Abstract
Recent data reveal that a substantial fraction of transcripts generated by RNA polymerases I, II, and III are rapidly degraded in the nucleus by the combined action of the exosome and a noncanonical poly(A) polymerase activity. This work identifies a domain within the yeast nucleolus that is enriched in polyadenylated RNAs in the absence of the nuclear exosome RNase Rrp6 or the exosome cofactor Mtr4. In normal yeast cells, poly(A)+ RNA was undetectable in the nucleolus but the depletion of either Rrp6 or Mtr4 led to the accumulation of polyadenylated RNAs in a discrete subnucleolar region. This nucleolar poly(A) domain is enriched for the U14 snoRNA and the snoRNP protein Nop1 but is distinct from the nucleolar body that functions in snoRNA maturation. In strains lacking both Rrp6 and the poly(A) polymerase Trf4, the accumulation of poly(A)+ RNA was suppressed, suggesting the involvement of the Trf4-Air1/2-Mtr4 polyadenylation (TRAMP) complex. The accumulation of polyadenylated snoRNAs in a discrete nucleolar domain may promote their recognition as substrates for the exosome.
A feature of RNAs in all organisms is that they are transcribed as precursors that undergo subsequent processing events to generate the mature functional forms. This was first found for the abundant cytoplasmic rRNAs, mRNAs, and tRNAs and was subsequently shown for the small nuclear RNAs (snRNAs), which participate in pre-mRNA splicing, and the small nucleolar RNAs (snoRNAs), which participate in rRNA processing and modification. Common steps in RNA maturation include endonuclease and exonuclease digestion, splicing, nucleotide modification, 3′ poly(A) tail addition, and 5′ capping. A major RNA processing activity is the multisubunit exosome complex (for reviews, see references 5, 11, and 34). The exosome contains multiple exoribonucleases that progressively digest RNAs from the 3′ end. The exosome functions in both the accurate processing of some RNA species and the complete degradation of defective RNAs (35).
The exosome was initially identified and characterized in the budding yeast Saccharomyces cerevisiae but is conserved in all eukaryotes analyzed, and a closely related complex is found in Archaea (20). Homologues of the yeast exosome proteins are present in humans (19), and at least four of them are functional when expressed in yeast, complementing mutations in the corresponding yeast genes (24). The yeast exosome is present in the nucleus, nucleolus, and cytoplasm and is comprised of 10 “core” subunits, all of which are essential for viability (reviewed in reference 11). An additional exonuclease, Rrp6, associates exclusively with the nuclear exosome in yeast (2, 4). The exosome complex purified from yeast lysates exhibited little activity (23), suggesting that its activation depends on additional factors that are not permanently associated with the complex. Consistent with this model, the exosome cofactor Mtr4 (a putative RNA helicase) was shown to associate with a poly(A) polymerase, Trf4, and one of two redundant zinc knuckle proteins, Air1/2, forming a novel nuclear Trf4-Air1/2-Mtr4 polyadenylation (TRAMP) complex that can activate the exonuclease activities (13, 18, 31, 38).
The addition of poly(A) tails appears to be a general mechanism to target transcripts made by all three nuclear RNA polymerases for exosome-dependent degradation (38). Previous reports indicate that yeast strains lacking Rrp6 accumulate polyadenylated precursors to both snoRNAs (1, 22, 33) and rRNA (8, 12, 17). These polyadenylated species are thought to generally represent nonfunctional transcripts that have been targeted for degradation. In addition, other polyadenylated transcripts of unknown function, which are associated with promoter regions or result from intergenic transcription, accumulate in strains lacking Rrp6 (6, 38).
In the present work, we analyze the subcellular distribution of polyadenylated RNAs in yeast strains lacking Rrp6 and we report the presence of a discrete compartment within the nucleolus that accumulates poly(A)+ RNA and U14 snoRNA. This type of compartmentalization may contribute to promoting the recognition of nucleolar polyadenylated RNAs as substrates for the exosome.
MATERIALS AND METHODS
Strains and plasmids.
The strains of S. cerevisiae used in this study are listed in Table 1. All strains were routinely grown to mid-log phase in yeast extract-peptone-dextrose (YPD) medium containing either 2% glucose or 2% galactose. Plasmids were introduced by the standard polyethylene glycol-lithium procedure, as described previously (29). Transformed strains were grown in minimal YNB medium containing 0.67% yeast nitrogen base supplemented with the appropriate amino acids and 2% glucose. The following expression plasmids were used in this study: Nop1-green fluorescent protein (GFP) (30), Tgs1-GFP (25), Nup49-GFP (3), and GFP-Rrp43 (39).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| D271 | MATaade2-1 his3 leu2 trp1 ura3 | 35a |
| YCBA63 | MATα, like D271 but HIS3-GAL::protA-RRP41 | 29 |
| YCBA81 | Like D271 but HIS5sp-GAL-3HA-DOB1 | 29 |
| YCA12 | MATaade2-1 his3Δ200 leu2-3,112 trp1-1 ura3-1 can1-100 Kl TRP1::rrp6 | 2 |
| D162 | MATα ura3-52 leu2Δ1 his3Δ200 rat1.1 | 2a |
| pap1-5 | MATaade2 his3 trp1 ura3 leu2 LEU2::pap1 (ppap1-5) | 21 |
| YCA42 | Like pap1-5 strain but rrp6Δ::URA3Kl | D. Tollervey, unpublished strain |
| YCT56 | Like pap1-5 strain but HIS3-GAL10::protA-RRP41 | D. Tollervey, unpublished strain |
| trf4Δ | MATahis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 trf4Δ::kanMX | 12 |
| rrp6Δ trf4Δ | MATahis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 trf4Δ::kanMX rrp6Δ::natMX | 12 |
RNA extraction and Northern blotting.
Yeast RNA extraction, Northern blotting, and deadenylation analysis were performed as described previously (18, 22). The sequences of oligonucleotides used as probes were previously reported.
Fluorescence in situ hybridization.
Yeast cells were processed for fluorescence in situ hybridization (FISH) as described previously (36, 32), except that a treatment with 0.1% sodium borohydride for 10 min was introduced after rehydration but before hybridization. Hybridization stringency conditions were 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 10% formamide for poly(A)+ RNA and U3 snoRNA and 2× SSC and 50% formamide for actin mRNA and U14 snoRNA. As a control, cells were treated with 1.5 U RNase A (Roche) in 2× SSC at 37°C for 60 min prior to the in situ hybridization with each probe. When needed, cells were hybridized with the oligo(dT) probe, fixed in 4% formaldehyde/2× SSC for 5 min, washed two times for 10 min in 2× SSC, and then hybridized with another probe. Samples were mounted in 90% glycerol and 1 mg/ml p-phenylenediamine in phosphate-buffered saline (PBS).
Poly(A)+ RNA was detected by using a Cy3-labeled oligo(dT)50 probe (MWG-Biotech AG). ACT1 mRNA was detected with a mixture of oligonucleotides (MWG-Biotech AG) with the sequences 5′ CRTTCTGGAGGAGCRATGATCTTGACRTTCATGGAAGRTGGAGCCAAAGRG 3′, 5′ CRTGGGAACATGGTGGTACCACCGGACATAACGATGTTACCGTATAATTRC 3′, 5′ CRGAAGAGTACARGGACAAAACGGCRTGGATGGAARCGTAGAAGGCTGRA 3′, and 5′ ARCCATACCGACCRTGATACCTTGGRGTCTTGGTCTRCCGACGATAGARG 3′.
These probes were synthesized with modified nucleotides to incorporate primary amines (amino-allyl timine is represented by R) and then conjugated with Cy3. The U3 probe was described previously (37). The probe to detect mature U14 snoRNA was in vitro transcribed in the presence of digoxigenin-UTP. The following oligonucleotide was synthesized to detect 3′-extended U14: 5AAA GTA AAC ACA 5GA TAC TAC AGT 5TA CGA TCA CTC AG5 CAT CCT AGG AA5 (“5” represents dT-digoxigenin); the underlined portion of the sequence is complementary to the 3′ end of mature U14 snoRNA, and the remaining portion of the sequence is specific for 3′-extended forms.
After hybridization with digoxigenin-conjugated probes, cells were treated with 4% formaldehyde/2× SSC for 5 min and washed first in 2× SSC and then in PBS-0.1% Triton X-100. Subsequently, cells were incubated for 10 min in PBS containing 1% skim milk and 1% RNase-free bovine serum albumin. Cells were then incubated for 1 h with anti-digoxigenin antibody conjugated to fluorescein isothiocyanate (Jackson ImmunoResearch Laboratories), followed by anti-fluorescein isothiocyanate antibody conjugated to Alexa-488 (Molecular Probes).
Fluorescence microscopy.
Images were acquired with a Zeiss LSM 510 confocal microscope (Carl Zeiss, Jena, Germany) using either the Plan Neofluar 100×/1.3 or the Plan Apochromat 63×/1.4 objective. Green fluorescence was detected by using the 488-nm laser line of an Ar laser (25-mW nominal output) and an LP 505 filter. Red fluorescence was detected by using a 543-nm HeNe laser (1 mW) and an LP 560 filter. Alternatively, yeast cells were imaged on a Zeiss Axiovert 200 microscope equipped with a CoolSNAP HQ charge-coupled device (Roper Scientific Photometrics, Tucson, AZ).
FRAP analysis.
To prepare yeast cells for fluorescence recovery after photobleaching (FRAP) experiments, 1 ml of each cell culture grown to mid-log phase was harvested and resuspended in 100 μl of medium; 5 μl of this cell suspension was added to a coverslip coated with 40% gelatin in medium. Live cells were imaged at 23°C, 30°C, or 37°C maintained by a heating/cooling frame (LaCon, Staig, Germany) in conjunction with an objective heater (PeCon, Erbach, Germany). Each FRAP analysis started with three image scans, followed by a single bleach pulse of approximately 70 ms on a defined region. A series of 97 single-section images were then collected at 155-ms intervals, with the first image acquired 2 ms after the end of bleaching. The image size was 512 by 100 pixels, and the pixel width was 14.3 nm. For imaging, the laser power was attenuated to 1% of the bleach intensity. The pinhole was set to 1 airy unit. For each time series, normalization was carried out by measuring the fluorescence intensity in the bleached, unbleached, and background areas. The fluorescence intensity measured in the bleached area during recovery was divided by the initial value of the fluorescence in that same area: R1 = FB(t)/FB(ini), where FB(t) is the fluorescence in the bleached area at time t, and FB(ini) is the initial fluorescence in that same area. Each time an image is acquired, some fluorescence is lost due to the laser scan; to compensate for this loss, each R1 value was multiplied by the following factor A: A = FT(AB)/FT(t), where FT(AB) is the total postbleach nucleolar fluorescence (bleached and nonbleached areas) and FT(t) is the fluorescence in the bleach and nonbleached area at time t. Since a global loss of fluorescence was also observed after the bleach, we had to compensate by multiplying the R1 value by factor B: B = FB(ini)/[FB(final) × FT(AB)/FT(final)], where FB(final) is the fluorescence in the bleached area in the last image acquired and FT(final) is the total nucleolar fluorescence (bleached and nonbleached areas) in the last image acquired. Finally, in order to estimate whether a fraction of the molecules in the bleached area was immobilized, we calculated the factor C, which yields values lower than 1 in the presence of an immobile fraction: C = [FB(final)/FNB(final)]/[FB(ini)/FNB(ini)], where FB(ini) and FB(final) represent the fluorescence in the bleached area in the first and last images acquired, respectively, and FNB(ini) and FNB(final) are the fluorescence values in the nonbleached area in the first and last images acquired, respectively. The final formula, which is the product of R1, A, B, and C, represents the normalized fluorescence recovery in the bleached area over time: R = [FB(t) × FT(final) × FNB(ini)]/[FT(t) × FNB(final) × FB(ini)].
RESULTS
Subcellular distribution of poly(A)+ RNA in yeast cells lacking Rrp6.
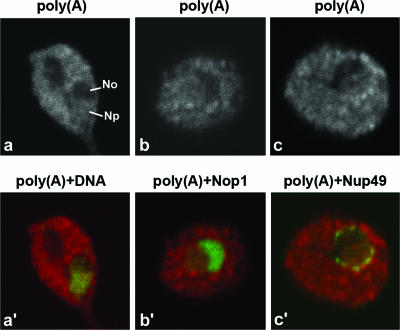
The distribution of polyadenylated RNAs was initially visualized in wild-type yeast cells by FISH using a Cy3-labeled oligo(dT)50 probe. Following FISH, the cells were incubated with the DNA stain DAPI (4′,6′-diamidino-2-phenylindole) to identify the nucleoplasm (Fig. 1a and a′). Additionally, FISH was performed in cells also expressing the nucleolar marker Nop1-GFP (Fig. 1b and b′) or the nuclear pore component Nup49-GFP to visualize the nuclear rim (Fig. 1c and c′). Poly(A)+ RNA was detected predominantly in the cytoplasm, with less intense staining in the nucleoplasm. A region of higher poly(A) staining intensity was often visible around the nuclear rim (Fig. 1c and c′), whereas no labeling was detected in the region labeled by Nop1-GFP (Fig. 1b and b′). Thus, poly(A)+ RNA is detected in the nucleoplasm but not the nucleolus in wild-type cells growing at 23°C.
FIG. 1.
Poly(A)+ RNA is normally not detected in the nucleolus. (a, b, and c) Wild-type yeast cells were grown at 23°C in YPD medium to mid-log phase. Poly(A)+ RNA was detected by FISH using a Cy3-labeled oligo(dT)50 probe. Cells were further labeled with the DNA stain DAPI [in panel a′, poly(A)+ RNA is visualized in red and DNA is visualized in green]. Alternatively, FISH was performed in cells expressing Nop1-GFP [in panel b′, poly(A)+ RNA is visualized in red and GFP is visualized in green] or Nup49-GFP [in panel c′, poly(A)+ RNA is visualized in red and GFP is visualized in green]. Np, nucleoplasm; No, nucleolus.
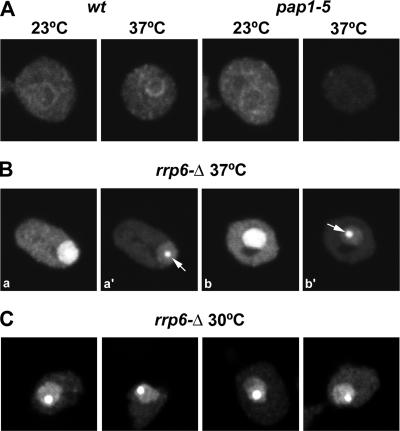
Eukaryotic mRNAs have characteristic 3′ poly(A) tails that are synthesized by the poly(A) polymerase Pap1 in yeast (26). In wild-type cells, the distribution of poly(A)+ RNA remains similar after incubation at 37°C (Fig. 2A). In contrast, strains carrying the temperature-sensitive lethal pap1-5 mutation (21) showed a drastic reduction in the poly(A) signal at the nonpermissive temperature (37°C). This result suggests that the poly(A)+ RNA detected in both the cytoplasm and nucleoplasm of wild-type cells corresponds predominantly to mRNAs.
FIG. 2.
Poly(A)+ RNA accumulates in a discrete nuclear focus in the rrp6Δ mutant. (A to C) Poly(A)+ RNA was detected by FISH in cells that were either grown at 23°C and incubated for 1 h at 37°C or grown at 30°C, as indicated. For a comparison of fluorescence signals, the images depicted in panel B, parts a, and b, were acquired with the same settings as those used for panel A. Panels B, parts a and a′, and B, parts b and b′, show different exposures of the same cell. Arrows point to the poly(A) domain. wt, wild type.
We next analyzed the localization of poly(A)+ RNA in a strain lacking the nuclear exosome component Rrp6, which is viable but lethally temperature sensitive. The basis of the temperature-sensitive lethality of the cells lacking Rrp6 (rrp6Δ mutation) has long been unclear, since the phenotypes in RNA maturation appeared nonconditional, with almost identical processing defects seen during growth at 23°C and at 30°C and following transfer to 37°C.
The rrp6Δ strain showed a striking accumulation of nuclear poly(A)+ RNA 1 h after transfer to 37°C (Fig. 2B). To allow the direct comparison of fluorescence intensities, images from the wild-type (Fig. 2A) and rrp6Δ mutant (Fig. 2B, panels a and b) were acquired by using the same microscope settings. The abundant accumulation of nuclear poly(A)+ RNA in the rrp6Δ strain saturates the image (Fig. 2B, panels a and b). Following the adjustment of the acquisition settings, we observed that the poly(A)+ RNA was not homogeneously accumulated throughout the nucleus but was concentrated in a discrete region that we shall refer to as the poly(A) domain (Fig. 2B, panels a′ and b′). At 30°C, rrp6Δ strains are viable but impaired in growth and, as shown in Fig. 2C, the poly(A) domain was also present under these conditions.
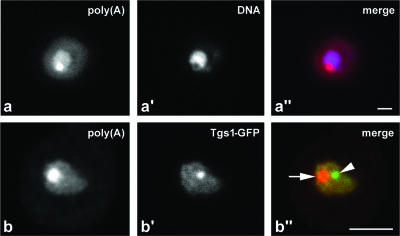
Comparison to DAPI staining (Fig. 3a to a″) showed that the nuclear poly(A) domain is excluded from the nucleoplasm, suggesting a nucleolar localization. To better characterize this localization, the rrp6Δ strain was transformed with a plasmid expressing Tgs1-GFP. Tgs1 is an RNA methyltransferase that converts the monomethyl cap (m7G) present on snRNA and snoRNA precursors into the 2,2,7-trimethylguanosine (m3G) cap structure that is a characteristic feature of the mature RNAs (25). In wild-type cells, Tgs1-GFP is detected throughout the nucleolus but concentrated in the “nucleolar body” (NB). This NB is a structure implicated in snoRNA maturation, which may be related to the mammalian Cajal body (36, 37). In the rrp6Δ strain incubated at 37°C, Tgs1-GFP was localized to the nucleolus and NB (Fig. 3b′) but did not colocalize with the poly(A) domain (Fig. 3b and b″). We conclude that the absence of Rrp6 leads to the accumulation of polyadenylated RNA in a discrete domain within the nucleolus, which is distinct from the NB.
FIG. 3.
The poly(A) domain localizes in the nucleolus and is distinct from the NB. Cells lacking Rrp6, rrp6Δ cells, were grown at 23°C and incubated for 1 h at 37°C. Poly(A)+ RNA was detected by FISH. Panels a to a″ depict cells double labeled with the DNA stain DAPI; in panel a″, poly(A)+ RNA is visualized in red and DNA is visualized in blue. Panels b to b″ show cells expressing Tgs1-GFP; in panel b″, poly(A) is visualized in red and Tgs1-GFP is visualized in green. The arrow points to the poly(A) domain, and the arrowhead points to the NB. Bar, 2 μm.
The nucleolar poly(A) domain accumulates Nop1 and U14 snoRNA.
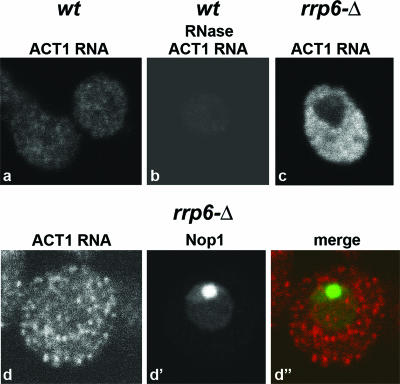
Previous studies suggested that mRNAs may transit through the nucleolus in yeast (reviewed in reference 28), and we therefore assessed whether the poly(A) domain observed in rrp6Δ cells contains mRNA. FISH was performed with Cy3-labeled probes complementary to exonic sequences of the ACT1 (actin) mRNA. As shown in Fig. 4, staining was detected predominantly in the cytoplasm, with no nuclear or nucleolar accumulation in the rrp6Δ strain. This suggests that RNAs other than mRNAs are present in the nucleolar poly(A) domain.
FIG. 4.
Localization of actin mRNA (ACT1 RNA) in wild-type and rrp6Δ yeast strains. Cells were grown at 23°C and incubated for 1 h at 37°C. FISH was performed using oligonucleotide probes complementary to the coding sequence of actin mRNA. For a direct comparison of staining intensities, images were acquired with the same confocal settings. wt, wild type.
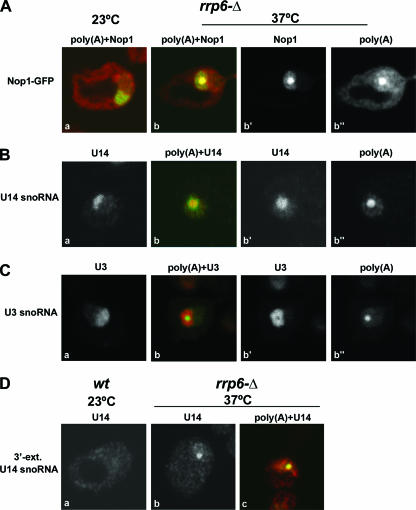
Accumulation of polyadenylated precursors to box C/D snoRNAs was previously observed in strains lacking Rrp6 (1, 18, 22, 33, 38), suggesting that these might contribute to the nucleolar poly(A) signal. All box C/D snoRNAs associate with Nop1/fibrillarin in snoRNP complexes (reviewed in reference 9), and we therefore analyzed the distribution of a Nop1-GFP fusion. In wild-type cells, Nop1-GFP distributes uniformly throughout the nucleolus (37) and a similar distribution was observed in rrp6Δ cells grown at 23°C (Fig. 5A, panel a). However, following the transfer of the rrp6Δ strain to 37°C for 1 h, Nop1-GFP became concentrated in a subnucleolar region that precisely coincided with the oligo(dT) staining (Fig. 5A, panels b to b″). The localization of Nop1 supported the model that the poly(A)+ RNAs accumulated in the nucleolus of rrp6Δ cells include polyadenylated snoRNAs. We therefore analyzed the localization of the box C/D snoRNAs U14 and U3. Previous studies revealed that polyadenylated U14 snoRNA accumulates in the rrp6Δ strain, whereas the polyadenylation of U3 was not observed (1). FISH analysis of rrp6Δ cells growing at 23°C with probes targeting either U14 (Fig. 5B, panel a) or U3 (Fig. 5C, panel a) snoRNAs revealed uniform staining of the nucleolus. In rrp6Δ cells incubated at 37°C the U14 signal was enriched in a focus that colocalized with the poly(A) domain (Fig. 5B, panels b to b″). In contrast, the U3 signal remained distributed throughout the nucleolus and was apparently excluded from the poly(A) domain (Fig. 5C, panels b to b″). As strains lacking Rrp6 were previously shown to accumulate 3′-extended box C/D snoRNA species (1, 18, 22, 33, 38), we performed FISH with an oligonucleotide probe that detects 3′-extended forms of U14 (Fig. 5D). The probe contains 18 nucleotides complementary to the 3′ end of mature U14 and 29 nucleotides complementary to 3′-extended intermediates. This probe produced no detectable signal in wild-type cells (Fig. 5D, panel a), probably because the snoRNA precursors undergo very rapid maturation. However, a nucleolar focus was clearly visible in the rrp6Δ mutant incubated at 37°C (Fig. 5D, panels b and c).
FIG. 5.
The nucleolar poly(A) domain accumulates Nop1 and 3′-extended U14 snoRNA. Cells lacking Rrp6, rrp6Δ cells, were grown at 23°C and incubated for 1 h at 37°C (A to C). Panels b to b″ show the same cell. (A) Detection of poly(A)+ RNA in cells expressing Nop1-GFP [poly(A)+ RNA is visualized in red, and GFP is visualized in green]. (B) Double labeling for poly(A)+ RNA and U14 snoRNA [poly(A)+ RNA is visualized in red, and U14 is visualized in green]. (C) Double labeling for poly(A)+ RNA and U3 snoRNA [poly(A)+ RNA is visualized in green, and U3 is visualized in red]. (D) FISH was performed using a probe for 3′-extended U14 snoRNA. Wild-type (wt) and rrp6Δ strains were grown at 23°C and incubated for 1 h at 37°C. Note that panels a, b, and c show distinct cells; the cell in panel c was double labeled for poly(A)+ RNA and 3′-extended (ext.) U14 snoRNA [poly(A)+ RNA is visualized in red, and U14 is visualized in green].
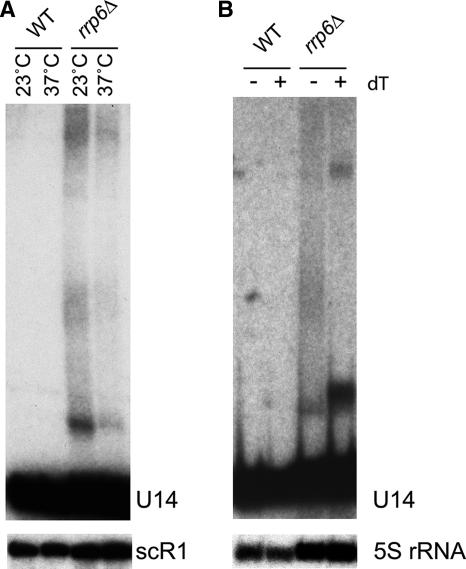
Northern blot analysis (Fig. 6) confirmed that 3′-extended and polyadenylated U14 species are present in the rrp6Δ strain used for these analyses. In the rrp6Δ strain, but not in the wild type, long heterogeneous forms of U14 were readily detected (Fig. 6A) both at 23°C and following transfer to 37°C for 1 h. Treatment of the RNAs extracted after 1 h at 37°C with RNase H and oligo(dT) to remove poly(A) tracts collapsed the 3′-extended U14 population into more discrete bands (Fig. 6B), showing that the heterogeneity largely arose from a distribution of poly(A) tail lengths. Since the 3′-extended forms of U14 in the rrp6Δ strain are predominantly polyadenylated, we conclude that the 3′-extended forms of U14 detected in the nucleolar poly(A) domain by in situ hybridization are also polyadenylated.
FIG. 6.
The U14 snoRNA is 3′ extended and polyadenylated in the rrp6Δ strain at the nonpermissive temperature. Total RNA was extracted from the wild type (WT) and rrp6Δ strains and resolved in 8% acrylamide-urea gels. (A) Northern hybridization with probes for U14 snoRNA (oligonucleotide 202) and scR1 (18, 22). Cells were grown at 23°C and incubated for 1 h at 37°C, as indicated. (B) Total RNA recovered from cells incubated for 1 h at 37°C was deadenylated by treatment with RNase H and oligo(dT) prior to gel separation. Samples were hybridized with probes for U14 snoRNA (oligonucleotide 202) and 5S rRNA (oligonucleotide sequence CTACTCGGTCAGGCTC). −, absence of; +, presence of.
Taken together, these results suggest that the nucleolar poly(A) domain represents a compartment within the nucleolus that is enriched in polyadenylated RNAs, including precursors to the U14 box C/D snoRNA. However, the lack of a clear temperature effect in the Northern analyses shows that the appearance of the focus is not due to the accumulation of polyadenylated U14 only at 37°C.
Formation of the nucleolar poly(A) domain requires both Trf4 and Pap1.
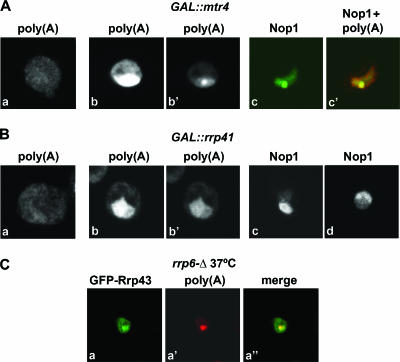
The yeast TRAMP complex contains a poly(A) polymerase, either Trf4 or Trf5, a zinc knuckle protein, either Air1 or Air2, and the RNA helicase Mtr4/Dob1 (18, 31, 38). In vivo, this complex is required for the polyadenylation and degradation of exosome substrates, including rRNA and snoRNA precursors (13, 18, 31, 38). The genetic depletion of Mtr4 leads to the hyperadenylation of several pre-RNA species, probably because polyadenylation has been uncoupled from degradation by the exosome (12). To determine whether Mtr4 is required for the localization of RNAs to the nucleolar poly(A) domain, we analyzed a strain in which Mtr4 was under GAL regulation. Cells depleted of Mtr4 by growth on glucose medium showed strong nuclear accumulation of poly(A)+ RNA (Fig. 7A, compare panels a and b). After a shorter exposure, the poly(A) signal appears concentrated in a focus (Fig. 7A, panel b′) that localizes to the nucleolus and is highly enriched in Nop1 (Fig. 7A, panels c and c′). Consistent with these observations, mtr4 was originally isolated as a temperature-sensitive mutant, which accumulated poly(A)+ RNA as a single dot in the nucleus at 37°C (14).
FIG. 7.
Depletion of Mtrf4 induces formation of the poly(A) domain. (A to B) The GAL::mtr4 and GAL::rrp41 strains were grown in YPD-2% galactose at 23°C and then transferred to glucose medium at 23°C for 20 h. Poly(A)+ RNA was detected by FISH, and Nop1 was detected by immunofluorescence. Panels A, part a, and B, part a, show cells grown in galactose medium. All other panels depict cells incubated in glucose medium. Panels A, parts b and b′, and B, parts b and b′, show different exposures of the same cell. Panel B, part d, shows cells that were grown in YPD-2% galactose at 23°C, transferred to glucose medium at 23°C for 20 h, and further incubated at 37°C for 1 h. (C) A GFP-tagged version of exosome component Rrp43 was expressed in the rrp6Δ strain. The resulting cells were grown at 23°C and shifted to 37°C for 1 h. Poly(A)+ RNA was detected by FISH.
For comparison, we analyzed a strain in which the core exosome component Rrp41 was under GAL regulation. Cells depleted of Rrp41 by growth on glucose medium also showed strong nuclear accumulation of poly(A)+ RNA (Fig. 7B, compare panels a and b), but the poly(A)+ staining failed to form any clear focus (Fig. 7B, panel b′). Nop1 remained uniformly distributed throughout the nucleolus following the depletion of Rrp41 (Fig. 7B, panel c). The same result was obtained after incubation at 37°C for 1 h (Fig. 7B, panel d), showing that the formation of the poly(A) domain is not induced simply by the incubation of cells at 37°C. Thus, in contrast to the depletion of Rrp6 and Mtr4, the depletion of Rrp41 does not induce the formation of the nucleolar poly(A) domain.
Taken together, the data suggest that polyadenylated RNAs accumulate at a subnucleolar structure prior to degradation by the nuclear exosome and we speculated that this structure might also be the site of exosome activity. We therefore assessed whether the poly(A) domain also contained components of the exosome. A GFP-tagged version of exosome component Rrp43 was expressed in the rrp6Δ strain, and the resulting cells were grown at 23°C and shifted to 37°C for 1 h (Fig. 7C). GFP-Rrp43 was distributed throughout the nucleus, but in a fraction (∼50%) of cells, the protein was enriched in a focus that colocalized with the poly(A) domain (Fig. 7C).
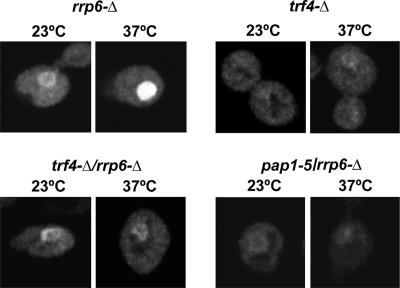
To investigate the poly(A) polymerase(s) responsible for the polyadenylation of the RNA present in the nucleolar poly(A) domain, we analyzed rrp6Δ mutants that lacked Trf4 or that carried a point mutation in the canonical poly(A) polymerase Pap1 (Fig. 8). In the rrp6Δ trf4Δ strain, the poly(A)+ RNA staining intensity was lower than that in the rrp6Δ control 1 h after transfer to 37°C. Some nuclear poly(A)+ RNA was accumulated but failed to form a discrete focus. This result indicates that Trf4 plays a major role in generating the polyadenylated RNA. In the rrp6Δ pap1-5 (21) strain grown at 23°C, the poly(A)+ RNA staining intensity was lower than that in the rrp6Δ control (Fig. 8) and, 1 h after transfer to 37°C, only a low level of nuclear poly(A) staining was detected. This result suggests that Pap1 is also required for the polyadenylation of the nucleolar-localized RNAs in the rrp6Δ strain at 37°C. However, Pap1 is involved in the synthesis of many or all mRNAs and we cannot exclude the possibility that its inactivation results in indirect effects, conceivably reducing the abundance of the mRNAs encoding Trf4 or other factors involved in polyadenylation.
FIG. 8.
Trf4 and Pap1 are required for formation of the poly(A) domain in the rrp6Δ mutant. All strains were grown at 23°C in YPD medium to early/mid-log phase. Poly(A)+ RNA was detected by FISH in cells that were either grown at 23°C or incubated for 1 h at 37°C, as indicated.
Nop1-GFP dynamics are similar in the nucleolus and poly(A) focus.
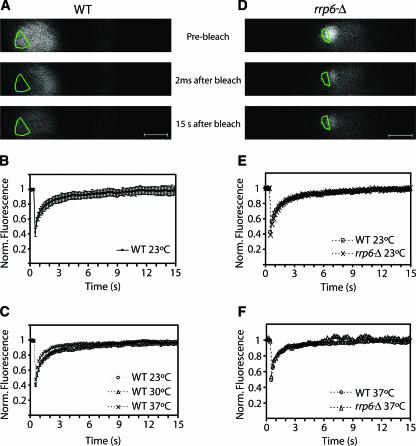
To assess the stability of the poly(A) domain, we compared the dynamics of Nop1-GFP in the nucleolus and poly(A) domain by FRAP in wild-type and rrp6Δ strains (Fig. 9). In a FRAP experiment, the GFP signal is irreversibly bleached in a defined target region by a short, high-intensity laser pulse. Subsequent diffusion of surrounding nonbleached fluorescent molecules into the bleached area leads to a recovery of the signal, which is recorded by time-lapse confocal microscopy. In wild-type cells, Nop1-GFP appeared homogeneously distributed throughout the typical crescent-shaped nucleolus. Photobleaching was performed over a region corresponding to approximately one-third of the nucleolus (Fig. 9A). Complete fluorescence recovery was observed within 15 s after the bleaching, irrespective of whether the cells were incubated at 23, 30, or 37°C (Fig. 9B and C). Thus, Nop1-GFP rapidly associates and dissociates with the yeast nucleolus, as previously described for its mammalian counterpart fibrillarin (27).
FIG. 9.
Dynamics of Nop1-GFP in the nucleolus and in the poly(A) domain. Wild-type and rrp6Δ yeast cells expressing Nop1-GFP were grown at 23°C and incubated for 1 h at 30 or 37°C. During the FRAP experiments, the cells were kept in the microscope stage at 23, 30, or 37°C. A sequence of 100 images was taken at 6.4 frames per second. Shown are images taken before bleaching, immediately after bleaching, and after recovery for 15 s (A and D). The bleached region is outlined. This region comprised either about one-third of the nucleolus or about one-half of the nucleolar poly(A) domain. The fluorescence intensity in the bleached region was measured and expressed as the normalized fluorescence R. The recovery curves correspond to a pool of three independent experiments, with at least 10 different cells analyzed per experiment. Error bars represent standard deviations. Scale bar = 1 μm. (B) Fluorescence recovery for wild-type cells at 23°C. (C) Comparison of fluorescence recovery rates of wild-type cells at 23, 30, and 37°C. (E and F) Comparison of fluorescence recovery rates in wild-type and rrp6Δ cells incubated at 23°C and 37°C, respectively.
At the permissive temperature (23°C), the rrp6Δ strain showed Nop1-GFP distribution and recovery kinetics that were similar to those of the wild type (Fig. 9E). One hour after transfer to 37°C, Nop1-GFP was accumulated within the nucleolar poly(A) domain. A region encompassing approximately half of this domain was bleached, and fluorescence recovery was monitored (Fig. 9D). The kinetics were similar to those observed in the nucleoli of wild-type cells incubated at 37°C (Fig. 9F). We conclude that although Nop1 is enriched in an apparently discrete domain, the protein is rapidly and continuously exchanged between this compartment and the surrounding nucleolus. Moreover, the mobility rates of Nop1 are similar whether it roams through the poly(A) domain or through the nucleolus.
DISCUSSION
The snoRNAs represent an abundant group of non-protein-encoding RNAs that function in the biogenesis of rRNA and snRNAs (reviewed in reference 15). As most classes of cellular RNAs, mature snoRNAs are generated by a complex processing pathway. In strains that are defective for the exosome, 3′-extended forms of many snoRNAs are detected, although it is currently unclear to what extent these represent maturation intermediates or RNAs that have been identified by surveillance activities and targeted for degradation by the exosome (1, 18, 34). Many of these extended snoRNAs are polyadenylated, suggesting that the addition of the poly(A) tails would normally promote their degradation (18, 31, 38). This implies that a significant fraction of the snoRNA precursor population may normally be degraded by the exosome. Here we report that in yeast cells lacking the exosome-associated exonuclease Rrp6, polyadenylated RNAs, including 3′-extended and polyadenylated forms of the U14 snoRNA, accumulate in a discrete compartment within the nucleolus.
Because most eukaryotic mRNAs have a 3′ poly(A) tail, the detection of poly(A)+ RNA by in situ hybridization with oligo(dT) probes has generally been considered to reflect the distribution of cellular mRNA. Our data for the wild-type yeast strains were indeed consistent with this interpretation. Poly(A) staining was detected predominantly in the cytoplasm and was greatly reduced in a strain carrying a mutation in Pap1, the polymerase responsible for the synthesis of poly(A) tails on nuclear pre-mRNAs (21). Poly(A) staining was also observed in the nucleoplasm, albeit at lower levels than in the cytoplasm, but the staining was not detected in the nucleolus (Fig. 1). This indicates that polyadenylated mRNAs are detectable in the nucleoplasm, whereas the nucleolus contains little polyadenylated RNA in a wild-type cell. We observed a region of higher poly(A) staining intensity in close proximity to the nuclear rim, which was identified by a GFP-tagged nucleoporin, possibly reflecting a local concentration of mRNAs in association with the nucleocytoplasmic transport machinery.
A substantially different pattern of poly(A) staining was observed in a strain lacking Rrp6. The poly(A)+ RNA signal was greatly increased in the nucleus (Fig. 2) and was particularly enriched in a subnucleolar focus, which we refer to as the nucleolar poly(A) domain (Fig. 3). Microarray analyses have not revealed dramatic increases in the levels of most mRNAs in the rrp6Δ strain at 37°C (10, 16, 17), and visualization of the ACT1 mRNA in the Rrp6-depleted strain revealed no accumulation in the nucleus or in the nucleolus (Fig. 4). This result indicates that the polyadenylated RNA species that are being detected in the nucleolar poly(A) domain are unlikely to correspond to mRNAs.
As shown in Fig. 5, the distributions of the U14 snoRNA and the snoRNA-associated protein Nop1 were drastically altered in the rrp6Δ strain at the nonpermissive temperature. Nop1 and U14 showed a uniform nucleolar staining at 23°C but became highly enriched in a focus that precisely colocalized with the nucleolar poly(A) domain following transfer to 37°C. Since strains lacking Rrp6 accumulate 3′-extended and polyadenylated snoRNAs, including U14 (Fig. 5D and 6), our results strongly suggest that the nucleolar poly(A) domain contains polyadenylated snoRNAs. This idea is supported by the finding that U3 snoRNA, which was not found to be polyadenylated in rrp6Δ strains (1), appeared to be excluded from the nucleolar poly(A) domain (Fig. 5C). The maturation of snoRNPs, including U14, involves passage through an NB that may be related to the human Cajal body (37). However, an analysis of the localization of the snoRNA cap-trimethylase Tgs1 showed that the nucleolar poly(A) domain and NB are physically distinct (Fig. 3).
The use of a probe specific for the 3′-extended form of U14 snoRNA showed that the accumulated poly(A)+ forms are indeed present in the poly(A) domain. However, the Northern analyses show that these represent only a small fraction of the total U14 population, indicating that mature, presumably functional U14 can also localize to the poly(A) domain. Analyses of strains carrying point mutations in Sda1 identified a subnucleolar structure termed the No-body (7). Sda1 is associated with a late pre-60S ribosomal subunit and is required for its export to the cytoplasm. Following the inhibition of 60S ribosomal subunit export in an sda1-2 mutant strain, the NB is very strongly and rapidly enriched for pre-60S particles, which are substrates for polyadenylation by Trf4 and degradation by the exosome. However, pre-40S particles that are not known to be defective in sda1-2 mutants are also accumulated, presumably transiently, in the NBs. This result suggests that the production of abundant degradation substrates induces the formation of visible surveillance centers. These detectably contain both the defective RNA-protein complexes and at least some “normal” complexes that will presumably pass the surveillance steps and be released. The relationship between the NB and the nucleolar poly(A) domain reported here remains to be determined. It is, however, unlikely that they are simply identical, since the loss of Rrp6 promoted the formation of the nucleolar poly(A) domain but inhibited NB formation (7).
The degradation of nuclear RNAs by the exosome is activated by the TRAMP complex (18, 31, 38), and our results show that TRAMP is required for the formation of the nucleolar poly(A) domain (Fig. 7 and 8). The poly(A) polymerase activity of Trf4 is distinct from that of Pap1, which is responsible for the synthesis of poly(A) tails of nuclear mRNA precursors. Within the mRNA 3′ synthesis machinery, Pap1 is highly processive and rapidly adds 60 to 90 adenine residues to mRNAs. In contrast, Trf4 exhibits a slow and distributive polyadenylation activity in vitro (18). In wild-type cells, the poly(A) tails synthesized by Trf4 may never normally be extended to more than a few nucleotides, due to rapid degradation by the exosome (18), whereas in situ detection by the (dT)50 probe presumably requires the presence of long A tails. In the absence of exosome activity, the short oligo(A) tails may act as primers for polyadenylation by Pap1 (18). Consistent with this model, reduced polyadenylation of both snoRNA and rRNA precursors was reported in a pap1-1 rrp6Δ strain relative to the rrp6Δ single mutant (17, 33). This result may explain the great reduction in the nucleolar poly(A) domain seen in the pap1-5 rrp6Δ double-mutant strain at 37°C (Fig. 8). However, it is also possible that the effects of pap1 mutations are indirect consequences of defective mRNA polyadenylation and subsequent mRNA instability.
Rrp6 and Mtr4 are implicated in the surveillance and degradation of many different nuclear RNA species (5, 6, 13, 17, 18, 31, 38). The lack of nucleolar poly(A) staining in normal cells is consistent with the view that polyadenylated RNAs are normally very short-lived intermediates that are rapidly degraded by the exosome. The absence of Rrp6 results in the accumulation of polyadenylated forms of snoRNAs, snRNAs, pre-rRNA, rRNA, intergenic cryptic unstable transcripts, and other transcripts, each of which presumably contributes to the strong nuclear accumulation of poly(A)+ RNAs. It remains to be established whether all of these species are enriched in the nucleolar poly(A) domain. Since an exosome component, Rrp43, is also detected to be enriched in the poly(A) domain (Fig. 7C), it is most likely that this domain corresponds to a dedicated compartment where polyadenylation and degradation of nucleolar RNAs takes place. We speculate that the compartmentalization of polyadenylated RNAs may promote their efficient recognition as substrates for degradation by the exosome.
Acknowledgments
We thank Edouard Bertrand for the gift of plasmids and probes and for helpful advice on yeast immunofluorescence and in situ hybridization procedures. We are also grateful to E. Hurt for kindly providing Nup49-GFP and D. Goldfarb for GFP-Rrp43.
T.C. was the recipient of a fellowship from Fundação para a Ciência e Tecnologia, Portugal. This work was supported by the European Commission QLG2-CT-2001-01554. D.T. and L.M. were supported by the Wellcome Trust.
Footnotes
Published ahead of print on 2 April 2007.
REFERENCES
- 1.Allmang, C., J. Kufel, G. Chanfreau, P. Mitchell, E. Petfalski, and D. Tollervey. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18:5399-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allmang, C., E. Petfalski, A. Podtelejnikov, M. Mann, D. Tollervey, and P. Mitchell. 1999. The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 13:2148-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Amberg, D. C., A. L. Goldstein, and C. N. Cole. 1992. Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev. 6:1173-1189. [DOI] [PubMed] [Google Scholar]
- 3.Belgareh, N., and V. Doye. 1997. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J. Cell Biol. 136:747-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briggs, M. W., K. T. Burkard, and J. S. Butler. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 273:13255-13263. [DOI] [PubMed] [Google Scholar]
- 5.Butler, J. S. 2002. The yin and yang of the exosome. Trends Cell Biol. 12:90-96. [DOI] [PubMed] [Google Scholar]
- 6.Davis, C. A., and M. J. Ares. 2006. Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 103:3262-3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dez, C., J. Houseley, and D. Tollervey. 2006. Surveillance of nuclear-restricted pre-ribosomes within a subnucleolar region of Saccharomyces cerevisiae. EMBO J. 25:1534-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang, F., J. Hoskins, and J. S. Butler. 2004. 5-Fluorouracil enhances exosome-dependent accumulation of polyadenylated rRNAs. Mol. Cell. Biol. 24:10766-10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henras, A. K., C. Dez, and Y. Henry. 2004. RNA structure and function in C/D and H/ACA s(no)RNPs. Curr. Opin. Struct. Biol. 14:335-343. [DOI] [PubMed] [Google Scholar]
- 10.Houalla, R., F. Devaux, A. Fatica, J. Kufel, D. Barrass, C. Torchet, and D. Tollervey. 2006. Microarray detection of novel nuclear RNA substrates for the exosome. Yeast 23:439-454. [DOI] [PubMed] [Google Scholar]
- 11.Houseley, J., J. LaCava, and D. Tollervey. 2006. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 7:529-539. [DOI] [PubMed] [Google Scholar]
- 12.Houseley, J., and D. Tollervey. 2006. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep. 7:205-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadaba, S., A. Krueger, T. Trice, A. M. Krecic, A. G. Hinnebusch, and J. Anderson. 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18:1227-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kadowaki, T., S. Chen, M. Hitomi, E. Jacobs, C. Kumagai, S. Liang, R. Schneiter, D. Singleton, J. Wisniewska, and A. M. Tartakoff. 1994. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol. 126:649-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 16.Kuai, L., B. Das, and F. Sherman. 2005. A nuclear degradation pathway controls the abundance of normal mRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 102:13962-13967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuai, L., F. Fang, J. S. Butler, and F. Sherman. 2004. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101:8581-8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaCava, J., J. Houseley, C. Saveanu, E. Petfalski, E. Thompson, A. Jacquier, and D. Tollervey. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121:713-724. [DOI] [PubMed] [Google Scholar]
- 19.Lejeune, F., X. Li, and L. E. Maquat. 2003. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12:675-687. [DOI] [PubMed] [Google Scholar]
- 20.Lorentzen, E., P. Walter, S. Fribourg, E. Evguenieva-Hackenberg, G. Klug, and E. Conti. 2005. The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 12:575-581. [DOI] [PubMed] [Google Scholar]
- 21.Minvielle-Sebastia, L., P. J. Preker, and W. Keller. 1994. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′ end processing factor. Science 266:1702-1705. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell, P., E. Petfalski, R. Houalla, A. Podtelejnikov, M. Mann, and D. Tollervey. 2003. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell. Biol. 23:6982-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchell, P., E. Petfalski, A. Shevchenko, M. Mann, and D. Tollervey. 1997. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91:457-466. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell, P., and D. Tollervey. 2000. Musing on the structural organization of the exosome complex. Nat. Struct. Biol. 7:843-846. [DOI] [PubMed] [Google Scholar]
- 25.Mouaikel, J., C. Verheggen, E. Bertrand, J. Tazi, and R. Bordonne. 2002. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell 9:891-901. [DOI] [PubMed] [Google Scholar]
- 26.Patel, D., and J. S. Butler. 1992. Conditional defect in mRNA 3′ end processing caused by a mutation in the gene for poly(A) polymerase. Mol. Cell. Biol. 12:3297-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 28.Schneiter, R., T. Kadowaki, and A. M. Tartakoff. 1995. mRNA transport in yeast: time to reinvestigate the functions of the nucleolus. Mol. Biol. Cell 6:357-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torchet, C., C. Bousquet-Antonelli, L. Milligan, E. Thompson, J. Kufel, and D. Tollervey. 2002. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell 9:1285-1296. [DOI] [PubMed] [Google Scholar]
- 30.Trumtel, S., I. Leger-Silvestre, P. E. Gleizes, F. Teulieres, and N. Gas. 2000. Assembly and functional organization of the nucleolus: ultrastructural analysis of Saccharomyces cerevisiae mutants. Mol. Biol. Cell 11:2175-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanácová, S., J. Wolf, G. Martin, D. Blank, S. Dettwiler, A. Friedlein, H. Langen, G. Keith, and W. Keller. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 3:e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van de Corput, M. P., and F. G. Grosveld. 2001. Fluorescence in situ hybridization analysis of transcript dynamics in cells. Methods 25:111-118. [DOI] [PubMed] [Google Scholar]
- 33.van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Hoof, A., and R. Parker. 1999. The exosome: a proteasome for RNA? Cell 99:347-350. [DOI] [PubMed] [Google Scholar]
- 35.Vasudevan, S., and S. W. Peltz. 2003. Nuclear mRNA surveillance. Curr. Opin. Cell Biol. 15:332-337. [DOI] [PubMed] [Google Scholar]
- 35a.Venema, J., and D. Tollervey. 1996. RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J. 15:5701-5714. [PMC free article] [PubMed] [Google Scholar]
- 36.Verheggen, C., D. L. Lafontaine, D. Samarsky, J. Mouaikel, J. M. Blanchard, R. Bordonne, and E. Bertrand. 2002. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J. 21:2736-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verheggen, C., J. Mouaikel, M. Thiry, J. M. Blanchard, D. Tollervey, R. Bordonne, D. L. Lafontaine, and E. Bertrand. 2001. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 20:5480-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wyers, F., M. Rougemaille, G. Badis, J. C. Rousselle, M. E. Dufour, J. Boulay, B. Regnault, F. Devaux, A. Namane, B. Seraphin, D. Libri, and A. Jacquier. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121:725-737. [DOI] [PubMed] [Google Scholar]
- 39.Zanchin, N. I., and D. S. Goldfarb. 1999. Nip7p interacts with Nop8p, an essential nucleolar protein required for 60S ribosome biogenesis, and the exosome subunit Rrp43p. Mol. Cell. Biol. 19:1518-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]