Abstract
p53 is extensively posttranslationally modified in response to various types of cellular stress. Such modifications have been implicated in the regulation of p53 protein levels as well as its DNA binding and transcriptional activities. Treatment of cells with doxorubicin causes phosphorylation and acetylation of p53, transcriptional upregulation of p21 and other target genes, and growth arrest. In contrast, downregulation of Mdm2 by a small interfering RNA (siRNA) approach led to increased levels of p53 lacking phosphorylation at serine 15 and acetylation at lysine 382. Levels of binding of p53 to the p21 promoter were comparable following treatment with doxorubicin or Mdm2 siRNA. Moreover, p53 was transcriptionally active and capable of inducing or repressing a variety of its target genes. Surprisingly, p53 upregulated by Mdm2 siRNA had no effect on cell cycle progression. Although comparable in level to that achieved by treatment with the p53 activators actinomycin D and nutlin-3, the increases in p53 and p21 after downregulation of Mdm2 were not sufficient to trigger cell cycle arrest. This version of p21 was capable of interacting with cyclin-dependent kinase 2 (Cdk2) but failed to inhibit its activity. Taken together, these results argue that Mdm2 is needed for full inhibition of Cdk2 activity by p21, thereby positively contributing to p53-dependent cell cycle arrest.
The tumor suppressor p53 is maintained at low levels in unstressed cells due to continuous degradation through the proteasome pathway (30). Mdm2, the product of a p53-induced target gene, mediates this process by ubiquitinating p53, thereby creating a negative-feedback loop (27, 37). In response to numerous stimuli, p53 becomes stabilized, accumulates in the nucleus, and regulates gene expression (2). Stabilization of p53 is thought to be triggered primarily through posttranslational modifications that affect its interaction with Mdm2 (10, 41). Activation of multiple pathways by different stimuli leads to p53 modification on at least 20 residues, including phosphorylation, acetylation, sumoylation, and glycosylation (1). Phosphorylation of Mdm2 following cellular stress has also been shown to inhibit its interaction with p53 and to play a role in p53 accumulation (31). In addition to its role in protein stabilization, posttranslational modification of p53 has also been implicated in the regulation of its transcriptional activity. The C terminus of p53 has been suggested to be a regulator of DNA binding (21). Modifications within this domain, including phosphorylation at serines 315, 378, and 392 or acetylation at lysines 373 and 382, have been shown to enhance the sequence-specific DNA binding of p53 (17, 21, 29, 43). Phosphorylation of serine 15 has been reported to stimulate the interaction of p53 with the acetyltransferases p300 and CBP (28).
Following cellular stress, p53 induces the expression of the cyclin-dependent kinase (cdk) inhibitor p21, among other target genes. p21 is a key mediator of arrest at the G1 phase of the cell cycle and contributes to the G2 arrest (6, 47). Moreover, p21 has been shown to be both sufficient and required for proper G1 arrest (5, 12, 38, 47) through binding and inhibition of cdk complexes (6, 18).
Here, the activities of p53 stabilized by downregulation of Mdm2 were investigated using a small interfering RNA (siRNA) approach. Mdm2 ablation caused an upregulation of p53 lacking a subset of posttranslational modifications. Nevertheless, this p53 was competent for specific DNA binding and transcriptional regulation. Intriguingly, p21 expression was induced but cells failed to become arrested. This p21 interacted with cyclin-dependent kinase 2 but failed to inhibit its activity. This suggests a role for Mdm2 in the ability of p53 and p21 to induce cell cycle arrest.
MATERIALS AND METHODS
Cell lines and drug treatments.
U2OS, HT1080, G361, WI-38, and HCT116 cell lines were purchased from ATCC. Cells were grown in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Control and p53 short hairpin RNA (shRNA) stable cell lines were generated by cotransfection of either TranSilent control or p53 shRNA vectors (Panomics) and pBabe-Puro (D. J. Lukin and J. J. Manfredi, unpublished data). A p21 shRNA stable cell line, p53−/− mouse embryonic fibroblasts (MEFs), and 293T cells were generous gifts from A.W. Lin, G. Lozano, and P. Frenette, respectively. Doxorubicin and actinomycin D were purchased from Sigma. Nutlin-3 (racemic mix) was purchased from Calbiochem.
siRNA transfections and immunoblotting.
An Mdm2 pool of RNA oligonucleotides was purchased from Dharmacon and used according to the manufacturer's instructions at a final concentration of 100 nM. Control siRNA r(AUGAACGUGAAUUGCUCAAUU) was purchased from QIAGEN. U2OS, HT1080, and WI-38 cells were transfected using Oligofectamine and G361 cells using Lipofectamine 2000 (Invitrogen). When indicated, drugs were added to cells 5 h after transfection, at the time when complete growth medium was added.
Cells were washed and harvested in phosphate-buffered saline (PBS) and then lysed in 50 mM HEPES (pH 7.5)-1% Triton X-100-150 mM NaCl-1 mM MgCl2-1 mM phenylmethylsulfonyl fluoride-5 μg/ml leupeptin-50 μg/ml aprotinin. Equal amounts of proteins, typically 50 μg, were electrophoresed by use of sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, transferred to nitrocellulose membranes (Bio-Rad), and blotted with anti-p53 (DO-1), anti-p21 (C-19), and anti-cyclin-dependent kinase 2 (anti-Cdk2) (M2) from Santa Cruz Biotechnology Inc. Anti-Mdm2 (Ab1) and antiactin (Ab-1) were from Calbiochem. Anti-pRb (4H1) was purchased from Cell Signaling Technology. Anti-p21 (SMX30; BD PharMingen) was used for detection of mouse and human p21. To analyze p53 phosphorylation and acetylation status, cells were treated and lysed as described in the presence of the following phosphatase and acetylase inhibitors: 0.1 mM sodium orthovanadate, 1 mM sodium fluoride, 15 mM p-nitrophenylphosphate (PNPP), and 5 μM trichostatin A. p53 polyclonal phosphospecific antibodies Ser6, Ser15, and Ser37 were from Cell Signaling Technology. Anti-p53 acetylated Lys382 was from Trevigen, Inc. Anti-mouse and -rabbit antibodies were from ICN Biomedicals Inc. The signal was detected using enhanced chemiluminescence (ECL) detection reagent from Amersham Life Sciences.
Indirect immunofluorescence.
Cells were grown on glass coverslips and transfected or treated as indicated. Coverslips were washed with PBS, and cells were fixed in ice-cold 100% methanol for 20 min at −20°C followed by cold 100% acetone for 15 to 20 s. Coverslips were then stored at 4°C in 50% glycerol-PBS until processed. Coverslips were washed three times with PBS, blocked for 10 min at room temperature in 5% normal goat serum (NGS) in PBS, washed with PBS, and incubated with primary antibodies in 5% NGS-PBS for 1 h at room temperature. After three washes in PBS, coverslips were incubated for 30 min at room temperature with secondary antibodies in 5% NGS-PBS, washed three times for 5 min in PBS, and mounted in 50% glycerol-0.1 mg/ml p-phenylenediamine-0.1 μg/ml DAPI (4′,6′-diamidino-2-phenylindole). The following primary and secondary antibodies were used: anti-p53 (DO-1 or FL393; Santa Cruz) (1:500), anti-phosphoserine 15 (polyclonal; Cell Signaling) (1:750), anti-phosphorylated H2AX (γH2AX; UPSTATE) (1:500), anti-Mdm2 (Ab-1; Calbiochem) (1:500), anti-p21 (SMX30; BD PharMingen) (1:500), and Alexa 594- or 488-conjugated anti-mouse and -rabbit antibodies (Molecular Probes) (1:500). Fluorescence was captured with a Nikon E-700 epifluorescence photomicroscope (Tokyo, Japan) using a Diagnostic Instruments Inc. RT-SE SPOT digital camera and Adobe Photoshop 5.0 software.
Oligonucleotide pull-down assay.
Cells were lysed in the presence of phosphatase and acetylase inhibitors as described for immunoblotting. The lysates were centrifuged for 3 min at 14,000 rpm, and NaCl, dithiothreitol (DTT), EDTA, MgCl2, and glycerol were added to the supernatants to achieve final concentrations of 500 mM, 1 mM, 0.2 mM, 1.5 mM, and 20%, respectively. Extracts were aliquoted, frozen in a dry ice-ethanol bath, and stored at −70°C. Cell extracts (150 to 300 μg) were incubated with 0.5 to 2 μg of biotinylated p21 5′ or control oligonucleotides (QIAGEN) in a total volume of 600 μl of DNA binding buffer (20 mM HEPES [pH 7.5], 83 mM NaCl, 0.1 mM EDTA, 12% glycerol, 2 mM MgCl2, 2 mM spermidine, 0.7 mM DTT, and 10 μg salmon sperm DNA). Protein levels were normalized by the addition of bovine serum albumin (Sigma). After a 20-min incubation at room temperature, 30 to 50 μl of streptavidin-conjugated beads (ImmunoPure streptavidin; Pierce Chemical) was added, and samples were rocked for 1 h at 4°C. Beads were washed three times with DNA binding buffer without DNA, resuspended in 1× Laemmli sample buffer, heated to 95°C for 3 min, and analyzed by immunoblotting.
The sequences of the biotinylated oligonucleotides (Operon Biotechnologies, Inc.) were as follows: for p21 5′, 5′-AATTCGGTACCGAACATGTCCCAACATGTTGGCTAGCG-3′ (where the underlined sequence corresponds to the p53 response element); and for the control, 5′-AATTCGGTACCTCGAAGAAGACGTGCAGGGACCCGCTAGCG-3′.
Chromatin immunoprecipitation (ChIP) assay.
Assays were performed essentially as described by Espinosa et al. (14) using anti-p53 antibody (DO-1; Santa Cruz). Primers for radioactive PCR amplification were as described by St. Clair et al. (42).
RNA extraction, RT-PCR, and real-time PCR.
RNA was extracted using an RNeasy kit (QIAGEN). Immediately after extraction, 1 to 2 μg of RNA was used for cDNA synthesis with SuperScript II (Invitrogen) according to the manufacturer's instructions. Reverse transcription-PCR (RT-PCR) analysis was performed as described by St. Clair et al. (42).
Quantitative real-time PCR was performed using the following PCR primers (Operon Biotechnologies, Inc.) and an ABI PRISM 7900HT sequence detection system (Applied Biosystems, Foster City, CA): for p21, 5′-ACTCTCAGGGTCGAAAACGG-3′-5′-CCTCGCGCTTCCAGGACTG-3′; for Mdm2, 5′-CTGTGTTCAGTGGCGATTGG-3′-5′-AGGGTCTCTTGTTCCGAAGC-3′; for 14 3 3σ, 5′-TCTGATCCAGAAGGCCAAGC-3′-5′-GCCCACCACGTTCTTATAGG-3′; for Survivin, 5′-TCCGGTTGCGCTTTCCT-3′-5′-TCTTCTTATTGTTGGTTTCCTTTGC-3′; for PUMA, 5′-AGAGGGAGGAGTCTGGGAGTG-3′-5′-GCAGCGCATATACAGTATCTTACAGG-3′; for Noxa, 5′-TGGAAGTCGAGTGTGCTACTCAACT-3′-5′-AGATTCAGAAGTTTCTGCCGGAA-3′; for PIG3, 5′-CAAATGGCA GAAAAGCTTGGAG-3′-5′-GGCAGTTGACGTTCTTCTCC-3′; and for GAPDH (glyceraldehyde-3-phosphate dehydrogenase), 5′-CAATGACCCCTTCATTGACC-3′-5′-GATCTCGCTCCTGGAAGATG-3′.
The reactions were performed in quadruplicates. For each gene, the median was considered and normalized using the median of the GAPDH replicates. Severalfold induction values were calculated using the following equation: 2[(GAPDH − gene X)treatment + (gene X − GAPDH)control].
Luciferase reporter assay.
U2OS and HCT116 cells were seeded in 24-well dishes and transfected using Lipofectamine 2000 (Invitrogen) with 200 ng of luciferase reporter plasmid (the p21 5′ response element cloned upstream of the minimal E1b-TATA promoter in the pGL3 reporter vector), 5 ng of pRL-Renilla, and control or Mdm2 siRNA oligonucleotides (100 nM). At 24 h after transfection, cells were treated with 0.5 μg/ml doxorubicin when indicated. At 48 h after transfection, luciferase and Renilla activities were measured using a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions.
Flow cytometry and BrdU incorporation assay.
Cells were pulsed with bromodeoxyuridine (BrdU) that was added to the culture medium at a 10 μM final concentration for 1 h before harvesting. Cells were trypsinized, collected by centrifugation, and fixed in 70% ethanol at −20°C. DNA was denatured by incubation for 30 min with 2 N HCl-0.5% Triton X-100 followed by neutralization with 0.1 M Na2B4O7·10 H2O (pH 8.5). Cells were then stained with fluorescein isothiocyanate-conjugated anti-BrdU antibody (catalog no. 347583; Becton Dickinson) for 1 h at room temperature in the dark, centrifuged, resuspended in PBS with 20 μg/ml propidium iodide and 1 mg/ml RNase A, and kept in the dark for 30 min. Cell cycle distribution and BrdU incorporation were analyzed using a FACSCalibur flow cytometer (Becton Dickinson) and CellQuest software (BD Biosciences).
Cdk2-p21 coimmunoprecipitation.
Following treatments as indicated, cells were washed and harvested in PBS and then lysed as described for immunoblotting with protease and phosphatase inhibitors. A 500-μg volume of proteins was mixed with 3 μl of anti-Cdk2 polyclonal antibody (M2; Santa Cruz) and 30 μl of a 50% slurry of protein A-Sepharose (Amersham Biosciences) and rocked at 4°C for 2 h. The beads were washed three times with RIPA (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 5 mM EDTA), resuspended in 30 μl of 2× sample buffer, and heated to 95°C for 5 min. Proteins were resolved in 12% to 15% polyacrylamide gels and analyzed by immunoblotting using anti-Cdk2 (M2) and anti-p21 (SMX30) antibodies (BD PharMingen).
Cdk2 kinase assay.
Cells were washed with PBS and lysed in HB buffer (25 mM MOPS [morpholinepropanesulfonic acid] [pH 7.2], 15 mM MgCl2, 15 mM EGTA, 1 mM DTT, 1% Triton X-100) with protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 5 μg/ml leupeptin, and 50 μg/ml aprotinin) and phosphatase inhibitors (60 mM β-glycerophosphate, 15 mM PNPP, and 0.1 mM sodium orthovanadate). After centrifugation, proteins were quantitated using a Bradford assay (Bio-Rad). Proteins (400 to 1,000 μg) were immunoprecipitated with 5 to 10 μl of anti-Cdk2 antibodies (M2) and 40 to 50 μl of a 50% slurry of protein A-Sepharose (Amersham Biosciences) and rocked at 4°C for 1 to 2 h. The beads were washed three times with 1 ml of HB buffer with protease and phosphatase inhibitors followed by a brief spin and were resuspended in 15 μl of HB buffer. To each sample was added 15 μl of 2× kinase reaction buffer (HB buffer containing 200 μM ATP, 1 mg/ml H1 histone, and 1 μl of [γ-32P]ATP [Perkin-Elmer]) (6,000 Ci/mmol [10 μCi/μl] per 10 μl of 2× reaction buffer). Samples were incubated for 15 to 20 min at 37°C in a block with agitation. The reaction was stopped by addition of 7.5 μl of 5× sample buffer (60 mM Tris-HCl [pH 6.8], 25% glycerol, 2% SDS, 14.4 mM β-mercaptoethanol, 0.1% bromophenol blue) and 5 min incubation at 95°C. Samples were electrophoresed on 12% denaturing gels. Gels were Coomassie stained, dried on Whatman paper, and exposed to Kodak XAR film. Quantification was performed using Kodak phosphorimager screens and Personal Molecular Imager FX and Quantity One software (Bio-Rad).
Retroviral infection.
Ecotropic retroviruses were generated by transient transfection of 293T cells with 5 μg of pCL-Eco and 5 μg of pBabe-puro (pBP) or pBP expressing human p53 (pBP53) or mouse p21 (pBP21) by use of a standard calcium phosphate precipitation protocol. Retroviral supernatants were collected 48 h after transfection and filtered through a 0.45 μm filter. p53−/− MEFs were incubated with 2 ml of freshly prepared retroviral supernatants in the presence of 8 μg/ml Polybrene at 37°C for 2 h followed by addition of growth medium to achieve a final volume of 7 ml. The infection was repeated the following day. The cells were harvested 48 h after the initial infection and processed as described for the immunoblotting and cdk2 kinase assays. pCL-Eco and pBP21 were generous gifts from M. O'Connell and S. A. Aaronson, respectively.
RESULTS
Downregulation of Mdm2 increases p53 levels comparably to treatment with doxorubicin but without inducing posttranslational modifications.
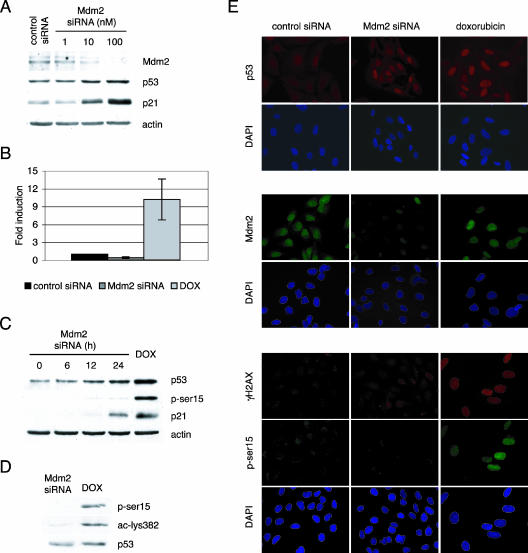
Transient transfection of U2OS cells with a pool of siRNA oligonucleotides targeting Mdm2 caused downregulation of Mdm2 protein levels that was detected by immunoblotting (Fig. 1A). Abrogation of Mdm2 expression led to upregulation of both p53 and p21 proteins (Fig. 1A). This siRNA approach resulted in a 50% to 80% reduction in the mRNA levels of Mdm2 (Fig. 1B). The level of p53 achieved following Mdm2 siRNA treatment closely resembled that obtained after DNA damage of the cells with doxorubicin (Fig. 1C). However, a comparison of similar levels of p53 showed that, in contrast to cells treated with doxorubicin, p53 upregulation by Mdm2 siRNA did not result in significant phosphorylation at serine 15 or acetylation at lysine 382 (Fig. 1C and D). A time course analysis showed that although the transfection procedure might be sensed by the cells as a type of cellular stress, this was not sufficient to induce modification of p53, not even at early time points (Fig. 1C). These findings were confirmed by immunohistochemistry (Fig. 1E). The siRNA approach downregulated Mdm2 protein levels, leading to an increase in p53 expression comparable to that seen upon treatment with doxorubicin but lacking the phosphorylation on serine 15 (Fig. 1E). In addition, the siRNA transfection did not seem to cause DNA damage, as suggested by the absence of phosphorylation of histone H2AX, although it was readily detectable by immunofluorescence after treatment with doxorubicin (Fig. 1E).
FIG. 1.
Downregulation of Mdm2 by siRNA causes p53 and p21 upregulation in the absence of DNA damage and posttranslational modifications. Mdm2 protein levels were downregulated in U2OS cells by use of siRNA oligonucleotides. (A and B) At 48 h after transfection, Mdm2, p53, p21, and actin levels in the cellular extracts were assayed by immunoblotting (A) and Mdm2 mRNA levels were quantitated by quantitative RT-PCR (B). (C and D) p53 stabilized by Mdm2 downregulation or 0.1 μg/ml doxorubicin (DOX) was analyzed for posttranslational modifications by immunoblotting. (E) At 48 h after transfection with control or Mdm2 siRNA or treatment with 0.1 μg/ml doxorubicin, U2OS cells were fixed and p53, Mdm2, γH2AX, and phosphorylated p53 (p-ser15) were localized by indirect immunofluorescence.
Posttranslational modifications are not required for binding of p53 to its response elements in the p21 promoter in vitro and in cells.
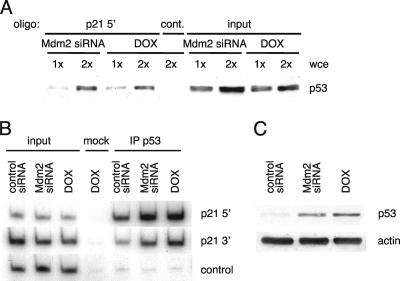
To determine whether the absence of posttranslational modifications affected p53 sequence-specific binding, p53 proteins from U2OS cells transfected with Mdm2 siRNA or treated with doxorubicin were compared with respect to their DNA binding activities. Cell extracts were incubated with a biotinylated probe containing the p21 5′ response element (−2262 to −2281 of the human p21 promoter) or a nonrelated control sequence. The oligonucleotides were pulled down with streptavidin beads, and the presence of p53 in the precipitated complexes was assayed by immunoblotting. The binding to the probe of p53 from Mdm2 siRNA-transfected cells was indistinguishable from that of p53 from doxorubicin-treated cells (Fig. 2A). This result suggested that DNA damage-associated posttranslational modification of p53 does not contribute to in vitro DNA binding.
FIG. 2.
p53 upregulated by Mdm2 siRNA binds to the response elements in the p21 promoter both in vitro and in cells. U2OS cells were transfected with Mdm2 siRNA oligonucleotides or treated with 0.1 μg/ml doxorubicin (DOX) for 24 h. (A) Volumes of the different extracts containing similar amounts of p53 were used in oligonucleotide pull-down assays to measure binding to the p21 5′ biotinylated probe or a control probe (cont.). 1x and 2x represent two levels of whole cell extracts (wce), and “input” corresponds to 10% of those amounts. The presence of p53 in the precipitated complexes was detected by immunoblotting. (B) Cells were cross-linked, and extracts were subjected to ChIP analysis. Radioactive PCR was performed to detect the occupancy by p53 of the response elements in the p21 promoter. A nonrelated genomic region was amplified as a control. (C) p53 levels in the extracts used for ChIP were assayed by immunoblotting.
p53 binding to DNA was next compared in the cellular context by use of a ChIP assay. Immunoprecipitation of p53 from cells transfected with Mdm2 siRNA or treated with doxorubicin followed by PCR analysis showed a strong and similar association of p53 to both response elements in the p21 promoter that was not affected by DNA damage and p53 modification (Fig. 2B). p53 was present in the p21 promoter before treatment (control siRNA), and the increase in its binding following stabilization by Mdm2 siRNA or doxorubicin was comparable to that previously seen, as was the increase in p53 protein levels (Fig. 2C). This result further supported the idea that posttranslational modification of p53 does not affect its sequence-specific DNA binding to the p21 promoter.
Upregulated p53 in response to Mdm2 siRNA induces expression of p21 protein and mRNA levels.
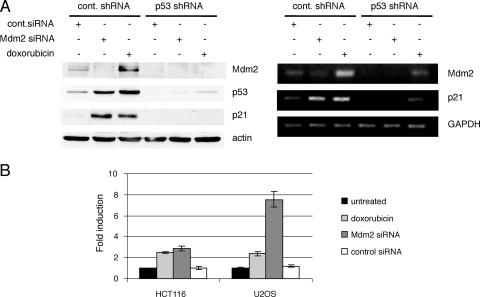
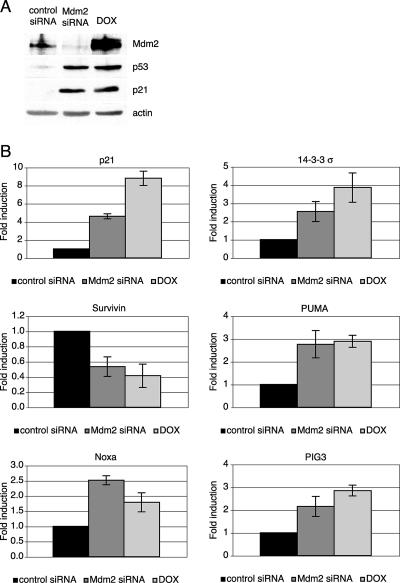
p21 has been reported to be degraded by both ubiquitin-dependent and -independent mechanisms. Mdm2 has been implicated in this pathway and shown to interact with p21 and negatively regulate its turnover (24, 56). Consistent with this, downregulation of Mdm2 by siRNA caused an increase in p21 protein levels. The upregulation was not observed in U2OS cells stably transfected with a plasmid expressing a p53 shRNA compared to the results seen with a control shRNA clone (Fig. 3A, left panel). This result suggested that p21 upregulation was at least in part dependent on the presence of p53. To determine whether the increase in p21 levels was a consequence of Mdm2 regulation of p21 stability or a direct transcriptional effect of p53, cells were transfected with control or Mdm2 siRNA or treated with doxorubicin and p21 mRNA levels were examined. RT-PCR analysis showed that p21 transcription was induced to similar extents in the control cells following both Mdm2 siRNA and doxorubicin treatment but was absent in the p53 shRNA clone (Fig. 3A, right panel). This result indicated that p53 upregulated by Mdm2 siRNA is transcriptionally active on the p21 promoter, leading to an increase in p21 protein levels. In addition, a reporter plasmid containing the p21 5′ response element upstream of the minimal adenovirus E1b promoter was activated in cells transfected with Mdm2, but not in those transfected with control siRNA, as well as in cells treated with doxorubicin (Fig. 3B). Finally, more careful examination of p53 transcriptional activity by use of quantitative real-time PCR revealed a similar ability of p53 to transactivate or repress multiple target genes (Fig. 4B). This demonstrated that p53 upregulated by ablation of Mdm2 was transcriptionally active. Despite the different mRNA levels, p21 protein levels were nearly identical following either treatment, suggesting that these might be the result of the combined effects on transcription and protein stability (Fig. 4A).
FIG. 3.
Upregulation of p53 by Mdm2 siRNA induces expression of p21 protein and mRNA to a level comparable to that seen with treatment with doxorubicin. (A) Clones of U2OS cells stably transfected with control (cont.) or p53 shRNA were transiently transfected with control or Mdm2 siRNA or treated with 0.1 μg/ml doxorubicin as indicated. Cell lysates were immunoblotted (left panel), and corresponding dishes were subjected to RT-PCR analysis (right panel). (B) HCT116 and U2OS cells were cotransfected with 200 ng of a luciferase reporter construct containing the p21 5′ response element, control or Mdm2 siRNA, and 5 ng of pRL-Renilla for internal control. At 24 h after transfection, cells were treated with 0.5 μg/ml doxorubicin when indicated. At 48 h after transfection, cells were lysed and assayed for luciferase and Renilla activities. The indicated values represent the averages of data obtained in three independent experiments, each performed in duplicate. Error bars represent standard errors of the means.
FIG. 4.
p53 upregulated by Mdm2 siRNA regulates transcription of various target genes. U2OS cells were transfected with control or Mdm2 siRNA or treated with doxorubicin (DOX) as indicated. At 48 h after treatment, cell lysates were immunoblotted (A) and mRNA levels from corresponding dishes were measured by quantitative RT-PCR (B).
Taken together, these results supported the notion that downregulation of Mdm2 by siRNA causes an increase in p53 levels that, devoid of significant modification, is capable of resulting in the binding of p53 to the p21 promoter in cells. Further, p21 is transcriptionally upregulated by p53 and the induced p21 protein level is comparable to that observed following treatment with doxorubicin.
Despite induction of p21, upregulation of p53 by Mdm2 siRNA fails to trigger cell cycle arrest compared to treatment with doxorubicin.
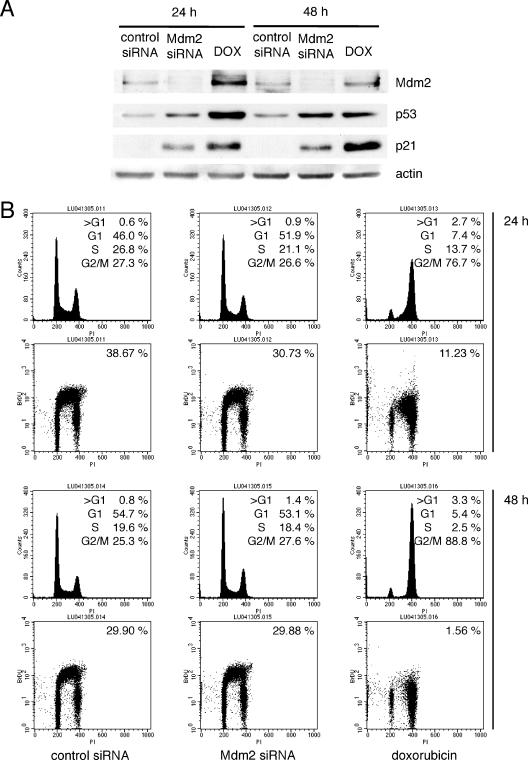
The biological effect of the upregulation of p53 by Mdm2 siRNA was next examined. U2OS cells were transfected with control or Mdm2 siRNA or treated with doxorubicin for 24 and 48 h, and cell cycle profiles and BrdU incorporation were analyzed. Treatment of cells with doxorubicin caused a cell cycle arrest predominantly at the G2 phase, and the percentage of cells in the S phase incorporating BrdU was reduced from 39% to 11% and 2% at 24 and 48 h, respectively. Surprisingly, a comparison of cells transfected with control or Mdm2 siRNA showed that, despite upregulation of p53 and p21, there were no substantial changes in their cell cycle profiles or in the fraction of cells incorporating BrdU (Fig. 5).
FIG. 5.
p53 upregulated by Mdm2 siRNA fails to induce cell cycle arrest in U2OS cells. U2OS cells were transfected with control or Mdm2 siRNA or treated with doxorubicin (DOX) as indicated. At 24 and 48 h after transfection, cell lysates were immunoblotted (A) and corresponding dishes were pulsed with BrdU and subjected to staining and flow cytometry (B). The percentages of BrdU-positive cells are indicated at the top right corner of the panels.
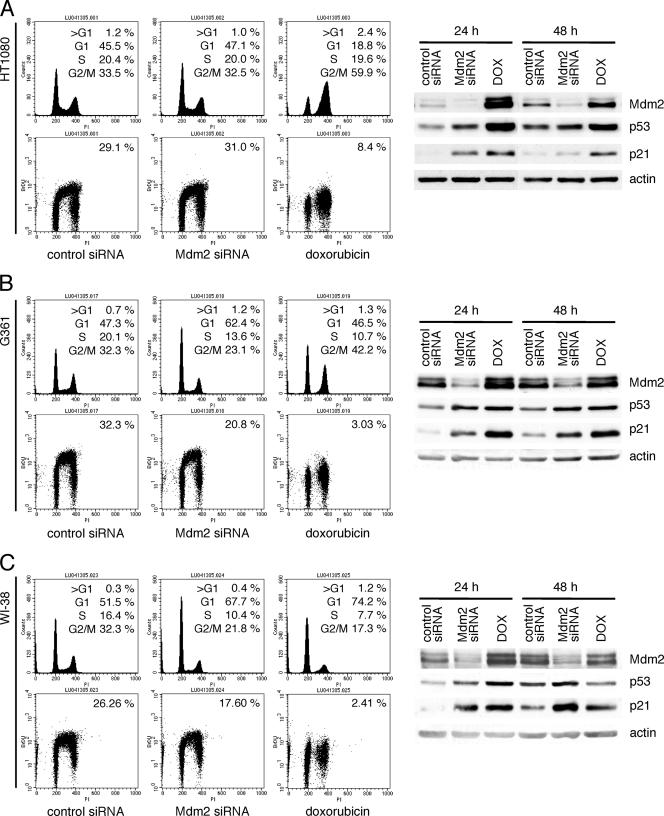
To further explore this result, similar experiments were performed using two other cancer cell lines (HT1080 fibrosarcoma cells and G361 melanoma cells) and primary lung fibroblasts (WI-38 cells). In all cell lines examined, downregulation of Mdm2 by siRNA transfection led to upregulation of p53 and p21 at levels that were similar to those achieved following treatment with doxorubicin (Fig. 6, right panels). However, consistent with the result obtained using U2OS cells, Mdm2 downregulation had no or a modest effect on the cell cycle profile of these cells compared to doxorubicin treatment. HT1080 showed no differences in the cell cycle distribution results between cells transfected with control or Mdm2 siRNA. G361 and WI-38 showed a slight arrest in the G1 phase of the cell cycle following Mdm2 downregulation, with only a 35% reduction in the fraction of cells incorporating BrdU, in contrast to a greater than 90% reduction when the cells were treated with doxorubicin (Fig. 6, left panels). These results suggested that an increase in p53 and p21 levels is not sufficient to induce growth arrest in these cells.
FIG. 6.
p53 upregulated by Mdm2 siRNA fails to induce cell cycle arrest in HT1080, G361, and WI-38 cells. HT1080, G361, and WI-38 cells were transfected with control or Mdm2 siRNA or treated with 0.1 μg/ml doxorubicin (DOX) as indicated. At 24 and 48 h after transfection, cell lysates were immunoblotted, and at 24 h, corresponding dishes were pulsed with BrdU and subjected to staining and flow cytometry.
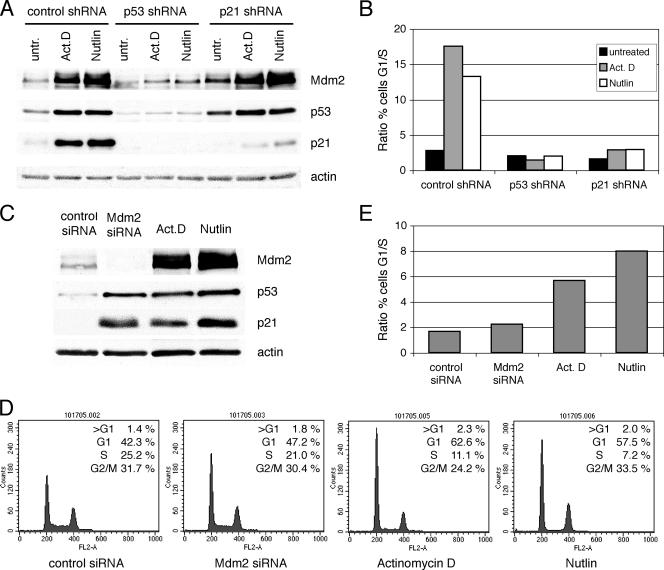
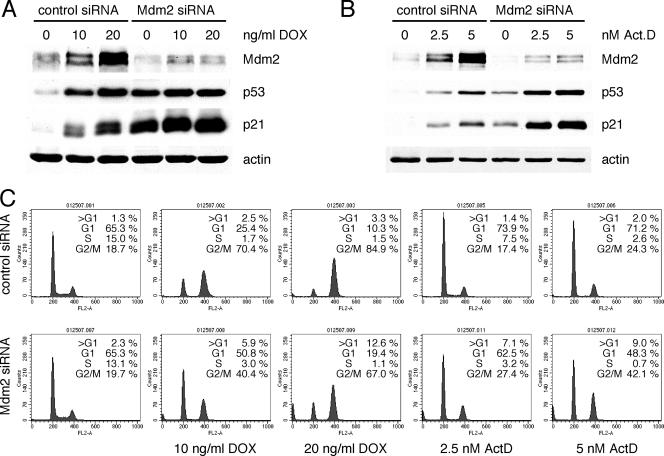
Induction of p53 and p21 to levels comparable to those seen with actinomycin D or nutlin is not sufficient to trigger cell cycle arrest in U2OS cells following Mdm2 siRNA treatment.
The cellular response to doxorubicin is known to have both p53-dependent and p53-independent components (36). It remained then formally possible that the cell cycle arrest induced by doxorubicin did not involve p53. To better address the ability of p53 and p21 to induce growth arrest when upregulated following ablation of Mdm2, transfection of cells with Mdm2 siRNA was compared to treatment with actinomycin D and nutlin-3. Low doses of actinomycin D have been shown to induce p53-dependent growth arrest (7). Nutlin-3 is a low-molecular-weight compound that activates p53 by disrupting its interaction with Mdm2 (45). U2OS stable clones expressing control shRNA were treated with actinomycin D and nutlin-3, and their response was compared to that of p53 or p21 shRNA clones (Fig. 7A). The G1 and G2 arrest caused by actinomycin D or nutlin-3 was severely impaired when p53 and p21 were downregulated, confirming their requirement (Fig. 7B). Next, p53 and p21 levels achieved by siRNA-mediated ablation of Mdm2 were compared to those obtained following treatment with actinomycin D and nutlin-3. Under all three sets of conditions, nearly identical p53 and p21 protein levels were observed (Fig. 7C). Surprisingly, in contrast to the drug-treated cells, those transfected with Mdm2 siRNA failed to undergo cell cycle arrest (Fig. 7D and E). These results showed that p53 and p21 levels that are capable of, and required for, triggering growth arrest in cells treated with actinomycin D or nutlin-3 fail to do so when they are induced by downregulation of Mdm2.
FIG. 7.
Induction of p53 and p21 to levels comparable to those seen with actinomycin D (Act.D) or nutlin-3 is not sufficient to trigger cell cycle arrest in U2OS cells following Mdm2 siRNA treatment. (A) Stable U2OS clones expressing control, p53, or p21 shRNA were treated with 5 nM actinomycin D or 2.5 μM nutlin-3. At 48 h after treatment, cell lysates were immunoblotted. untr., untreated. (B) Cells were treated as described above, and cell cycle profiles were analyzed by flow cytometry. The histogram shows the ratio of the percentage of cells in G1 phase to that of cells in S phase. (C) U2OS cells were transfected with control or Mdm2 siRNA oligonucleotides or were treated with actinomycin D or nutlin-3. At 48 h after treatment, cell lysates were immunoblotted. (D and E) Cells from corresponding dishes were stained with propidium iodide and analyzed by flow cytometry (D); the results are represented in panel E.
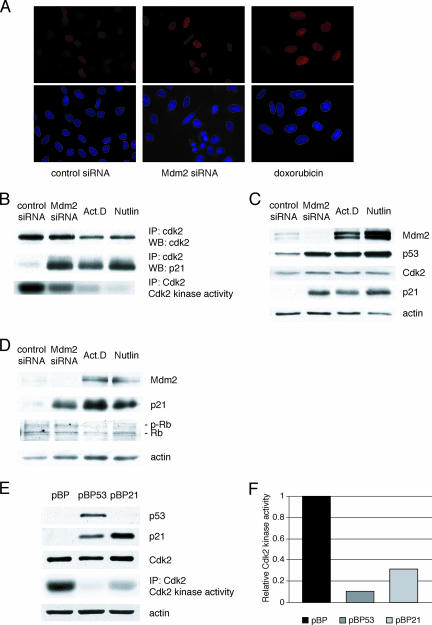
p21 upregulated following Mdm2 siRNA localizes to the nucleus and binds to Cdk2 but does not inhibit Cdk2 kinase activity.
Among the several target genes induced by p53 upon cellular stress, p21 is the best-characterized mediator of the G1 arrest and, to a lesser extent, the G2 arrest. To try to understand its inability to induce cell cycle arrest following Mdm2 downregulation, the cellular localization of p21 was first examined. U2OS cells were treated with control or Mdm2 siRNA or with doxorubicin, and the p21 protein was visualized by indirect immunofluorescence. p21 was upregulated and localized to the nucleus in cells treated with Mdm2 siRNA or doxorubicin, indicating that cellular localization was not the cause for its dysfunctionality (Fig. 8A). Next, the ability of p21 to bind to the cyclin-dependent kinase Cdk2 was investigated. Comparable amounts of p21 were found associated with Cdk2 in coimmunoprecipitation assays using extracts from cells treated with Mdm2 siRNA, actinomycin D, or nutlin-3 (Fig. 8B). This observation suggested that the inability of p21 to induce growth arrest in cells following Mdm2 downregulation could not be explained by an impaired association with Cdk2. However, when the Cdk2 kinase activity in the same extracts was assayed, it was found to be significantly decreased in extracts from cells treated with actinomycin D or nutlin-3 compared to that seen with extracts from cells treated with Mdm2 siRNA (Fig. 8B). Consistent with this, there was a reduction of the phosphorylated form of pRb in the extracts from the drug-treated cells (Fig. 8D). These results would argue that p21 upregulated by ablation of Mdm2 seems to require some additional activation in order to efficiently inhibit Cdk2 kinase activity once they are associated. Whether this activation involves posttranslational modification or interacting protein(s) remains to be explored.
FIG. 8.
p21 upregulated following Mdm2 siRNA treatment localizes to the nucleus and binds to Cdk2 but does not inhibit Cdk2 kinase activity. (A) U2OS cells were grown on coverslips and transfected with control or Mdm2 siRNA or treated with doxorubicin as indicated. At 48 h after treatment, p21 was localized by indirect immunofluorescence. (B) U2OS cells were transfected with control or Mdm2 siRNA or treated with 5 nM actinomycin D (Act.D) or 2.5 μM nutlin-3. At 48 h after treatment, cell extracts were immunoprecipitated with anti-Cdk2 antibody. p21 and Cdk2 were detected in the immunoprecipitates (IP) by immunoblotting (WB). Cdk2 in cell extracts from corresponding dishes was immunoprecipitated and assayed for histone H1 kinase activity. (C) Protein levels in extracts used as described for panel B were assayed by immunoblotting. (D) U2OS cells were treated as described for panel B, and Mdm2, p21, pRb, and actin levels in the cellular extracts were assayed by immunoblotting. (E) p53−/− MEFs were infected with control or p53- or p21-encoding retroviruses. At 48 h after infection, cell lysates were immunoblotted. Cdk2 kinase activity from corresponding dishes was assayed as described, and the results are quantitated in panel F.
Finally, ectopic expression of p21 in p53-null mouse embryo fibroblasts by retroviral infection produced p21 levels that were higher than those obtained by induction of endogenous p21 upon retroviral expression of p53. However, a greater reduction of Cdk2 kinase activity was observed in response to the p53-expressing retrovirus. This is consistent with the notion that that other downstream effectors of the p53 response contribute to Cdk2 inhibition (Fig. 8E and F).
Cellular stress induces cell cycle arrest despite downregulation of Mdm2.
The role of Mdm2 in the p53 response to cellular stress was examined next. U2OS cells were simultaneously transfected with control or Mdm2 siRNA oligonucleotides and treated with doxorubicin or actinomycin D. As a result of p53 activation, both drugs caused an upregulation of Mdm2 that was significantly reduced in the cells transfected with Mdm2 siRNA (Fig. 9A and B). The ablation of the induced Mdm2, however, was not complete. The p53 levels achieved by treatment with doxorubicin or actinomycin D were slightly greater in the cells transfected with Mdm2 siRNA than in the control cells. The induction of p21 was significantly more pronounced in the Mdm2-ablated cells. Mdm2 has been shown to modulate p21 levels, but little is know concerning whether this regulation is maintained or affected during the cellular response to stress. It is therefore possible that the combination of Mdm2 downregulation and treatment with p53-activating drugs could trigger a more robust induction of p21 due to both increased transcription and protein stability. Doxorubicin and actinomycin D caused growth arrest in the control cells, as shown previously. Both drugs were also able to induce cell cycle arrest in the cells in which Mdm2 was downregulated by siRNA (Fig. 9C). Similar results were obtained using two independent Mdm2 siRNA duplexes, although the downregulation was not as efficient as with the pool of oligonucleotides (data not shown). One explanation for the results obtained using combined treatments of Mdm2 siRNA and doxorubicin or actinomycin is that residual amounts of Mdm2 were sufficient to allow p21 to be fully active. Alternatively, it is possible that the increased p21 levels in the Mdm2-ablated cells were able to overcome a requirement for an additional activation.
FIG. 9.
Cellular stress induces cell cycle arrest despite downregulation of Mdm2. (A and B) U2OS cells were transfected with the indicated siRNA oligonucleotides and treated with doxorubicin (DOX) (A) or actinomycin D (Act.D) (B) as described in Material and Methods. At 48 h after treatment, cell lysates were immunoblotted. (C) U2OS cells from corresponding dishes were stained with propidium iodide and analyzed by flow cytometry.
DISCUSSION
The p53 pathway is inoperative in most cancer cells, with p53 being frequently inactivated by mutation. In tumor cells that retain wild-type p53, the pathway is inactivated by other mechanisms, including Mdm2 overexpression or loss of p14ARF (46). Because of this, restoration of the p53 pathway has been the focus of intensive research. In the case of wild-type p53-expressing tumors, disruption of the negative regulation of p53 by Mdm2 represents an attractive approach for therapeutic treatment. Several strategies have been devised and investigated to this purpose, including the use of small compounds and p14ARF-derived peptides that disrupt the p53-Mdm2 interaction and the use of antisense or siRNA oligonucleotides that downregulate Mdm2 expression levels (32, 44, 55). In addition, some of these tools have been used to investigate the role of posttranslational modification in p53 activities. With respect to Mdm2 downregulation, most published reports have used an antisense approach with modified backbones, and many of those studies presented conflicting results involving nonspecific effects caused by oligonucleotides (48, 50), p53-independent effects of Mdm2 (49, 53), or nonphysiological levels of p53 (48, 51). The goal of the present study was to evaluate the growth-suppressive ability of p53 and p21 when they are upregulated at physiological levels by siRNA-mediated downregulation of Mdm2 in the absence of DNA damage or other types of cellular stress.
Abrogation of Mdm2 expression by siRNA oligonucleotides led to stabilization of p53 without inducing DNA damage and the associated modifications of p53 at serine 15 or lysine 382 (Fig. 1). p53 was nevertheless capable of binding to DNA in a sequence-specific manner, both in vitro and in cells, in a manner comparable to that of p53 activated by treatment with the DNA-damaging agent doxorubicin (Fig. 2). Following Mdm2 downregulation, p21 was transcriptionally upregulated in p53-expressing cells but not in cells in which p53 expression had been ablated by shRNA (Fig. 4A). This observation is in contrast with some previously reported studies in which treatment of cells with anti-Mdm2 antisense oligonucleotides induced p21 mRNA levels in prostate cancer cells regardless of their p53 status. This suggested that downregulation of Mdm2 affected p21 transcriptional activation in a yet-unknown p53-independent manner (55). Subsequent studies provided evidence of an interaction between Mdm2 and p21 in cells and resulted in reports that Mdm2 reduced p21 protein stability in a ubiquitin- and p53-independent way (56). Although these results can account for the upregulation of p21 protein levels in p53 mutant-null cells, the mechanism(s) by which Mdm2 downregulation would lead to induction of p21 mRNA in these cells still remains to be elucidated.
In multiple cancer and normal cell lines, downregulation of Mdm2 protein levels caused accumulation of p53. The p53 target gene p21 was equally upregulated, presumably by a combined effect of p53-mediated transcriptional induction and increased stability in the absence of Mdm2 (Fig. 1, 5, and 6). Surprisingly, cells treated with Mdm2 siRNA, in spite of attaining p53 and p21 levels similar to those obtained by treatment with different drugs such as doxorubicin, actinomycin D, or nutlin-3, failed to undergo cell cycle arrest (Fig. 5 to 7). It has been reported that in the absence of Mdm2, the p53 response can be switched from growth arrest to apoptosis (13). Under these conditions, however, it is unlikely that the observed impaired growth arrest could result from early apoptosis of the cells that were responsive to the upregulation of p53 following Mdm2 ablation. There were no indications of cell death by microscopic examination of cell morphology or measurements of cell number. Importantly, the absence of floating cells and lack of a hypodiploid fraction detectable by flow cytometric analysis at 24 and 48 h (Fig. 5 to 7 and 9 and data not shown) also support the idea of the absence of apoptosis. In fact, a slight increase in cells with <2N DNA content was observed only when Mdm2 siRNA treatment was combined with doxorubicin or actinomycin D treatment (Fig. 9C).
Transfection of cells with Mdm2 siRNA oligonucleotides did not appear to cause DNA damage or activate the ATM pathway, since γH2AX reactivity was not detected (Fig. 1E). In addition, p53 upregulated by Mdm2 siRNA showed no detectable phosphorylation at serine 15 or acetylation at lysine 382, two of the best-characterized posttranslational modification sites of p53 (Fig. 1C to E). Therefore, a possible explanation for the impaired growth arrest could be that p53 requires posttranscriptional modifications in order to be fully active. Several reports, however, argue against this hypothesis, as the use of nutlins or overexpression of p14ARF activates p53 without inducing phosphorylation at several key residues (23, 44). Another possible reason could be that a p53-independent pathway cooperates with the p53 pathway to block cell cycle progression. This could be the case in cells treated with doxorubicin, but the cell cycle arrest observed following treatment with nutlin-3 or low doses of actinomycin D proved to be strictly p53 and p21 dependent (Fig. 7). Taken together, these results would suggest that upregulation of p53 and p21 at physiological levels is required but not sufficient to trigger growth arrest. The possibility that p21 might be inoperative due to mislocalization to the cytoplasm was ruled out, as it was detected in the nucleus by immunofluorescence assays (Fig. 8A). In addition, p21 was able to interact with Cdk2, but for reasons that remain to be explored, it appeared to be unable to significantly inhibit its kinase activity (Fig. 8B and C). Interestingly, downregulation of Mdm2 led to a reduction in Cdk2 activity that was intermediate between the activity levels seen with control cells and those seen with cells treated with actinomycin D or nutlin-3. Yet this partial inhibition of Cdk2 was not sufficient to prevent cell cycle progression. This observation suggests that there is a threshold for the reduction in kinase activity that is needed to cause growth arrest.
It is possible to speculate that following Mdm2 siRNA treatment, resultant differences, including posttranslational modification or levels of interacting proteins, might affect the ability of p21 to inhibit Cdk2. To test this possibility, the p53 response to cellular stress was tested in cells in which Mdm2 was downregulated by siRNA transfection. Doxorubicin or actinomycin D treatment was able to cause cell cycle arrest even in the presence of reduced Mdm2 levels (Fig. 9C). This result could be due to residual amounts of Mdm2 exerting an effect or, alternatively, to the enhanced upregulation of p21 that is observed.
Early studies of p21 and cdk-cyclin complexes resulted in reports that most of the active Cdk2/cyclin complexes found in normal diploid fibroblasts contained p21 (19, 54). In vitro studies using recombinant proteins suggested that multiple p21 molecules bound to a Cdk2/cyclin complex and led the authors to propose a model in which p21 could associate with the Cdk2 complex through noninhibitory and inhibitory binding sites (19). In contrast to this result, analysis of the crystal structure of a complex of an N-terminal peptide of p27, Cdk2, and cyclin A suggested that the presence of a single p27 molecule should be sufficient to inhibit Cdk2 activity (39). However, p21 differs from the other cyclin-dependent kinase inhibitors p27 and p57 in that its C terminus contains a second Cy region and is capable of binding to several factors (8). In addition, in vitro studies using the N terminus of p21 indicated that the nature of the interaction of p21 with the Cdk2/cyclin E or Cdk2/cyclin A complexes is different. This was shown by detection of differential sensitivities to amino acid substitutions in p21, deletion of its N-terminal Cy domain, and p21 monoclonal antibodies (52). Several reports support the idea that p21 can associate with Cdk2/cyclin complexes without inhibiting its activity and that interacting proteins might modulate p21 function. A study of growth inhibition caused by the presence of transforming growth factor β1 showed that an increased association of p21 to Cdk2 correlated with inhibition of its kinase activity in normal prostate cells but not cancer cells, making the latter insensitive to the treatment (9). Two independent publications reported that the human papillomavirus 16 E7 oncoprotein can interact with the C terminus of p21 and, without displacing it from the Cdk2/cyclin complex, block its ability to inhibit Cdk2 kinase activity (16, 25). A similar function was also attributed to the SET/TAF-1/I2PP2A oncoprotein, which associated with p21 through its C terminus and reversed the inhibition by p21 of Cdk2/cyclin E complexes but not Cdk2/cyclin A complexes (15). Along the same line, but causing the opposite effect, the p21-interacting protein TOK-1 was isolated in a two-hybrid screening. One of the TOK-1-characterized isoforms colocalized with p21 in nuclei, bound to its C terminus, and enhanced the inhibition of Cdk2 by p21 (34). Examination of p21/Cdk2/cyclin complexes in cells treated with Mdm2 siRNA or genotoxic drugs might provide some insight into the mechanisms that modulate p21 activity in the absence and presence of cellular stress.
Because it is overexpressed in several types of tumors and also due to the fact that it is the main negative regulator of the tumor suppressor p53, Mdm2 is considered to be an oncogene (22). In addition to p53, Mdm2 promotes the degradation of several growth-suppressive proteins, and multiple ways have been reported by which Mdm2 can counteract both p53-dependent and-independent pathways that trigger cell cycle arrest (3, 24, 40). The Mdm2 protein, however, has another face. In some cell lines, Mdm2 overexpression has been reported to induce G1 arrest, an activity that appears to be impaired in most cancer cell lines (4, 11, 57). Stable expression of excess Mdm2 in H1299 cells that express tetracycline-regulated p53 led to an increase in the fraction of cells arrested in G2 upon induction of p53 (33). Downregulation of Mdm2 by siRNA is likely to affect numerous proteins in addition to p53 and p21. In fact, whereas after DNA damage Mdm2 promotes MdmX degradation (26, 35), downregulation of Mdm2 by siRNA caused an upregulation of MdmX protein levels (data not shown). Increased MdmX levels are expected to negatively affect p53 transcriptional activity. This could account for the different efficiencies of p53 in regulating transcription observed in cells treated with Mdm2 siRNA compared to that resulting from doxorubicin treatment. These differences were not of great magnitude and, in the case of p21, were compensated by its increased protein stability. However, with a continuously growing number of p53 target genes involved in cell cycle arrest, it is possible to imagine that increased MdmX levels might have a more profound impact on the p53 transcriptional regulation of some of them. Finally, it is also possible that, in addition to MdmX, other targets of degradation by Mdm2 will be identified that might contribute to cell cycle arrest. For instance, a factor that modulates the ability of p21 to inhibit Cdk2 kinase activity could be one of such Mdm2 targets.
The multiplicity and variety of Mdm2 functions are most probably obscured by cell type-specific effects, most notably by potential different functions of the many splice variants that are generated from the mdm2 gene in different cell lines (20). A more careful and detailed examination of Mdm2 multiple activities in both its full-length and its spliced forms is required. Finally, a detailed examination of p21, in terms of posttranslational modifications and/or interacting partners, needs to be undertaken. This will provide molecular details of the mechanisms that allow p21 to trigger cell cycle arrest when upregulated at physiological levels.
Acknowledgments
We thank A. W. Lin (Roswell Park Cancer Institute) for the p21 shRNA stable cell line and G. Lozano (M. D. Anderson Cancer Center) for the p53-null mouse embryo fibroblasts. We are grateful to Matthew O'Connell and Zhen-Qiang Pan for their technical advice and helpful discussions and for sharing reagents. Present and past members of the Manfredi laboratory Lois Resnick-Silverman, Dana Lukin, Anthony Mastropietro, Selvon St. Clair, Wendy Liu, Sejal Patel, Shohreh Varmeh-Ziaie, Pierre-Jacques Hamard, Luis Carvajal, and Kester Haye are thanked for their help and support. Quantitative PCR was performed at the Mount Sinai shared research facility.
These studies were supported by a grant from the National Cancer Institute (R01 CA86001).
Footnotes
Published ahead of print on 19 March 2007.
REFERENCES
- 1.Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764-2772. [DOI] [PubMed] [Google Scholar]
- 2.Ashcroft, M., Y. Taya, and K. H. Vousden. 2000. Stress signals utilize multiple pathways to stabilize p53. Mol. Cell. Biol. 20:3224-3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyd, M. T., N. Vlatkovic, and D. S. Haines. 2000. A novel cellular protein (MTBP) binds to MDM2 and induces a G1 arrest that is suppressed by MDM2. J. Biol. Chem. 275:31883-31890. [DOI] [PubMed] [Google Scholar]
- 4.Brown, D. R., C. A. Thomas, and S. P. Deb. 1998. The human oncoprotein MDM2 arrests the cell cycle: elimination of its cell-cycle-inhibitory function induces tumorigenesis. EMBO J. 17:2513-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brugarolas, J., C. Chandrasekaran, J. I. Gordon, D. Beach, T. Jacks, and G. J. Hannon. 1995. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377:552-557. [DOI] [PubMed] [Google Scholar]
- 6.Bunz, F., A. Dutriaux, C. Lengauer, T. Waldman, S. Zhou, J. P. Brown, J. M. Sedivy, K. W. Kinzler, and B. Vogelstein. 1998. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282:1497-1501. [DOI] [PubMed] [Google Scholar]
- 7.Chang, D., F. Chen, F. Zhang, B. C. McKay, and M. Ljungman. 1999. Dose-dependent effects of DNA-damaging agents on p53-mediated cell cycle arrest. Cell Growth Differ. 10:155-162. [PubMed] [Google Scholar]
- 8.Child, E. S., and D. J. Mann. 2006. The intricacies of p21 phosphorylation: protein/protein interactions, subcellular localization and stability. Cell Cycle 5:1313-1319. [DOI] [PubMed] [Google Scholar]
- 9.Cipriano, S. C., and Y. Q. Chen. 1998. Insensitivity to growth inhibition by TGF-β1 correlates with a lack of inhibition of the CDK2 activity in prostate carcinoma cells. Oncogene 17:1549-1556. [DOI] [PubMed] [Google Scholar]
- 10.Craig, A. L., L. Burch, B. Vojtesek, J. Mikutowska, A. Thompson, and T. R. Hupp. 1999. Novel phosphorylation sites of human tumour suppressor protein p53 at Ser20 and Thr18 that disrupt the binding of mdm2 (mouse double minute 2) protein are modified in human cancers. Biochem. J. 342(Pt. 1):133-141. [PMC free article] [PubMed] [Google Scholar]
- 11.Dang, J., M. L. Kuo, C. M. Eischen, L. Stepanova, C. J. Sherr, and M. F. Roussel. 2002. The RING domain of Mdm2 can inhibit cell proliferation. Cancer Res. 62:1222-1230. [PubMed] [Google Scholar]
- 12.Deng, C., P. Zhang, J. W. Harper, S. J. Elledge, and P. Leder. 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82:675-684. [DOI] [PubMed] [Google Scholar]
- 13.de Rozieres, S., R. Maya, M. Oren, and G. Lozano. 2000. The loss of mdm2 induces p53-mediated apoptosis. Oncogene 19:1691-1697. [DOI] [PubMed] [Google Scholar]
- 14.Espinosa, J. M., R. E. Verdun, and B. M. Emerson. 2003. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol. Cell 12:1015-1027. [DOI] [PubMed] [Google Scholar]
- 15.Estanyol, J. M., M. Jaumot, O. Casanovas, A. Rodriguez-Vilarrupla, N. Agell, and O. Bachs. 1999. The protein SET regulates the inhibitory effect of p21(Cip1) on cyclin E-cyclin-dependent kinase 2 activity. J. Biol. Chem. 274:33161-33165. [DOI] [PubMed] [Google Scholar]
- 16.Funk, J. O., S. Waga, J. B. Harry, E. Espling, B. Stillman, and D. A. Galloway. 1997. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 11:2090-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 18.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 19.Harper, J. W., S. J. Elledge, K. Keyomarsi, B. Dynlacht, L. H. Tsai, P. Zhang, S. Dobrowolski, C. Bai, L. Connell-Crowley, E. Swindell, et al. 1995. Inhibition of cyclin-dependent kinases by p21. Mol. Biol. Cell 6:387-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris, L. C. 2005. MDM2 splice variants and their therapeutic implications. Curr. Cancer Drug Targets 5:21-26. [DOI] [PubMed] [Google Scholar]
- 21.Hupp, T. R., D. W. Meek, C. A. Midgley, and D. P. Lane. 1992. Regulation of the specific DNA binding function of p53. Cell 71:875-886. [DOI] [PubMed] [Google Scholar]
- 22.Iwakuma, T., and G. Lozano. 2003. MDM2, an introduction. Mol. Cancer Res. 1:993-1000. [PubMed] [Google Scholar]
- 23.Jackson, M. W., M. K. Agarwal, M. L. Agarwal, A. Agarwal, P. Stanhope-Baker, B. R. Williams, and G. R. Stark. 2004. Limited role of N-terminal phosphoserine residues in the activation of transcription by p53. Oncogene 23:4477-4487. [DOI] [PubMed] [Google Scholar]
- 24.Jin, Y., H. Lee, S. X. Zeng, M. S. Dai, and H. Lu. 2003. MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 22:6365-6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones, D. L., R. M. Alani, and K. Munger. 1997. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 11:2101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawai, H., D. Wiederschain, H. Kitao, J. Stuart, K. K. Tsai, and Z. M. Yuan. 2003. DNA damage-induced MDMX degradation is mediated by MDM2. J. Biol. Chem. 278:45946-45953. [DOI] [PubMed] [Google Scholar]
- 27.Kubbutat, M. H., R. L. Ludwig, M. Ashcroft, and K. H. Vousden. 1998. Regulation of Mdm2-directed degradation by the C terminus of p53. Mol. Cell. Biol. 18:5690-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert, P. F., F. Kashanchi, M. F. Radonovich, R. Shiekhattar, and J. N. Brady. 1998. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 273:33048-33053. [DOI] [PubMed] [Google Scholar]
- 29.Lu, H., R. P. Fisher, P. Bailey, and A. J. Levine. 1997. The CDK7-cycH-p36 complex of transcription factor IIH phosphorylates p53, enhancing its sequence-specific DNA binding activity in vitro. Mol. Cell. Biol. 17:5923-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maki, C. G., J. M. Huibregtse, and P. M. Howley. 1996. In vivo ubiquitination and proteasome-mediated degradation of p53(1). Cancer Res. 56:2649-2654. [PubMed] [Google Scholar]
- 31.Meek, D. W., and U. Knippschild. 2003. Posttranslational modification of MDM2. Mol. Cancer Res. 1:1017-1026. [PubMed] [Google Scholar]
- 32.Midgley, C. A., J. M. Desterro, M. K. Saville, S. Howard, A. Sparks, R. T. Hay, and D. P. Lane. 2000. An N-terminal p14ARF peptide blocks Mdm2-dependent ubiquitination in vitro and can activate p53 in vivo. Oncogene 19:2312-2323. [DOI] [PubMed] [Google Scholar]
- 33.Ohkubo, S., T. Tanaka, Y. Taya, K. Kitazato, and C. Prives. 2006. Excess HDM2 impacts cell cycle and apoptosis and has a selective effect on p53 dependent transcription. J. Biol. Chem. 281:16943-16950. [DOI] [PubMed] [Google Scholar]
- 34.Ono, T., H. Kitaura, H. Ugai, T. Murata, K. K. Yokoyama, S. M. Iguchi-Ariga, and H. Ariga. 2000. TOK-1, a novel p21Cip1-binding protein that cooperatively enhances p21-dependent inhibitory activity toward CDK2 kinase. J. Biol. Chem. 275:31145-31154. [DOI] [PubMed] [Google Scholar]
- 35.Pan, Y., and J. Chen. 2003. MDM2 promotes ubiquitination and degradation of MDMX. Mol. Cell. Biol. 23:5113-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Passalaris, T. M., J. A. Benanti, L. Gewin, T. Kiyono, and D. A. Galloway. 1999. The G2 checkpoint is maintained by redundant pathways. Mol. Cell. Biol. 19:5872-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez, M. S., J. M. Desterro, S. Lain, D. P. Lane, and R. T. Hay. 2000. Multiple C-terminal lysine residues target p53 for ubiquitin-proteasome-mediated degradation. Mol. Cell. Biol. 20:8458-8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rousseau, D., D. Cannella, J. Boulaire, P. Fitzgerald, A. Fotedar, and R. Fotedar. 1999. Growth inhibition by CDK-cyclin and PCNA binding domains of p21 occurs by distinct mechanisms and is regulated by ubiquitin-proteasome pathway. Oncogene 18:4313-4325. [DOI] [PubMed] [Google Scholar]
- 39.Russo, A. A., P. D. Jeffrey, A. K. Patten, J. Massague, and N. P. Pavletich. 1996. Crystal structure of the p27Kip1 cyclin-dependent-kinase inhibitor bound to the cyclin A-Cdk2 complex. Nature 382:325-331. [DOI] [PubMed] [Google Scholar]
- 40.Sdek, P., H. Ying, D. L. Chang, W. Qiu, H. Zheng, R. Touitou, M. J. Allday, and Z. X. Xiao. 2005. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol. Cell 20:699-708. [DOI] [PubMed] [Google Scholar]
- 41.Shieh, S. Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 42.St. Clair, S., L. Giono, S. Varmeh-Ziaie, L. Resnick-Silverman, W. J. Liu, A. Padi, J. Dastidar, A. DaCosta, M. Mattia, and J. J. Manfredi. 2004. DNA damage-induced downregulation of Cdc25C is mediated by p53 via two independent mechanisms: one involves direct binding to the cdc25C promoter. Mol. Cell 16:725-736. [DOI] [PubMed] [Google Scholar]
- 43.Takenaka, I., F. Morin, B. R. Seizinger, and N. Kley. 1995. Regulation of the sequence-specific DNA binding function of p53 by protein kinase C and protein phosphatases. J. Biol. Chem. 270:5405-5411. [DOI] [PubMed] [Google Scholar]
- 44.Thompson, T., C. Tovar, H. Yang, D. Carvajal, B. T. Vu, Q. Xu, G. M. Wahl, D. C. Heimbrook, and L. T. Vassilev. 2004. Phosphorylation of p53 on key serines is dispensable for transcriptional activation and apoptosis. J. Biol. Chem. 279:53015-53022. [DOI] [PubMed] [Google Scholar]
- 45.Vassilev, L. T., B. T. Vu, B. Graves, D. Carvajal, F. Podlaski, Z. Filipovic, N. Kong, U. Kammlott, C. Lukacs, C. Klein, N. Fotouhi, and E. A. Liu. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844-848. [DOI] [PubMed] [Google Scholar]
- 46.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 47.Waldman, T., K. W. Kinzler, and B. Vogelstein. 1995. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 55:5187-5190. [PubMed] [Google Scholar]
- 48.Wang, H., L. Nan, D. Yu, S. Agrawal, and R. Zhang. 2001. Antisense anti-MDM2 oligonucleotides as a novel therapeutic approach to human breast cancer: in vitro and in vivo activities and mechanisms. Clin. Cancer Res. 7:3613-3624. [PubMed] [Google Scholar]
- 49.Wang, H., L. Nan, D. Yu, J. R. Lindsey, S. Agrawal, and R. Zhang. 2002. Anti-tumor efficacy of a novel antisense anti-MDM2 mixed-backbone oligonucleotide in human colon cancer models: p53-dependent and p53-independent mechanisms. Mol. Med. 8:185-199. [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, H., S. Wang, L. Nan, D. Yu, S. Agrawal, and R. Zhang. 2002. Antisense anti-MDM2 mixed-backbone oligonucleotides enhance therapeutic efficacy of topoisomerase I inhibitor irinotecan in nude mice bearing human cancer xenografts: in vivo activity and mechanisms. Int. J. Oncol. 20:745-752. [PubMed] [Google Scholar]
- 51.Wang, H., D. Yu, S. Agrawal, and R. Zhang. 2003. Experimental therapy of human prostate cancer by inhibiting MDM2 expression with novel mixed-backbone antisense oligonucleotides: in vitro and in vivo activities and mechanisms. Prostate 54:194-205. [DOI] [PubMed] [Google Scholar]
- 52.Wohlschlegel, J. A., B. T. Dwyer, D. Y. Takeda, and A. Dutta. 2001. Mutational analysis of the Cy motif from p21 reveals sequence degeneracy and specificity for different cyclin-dependent kinases. Mol. Cell. Biol. 21:4868-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Würl, P., F. Bartel, A. Meye, M. Kappler, M. Bache, H. Schmidt, M. Schonfelder, and H. Taubert. 2002. Growth reduction of a xenotransplanted human soft tissue sarcoma by MDM2 antisense therapy via implanted osmotic minipumps. Int. J. Oncol. 20:1087-1093. [PubMed] [Google Scholar]
- 54.Zhang, H., G. J. Hannon, and D. Beach. 1994. p21-containing cyclin kinases exist in both active and inactive states. Genes Dev. 8:1750-1758. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, Z., M. Li, H. Wang, S. Agrawal, and R. Zhang. 2003. Antisense therapy targeting MDM2 oncogene in prostate cancer: effects on proliferation, apoptosis, multiple gene expression, and chemotherapy. Proc. Natl. Acad. Sci. USA 100:11636-11641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, Z., H. Wang, M. Li, S. Agrawal, X. Chen, and R. Zhang. 2004. MDM2 is a negative regulator of p21WAF1/CIP1, independent of p53. J. Biol. Chem. 279:16000-16006. [DOI] [PubMed] [Google Scholar]
- 57.Zhou, R., R. Frum, S. Deb, and S. P. Deb. 2005. The growth arrest function of the human oncoprotein mouse double minute-2 is disabled by downstream mutation in cancer cells. Cancer Res. 65:1839-1848. [DOI] [PubMed] [Google Scholar]