Abstract
One outcome of T-cell receptor (TCR) signaling is increased affinity and avidity of integrins for their ligands. This occurs through a process known as inside-out signaling, which has been shown to require several molecular components including the adapter proteins ADAP (adhesion and degranulation-promoting adapter protein) and SKAP-55 (55-kDa src kinase-associated phosphoprotein) and the small GTPase Rap1. Herein, we provide evidence linking ADAP and SKAP-55 to RIAM, a recently described adapter protein that binds selectively to active Rap1. We identified RIAM as a key component linking the ADAP/SKAP-55 module to the small GTPase Rap1, facilitating TCR-mediated integrin activation. We show that RIAM constitutively interacts with SKAP-55 in both a heterologous transfection system and primary T cells and map the region essential for this interaction. Additionally, we find that the SKAP-55/RIAM complex is essential both for TCR-mediated adhesion and for efficient conjugate formation between T cells and antigen-presenting cells. Mechanistic studies revealed that the ADAP/SKAP-55 module relocalized RIAM and Rap1 to the plasma membrane following TCR activation to facilitate integrin activation. These results describe for the first time a link between ADAP/SKAP-55 and the Rap1/RIAM complex and provide a potential new mechanism for TCR-mediated integrin activation.
Integrins are cell surface receptors that mediate cell-cell interactions and cell-matrix interactions. These receptors are critical for T-cell migration to peripheral lymph nodes and inflammatory sites and are necessary for productive interactions between T cells and antigen-presenting cells (APCs) (22). The major integrins expressed on T cells are the β2 integrin LFA-1 (lymphocyte function-associated antigen 1; αLβ2) and the β1 family of integrins (very late antigen [VLA]; α4β1, α5β1, and α6β1). LFA-1 binds to intercellular adhesion molecule 1 (ICAM-1), ICAM-2, and ICAM-3, whereas the β1 family binds to vascular cell adhesion molecule 1 or the extracellular matrix protein fibronectin (12, 36). On resting T cells, integrins are maintained in an inactive state and adopt a low-affinity conformation for their ligands. T-cell receptor (TCR) engagement by a peptide-major histocompatibility complex (MHC) complex induces integrin clustering as well as a conformational change leading to a higher-affinity state, thereby markedly enhancing the avidity of integrins for their ligands (1, 22, 32). This activation-induced modulation of integrin activation via antigen receptors (or other cell surface receptors) occurs through a process known as inside-out signaling (9, 22).
The molecular basis for inside-out signaling remains poorly understood; however, several proteins in the TCR signaling pathway are known to be involved (22, 39). Following engagement of the TCR and activation of src and syk family protein tyrosine kinases, the transmembrane adapter LAT (linker for activation of T cells) and the cytosolic adapter protein SLP-76 (76-kDa src homology 2 domain-containing leukocyte phosphoprotein) form a scaffold to create a critical signaling complex (25, 42). Elements of this complex include ADAP (adhesion- and degranulation-promoting adapter protein) and SKAP-55 (55-kDa src kinase-associated phosphoprotein), two adapter proteins constitutively associated through the interaction of the SH3 (src homology 3) domain of SKAP-55 with the proline-rich domain of ADAP (29, 30). The phosphorylation of ADAP through the src kinase Fyn (3), following TCR stimulation, allows ADAP to interact with the SH2 domain of SLP-76 and recruits the ADAP/SKAP-55 complex to the plasma membrane via the SLP-76/GADS/LAT module (8, 33). T cells from ADAP-deficient mice show impaired LFA-1- and VLA-4-mediated adhesion, whereas the knockdown of SKAP-55 by small interfering RNA (siRNA) decreases T-cell-APC conjugation and TCR-mediated activation of LFA-1 and VLA-4 (10, 15, 23, 35).
Another molecule essential for inside-out signaling from the TCR to integrins is the GTPase Rap1 (4). Like other low-molecular-weight GTP-binding proteins, Rap1 cycles between an inactive GDP-bound form and an active form associated with GTP. Activation of Rap1 has been shown to regulate T-cell adhesion and spreading. In T cells, constitutively active Rap1 increases adhesion to integrin ligands, including ICAM-1 and fibronectin, whereas a dominant-negative Rap1 mutant blocks TCR-induced integrin activation (16, 17, 37). The mechanism for how Rap1 regulates integrin activation is not completely understood; however, recent studies have identified several effectors that bind active Rap1, including RAPL (regulator for cell adhesion and polarization enriched in lymphoid tissues), PKD1 (protein kinase D1), and RIAM (Rap1-GTP interacting adapter molecule), which appear essential for linking Rap1 to the integrins. RAPL and PKD1 are known to regulate the subcellular localization of Rap1 at the plasma membrane via their associations with the αL-integrin subunit of LFA-1 (RAPL) and the β1-integrin subunit of VLA-4 (PKD1) (19, 31). RIAM has been shown to regulate VLA-4 integrin (α4β1) and LFA-1 integrin (αLβ2) activation in Jurkat T cells through its association with Rap1 upon TCR ligation (27). Additionally, RIAM has been linked to the regulation of actin dynamics in T cells through its interaction with Ena/VASP (vasodilator-stimulated phosphoprotein) family proteins and profilin (14, 27). Recently, RIAM has also been shown to regulate one of the major platelet integrins, αIIbβ3, through talin-mediated integrin activation (11).
Although the critical role of Rap1 in TCR-induced integrin activation is now appreciated, exactly how TCR stimulation links to Rap1 activation remains unclear. One clue to this process followed from the observation that the interaction between ADAP and SKAP-55 is critical for optimal TCR-mediated integrin activation. We have recently found that the ADAP/SKAP-55 module is required for the recruitment of Rap1 to the plasma membrane (23). How the ADAP/SKAP-55 complex is linked to Rap1 is the subject of this study. In this report we describe, for the first time, an interaction among ADAP, SKAP-55, and RIAM and demonstrate that this larger complex is critical for efficient translocation of active Rap1 to the membrane, facilitating TCR-mediated adhesion and conjugate formation between T cells and APCs.
MATERIALS AND METHODS
Constructs.
(i) RIAM cloning.
A cDNA encoding a full-length open reading frame of RIAM was obtained by reverse transcription-PCR (RT-PCR), using the SuperScript One-Step RT-PCR System with Platinum Taq (Invitrogen) from total RNA of murine T-cell blasts. The following primers were used: EcoRV-EcoRI-RIAM-Met-sense, 5′-CCGGATATCGCGAATTCTGGTCGACGTCAAGATGGGTGAATCAAATGAA-3′, and BamHI-RIAM-stop-antisense, 5′-CGCGGATCCAGAGCATCCCAAATACTAAC-3′. The digested PCR product was cloned into pFlag-CMV-4 (Sigma) and pEGFP-C3 (Clontech).
(ii) RIAM fragment cloning.
The murine RIAM domains were described previously (14). We designed our RIAM fragments to include the entirety of the appropriate domains and therefore included a small number of amino acids both amino and carboxyl terminal to the regions defined by Jenzora et al. (14). For ease of description, however, we designated these domains as the “RIAM Ras association (RA) domain” and “RIAM pleckstrin homology (PH) domain,” recognizing that each domain is extended slightly. These RIAM fragment domains were amplified by PCR using full-length RIAM as a template. The following primers were used: EcoRI-ProI-sense, 5′-TTGAATTCTATGGGTGAATCAAATGAAGA-3′; BamHI-ProI-antisense, 5′-GGATCCCTAGGCTTCTTCTTCTTCTTTA-3′; EcoRI-RA-PH-sense, 5′-TTGAATTCTAAAGAAGAAGAAGAAGCCAAAG-3′; BamHI-RA-PH-antisense, 5′-GGATCCCTATCCAGCTCTTGCCACAG-3′; EcoRI-RA-antisense, 5′-GGATCCCTATAAGTATTCTTTGTTTTTAGCGTTC-3′; EcoRI-PH-sense, 5′-TTGAATTCTATGGAAAGCTTCTGCGGCACATCCA-3′; EcoRI-ProII-sense, 5′-TTGAATTCTGCTGTGGCAAGAGCTGG-3′; and BamHI-ProII-antisense, 5′-GGATCCCTAGGGTATATTGCCTCTTTTC-3′. The PCR products were digested with EcoRI and BamHI and cloned into pFlag-CMV-4 and pEGFP-C1 vectors (Sigma and Clontech), respectively.
(iii) ADAP cloning.
A cDNA encoding a full-length open reading frame of ADAP (120 kDa) was obtained by RT-PCR from total RNA of murine T-cell blasts. The following primers were used: KpnI-ADAP-Met-sense, 5′-CGGTACCCCTCATGGCGAAGTTCAACACG-3′, and XhoI-ADAP-stop-antisense, 5′-CTCGAGCAGTGGAATGAACAGAGCAG-3′. The digested PCR product was cloned into pcDNA3.1 His (Invitrogen). Human cloning of Flag-tagged ADAP cDNA into pEF-BOS expression vector has been described previously (23).
(iv) SKAP-55 cloning.
Cloning of full-length SKAP-55 fused with either a hemagglutinin (HA) or a Flag tag into the pEF-BOS vector has been reported previously (30).
(v) SLP-76 cloning.
Cloning of full-length Slp-76 fused with a Flag tag into the pEF-BOS vector has been reported previously (43).
(vi) Rap1 mutant.
The cDNA of the Rap1Q63E mutant was kindly provided by Alfred Wittinghofer (Max-Planck-Institut für Molekulare Physiologie, Dortmund, Germany) and was used as a template to be reamplified with the following primers: EcoRI-Q63E(Rap1)-sense, 5′-GTAATCTTGGAATTCTGATGCGTGAGTACAAGCTAGTGGTC-3′, and BamHI-Q63E(Rap1)-antisense, 5′-GTGGATCCCTCGAGAGCTGCTGCTGACTATGGG-3′. The cDNA was subsequently cloned into the pEGFP-C3 vector (Clontech). pGEX RalGDS-RBD was kindly provided by Johannes L. Bos (Utrecht Medical Center, Utrecht, The Netherlands). The constructs for pDsRedC1-tagged Rap1G12V were kindly provided by Ignacio Rubio (University of Jena, Jena, Germany).
(vii) siRNA of SKAP-55.
For siRNA of SKAP-55, the oligonucleotides SK4 (5′-GAAAGAATCCTGCTTTGAA-3′) and SK1 (5′-GCGATTAGAGATCATACTA-3′) cloned into the pSuper vector (OligoEngine) were used (23). The same oligonucleotides, SK4 and SK1, were also cloned into pCMS3-EGFP vector (23).
Cell culture and transfection.
The Jurkat (JE6.1; ATCC) T-cell line and the Raji B-cell line were cultured in RPMI 1640 (Gibco BRL) supplemented with 10% fetal calf serum (FCS) at 37°C with 5% CO2. For immunofluorescence, Jurkat T cells were electroporated with 20 μg of plasmid DNA using the BTX electroporator (310 V and 10 ms). For adhesion assays, Jurkat cells were electroporated using 30 μg of plasmid DNA, as previously described (23). For siRNA of SKAP-55, Jurkat T cells were transfected with 40 μg of the indicated vector and cultured for 48 h before use. The 293T cell line was cultured in Dulbecco modified Eagle medium (Gibco BRL) supplemented with 10% FCS and transfected using the liposomal transfection reagent Lipofectamine (Invitrogen). Primary human T cells were prepared from a healthy donor by standard separation techniques using AutoMACS (Miltenyi Biotech). Total mouse splenocytes were stimulated with phorbol myristate acetate (10−7 M)-ionomycin (10−6 M) (Sigma) and cultured in RPMI supplemented with 10% FCS and 0.01 M β-mercaptoethanol for 6 days in the presence of interleukin-2 (40 U/ml) to generate T-cell blasts.
Genetically altered mice.
Mice made deficient in ADAP by gene targeting have been described previously (35). Splenic T cells were prepared from wild-type or ADAP-deficient animals by standard separation techniques using AutoMACS (Miltenyi Biotech) per the manufacturer's instructions.
Immunoprecipitation and Western blot analysis.
Flag-RIAM or fragments and HA/Flag-SKAP-55, His/Flag-ADAP, or GFP (green fluorescent protein)/HA/myc-Rap1 and its mutants were coexpressed in 293T cells. One day after transfection, cells were lysed in IPP150 lysis buffer (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.1% NP-40 containing 2.5 mM MgCl2, when Rap1 or mutants were coexpressed) supplemented with one protease inhibitor cocktail tablet (EDTA free; Roche). The lysate was centrifuged at 25,000 × g for 10 min, and the supernatant was rotated for 2 h at 4°C with anti-Flag M2 affinity gel (Sigma) or anti-HA (Roche). For the HA immunoprecipitation, protein G-agarose beads (Santa Cruz) were added for 1 h. The beads were then washed and recovered in sample buffer and loaded onto a 4 to 15% gradient sodium dodecyl sulfate (SDS)-polyacrylamide gel (Bio-Rad). Anti-M2 (Sigma), anti-GFP (full length; Clontech), and anti-HA (Roche) were used to develop the Western blots. For immunoprecipitation of endogenous proteins, murine T-cell blasts (1.5 × 107) were rested in phosphate-buffered saline (PBS) or stimulated for 10 min with 500A2 (anti-CD3; BD Pharmingen) or with 2.5 mM pervanadate (PV) for 3 min. Cells were lysed in IPP150 containing 2.5 mM MgCl2, supplemented with a protease inhibitor cocktail tablet (EDTA free; Roche). Jurkat T cells (1.0 × 107) or primary human T cells (2.0 × 107) were left untreated or stimulated for 5 min with monoclonal antibody (MAb) C305 (41) (provided by Arthur Weiss, University of California, San Francisco, San Francisco, CA) or MEM92 (provided by Vaclav Horejsi, Academy of Sciences of the Czech Republic, Prague, Czech Republic) and lysed as described above. The protein concentration of the lysates was determined using the Bradford assay (Roth), and a total of 500 μg protein was used for immunoprecipitation studies. Either the polyclonal rabbit antiserum to RIAM (14) or the sheep antiserum to ADAP (33) was used for immunoprecipitation. Immune complexes were analyzed as described above using anti-SKAP-55 (BD Pharmingen), anti-SKAP-55 MAb (23), anti-RIAM (14), anti-ADAP (Upstate), anti-ADAP (33), or anti-Rap1 (Santa Cruz) for Western blot analysis. For proper stimulation of T cells, lysates were also analyzed for phospho-extracellular signal-regulated kinase 1/2 (phospho-ERK1/2) and ERK1/2 (Cell Signaling).
Immunofluorescence, confocal microscopy, and quantification.
Transfected Jurkat T cells were plated onto glass coverslips coated with poly-l-lysine (Sigma) for 45 min at room temperature or for 20 min at 37°C with Raji cells preincubated with 0.5 μg/ml of staphylococcal enterotoxin E (SEE; Toxin Technologies) as well as loaded with the cell tracker Blue CMAC to generate conjugates. Cells were fixed in 3.7% paraformaldehyde, washed in 50 mM NH4Cl, and stained for phalloidin or HA-SKAP-55 and mounted in a medium containing N-propyl gallate (Sigma). Phalloidin-Alexa Fluor 594 and 488, rabbit anti-rat immunoglobulin G (IgG)-Alexa Fluor 594, and the cell tracker Blue CMAC were obtained from Molecular Probes. Cells were analyzed using a Nikon Eclipse E800 microscope. Image analysis was performed using IPLab version 3.9.3 r4 and Adobe Photoshop 6.0 software. For each study, more than 30 transfected cells were analyzed, and each experiment was repeated at least three times. To examine the effect of SKAP-55 on Rap1 localization, Jurkat T cells were transfected with either pCMS3E-C (scrambled) or pCMS3E-SK (an siRNA against SKAP-55) in combination with pDsRed-tagged RapG12V. After 48 h, resting or stimulated (5 min with anti-TCR MAb C305) Jurkat T cells were seeded onto poly-l-lysine-coated slides, fixed with 3.5% paraformaldehyde in PBS, and mounted with Mowiol (Calbiochem). Cells were imaged with a Leica TCS SP2 laser scanning confocal system and analyzed using Leica software. Corel Photopaint histogram tools were used to obtain the number of red pixels per cell of Rap1G12V (pDsRedC1 Rap1G12V) colocalized at the plasma membrane area compared to the total amount of pDsRedC1 Rap1G12V (Rap1 pixels) within the cell.
Purified T cells (1 × 107 cells) from wild-type or ADAP-knockout mice were preincubated with biotinylated CD3 MAb (2C11; BD Pharmingen) for 30 min at 4°C and cross-linked with streptavidin (Dianova) for 3 min at 37°C. T cells were seeded onto poly-l-lysine-coated slides, fixed with 3.5% paraformaldehyde in PBS, permeabilized with 0.3% Triton X-100, and blocked with 5% horse serum in PBS. Endogenous RIAM was detected using a polyclonal rabbit antiserum to RIAM (14) and visualized with goat anti-rabbit antibody-fluorescein isothiocyanate and tetramethyl rhodamine isocyanate-phalloidin (Sigma). Cells were imaged with a Leica TCS SP2 laser scanning confocal system and analyzed using Leica software.
Flow cytometry, TCR-mediated CD69 expression, and calcium mobilization.
For flow cytometry analysis of the β2 subunit of LFA-1 (MEM48), the β1 subunit of VLA-4 (MEM101A), CD3 (OKT3; ATCC), or MHC class I (W6/32; ATCC), MAbs in combination with Cy5-conjugated anti-mouse IgG (Dianova) were used, and samples were analyzed using a FACSCalibur flow cytometer and CellquestPro software (BD Bioscience). MEM48 and MEM101A were provided by Vaclav Horejsi (Academy of Sciences of the Czech Republic, Prague, Czech Republic). The measurement of CD69 upregulation and calcium mobilization have been described elsewhere (38). For TCR-induced CD69 expression, cells were cultured for 18 h in wells containing RPMI supplemented with 10% FCS alone (medium) or medium containing 20-ng/ml phorbol myristate acetate or in wells that had earlier been coated with the anti-TCR MAb C305. After overnight culture, cells were stained with an anti-CD69 antibody (BD Pharmingen) in combination with Cy5-conjugated anti-mouse IgG, and surface expression was analyzed by fluorescence-activated cell sorting (FACS). For calcium mobilization, cells were loaded with Indo-1 AM (Molecular Probes) at 37°C for 45 min in RPMI 1640 without phenol red (Gibco BRL) supplemented with 10% FCS. Cells were washed and reincubated for an additional 30 min at 37°C in the same RPMI medium. Baseline calcium levels were measured for 30 s; cells were stimulated with the C305 antibody followed by ionomycin (1 μg/ml) treatment. Data for calcium mobilization were acquired on a FACSort LSR flow cytometer (BD Bioscience), and ratiometric analysis was performed with Flow Jo software.
Adhesion assay.
Adhesion assays were performed as previously described (23, 24, 34). Briefly, Jurkat T cells (1 × 106 cells per dish) were incubated with OKT3 for 30 min at 37°C prior to the adhesion on fibronectin- or ICAM-1-coated dishes. The bound cell fraction was determined by counting four independent fields by microscopy using an ocular counting reticule. The induction (n-fold) of cell binding to fibronectin was calculated as a ratio of the mean of stimulated to the mean of unstimulated vector-transfected cells for each experiment. The expression of GFP or individual GFP fusion proteins of RIAM was analyzed by Western blotting or flow cytometric analysis.
Rap1 activity assay.
Rap1 activity was assessed as previously described (23, 24, 34). Briefly, Jurkat T cells (5 × 106 cells) were either left untreated or stimulated for 5 min with the anti-TCR MAb C305 and then lysed. Activated Rap1 was isolated using a glutathione S-transferase-RalGDS-Rap1 binding fusion protein (5), and bound Rap1 was quantified by Western blotting using anti-Rap1. The expression of GFP or individual GFP fusion proteins of RIAM was analyzed by Western blot analysis.
Conjugate assay.
Raji B cells were pulsed for 16 h with 2-μg/ml SEE or left untreated and stained with 0.5 μM of the red cell-labeling agent DDAO-SE (Molecular Probes). Jurkat T cells were incubated with an equal number of B cells for 30 min at 37°C. Nonspecific aggregates were disrupted by vortexing, and then cells were fixed by adding 3.5% paraformaldehyde and analyzed using a FACSCalibur flow cytometer (BD Bioscience). The percent conjugation was defined as the number of double-positive events in the upper right quadrant divided by the total number of GFP-expressing cells multiplied by 100.
Isolation of plasma membrane fractions.
Isolation of cytosolic and plasma membrane fractions has been described previously (23, 24, 34). Briefly, either resting or stimulated (MAb C305 for 5 min) Jurkat T cells (2 × 107) or purified splenic T cells (6 × 107) from mice (which for stimulation were preincubated with biotinylated CD3 MAb [2C11; BD Pharmingen] for 30 min at 4°C and cross-linked with streptavidin [Dianova] for 3 min at 37°C) were washed in RPMI and resuspended on ice in a hypotonic buffer. Cells were sheared; nuclei and unbroken cells were removed by low-speed centrifugation. The remaining supernatant was recentrifuged, and the cytosolic fraction (supernatant) was collected. The remaining pellet (membrane fraction) was washed twice with hypotonic buffer (washing fraction) and finally resuspended on ice in lysis buffer as described above. To ensure equal loading, the protein concentrations of the cytosolic and membrane fractions were determined by the Bradford assay. Western blot analysis revealed that the membrane fraction was strongly enriched for the plasma membrane markers Na+/K+ ATPase and LAT (an integral membrane protein) and contained only minor amounts of EEA1-positive membranes (EEA1 is an endosomal marker), whereas most of the EEA1-positive membranes were found in the washing fraction (data not shown).
RESULTS
RIAM interacts and colocalizes with SKAP-55.
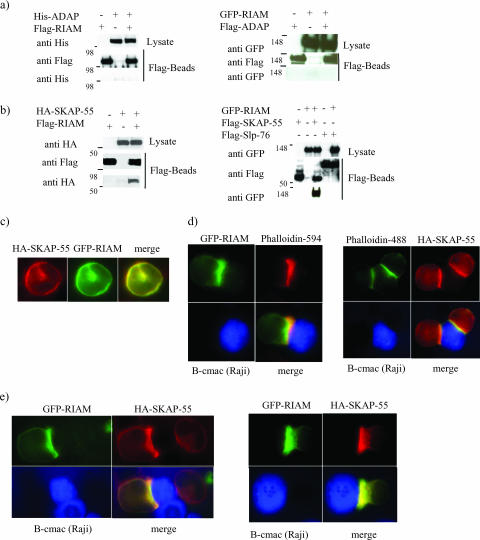
Both ADAP and RIAM have been shown to interact with the actin-binding proteins VASP and profilin, and both play critical roles in TCR-mediated integrin activation. Therefore, we speculated that the two adapter proteins may function in overlapping pathways to regulate integrin function in T cells. To address this notion, we tested if RIAM associates directly with ADAP. 293T cells were cotransfected with His-tagged ADAP and Flag-tagged RIAM; however, anti-Flag immunoprecipitates from the transfectants failed to show an association between the two proteins (Fig. 1a, left). To confirm this finding, we cotransfected 293T cells with Flag-ADAP and GFP-RIAM and again performed anti-Flag immunoprecipitation. As shown (Fig. 1a, right), we again found no evidence of association between the two proteins.
FIG. 1.
The adapter protein RIAM interacts with SKAP-55 in transfected 293T cells and colocalizes with SKAP-55 in the Jurkat T-cell line. (a) (Left) His-ADAP and Flag-RIAM were cotransfected into 293T cells. Cell lysates were immunoprecipitated with anti-Flag and then separated by SDS-polyacrylamide gel electrophoresis. Coprecipitated RIAM and ADAP were examined by immunoblot analysis with anti-His and anti-Flag. (Right) Flag-ADAP and GFP-RIAM were cotransfected into 293T cells. Cell lysates were immunoprecipitated with anti-Flag and then separated by SDS-polyacrylamide gel electrophoresis. Coprecipitated RIAM and ADAP were examined by immunoblot analysis with anti-GFP and anti-Flag. (b) (Left) HA-SKAP-55 and Flag-RIAM were cotransfected into 293T cells. Cell lysates were immunoprecipitated with anti-Flag. Coprecipitated RIAM and SKAP-55 were examined by immunoblot analysis with anti-HA and anti-Flag. (Right) Flag-SKAP-55 and GFP-RIAM or Flag-SLP-76 and GFP-RIAM were cotransfected into 293T cells. The anti-Flag immunoprecipitation reveals only an interaction between SKAP-55 and RIAM. (c) Jurkat T cells were cotransfected with GFP-RIAM and HA-SKAP-55 constructs. Fixed cells were labeled with an anti-HA antibody followed by anti-rat antibody-Alexa Fluor 594 staining (red). (d) The Jurkat T cells transfected with either GFP-RIAM or HA-SKAP-55 were used to form conjugates with SEE-pulsed Raji B cells. The Raji cells were also labeled in blue with the cell tracker Blue CMAC. Cells were then fixed, permeabilized, and stained with phalloidin 594 (red) or phalloidin 488 (green). Conjugates were analyzed for accumulation of HA-SKAP-55 or GFP-RIAM at the T-B interface. (e) Jurkat T cells were cotransfected with GFP-RIAM and HA-SKAP-55. Conjugates were formed and analyzed for HA-SKAP-55 and GFP-RIAM colocalization. Representative conjugates are shown. Each study was repeated at least three times, and in each study, more than 30 conjugates were examined per condition.
Because ADAP is known to function via its association with SKAP-55, we questioned if SKAP-55 may potentially serve as a link to RIAM. When 293T cells were transfected with cDNA encoding Flag-RIAM along with a construct encoding HA-SKAP-55, anti-Flag immunoprecipitation revealed a strong interaction between the two proteins (Fig. 1b, left). Similarly, Flag-SKAP-55 associates with GFP-RIAM after anti-Flag immunoprecipitation in transfected 293T cells (Fig. 1b, right). As a specificity control, we expressed Flag-SLP-76 along with GFP-RIAM and found no interaction (Fig. 1b, right).
To extend these findings to T cells and to determine if the interaction between SKAP-55 and RIAM is inducible, we studied colocalization in transfected Jurkat cells. For these experiments, we expressed RIAM fused to a GFP along with HA-tagged SKAP-55. Both proteins were found diffusely in the cytosol in unstimulated Jurkat T cells (Fig. 1c). SKAP-55 has been shown previously to be recruited to the contact site when the TCR is engaged (10, 35, 40). To determine whether RIAM is similarly relocalized upon T-cell activation, we formed conjugates between transfected Jurkat cells and Raji B cells pulsed with SEE. GFP-RIAM was recruited to the contact site in >90% of all conjugates, similar to what we observed when SKAP-55 localization was studied (Fig. 1d). To determine more directly if SKAP-55 and RIAM relocalize to the same site, we cotransfected Jurkat cells with the two constructs and generated conjugates. Both proteins were concentrated at the contact site and in the merged image appeared to localize together, again in >90% of conjugates (Fig. 1e). Collectively, these results suggest that RIAM and SKAP-55 interact in T cells and colocalize at the interface of the T-cell-APC contact site.
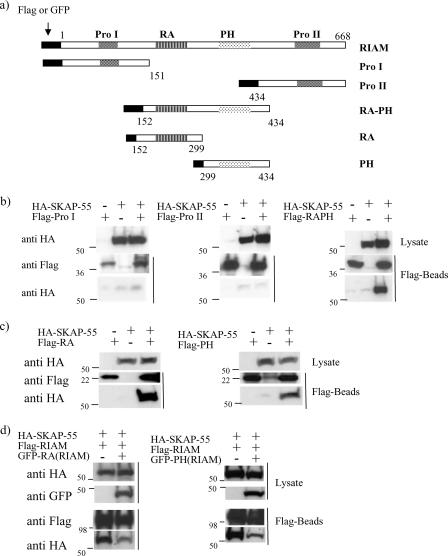
Mapping the region of RIAM responsible for SKAP-55 binding.
RIAM contains a central region with an RA domain and a PH domain flanked by two proline-rich regions (Fig. 2a). To determine which region of RIAM is involved in SKAP-55 binding, we generated cDNAs encoding Flag-tagged fusion proteins of the various RIAM fragments that include each domain (Fig. 2a). These cDNAs were cotransfected into 293T cells along with cDNA encoding HA-SKAP-55. Cell lysates were prepared and subjected to anti-Flag immunoprecipitation. The central region of RIAM (containing the RA and PH domains) interacted with SKAP-55 (Fig. 2b). In contrast, the two proline-rich regions failed to demonstrate an association (Fig. 2b). Further analysis of the central region of RIAM revealed that the RIAM RA as well as the RIAM PH domain bound independently to SKAP-55 (Fig. 2c). Consistently, we found that the SKAP-55/RIAM interaction could be blocked by either the isolated RA or PH domain of RIAM (Fig. 2d), indicating that the central region of RIAM is necessary and sufficient for SKAP-55 binding, and yet the interaction likely involves multiple contact sites.
FIG. 2.
Mapping the RIAM domains involved in SKAP-55 binding. (a) Schematic representation of the RIAM constructs used in these experiments. (b) Each domain of RIAM fused to a Flag tag was cotransfected with HA-SKAP-55 in 293T cells. Cell lysates were immunoprecipitated with anti-Flag and then separated by SDS-polyacrylamide gel electrophoresis. Coprecipitated RIAM domains and SKAP-55 were examined by immunoblot analysis with anti-HA and anti-Flag. (c) Flag-RIAM RA or Flag-RIAM PH was cotransfected along with HA-SKAP-55. Cell lysates were immunoprecipitated with anti-Flag. Coprecipitated domains of RIAM and SKAP-55 were examined by immunoblot analysis with anti-HA and anti-Flag. (d) The HA-SKAP-55 and Flag-RIAM were cotransfected with the GFP-RIAM RA or GFP-RIAM PH domain in 293T cells.
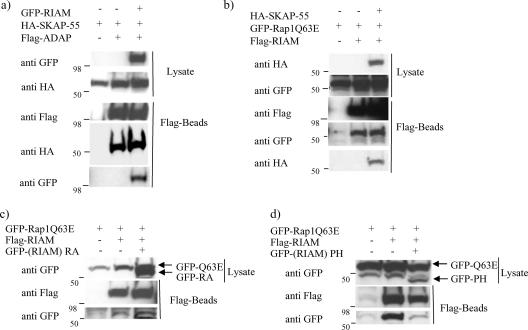
The RIAM/SKAP-55 complex associates with ADAP and the active form of Rap1.
As SKAP-55 and ADAP are known to interact constitutively, we examined whether RIAM overexpression impacts the binding of SKAP-55 to ADAP by expressing GFP-RIAM, HA-SKAP-55, and Flag-ADAP in 293T cells and then coimmunoprecipitating them with anti-Flag. Even when highly expressed, RIAM did not interfere with the interaction between SKAP-55 and ADAP (Fig. 3a). In fact, we found RIAM present in the ADAP immunoprecipitates, indicating that although ADAP and RIAM do not associate directly (Fig. 1), a trimolecular complex including ADAP, SKAP-55, and RIAM exists within the cell. We then addressed whether active Rap1, which has been shown to associate with RIAM, still binds RIAM when RIAM is associated with SKAP-55. For these experiments, we cotransfected HA-SKAP-55, Flag-RIAM, and GFP-tagged active Rap1 (the Q63E mutant) into 293T cells and then immunoprecipitated them with anti-Flag. We found that active Rap1 binds RIAM to the same extent in the presence and in the absence of SKAP-55. Additionally, we found that SKAP-55 was present in the Rap1/RIAM complex (Fig. 3b). To ensure that RIAM was the only molecular link between ADAP/SKAP-55 and Rap1, we addressed whether ADAP or SKAP-55 was able to interact individually with Rap1 and found no such association (data not shown). Since it has been shown previously that Rap1 binds RIAM only when Rap1 is in its active conformation (27), we speculate that one consequence of TCR engagement is the formation of a multimolecular complex including ADAP/SKAP-55/RIAM and active Rap1.
FIG. 3.
The SKAP-55/RIAM complex interacts with ADAP and the active form of Rap1. (a) Flag-ADAP, HA-SKAP-55, and GFP-RIAM were transfected in 293T cells. Cell lysates were immunoprecipitated with anti-Flag and then examined by immunoblot analysis using anti-GFP, anti-HA, and anti-Flag. (b) GFP-Rap1Q63E, HA-SKAP-55, and Flag-RIAM were transfected in 293T cells. Cell lysates were immunoprecipitated with anti-Flag. RIAM coprecipitates were examined by immunoblot analysis with anti-GFP, anti-HA, and anti-Flag. (c) Cell lysates containing GFP-Rap1Q63E, Flag-RIAM, and GFP-RIAM RA domain were immunoprecipitated with anti-Flag. The immunoblots were revealed using anti-GFP and anti-Flag. (d) Cell lysates containing GFP-Rap1Q63E, Flag-RIAM, and GFP-RIAM PH domain were immunoprecipitated with anti-Flag. The immunoblots were analyzed using anti-GFP and anti-Flag.
Work by others has shown that the central region of RIAM (consisting of the RA and PH domains) is involved in the recruitment of active Rap1 (27). To determine if the isolated RA or PH domain of RIAM (either of which interferes with RIAM/SKAP-55 binding [Fig. 2d]) is also able to block formation of the RIAM/Rap1 complex, 293T cells were cotransfected with RIAM and active Rap1 in the absence or presence of the RIAM RA or RIAM PH domain. The RA domain of RIAM failed to diminish the interaction between RIAM and active Rap1 (Fig. 3c). In contrast, however, the RIAM PH domain did interfere with complex formation between active Rap1 and RIAM (Fig. 3d).
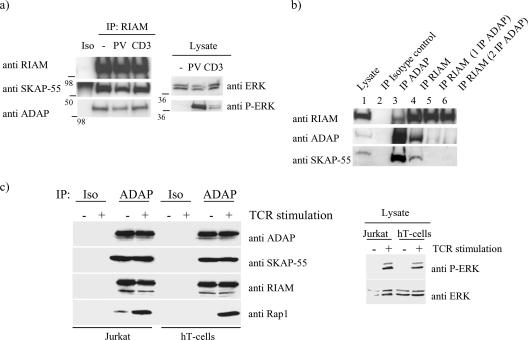
The endogenous ADAP/SKAP-55 complex interacts constitutively with RIAM in primary T cells.
We next asked if the RIAM/SKAP-55 association occurs in primary T cells. For these studies, T-cell blasts were left resting or were activated by cross-linking the TCR or stimulated with the phosphatase inhibitor PV. To ensure adequate T-cell activation, mitogen-associated protein kinase ERK1/2 phosphorylation was monitored (Fig. 4a). When cellular lysates were subjected to immunoprecipitation with anti-RIAM, we found that SKAP-55 associated with RIAM in both resting and activated cells (Fig. 4a). Additionally, we found ADAP in the RIAM immunoprecipitations, suggesting the presence of the trimolecular complex in both resting and activated T cells. To distinguish between an ADAP/SKAP-55/RIAM module and the presence of two different pools of SKAP-55, one associated with ADAP and the other with RIAM, we cleared ADAP from the cell lysate by immunoprecipitation and then asked if there remained a residual SKAP-55/RIAM association. ADAP immunoprecipitation followed by a RIAM immunoprecipitation, while still bringing down substantial amounts of RIAM, failed to coprecipitate SKAP-55 (Fig. 4b). These results suggest that RIAM is constitutively associated with SKAP-55 but only within a trimolecular complex including ADAP.
FIG. 4.
RIAM immunoprecipitates SKAP-55 and ADAP in primary T cells. (a) Murine T-cell blasts were rested or activated by cross-linking the TCR with an anti-CD3 antibody or with the phosphatase inhibitor PV. Cell lysates were immunoprecipitated with an isotype control or with an anti-RIAM polyclonal antibody and immunoblotted with anti-RIAM, anti-SKAP-55, and anti-ADAP. Cell lysates were also immunoblotted for the presence of ERK and phospho-ERK. (b) Murine T-cell BLAST lysates were immunoprecipitated with anti-RIAM or anti-ADAP. ADAP immunoprecipitations (one for lane 5 and two for lane 6) were performed before the RIAM immunoprecipitation. The immunoblots were analyzed using anti-RIAM, anti-SKAP-55, and anti-ADAP. (c) Jurkat T cells or primary human T cells (hT-cells) were stimulated for 5 min with MAb to CD3 (+) (C305 or MEM92) or left untreated (−). Cells were lysed, and immunoprecipitations were performed using anti-ADAP or an isotype control. Precipitates were analyzed by Western blot analysis for the presence of ADAP, SKAP-55, RIAM, or Rap1. Successful stimulation of T cells was assessed by analyzing the phosphorylation status of ERK. IP, immunoprecipitation; P-ERK, phospho-ERK; Iso, isotype control.
To address whether RIAM provides a link between the ADAP/SKAP-55 complex and the GTPase Rap1, we immunoprecipitated ADAP from lysates of resting or TCR-stimulated Jurkat T cells or purified primary human T cells. T-cell activation was monitored by inducible phosphorylation of ERK1/2. We confirmed the presence of a trimolecular complex including ADAP, SKAP-55, and RIAM in both resting and activated T cells (Fig. 4c). Rap1 was found to be associated with the ADAP/SKAP-55/RIAM complex but only after TCR engagement. These data indicate that the two cytosolic adapter proteins ADAP and SKAP-55 form a constitutive interaction with RIAM and provide a scaffold for the recruitment of the Rap1 effector protein following TCR ligation.
SKAP-55/RIAM complex disruption leads to decreased TCR-mediated adhesion.
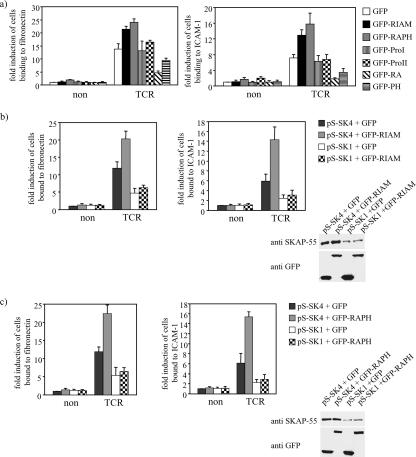
To investigate the functional importance of the SKAP-55/RIAM interaction in TCR-mediated inside-out signaling, we transiently expressed GFP-RIAM in Jurkat T cells and then assessed the transfectants for their ability to adhere to fibronectin or to ICAM-1 after triggering of the TCR. Confirming previous work by others (27), overexpression of RIAM enhanced adhesion of T cells to fibronectin or ICAM-1 (Fig. 5a). We then explored the structural features of RIAM sufficient for this effect by transfecting cDNAs encoding various RIAM domains. The transfection efficiency was analyzed 24 h posttransfection by anti-GFP Western blot analysis or FACS analysis (data not shown). We observed an increase in Jurkat cell adhesion to fibronectin as well as to ICAM-1 when the central region of RIAM, containing the RA and PH domains, was expressed. In contrast, the two proline-rich regions of RIAM (which cannot bind SKAP-55) failed to show an effect on cell adhesion.
FIG. 5.
Disruption of the SKAP-55/RIAM complex interferes with the TCR-dependent β1- and β2-integrin activation. (a) Jurkat T cells were transfected with cDNAs encoding GFP, GFP-RIAM, GFP-RAPH, GFP-ProI, or GFP-ProII. Transfectants either were left untreated (non) or were stimulated with anti-CD3 OKT3 (TCR) for 30 min and subsequently analyzed for their ability to adhere to fibronectin or to ICAM-1. Adhesion data represent the means ± the standard errors of the means of three independently performed experiments. (b) Jurkat T cells transfected with pS-SK1 (siRNA directed against SKAP-55) or pS-SK4 (scrambled siRNA) were cotransfected with either GFP alone or GFP-RIAM and analyzed for adhesion to fibronectin or ICAM-1 under resting (non) or TCR-stimulated (TCR) conditions. (c) Jurkat T cells transfected with pS-SK1 or pS-SK4 were transfected with GFP or GFP-RAPH and then analyzed for adhesion to fibronectin or ICAM-1 under resting (non) or TCR-stimulated (TCR) conditions.
To address if the interaction between RIAM and SKAP-55 is essential for the effect of RIAM on cell adhesion, we tested the impact of overexpressing the RIAM RA or the RIAM PH domain in the adhesion assay. Expression of either domain impaired adhesion of Jurkat cells to fibronectin and ICAM-1 following TCR engagement (Fig. 5b). This result is not due to decreased activation of Rap1, since expression of the isolated RIAM domains did not affect this event (data not shown). Similarly, expression of full-length RIAM or its isolated domains did not alter the expression levels of MHC class II, TCR, LFA-1, and VLA-4 or upregulation of the activation marker CD69 following TCR engagement or calcium mobilization (data not shown).
To confirm that RIAM (or the isolated RAPH domain) requires an interaction with SKAP-55 to regulate TCR-mediated integrin activation, we reduced expression of SKAP-55 in Jurkat T cells using an interfering RNA (RNAi)-based approach (23) and coexpressed GFP-RIAM or GFP-RAPH in parallel (Fig. 5b and c). The transfectants were assessed for their ability to adhere to fibronectin or to ICAM-1 after triggering of the TCR. The studies shown in Fig. 5b and c demonstrate that loss of SKAP-55 expression precludes the enhanced adhesion of T cells mediated by RIAM or the RIAM RAPH domain. Collectively these data demonstrate that the SKAP-55/RIAM complex is critical for optimizing the TCR-mediated adhesion.
SKAP-55/RIAM complex disruption leads to decreased T-cell conjugate formation.
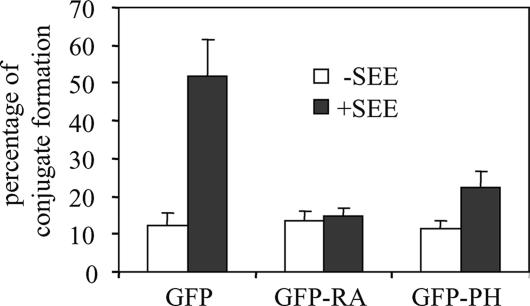
SKAP-55 has also been identified as one key regulator of T-cell-APC conjugate formation (40). To investigate whether SKAP-55 must also associate with RIAM to support conjugates, we disrupted this complex and then measured the impact on conjugate formation. For these experiments, we transiently expressed GFP, GFP-RA, or GFP-PH in Jurkat T cells and tested the ability of the transfectants to form conjugates with Raji B cells pulsed with SEE. Expression of either RIAM domain that blocked interactions with SKAP-55 abrogated conjugate formation (Fig. 6). These results demonstrate that the SKAP-55/RIAM interaction is essential for stable TCR-stimulated T-cell-APC conjugates.
FIG. 6.
Impaired conjugate formation between B cells and Jurkat cells expressing the RIAM RA or RIAM PH domain. Jurkat T cells transiently transfected with either GFP, GFP-RA, or GFP-PH were incubated with unpulsed or SEE-pulsed DDAO-SE-labeled Raji B cells. The percentage of conjugate formation between Jurkat T cells and Raji B cells was determined by FACS analysis. A histogram representative of a set of two-dimensional plots is shown. Data are the average means and standard errors of the means from three independent experiments.
The ADAP/SKAP-55 complex facilitates TCR-mediated plasma membrane recruitment of RIAM and Rap1.
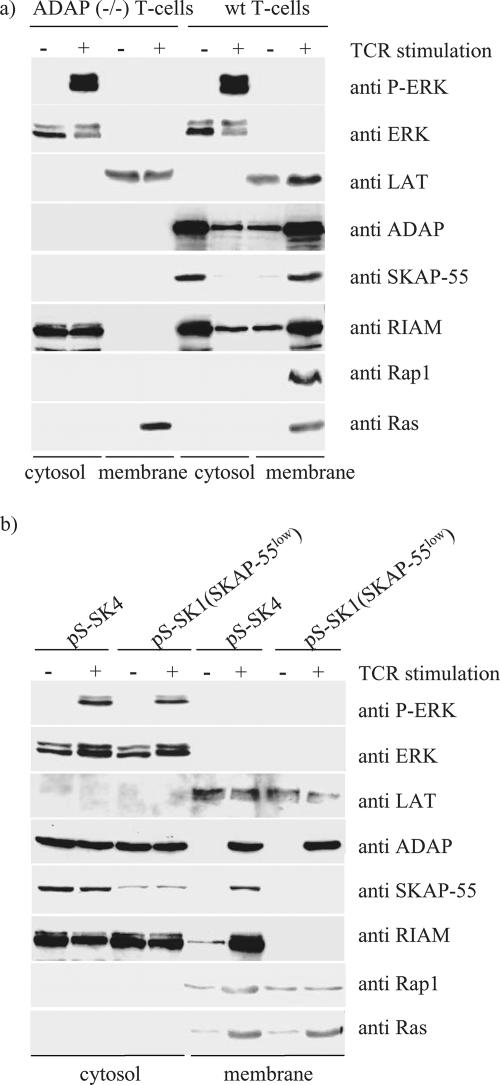
We have shown previously that the ADAP/SKAP-55 module is required for plasma membrane recruitment of Rap1 following TCR activation (23). Based on the data presented above, we hypothesized that ADAP and SKAP-55 would be required also for relocalization of RIAM in stimulated T cells. To test this notion, we examined unstimulated or TCR-stimulated ADAP-deficient T lymphocytes for the presence of RIAM at the membrane. In contrast to wild-type T cells, those cells lacking ADAP demonstrated a marked reduction in plasma membrane recruitment of RIAM upon TCR activation (Fig. 7a). As expected, the TCR-mediated increase in membrane targeting of Rap1 seen in ADAP-deficient T cells was also attenuated.
FIG. 7.
Disruption of the ADAP/SKAP-55 module interferes with the plasma membrane localization of RIAM and Rap1. (a) Purified splenic T cells prepared from ADAP-deficient [ADAP (−/−)] or wild-type (wt) mice were preincubated with biotinylated anti-CD3 MAb (2C11) and then cross-linked with streptavidin for 3 min at 37°C. Subsequently, membrane and cytosolic fractions were prepared from unstimulated (−) or stimulated (+) cells. The individual samples were analyzed by Western blotting using anti-ADAP, anti-SKAP-55, anti-RIAM, anti-Rap1, and anti-Ras. To control the fractionation efficiency and proper TCR stimulation, fractions were assessed for the presence of LAT (plasma membrane), ERK (cytosol), or phospho-ERK (stimulation) (P-ERK). (b) Jurkat T cells were transiently transfected with either pS-SK4 (scrambled) or pS-SK1 (siRNA of SKAP-55) or were either left unstimulated (−) or stimulated for 5 min with the anti-TCR MAb C305. Cytosolic and membrane fractions were analyzed as described above.
We and others have shown previously (13, 23) that ADAP expression appears to be required for stable expression of SKAP-55. Thus, in our experiment using ADAP-deficient cells, it was impossible to determine if failed RIAM and Rap1 membrane recruitment was due to loss of ADAP or SKAP-55. To test more directly the importance of SKAP-55 in this process, we made further use of Jurkat cells transfected with the RNAi targeting SKAP-55. As shown, “knockdown” of SKAP-55 markedly attenuates TCR-mediated membrane targeting of both Rap1 and RIAM, while TCR-mediated membrane recruitment of ADAP and Ras is unaffected (Fig. 7b).
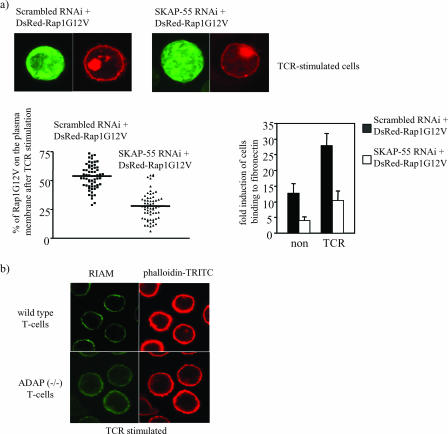
Additionally, to address whether Rap1 still requires the ADAP/SKAP-55/RIAM module for membrane translocation, we overexpressed a constitutively active mutant of Rap1 (Rap1G12V) in Jurkat T cells in which expression of SKAP-55 was downregulated by RNAi. As shown in Fig. 8a, “knockdown” of SKAP-55 expression strongly antagonized the adhesion-promoting effect of Rap1G12V. Additionally, under these same conditions, TCR-mediated membrane recruitment of Rap1G12V was markedly decreased (Fig. 8a).
FIG. 8.
Disruption of the ADAP/SKAP-55 module interferes with plasma membrane localization of both Rap1G12V and RIAM. (a) Jurkat T cells were transfected with constructs encoding a scrambled RNAi or an RNAi directed against SKAP-55 along with a construct encoding DsRed-tagged Rap1G12V. Cells were stimulated for 5 min through the TCR and were then imaged by confocal laser scanning microscopy. Rap1G12V translocation to the membrane was analyzed by determining the intensity of DsRed staining using Corel software (60 cells examined for each experiment). The percentage of total pixels representing Rap1G12V at the membrane for each cell is shown in the scattergram (left). This study is representative of three independent experiments. In parallel, transfectants were analyzed for their capability to adhere to fibronectin. The adhesion results are shown in the bar graph (right) and represent the means ± the standard errors of the means of three independent experiments. (b) Purified splenic T cells prepared from ADAP-deficient [ADAP (−/−)] or wild-type mice were preincubated with biotinylated anti-CD3 MAb and then cross-linked with streptavidin for 3 min at 37°C. Cells were fixed, permeabilized, and stained with phalloidin-tetramethyl rhodamine isocyanate (TRITC) (red) and RIAM in combination with anti-rabbit IgG-fluorescein isothiocyanate.
Finally, to extend these findings to primary T cells, we again made use of cells from the ADAP-deficient mice, comparing RIAM localization in these cells with that seen in T cells from wild-type animals. Cells were stimulated via the TCR and assessed for RIAM localization. Whereas in wild-type cells TCR stimulation results in obvious translocation of RIAM to the membrane, the response in ADAP-deficient T cells is much less pronounced, with most of the RIAM appearing diffusely in the cytosol (Fig. 8b). Collectively, these data reinforce the notion that one important role of the ADAP/SKAP-55 module is to relocalize RIAM and active Rap1 to the plasma membrane following TCR ligation, thus facilitating integrin activation.
DISCUSSION
Recent work from several groups has identified the important role played by various adapter proteins in the regulation of integrin activation upon engagement of the TCR (39). Both gain- and loss-of-function studies of primary T cells have demonstrated that ADAP and SKAP-55 are two such critical adapters for TCR-mediated inside-out-signaling of integrins (10, 35, 40). It appears that the constitutive association of ADAP with SKAP-55 is critical for their effect on integrin function (23). One role for the ADAP/SKAP-55 complex appears to be recruitment of activated Rap1, a low-molecular-weight GTPase critical for integrin activation, to the plasma membrane. It remained unclear, however, how the ADAP/SKAP-55 complex intersected with the Rap1 signaling pathway. In this study, we revealed a potential molecular mechanism by showing that in addition to binding ADAP, SKAP-55 also interacts with RIAM, an adapter protein known to associate with Rap1 when the GTPase is in its active conformation.
Studies of transfected heterologous cells, the Jurkat T-cell line, and primary cells confirmed a trimolecular complex consisting of ADAP, SKAP-55, and RIAM. Dissection of this complex further identified SKAP-55 as the key RIAM binder, and mapping studies demonstrated that the region essential for this interaction involved the central domain of RIAM (RAPH domain). Furthermore, the SKAP-55/RIAM complex associates with active Rap1, suggesting the possibility that one essential function of ADAP and SKAP-55 is to provide a scaffold, indirectly recruiting active Rap1. Concordant with this model is our observation that expression of RIAM domains that interfere with the SKAP-55/RIAM interaction diminished TCR-mediated T-cell adhesion to fibronectin and ICAM-1 as well as the ability of T cells to form conjugates with APCs. Moreover, we found that the ADAP/SKAP-55 module is required for the plasma membrane recruitment of RIAM after TCR activation and that one role of the ADAP/SKAP-55/RIAM module is to relocalize Rap1 to the plasma membrane following TCR activation.
An important next question to consider is how the association between the ADAP/SKAP-55 and RIAM/Rap1 modules functions to couple TCR engagement to increased integrin function. Both ADAP and RIAM are known to interact with actin-binding proteins including Ena/VASP family members and profilin (7, 14, 26, 27). Interestingly, however, we found that a mutant of RIAM, which no longer is able to bind these molecules (the RIAM RAPH domain construct), still increased TCR-stimulated Jurkat cell adhesion. This result is consistent with prior work by Lafuente et al. (27), who showed that RIAM function remains intact even when the RIAM/VASP interaction is blocked by expression of ActA repeats. A potential explanation for why RIAM can still mediate cell adhesion even when it is unable to interact with these actin-remodeling proteins is that ADAP can bind directly to Ena/VASP family members (14, 27), thus potentially compensating for RIAM deficiency. Similarly, although there is a striking integrin defect in ADAP-deficient T cells, these cells still polymerize actin when they encounter anti-TCR-coated beads (35), perhaps due to compensation by RIAM. It will be important to generate cells in which both ADAP and RIAM are absent to determine the effect of this combined deficiency on actin dynamics.
We were intrigued by the finding that the PH domain of RIAM contributes to such a great extent to SKAP-55 binding. Although PH domains are best known for phosphoinositide interactions, recent studies have indicated that the specificity of membrane targeting by PH domains may be conferred by protein interactions, either alone or together with phospholipids (21, 28). In vitro binding studies have revealed that the PH domain of RIAM displays highest affinity for phosphatidylinositol monophosphates (14). However, we found that the tandem RA and PH domains of RIAM are not sufficient for membrane targeting, as the deficiency of SKAP-55 results in retention of RIAM in the cytosol following TCR engagement (Fig. 7). Thus, a GTP-Rap1/RIAM module alone is not sufficient for membrane targeting after TCR stimulation, as a requirement for both ADAP and SKAP-55 remains.
Although apparently necessary for the recruitment of active Rap1 to the plasma membrane, the ADAP/SKAP-55/RIAM module is not needed for activation of the GTPase following TCR engagement. This finding was shown in our previous study using primary T cells obtained from ADAP-deficient mice and in Jurkat T cells in which SKAP-55 was knocked down using siRNAs (23). Previous work by others has shown that RIAM is required for localization of Rap1-GTP at the plasma membrane (27). In our current study, expression of the isolated RA or PH domain of RIAM (either of which abrogates the SKAP-55/RIAM interaction) had no effect on TCR-mediated activation of Rap1. However, “knockdown” of SKAP-55 in Jurkat cells markedly reduced the inducible relocalization of active Rap1 to the plasma membrane from the cytosol. Collectively, these results demonstrate that the role of the ADAP/SKAP-55/RIAM module is not to activate Rap1 but rather to promote its translocation to the plasma membrane upon TCR engagement.
An important question to consider is how Rap1 regulates integrin function in T cells. One attractive candidate is RAPL, a Rap1-binding effector protein that mediates Rap1-dependent integrin activation (18-20). Although the mechanism by which RAPL functions in this pathway is not completely clear, RAPL has been shown to coimmunoprecipitate with LFA-1 in the presence of active Rap1, providing an indirect link between Rap1 and an important T-cell integrin (19). It remains unclear, however, whether RIAM works synergistically with RAPL to bring active Rap1 to the plasma membrane or whether these two proteins function in independent pathways. Another potential effector of active Rap1 is PKD1, which has recently been implicated in integrin function in T cells as having an adapter-like role linking active Rap1 to β1 integrins (31). Further studies are required to determine how RAPL and PKD1 intersect with the ADAP/SKAP-55/RIAM signaling cascade to facilitate integrin activation.
In conclusion, our work suggests one means by which TCR signaling may utilize a network of adapter molecules to bring active Rap1 to its site of action. Our model suggests that TCR engagement activates Rap1 in an ADAP/SKAP-55/RIAM-independent fashion. Previous work by others suggests a number of possible links between the TCR and Rap1 activation, including several guanine nucleotide exchange factors such as C3G and CalDAG-GEFI, which are activated downstream of Fyn and phospholipase Cγ, respectively (2, 6). However, our data indicate that, although not required for its activation, Rap1 cannot be recruited to its site of action without the ADAP/SKAP-55/RIAM complex. Additional studies are now required to determine the mechanism(s) by which other key effector molecules, for example, RAPL and PKD1, function in this pathway to optimize TCR-mediated integrin function.
Acknowledgments
We thank Arthur Weiss, Alfred Wittinghofer, Johannes L. Bos, Ignacio Rubio, and Vaclav Horejsi for providing reagents and Martha Jordan and Marisa Juntilla for critical reading of the manuscript. We thank A. Nehring and J. Hoppe for excellent technical assistance.
This work was supported by long-term postdoctoral fellowships from the European Molecular Biology Organization (G.M.), by Deutsche Forschungsgemeinschaft grants KL1292/5-1 and GRK1167 (S.K. and B.S.), and by the U.S. National Institutes of Health (G.K.).
Footnotes
Published ahead of print on 2 April 2007.
REFERENCES
- 1.Bazzoni, G., and M. E. Hemler. 1998. Are changes in integrin affinity and conformation overemphasized? Trends Biochem. Sci. 23:30-34. [DOI] [PubMed] [Google Scholar]
- 2.Bivona, T. G., H. H. Wiener, I. M. Ahearn, J. Silletti, V. K. Chiu, and M. R. Philips. 2004. Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J. Cell Biol. 164:461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boerth, N. J., B. A. Judd, and G. A. Koretzky. 2000. Functional association between SLAP-130 and SLP-76 in Jurkat T cells. J. Biol. Chem. 275:5143-51452. [DOI] [PubMed] [Google Scholar]
- 4.Bos, J. L. 2005. Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 17:123-128. [DOI] [PubMed] [Google Scholar]
- 5.Bos, J. L., B. Franke, L. M'Rabet, K. Reedquist, and F. Zwartkruis. 1997. In search of a function for the Ras-like GTPase Rap1. FEBS Lett. 410:59-62. [DOI] [PubMed] [Google Scholar]
- 6.Boussiotis, V. A., G. J. Freeman, A. Berezovskaya, D. L. Barber, and L. M. Nadler. 1997. Maintenance of human T cell anergy: blocking of IL-2 gene transcription by activated Rap1. Science 278:124-128. [DOI] [PubMed] [Google Scholar]
- 7.Coppolino, M. G., M. Krause, P. Hagendorff, D. A. Monner, W. Trimble, S. Grinstein, J. Wehland, and A. S. Sechi. 2001. Evidence for a molecular complex consisting of Fyb/SLAP, SLP-76, Nck, VASP and WASP that links the actin cytoskeleton to Fcgamma receptor signalling during phagocytosis. J. Cell Sci. 114:4307-4318. [DOI] [PubMed] [Google Scholar]
- 8.da Silva, A. J., Z. Li, C. de Vera, E. Canto, P. Findell, and C. E. Rudd. 1997. Cloning of a novel T-cell protein FYB that binds FYN and SH2-domain-containing leukocyte protein 76 and modulates interleukin 2 production. Proc. Natl. Acad. Sci. USA 94:7493-7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dustin, M. L., and T. A. Springer. 1989. T-cell receptor cross-linking transiently stimulates adhesiveness through LFA-1. Nature 341:619-624. [DOI] [PubMed] [Google Scholar]
- 10.Griffiths, E. K., C. Krawczyk, Y. Y. Kong, M. Raab, S. J. Hyduk, D. Bouchard, V. S. Chan, I. Kozieradzki, A. J. Oliveira-Dos-Santos, A. Wakeham, P. S. Ohashi, M. I. Cybulsky, C. E. Rudd, and J. M. Penninger. 2001. Positive regulation of T cell activation and integrin adhesion by the adapter Fyb/Slap. Science 293:2260-2263. [DOI] [PubMed] [Google Scholar]
- 11.Han, J., C. J. Lim, N. Watanabe, A. Soriani, B. Ratnikov, D. A. Calderwood, W. Puzon-McLaughlin, E. M. Lafuente, V. A. Boussiotis, S. J. Shattil, and M. H. Ginsberg. 2006. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr. Biol. 16:1796-1806. [DOI] [PubMed] [Google Scholar]
- 12.Hogg, N., M. Laschinger, K. Giles, and A. McDowall. 2003. T-cell integrins: more than just sticking points. J. Cell Sci. 116:4695-4705. [DOI] [PubMed] [Google Scholar]
- 13.Huang, Y., D. D. Norton, P. Precht, J. L. Martindale, J. K. Burkhardt, and R. L. Wange. 2005. Deficiency of ADAP/Fyb/SLAP-130 destabilizes SKAP55 in Jurkat T cells. J. Biol. Chem. 280:23576-23583. [DOI] [PubMed] [Google Scholar]
- 14.Jenzora, A., B. Behrendt, J. V. Small, J. Wehland, and T. E. Stradal. 2005. PREL1 provides a link from Ras signalling to the actin cytoskeleton via Ena/VASP proteins. FEBS Lett. 579:455-463. [DOI] [PubMed] [Google Scholar]
- 15.Jo, E. K., H. Wang, and C. E. Rudd. 2005. An essential role for SKAP-55 in LFA-1 clustering on T cells that cannot be substituted by SKAP-55R. J. Exp. Med. 201:1733-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katagiri, K., M. Hattori, N. Minato, S. Irie, K. Takatsu, and T. Kinashi. 2000. Rap1 is a potent activation signal for leukocyte function-associated antigen 1 distinct from protein kinase C and phosphatidylinositol-3-OH kinase. Mol. Cell. Biol. 20:1956-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katagiri, K., M. Hattori, N. Minato, and T. Kinashi. 2002. Rap1 functions as a key regulator of T-cell and antigen-presenting cell interactions and modulates T-cell responses. Mol. Cell. Biol. 22:1001-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katagiri, K., M. Imamura, and T. Kinashi. 2006. Spatiotemporal regulation of the kinase Mst1 by binding protein RAPL is critical for lymphocyte polarity and adhesion. Nat. Immunol. 7:919-928. [DOI] [PubMed] [Google Scholar]
- 19.Katagiri, K., A. Maeda, M. Shimonaka, and T. Kinashi. 2003. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat. Immunol. 4:741-748. [DOI] [PubMed] [Google Scholar]
- 20.Katagiri, K., N. Ohnishi, K. Kabashima, T. Iyoda, N. Takeda, Y. Shinkai, K. Inaba, and T. Kinashi. 2004. Crucial functions of the Rap1 effector molecule RAPL in lymphocyte and dendritic cell trafficking. Nat. Immunol. 5:1045-1051. [DOI] [PubMed] [Google Scholar]
- 21.Kavran, J. M., D. E. Klein, A. Lee, M. Falasca, S. J. Isakoff, E. Y. Skolnik, and M. A. Lemmon. 1998. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 273:30497-30508. [DOI] [PubMed] [Google Scholar]
- 22.Kinashi, T. 2005. Intracellular signalling controlling integrin activation in lymphocytes. Nat. Rev. Immunol. 5:546-559. [DOI] [PubMed] [Google Scholar]
- 23.Kliche, S., D. Breitling, M. Togni, R. Pusch, K. Heuer, X. Wang, C. Freund, A. Kasirer-Friede, G. Menasche, G. A. Koretzky, and B. Schraven. 2006. The ADAP/SKAP55 signaling module regulates T-cell receptor-mediated integrin activation through plasma membrane targeting of Rap1. Mol. Cell. Biol. 26:7130-7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolanus, W., W. Nagel, B. Schiller, L. Zeitlmann, S. Godar, H. Stockinger, and B. Seed. 1996. Alpha L beta 2 integrin/LFA-1 binding to ICAM-1 induced by cytohesin-1, a cytoplasmic regulatory molecule. Cell 86:233-242. [DOI] [PubMed] [Google Scholar]
- 25.Koretzky, G. A., and P. S. Myung. 2001. Positive and negative regulation of T-cell activation by adaptor proteins. Nat. Rev. Immunol. 1:95-107. [DOI] [PubMed] [Google Scholar]
- 26.Krause, M., A. S. Sechi, M. Konradt, D. Monner, F. B. Gertler, and J. Wehland. 2000. Fyn-binding protein (Fyb)/SLP-76-associated protein (SLAP), Ena/vasodilator-stimulated phosphoprotein (VASP) proteins and the Arp2/3 complex link T cell receptor (TCR) signaling to the actin cytoskeleton. J. Cell Biol. 149:181-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lafuente, E. M., A. A. van Puijenbroek, M. Krause, C. V. Carman, G. J. Freeman, A. Berezovskaya, E. Constantine, T. A. Springer, F. B. Gertler, and V. A. Boussiotis. 2004. RIAM, an Ena/VASP and profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev. Cell 7:585-595. [DOI] [PubMed] [Google Scholar]
- 28.Lemmon, M. A. 2004. Pleckstrin homology domains: not just for phosphoinositides. Biochem. Soc. Trans. 32:707-711. [DOI] [PubMed] [Google Scholar]
- 29.Liu, J., H. Kang, M. Raab, A. J. da Silva, S. K. Kraeft, and C. E. Rudd. 1998. FYB (FYN binding protein) serves as a binding partner for lymphoid protein and FYN kinase substrate SKAP55 and a SKAP55-related protein in T cells. Proc. Natl. Acad. Sci. USA 95:8779-8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marie-Cardine, A., L. R. Hendricks-Taylor, N. J. Boerth, H. Zhao, B. Schraven, and G. A. Koretzky. 1998. Molecular interaction between the Fyn-associated protein SKAP55 and the SLP-76-associated phosphoprotein SLAP-130. J. Biol. Chem. 273:25789-25795. [DOI] [PubMed] [Google Scholar]
- 31.Medeiros, R. B., D. M. Dickey, H. Chung, A. C. Quale, L. R. Nagarajan, D. D. Billadeau, and Y. Shimizu. 2005. Protein kinase D1 and the beta 1 integrin cytoplasmic domain control beta 1 integrin function via regulation of Rap1 activation. Immunity 23:213-226. [DOI] [PubMed] [Google Scholar]
- 32.Mueller, K. L., M. A. Daniels, A. Felthauser, C. Kao, S. C. Jameson, and Y. Shimizu. 2004. Cutting edge: LFA-1 integrin-dependent T cell adhesion is regulated by both ag specificity and sensitivity. J. Immunol. 173:2222-2226. [DOI] [PubMed] [Google Scholar]
- 33.Musci, M. A., L. R. Hendricks-Taylor, D. G. Motto, M. Paskind, J. Kamens, C. W. Turck, and G. A. Koretzky. 1997. Molecular cloning of SLAP-130, an SLP-76-associated substrate of the T cell antigen receptor-stimulated protein tyrosine kinases. J. Biol. Chem. 272:11674-11677. [DOI] [PubMed] [Google Scholar]
- 34.Nagel, W., L. Zeitlmann, P. Schilcher, C. Geiger, J. Kolanus, and W. Kolanus. 1998. Phosphoinositide 3-OH kinase activates the beta2 integrin adhesion pathway and induces membrane recruitment of cytohesin-1. J. Biol. Chem. 273:14853-14861. [DOI] [PubMed] [Google Scholar]
- 35.Peterson, E. J., M. L. Woods, S. A. Dmowski, G. Derimanov, M. S. Jordan, J. N. Wu, P. S. Myung, Q. H. Liu, J. T. Pribila, B. D. Freedman, Y. Shimizu, and G. A. Koretzky. 2001. Coupling of the TCR to integrin activation by Slap-130/Fyb. Science 293:2263-2265. [DOI] [PubMed] [Google Scholar]
- 36.Pribila, J. T., A. C. Quale, K. L. Mueller, and Y. Shimizu. 2004. Integrins and T cell-mediated immunity. Annu. Rev. Immunol. 22:157-180. [DOI] [PubMed] [Google Scholar]
- 37.Sebzda, E., M. Bracke, T. Tugal, N. Hogg, and D. A. Cantrell. 2002. Rap1A positively regulates T cells via integrin activation rather than inhibiting lymphocyte signaling. Nat. Immunol. 3:251-258. [DOI] [PubMed] [Google Scholar]
- 38.Singer, A. L., S. C. Bunnell, A. E. Obstfeld, M. S. Jordan, J. N. Wu, P. S. Myung, L. E. Samelson, and G. A. Koretzky. 2004. Roles of the proline-rich domain in SLP-76 subcellular localization and T cell function. J. Biol. Chem. 279:15481-15490. [DOI] [PubMed] [Google Scholar]
- 39.Togni, M., J. Lindquist, A. Gerber, U. Kolsch, A. Hamm-Baarke, S. Kliche, and B. Schraven. 2004. The role of adaptor proteins in lymphocyte activation. Mol. Immunol. 41:615-630. [DOI] [PubMed] [Google Scholar]
- 40.Wang, H., E. Y. Moon, A. Azouz, X. Wu, A. Smith, H. Schneider, N. Hogg, and C. E. Rudd. 2003. SKAP-55 regulates integrin adhesion and formation of T cell-APC conjugates. Nat. Immunol. 4:366-374. [DOI] [PubMed] [Google Scholar]
- 41.Weiss, A., and J. D. Stobo. 1984. Requirement for the coexpression of T3 and the T cell antigen receptor on a malignant human T cell line. J. Exp. Med. 160:1284-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, J. N., and G. A. Koretzky. 2004. The SLP-76 family of adapter proteins. Semin. Immunol. 16:379-393. [DOI] [PubMed] [Google Scholar]
- 43.Zhong, X. P., E. A. Hainey, B. A. Olenchock, H. Zhao, M. K. Topham, and G. A. Koretzky. 2002. Regulation of T cell receptor-induced activation of the Ras-ERK pathway by diacylglycerol kinase zeta. J. Biol. Chem. 277:31089-31098. [DOI] [PubMed] [Google Scholar]