Abstract
Gene expression in the gut is segmentally regulated, but little is known of the molecular origin of patterning. Analysis of gene expression in colons from mice lacking the methyl-CpG binding repressor MBD2 revealed frequent activation of genes that are normally only expressed in the exocrine pancreas and duodenum. Reduced DNA methylation activated the same gene set in the colon. No significant differences in DNA methylation between the colon and duodenum were detected, but MBD2 was significantly more abundant in the colon. The relevance of MBD2 concentration was tested in a human colon cancer cell line. Depletion of MBD2 was again found to activate exocrine pancreatic genes. Gene activation in this cell culture model was accompanied by loss of promoter-bound MBD2 and increased histone acetylation. The results suggest that modulation of MBD2 during gut development establishes a region-specific gene expression pattern that is essential for establishing correct segmental character.
CpG methylation plays a role in long-term gene silencing such as X inactivation and imprinting, but its role in differentiation of specialized cells and organogenesis is less well documented. An understanding of the involvement of DNA methylation in development can be gained by functional studies of methyl-CpG-binding proteins (MBPs), which mediate downstream effects by binding to methylated DNA and recruiting protein partners (12, 17). MBPs fall into two families: the MBD proteins, MBD1, MBD2, MBD4, and MeCP2, which share a related ∼80-amino-acid methyl-CpG binding domain (10), and the Kaiso-like proteins, Kaiso, ZBTB4, and ZBTB38, which bind DNA methylated at CpGs via a conserved zinc-finger motif (9, 27). Biochemical analysis of MeCP2, MBD1, MBD2, and Kaiso has provided evidence that each can associate with a specific corepressor complex and bring about transcriptional silencing in model gene expression systems.
The present study concerns MBD2, which is a transcriptional repressor that can be isolated in a complex with components of the NuRD complex, including the SNF2-like motor protein Mi-2 and histone deacetylases (8, 22, 26, 34). MBD2 is the only member of the MBD protein family to be represented by invertebrate orthologs (12). Evidence for the involvement of MBD2 in differentiation and development has come from mouse genetics. Mice in which the Mbd2 gene is deleted are viable and fertile (11), but they mis-express the interleukin 4 and interferon γ genes in T helper (TH) cells (13). Expression of the interferon γ and interleukin 4 genes is mutually exclusive in the TH1 and TH2 derivatives of wild-type naive TH cells, but Mbd2−/− TH cells often expressed both genes simultaneously. Failure to adequately repress these genes in differentiated TH1 and TH2 cells alters the immune response in MBD2-deficient animals (14). Also, aberrant derepression of human fetal globin genes in Mbd2-null mice bearing a transgene that contains the human β-like globin gene cluster has also been reported (31). In addition to these examples of gene misregulation, it has been found that depletion of MBD2 greatly reduces the number of intestinal adenomas in tumor-prone ApcMin mice (32). Although the molecular basis of this effect is unknown, it may be relevant that MBD2 is required for silencing of specific tumor-suppressor genes in human colorectal carcinoma cells and in other human cancer cell lines (3, 24). MBD2-mediated repression of tumor suppressor genes may therefore be a prerequisite for tumor formation, although other explanations cannot currently be ruled out.
In the present study we investigated gene expression in the colons of Mbd2−/− mice. We found that a set of genes coding for digestive enzymes normally expressed only in duodenum and pancreas are highly overexpressed in the Mbd2-null colon, the level of overexpression being variable between individual mice. DNA methylation profiles were indistinguishable between wild-type duodenum and colon, but MBD2 was ∼5-fold more abundant in colon. The MBD2-deficient phenotype could be mimicked in human colorectal carcinoma cells by depletion of MBD2 or DNA methylation, leading to overexpression and altered chromatin structure at these genes. Our results establish that a stable gene expression pattern in the mature gut is dependent on DNA methylation and MBD2.
MATERIALS AND METHODS
Mice and tissue samples.
Mbd2−/− mice (11) are kept as inbred homozygous lines on a C57BL/6 and a BALB/c background. Dnmt1c/+ mice (23) are kept on a C57BL/6 background.
Cell lines.
HCT116 cells were obtained from the American Type Culture Collection. DKO cells were a gift from B. Vogelstein. Both cell lines were maintained in RPMI medium containing 10% (vol/vol) fetal calf serum. For HDAC inhibition, cells were treated with 500 nM trichostatin A (Sigma) for 18 h.
MBD2 siRNA and shRNA.
Two different predesigned small interfering RNAs (siRNAs) directed against MBD2 (ID no. 14015 and ID no. 13925) were purchased from Ambion. Both siRNAs (150 pmol each) were cotransfected three times at 24-h intervals into HCT116 cells using Oligofectamine (Invitrogen). Silencer negative control siRNA#1 (Ambion) was used as a control. To generate the small hairpin RNA (shRNA) vector, a 64-bp hairpin sequence directed against MBD2 was cloned into the pSuper vector with puromycin resistance. Stable cell clones were obtained after puromycin selection.
Real-time RT-PCR.
Total RNA was extracted from mouse intestines using RNA-Bee (Tel-Test) and cultured cells using TriReagent (Sigma) according to the manufacturer's instructions. After DNase treatment with DNA-Free (Ambion), cDNA was transcribed using Moloney murine leukemia virus reverse transcriptase (Promega). Real-time PCR was carried out with iQ SYBR Green Supermix (Bio-Rad) on an iCycler (Bio-Rad) according to the manufacturer's instructions. The primer sequences used for reverse transcription-PCR (RT-PCR) can be obtained on request.
Western blotting and immunohistochemistry.
Approximately 100 mg of snap-frozen mouse intestine was placed into 600 μl of Laemmli buffer (60 mM Tris-HCl, 100 mM dithiothreitol, 10% glycerol, 2% sodium dodecyl sulfate [SDS]), sonicated for 15 s, and boiled for 6 min. After spinning for 4 min at 14,000 rpm (4°C), the supernatant was run on a polyacrylamide gel. Western blot analyses with protein extracts from mouse tissue or cultured human cells were performed using standard protocols. Rabbit R593 anti-MBD2 antiserum (26) was used to probe the blots. Immunohistochemistry was performed on 5-μm cryosections fixed in 4% paraformaldehyde. After the sections were boiled in 10 mM citric acid (pH 6.0) for 5 min, staining was performed using the Mouse-on-Mouse peroxidase kit (Vector Laboratories) according to the manufacturer's instructions. A mouse monoclonal antibody (anti-human spasmolytic polypeptide, catalog no. NCL-HSP; Novocastra) was used to detect TFF2 (spasmolytic polypeptide).
Chromatin immunoprecipitation (ChIP).
Cells were grown until confluent, cross-linked in 1% formaldehyde in phosphate-buffered saline (PBS) for 10 min, washed in PBS, and lysed in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.0]). The chromatin was sonicated for 3 min at 35% amplitude using a digital sonifier (Branson) and then diluted 1:10 in immunoprecipitation dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl [pH 8.0]). After the samples were precleared for 2 h with salmon sperm DNA-bovine serum albumin-protein G-Sepharose beads, 15 μg of affinity-purified anti-MBD2 antiserum (S923 [26]), anti-acetyl histone H4 antibody (catalog no. 06 to 598, Upstate), or unrelated control antibodies were added to each sample, followed by rotation overnight at 4°C. To collect the immunocomplexes, 100 μl of protein G-Sepharose was added to the samples for 1 h at 4°C. The beads were then washed once in buffer 1 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl), four times in buffer 2 (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 500 mM NaCl, 20 mM Tris-HCl), once in buffer 3 (250 mM LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl), and twice in TE buffer (10 mM Tris-HCl, 1 mM EDTA). Immunocomplexes were then eluted with 2 × 100 μl of extraction buffer (1% SDS, 100 mM NaHCO3), and cross-links were reversed by adding 5 M NaCl to a final concentration of 300 mM, followed by incubation at 65°C overnight. The primer sequences for amplification of the TFF2 promoter can be obtained on request.
Bisulfite genomic sequencing.
DNA was phenol-chloroform extracted from cultured cells or mouse intestine after proteinase K digestion. DNA (2 μg) was bisulfite modified and PCR amplified (primer sequences are available on request). PCR products were gel purified and cloned using the TOPO TA cloning kit (Invitrogen). Colonies for each PCR (10-14) were picked for sequencing.
Global DNA methylation analysis.
Decreasing amounts of DNA were slot blotted onto a nitrocellulose membrane and then incubated with recombinant protein consisting of four sequential hemagglutinin (HA)-tagged methyl-CpG-binding domains of MBD1, which can bind to methylated but not unmethylated DNA (16). Incubation with an anti-HA antibody allowed visualization and quantification of the amount of methylated DNA in the sample. M.Sss1-methylated DNA and DNA isolated from bacteriophage lambda were used as positive and negative controls, respectively, for CpG methylation.
RESULTS
ExPa genes are activated in colons of Mbd2-null and Dnmt1-deficient mice.
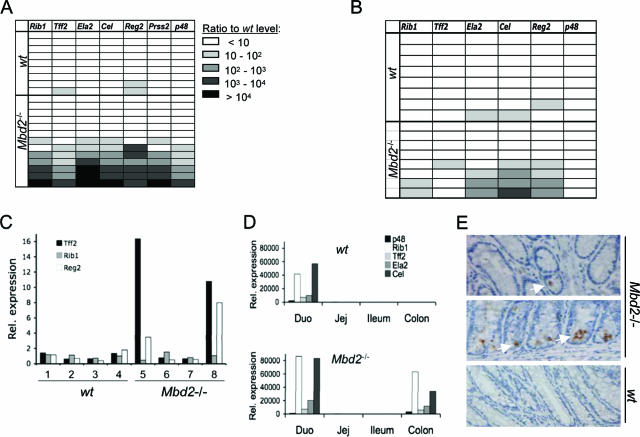
As part of our investigations concerning the resistance of Mbd2-null mice to intestinal tumorigenesis (32), we examined gene expression patterns in colons from wild-type and Mbd2-null mice by using Affymetrix gene expression microarrays. A group of genes stood out as highly overexpressed (>30-fold) specifically in Mbd2-null colons (Table 1; see the Affymetrix data set in the supplemental material). We noted that other members of the MBD family—MBD1, MBD4, and MeCP2—were not transcriptionally upregulated in the absence of MBD2 (see the Affymetrix data set in the supplemental material). The overexpressed genes predominantly encode digestive enzymes that are expressed in the exocrine pancreas and duodenum and secreted into the duodenum, where they enzymatically degrade nutrients. These genes are normally silent in the colon, whose main function is water and salt resorption (7). To verify the microarray results, we analyzed expression of six exocrine pancreatic (ExPa) genes in the colons of 9 wild-type and 13 Mbd2-null BALB/c mice using quantitative RT-PCR (Fig. 1A). We excluded amylase 1 from the analysis because it is also expressed at significant levels in wild-type colon (data not shown). Surprisingly, significant overexpression of these genes was seen in only 7 of 13 Mbd2-deficient colons: an apparent penetrance of ∼50%. Levels of overexpression also varied widely, with some animals expressing >10,000-fold more than the basal levels in wild-type colon. The results demonstrate a continuum of overexpression in the absence of MBD2.
TABLE 1.
ExPa genes found to be strongly upregulated in the Mbd2-null colon by Affymetrix gene expression microarray
| Gene | Description | UniGene accession no. | GenBank accession no. | Fold upregulation in Mbd2−/− colon |
|---|---|---|---|---|
| Amy1 | Amylase 1 | Mm.378882 | X02578 | 2,084 |
| Prss2 | Serine protease 2 | Mm.276926 | X04574 | 267 |
| Reg2 | Regenerating islet-derived 2 | Mm.46360 | D14011 | 99 |
| Ela2 | Elastase 2 | Mm.45316 | X04573 | 71 |
| Rib1 | Pancreatic RNase 1 | X60103 | 60 | |
| Tff2 | Trefoil factor 2 | Mm.1825 | U78770 | 52 |
| Cel | Carboxyl ester lipase | Mm.236017 | U37386 | 32 |
FIG. 1.
Expression of ExPa genes in wild-type and Mbd2−/− mice. (A) Real-time quantitative RT-PCR analysis of ExPa gene expression in the colon of nine wild-type and 13 Mbd2-null BALB/c mice. Each horizontal row corresponds to one mouse. Values represent the fold overexpression of genes normalized to average expression in wild-type samples. (B) The partial penetrance of ExPa gene overexpression is independent of the mouse strain. Detection of ExPa gene expression levels in the colon of eight wild-type and eight Mbd2-null C57BL/6 mice by real-time RT-PCR as in panel A. (C) ExPa gene expression levels in colons isolated from 4-day-old BALB/c wild-type and Mbd2-null mice. The expression levels were analyzed by real-time RT-PCR and were normalized to the average expression in wild-type samples. (D) Real-time RT-PCR analysis for expression patterns of ExPa genes in four intestinal segments (Duo, duodenum; Jej, jejunum) of wild-type and Mbd2-null mice. Overexpression levels are calculated compared to the levels in the wild-type colon. (E) Immunohistochemical detection of TFF2 in colonic epithelium of Mbd2-null mice (top and middle panels) and wild-type mice (bottom panel). Positively stained enterocytes (brown spots; white arrows show examples) and goblet cells are only seen in mutant crypts.
Several considerations argue that phenotypic variability is not genetic in origin. First, these mice were back-crossed for eight generations onto a BALB/c genetic background. Second, incomplete penetrance was independent of genetic background, since four of eight Mbd2-null mutants that had been back-crossed onto a C57BL/6 background (eight generations) showed derepression of ExPa genes (Fig. 1B). Third, significant heterogeneity arose between siblings, since only two of four adult littermates (one male and one female) showed expression of the ExPa genes in colon (data not shown). We also considered the possibility that the activation of ExPa genes correlated positively or negatively with postnatal age, but no correlation was seen between ExPa gene overexpression and postweaning age (data not shown). Importantly, two of four newborn (postnatal day 4) Mbd2-null mice showed upregulation of ExPa enzymes in the colon (Fig. 1C), indicating that the incomplete penetrance of ExPa gene expression is present soon after birth in the Mbd2-null mouse colon.
Differentiation of the exocrine pancreas is under the control of the transcription factor complex Ptf1, which comprises two widely expressed proteins, p64 and p75, plus the highly tissue specific factor p48 (19, 20, 30). Mice carrying a disrupted p48 gene specifically lack an exocrine pancreas (30). We sought to determine whether the overexpression of ExPa genes correlated with p48 mis-expression in MBD2-deficient colons. The results showed a strong correlation, since p48 expression was confined to colons that also overexpressed the pancreas-specific gene set (Fig. 1A). Derepression of p48 may therefore trigger the overexpression phenotype. We also tested the expression of Cdx1 and Cdx2, which are required for anterior-posterior patterning and the development of the intestinal epithelium. No significant difference in mRNA levels between wild-type and Mbd2-null colons was observed (data not shown).
To determine which regions of the Mbd2-null intestine overexpress ExPa genes, we isolated RNA from the colon and the three segments comprising the small intestine: the duodenum, the jejunum, and the ileum. Real-time RT-PCR revealed that ExPa genes are, as expected, expressed in the duodenum in wild-type mice (Fig. 1D). In Mbd2-null mice, aberrant expression of digestive enzymes was only observed in the colon, since the jejunum and ileum were not affected. At their highest, the expression levels of p48, Tff2, and Ela2 in the colons of Mbd2-deficient mice were comparable to levels seen in the duodenum. We next sought to determine whether variable ExPa gene expression in Mbd2-null mice was caused by different numbers of expressing cells per colon or different levels of expression in a constant set of cells. Strong cytoplasmic immunostaining for TFF2 was observed in goblet cells and enterocytes in two of five Mbd2-null mouse colons (Fig. 1E and data not shown). One ExPa-expressing colon showed TFF2 expression in multiple cells in adjacent crypts (Fig. 1E, middle panel), whereas in the other mouse strong staining was only seen in single cells in a small fraction of crypts (Fig. 1E, top panel). No TFF2-positive crypt cells were seen in five wild-type mouse colons (Fig. 1E, bottom panel, and data not shown). Variability of expression levels in Mbd2-deficient mice may therefore be due to different numbers of crypt cells that escape ExPa gene repression.
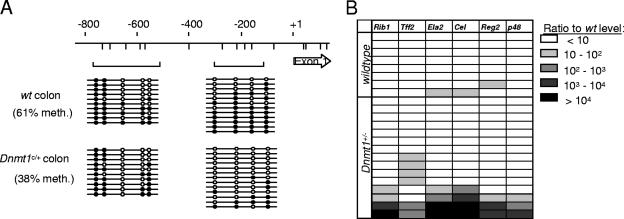
Since MBD2 is reported to read the DNA methylation signal, we speculated that reduced levels of DNA methylation would also induce inappropriate expression of the same genes. Absence of the maintenance DNA methyltransferase DNMT1 is lethal in mice (23), but phenotypic effects on cancer susceptibility have been observed in Dnmt1+/− heterozygotes, suggesting that reducing the amount of enzyme by half affects DNA methylation (21). To test for the effects of heterozygosity of the Dnmt1c allele on DNA methylation levels, we carried out bisulfite genomic sequencing of a region upstream of the Tff2 promoter. Average DNA methylation across the region was significantly less in Dnmt1c/+ heterozygotes than in wild-type mice (61% versus 38%; Fig. 2A). When the expression of ExPa genes was examined in Dnmt1c/+ mice, we observed overexpression of the ExPa genes in 4 of 15 mice (Fig. 2B). As in the case of Mbd2-null mice, Dnmt1 deficiency increased the probability that colon cells escape the repression of ExPa genes that normally prevails in this organ.
FIG. 2.
Aberrant expression of ExPa genes in the colons of Dnmt1c/+ mice. (A) Bisulfite genomic sequencing of the mouse Tff2 promoter region in the colon of wild-type and Dnmt1c/+ C57BL/6 mice. CpGs are represented by vertical lines. Brackets show PCR products amplified after bisulfite treatment. Empty circles represent unmethylated cytosine residues, and filled circles represent methylated cytosine residues. (B) Real-time RT-PCR analysis of ExPa gene expression in wild-type and Dnmt1c/+ C57BL/6 mouse colons. Each horizontal row corresponds to one mouse. Values correspond to the fold overexpression of genes normalized to average expression in wild-type samples.
MBD2 abundance controls ExPa gene expression.
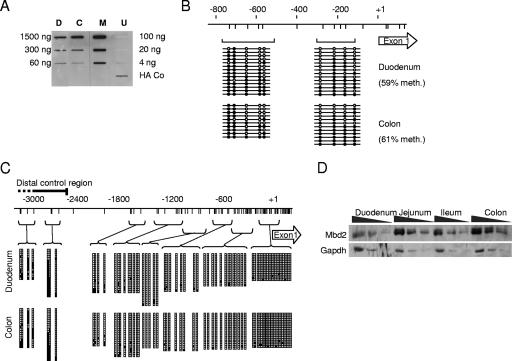
The genetic results show that deficiency of either DNA methylation or of a protein that binds to methylated DNA causes gene activation specifically in the colon. Targeting of transcriptional silencing to the colon may be due to higher levels of CpG methylation or a higher abundance of MBD2 in distal regions of the intestinal tract. We first examined global DNA methylation levels using as an assay the binding of a poly-MBD domain reagent that provides a semiquantitative measure of CpG methylation (16). No difference was observed between colon and duodenum (Fig. 3A). Detailed analysis of two regions of the Tff2 gene promoter also showed levels of CpG methylation that were not significantly different between the colon and the duodenum (61% versus 59% methylation, respectively, across a total of 16 CpG sites; Fig. 3B). Analysis of the p48 gene established the presence of a CpG island that is nonmethylated in duodenum, colon, and leukocyte DNA. We tested CpG methylation in the upstream “negative regulatory domain” of p48, including a conserved region 2 to 4 kb upstream of the transcription start site that has been implicated in silencing of p48 (18, 30). CpG methylation was detected, but again, no consistent differences between the colon and the duodenum were observed (Fig. 3C).
FIG. 3.
Comparison of MBD2 and DNA methylation levels in the duodenum and the colon. (A) Analysis of global methylation levels in duodenum (D) and colon (C) as determined by an affinity labeling assay with a HA-tagged poly-MBD domain reagent (16). M.Sss1-methylated (M) and nonmethylated bacteriophage lambda DNA (U) were positive and negative controls, respectively. The single HA-Co slot contained immobilized HA-tagged protein as a positive control for the anti-HA monoclonal antibody. (B) Bisulfite genomic sequencing of the Tff2 upstream region in duodenum and colon. The map shows the promoter region and first exon (open arrow). CpGs are represented by vertical lines. Brackets show PCR products amplified after bisulfite treatment. Empty circles represent unmethylated cytosine residues, and filled circles represent methylated cytosine residues. Each line corresponds to a sequenced DNA strand. (C) Bisulfite genomic sequencing of the p48 promoter in the colon and duodenum of wild-type mice. CpGs are represented by vertical lines. Horizontal lines linked to brackets show PCR products amplified after bisulfite treatment. Empty squares represent unmethylated cytosine residues, and filled squares represent methylated cytosine residues. Each row represents a single sequenced DNA molecule, and column height corresponds to the number of strands sequenced (i.e., 14 to 24) for each PCR product. (D) Western blot analysis for MBD2 and GAPDH in protein extracts from wild-type duodenum, jejunum, ileum, and colon. A twofold dilution series for each protein extract is shown.
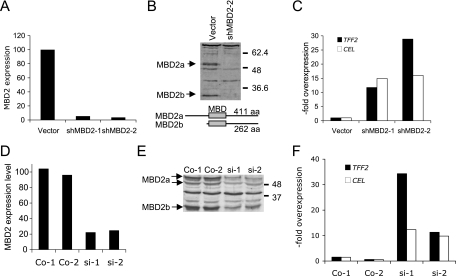
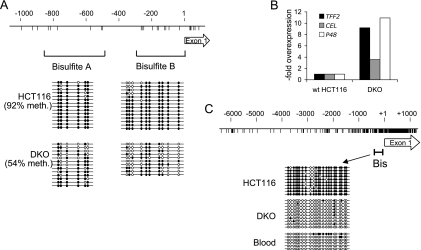
In contrast to the apparent constancy of DNA methylation, the concentration of MBD2 was ∼5-fold higher in the colon than in the duodenum as determined by Western blot analysis when normalized against GAPDH (glyceraldehyde-3-phosphate dehydrogenase) protein (Fig. 3D). Levels of MBD2 in the ileum and jejunum, where ExPa genes are also silent, were likewise higher than in the duodenum and were similar to colon levels (Fig. 3D). We only detected the larger MBD2a isoform with antibodies that detect both MBD2a and MBD2b, suggesting that MBD2b is a rare form in this tissue (data not shown). The relative abundance of MBD2 in the colon is compatible with the hypothesis that various concentrations of this CpG methylation-dependent repressor contribute to the control of differential gene expression in colon and duodenum. To test this idea, we sought to determine whether expression of the ExPa genes is affected by artificial modulation of MBD2 concentrations in a model system: the human colon cancer cell line HCT116. We depleted MBD2 by stably expressing an shRNA directed against MBD2 mRNA. Real-time RT-PCR showed an mRNA knockdown of more than 90% in two shRNA-expressing clones (Fig. 4A), and Western blot analysis confirmed almost complete depletion of both MBD2a and MBD2b isoforms (Fig. 4B). This treatment caused >10-fold overexpression of two digestive enzymes, encoded by the genes TFF2 and CEL, in both MBD2-depleted cell clones (Fig. 4C). A more modest downregulation of MBD2 that paralleled the ∼5-fold difference between mouse duodenum and colon was achieved by transfecting siRNAs targeted against MBD2 mRNA (Fig. 4D and E) and once again led to >10-fold upregulation of TFF2 and CEL gene expression (Fig. 4F). Therefore, the modulation of MBD2 levels is sufficient to regulate the expression of these genes in a human colon cell line. We also tested the effects of DNA hypomethylation on these genes, using a DNMT1−/−, DNMT3b−/− double mutant derivative of HCT116 cells (28). The double-knockout (DKO) cells carry mutations in the DNA methyltransferases DNMT1 and DNMT3b and show reduced DNA methylation levels, as monitored by bisulfite sequencing (Fig. 5A). We detected ∼10-fold upregulation of TFF2 and P48 and ∼4-fold overexpression of CEL in DKO cells (Fig. 5B), indicating that DNA methylation, like MBD2, plays a role in silencing these genes.
FIG. 4.
Depletion of MBD2 induces ExPa gene expression in a human colon cell line. (A) Real-time RT-PCR analysis for MBD2 expression levels in a mock-transfected HCT116 cell clone (vector) and two stable derivate cell lines expressing shRNA directed against MBD2 (shMBD2-1 and shMBD2-2). (B) Western blot analysis for MBD2 in cell extracts from mock-transfected cells (vector) and a cell clone expressing shRNA for MBD2 (shMBD2-2). The structure and length of the MBD2a and MBD2b isoforms is indicated below. (C) Real-time RT-PCR analysis of ExPa gene expression in control cells and cells stably expressing shRNA against MBD2. Values indicate the overexpression normalized to the vector control. (D) Moderate (four- to fivefold) reduction of MBD2 mRNA levels by siRNA knockdown in two transfected cell lines si-1 and si-2) and two mock-transfected control cell lines analyzed by real-time RT-PCR. (E) Western blot analysis of MBD2 isoform confirms partial depletion in the two siRNA-transfected cell lines. Wild-type levels of MBD2 are seen in control cell lines (Co-1 and Co-2). (F) Real-time RT-PCR analysis of TFF2 and CEL gene expression levels in control and MBD2-depleted HCT116 cells.
FIG. 5.
Activation of ExPa gene expression by depletion of DNA methylation in HCT116 cells. (A) Bisulfite genomic sequencing of the TFF2 promoter shows reduced DNA methylation in methylation-deficient HCT116 cells carrying mutations in DNMT1 and DNMT3 (DKO cells) compared to wild-type HCT116 cells. CpGs are represented by vertical lines. Dotted lines show PCR products amplified after bisulfite treatment. Empty circles represent unmethylated cytosine residues, and filled circles represent methylated cytosine residues. (B) Real-time RT-PCR for ExPa genes in wild-type HCT116 and DKO cells. (C) Bisulfite genomic sequencing of the p48 promoter CpG island shows dense CpG methylation in HCT116 cells but not in DKO cells or human blood. CpGs are represented by vertical lines. The bracket (Bis) shows PCR products amplified after bisulfite treatment.
We noticed that P48, RIB1, and ELA2 were not upregulated in MBD2-deficient HCT116 cells in contrast to Mbd2-null mouse colon. Since P48 is an upstream activator of ExPa genes, we investigated the reason for its silence. Bisulfite sequencing established that the promoter CpG island of human P48 is densely methylated in these cells, although this region is nonmethylated in human blood DNA (Fig. 5C). Most CpG islands are nonmethylated regardless of gene expression (1), but their aberrant methylation, particularly at tissue specifically expressed genes, is a common feature of cancer cells (15) and other permanent cell lines (2, 5), where it effectively silences the associated genes. The inference that P48 is silenced by aberrant methylation in HCT116 cells is supported by its induction in a DNA methylation-deficient derivative of the same line (Fig. 5B). Parenthetically, it is apparent from these results that P48 silencing due to aberrant methylation of its CpG island promoter in HCT116 cells is not primarily mediated by MBD2.
MBD2 binding and deacetylation of repressed exocrine pancreatic genes.
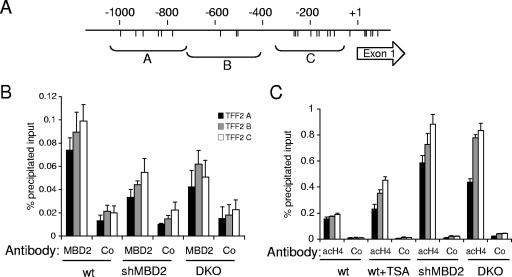
To test whether TFF2 is a direct MBD2 target, we carried out ChIP in wild-type, MBD2-depleted, and DNA methylation-deficient HCT116 cells. ChIP of genomic DNA corresponding to three primer pairs covering ∼1 kb upstream of the transcription start site was assayed by quantitative PCR. MBD2 was enriched at the TFF2 promoter in wild-type HCT116 cells, but binding to a domain extending 1 kb upstream of the transcription start site (Fig. 6A) was reduced by both MBD2-shRNA treatment and in DKO derivative cells (Fig. 6B). A similar reduction in MBD2 occupancy was observed at the P48 promoter in MBD2-deficient and DKO HCT116 cells (data not shown). Since MBD2 is known to recruit HDACs associated with the NuRD complex, we sought to determine whether reduced MBD2 binding was accompanied by an increase in histone acetylation levels at the TFF2 promoter. ChIP experiments revealed low levels of acetylated histone H4 in untreated HCT116 cells. These levels increased significantly after treatment with the histone deacetylase inhibitor trichostatin A but to a greater extent after MBD2 depletion by shRNA and in methylation-deficient DKO cells (Fig. 6C). Bisulfite genomic sequencing of the TFF2 promoter confirmed high levels of DNA methylation (>90%) in wild-type HCT116 cells but lower levels in DKO cells (Fig. 5A). The results demonstrate DNA methylation-dependent binding of MBD2 to the TFF2 promoter and show elevated histone acetylation in this region when MBD2 is depleted.
FIG. 6.
MBD2 binds to the TFF2 in a DNA methylation-dependent manner and reduces histone H4 acetylation levels. (A) Diagram indicating the location of CpG sites (vertical lines) and the TFF2 upstream fragments amplified by primer pairs A, B, and C. (B) ChIP shows reduced binding of MBD2 in regions A to C of the TFF2 promoter after either MBD2 depletion by RNA interference or depletion of DNA methylation in DNA methyltransferase mutant DKO cells. (C) Increased histone H4 acetylation after depletion of MBD2 or DNA methylation. ChIP analysis for panacetylated histone H4 at the TFF2 promoter was carried out with chromatin from wild-type and TSA-treated HCT116 cells, HCT116 cells expressing shRNA against MBD2, and DKO cells. Negative control immunoprecipitations in panels B and C (Co) were performed with an unrelated antibody (anti-α-amylase).
DISCUSSION
Our results identify MBD2 as a key regulator of a gene set that is normally expressed in the duodenum and exocrine pancreas but not in the colon. Silencing of these ExPa genes in the colon depends on MBD2, since in its absence about half of all mice show expression in this intestinal segment. We found no evidence that this effect is due to modulation of DNA methylation, since neither whole genomic DNA nor upstream regulatory regions of the p48 and Tff2 genes showed obvious differences in CpG methylation between the colon and the duodenum. These results imply that differential DNA methylation is not responsible for the difference, although we cannot altogether exclude the involvement of variable DNA methylation at specific genes or in a small subset of intestinal cells. The concentration of MBD2, on the other hand, was significantly higher in the colon than in the duodenum, raising the possibility that modulation of this repressor could regulate ExPa gene expression against a background of constant DNA methylation levels. We tested this idea in the human HCT116 colon cell line and found that depletion of MBD2 indeed caused activation of several ExPa genes. Since MBD2 is part of a NuRD complex, which contains histone deacetylases (8, 22), chromatin hyperacetylation is the expected result of MBD2 loss. Accordingly, loss of MBD2 was accompanied by significantly increased histone acetylation at the TFF2 gene. The results suggest that the high concentration of colonic MBD2 imposes silence on the ExPa genes in this intestinal region, even though the DNA methylation levels appear to be no different from those in the duodenum. The levels of MBD2 are also high in the jejunum and ileum, where ExPa genes remain silent, but we did not detect derepression of these genes in the absence of MBD2. The results indicate that MBD2 is not essential for ExPa gene silencing in these midintestinal regions.
Gene expression analysis along the length of the mouse intestine has shown that fewer specialized genes are expressed in the colon than in more proximal regions of the intestinal tract (4). Little is known about the molecular basis of segmental gene expression along the intestinal anterior-posterior axis. One hypothetical possibility is that expression patterns are regulated by the abundance of specific transcriptional activators in each segment. An alternative, but not mutually exclusive, possibility is that differential silencing of gene expression contributes to patterning through segmental variation in the abundance of transcriptional repressors. Previous work has indicated that variations in the expression of histone deacetylases, which are components of many corepressors, are crucial for embryonic development of the gastrointestinal tract (33). The observed anterior-posterior decline in transcriptional complexity along the intestine (4) could therefore be caused by increased cellular concentrations of broad specificity repressors in posterior intestinal segments.
We were initially prompted to study ExPa gene expression through an interest in the resistance of Mbd2-null mice to intestinal tumorigenesis in the Min mouse model (32). It is unlikely that our findings are relevant to this phenotype, however, for two major reasons. First, most adenomatous polyps in the Min mouse arise in the proximal intestinal tract (25), where we detected no altered expression of the ExPa genes. The location of ExPa gene mis-expression and the sites of tumorigenesis therefore do not match. Second, ExPa gene expression is not seen in all mice, whereas the tumor resistance phenotype appears fully penetrant. If failure to overexpress the ExPa genes abolished tumor resistance in mice, then a subset of animals (about half) with normal tumor susceptibility would have been detected. The absence of such a subset suggests that overexpression of ExPa genes is not causally related to tumor resistance. It is possible that other genes that are more subtly misregulated in the Mbd2-null intestine are relevant to the latter phenotype.
A puzzling feature of the colon phenotype is its incomplete penetrance. A continuum of ExPa gene expression levels is observed, ranging from very high (equivalent to duodenum) to no detectable expression in about half of the animals. It is common for transcriptional repressors to contribute to silencing as part of a redundant set of silencing mechanisms, and this can cause partial gene activation in a stochastic manner when one component is absent. Leakproof inactivation of genes on one X chromosome in female mammals, for example, relies on layered epigenetic silencing mechanisms, each of which reduces the probability per cell of gene reactivation (6). Indeed, MBD2 itself is not solely responsible for silencing of the interleukin-4 gene in TH cells, since not all MBD2-deficient TH cells express the gene (13). In spite of these precedents at the cellular level, variability of gene reactivation between members of an inbred population of mice is unexpected. Our studies make it unlikely that age, gender, or genetic background are critical factors affecting penetrance (see Results). Instead, they suggest that stochastic variation arises during development. We speculate that stem cells at the base of the colon crypts are variably dependent on MBD2 for silencing the ExPa genes in their differentiated descendants. MBD2 dependence or independence would be a stable feature that affects a particular crypt for the lifetime of its parent stem cell.
Mbd2-null animals that express ExPa genes showed coincident upregulation of p48. The p48 polypeptide is a component of Ptf1, a trimeric transcription factor that is essential for the development and maintenance of the differentiated state of the exocrine pancreas. A CpG island is located near the transcription start site of p48, which, like many CpG islands at highly tissue specifically expressed genes, is nonmethylated regardless of gene activity, except in cultured cell lines where aberrant methylation occurs. In view of its pivotal role in differentiation of the exocrine pancreas, we suspected that inappropriate p48 expression in the Mbd2-null colon is a trigger for ExPa gene expression. Under a simple hypothetical pathway, MBD2 would directly mediate repression of p48 in the colon. Although the p48 promoter is associated with a constitutively nonmethylated CpG island, which therefore should not bind MBD2, studies of the sequence requirements of p48 gene expression in mouse acinar and nonpancreatic cells have implicated a region extending up to 7 kb upstream of the p48 transcription start site in both positive and negative regulation of gene activity (18, 30). This region is not within the CpG island region and, accordingly, bisulfite sequence analysis in duodenum and colon detects CpG methylation. Also, MBD2 is bound to the equivalent region of human P48 by ChIP in HCT116 cells (data not shown). Taken together, our results are compatible with the hypothesis that the p48 gene is under direct regulatory control by MBD2.
MBD2 deficiency in HCT116 cells, which have silenced P48 via aberrant CpG island methylation, induced TFF2 and CEL, but not RIB1 or ELA2. Both RIB1 and ELA2 possess CpG-deficient promoters that do not correspond to CpG islands. The different behaviors of these genes in HCT116 cells and mouse colon could be explained if TFF2 and CEL are direct targets for MBD2 repression, whereas the induction of RIB1 and ELA2 requires the upstream expression of P48. In line with this hypothesis, the PTF1 transcriptional activator complex, which includes P48, has been shown to bind an enhancer of the ELA2 gene (29). Also, we show that MBD2 is associated with the TFF2 promoter region in HCT116 cells, and its absence is accompanied by alterations in the structure of TFF2 chromatin. Interestingly, a transgenic mouse with a reporter gene under the control of the multimerized Ela2 enhancer showed expression in the pancreas, duodenum and colon (29). It is conceivable that the absence of appropriate DNA methylation in the transgene prevented recruitment of MBD2, thereby permitting expression in the colon.
Supplementary Material
Acknowledgments
We are grateful to Jim Selfridge for help with mouse experiments, Karen Wilson and Dina De Sousa for technical assistance, and Aimée Deaton for comments on the manuscript.
This study was supported by supported by Programme Grants from Cancer Research UK and the Wellcome Trust.
Footnotes
Published ahead of print on 12 March 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Antequera, F. 2003. Structure, function and evolution of CpG island promoters. Cell Mol. Life Sci. 60:1647-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antequera, F., J. Boyes, and A. Bird. 1990. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell 62:503-514. [DOI] [PubMed] [Google Scholar]
- 3.Auriol, E., L. M. Billard, F. Magdinier, and R. Dante. 2005. Specific binding of the methyl binding domain protein 2 at the BRCA1-NBR2 locus. Nucleic Acids Res. 33:4243-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bates, M. D., C. R. Erwin, L. P. Sanford, D. Wiginton, J. A. Bezerra, L. C. Schatzman, A. G. Jegga, C. Ley-Ebert, S. S. Williams, K. A. Steinbrecher, B. W. Warner, M. B. Cohen, and B. J. Aronow. 2002. Novel genes and functional relationships in the adult mouse gastrointestinal tract identified by microarray analysis. Gastroenterology 122:1467-1482. [DOI] [PubMed] [Google Scholar]
- 5.Bird, A. P. 1987. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 3:342-347. [Google Scholar]
- 6.Csankovszki, G., A. Nagy, and R. Jaenisch. 2001. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J. Cell Biol. 153:773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Santa Barbara, P., G. R. van den Brink, and D. J. Roberts. 2003. Development and differentiation of the intestinal epithelium. Cell Mol. Life Sci. 60:1322-1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, Q., and Y. Zhang. 2001. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 15:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filion, G. J., S. Zhenilo, S. Salozhin, D. Yamada, E. Prokhortchouk, and P. A. Defossez. 2006. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol. Cell. Biol. 26:169-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hendrich, B., and A. Bird. 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 18:6538-6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hendrich, B., J. Guy, B. Ramsahoye, V. A. Wilson, and A. Bird. 2001. Closely related proteins Mbd2 and Mbd3 play distinctive but interacting roles in mouse development. Genes Dev. 15:710-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hendrich, B., and S. Tweedie. 2003. The methyl-CpG binding domain and the evolving role of DNA methylation in animals. Trends Genet. 19:269-277. [DOI] [PubMed] [Google Scholar]
- 13.Hutchins, A., A. Mullen, H. Lee, K. Barner, F. High, B. Hendrich, A. Bird, and S. Reiner. 2002. Gene silencing quantitatively controls the function of a developmental transactivator. Mol. Cell 10:81-91. [DOI] [PubMed] [Google Scholar]
- 14.Hutchins, A. S., D. Artis, B. D. Hendrich, A. P. Bird, P. Scott, and S. L. Reiner. 2005. Cutting edge: a critical role for gene silencing in preventing excessive type 1 immunity. J. Immunol. 175:5606-5610. [DOI] [PubMed] [Google Scholar]
- 15.Jones, P. A., and S. B. Baylin. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3:415-428. [DOI] [PubMed] [Google Scholar]
- 16.Jorgensen, H. F., K. Adie, P. Chaubert, and A. P. Bird. 2006. Engineering a high-affinity methyl-CpG-binding protein. Nucleic Acids Res. 34:e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klose, R. J., and A. P. Bird. 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem. Sci. 31:89-97. [DOI] [PubMed] [Google Scholar]
- 18.Knofler, M., A. Krapp, O. Hagenbuchle, and P. K. Wellauer. 1996. Constitutive expression of the gene for the cell-specific p48 DNA-binding subunit of pancreas transcription factor 1 in cultured cells is under control of binding sites for transcription factors Sp1 and αCbf. J. Biol. Chem. 271:21993-22002. [DOI] [PubMed] [Google Scholar]
- 19.Krapp, A., M. Knofler, S. Frutiger, G. J. Hughes, O. Hagenbuchle, and P. K. Wellauer. 1996. The p48 DNA-binding subunit of transcription factor PTF1 is a new exocrine pancreas-specific basic helix-loop-helix protein. EMBO J. 15:4317-4329. [PMC free article] [PubMed] [Google Scholar]
- 20.Krapp, A., M. Knofler, B. Ledermann, K. Burki, C. Berney, N. Zoerkler, O. Hagenbuchle, and P. K. Wellauer. 1998. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 12:3752-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laird, P. W., Jackson-Grusby, A. Fazeli, S. L. Dickinson, W. E. Jung, E. Li, R. A. Weinberg, and R. Jaenisch. 1995. Suppression of intestinal neoplasia by DNA hypomethylation. Cell 81:197-205. [DOI] [PubMed] [Google Scholar]
- 22.Le Guezennec, X., M. Vermeulen, A. B. Brinkman, W. A. Hoeijmakers, A. Cohen, E. Lasonder, and H. G. Stunnenberg. 2006. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol. Cell. Biol. 26:843-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 24.Lin, X., and W. G. Nelson. 2003. Methyl-CpG-binding domain protein-2 mediates transcriptional repression associated with hypermethylated GSTP1 CpG islands in MCF-7 breast cancer cells. Cancer Res. 63:498-504. [PubMed] [Google Scholar]
- 25.Moser, A. R., W. F. Dove, K. A. Roth, and J. I. Gordon. 1992. The Min (multiple intestinal neoplasia) mutation: its effect on gut epithelial cell differentiation and interaction with a modifier system. J. Cell Biol. 116:1517-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ng, H.-H., Y. Zhang, B. Hendrich, C. A. Johnson, B. M. Burner, H. Erdjument-Bromage, P. Tempst, D. Reinberg, and A. Bird. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23:58-61. [DOI] [PubMed] [Google Scholar]
- 27.Prokhortchouk, A., B. Hendrich, H. Jorgensen, A. Ruzov, M. Wilm, G. Georgiev, A. Bird, and E. Prokhortchouk. 2001. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev. 15:1613-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rhee, I., K. E. Bachman, B. H. Park, K. W. Jair, R. W. Yen, K. E. Schuebel, H. Cui, A. P. Feinberg, C. Lengauer, K. W. Kinzler, S. B. Baylin, and B. Vogelstein. 2002. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature 416:552-556. [DOI] [PubMed] [Google Scholar]
- 29.Rose, S. D., F. Kruse, G. H. Swift, R. J. MacDonald, and R. E. Hammer. 1994. A single element of the elastase I enhancer is sufficient to direct transcription selectively to the pancreas and gut. Mol. Cell. Biol. 14:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose, S. D., G. H. Swift, M. J. Peyton, R. E. Hammer, and R. J. MacDonald. 2001. The role of PTF1-P48 in pancreatic acinar gene expression. J. Biol. Chem. 276:44018-44026. [DOI] [PubMed] [Google Scholar]
- 31.Rupon, J. W., S. Z. Wang, K. Gaensler, J. Lloyd, and G. D. Ginder. 2006. Methyl binding domain protein 2 mediates γ-globin gene silencing in adult human betaYAC transgenic mice. Proc. Natl. Acad. Sci. USA 103:6617-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sansom, O. J., J. Berger, S. M. Bishop, B. Hendrich, A. Bird, and A. R. Clarke. 2003. Deficiency of Mbd2 suppresses intestinal tumorigenesis. Nat. Genet. 34:145-147. [DOI] [PubMed] [Google Scholar]
- 33.Tou, L., Q. Liu, and R. A. Shivdasani. 2004. Regulation of mammalian epithelial differentiation and intestine development by class I histone deacetylases. Mol. Cell. Biol. 24:3132-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wade, P. A., A. Gegonne, P. L. Jones, E. Ballestar, F. Aubry, and A. P. Wolffe. 1999. Mi-2 complex couples DNA methylation to chromatin remodeling and histone deacetylation. Nat. Genet. 23:62-66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.