Abstract
Histone methylation is an important posttranslational modification that contributes to chromatin-based processes including transcriptional regulation, DNA repair, and epigenetic inheritance. In the budding yeast Saccharomyces cerevisiae, histone lysine methylation occurs on histone H3 lysines 4, 36, and 79, and its deposition is coupled mainly to transcription. Until recently, histone methylation was considered to be irreversible, but the identification of histone demethylase enzymes has revealed that this modification can be dynamically regulated. In budding yeast, there are five proteins that contain the JmjC domain, a signature motif found in a large family of histone demethylases spanning many organisms. One JmjC-domain-containing protein in budding yeast, Jhd1, has recently been identified as being a histone demethylase that targets H3K36 modified in the di- and monomethyl state. Here, we identify a second JmjC-domain-containing histone demethylase, Rph1, which can specifically demethylate H3K36 tri- and dimethyl modification states. Surprisingly, Rph1 can remove H3K9 methylation, a histone modification not found in budding yeast chromatin. The capacity of Rph1 to demethylate H3K9 provides the first indication that S. cerevisiae may have once encoded an H3K9 methylation system and suggests that Rph1 is a functional vestige of this modification system.
Posttranslational modification of histone molecules within chromatin contributes epigenetic information to the underlying DNA-based genetic code (17). Recently, the histone lysine (K) methylation system has attracted a significant amount of interest due to its widespread roles in transcriptional regulation, DNA repair, and epigenetic inheritance (24). In higher eukaryotes, histone lysine methylation occurs on histone H3K4, K9, K27, K36, and K79 and histone H4K20. In general, histone H3K4, K36, and K79 methylation is associated with actively transcribed genes, whereas H3K9, K27, and H4K20 methylation is associated with silenced regions (24). Budding yeast has a less complex histone methylation system that encodes three histone lysine methyltransferase enzymes, Set1, Set2, and Dot1, that modify H3K4, K36, and K79, respectively (25). Histone lysine methylation in budding yeast is tightly coupled to the process of transcription, and the deposition of these modifications occurs mainly during the initiation and elongation phases of RNA polymerase II-based transcription (25). In particular, H3K36 methylation is tightly coupled to the process of active transcriptional elongation and forms an increasing concentration gradient from the 5′ to the 3′ end of the gene. H3K36 profiles are dictated by the preferential association of Set2 with the elongating form of RNA polymerase II, which is phosphorylated on Ser2 of the C-terminal domain (19, 21, 23, 31, 43). The functional outcome of histone methylation is often elicited through effector proteins that specifically recognize and interpret these histone modifications. In budding yeast, H3K36 methylation is recognized by the chromodomain protein EAF3, which is a stable component of the Sin3 corepressor complex. EAF3 acts as an effector protein by recruiting the Sin3 corepressor complex to the body of yeast genes, where it inhibits intragenic transcription (4, 13, 14). Under standard laboratory growth conditions, budding yeast lacking H3K36 methylation shows no obvious cellular defects. However, the widespread involvement of this modification in transcriptional elongation suggests that H3K36 methylation may have important roles in transcriptional fidelity under certain environmental or growth conditions.
Until recently, histone methylation was considered to be a static modification, but the identification of histone demethylase enzymes has revealed that this modification can be dynamically regulated (6, 15, 16, 32, 36, 41, 44). Thus far, two histone demethylase enzyme families have been identified, the LSD1 family and the JmjC-domain-containing family. These enzymes are potentially important chromatin regulators, given their capacity to modify epigenetic information through the direct removal of histone lysine methylation marks. Functional characterization of existing histone demethylase enzymes has revealed that individual enzymes recognize specific lysine residues and can distinguish between the monomethylation (me1), dimethylation (me2), and trimethylation (me3) states of their target substrates (16). The budding yeast genome is predicted to encode five JmjC-domain-containing proteins but has no apparent LSD1 homologue. JmjC-domain-containing proteins achieve histone demethylation by an oxidative mechanism requiring iron and alpha-ketoglutarate as cofactors and are capable of removing all three histone lysine methylation states (16). Jhd1 is the only active JmjC-domain-containing histone demethylase identified in budding yeast, and it targets the demethylation of H3K36me2 and H3K36me1 (36). Bioinformatic analysis indicates that other JmjC-domain-containing proteins in budding yeast may be enzymatically active based on the conservation of important cofactor binding residues and therefore may constitute novel histone demethylases (15).
Here, we characterize a second budding yeast JmjC-domain-containing protein, Rph1, and reveal that it is an H3K36 demethylase capable of removing the me3 modification state. Biochemical analysis of Rph1 demonstrates that this enzyme is also capable of removing H3K9 methylation despite the fact that S. cerevisiae chromatin lacks this modification. These observations reveal that H3K36me3 is a reversible modification in budding yeast and suggest that Rph1-mediated demethylation of H3K9 may be a functional vestige of an extinct H3K9 methylation system in S. cerevisiae.
MATERIALS AND METHODS
Expression plasmids and recombinant protein.
For recombinant protein expression, RPH1 was PCR amplified from yeast genomic DNA and cloned into the NcoI and NotI sites of pET28a (Novagen) encoding a C-terminal His tag. The H235A mutation in the predicted iron binding site was generated by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene). Deletion constructs were also generated by PCR and cloned into pET28a containing a C-terminal His tag. Recombinant protein was purified under native conditions according to the manufacturer's recommendations by using Ni-nitrilotriacetic acid (QIAGEN) affinity chromatography. For the expression of Rph1 in yeast, full-length Rph1 and Rph1 deletions were cloned into pAD4M (2μm Ampr LEU2 ADH1 promoter/terminator) containing an N-terminal Flag tag. In all cases, the sequences of PCR-amplified clones were confirmed by sequencing.
Demethylation assay and mass spectrometry.
All histone substrates were radioactively labeled as described previously (16, 36) using equal counts of labeled substrate for histone demethylation reactions. Histone demethylation and mass spectrometry assays were carried out as described previously (36). H3K36 peptide substrates used in mass spectrometry analyses encompass amino acids 28 to 45, containing a trimethyl modification, amino acids 32 to 42, containing a dimethyl modification, and amino acids 21 to 45, containing a monomethyl modification (Upstate Biotechnology). The H3K9me3 peptide substrate used in mass spectrometry analyses encompass amino acids 1 to 18 of histone H3 (Upstate Biotechnology). For Western blot analysis of histones after demethylase assays, the following antibodies were used at dilutions ranging from 1:200 to 1:1,000: Ab8898 for H3K9me3 (Abcam), Ab9050 for H3K36me3 (Abcam), and Ab9048 for H3K36me2 and H3K36me1 (Abcam).
Strains.
All strains except those in the telomeric silencing assay are of the BY4741 background. Strains used in the telomeric silencing assay are isogenic to strain YCB647 (35). The rph1Δ strain was generated by homologous recombination using a PCR-amplified natMX knockout cassette (10). Rph1 was Flag tagged by amplification of a p3Flag-KanMX cassette (9) using primer A (CCGCAGGACGGGAAAGCGGCCATTAATCAACAGAGTACACCTTTAAACAGGGAACAAAAGCTGGAG) and primer B (GCCTTCAAAATGAGAGATCTCGGTAAACAACTGGCAATGGTGAGTCACTATAGGGCGAATTGGGT) and introduced into strain BY4741 by homologous recombination.
Size exclusion chromatography and sucrose gradient analysis.
Whole-cell yeast extract or recombinant Rph1 (rRph1) was fractionated over a 24-ml Superose 6 size exclusion column (Amersham Biosciences) equilibrated with BC400 (40 mM HEPES [pH 7.9], 400 mM KCl, 0.5 mM dithiothreitol, 10% glycerol, 0.2 mM phenylmethylsulfonyl fluoride) with the aid of an AKTA purifier (Amersham biosciences) at a flow rate of 0.2 ml/min, and 250-μl fractions were collected. Every other fraction was analyzed for Rph1 by Western blotting or Coomassie staining. Sucrose gradients were formed at 4°C in 13-ml SW40 tubes using a manual two-chamber gradient former. Chamber 1 was loaded with buffer A (300 mM KCl, 20 mM HEPES [pH 7.9], 10% glycerol, 10 mM beta-mercaptoethanol) containing 5% sucrose, and chamber 2 was loaded with buffer A containing 20% sucrose. Rph1 and protein molecular weight markers were applied to the 5 to 20% sucrose gradient and centrifuged at 40,000 rpm in an SW40 rotor for 19 h at 4°C. Fractions (500 μl) were collected manually from the top of the gradient using a peristaltic pump fitted with a capillary tube. Each fraction was trichloroacetic acid precipitated and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie staining.
Native molecular weight and frictional coefficient calculations.
To determine the native molecular weight (Mr) and frictional coefficient (f/f0) of rRph1, the values obtained for radius and sedimentation in Fig. 4 were applied to equations 1 and 2 (34):
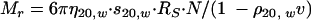 |
(1) |
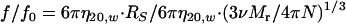 |
(2) |
where RS is Stoke's radius (cm), s20,w is the sedimentation velocity (S × 10−13), η20,w is the viscosity of water at 20°C (0.01002 g·s−1 cm−1), N is Avogadro's number (6.022 × 1023·mol−1), ρ20,w is the density of water at 20°C (0.9981 g·cm3), and ν is the partial specific volume (used 0.725 cm3/g).
FIG. 4.
Rph1 demethylates H3K36 in vivo. (A) Schematic representation of Flag-tagged Rph1 and Rph1 H235A yeast overexpression constructs. (B) Western blot analysis of yeast whole-cell extracts expressing Flag-Rph1 proteins. The asterisk denotes a cross-reactive band in the yeast extract, and the arrows indicate the overexpressed Rph1 proteins. (C) Yeast strains overexpressing Rph1 and Rph1 H235A constructs displayed in A were analyzed for growth (right). The left panel indicates the identity of each strain. Full-length Rph1 caused a severe defect in growth that was not alleviated by mutation of the catalytic domain. Deletion of the ZF domain rescued the growth defect, but in all cases, mutation of the catalytic domain had no effect on cell growth. (D) H3K36 methylation levels were analyzed in cells overexpressing Rph1 ΔZF and Rph1 JmjN/JmjC proteins (schematic representation on the left) using H3K36 modification-specific antibodies. An arrow indicates the H3K36 modification-specific band. The overexpression of both proteins caused a reduction in the levels of H3K36me3 and an increase in the levels of H3K36me2 and H3K36me1. Mutation of the catalytic domain abolished this effect, verifying that demethylation was a direct consequence of Rph1 catalytic function. The specificity of the H3K36 signal was verified by Western blot analysis using a set2Δ strain that lacks all H3K36 methylation states. WT, wild type.
Transfection and immunofluorescence microscopy.
NIH 3T3 cells were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and penicillin-streptomycin. For immunofluorescence, cells grown on coverslips in six-well plates were transfected with 2 to 6 μg of Flag-Rph1 expression plasmid using Fugene 6 transfection reagent (Roche). Cells were fixed 24 h posttransfection for 20 min in 4% paraformaldehyde, washed three times with phosphate-buffered saline (PBS), and subsequently permeabilized for 20 min in 0.5% Triton X-100-PBS. Permeabilized cells were washed two times in PBS and blocked in 3% bovine serum albumin-PBS for 30 min. Cells were incubated with primary antibody in a humidified chamber for 1 to 3 h using histone modification antibodies at a dilution of 1:100 and the Flag monoclonal M2 antibody (Sigma) at a dilution of 1:1,000. After primary antibody incubation, cells were washed three times and incubated with fluorescein isothiocyanate or rhodamine-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories). Cells were washed twice with PBS, stained with 4,6-diamidino-2-phenylindole dihydrochloride (DAPI), and mounted on glass slides in fluorescent mounting medium (DAKO). Slides were analyzed on a AxioSkop fluorescent microscope (Zeiss).
RESULTS
Rph1 is an H3K36me3 demethylase.
We and others recently identified and characterized mammalian JHDM3/JMJD2 histone demethylases that target H3K9/K36 methylation (6, 8, 16, 41). These proteins contain N-terminal JmjN and JmjC domains that are required for enzymatic activity. Bioinformatic analysis has identified an S. cerevisiae protein, Rph1, which has a high level of similarity to the JHDM3/JMJD2 proteins within its JmjN and JmjC domains. Residues predicted to function as cofactor binding sites are completely conserved between the JmjC domain of Rph1 and the JHDM3/JMJD2 proteins, suggesting that Rph1 could potentially encode a novel yeast histone demethylase (15). Interestingly, very little similarity exists between mammalian JHDM3/JMJD2 proteins and Rph1 outside of the JmjN and JmjC domains, suggesting that Rph1 may have unique substrate specificity and function in yeast.
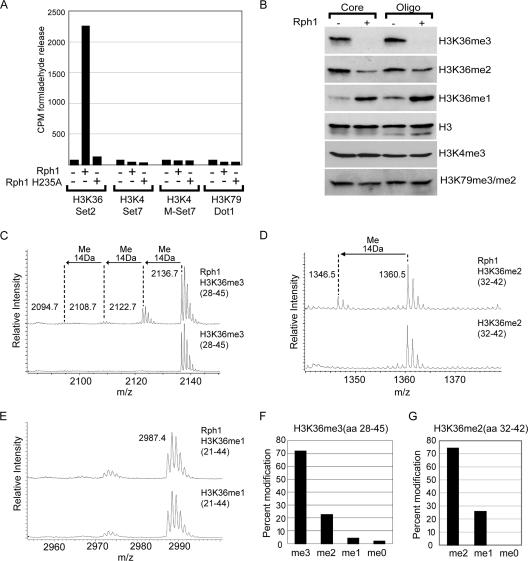
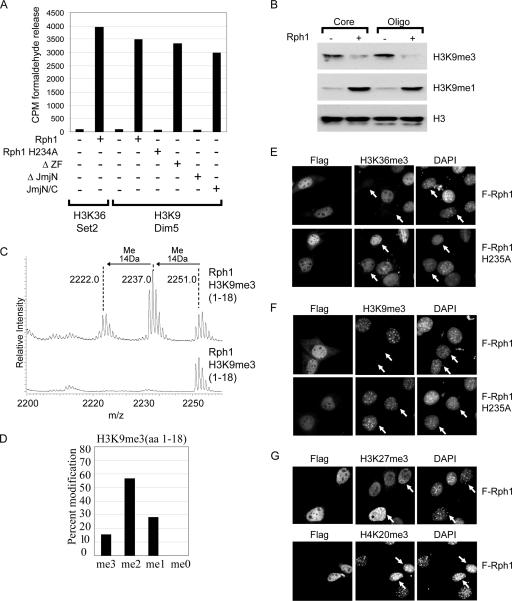
To test whether Rph1 is a histone demethylase, rRph1 or rRph1 with a replacement in a predicted iron binding residue (H235A) was used in a histone demethylase assay containing radioactively labeled methyl groups on histone H3 at positions K4, K36, and K79 (Fig. 1A). Histone demethylase activity was monitored by the release of the labeled reaction product formaldehyde. Demethylase activity was observed only when H3K36-labeled substrate was present in the reaction mixture, suggesting that Rph1 is an H3K36-specific histone demethylase (Fig. 1A). Mutation of a predicted iron binding residue within Rph1 completely abolished enzymatic activity, verifying that Rph1 relies on the JmjC domain for catalysis. Because histone lysine methylation can occur in three modification states, we sought to identify which H3K36 modification states are targeted by Rph1. Rph1 was incubated with core histones or oligonucleosomes, and the resulting methylation states were analyzed by Western blotting using modification-specific antibodies (Fig. 1B). Rph1-mediated demethylation culminated in a reduction of H3K36me3 and an accumulation of H3K36me1 but did not affect H3K4me3 or H3K79me3 methylation. Interestingly, this property of Rph1 differs from that of mammalian JHDM3/JMJD2 proteins, which are incapable of efficiently demethylating oligonucleosomal substrates (16).
FIG. 1.
Rph1 is an H3K36 demethylase capable of removing trimethyl lysine. (A) Labeled histone substrates corresponding to known histone methylation sites in budding yeast were incubated with rRph1 or Rph1 H235A. DOT1L, Set2, SET7, and mutant SET7 (M-SET7) methyltransferase enzymes were used to label histone substrates. Mutant SET7 was used to produce substrates containing higher H3K4 methylation states, as wild-type SET7 produces only monomethyl H3K4 (42). Histone demethylase activity was monitored by the release of labeled formaldehyde. Wild-type Rph1, but not the mutant Rph1 H235A, specifically demethylates H3K36-labeled substrate. CPM, counts per minute. (B) Core histones and oligonucleosomes (Oligo) were incubated with Rph1, and histone methylation levels were analyzed by Western blotting using H3K36, H3K4, and H3K79 methylation-specific antibodies. Rph1 demethylates H3K36me3 and H3K36me2, resulting in an accumulation of H3K36me1. (C to E) H3K36me3, H3K36me2, and H3K36me1 peptides were incubated with Rph1 in demethylase assays followed by mass spectrometry analysis. Rph1 specifically demethylates the H3K36me3 and H3K36me2 modification states. (F and G) Bar graphs representing the level of each modification state following Rph1-mediated demethylation of H3K36me3 and H3K36me2 peptides. aa, amino acids.
Rph1 activity towards purified histone substrates clearly demonstrates that Rph1 is an H3K36 demethylase. To fully define Rph1 substrate specificity, mass spectrometry was used to analyze the modification state of histone H3 peptides containing K36me3, K36me2, and K36me1 following demethylation by Rph1 (Fig. 1C, D, E, F, and G). In agreement with the Rph1-mediated demethylation of H3K36me3 observed by histone Western blotting, mass spectrometric analysis revealed that Rph1 efficiently demethylates H3K36me3, leading to processive reduction to the me2, me1, and me0 modification states (Fig. 1C). Rph1 is also capable of initiating demethylation on H3K36me2 substrates but is unable to demethylate the H3K36me1 modification state (Fig. 1D and E). Together, these data reveal the first yeast histone demethylase capable of removing the me3 modification state and demonstrate that Rph1 targets the demethylation of H3K36me3 and H3K36me2.
Rph1 requires both the JmjN and JmjC domains to catalyze histone demethylation.
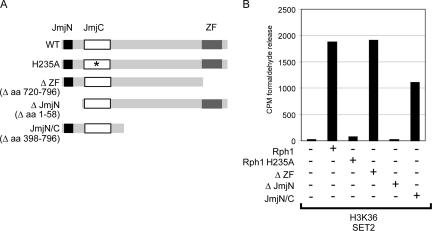
The Rph1 protein has three curated protein domains including a JmjN domain, a JmjC domain, and a zinc finger (ZF) domain. Mutation of a predicted iron binding site within the Rph1 JmjC domain abrogates demethylase activity, demonstrating that the JmjC domain is the catalytic core of the enzyme. Characterization of other JmjC-domain-containing proteins has revealed that additional domains can contribute to demethylase activity (8, 16, 36, 44). To understand which Rph1 domains are required for histone demethylation, a series of deletion proteins (Fig. 2A) were generated and analyzed for H3K36 demethylase activity using the formaldehyde release assay (Fig. 2B). A unique feature of Rph1 is its C-terminal ZF DNA binding domain, which is absent from the related mammalian JHDM3/JMJD2 histone demethylases (12). To determine whether this domain contributes to demethylase activity, the ZF was deleted, and the activity of the recombinant protein was analyzed by formaldehyde release (Fig. 2B). Removal of the ZF domain had no effect on enzymatic activity, suggesting that this domain may have alternative roles in vivo. In contrast, deletion of the JmjN domain completely abrogated H3K36 demethylase activity (Fig. 2B). Recently, the crystal structure of the human JHDM3A/JMJD2A protein was solved, revealing that the JmjN domain folds into the JmjC domain, creating a single structural entity that is enzymatically active (5). Given that Rph1 also relies on its JmjN domain for enzymatic activity, it seems likely that this domain contributes to the structure of the functional yeast enzyme. To determine whether the JmjN/JmjC domain alone is enzymatically active, a protein encompassing only these domains was generated and used in a histone demethylase assay. Although this protein showed a slight reduction in H3K36 demethylase activity, it was still capable of removing H3K36 methylation, demonstrating that the JmjN/JmjC domain is sufficient for demethylase activity (Fig. 2B). Together, these data show that Rph1 demethylase activity relies on the function of the JmjN and JmJC domains and indicate that the ZF domain may have alternative roles in vivo, perhaps involving protein targeting.
FIG. 2.
Rph1 requires the JmjN/JmjC domains but not the ZF domain for demethylase activity. (A) Schematic representation of Rph1 indicating the three curated domains within Rph1. The JmjN, JmjC, and ZF domains were individually deleted or mutated to examine the domain requirements and to map the smallest catalytically active fragment of Rph1. WT, wild type; aa, amino acids. (B) The mutant Rph1 proteins displayed in A were used in a histone demethylase assay containing H3K36-labeled substrate, and histone demethylase activity was monitored by formaldehyde release. The JmjN and JmjC domains of Rph1 are sufficient for H3K36 demethylation. CPM, counts per minute.
Deletion of RPH1 causes no overt cellular phenotype.
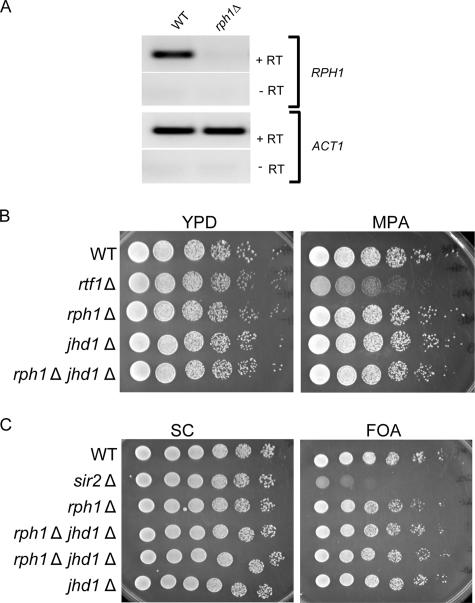
To analyze the role of Rph1 in the regulation of H3K36 in vivo, the RPH1 locus was disrupted by homologous recombination, and the absence of the Rph1 transcript was verified by reverse transcription (RT)-PCR (Fig. 3A). The Rph1-deficient strain was morphologically wild type, and analysis of H3K36 methylation levels by Western blotting with modification-specific antibodies revealed no global changes in H3K36 methylation (data not shown). Given that H3K36 methylation has been previously linked to transcriptional elongation, we tested whether a deletion of RPH1 causes sensitivity to mycophenoic acid (MPA), a drug that affects transcriptional elongation. Results shown in Fig. 3B indicate that the deletion of Rph1 does not confer sensitivity to MPA, nor does it cause defects in telomeric silencing (Fig. 3C). In addition, we have tested a number of conditions used for phenotypic analysis (Table 1) and observed no apparent phenotype. Yeast Jhd1 is also an H3K36 demethylase, but in contrast to Rph1, it specifically demethylates H3K36me2/me1 modification states. To examine whether there are synthetic effects in yeast lacking both Rph1 and Jhd1, a double mutant strain was generated and subjected to the same phenotypic analysis as the Rph1-deficient strain (Table 1). The double mutant strain failed to display any synthetic effects and grew normally under all conditions tested. Together, these data indicate that Rph1 and Jhd1 do not play an essential global role in regulating cellular processes including transcription, DNA replication, and heterochromatin function. This does not, however, rule out the possibility that Rph1 or Jhd1 contributes to these functions in a more subtle manner not realized using standard phenotypic analyses.
FIG. 3.
Deletion of RPH1 causes no overt phenotypes. (A) The RPH1 gene was disrupted by homologous recombination, and loss of the transcript was verified by RT-PCR. The top panel shows RPH1, and the bottom panel shows actin (ACT1) RT-PCR products. (B) Wild type (WT), rph1Δ, jhd1Δ, rph1Δ jhd1Δ, and rtf1Δ strains were spotted onto a yeast extract-peptone-dextrose (YPD) plate and a yeast extract-peptone-dextrose plate containing 50 μg/ml MPA in fivefold serial dilutions. Growth was analyzed following incubation for 2 days at 30°C. (C) Wild-type (WT), sir2Δ, rph1Δ, rph1Δ jhd1Δ, and jhd1Δ strains were spotted onto a synthetic complete (SC) plate and a synthetic complete plate containing 50 mg/ml 5-fluoroorotic acid (FOA) in fivefold serial dilutions. Growth was analyzed following incubation for 2 days at 30°C.
TABLE 1.
Phenotype analysis of the rph1Δ strain
| Phenotype | Functional implicationa | Control (reference) |
|---|---|---|
| Slow growth | General protein defects indicating important genes | |
| Heat sensitivity | General protein defects indicating important genes | spt4Δ (1) |
| MPA sensitivity | Transcription elongation | rtf1Δ (7) |
| Galactose fermentation | Transcriptional activation | snf2Δ (28) |
| Raffinose fermentation | Transcriptional derepression | snf2Δ (28) |
| Inositol auxotrophy | Inositol biosynthesis; transcriptional activation | spt7Δ (29) |
| Hydroxyurea sensitivity | DNA replication | htz1Δ (26) |
| Caffeine sensitivity | Mitogen-activated protein kinase pathway; chromatin remodeling | htz1Δ (26) |
| Telomeric silencing defect | Heterochromatin silencing | sir2Δ (35) |
See reference 11.
Rph1 can demethylate H3K36 in vivo.
To verify that Rph1 can target H3K36 demethylation in vivo, Flag-tagged Rph1 was overexpressed in wild-type cells (Fig. 4A and B). Interestingly, the overexpression of Rph1 resulted in a severe inhibition of cell growth, suggesting that elevated levels of Rph1 have a detrimental effect on cell function (Fig. 4C). To determine if the growth defect was a result of Rph1 demethylase activity, a catalytically inactive Rph1 was overexpressed, and growth was analyzed (Fig. 4A, B, and C). Like the wild-type Rph1 protein, the overexpression of the mutant protein resulted in a growth defect, indicating that the effect of Rph1 on cell growth is independent of demethylase activity (Fig. 4C). Rph1 has previously been shown to function as a transcriptional repressor, suggesting that the growth defect may be related to the silencing of genes involved in cell division or other growth-related pathways. The slow growth and low levels of protein expression in cells expressing full-length Rph1 made it impossible to reproducibly observe changes in H3K36 methylation. To try to separate the growth suppression and catalytic activities of Rph1, a protein lacking the C-terminal ZF domain was overexpressed (Fig. 4A and B). Deletion of the ZF domain completely abrogated the growth defect, indicating that the DNA binding ZF is important for growth suppression, perhaps functioning as a targeting mechanism for Rph1-mediated repression (12) (Fig. 4C). Cells expressing Rph1 that lack the ZF domain grew normally and expressed high levels of protein, making it possible to analyze the H3K36 methylation levels by Western blotting using modification-specific antibodies (Fig. 4D). Consistent with the observation that Rph1 is an H3K36me3 demethylase in vitro, the overexpression of Rph1 lacking the ZF caused a decrease in H3K36me3 methylation levels in vivo (Fig. 4D, compare lanes 1 and 2). Demethylase activity was dependent on an intact JmjC domain, as a point mutation in the catalytic domain abrogated this effect (Fig. 4D). In agreement with domain-mapping studies in vitro, the overexpression of the JmjN/JmjC domain alone was sufficient to catalyze H3K36me3 demethylation, and this function relied on an intact JmjC domain (Fig. 4D, lanes 4 and 5). Together, these data reveal that Rph1 functions to demethylate H3K36 in vivo and that elevated levels of Rph1 lead to growth defects that are independent of demethylase activity.
Rph1 is not stably associated with other proteins in vivo.
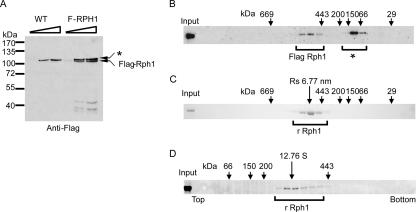
Many chromatin remodeling and chromatin-modifying enzymes are found in high-molecular-weight complexes containing auxiliary proteins that are required to regulate enzymatic function and target the enzyme to defined genomic regions (2-4, 13, 14, 20, 22, 27, 40). To identify potential Rph1 functional protein partners, we performed TAP-Tag purification, which failed to reveal any stably associated proteins (data not shown). To verify that Rph1 is not a component of a high-molecular-weight protein complex, extract from a Flag-tagged Rph1 strain (Fig. 5A) was fractionated by size exclusion chromatography (Fig. 5B). Rph1-containing fractions were identified by Western blot analysis using Flag-specific antibodies (Fig. 5B). Rph1 eluted from the size exclusion column with an apparent native molecular mass of greater that 440 kDa, which is much larger than its theoretical molecular mass of 90.2 kDa based on the amino acid composition (Fig. 5B). Rph1 affinity purification failed to reveal associated proteins, but size exclusion analysis suggests that the native molecular weight of Rph1 is larger than that expected for Rph1 alone. To determine whether the high apparent native molecular weight of Rph1 in size exclusion fractionations was due to an association with other proteins, rRph1 was separated over the same size exclusion column and analyzed by Coomassie staining (Fig. 5C). Surprisingly the recombinant protein also eluted from the size exclusion column with a native molecular mass of greater than 440 kDa (Fig. 5C). This observation indicates that the high apparent native molecular weight of Rph1 in yeast extracts is not due to additional stably associated proteins but instead is an intrinsic property of Rph1 alone. Given that size exclusion chromatography separates proteins based on radius and not molecular weight, the aberrant size of Rph1 in these experiments could be due to an abnormally elongated Rph1 molecule or the result of a homogenous multimeric Rph1 complex. Over four decades ago, Siegel and Monty derived a series of formulae that combine biophysical properties obtained from size exclusion chromatography and sedimentation analysis to accurately determine the native molecular weight of proteins and protein complexes (34). Using those formulae, the experimentally determined radius and sedimentation coefficient can be exploited to determine whether a given protein species has an abnormal elution profile due to a highly elongated shape or multimerization. The Stokes radius of rRph1 calculated from size exclusion chromatography was ∼6.77 nm (Fig. 5C). To determine the sedimentation coefficient, rRph1 was analyzed by sucrose gradient sedimentation. The sedimentation coefficient (s20,w) of rRph1 calculated from the sucrose gradient was ∼12.76 S (Fig. 5D). By applying values obtained from the size exclusion and sedimentation analysis to the Siegel and Monty formulas, the derived native molecular mass of Rph1 was calculated to be 355.43 kDa, and the frictional ratio (f/f0) was 1.45. This analysis suggests that Rph1 is not an elongated molecule but instead consists of four 90.2-kDa (theoretical mass) Rph1 subunits. It is surprising that Rph1 does not form a stable heterogeneous protein complex in budding yeast given that many other chromatin-modifying enzymes are found in high-molecular-weight complexes that have accessory proteins involved in targeting the enzymatic activity to chromatin. One explanation for the apparent absence of a stable Rph1 complex could be the intrinsic ability of Rph1 to directly bind DNA through its C-terminal ZF domain (12). The DNA binding properties of Rph1 may allow it to function independently of associated factors in recognizing target sites in chromatin and permit more transient interactions with additional protein factors while antagonizing H3K36 methylation.
FIG. 5.
Rph1 is not stably associated with other proteins in yeast extracts. (A) Endogenous Rph1 was Flag tagged, and the wild-type (WT) and Flag-tagged strains were analyzed by Western blotting using Flag-specific antibodies. A signal corresponding to Flag-Rph1 was evident only in the tagged strain. The asterisk indicates a cross-reactive band observed in budding yeast extract. (B) Flag-RHP1 yeast extract was fractionated by size exclusion chromatography, and the Rph1-containing fractions were identified by Flag-specific Western blotting. The * indicates a cross-reacting band found in yeast extracts. Size exclusion chromatography molecular mass markers are indicated above the panel. Rph1 eluted from the size exclusion column with an apparent molecular mass of greater than 440 kDa. (C) Recombinant Rph1 was fractionated by size exclusion chromatography, and the Rph1-containing fractions were identified by Coomassie staining. Size exclusion chromatography molecular mass markers are indicated above the panel, and the calculated radius of Rph1 is given above in nm. rRph1 eluted from the size exclusion column at the same position as endogenous Rph1. (D) Recombinant Rph1 was fractionated over a 5 to 20% sucrose gradient, and Rph1-containing fractions were identified by Coomassie staining. Molecular mass standards are indicted above the panel, and the calculated sedimentation coefficient in S is given above.
Rph1 demethylates H3K9 despite the absence of this modification in budding yeast.
In S. cerevisiae, histone lysine methylation is limited to positions K4, K36, and K79 of histone H3. Interestingly, the JHDM3/JMJD2 proteins in mammals, which are related to Rph1, are capable of removing both H3K36 and K9 methylation (6, 8, 16, 41). An H3K9 methylation system is absent from budding yeast, suggesting that the capacity of the mammalian JHDM3/JMJD2 demethylase enzymes to target H3K9 methylation may have been adaptively acquired during the evolution of a more complex chromatin modification system. To analyze whether Rph1 specifically catalyzes H3K36 demethylation, the recombinant enzyme was incubated with histone substrates radioactively labeled on H3K9, and demethylase activity was monitored by formaldehyde release (Fig. 6A). Surprisingly, Rph1 efficiently demethylated the H3K9-modified substrate requiring both the JmjN/JmjC domain but not the ZF motif for enzymatic activity (Fig. 6A). To verify the Rph1 H3K9 demethylase activity observed by formaldehyde release, Rph1 was incubated with histone substrates, and H3K9 methylation was assessed by Western blot analysis using modification-specific antibodies (Fig. 6B). Like its mammalian counterparts, Rph1 also targets the demethylation of H3K9me3, resulting in an accumulation of the me1 modification state. In contrast to the mammalian JHDM3 enzymes, Rph1 is capable of demethylating both core histone and oligonucleosomal substrates (Fig. 6B) (16). The capacity of Rph1 to demethylate H3K9me3 was also demonstrated by incubating Rph1 with an H3K9me3 peptide substrate and analyzing the resulting modification states by mass spectrometry (Fig. 6C and D). Rph1 demethylated the H3K9me3 substrate, resulting in the accumulation of me2 and me1 modified peptides (Fig. 6C and D).
FIG. 6.
Rph1 removes H3K9 methylation both in vitro and in vivo. (A) Histone substrates were radioactively labeled on H3K36 and H3K9 and incubated with the Rph1 proteins detailed in Fig. 2A. Demethylase activity was monitored by the release of radioactive formaldehyde. Rph1 efficiently demethylated both H3K36 and H3K9 substrates requiring an intact JmjN/JmjC domain for enzymatic activity. CPM, counts per minute. (B) Core histones and oligonucleosomes (Oligo) were incubated with Rph1, and histone methylation levels were analyzed by Western blotting with H3K9 modification-specific antibodies. Rph1 demethylates H3K9me3, resulting in an accumulation of K9me1. (C) An H3K9me3 peptide was incubated with Rph1, and the resulting modification state was analyzed by mass spectrometry. Rph1 can demethylate H3K9me3, leading to an accumulation of H3K9me2 and H3K9me1 modification states. (D) Bar graph representing the percentage of each modification state after Rph1-mediated demethylation of the H3K9me3 peptide. (E and F) Flag-tagged Rph1 and Rph1 H235A were expressed in NIH 3T3 cells, and the levels of H3K36me3 (E) and H3K9me3 (F) were analyzed by indirect immunofluorescence using histone methylation-specific antibodies. Rph1 localized to the nucleus and demethylated both H3K36me3 and H3K9me3 (top panels). Demethylase activity required an intact JmjC domain, as a mutation of the catalytic domain abrogated this effect (bottom panels). (G) Expression of Rph1 in NIH 3T3 cells does not cause demethylation of other repressive histone methylation marks including H3K27me3 (top) or H4K20me3 (bottom) as assessed by indirect immunofluorescence with methylation specific antibodies.
The surprising observation that Rph1 demethylates both H3K9 and K36 methylation indicates that the H3K9 demethylase activity of mammalian JHDM3/JMJD2 enzymes is not simply a feature acquired through adaptation but is likely a feature of the ancestral enzyme. To determine whether Rph1 can remove H3K36 and H3K9 methylation in cellular chromatin containing both of these modifications, an expression vector was generated to overexpress Flag-tagged Rph1 in mammalian cells. Consistent with a role in counteracting histone methylation, Rph1 localized predominantly to the nucleus when expressed in mouse NIH 3T3 cells (Fig. 6E and F). In cells expressing Rph1, both H3K36me3 methylation and H3K9me3 methylation were dramatically reduced as assessed by immunofluorescence using modification-specific antibodies (Fig. 6E and F, top panels). Demethylation by Rph1 in mammalian cells was dependent on an intact JmjC domain, as a mutation in the catalytic domain abolished demethylase activity (Fig. 6E and F, bottom panels). This effect on H3K9me3 and H3K36me3 methylation was specific, as other histone methylation marks associated with silencing in mammals, including H3K27me3 and H4K20me3, were unaffected (Fig. 6G). These observations strongly suggest that the bifunctional substrate specificity of the mammalian JHDM3 enzymes is not an acquired feature but instead is inherited from the ancestral form of the protein. Furthermore, this suggests that the budding yeast genome may have encoded an H3K9 methylation system that was lost at some point during evolution of the current budding yeast chromatin modification system. The fact that Rph1 has the capacity to target H3K9 methylation may represent a functional vestige of this H3K9 modification system in budding yeast. The activity of Rph1 towards H3K9 methylation has presumably been retained in the absence of this modification through selective pressure to preserve the structurally linked H3K36 demethylase activity.
DISCUSSION
The identification of demethylase enzymes has revealed that histone methylation can be dynamically regulated in a manner similar to that of histone acetylation and phosphorylation. In S. cerevisiae, the enzymes that place histone methylation marks are well characterized and coordinate mainly the addition of these modifications during the process of active transcription (25). Previously, only one histone demethylase enzyme, Jhd1, was identified in budding yeast. Jhd1 is a JmjC-domain-containing protein that catalyzes the demethylation of H3K36me2 and H3K36me1 modification states (36). Given that Jhd1 does not target H3K36me3 in yeast, it remained possible that this methylation state was irreversible.
Here, we identify Rph1 as being a histone demethylase with activity towards histone H3K36me3 and H3K36me2 modification states. Deletion of RPH1 does not affect global histone H3K36 methylation profiles, and deletion strains are viable, displaying no obvious morphological or cellular defects. This observation is not surprising given that a deletion of SET2, the sole H3K36 methyltransferase in budding yeast, causes no obvious cellular defects and has subtle effects on gene expression. The overexpression of Rph1 leads to a cellular growth defect, but this property appears to be independent of H3K36 demethylase activity and instead relies on the C-terminal ZF DNA binding domain. It remains possible that the growth defect in Rph1-overexpressing cells is due to demethylase-independent repression of growth-related genes through the ZF DNA binding domain. The overexpression of the Rph1 JmjN/JmjC domains alone is sufficient to mediate the demethylation of H3K36, verifying that this portion of the protein is catalytically competent in vivo. In contrast to many other chromatin-modifying enzymes, Rph1 does not stably associate with other proteins but instead forms a homogenous complex comprised of four Rph1 subunits. Often, chromatin remodeling complexes rely on associated protein factors for enzyme targeting, but the fact that Rph1 has an intrinsic DNA binding domain may alleviate the requirement for genomic targeting by auxiliary protein factors in some instances. Removal of the ZF relieves growth defects in cells overexpressing Rph1, supporting the argument that this domain contributes to protein function and perhaps genomic targeting in vivo. Additional functional analyses will be required to define specific genomic targets of Rph1 and to understand how Rph1-mediated demethylation contributes to transcriptional regulation by Rph1.
The two characterized budding yeast histone demethylase enzymes, Jhd1 and Rph1, both target H3K36 methylation. Two of the three remaining JmjC-domain-containing proteins, Gis1 and Ecm5, have mutations in cofactor binding residues that ablate demethylase activity (Y. Tsukada, K. E. Gardner, and Y. Zhang, unpublished data). The remaining protein, Yjr119C, is an H3K4 demethylase that catalyzes the removal of the H3K4me3 modification state (our unpublished data). Therefore, it appears that JmjC-domain-containing proteins in budding yeast target the removal of H3K4 and H3K36 methylation but not H3K79 methylation. H3K4 and H3K36 methylation are placed by SET domain-containing histone methyltransferases. In contrast, H3K79 methylation is catalyzed by DOT1, which does not have a SET domain. The inability of JmjC-domain-containing proteins to remove H3K79 methylation strikingly parallels the fact that a unique enzyme is required to place this modification. Perhaps H3K79 methylation is also removed by a novel class of demethylase enzymes with unique enzymatic properties. Further biochemical and genetic analyses of H3K79 methylation in budding yeast will be instrumental in determining whether this modification is dynamically regulated and provide insight into potentially novel enzymes involved in metabolizing this modification.
The JmjC-domain-containing histone demethylase enzymes characterized thus far have a very defined substrate specificity towards the lysine modification site and state. The catalytic domain of Rph1 is homologous to the mammalian JHMD3/JMJD2 enzymes, which target both H3K36 and H3K9 demethylation. The capacity of mammalian enzymes to target H3K9 methylation, a modification which is absent from budding yeast chromatin, may have adaptively evolved in the presence of enzymes that place this modification. Surprisingly, the characterization of Rph1 substrate specificity revealed that Rph1 is also capable of demethylating H3K9 in vitro as well as on mammalian chromatin in vivo. This property of Rph1 is not simply due to promiscuous substrate specificity, as Rph1 does not affect other yeast or mammalian histone methylation sites. The capacity of Rph1 to demethylate this modification suggests that an H3K9 methylation system may have once existed in budding yeast. Despite the fact that H3K9 methylation is no longer found in budding yeast chromatin, the enzymatic activity of Rph1 towards this modification may have been inadvertently retained due to its bifunctional requirement as a regulator of H3K36 methylation. Other components of the H3K9 methylation system, including the H3K9 methyltransferase, may have been lost or become functionally inactive.
No SET-domain-containing protein has been shown to modify H3K9 in budding yeast. The SET-domain-containing protein Set3 is a structurally integral component of a high-molecular-weight histone deacetylase complex (30) that, much like Set2, is targeted to the body of active genes, where it regulates chromatin modification (39). Deletion of Set2 in a strain lacking any component of the Set3 complex results in synthetic growth defects, suggesting that these factors contribute to similar processes (18). It has recently been demonstrated that in addition to H3K36 methylation, H3K9 methylation is targeted to the body of actively transcribed genes in mammalian cells (37, 38), and at least one mammalian histone deacetylase complex also contains H3K9 methyltransferase activity (33). No histone methyltransferase activity has been identified for the budding yeast Set3 complex, and residues within the SET domain that are required for methyltransferase activity are substituted. The role of this complex in the transcribed regions of yeast genes raises the possibility that Set3 may have once played a role analogous to that of the methyltransferases that place H3K9 methylation in the body of mammalian genes. During the evolution of the yeast chromatin modification system, a loss of selective pressure for H3K9 methylation could have potentially allowed components of this system to functionally deteriorate, while an intact H3K9 methylation system in higher eukaryotes was retained. Perhaps Set3 remains as a relic of this modification system due to its essential structural role in the assembly of the Set3 protein complex and its role in histone deacetylation. It will be interesting to determine whether the SET domain of Set3 can be replaced with the SET domain from an active H3K9 methyltransferase to recapitulate H3K9 methylation profiles in budding yeast that are found in the body of transcribed genes in mammals. The revelation that Rph1 can demethylate H3K9 provides the first evidence for the possibility of an extinct H3K9 methylation system in budding yeast and suggests that Rph1 may represent a functional vestige of this system.
Acknowledgments
We thank Henrik Dolhman and Chris Brandl for providing reagents. We are grateful to Brian Strahl for providing the BY4741, set2Δ, spt4Δ, rtf1Δ, snf2Δ, spt7Δ, htz1Δ, and sir2Δ strains. We thank Emma Turnbull and Nara Lee for critical reading of the manuscript.
This work was supported by NIH grants GM68804 (to Y.Z.) and P30 CA08748 (to P.T.). Y.Z. is an Investigator of the Howard Hughes Medical Institute. R.J.K. is supported by the Canadian Institutes of Health Research.
Footnotes
Published ahead of print on 19 March 2007.
REFERENCES
- 1.Basrai, M. A., J. Kingsbury, D. Koshland, F. Spencer, and P. Hieter. 1996. Faithful chromosome transmission requires Spt4p, a putative regulator of chromatin structure in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:2838-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cairns, B. R. 2005. Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 15:185-190. [DOI] [PubMed] [Google Scholar]
- 3.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 4.Carrozza, M. J., B. Li, L. Florens, T. Suganuma, S. K. Swanson, K. K. Lee, W. J. Shia, S. Anderson, J. Yates, M. P. Washburn, and J. L. Workman. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581-592. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Z., J. Zang, J. Whetstine, X. Hong, F. Davrazou, T. G. Kutateladze, M. Simpson, Q. Mao, C. H. Pan, S. Dai, J. Hagman, K. Hansen, Y. Shi, and G. Zhang. 2006. Structural insights into histone demethylation by JMJD2 family members. Cell 125:691-702. [DOI] [PubMed] [Google Scholar]
- 6.Cloos, P. A., J. Christensen, K. Agger, A. Maiolica, J. Rappsilber, T. Antal, K. H. Hansen, and K. Helin. 2006. The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3. Nature 442:307-311. [DOI] [PubMed] [Google Scholar]
- 7.Desmoucelles, C., B. Pinson, C. Saint-Marc, and B. Daignan-Fornier. 2002. Screening the yeast “disruptome” for mutants affecting resistance to the immunosuppressive drug, mycophenolic acid. J. Biol. Chem. 277:27036-27044. [DOI] [PubMed] [Google Scholar]
- 8.Fodor, B. D., S. Kubicek, M. Yonezawa, R. J. O'Sullivan, R. Sengupta, L. Perez-Burgos, S. Opravil, K. Mechtler, G. Schotta, and T. Jenuwein. 2006. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 20:1557-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelbart, M. E., T. Rechsteiner, T. J. Richmond, and T. Tsukiyama. 2001. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol. Cell. Biol. 21:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 11.Hampsey, M. 1997. A review of phenotypes in Saccharomyces cerevisiae. Yeast 13:1099-1133. [DOI] [PubMed] [Google Scholar]
- 12.Jang, Y. K., L. Wang, and G. B. Sancar. 1999. RPH1 and GIS1 are damage-responsive repressors of PHR1. Mol. Cell. Biol. 19:7630-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joshi, A. A., and K. Struhl. 2005. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20:971-978. [DOI] [PubMed] [Google Scholar]
- 14.Keogh, M. C., S. K. Kurdistani, S. A. Morris, S. H. Ahn, V. Podolny, S. R. Collins, M. Schuldiner, K. Chin, T. Punna, N. J. Thompson, C. Boone, A. Emili, J. S. Weissman, T. R. Hughes, B. D. Strahl, M. Grunstein, J. F. Greenblatt, S. Buratowski, and N. J. Krogan. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123:593-605. [DOI] [PubMed] [Google Scholar]
- 15.Klose, R. J., E. M. Kallin, and Y. Zhang. 2006. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7:715-727. [DOI] [PubMed] [Google Scholar]
- 16.Klose, R. J., K. Yamane, Y. Bae, D. Zhang, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442:312-316. [DOI] [PubMed] [Google Scholar]
- 17.Kornberg, R. D., and Y. Lorch. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285-294. [DOI] [PubMed] [Google Scholar]
- 18.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney, V. Canadien, D. P. Richards, B. K. Beattie, A. Emili, C. Boone, A. Shilatifard, S. Buratowski, and J. Greenblatt. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuzmichev, A., K. Nishioka, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16:2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, B., L. Howe, S. Anderson, J. R. Yates III, and J. L. Workman. 2003. The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278:8897-8903. [DOI] [PubMed] [Google Scholar]
- 22.Li, H., S. Ilin, W. Wang, E. M. Duncan, J. Wysocka, C. D. Allis, and D. J. Patel. 2006. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 442:91-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, J., D. Moazed, and S. P. Gygi. 2002. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 277:49383-49388. [DOI] [PubMed] [Google Scholar]
- 24.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6:838-849. [DOI] [PubMed] [Google Scholar]
- 25.Millar, C. B., and M. Grunstein. 2006. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell Biol. 7:657-666. [DOI] [PubMed] [Google Scholar]
- 26.Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen, and C. Wu. 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303:343-348. [DOI] [PubMed] [Google Scholar]
- 27.Muller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta, B. Wild, E. L. Miller, M. B. O'Connor, R. E. Kingston, and J. A. Simon. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111:197-208. [DOI] [PubMed] [Google Scholar]
- 28.Neigeborn, L., and M. Carlson. 1984. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108:845-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patton-Vogt, J. L., and S. A. Henry. 1998. GIT1, a gene encoding a novel transporter for glycerophosphoinositol in Saccharomyces cerevisiae. Genetics 149:1707-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pijnappel, W. W., D. Schaft, A. Roguev, A. Shevchenko, H. Tekotte, M. Wilm, G. Rigaut, B. Seraphin, R. Aasland, and A. F. Stewart. 2001. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15:2991-3004.11711434 [Google Scholar]
- 31.Schaft, D., A. Roguev, K. M. Kotovic, A. Shevchenko, M. Sarov, A. Shevchenko, K. M. Neugebauer, and A. F. Stewart. 2003. The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 31:2475-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero, and Y. Shi. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 33.Shi, Y., J.-I. Sawada, G. Sui, E. B. Affar, J. R. Whetstine, F. Lan, H. Ogawa, M. P.-S. Luke, Y. Nakatani, and Y. Shi. 2003. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature 422:735-738. [DOI] [PubMed] [Google Scholar]
- 34.Siegel, L. M., and K. J. Monty. 1966. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta 112:346-362. [DOI] [PubMed] [Google Scholar]
- 35.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukada, Y., J. Fang, H. Erdjument-Bromage, M. E. Warren, C. H. Borchers, P. Tempst, and Y. Zhang. 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439:811-816. [DOI] [PubMed] [Google Scholar]
- 37.Vakoc, C. R., S. A. Mandat, B. A. Olenchock, and G. A. Blobel. 2005. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell 19:381-391. [DOI] [PubMed] [Google Scholar]
- 38.Vakoc, C. R., M. M. Sachdeva, H. Wang, and G. A. Blobel. 2006. A profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell. Biol. 26:9185-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, A., S. K. Kurdistani, and M. Grunstein. 2002. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298:1412-1414. [DOI] [PubMed] [Google Scholar]
- 40.Wang, H., W. An, R. Cao, L. Xia, H. Erdjument-Bromage, B. Chatton, P. Tempst, R. G. Roeder, and Y. Zhang. 2003. mAM facilitates conversion by ESET of dimethyl to trimethyl lysine 9 of histone H3 to cause transcriptional repression. Mol. Cell 12:475-487. [DOI] [PubMed] [Google Scholar]
- 41.Whetstine, J. R., A. Nottke, F. Lan, M. Huarte, S. Smolikov, Z. Chen, E. Spooner, E. Li, G. Zhang, M. Colaiacovo, and Y. Shi. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125:467-481. [DOI] [PubMed] [Google Scholar]
- 42.Xiao, B., C. Jing, J. R. Wilson, P. A. Walker, N. Vasisht, G. Kelly, S. Howell, I. A. Taylor, G. M. Blackburn, and S. J. Gamblin. 2003. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature 421:652-656. [DOI] [PubMed] [Google Scholar]
- 43.Xiao, T., H. Hall, K. O. Kizer, Y. Shibata, M. C. Hall, C. H. Borchers, and B. D. Strahl. 2003. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamane, K., C. Toumazou, Y. Tsukada, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125:483-495. [DOI] [PubMed] [Google Scholar]