Abstract
Cardiac hypertrophy is associated with a dramatic change in the gene expression profile of cardiac myocytes. Many genes important during development of the fetal heart but repressed in the adult tissue are reexpressed, resulting in gross physiological changes that lead to arrhythmias, cardiac failure, and sudden death. One transcription factor thought to be important in repressing the expression of fetal genes in the adult heart is the transcriptional repressor REST (repressor element 1-silencing transcription factor). Although REST has been shown to repress several fetal cardiac genes and inhibition of REST function is sufficient to induce cardiac hypertrophy, the molecular mechanisms employed in this repression are not known. Here we show that continued REST expression prevents increases in the levels of the BNP (Nppb) and ANP (Nppa) genes, encoding brain and atrial natriuretic peptides, in adult rat ventricular myocytes in response to endothelin-1 and that inhibition of REST results in increased expression of these genes in H9c2 cells. Increased expression of Nppb and Nppa correlates with increased histone H4 acetylation and histone H3 lysine 4 methylation of promoter-proximal regions of these genes. Furthermore, using deletions of individual REST repression domains, we show that the combined activities of two domains of REST are required to efficiently repress transcription of the Nppb gene; however, a single repression domain is sufficient to repress the Nppa gene. These data provide some of the first insights into the molecular mechanism that may be important for the changes in gene expression profile seen in cardiac hypertrophy.
The repressor element 1-silencing transcription factor (REST) was originally identified as an important transcription factor regulating the expression of neuron-specific genes (12, 53) but has since been shown to be a key transcriptional regulator in heart development (28) and vascular smooth muscle growth (11). Disruption of REST function by expression of a dominant-negative form specifically in the heart results in cardiomyopathy, arrhythmias, and sudden death (28). These effects are thought to result from the reexpression of fetal cardiac genes and have led to the proposition that REST represses the fetal cardiac gene program in the adult heart (28). In vascular smooth muscle, loss of REST has been implicated in neointimal hyperplasia, and inhibition of REST results in increased smooth muscle proliferation (11). Several genes that are repressed by REST in myocytes have been identified, including the genes encoding the brain and atrial natriuretic peptides (Nppb and Nppa, encoding BNP and ANP, respectively), α-skeletal actin (Acta1), potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channels 2 and 4 (Hcn2 and Hcn4), and voltage-gated calcium channel subunit alpha Cav3.2 (Cacna1h) (26, 43). Levels of BNP and ANP are particularly important, since increased levels of these peptides in circulation are clinical indicators of the severity of hypertrophy (8, 29, 38, 61). In adult ventricular myocytes, expression of both BNP and ANP is increased in cardiac hypertrophy, and as they are secreted via the constitutive secretory pathway, increased expression results in increased levels of circulating peptides (38, 52). Binding sites for REST (RE1 sites) have been identified in both Nppb and Nppa gene regulatory regions, and a role for REST in repression of these genes has been identified in ventricular myocytes (27, 28, 43). Since removal of REST function within the heart in transgenic mice results in increased ANP and BNP expression and cardiac hypertrophy, it has been proposed that repression of these genes by REST is an important component of normal heart function (28). The molecular mechanisms involved in REST repression of Nppb and Nppa genes, however, are not known.
REST is able to recruit two independent corepressor complexes through N-terminal and C-terminal repression domains (2, 15, 21, 42, 49, 59). Via its N-terminal repression domain, REST interacts with the mSin3 corepressor complex, and repression via the N terminus is associated with class I (15, 21, 42, 49) and class II (40) histone deacetylase (HDAC) activity. The C-terminal repression domain of REST interacts with the corepressor CoREST, which, like mSin3, is part of a larger complex (2, 16, 22, 65). The CoREST corepressor complex contains HDAC1, HDAC2, and lysine-specific histone demethylase 1 (LSD1, also known as BHC110), which represses transcription by demethylating histone H3 lysine 4 (H3K4) (16, 22, 54, 65). The significance of, and the requirement for, two independent repression domains in REST is not entirely clear. When fused to a Gal4 DNA binding domain, both the N- and C-terminal repression domains are able to independently repress transcription of a reporter gene containing Gal4 binding sites (59), and deletion of either domain from the full-length protein results in some loss of repressor activity, but repressor activity is lost completely only with the removal of both domains (4). REST is able to recruit both mSin3 and CoREST to the Scn2a2 (Nav1.2) RE1 site in L6 and JTC-19 cells (4, 6); however, the mechanisms of REST repression appear to be gene and cell type dependent. Scn2a2 expression was derepressed by the HDAC inhibitor trichostatin A (TSA) in HEK293 and JTC-19 cells but not in Rat-1 and Neuro-2a cells (4, 6, 34, 49). Additionally, inhibition of CoREST recruitment is sufficient to inhibit Scn2a2 but not Stmn2 (SCG10) gene expression in Rat-1 cells (34). Most of the studies of REST have focused on silencing of RE1 genes in nonneuronal cells or repression of RE1 genes in neurons (5, 9, 10, 34, 39, 45, 67). In cardiac myocytes, REST repression of Nppa is associated with decreased histone acetylation, though whether this is due to recruitment of HDAC activity by the N- or C-terminal repression domains is not clear (27). In response to the hypertrophy-inducing stimulus endothelin-1 (ET-1), adult rat ventricular myocytes show increased expression of Nppb and Nppa mRNA.
Here we show that continued expression of REST using adenovirus delivery is sufficient to prevent ET-1-induced increases in expression of Nppb and Nppa. In a rat ventricular myocyte cell line (H9c2), REST is recruited to and represses the expression of the Nppb and Nppa genes, and inhibition of REST function results in increased Nppb and Nppa expression. Both ET-1 treatment of adult ventricular myocytes and inhibition of REST in H9c2 cells are associated with increases in both histone H4 acetylation and dimethyl H3K4. We show that recruitment of HDAC activity by REST is not sufficient to repress Nppb or Nppa, and we provide evidence that demethylation of H3K4 is a vital component of REST repression. Expression of ectopic REST lacking either the N- or the C-terminal repression domain leads to partial derepression of Nppb transcription and associated chromatin changes, suggesting that both domains are required for repression of Nppb in these cells. Conversely, our data suggest that either domain alone is able to repress Nppa transcription and maintain a repressed chromatin state at the Nppa promoter. This is the first study to demonstrate a functional requirement for both repression domains of REST to regulate the levels of an actively transcribed gene, and it provides insights into the chromatin changes that may be important in cardiac hypertrophy.
MATERIALS AND METHODS
Myocyte isolation.
Male Wistar rats (250 to 300 g) were killed humanely by cervical dislocation following stunning. Hearts were removed in accordance with the Home Office Guidance on the Operation of the Animals (Scientific Procedures) Act of 1986, and ventricular myocytes were isolated as described by Harrison et al. (17). Briefly, isolated hearts were perfused first with a HEPES-based isolation solution containing 5 mM HEPES, 130 mM NaCl, 5.4 mM KCl, 1.4 mM MgCl2, 0.4 mM NaH2PO4, 10 mM glucose, 20 mM taurine, 10 mM creatine, and 750 μM Ca2+ (pH 7.4) for 5 min and then for a further 4 min with the same buffer lacking calcium with 0.1 mM EGTA. After a 10-min perfusion with 1 mg/ml collagenase (type I; Worthington) and 0.8 mg/ml protease (XIV; Sigma), ventricles were isolated, finely chopped, and gently shaken at 37°C for 30 min in an isolation solution containing collagenase and 1% bovine serum albumin. Single cells were harvested by filtration, pelleted, and resuspended in a calcium-containing isolation solution before being plated onto laminin (20 μg/ml; Invitrogen)-coated plates. Cells were cultured at 37°C under 5% CO2 in Dulbecco's modified Eagle's medium (Life Technologies, Inc.) supplemented with 10% (vol/vol) fetal calf serum, 2 mM l-glutamine, 6 g/liter penicillin, 10 g/liter streptomycin, and 1% nonessential amino acids (Sigma). Four hours after plating, fresh medium containing 1% fetal calf serum with or without 10 nM ET-1 and/or adenovirus was added to the cells, which were harvested after a further 24 h.
Cell culture.
H9c2 cells obtained from the European Collection of Cell Cultures were cultured in Dulbecco's modified Eagle's medium (Life Technologies, Inc.) supplemented with 10% (vol/vol) fetal calf serum, 2 mM glutamine, streptomycin (10 g/liter), and penicillin (10 g/liter) at 37°C under 5% CO2. Cells were incubated with 300 nM TSA (Wako) or 1 mM 5′-deoxy-5′-methyl-thioadenosine (MTA; Sigma) for 48 h prior to isolation of RNA and reverse transcription-PCR (RT-PCR) analysis.
Immunohistochemistry.
H9c2 cells were fixed and stained using the REST antiserum R2174 (62) (1:1,000) or preimmune serum. Staining was visualized by fluorescence using a fluorescein isothiocyanate (FITC)-conjugated anti-rabbit secondary antibody (1:100; Santa Cruz). Nuclei were counterstained using 4′,6′-diamidino-2-phenylindole (DAPI).
RT-PCR.
Total RNA was isolated from H9c2 cells using Tri reagent (Sigma) and reverse transcribed using oligo(dT), random primers, and the Moloney murine leukemia virus reverse transcriptase RNase H(−) point mutant (Promega). PCR was performed using the following oligonucleotide primers: for cyclophilin, sense primer 5′-ACCCCACCGTGTTCTTCGAC and antisense primer 5′-TGGACTTGCCACCAGTGCCA; for Nppb, sense primer 5′-GACTCCGGCTTCTGATCTG and antisense primer 5′-ACTGTGGCAAGTTTGTGCTG; for. Nppa, sense primer 5′-CTGCTTTCTGAAAGGGGTGA and antisense primer 5′-CGGTGTGTCACACAGCTTGG; for Rest (rat), sense primer 5′-ACTTTGTCCTTACTCAAGTTCTCAG and antisense primer 5′-ATGGCGGGTTACTTCATGTT; for REST (human), sense primer 5′-CGAACTCACACAGGAGAACG and antisense primer 5′-GAGGCCACATAATTGCACTG. Quantitative PCR was performed using SYBR green I incorporation, measured using an iCycler iQ system (Bio-Rad). All reactions were performed in duplicate alongside negative controls containing RNA as a template.
Adenovirus construction and amplification.
Adenoviruses containing either full-length REST or the DNA binding domain (amino acids 234 to 437) of REST (dominant-negative REST [DN-REST]) have been described previously (11, 62). A REST sequence encoding either amino acids 1 to 1097 (REST), 73 to 1097 (C-REST), or 1 to 1017 (N-REST) was amplified by PCR and cloned into pAdTrack-CMV. The resulting plasmids were linearized with PmeI and electroporated into competent Escherichia coli BJ5183 containing the pAdEasy-1 plasmid (19). Recombinants were selected for by growth on kanamycin. The resulting adenovirus plasmids were purified, linearized with PacI, and transfected into packaging HEK293 cells to produce adenovirus particles. Viral amplification was achieved via four rounds of infection, and the virus was purified through cesium chloride gradients using standard protocols. H9c2 cells were infected using approximately 1 × 1012 virus particles/ml and were harvested for analysis after 48 h.
Gel shift assays.
DNA fragments containing the rat Nppb and Nppa RE1 sites were amplified using the following primers: for Nppb, sense primer 5′-CGCGAAGCTTGGCAGGGTATCAGAGTGGTT and antisense primer 5′-CGCGAAGCTTAGTTAGCACCCACCATCACC; for Nppa, sense primer 5′-CGCGAAGCTTGGCCTTACCTCTCCCACTCT and antisense primer 5′-CGCGAAGCTTGGTGACAGAAAGGAGCCAAA. PCR products were digested with HindIII, and [α-32P]dATP was incorporated by Klenow fill-in. Probes were run on a 4% polyacrylamide gel and purified. Nuclear protein was prepared from H9c2 cells using the procedure described by Andrews and Faller (3) and was quantified using a DC protein assay kit (Bio-Rad). Protein (20 μg for uninfected and 6 μg for infected H9c2 cells) was preincubated on ice, with or without competitor DNA, for 20 min in 19 μl of a solution containing 20 mM HEPES (pH 7.9), 100 mM KCl, 5 mM MgCl2, 8% (vol/vol) glycerol, and 1 μg of calf thymus DNA. Approximately 10,000 to 20,000 cpm of radioactive probe was added to each reaction mixture and incubated for 20 min at room temperature. For supershift experiments, 2 μg of anti-REST (P18; Santa Cruz) or anti-Sp1 (H-225; Santa Cruz) antibody was added, and reaction mixtures were incubated for a further 30 min at room temperature. Reactions were run on 0.5× Tris-borate-EDTA polyacrylamide gels, fixed, dried, and exposed to Biomax X-ray film (Kodak) for 16 h. Complementary oligonucleotides for the following sites (with sequences given in parentheses) were annealed and used for competition experiments: Chrm4 RE1 (5′-GTACGGAGCTGTCCGAGGTGCTGAATCTGCCT), Nppb RE1 (5′-GGTGATCAGAACCATGGACACTACCA), Nppa RE1 (5′-AAACTTCAGCACCACGGACAGACGCTG), and Sp1 (5′-AATTCCCCGAGGGGCGCCTAGTCCCCATG).
Chromatin immunoprecipitation (ChIP).
Anti-REST (R2174 [62]), anti-Myc (Sigma), anti-acetylated H4 (Abcam), and anti-dimethyl H3K4 (Abcam) antibodies were used to immunoprecipitate cross-linked chromatin. H9c2 cells were cross-linked with 0.37% formaldehyde for 10 min at room temperature. Glycine was added to a final concentration of 0.125 M to terminate the cross-linking reaction. Cells were harvested, washed with phosphate-buffered saline, resuspended in cell lysis buffer (10 mM Tris-HCl [pH 8.0], 10 mM NaCl, 0.2% NP-40, 10 mM sodium butyrate, 50 μg/ml phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml leupeptin), and incubated on ice for 10 min. After centrifugation, nuclei were resuspended in 1.2 ml of nuclear lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% sodium dodecyl sulfate, 10 mM sodium butyrate, 50 μg/ml PMSF, 1 μg/ml leupeptin) and incubated on ice for 10 min before addition of 0.72 ml of immunoprecipitation dilution buffer (20 mM Tris-HCl [pH 8.1], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 0.01% sodium dodecyl sulfate, 10 mM sodium butyrate, 50 μg/ml PMSF, 1 μg/ml leupeptin). Fixed chromatin was fragmented by sonication to an average size of 500 to 700 bp. Sonicated extracts were precleared with protein G-Sepharose and chromatin immunoprecipitated using 4 μg of antibody or 10 μl of crude serum. Cross-links were reversed, and protein was removed by digestion with proteinase K and phenol extraction. Purified DNA was resuspended in 50 μl of Tris-EDTA, pH 8, and 2 μl was used for quantitative PCR. Products were quantified using SYBR green I incorporation, measured using an iCycler iQ system (Bio-Rad) with the following primers: for the Nppa promoter, sense primer 5′-GTTGGCTTCCTGGCTGACT and antisense primer 5′-CACCCCCACCCTAGATGTC; for Nppa RE1, sense primer 5′-TTTGGCTCCTTTCTGTCACC and antisense primer 5′-CACACACACACACACACACG; for Nppb RE1, sense primer 5′-TTACAGGTTCGAGGACACTC and antisense primer 5′-GCTTATGGGGTGACTCATC; for the control, sense primer 5′-TCCTCACCTGGACACCCTAC and antisense primer 5′-ACGTAGCAGAGCCAGTAGCC. A minimum of three independent experiments were performed, and significance was analyzed using a Student t test.
RESULTS
REST inhibits increased Nppb and Nppa expression induced by a hypertrophic stimulus.
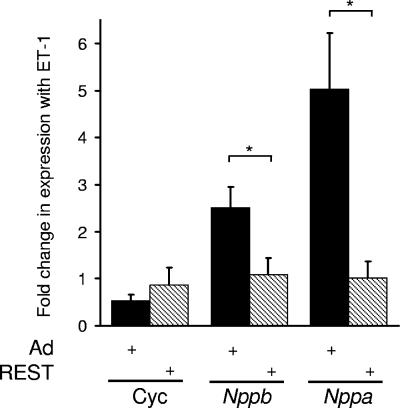
Inhibition of REST and relief of target gene repression has previously been implicated in the development of cardiac hypertrophy (27, 28, 40). Hypertrophy-inducing agents, such as ET-1, are thought to act, at least in part, by inhibiting REST function. In support of such notions, expression of a dominant-negative REST in rat ventricular myocytes results in increased expression of Nppb and Nppa mRNA and reduces the increase in levels of these genes in response to ET-1 (28). To complement such observations, we examined the effects of ectopic REST expression in rat ventricular myocytes on Nppb and Nppa expression in response to ET-1. In the presence of a control adenovirus, ET-1 resulted in 2.5- ± 0.4-fold and 5.0- ± 1.2-fold increases in Nppb and Nppa expression, respectively (Fig. 1). In the presence of an adenovirus expressing the full-length REST protein, the increases in Nppb and Nppa expression in response to ET-1 were abolished (Fig. 1). Neither REST nor ET-1 resulted in changes to a control gene, cyclophilin (Fig. 1), and infection of H9c2 cells with a control adenovirus did not affect basal or ET-1-induced Nppb and Nppa expression (data not shown). Together with published observations, these new data provide strong support for the notion that inhibition of REST is important in driving the gene expression changes seen in cardiac hypertrophy.
FIG. 1.
Ectopic REST expression prevents increased Nppb and Nppa mRNA levels in adult rat ventricular myocytes in response to ET-1. Data are changes in cyclophilin (Cyc), Nppb, and Nppa mRNA levels in cells infected with a control adenovirus (Ad) or REST-expressing adenovirus treated with ET-1 relative to untreated cells (means ± standard errors of the means; n = 4). *, P < 0.05.
REST represses Nppb and Nppa transcription in H9c2 cardiomyocytes.
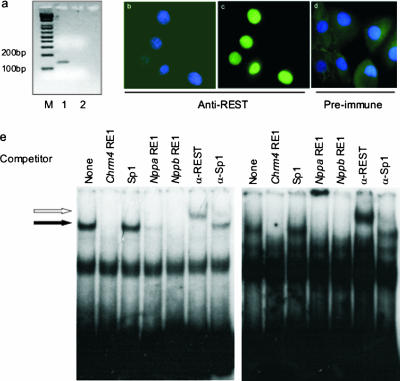
In order to examine the molecular mechanism by which REST regulates heart-specific gene expression, we used H9c2 cells, which are derived from embryonic rat ventricular heart tissue (25). Although it has been shown that rat cardiac myocytes express REST in vivo (27, 28), the expression of REST in H9c2 cells has not been documented. Using RT-PCR, we obtained a specific product derived from REST mRNA in H9c2 cells (Fig. 2a). Moreover, immunohistochemistry using an anti-REST antiserum but not preimmune serum detected REST protein in H9c2 nuclei (Fig. 2b to d). To determine if the REST protein is functional, we used a gel shift assay. Nuclear extracts prepared from H9c2 cells produced a specific DNA-protein complex with RE1 sites of the Nppb (Fig. 2e, left) and Nppa (Fig. 2e, right) genes. The RE1 sequences of both genes produced a specific complex (Fig. 2e) which was competed by excess unlabeled rat Chrm4, Nppa, and Nppb RE1 sequences but not by the binding site of the transcription factor Sp1. Further, this complex was specifically recognized by an anti-REST antibody, producing a more slowly migrating complex (Fig. 2e), but not by an antibody to Sp1. Thus, H9c2 cells express REST protein within the nucleus that is able to bind the Nppb and Nppa RE1 sites in vitro.
FIG. 2.
H9c2 cells express REST. (a) PCR from H9c2 RNA either treated with reverse transcriptase (lane 1) or left untreated (lane 2). M, 1-kb Plus DNA ladder (Life Technologies). (b to d) Immunohistochemistry of H9c2 cells with anti-REST serum (b and c) or preimmune serum (d). DAPI-stained nuclei appear blue, and REST protein detected by FITC staining appears green. The DAPI and FITC images for anti-REST serum are shown separately (panels b and c, respectively), whereas the two images for preimmune serum have been overlaid (d). (e) Gel mobility shift assay of the Nppb (left panel) and Nppa (right panel) gene RE1 sites with 20 μg of nuclear extracts from H9c2 cells in the presence of no competitor (None), an RE1 site from the Chrm4 gene (Chrm4 RE1), an unrelated binding site (Sp1), or RE1 sites from the Nppa and Nppb genes (Nppa RE1 and Nppb RE1, respectively). Specific DNA-protein complexes are indicated by the filled arrow. REST protein was identified in the complex using an anti-REST antibody (α-REST), indicated by the open arrow, and an anti-Sp1 antibody (α-Sp1) was used to control for specificity.
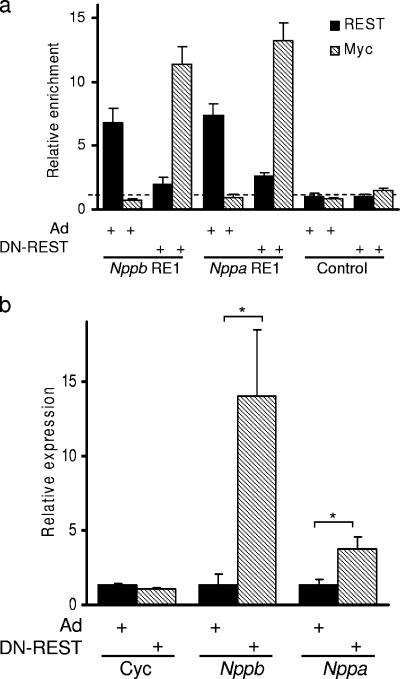
For REST to regulate Nppb and Nppa expression in H9c2 cells, we would expect it to be recruited to the RE1 sites of these genes in vivo. To investigate this, we examined the ability of an anti-REST antibody to precipitate Nppb and Nppa RE1 sequences from H9c2 derived chromatin (ChIP assay). We observed that both Nppb and Nppa RE1 sequences were specifically enriched by an anti-REST antibody in uninfected (data not shown) and control adenovirus-infected (Fig. 3a) cells. Precipitation of the Nppb and Nppa RE1 sequences by the anti-REST antibody was specific; a control sequence that is not associated with an RE1 site was not precipitated (Fig. 3a).
FIG. 3.
REST represses Nppb and Nppa transcription in H9c2 cells. (a) Quantification of Nppb RE1, Nppa RE1, and control sequences precipitated by anti-REST (solid bars) and anti-Myc (hatched bars) antibodies from control adenovirus (Ad)- and DN-REST-infected H9c2 cells (means ± standard errors of the means; n = 4). Dashed line represents level of background precipitation. (b) Quantitative RT-PCR of cyclophilin (Cyc), Nppb, and Nppa mRNA levels in H9c2 cells infected with a control adenovirus (Ad) (solid bars) or DN-REST (hatched bars) relative to those in uninfected control cells (means ± standard error of the means; n = 3). *, P < 0.05.
Having established that REST is recruited to both the Nppb and Nppa RE1 sites in H9c2 cells, we next wished to determine the functional effects of REST on Nppb and Nppa transcription levels. To this end we used an adenovirus construct to introduce a myc-tagged dominant-negative form of REST (DN-REST [62]) into more than 90% of the cell population in order to displace endogenous REST from its genomic sites. By use of the ChIP assay, Nppb and Nppa RE1 sequences were efficiently precipitated by a REST antiserum, but not by an anti-myc antibody, in cells infected with a control adenovirus (Fig. 3a). Precipitation from control virus-infected cells was not different from that from uninfected cells (6.8- ± 0.74- compared with 6.6- ± 1.09-fold for Nppb and 7.3- ± 2.43-fold compared with 7.5- ± 0.98-fold for Nppa). In DN-REST-infected cells, Nppb and Nppa RE1 sequences were precipitated by the anti-myc antibody but not by the anti-REST antibody (which recognizes a C-terminal epitope not present in DN-REST), indicating that endogenous REST is not present at these sites but has been displaced by the introduced DN-REST (Fig. 3a). The non-RE1 sequence was not precipitated by the anti-REST antibody or the anti-myc antibody (Fig. 3a). These data demonstrate that endogenous REST is recruited to the Nppb and Nppa RE1 sites and that REST can be displaced by introduction of a dominant-negative form, which contains only the DNA binding domain. To determine the functional consequence of replacing endogenous REST with DN-REST, which lacks all the repressor function of the full-length protein (59), we used RT-PCR of mRNA derived from uninfected, control-infected, and DN-REST-infected H9c2 cells to quantify Nppb and Nppa expression levels. Infection of H9c2 cells with a control virus did not alter the Nppb or Nppa mRNA level (Fig. 3b), while introduction of DN-REST resulted in increased expression of Nppb and Nppa genes but not of cyclophilin, a gene lacking an RE1 site (Fig. 3b). Nppb transcript levels were induced to a much greater level than Nppa levels in these cells (10.3- and 2.8-fold, respectively), and these levels are similar to the induction of Nppb and Nppa seen in response to DN-REST in neonatal cardiomyocytes (12.0- and 2.5-fold, respectively [28]), though they show some differences from changes induced by ET-1 (Fig. 1) (28). A greater level of Nppb induction probably reflects either the presence of transcriptional activators specific for Nppb or the presence of other repressors specific for Nppa, or both. However, we cannot be certain that, although the DN-REST does not contain any repressor activity (59), it does not block access to a transcriptional activator binding to an overlapping site within the Nppa gene. Together these data show that REST is recruited to the RE1 sites of Nppb and Nppa and represses transcription of these genes in H9c2 cells.
REST repression is mediated in part by histone deacetylation.
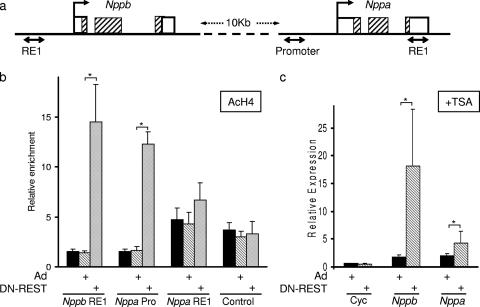
Changes in gene expression are often brought about via covalent modifications of histones, resulting in altered chromatin structure and a change in the ability to recruit other transcriptional regulatory proteins. We investigated changes in histone modifications that are associated with derepression of Nppb and Nppa transcription mediated by DN-REST. The Nppb RE1 site is located approximately 500 bp 5′ to the transcription start site (43) (Fig. 4a), and thus we were able to use the Nppb RE1 primers to interrogate histone modifications at the promoter of the Nppb gene. The Nppa RE1 site, however, is located in the 3′ untranslated region (27) (Fig. 4a), and in order to interrogate the histone modifications at the Nppa promoter, we designed specific primers located 500 bp from the Nppa transcription start site. Since REST is known to repress transcription via the recruitment of HDACs (15, 21, 42, 49), and HDAC activity is thought to play a role in Nppa repression (27, 28, 40), we used an antibody to acetylated histone H4 (AcH4) in a ChIP assay to examine the level of histone H4 acetylation at the Nppb and Nppa genes in the presence and absence of REST in H9c2 cells. Displacement of REST by DN-REST resulted in a significant increase in H4 acetylation at both the Nppb and Nppa promoter regions (10.4- and 7.3-fold, respectively) (Fig. 4b). Interestingly the Nppa RE1 region showed only a modest (1.6-fold) increase in H4 acetylation levels, though the basal level of acetylation did appear greater than that for either the Nppb or Nppa promoter region (Fig. 4b). Control DNA did not show any change in H4 acetylation levels in response to DN-REST (Fig. 4b). These data indicate that removal of REST from the Nppb and Nppa genes results in increased histone acetylation at the promoters, and they suggest that HDACs recruited by REST play a role in repressing Nppb and Nppa transcription by antagonizing histone acetyltransferases. Although it has previously been reported that HDAC activity is involved in Nppa repression (27, 28, 40), it is not known if other enzyme activities recruited by REST are also required. To test if HDAC activity alone was sufficient for the observed repression of Nppb and Nppa by REST, we examined the effects on Nppb and Nppa expression of removing REST in the presence of TSA. If REST-mediated repression occurs solely via HDAC activity, then treatment with TSA should result in derepression of Nppb and Nppa, and subsequent displacement of REST would have no additional effect. Conversely, any repression mediated by REST in the presence of TSA must involve mechanisms other than recruitment of HDAC activity. Treatment of H9c2 cells with TSA alone resulted in some derepression of Nppb and Nppa transcription; however, displacement of REST in the continued presence of TSA resulted in further increases in Nppb and Nppa transcript levels (10.2- and 2.1-fold, respectively, above that with TSA alone) (Fig. 4c). These data indicate that REST represses Nppb and Nppa transcription using mechanisms additional to HDAC activity.
FIG. 4.
Derepression of Nppb and Nppa in H9c2 cells is associated with increased histone acetylation. (a) Diagram indicating the locations of Nppb RE1 and Nppa RE1 sites in relation to the Nppb and Nppa genes on rat chromosome 5. Sequences amplified by PCR for ChIP experiments are represented by three double-headed arrows. The transcription initiation sites are indicated by arrows. Open boxes, noncoding sequences; hatched boxes, coding sequences. (b) Quantification of Nppb RE1, Nppa promoter (Nppa Pro), Nppa RE1, and control sequences precipitated by an acetylated histone H4 antiserum in uninfected H9c2 cells (solid bars), H9c2 cells infected with a control adenovirus (Ad) (hatched bars), or H9c2 cells infected with DN-REST (shaded bars) (means ± standard errors of the means; n = 5). *, P < 0.05. (c) Quantitative RT-PCR of cyclophilin (Cyc), Nppb, and Nppa mRNA levels in H9c2 cells infected with a control adenovirus (Ad) (solid bars) or with an adenovirus containing DN-REST (hatched bars) in the presence of TSA, expressed relative to levels in vehicle-treated control cells (means ± standard errors of the means; n = 4). *, P < 0.05.
A role for histone demethylation in repression of Nppb and Nppa.
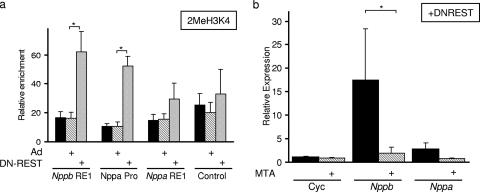
In addition to recruiting HDACs, REST also recruits, via its C terminus, a corepressor complex that contains CoREST and a lysine demethylase, LSD1 (4, 16, 54, 65). LSD1 acts to demethylate H3K4 and by doing so represses transcription (54). We investigated a potential role for H3K4 demethylation as a mechanism for REST-mediated repression of the Nppb and Nppa genes. Using an anti-dimethyl H3K4 antibody, we performed ChIP on control, adenovirus-infected, and DN-REST-infected H9c2 cells. Displacement of REST by DN-REST resulted in an increase in dimethylated H3K4 levels at both the Nppb and Nppa promoters (3.82- and 4.84-fold, respectively). Levels of dimethyl H3K4 were modestly increased at the Nppa RE1 site (1.89-fold) and were unchanged at the control DNA (Fig. 5a). Thus, displacement of REST results in an increase in dimethyl H3K4 levels at the Nppb and Nppa genes, indicating that REST recruits H3K4 demethylase activity to the Nppb and Nppa genes. To test if the increased methylation of H3K4 observed is important for the increase in transcription, we took advantage of the fact that methyltransferases can be inhibited by MTA. In the absence of MTA, displacement of REST results in increased dimethylated H3K4 levels and increased Nppb and Nppa transcription (Fig. 5a and b). This induction of Nppb and Nppa transcription was inhibited by MTA, indicating that the increased methylation seen upon REST displacement is important for derepression (Fig. 5b). MTA did not alter the expression of a gene that is not regulated by REST, cyclophilin (Fig. 5b). Together these data indicate that lysine demethylase activity is important for REST-mediated repression of Nppb and Nppa.
FIG. 5.
Derepression of Nppb and Nppa in H9c2 cells is associated with increased dimethyl H3K4 levels. (a) Quantification of Nppb RE1, Nppa promoter (Nppa Pro), Nppa RE1, and control sequences precipitated by an anti-dimethyl H3K4 antibody in uninfected H9c2 cells (solid bars), H9c2 cells infected with a control adenovirus (Ad) (hatched bars), or H9c2 cells infected with an adenovirus containing DN-REST (shaded bars) (means ± standard errors of the means). *, P < 0.05. (b) Quantitative RT-PCR of cyclophilin (Cyc), Nppb, and Nppa mRNA levels in DN-REST-expressing H9c2 cells in the presence (solid bars) or absence (hatched bars) of MTA expressed relative to those in vehicle-treated control cells (means ± standard errors of the means; n = 5). *, P < 0.05.
Both the N- and C-terminal domains of REST contribute to repression.
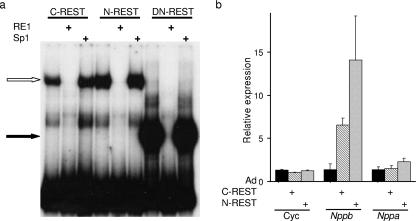
REST has two functional repression domains that recruit distinct corepressor complexes (15, 21, 42, 47, 49). In order to investigate the relative contribution of each repression domain to Nppb and Nppa transcriptional regulation in H9c2 cells, we used adenovirus constructs expressing either a REST protein lacking the N-terminal mSin3 interaction domain (C-REST) or a REST protein lacking the C-terminal CoREST interaction domain (N-REST). Each of these proteins is unable to repress transcription via the N- or C-terminal repression domain, respectively, while the other repression domain should remain functional (4). A gel shift assay was performed to ensure that C- and N-REST produced functional proteins able to bind an RE1 site. H9c2 nuclear extracts from C- and N-REST-adenovirus-infected cells produced specific RE1 binding proteins (Fig. 6a). We examined the effects of these REST mutants on Nppb and Nppa transcription by using RT-PCR. Expression of either C- or N-REST resulted in derepression of Nppb expression (4.8- and 10.3-fold, respectively) but did not affect the levels of a control gene, cyclophilin (Fig. 6b). Interestingly, neither expression of C-REST nor expression of N-REST resulted in a significant derepression of Nppa expression (Fig. 6b), despite our previous observations that expression of DN-REST resulted in a 2.8-fold increase in Nppa transcript levels (Fig. 3b). Therefore, a singular repression domain of REST is sufficient to repress Nppa, but both repressor domains are not required to repress Nppb transcription. This apparent difference in Nppb and Nppa responses to REST is likely to be linked to the fact that the Nppb promoter is more active than the Nppa promoter in the absence of REST repression in H9c2 cells (Fig. 3b). Two possible explanations for this are the presence of a transcriptional activator specific for Nppb and/or the presence of a transcriptional repressor specific for Nppa in H9c2 cells.
FIG. 6.
Both REST repression domains are required for Nppb but not Nppa repression in H9c2 cells. (a) Gel mobility shift assay of the Nppb gene RE1 site with 6 μg of nuclear extracts from C-REST-, N-REST-, and DN-REST-infected H9c2 cells in the presence of no competitor, an RE1 site from the Chrm4 gene (RE1), or an unrelated binding site (Sp1). Specific DNA-protein complexes are indicated by a open (C-REST and N-REST) or filled (DN-REST) arrow. (b) Quantitative RT-PCR of cyclophilin (Cyc), Nppb, and Nppa mRNA levels in H9c2 cells infected with a control adenovirus (Ad) (solid bars), C-REST (hatched bars), or N-REST (shaded bars) relative to those in uninfected control cells (means ± standard errors of the means; n = 3).
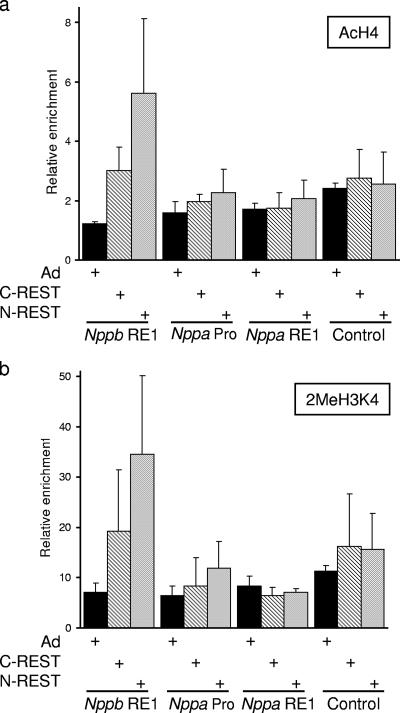
To determine the change in chromatin associated with derepression by C- and N-REST, we examined the levels of AcH4 and dimethyl H3K4 in the presence of endogenous REST, C-REST, or N-REST at the Nppb and Nppa genes. Expression of either C- or N-REST resulted in an increase in AcH4 levels at the Nppb RE1; a greater increase was observed with N-REST (4.6-fold compared with 2.4-fold for C-REST [Fig. 7a]). This suggests that HDAC recruitment via the C terminus is functionally important in regulating Nppb levels. Expression of either C- or N-REST also resulted in increased dimethyl H3K4 levels at the Nppb RE1, and again a greater increase was observed with N-REST (4.9-fold compared with 2.7-fold for C-REST [Fig. 7b]). The C terminus of REST is known to recruit a corepressor complex containing histone demethylase activity (16, 54); thus, loss of this activity would be expected to result in increased dimethyl H3K4 levels. The mSin3 complex recruited by the N-terminal repression domain has not, however, been associated with histone demethylase activity, yet expression of C-REST results in increased dimethyl H3K4 levels (Fig. 7b). It has been shown that the demethylase activity of LSD1 is impaired on acetylated histones and is stimulated by HDAC activity (30). Thus, the increased methylation resulting from the loss of the mSin3 complex may be the result of increased acetylation levels inhibiting REST-recruited demethylase activity. Other possible explanations for this observation are that the mSin3 complex is associated with a previously unidentified histone demethylase activity or that increased histone acetylation resulting from the loss of HDAC activity results in the increased recruitment of an H3K4 histone methylase that antagonizes REST-recruited demethylase activity. These data show that both acetylation and methylation levels of the Nppb gene are affected in response to C- or N-REST, suggesting that the two repression domains are interdependent for effective Nppb repression. We observed no significant changes in acetylation or methylation at the Nppa promoter or RE1 region in response to C- or N-REST (Fig. 7a and b). Such an absence of changes in histone modifications is consistent with our findings that neither of these constructs resulted in derepression of Nppa (Fig. 6).
FIG. 7.
Chromatin changes associated with C-REST and N-REST in H9c2 cells. (a and b) Quantification of Nppb RE1, Nppa promoter (Nppa Pro), Nppa RE1, and control sequences precipitated by anti-acetylated H4 (a) or anti-dimethyl H3K4 (b) in H9c2 cells infected with a control adenovirus (Ad), C-REST, or N-REST (means ± standard errors of the means; n = 3).
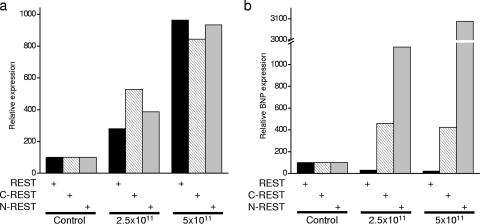
Because we have introduced ectopic REST constructs that are able to interact with corepressor complexes, it is possible that some of our data may be the result of squelching (14, 32). Squelching occurs when high concentrations of a particular domain compete with DNA-bound transcription factors for corepressor or coactivator recruitment. In our experiments, both C- and N-REST have domains that recruit corepressor complexes. If the concentration of free C- or N-REST is high, then it will compete with DNA-bound C- or N-REST for these complexes, thus inhibiting their ability to repress transcription. In order to test this possibility, we titrated ectopic REST, C-REST, and N-REST into H9c2 cells and examined the effects on Nppb expression. Using adenovirus delivery, we were able to express REST, C-REST, and N-REST to comparable levels at both 2.5 × 1011 and 5 × 1011 PFU/ml (Fig. 8a). Expression of REST at either level resulted in further Nppb repression, while C- and N-REST resulted in derepression of Nppb (Fig. 8b). The fact that full-length REST, expressed at levels comparable to those of C- and N-REST, resulted in further repression of Nppb indicates that our data are not the result of squelching. Rather, our data suggest that the functions of the two repression domains of REST are interdependent; they provide the efficient repression of an activated promoter only when working in combination. However, these repression domains provide sufficient function independently to repress the activity of a weak promoter, perhaps working in combination with a second transcriptional repressor.
FIG. 8.
Derepression of Nppb in H9c2 cells is not due to squelching. (a) Quantitative RT-PCR of ectopically expressed REST, C-REST, or N-REST introduced using 2.5 × 1011 or 5 × 1011 PFU/ml of adenovirus. (b) Quantitative RT-PCR of Nppb mRNA in the presence of ectopically expressed REST, C-REST, or N-REST introduced using 2.5 × 1011 or 5 × 1011 PFU/ml of adenovirus.
Chromatin changes in response to a hypertrophic stimulus.
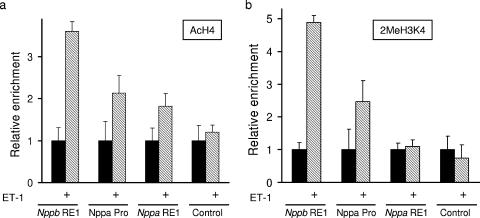
To correlate the changes in chromatin seen with a loss of REST activity in H9c2 cells to changes in chromatin in response to hypertrophic stimuli, we treated primary adult rat ventricular myocytes with ET-1 and examined the levels of H4 acetylation and H3K4 methylation. In the presence of ET-1, the Nppb RE1 and Nppa promoter regions showed increases in H4 acetylation and H3K4 methylation, while no changes in these chromatin marks were observed for the control, non-RE1 region (Fig. 9). Such data are consistent with the changes observed in H9c2 cells in response to DN-REST (Fig. 4b and 5a), suggesting that these chromatin changes are an important component of the gene expression changes in hypertrophy. Together, our data for H9c2 cells and adult rat ventricular myocytes suggest that REST is important for maintaining appropriate Nppb and Nppa gene expression in normal hearts and that the antagonism of REST-recruited chromatin-modifying enzymes is important in the development of cardiac hypertrophy.
FIG. 9.
Chromatin changes in adult rat ventricular myocytes in response to ET-1. (a and b) Quantification of Nppb RE1, Nppa promoter (Nppa Pro), Nppa RE1, and control sequences precipitated by anti-acetylated H4 (a) or anti-dimethyl H3K4 (b) in control (solid bars) and ET-1 treated (hatched bars) rat ventricular myocytes (means ± standard error of the means; n = 4).
DISCUSSION
Here we provide evidence that both repression domains of REST are required to maintain repression of Nppb transcription, while either of these domains is sufficient to retain repression of Nppa transcription, in the ventricular myocyte cell line, H9c2. The differences in the abilities of individual REST repression domains to repress Nppb and Nppa transcription most likely reflect the different set of transcriptional activators and/or repressors recruited to these promoters in H9c2 cells and ventricular myocytes. Studies of Nppb and Nppa gene regulation have identified several transcription factors that are important in controlling their expression (reviewed in references 13, 20, 35, and 51). In addition to an RE1 site, the promoters of both the Nppa and the Nppb gene contain several common transcription factor binding sites, which include GATA and Nkx2.5 binding elements (18, 36, 60). Interactions of GATA-4 and Nkx2.5 are important for both Nppb and Nppa transcription (13, 46, 56), and transcription of both genes is induced by ET-1, possibly via inhibition of REST (27, 28, 40) and stimulation of Ets factors (46). Although the exact mechanism of ET-1 inhibition of REST is not known, it may involve stimulation of calcium/calmodulin-dependent protein kinase and phosphorylation of the class II HDACs, HDAC4 and HDAC5. Once phosphorylated, HDAC4 and HDAC5 are exported from the nucleus, where they are unable to form part of a REST-corepressor complex (40).
In addition to their many similar features, there are also differences between Nppb and Nppa promoters. The Nppb promoter is more similar to promoters expressed in erythroid cells due to the presence of binding sites for the erythroid kruppel-like factor zinc finger proteins and AP1 motifs (36). The Nppa promoter contains several motifs that are also found in other cardiac genes, including the CArG serum response element and several E-boxes that recruit a dHAND/MEF2c complex (55, 66). One potential candidate for an Nppa-specific factor is the transcriptional repressor Jumonji, which is important for repressing Nppa expression in ventricular myocytes (24, 31). The continued presence of Jumonji in the absence of REST would provide one explanation of why the Nppa promoter appears less active than the Nppb promoter in H9c2 cells. Many of the transcription factors that regulate Nppb and Nppa do so in a combinatorial manner. For example, serum response factor activates both Nppb and Nppa transcription, but its function is inhibited by interactions with the homeobox-containing protein, HOP (55). GATA-4 associates with Nkx2.5 to enhance transcription, though this is antagonized by FOG-2 (33), while activation by MEF2 is antagonized by Twist (57). Some of the transcription factors that regulate the Nppb and Nppa promoters (for example, GATA-4) are known to recruit histone acetyltransferase activity, thus providing a direct antagonism of any REST repression mediated by HDAC activity.
Potential roles for the N- and C-terminal repression domains of REST have not been previously studied for cardiac cells; however, different roles have been suggested for these domains in the regulation of RE1-containing genes in other cell types. For example, REST repression of the Scn2a2 promoter in Rat-1 cells has been reported to involve CoREST but not HDAC recruitment (34), while in C6, JTC-19, and HEK293 cells, HDAC recruitment and activity are important (4, 6, 21). REST repression of the Chrm4, Gria2 (GluR2), and Grin1 (NMDAR1) genes also involves HDAC activity (6, 21, 42, 49, 62). Repression of the Stmn2 gene is achieved by HDAC activity in 3T3 NIH cells but requires a combination of HDAC activity and CoREST-recruited DNA methylase in Rat-1 cells (34, 42). In Neuro-2a cells, REST repression of the Chrm4 promoter is mediated via HDAC activity, but repression of the Scn2a2 promoter is not (49). Work with Rat-1 cells led to the proposal that recruitment of the CoREST complex by the C-terminal repression domain is important for long-term gene silencing, while recruitment of the mSin3 complex is important for transient repression (34). Here we show that recruitment of histone demethylase activity by the C-terminal domain does not mediate gene silencing of Nppb or Nppa in H9c2 cells but is also important for transient repression of Nppb transcription.
The changes in chromatin modifications that we see in response to displacement of functional REST occur at the proximal promoter regions (Fig. 4b and 5a). The Nppb RE1 site is only 500 bp from the Nppb transcriptional start site, and because the genomic fragments from our ChIP experiments have an average size of 600 bp, we are not able to resolve these locations. The Nppa RE1 site, however, is located in the 3′ untranslated region of the Nppa gene, 2 kb from the transcription start site (27), allowing us to resolve these locations in our ChIP assay. It has been suggested that REST is able to act over large distances, since RE1 sites have been identified within introns and in regions distal to promoters of genes (7, 23, 34). Our studies provide the first data to show that REST is able to influence the chromatin modifications of promoter regions located some distance from an RE1 site. The level of histone acetylation at the Nppb and Nppa promoters will be defined by combinatorial actions of histone acetyltransferase and HDAC activities recruited by the transcription factors present. Similar levels of AcH4 are present at the promoter regions of Nppb and Nppa in H9c2 cells (Fig. 4b); however, expression of DN-REST results in a greater increase in AcH4 levels at the Nppb promoter (10.4-fold compared with 7.3-fold for Nppa [Fig. 4b]). These data indicate that in H9c2 cells, more histone acetyltransferase activity is recruited to the Nppb than to the Nppa gene. Removal of either REST repression domain results in some increase in AcH4 levels at the Nppb but not the Nppa promoter (Fig. 7a), suggesting that a single HDAC-containing complex is sufficient to antagonize the acetyltransferases present at the Nppa gene but not at the Nppb gene. Perhaps more interestingly, only one of the repression domains of REST, the C-terminal domain, has been associated with histone demethylase activity (22, 54, 65), yet removal of either repression domain results in increased methylated H3K4 at the Nppb promoter (Fig. 7b). Though we cannot rule out the possibility of recruitment of histone demethylase activity as part of the mSin3 complex, the mSin3 complex has been widely studied, and no evidence for associated histone demethylation has been reported. Removal of histone acetylation by HDACs has been shown to be required to stimulate the demethylase activity of LSD1 (30). Thus, increased acetylation levels may impair LSD1 activity, resulting in increased H3K4 methylation levels. Another possibility is that increased histone acetylation seen at the Nppb promoter leads to increased recruitment of a histone methyltransferase protein via a protein containing a bromodomain (which recognizes acetylated lysines). Although such recruitment does not need to be direct, it is interesting that two histone methyltransferase proteins, MLL1 and ASH1, contain bromodomains and that MLL1 methylates H3K4 (37, 41). Furthermore, recruitment of MLL1 has been implicated in antagonizing REST function during neuronal differentiation (63). Increased histone acetylation is also important as a first step in the induction of the beta interferon gene and is required solely for the recruitment of the SWI/SNF complex, which subsequently remodels the local chromatin, resulting in increased transcription (1). However, increased acetylation also results in increased recruitment of REST via the chromatin-remodeling enzyme BRG1, suggesting that increased acetylation may also lead to increased repression (44).
In addition to recruitment of HDAC and histone demethylase activity, REST is able to repress expression by recruiting a histone methyltransferase, G9a, which methylates H3K9 (48, 58). It is unlikely that G9a activity is responsible for continued Nppa repression, since G9a is recruited by the C-terminal domain of REST (48); thus, G9a would not be recruited by our N-REST construct. REST has also been suggested to recruit RNA polymerase C-terminal domain phosphatases, which also act to repress transcription (64). It is not known if these C-terminal domain phosphatases are recruited directly by REST or which part of the REST protein is responsible for their recruitment. It is possible that our C-REST and N-REST proteins are able to recruit these C-terminal domain phosphatases, which are expressed in the heart (64), and their activity may contribute to continued Nppa repression (Fig. 6b). However, given that C-terminal domain phosphorylation of RNA polymerase is a fundamental event in gene transcription, we would expect any recruited C-terminal domain phosphatases to also repress Nppb transcription.
In summary, we provide evidence that both repression domains of REST are required for effective repression of Nppb transcription in H9c2 cells and that this is achieved by a combination of targeted histone deacetylation and histone demethylation. Conversely, either REST domain retains sufficient repression activity to maintain repression of Nppa, which is driven from a less active promoter. These data suggest that the actions of multiple chromatin-modifying complexes are important in maintaining appropriate cardiac gene expression.
Acknowledgments
This work was supported by the British Heart Foundation.
We thank Rory Johnson and Mariusz Mucha for critical reading of the manuscript.
Footnotes
Published ahead of print on 19 March 2007.
REFERENCES
- 1.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. [DOI] [PubMed] [Google Scholar]
- 2.Andres, M. E., C. Burger, M. J. Peral-Rubio, E. Battaglioli, M. E. Anderson, J. Grimes, J. Dallman, N. Ballas, and G. Mandel. 1999. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc. Natl. Acad. Sci. USA 96:9873-9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballas, N., E. Battaglioli, F. Atouf, M. E. Andres, J. Chenoweth, M. E. Anderson, C. Burger, M. Moniwa, J. R. Davie, W. J. Bowers, H. J. Federoff, D. W. Rose, M. G. Rosenfeld, P. Brehm, and G. Mandel. 2001. Regulation of neuronal traits by a novel transcriptional complex. Neuron 31:353-365. [DOI] [PubMed] [Google Scholar]
- 5.Ballas, N., C. Grunseich, D. D. Lu, J. C. Speh, and G. Mandel. 2005. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell 121:645-657. [DOI] [PubMed] [Google Scholar]
- 6.Belyaev, N. D., I. C. Wood, A. W. Bruce, M. Street, J. B. Trinh, and N. J. Buckley. 2004. Distinct RE-1 silencing transcription factor-containing complexes interact with different target genes. J. Biol. Chem. 279:556-561. [DOI] [PubMed] [Google Scholar]
- 7.Bruce, A. W., I. J. Donaldson, I. C. Wood, S. A. Yerbury, M. I. Sadowski, M. Chapman, B. Gottgens, and N. J. Buckley. 2004. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. USA 101:10458-10463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnett, J. C., Jr., P. C. Kao, D. C. Hu, D. W. Heser, D. Heublein, J. P. Granger, T. J. Opgenorth, and G. S. Reeder. 1986. Atrial natriuretic peptide elevation in congestive heart failure in the human. Science 231:1145-1147. [DOI] [PubMed] [Google Scholar]
- 9.Calderone, A., T. Jover, K. M. Noh, H. Tanaka, H. Yokota, Y. Lin, S. Y. Grooms, R. Regis, M. V. Bennett, and R. S. Zukin. 2003. Ischemic insults derepress the gene silencer REST in neurons destined to die. J. Neurosci. 23:2112-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, Z. F., A. J. Paquette, and D. J. Anderson. 1998. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 20:136-142. [DOI] [PubMed] [Google Scholar]
- 11.Cheong, A., A. J. Bingham, J. Li, B. Kumar, P. Sukumar, C. Munsch, N. J. Buckley, C. B. Neylon, K. E. Porter, D. J. Beech, and I. C. Wood. 2005. Downregulated REST transcription factor is a switch enabling critical potassium channel expression and cell proliferation. Mol. Cell 20:45-52. [DOI] [PubMed] [Google Scholar]
- 12.Chong, J. A., J. Tapia-Ramirez, S. Kim, J. J. Toledo-Aral, Y. Zheng, M. C. Boutros, Y. M. Altshuller, M. A. Frohman, S. D. Kraner, and G. Mandel. 1995. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell 80:949-957. [DOI] [PubMed] [Google Scholar]
- 13.Durocher, D., C. Grepin, and M. Nemer. 1998. Regulation of gene expression in the endocrine heart. Recent Prog. Horm. Res. 53:7-23. [PubMed] [Google Scholar]
- 14.Gill, G., and M. Ptashne. 1988. Negative effect of the transcriptional activator GAL4. Nature 334:721-724. [DOI] [PubMed] [Google Scholar]
- 15.Grimes, J. A., S. J. Nielsen, E. Battaglioli, E. A. Miska, J. C. Speh, D. L. Berry, F. Atouf, B. C. Holdener, G. Mandel, and T. Kouzarides. 2000. The co-repressor mSin3A is a functional component of the REST-CoREST repressor complex. J. Biol. Chem. 275:9461-9467. [DOI] [PubMed] [Google Scholar]
- 16.Hakimi, M. A., D. A. Bochar, J. Chenoweth, W. S. Lane, G. Mandel, and R. Shiekhattar. 2002. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. USA 99:7420-7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison, S. M., E. McCall, and M. R. Boyett. 1992. The relationship between contraction and intracellular sodium in rat and guinea-pig ventricular myocytes. J. Physiol. 449:517-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He, Q., M. Mendez, and M. C. LaPointe. 2002. Regulation of the human brain natriuretic peptide gene by GATA-4. Am. J. Physiol. Endocrinol. Metab. 283:E50-E57. [DOI] [PubMed] [Google Scholar]
- 19.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houweling, A. C., M. M. van Borren, A. F. Moorman, and V. M. Christoffels. 2005. Expression and regulation of the atrial natriuretic factor encoding gene Nppa during development and disease. Cardiovasc. Res. 67:583-593. [DOI] [PubMed] [Google Scholar]
- 21.Huang, Y., S. J. Myers, and R. Dingledine. 1999. Transcriptional repression by REST: recruitment of Sin3A and histone deacetylase to neuronal genes. Nat. Neurosci. 2:867-872. [DOI] [PubMed] [Google Scholar]
- 22.Humphrey, G. W., Y. Wang, V. R. Russanova, T. Hirai, J. Qin, Y. Nakatani, and B. H. Howard. 2001. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem. 276:6817-6824. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, R., R. J. Gamblin, L. Ooi, A. W. Bruce, I. J. Donaldson, D. R. Westhead, I. C. Wood, R. M. Jackson, and N. J. Buckley. 2006. Identification of the REST regulon reveals extensive transposable element-mediated binding site duplication. Nucleic Acids Res. 34:3862-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, T. G., J. Chen, J. Sadoshima, and Y. Lee. 2004. Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors. Mol. Cell. Biol. 24:10151-10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimes, B. W., and B. L. Brandt. 1976. Properties of a clonal muscle cell line from rat heart. Exp. Cell Res. 98:367-381. [DOI] [PubMed] [Google Scholar]
- 26.Kuratomi, S., A. Kuratomi, K. Kuwahara, T. M. Ishii, K. Nakao, Y. Saito, and M. Takano. 2007. NRSF regulates the developmental and hypertrophic changes of HCN4 transcription in rat cardiac myocytes. Biochem. Biophys. Res. Commun. 353:67-73. [DOI] [PubMed] [Google Scholar]
- 27.Kuwahara, K., Y. Saito, E. Ogawa, N. Takahashi, Y. Nakagawa, Y. Naruse, M. Harada, I. Hamanaka, T. Izumi, Y. Miyamoto, I. Kishimoto, R. Kawakami, M. Nakanishi, N. Mori, and K. Nakao. 2001. The neuron-restrictive silencer element-neuron-restrictive silencer factor system regulates basal and endothelin 1-inducible atrial natriuretic peptide gene expression in ventricular myocytes. Mol. Cell. Biol. 21:2085-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuwahara, K., Y. Saito, M. Takano, Y. Arai, S. Yasuno, Y. Nakagawa, N. Takahashi, Y. Adachi, G. Takemura, M. Horie, Y. Miyamoto, T. Morisaki, S. Kuratomi, A. Noma, H. Fujiwara, Y. Yoshimasa, H. Kinoshita, R. Kawakami, I. Kishimoto, M. Nakanishi, S. Usami, M. Harada, and K. Nakao. 2003. NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. EMBO J. 22:6310-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langenickel, T., I. Pagel, K. Hohnel, R. Dietz, and R. Willenbrock. 2000. Differential regulation of cardiac ANP and BNP mRNA in different stages of experimental heart failure. Am. J. Physiol. Heart Circ. Physiol. 278:H1500-H1506. [DOI] [PubMed] [Google Scholar]
- 30.Lee, M. G., C. Wynder, D. A. Bochar, M. A. Hakimi, N. Cooch, and R. Shiekhattar. 2006. Functional interplay between histone demethylase and deacetylase enzymes. Mol. Cell. Biol. 26:6395-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee, Y., A. J. Song, R. Baker, B. Micales, S. J. Conway, and G. E. Lyons. 2000. Jumonji, a nuclear protein that is necessary for normal heart development. Circ. Res. 86:932-938. [DOI] [PubMed] [Google Scholar]
- 32.Leichter, M., and G. Thiel. 1999. Transcriptional repression by the zinc finger protein REST is mediated by titratable nuclear factors. Eur. J. Neurosci. 11:1937-1946. [DOI] [PubMed] [Google Scholar]
- 33.Lu, J. R., T. A. McKinsey, H. Xu, D. Z. Wang, J. A. Richardson, and E. N. Olson. 1999. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol. Cell. Biol. 19:4495-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lunyak, V. V., R. Burgess, G. G. Prefontaine, C. Nelson, S. H. Sze, J. Chenoweth, P. Schwartz, P. A. Pevzner, C. Glass, G. Mandel, and M. G. Rosenfeld. 2002. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science 298:1747-1752. [DOI] [PubMed] [Google Scholar]
- 35.Ma, K. K., K. Banas, and A. J. de Bold. 2005. Determinants of inducible brain natriuretic peptide promoter activity. Regul. Pept. 128:169-176. [DOI] [PubMed] [Google Scholar]
- 36.McBride, K., and M. Nemer. 2001. Regulation of the ANF and BNP promoters by GATA factors: lessons learned for cardiac transcription. Can. J. Physiol. Pharmacol. 79:673-681. [PubMed] [Google Scholar]
- 37.Milne, T. A., S. D. Briggs, H. W. Brock, M. E. Martin, D. Gibbs, C. D. Allis, and J. L. Hess. 2002. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10:1107-1117. [DOI] [PubMed] [Google Scholar]
- 38.Mukoyama, M., K. Nakao, K. Obata, M. Jougasaki, M. Yoshimura, E. Morita, K. Hosoda, S. Suga, Y. Ogawa, H. Yasue, et al. 1991. Augmented secretion of brain natriuretic peptide in acute myocardial infarction. Biochem. Biophys. Res. Commun. 180:431-436. [DOI] [PubMed] [Google Scholar]
- 39.Nadeau, H., and H. A. Lester. 2002. NRSF causes cAMP-sensitive suppression of sodium current in cultured hippocampal neurons. J. Neurophysiol. 88:409-421. [DOI] [PubMed] [Google Scholar]
- 40.Nakagawa, Y., K. Kuwahara, M. Harada, N. Takahashi, S. Yasuno, Y. Adachi, R. Kawakami, M. Nakanishi, K. Tanimoto, S. Usami, H. Kinoshita, Y. Saito, and K. Nakao. 2006. Class II HDACs mediate CaMK-dependent signaling to NRSF in ventricular myocytes. J. Mol. Cell. Cardiol. 41:1010-1022. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura, T., T. Mori, S. Tada, W. Krajewski, T. Rozovskaia, R. Wassell, G. Dubois, A. Mazo, C. M. Croce, and E. Canaani. 2002. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol. Cell 10:1119-1128. [DOI] [PubMed] [Google Scholar]
- 42.Naruse, Y., T. Aoki, T. Kojima, and N. Mori. 1999. Neural restrictive silencer factor recruits mSin3 and histone deacetylase complex to repress neuron-specific target genes. Proc. Natl. Acad. Sci. USA 96:13691-13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogawa, E., Y. Saito, K. Kuwahara, M. Harada, Y. Miyamoto, I. Hamanaka, N. Kajiyama, N. Takahashi, T. Izumi, R. Kawakami, I. Kishimoto, Y. Naruse, N. Mori, and K. Nakao. 2002. Fibronectin signaling stimulates BNP gene transcription by inhibiting neuron-restrictive silencer element-dependent repression. Cardiovasc. Res. 53:451-459. [DOI] [PubMed] [Google Scholar]
- 44.Ooi, L., N. D. Belyaev, K. Miyake, I. C. Wood, and N. J. Buckley. 2006. BRG1 chromatin remodeling activity is required for efficient chromatin binding by repressor element 1-silencing transcription factor (REST) and facilitates REST-mediated repression. J. Biol. Chem. 281:38974-38980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paquette, A. J., S. E. Perez, and D. J. Anderson. 2000. Constitutive expression of the neuron-restrictive silencer factor (NRSF)/REST in differentiating neurons disrupts neuronal gene expression and causes axon pathfinding errors in vivo. Proc. Natl. Acad. Sci. USA 97:12318-12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pikkarainen, S., H. Tokola, R. Kerkela, T. Majalahti-Palviainen, O. Vuolteenaho, and H. Ruskoaho. 2003. Endothelin-1-specific activation of B-type natriuretic peptide gene via p38 mitogen-activated protein kinase and nuclear ETS factors. J. Biol. Chem. 278:3969-3975. [DOI] [PubMed] [Google Scholar]
- 47.Rivard, A., N. Principe, and V. Andres. 2000. Age-dependent increase in c-fos activity and cyclin A expression in vascular smooth muscle cells. A potential link between aging, smooth muscle cell proliferation and atherosclerosis. Cardiovasc. Res. 45:1026-1034. [DOI] [PubMed] [Google Scholar]
- 48.Roopra, A., R. Qazi, B. Schoenike, T. J. Daley, and J. F. Morrison. 2004. Localized domains of g9a-mediated histone methylation are required for silencing of neuronal genes. Mol. Cell 14:727-738. [DOI] [PubMed] [Google Scholar]
- 49.Roopra, A., L. Sharling, I. C. Wood, T. Briggs, U. Bachfischer, A. J. Paquette, and N. J. Buckley. 2000. Transcriptional repression by neuron-restrictive silencer factor is mediated via the Sin3-histone deacetylase complex. Mol. Cell. Biol. 20:2147-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosati, B., F. Grau, and D. McKinnon. 2006. Regional variation in mRNA transcript abundance within the ventricular wall. J. Mol. Cell. Cardiol. 40:295-302. [DOI] [PubMed] [Google Scholar]
- 51.Rosenzweig, A., T. D. Halazonetis, J. G. Seidman, and C. E. Seidman. 1991. Proximal regulatory domains of rat atrial natriuretic factor gene. Circulation 84:1256-1265. [DOI] [PubMed] [Google Scholar]
- 52.Saito, Y., K. Nakao, H. Arai, K. Nishimura, K. Okumura, K. Obata, G. Takemura, H. Fujiwara, A. Sugawara, T. Yamada, et al. 1989. Augmented expression of atrial natriuretic polypeptide gene in ventricle of human failing heart. J. Clin. Investig. 83:298-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schoenherr, C. J., and D. J. Anderson. 1995. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science 267:1360-1363. [DOI] [PubMed] [Google Scholar]
- 54.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero, and Y. Shi. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 55.Shin, C. H., Z. P. Liu, R. Passier, C. L. Zhang, D. Z. Wang, T. M. Harris, H. Yamagishi, J. A. Richardson, G. Childs, and E. N. Olson. 2002. Modulation of cardiac growth and development by HOP, an unusual homeodomain protein. Cell 110:725-735. [DOI] [PubMed] [Google Scholar]
- 56.Shiojima, I., I. Komuro, T. Oka, Y. Hiroi, T. Mizuno, E. Takimoto, K. Monzen, R. Aikawa, H. Akazawa, T. Yamazaki, S. Kudoh, and Y. Yazaki. 1999. Context-dependent transcriptional cooperation mediated by cardiac transcription factors Csx/Nkx-2.5 and GATA-4. J. Biol. Chem. 274:8231-8239. [DOI] [PubMed] [Google Scholar]
- 57.Spicer, D. B., J. Rhee, W. L. Cheung, and A. B. Lassar. 1996. Inhibition of myogenic bHLH and MEF2 transcription factors by the bHLH protein Twist. Science 272:1476-1480. [DOI] [PubMed] [Google Scholar]
- 58.Tachibana, M., K. Sugimoto, T. Fukushima, and Y. Shinkai. 2001. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 276:25309-25317. [DOI] [PubMed] [Google Scholar]
- 59.Tapia-Ramirez, J., B. J. Eggen, M. J. Peral-Rubio, J. J. Toledo-Aral, and G. Mandel. 1997. A single zinc finger motif in the silencing factor REST represses the neural-specific type II sodium channel promoter. Proc. Natl. Acad. Sci. USA 94:1177-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thuerauf, D. J., D. S. Hanford, and C. C. Glembotski. 1994. Regulation of rat brain natriuretic peptide transcription. A potential role for GATA-related transcription factors in myocardial cell gene expression. J. Biol. Chem. 269:17772-17775. [PubMed] [Google Scholar]
- 61.Wei, C. M., P. C. Kao, J. T. Lin, D. M. Heublein, H. V. Schaff, and J. C. Burnett, Jr. 1993. Circulating beta-atrial natriuretic factor in congestive heart failure in humans. Circulation 88:1016-1020. [DOI] [PubMed] [Google Scholar]
- 62.Wood, I. C., N. D. Belyaev, A. W. Bruce, C. Jones, M. Mistry, A. Roopra, and N. J. Buckley. 2003. Interaction of the repressor element 1-silencing transcription factor (REST) with target genes. J. Mol. Biol. 334:863-874. [DOI] [PubMed] [Google Scholar]
- 63.Wynder, C., M. A. Hakimi, J. A. Epstein, A. Shilatifard, and R. Shiekhattar. 2005. Recruitment of MLL by HMG-domain protein iBRAF promotes neural differentiation. Nat. Cell Biol. 7:1113-1117. [DOI] [PubMed] [Google Scholar]
- 64.Yeo, M., S. K. Lee, B. Lee, E. C. Ruiz, S. L. Pfaff, and G. N. Gill. 2005. Small CTD phosphatases function in silencing neuronal gene expression. Science 307:596-600. [DOI] [PubMed] [Google Scholar]
- 65.You, A., J. K. Tong, C. M. Grozinger, and S. L. Schreiber. 2001. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 98:1454-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zang, M. X., Y. Li, L. X. Xue, H. T. Jia, and H. Jing. 2004. Cooperative activation of atrial naturetic peptide promoter by dHAND and MEF2C. J. Cell. Biochem. 93:1255-1266. [DOI] [PubMed] [Google Scholar]
- 67.Zuccato, C., M. Tartari, A. Crotti, D. Goffredo, M. Valenza, L. Conti, T. Cataudella, B. R. Leavitt, M. R. Hayden, T. Timmusk, D. Rigamonti, and E. Cattaneo. 2003. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 35:76-83. [DOI] [PubMed] [Google Scholar]