 (D) ORFs. Inputs (In) and immunoprecipitated (Ip) DNAs were PCR amplified in the presence of [32P]dATP, and numbers indicate immunoprecipitated/input ratios. (E) Htz1 background levels were controlled by chromatin IP as in A using non-Htz1-tagged wild-type, tup1Δ, and cyc8Δ mutant strains. The primers used amplify the GAL1 UAS region. (F) Western blot analysis of total protein extracts obtained from the indicated HA-Htz1-expressing strains grown in glucose. HA-specific and anti-H3 antibodies were used. (G) Nucleosome positioning at the GAL1 promoter in wild-type and tup1Δ strains as assayed by MNase sensitivity; “n” indicates naked genomic DNA digested with MNase. Also shown is a schematic representation of the GAL1 promoter indicating the position of three nucleosomes (−1, −2, and +1). The UASGAL (UAS), the TATA element (T), and TSS (arrow) are also indicated. Histone Htz1 (H) and histone H3 (I) deposition at GAL1 in wild-type and hda1 and mig1 deletion yeast strains was monitored by chromatin IPs performed as described above (A and B).
The Tup1 requirement of Htz1 deposition is not restricted to the GAL1 promoter.
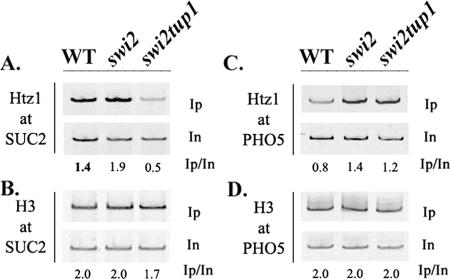
Examination of the generality of the involvement of Tup1 in Htz1 deposition is complicated by the fact that, in contrast to genes involved in galactose metabolism, most Tup1-regulated genes are constitutively expressed in a TUP1 deletion strain. This necessitates an experimental setting that could allow the testing of Htz1 deposition in a tup1Δ strain not only in the absence of active transcription (a situation that could be achieved by using a TATA-less promoter context) but also in the absence of subsequent promoter nucleosome remodeling or loss. Transcription of SUC2, another Tup1-repressible gene not related to galactose metabolism, depends on the Swi/Snf chromatin remodeling complex, and the absence of this complex abolishes SUC2 promoter nucleosome remodeling and transcription even in a mutant background lacking the Tup1-encoding gene (11). We examined the levels of Htz1 deposited at the SUC2 promoter in wild-type, swi2Δ, and swi2Δ tup1Δ double mutant strains grown in glucose medium. As indicated in Fig. 4A, the levels of Htz1 deposited at SUC2 in the swi2Δ mutant strain were comparable to those observed in the wild-type strain, but they were significantly higher than those observed in the swi2Δ tup1Δ double mutant strain. No difference in Htz1 deposition was observed at the PHO5 promoter (Fig. 4C), while levels of histone H3 at both promoters were comparable in wild-type, swi2Δ and swi2Δ tup1Δ strains (Fig. 4B and D). This experiment demonstrated that the Tup1 requirement for Htz1 deposition is also evident for SUC2 and may affect additional Tup1-regulated genes.
FIG. 4.
Htz1 deposition at the SUC2 promoter depends on the Tup1 corepressor. Chromatin IPs were performed in glucose-grown wild-type (WT) and swi2Δ and swi2Δtup1Δ mutant strains all expressing an HA epitope-tagged histone Htz1. Deposition of Htz1 or H3 in these strains was monitored for SUC2 (A and B) and PHO5 (C and D). Inputs (In) and immunoprecipitated (Ip) DNAs were PCR amplified in the presence of [32P]dATP, and numbers indicate immunoprecipitated/input ratios.
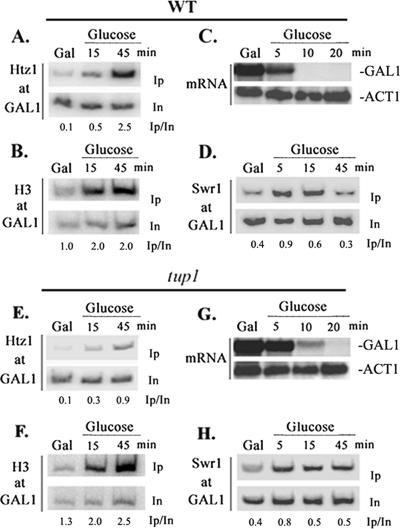
Tup1-dependent Htz1 deposition at GAL1 occurs rapidly upon reestablishment of repression.
We have established that Tup1 is required for Htz1 deposition at the GAL1 promoter in cells grown in steady state in glucose. This requirement could reflect either the maintenance of the Htz1-modified nucleosomal structure or the dynamics of its establishment. In that respect, Htz1 deposition should occur upon the reestablishment of the transcriptionally repressed state, since the high levels of Htz1 at GAL1 observed in glucose-grown cells are significantly decreased when growth occurs under induction conditions (1). Indeed, as indicated in Fig. 5A, increased levels of Htz1 at GAL1 were evident within 15 min after shifting cultures from galactose to glucose and reached the highest values after 45 min of growth in glucose. When the same chromatin samples were immunoprecipitated with antibodies raised against histone H3, we observed that histone H3 occupancy preceded that of Htz1 (15 versus 45 min) (Fig. 5B), possibly reflecting Htz1 deposition at a preformed canonical nucleosome. Finally, GAL1 mRNA analysis in the wild-type strain showed that upon shifting yeast cultures in glucose medium, GAL1 transcription ceased almost instantaneously, since mRNA levels drop with kinetics in concert with the average half-life of yeast mRNAs (within 5 to 10 min) (Fig. 5C), suggesting that the transcription turnoff preceded the Htz1 deposition. In accordance with the steady-state results (Fig. 3A), Htz1 deposition at the GAL1 promoter was inefficient when tup1Δ cells were shifted to glucose medium (Fig. 5E). Chromatin IPs using antibodies raised against histone H3 showed a histone H3 deposition kinetic similar to that observed in the wild-type strain (Fig. 5F), suggesting that nucleosome re-formation takes place normally upon transcriptional shutdown even in the absence of Tup1. Finally, upon shifting tup1Δ cells in glucose (Fig. 5G), GAL1 transcription ceased with only a slight delay (within 10 min), suggesting that the relatively inefficient Htz1 deposition at GAL1 in yeast cells lacking Tup1 is not a result of persistent transcription in this genetic background. The results presented above indicated that Tup1 is required for the establishment of Htz1-modified nucleosomes at the GAL1 promoter, which occurs rapidly upon transcriptional repression.
FIG. 5.
Htz1 deposition and SWR1-C recruitment at GAL1 occur shortly upon glucose repression. (Top) Histone Htz1 (A) and histone H3 (B) deposition at GAL1. Chromatin IPs were performed in parallel with mRNA analysis in a wild-type (WT) strain expressing HA epitope-tagged Htz1. Yeast cells grown in yeast extract-peptone-galactose medium (Gal) were transferred into yeast extract-peptone-glucose medium (Glucose) for the indicated time points. HA-specific antibody (A), H3-specific antibody (B), and GAL1 promoter primers were used for PCR amplification of input (In) and immunoprecipitated (Ip) DNA. Numbers indicate immunoprecipitated/input ratios. (C) GAL1 mRNA analysis in galactose- and glucose-grown cells. The wild-type strain described above was treated as described above (A), total RNA was extracted at the indicated time points, and it was fractionated in a formaldehyde agarose gel and hybridized with 32P-labeled GAL1 and ACT1 probes. (D) SWR1-C occupancy of GAL1. Chromatin IPs were performed using Myc-specific antibody and GAL1 primers as described above (A) in a wild-type strain expressing Myc epitope-tagged Swr1. Yeast cells were cultured in galactose and then transferred in glucose for the indicated time points. (Bottom) The chromatin IP experiments presented in panels E, F, and H as well as GAL1 mRNA analysis of G were performed in the tup1Δ mutant strain as described above (A, B, C, and D, respectively).
SWR1-C is transiently tethered at GAL1 even in a tup1Δ mutant strain.
It is known that the SWR1 complex is largely responsible for global genomic Htz1 deposition at promoter nucleosomes. Thus, the kinetics of Htz1 deposition at GAL1 allowed us to examine whether the recruitment of SWR1-C was quantitatively dependent on Tup1. For this purpose, we constructed yeast strains expressing an epitope-tagged version of Swr1, the essential SWR1-C ATPase subunit. First, we examined whether SWR1-C was tethered at GAL1 upon glucose repression. As demonstrated in the chromatin IP experiment shown in Fig. 5D, Swr1 cross-links with the GAL1 promoter DNA with relatively low efficiency in galactose-grown cells, but its occupancy increased rapidly within 5 to 15 min after the removal of galactose and the addition of glucose into the growth medium. Swr1 occupancy was transient and dropped thereafter, reaching levels similar to those observed in steady-state galactose-grown cells within 45 min. We concluded that SWR1-C recruitment at GAL1 is increased rapidly and transiently upon transcriptional repression. Given the Tup1 requirement for Htz1 deposition, we then examined whether the extent of tethering of SWR1-C at GAL1 was dependent on the Tup1 corepressor. Surprisingly, the transient increase in the SWR1-C occupancy of the GAL1 promoter that was observed in the wild-type strain was also qualitatively and quantitatively similar in the tup1Δ mutant strain (Fig. 5H). We concluded that Tup1 is not required for the recruitment of the SWR1-C at the GAL1 promoter.
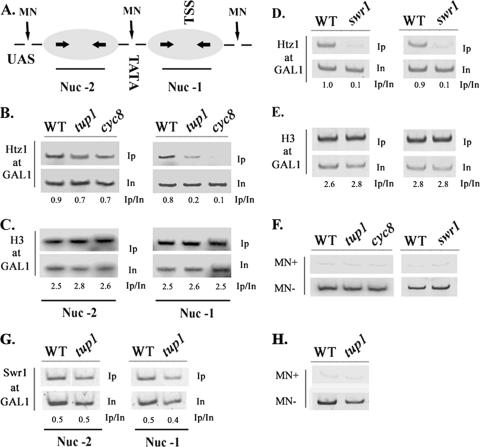
Tup1 is specifically required for Htz1 deposition at the promoter-proximal GAL1 nucleosome.
The fact that the normally recruited SWR1-C resulted in lower (but not absent) levels of Htz1 deposition at GAL1 in a tup1Δ strain could be due to distinct requirements of each promoter nucleosome for such deposition. In order to investigate this possibility, we first monitored the presence of Htz1 at the GAL1 promoter in individual nucleosomes. For this purpose, we applied a modified version of the standard chromatin IP protocol, where formaldehyde-fixed chromatin was digested by extensive micrococcal nuclease treatments and immunoprecipitated DNA was detected by PCR using primers corresponding to the promoter DNA wrapped around the two positioned nucleosomes (Fig. 6A and F [see the legend for details]). As shown in Fig. 6B (left), the presence of Htz1 in nucleosome −2 was detected at comparable levels in wild-type as well as in tup1Δ and cyc8Δ strains grown in glucose. By contrast, the presence of the histone variant in nucleosome −1 was dramatically compromised in either tup1Δ or cyc8Δ strains (Fig. 6B, right), and this did not reflect nucleosome loss since the levels of histone H3 at nucleosome −1 were not affected by the tup1Δ or the cyc8Δ mutation (Fig. 6C, right; see also the left panel for nucleosome −2). In contrast to the tup1Δ and cyc8Δ mutant strains, cells lacking the Swr1 ATPase subunit of SWR1-C (swr1Δ) failed to deposit Htz1 at both −2 and −1 nucleosomes of the GAL1 promoter (Fig. 6D, left and right panels, respectively), while the levels of histone H3 in this mutant background were normal at either nucleosomal position (Fig. 6E). In addition, SWR-C was recruited equally well to both nucleosomes upon repression on GAL1 (Fig. 6G and H). These observations indicated that the Tup1-Cyc8 corepressor complex is specifically required for SWR1-C-dependent Htz1 deposition at the TSS-positioned nucleosome −1 of the GAL1 promoter.
FIG. 6.
Tup1-dependent Htz1 deposition involves only the GAL1-proximal nucleosome. (A) Schematic representation of the GAL1 promoter nucleosomal organization. The positions of the distal (Nuc −2) and proximal (Nuc −1) nucleosomes relative to the UASGAL, TATA element, and TSS are indicated along with MNase-sensitive sites. Arrows within nucleosomes represent the positions of primers used for PCR amplification. (B) Histone Htz1 and histone H3 (C) depositions at the distal (Nuc −2) and proximal (Nuc −1) GAL1 nucleosomes were monitored in glucose grown wild-type (WT), tup1, and cyc8 strains following chromatin IPs and PCR amplification using nucleosome-specific primers. Histone Htz1 (D) and histone H3 (E) deposition at nucleosome −2 and nucleosome −1 was monitored in wild-type and swr1 strains by chromatin IPs and PCR amplification as described above. Numbers indicate immunoprecipitated/input ratios. (F) The extent of MNase digestion in the above-described experiments was confirmed by PCR amplification of MNase-digested (MN+) or untreated (MN−) input DNA obtained from the indicated strains using primers spanning the region of both nucleosome −2 and nucleosome −1. (G) Swr1 recruitment on either nucleosome (nucleosome −2 and nucleosome −1) was monitored by chromatin IP in WT and tup1 strains grown in glucose as described above (B). (H) MNase treatment was confirmed as described above (F).
DISCUSSION
We have attempted to identify elements that determine the deposition of histone Htz1 at the GAL1 promoter. In the course of these studies, it was revealed that the Tup1 corepressor is an important determinant for Htz1 deposition for one specific nucleosome, the promoter-proximal one, and that this epigenetic mark facilitated the recruitment of Mediator upon transcriptional activation. Moreover, we have demonstrated that the Tup1 contribution is evident during the reestablishment of the repressed state when the SWR1-C remodeling complex is transiently recruited to the promoter.
The involvement of Tup1 in Htz1 deposition on GAL1 became evident when decreased levels of Htz1 were measured in a tup1Δ background. Fine mapping of Htz1 deposited along the two promoter nucleosomes demonstrated that Tup1 is actually required for only the promoter-proximal nucleosome (nucleosome −1). By contrast, both proximal (nucleosome −1) and distal (nucleosome −2) nucleosomes depend on SWR1-C for Htz1 deposition. This fact establishes for the first time differences in the dynamics of SWR1-C-mediated Htz1 deposition between adjacent nucleosomes. Noticeably, the distal nucleosome is localized between the UAS and the TATA element, and thus, it does not preclude the binding of both Gal4 and TBP to the promoter, whereas the proximal nucleosome covers the transcription start point, and in principle, it should be removed for transcription initiation to occur (23). It is conceivable that the differential requirement for Htz1 deposition could reflect distinct functions of the two nucleosomes in the repression-activation dynamics, with nucleosome −1 being the target of Tup1-mediated repression.
In agreement with the above-mentioned notion, in yeast cells lacking Htz1, the binding of the Gal4 activator and of TBP as well as the recruitment of the SAGA coactivator complex at GAL1 were at levels that were indistinguishable from those of the wild-type strain. By contrast, recruitment of Mediator was compromised, at least at the early points of galactose induction. Furthermore, Tup1 not only was required for efficient deposition of Htz1 in nucleosome −1 but was also essential for the manifestation of the transcriptional defect in GAL1 induction when Htz1 was absent. It is well established that Tup1 represses transcription through a multitude of mechanisms, one of them being through interference with the function of Mediator, while another is mediated through chromatin (12, 45). We postulate that under repressive conditions, Tup1, on one hand, ensures that Mediator will not be recruited and that the repressive nature of nucleosome −1 will be maintained, and at the same time, it ensures the presence of Htz1 on this nucleosome. This epigenetic marking operates under transcription-inducing conditions and contributes to the rapid neutralization of Tup1-mediated repression, permitting the recruitment of Mediator. It should be noted that Tup1 is continuously bound to the GAL1 promoter, even under activation conditions (31). Tup1 neutralization could be based on the fact that Htz1-bearing nucleosomes facilitate activation through their susceptibility to loss, thereby helping to expose promoter DNA to general transcription factors (44). Based on these data, we propose that Htz1-containing nucleosomes deposited at GAL1 facilitate Mediator recruitment and transcription initiation, possibly through their susceptibility to loss upon transcriptional induction. A functional interplay between Htz1 and Mediator might be more general, since this transcriptional coactivator is associated with a number of environmentally reprogrammed genes (10), most of them highly enriched in Htz1-containing promoter nucleosomes (13, 32, 44).
How does Tup1 specify Htz1 deposition? Clues concerning the answer to this question came by examining the dynamics of the shift from inducing to repressing conditions. First, we observed rapid kinetics for the de novo deposition of Htz1 similar to those recently described for the PHO5 promoter (26). Second, we measured a transient recruitment of the SWR1 complex that correlated with the kinetics of Htz1 deposition. This recruitment was not dependent on the continuously bound Tup1, a fact consistent with the Tup1 independence for Htz1 deposition of nucleosome −2. Actually, the deposition reaction taking place at nucleosome −2 demonstrates that when tethered at GAL1, SWR1-C is competent for histone exchange, and thus, nucleosome −1 cannot serve as a proper substrate for SWR1-C in the absence of the corepressor. Tup1 might be essential for the proper re-formation and positioning of the proximal nucleosome upon glucose repression. However, both the extent of histone H3 deposition, indicative of nucleosome formation, and the steady-state MNase protection experiment indicated that even in the absence of the corepressor, GAL1 promoter nucleosomes are properly positioned at this level of resolution. Alternatively, it could be envisioned that contacts between the proximal GAL1 nucleosome and Tup1, which is known to interact with hypoacetylated H3 and H4 histone tails (7), and/or Tup1-directed histone-modifying activities may be important for the histone exchange reaction. However, in the absence of histone deacetylases such as Hda1, which is important for Tup1-mediated function (43), or even Rpd3, Htz1 was normally deposited at the GAL1 promoter. Finally, the artificial tethering of Tup1-Cyc8 to a test promoter was not sufficient to deposit Htz1 (data not shown). Given the above-described data, we envision that the transcriptionally poised state of GAL1 in a tup1Δ background somehow compromises the function of SWR1-C at nucleosome −1. Whatever the exact mechanisms, our data suggest that gene-specific transcriptional regulators such as Tup1 play essential roles in SWR1-C-mediated Htz1 deposition at promoters.
This mechanism of Htz1 deposition could involve the majority of the Tup1-repressed genes, a hypothesis that is weakly supported by the observed increase in Htz1 deposition in Tup1-unrelated chromatin (PHO5 and GAL1 ORF) in tup1Δ strains. Unfortunately, this supposition cannot be easily tested since, unlike genes belonging to the galactose utilization system, for the majority of Tup1-regulated genes, analysis of promoter chromatin is complicated by their constitutive derepression in tup1Δ strains. We circumvented this problem for the SUC2 promoter by using the swi2 genetic background that not only precludes transcription but, more importantly, also preserves the promoter chromatin structure (11). Although we cannot yet generalize these findings, the requirement of Tup1 for Htz1 deposition for an additional glucose-repressed promoter strongly argues for the uncovering of a more general mechanism.
The primary goal of our studies was to define gene-specific determinants for SWR1-C recruitment and Htz1 deposition. Although we identified Tup1 as being a gene-specific cofactor for such deposition, we failed to identify elements that specify SWR1-C recruitment. The timing of such recruitment may suggest the involvement of specific chromatin rearrangements that occur upon transcriptional repression but not on transcription per se, since Htz1 was also deposited in a transcriptionally inactive TATA-less GAL1 promoter allele (data not shown). Alternatively, one could postulate that SWR1-C has a general and not a directed affinity for specific architectural features of chromatin. These features might be as general as those that differentiate promoter from nonpromoter regions in yeast chromatin (37) or more specific than those specified by Reb1-dA:dT (32). Nevertheless, following its recruitment, the function of the Htz1 exchange complex at specific nucleosomes might then be dictated by promoter-specific factors such as the Tup1 corepressor.
Acknowledgments
We thank Niki Gounalaki for technical support, Manolis Papamichos-Chronakis for materials, and Irene Topalidou for advice and helpful suggestions.
This work was supported by PENED grant 2001/01/ED509 (General Secretary of Research and Technology, Greek Ministry of Development) to G.T.
Footnotes
Published ahead of print on 26 March 2007.
REFERENCES
- 1.Adam, M., F. Robert, M. Larochelle, and L. Gaudreau. 2001. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell. Biol. 21:6270-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 3.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 5.Camilloni, G., F. Della Seta, R. Negri, A. G. Ficca, and E. Di Mauro. 1986. Structure of RNA polymerase II promoters. Conformational alterations and template properties of circularized Saccharomyces cerevisiae GAL1-GAL10 divergent promoters. EMBO J. 5:763-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davie, J. K., D. G. Edmondson, C. B. Coco, and S. Y. Dent. 2003. Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J. Biol. Chem. 278:50158-50162. [DOI] [PubMed] [Google Scholar]
- 7.Davie, J. K., R. J. Trumbly, and S. Y. Dent. 2002. Histone-dependent association of Tup1-Ssn6 with repressed genes in vivo. Mol. Cell. Biol. 22:693-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan, J. Y., F. Gordon, K. Luger, J. C. Hansen, and D. J. Tremethick. 2002. The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat. Struct. Biol. 9:172-176. [DOI] [PubMed] [Google Scholar]
- 10.Fan, X., D. M. Chou, and K. Struhl. 2006. Activator-specific recruitment of Mediator in vivo. Nat. Struct. Mol. Biol. 13:117-120. [DOI] [PubMed] [Google Scholar]
- 11.Gavin, I. M., M. P. Kladde, and R. T. Simpson. 2000. Tup1p represses Mcm1p transcriptional activation and chromatin remodeling of an a-cell-specific gene. EMBO J. 19:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green, S. R., and A. D. Johnson. 2004. Promoter-dependent roles for the Srb10 cyclin-dependent kinase and the Hda1 deacetylase in Tup1-mediated repression in Saccharomyces cerevisiae. Mol. Biol. Cell 15:4191-4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guillemette, B., A. R. Bataille, N. Gevry, M. Adam, M. Blanchette, F. Robert, and L. Gaudreau. 2005. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3:e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han, S. J., J. S. Lee, J. S. Kang, and Y. J. Kim. 2001. Med9/Cse2 and Gal11 modules are required for transcriptional repression of distinct group of genes. J. Biol. Chem. 276:37020-37026. [DOI] [PubMed] [Google Scholar]
- 15.Jackson, J. D., and M. A. Gorovsky. 2000. Histone H2A.Z has a conserved function that is distinct from that of the major H2A sequence variants. Nucleic Acids Res. 28:3811-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keogh, M. C., T. A. Mennella, C. Sawa, S. Berthelet, N. J. Krogan, A. Wolek, V. Podolny, L. R. Carpenter, J. F. Greenblatt, K. Baetz, and S. Buratowski. 2006. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20:660-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 18.Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings, A. J. Link, H. D. Madhani, and J. Rine. 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2:E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan, H. Ding, R. A. Haw, J. Pootoolal, A. Tong, V. Canadien, D. P. Richards, X. Wu, A. Emili, T. R. Hughes, S. Buratowski, and J. F. Greenblatt. 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12:1565-1576. [DOI] [PubMed] [Google Scholar]
- 20.Ladurner, A. G., C. Inouye, R. Jain, and R. Tjian. 2003. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell 11:365-376. [DOI] [PubMed] [Google Scholar]
- 21.Li, B., S. G. Pattenden, D. Lee, J. Gutierrez, J. Chen, C. Seidel, J. Gerton, and J. L. Workman. 2005. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl. Acad. Sci. USA 102:18385-18390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieb, J. D., and N. D. Clarke. 2005. Control of transcription through intragenic patterns of nucleosome composition. Cell 123:1187-1190. [DOI] [PubMed] [Google Scholar]
- 23.Lomvardas, S., and D. Thanos. 2001. Nucleosome sliding via TBP DNA binding in vivo. Cell 106:685-696. [DOI] [PubMed] [Google Scholar]
- 24.Matangkasombut, O., and S. Buratowski. 2003. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell 11:353-363. [DOI] [PubMed] [Google Scholar]
- 25.Meneghini, M. D., M. Wu, and H. D. Madhani. 2003. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112:725-736. [DOI] [PubMed] [Google Scholar]
- 26.Millar, C. B., F. Xu, K. Zhang, and M. Grunstein. 2006. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20:711-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 28.Morrow, B. E., Q. Ju, and J. R. Warner. 1990. Purification and characterization of the yeast rDNA binding protein REB1. J. Biol. Chem. 265:20778-20783. [PubMed] [Google Scholar]
- 29.Papamichos-Chronakis, M., R. S. Conlan, N. Gounalaki, T. Copf, and D. Tzamarias. 2000. Hrs1/Med3 is a Cyc8-Tup1 corepressor target in the RNA polymerase II holoenzyme. J. Biol. Chem. 275:8397-8403. [DOI] [PubMed] [Google Scholar]
- 30.Papamichos-Chronakis, M., T. Gligoris, and D. Tzamarias. 2004. The Snf1 kinase controls glucose repression in yeast by modulating interactions between the Mig1 repressor and the Cyc8-Tup1 co-repressor. EMBO Rep. 5:368-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papamichos-Chronakis, M., T. Petrakis, E. Ktistaki, I. Topalidou, and D. Tzamarias. 2002. Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol. Cell 9:1297-1305. [DOI] [PubMed] [Google Scholar]
- 32.Raisner, R. M., P. D. Hartley, M. D. Meneghini, M. Z. Bao, C. L. Liu, S. L. Schreiber, O. J. Rando, and H. D. Madhani. 2005. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123:233-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rangasamy, D., L. Berven, P. Ridgway, and D. J. Tremethick. 2003. Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J. 22:1599-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 35.Santisteban, M. S., T. Kalashnikova, and M. M. Smith. 2000. Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103:411-422. [DOI] [PubMed] [Google Scholar]
- 36.Schwabish, M. A., and K. Struhl. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24:10111-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sekinger, E. A., Z. Moqtaderi, and K. Struhl. 2005. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell 18:735-748. [DOI] [PubMed] [Google Scholar]
- 38.Suto, R. K., M. J. Clarkson, D. J. Tremethick, and K. Luger. 2000. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 7:1121-1124. [DOI] [PubMed] [Google Scholar]
- 39.Syntichaki, P., I. Topalidou, and G. Thireos. 2000. The Gcn5 bromodomain co-ordinates nucleosome remodelling. Nature 404:414-417. [DOI] [PubMed] [Google Scholar]
- 40.Topalidou, I., M. Papamichos-Chronakis, and G. Thireos. 2003. Post-TATA binding protein recruitment clearance of Gcn5-dependent histone acetylation within promoter nucleosomes. Mol. Cell. Biol. 23:7809-7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Topalidou, I., M. Papamichos-Chronakis, G. Thireos, and D. Tzamarias. 2004. Spt3 and Mot1 cooperate in nucleosome remodeling independently of TBP recruitment. EMBO J. 23:1943-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tzamarias, D., and K. Struhl. 1994. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature 369:758-761. [DOI] [PubMed] [Google Scholar]
- 43.Wu, J., N. Suka, M. Carlson, and M. Grunstein. 2001. TUP1 utilizes histone H3/H2B-specific HDA1 deacetylase to repress gene activity in yeast. Mol. Cell 7:117-126. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, H., D. N. Roberts, and B. R. Cairns. 2005. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123:219-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang, Z., and J. C. Reese. 2004. Redundant mechanisms are used by Ssn6-Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J. Biol. Chem. 279:39240-39250. [DOI] [PubMed] [Google Scholar]