Abstract
The best-characterized biochemical feature of apoptosis is degradation of genomic DNA into oligonucleosomes. The endonuclease responsible for DNA degradation in caspase-dependent apoptosis is caspase-activated DNase. In caspase-independent apoptosis, different endonucleases may be activated according to the cell line and the original insult. Among the known effectors of caspase-independent cell death, L-DNase II (LEI [leukocyte elastase inhibitor]-derived DNase II) has been previously characterized by our laboratory. We have thus shown that this endonuclease derives from the serpin superfamily member LEI by posttranslational modification (A. Torriglia, P. Perani, J. Y. Brossas, E. Chaudun, J. Treton, Y. Courtois, and M. F. Counis, Mol. Cell. Biol. 18:3612-3619, 1998). In this work, we assessed the molecular mechanism involved in the change in the enzymatic activity of this molecule from an antiprotease to an endonuclease. We report that the cleavage of LEI by elastase at its reactive center loop abolishes its antiprotease activity and leads to a conformational modification that exposes an endonuclease active site and a nuclear localization signal. This represents a novel molecular mechanism for a complete functional conversion induced by changing the conformation of a serpin. We also show that this molecular transformation affects cellular fate and that both endonuclease activity and nuclear translocation of L-DNase II are needed to induce cell death.
Apoptosis is a form of cell death characterized by specific morphological features. In early stages of this process, DNA is cleaved into large fragments (50 to 300 kb) that are later degraded into oligonucleosomes (180 bp). Degradation of genomic DNA into oligonucleosomes is one of the most-studied features of apoptosis. In many cases, apoptosis is triggered by the activation of specific proteases called caspases. Upon the activation of caspases, DNA degradation is triggered by the activation of CAD (caspase-activated DNase). Nevertheless, in systems where caspases are not activated, other proteases, like serine proteases or cathepsins, and different endonucleases are activated, also leading to a cell death morphologically indistinguishable from caspase-dependent cell death (19). Indeed, many enzymes have been proposed to be responsible for DNA degradation, such as DNase I (29), DNase II (3), DNase γ (35), and NUC18/cyclophilin A (24). However, none of them appeared to fulfill the criteria for an apoptotic DNase completely. Besides, recent studies of apoptotic degradation in vivo and in vitro indicate that two independent systems are involved in DNA degradation during programmed cell death (26), a cell-autonomous system that functions in dying cells (involving the enzymes mentioned above) and another system that works after dying cells are engulfed by phagocytes.
In mammalian cells, caspase-independent apoptotic DNA degradation has been associated with two mitochondrial proteins, i.e., endonuclease G (20) and apoptosis-inducing factor (37). These proteins are translocated to the nucleus upon release from the mitochondria. They may be involved in a pathway alternative to CAD/ICAD leading to genomic DNA fragmentation in a caspase-independent manner.
Another effector of caspase-independent apoptosis-like cell death now recognized is LEI (leukocyte elastase inhibitor)/L-DNase II (LEI-derived DNase II) (34), a protein characterized in our laboratory (41). The activation of this DNase II (acid, cation-independent DNase) was first discovered in lens tissue during its terminal differentiation (39), which is an apoptosis-related cellular process (12). The activation of this enzyme is also seen in other physiological models, such as neural apoptosis during retinal development (38), as well as in different cell lines (5, 9).
Like most enzymes involved in apoptosis, L-DNase II is synthesized as a precursor. This precursor is LEI, a protein of the serpin superfamily, which has a protease inhibitor activity and a cytoplasmic localization. After cleavage by elastase (41) or apoptotic protease 24 (1) (but not caspases (40), LEI loses its antiprotease activity and is transformed into L-DNase II, a nuclear protein with endonuclease activity.
Proteins of the serpin superfamily have conformational features present in all of the crystal structures of serpins and serpin-protease complexes that have been reported to date (36, 42). The key feature of all serpins is the reactive-center loop (RCL). The relevant amino acids are tethered between β-sheets A and C, which exist in an exposed conformation. The specificity of the serpin is determined by the amino acid sequence encompassed by the RCL. With few exceptions, serpins inhibit serine proteases by an irreversible suicide-substrate inhibitory mechanism. Protease attacks on the P1 residue of the RCL, leading to a covalent ester linkage between the P1 residue of the RCL and the active site of the serine. This will cause cleavage of the P1-P1′ peptide bond of the serpin. Thus, the RCL inserts itself into sodium deoxycholate, β-sheet, thereby imparting enhanced stability to the complex, disrupting protease structure and rendering it inactive. This mechanism, called the stressed-to-relaxed transition, is associated with a change in the apparent molecular mass of the serpin. The preferred conformation of serpin is its relaxed form, which is more stable from a thermodynamic point of view. Here, we investigate the molecular basis of the change in LEI activity from a protease inhibitor to an endonuclease, as well as the nuclear translocation linked to this transformation. We show that the conformational modification typical of serpins is responsible for this transformation and that both endonuclease activity and nuclear translocation are key events in the induction of cell death (2).
MATERIALS AND METHODS
Antiprotease and endonuclease activities.
Site-directed mutagenesis was performed on a wild-type-LEI-pET 23d(+) construct with the QuikChange mutagenesis kit from Stratagene. An alanine residue at position P10 of the hinge region of the reactive loop of LEI (AP10T mutant) and histidine 368 (H368A mutant) were changed into threonine and alanine, respectively.
Recombinant proteins were produced in Escherichia coli strain BL21/pLysS and purified with His-select cartridges (Sigma).
Increasing concentrations of wild-type or mutant LEI (17.5 to 280 μg/ml) were incubated with elastase (0.1 μg/ml) in the presence of 2 mM pNaMAAPV, a synthetic substrate. The resulting color reaction was measured at 405 nm.
DNase activity was measured after overnight cleavage of LEI with equimolar quantities of elastase at 37°C (this step was omitted with 358Stop LEI). An 841-ng sample of wild-type, AP10T, H368A, or 358stop LEI was incubated with 2.5 μg of plasmid DNA in a time-dependent manner at 37°C in 20 mM Tris-EDTA, pH 5.5. Untreated wild-type LEI was incubated with DNA as a negative control.
Nuclear translocation.
BHK cells were grown as a monolayer at a density of 45,000 cells/cm2 in Dulbecco's modified Eagle's medium (Life Technology) supplemented with 10% fetal calf serum (FCS), 4 mM glutamine, 200 U/ml penicillin, and 0.2 mg/ml streptomycin (all from Life Technology) at 37°C in a humidified atmosphere containing 5% CO2.
Porcine LEI cDNA was subcloned into the XhoI/SalI restriction sites of plasmid pDsRed2-C1 (Clontech) and mutated by site-directed mutagenesis as before.
BHK cells were transfected with jetPEI (PolyPlus Laboratories) according to the manufacturer's protocol. The pCDNA3-1/NTGFP topo construct contained wild-type LEI. Apoptosis was induced with 40 μM 5-(N-ethyl-N-isopropyl)amiloride (EIPA), and pictures were taken 16 h after induction. All of the apoptotic cells present in the well (24-well plate) were morphologically identified. Cells displaying red and green (colocalized) fluorescence in the nucleus were considered to express LEI constructs displaying no impairment of nuclear translocation. Dissociation of fluorescence with an increase in green fluorescence in the nucleus was considered to indicate impairment of nuclear translocation.
Cell survival.
Porcine LEI cDNA was subcloned into the BamHI/XhoI restriction sites of plasmid pZeoSV2 (Invitrogen). BHK cells were seeded at a density of 20,000 cells/cm2 in 24-well plates. Two hours before transfection, the culture medium was replaced with fresh medium without FCS. The transfection medium contained 0.4 μg of pZeo-LEI and 3 μl of Lipofectin reagent (Life Technologies, Inc.) per well. The transfection process occurred at 37°C for 4 h and was stopped with Dulbecco modified Eagle medium containing 20% FCS.
Apoptosis was induced with 40 μM 5-(N,N-hexamethylene)amiloride (HMA). Survival was measured by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] method. At the end of cell treatment, culture medium was removed and 250 μl of MTT (1 mg/ml) was added to each well. The plate was kept in a CO2 incubator for 1 h. Cells were then lysed by the addition of 250 μl of isopropanol. The degree of MTT reduction in each sample was subsequently assessed by measuring absorbance at 570 versus 630 nm with a microplate reader (Bio-Rad).
Pull-down assay.
Recombinant proteins (wild-type LEI, K225A mutant LEI, and calmodulin) were produced in E. coli strain BL21/pLysS and loaded into His-select cartridges (Sigma) according to the manufacturer's protocol. HeLa cells were grown to confluence for 3 days in 75-cm2 flasks before lysis in RIPA buffer (50 mM Tris [pH 7.2], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 1% sodium dodecyl sulfate, 2 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 5 μg/ml pepstatin, 5 μg/ml aprotinin) and loaded onto the column. Bound material was quantified by the Bradford assay (Bio-Rad); 25 μg of each sample was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted for importin α (monoclonal clone IM-75; Sigma). Calmodulin was used as a negative control, and crude extract of HeLa cells (25 μg) was used as a positive control.
Protein Explorer.
Proteins were analyzed by using the Protein Explorer website (http://www.umass.edu/microbio/chime/pe_beta/pe/protexpl/frntdoor.htm). Upon introduction of the Protein Data Base (PDB) code of each protein, the molecular electrostatic potential was calculated with the Advanced Explorer tools.
RESULTS
Conformational change in LEI.
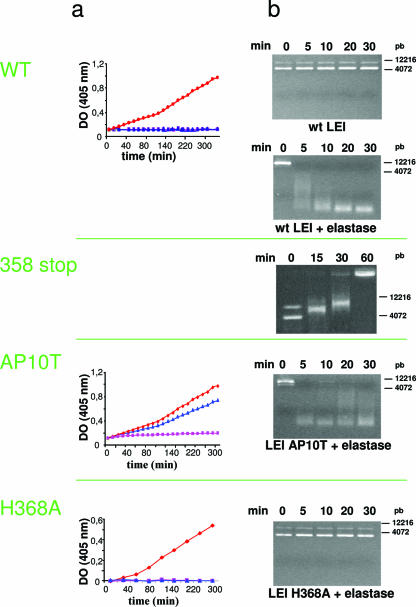
In previous studies, we have shown that limited cleavage transforms LEI into L-DNase II (41). However, the mechanism leading to the production of L-DNase II was not fully established. To assess this mechanism, we first tested whether the transition from LEI to L-DNase II involves proteolytic cleavage of the N- or C-terminal region. Previous published data show that LEI (42 kDa) is preferentially cleaved by elastase at amino acid 358, in the C-terminal region of the protein (Fig. 1), leading to a protein with an apparent molecular mass of 35 kDa (32). Even though this cleavage induces LEI transformation, it does not lead to release of the C-terminal peptide. Indeed, Edman degradation indicated the presence of this peptide in L-DNase II (41). Moreover, trypsin digestion of p42 and p35, followed by mass spectrometry of the peptides obtained, showed no significant difference between digests (data not shown), other than the changes expected from a cleavage described earlier (32). This suggested that, despite their different apparent molecular masses, the peptide contents of these proteins were comparable. We also introduced a stop codon at nucleotide 1074, corresponding to residue 358, which is the cleavage point of elastase in LEI (32). This produced an inactive endonuclease, although its ability to bind DNA was preserved (Fig. 2b, 358stop mutant). Indeed, a shift effect but no plasmid degradation was observed when the 358stop mutant was incubated with DNA for different times (compare with wild-type LEI before and after cleavage by elastase in Fig. 2b). This result suggested that the DNA binding site was in the peptide upstream of residue 358. It also might suggest that the C-terminal peptide supported the endonuclease active site.
FIG. 1.
Sequence alignment of horse, human, and pig LEI proteins. By convention, amino acid numbering refers to the PDB nomenclature of serpins that aligns all of the serpins with α1-antitrypsin. UniProtKB/Swiss-Prot entries: horse, LEI P05619; human, LEI P30740; pig, LEI P80229. The hinge region is light green, the inhibitory consensus pattern is dark green, and the P1-P1′ elastase cleavage site is blue. The RCL integrated into the main β-sheet of LEI after cleavage is in bold, and the single elastase recognition site is underlined. His 287, which is lost in human LEI and replaced with Ser, is cyan. The two cysteines of pig LEI are purple. The sequences obtained by Edman degradation are yellow (41). The bipartite NLS is red, and the endonuclease active site is orange. Red arrowheads indicate points of site-directed mutagenesis (1, main NLS mutant; 2, hinge region mutant; 3, endonuclease active-site mutant).
FIG. 2.
Antiprotease and endonuclease activities of three different LEI mutants. Site-directed mutagenesis was performed with a wild-type (WT or wt) LEI-pET 23d(+) construct and the QuikChange mutagenesis kit (Stratagene). An alanine residue of the hinge region of the RCL of LEI (AP10T mutant) and histidine 368 of the endonuclease active site (H368A mutant) were changed to threonine and alanine, respectively. Recombinant proteins were produced by E. coli strain BL21/pLysS and purified with His-select cartridges (Sigma). Increasing concentrations of wild-type or mutant LEI (17.5 to 280 μg/ml) (magenta and blue lines) were incubated with elastase (0.1 μg/ml) in the presence of a 2 mM concentration of a synthetic substrate, pNaMAAPV. The resulting colored reaction was measured (DO, optical density) at 405 nm (a). As a control, elastase and its substrate were incubated alone (red). In the AP10T mutant, the antiprotease activity is strongly affected compared to that of wild-type LEI. On the contrary, this activity is not affected in the H368A mutant. DNase activity (b) was measured after overnight cleavage of LEI with equimolar quantities of elastase at 37°C. Samples (841 ng) of wild-type, AP10T, and H368A LEI were incubated with 2.5 μg of plasmid DNA for different times at 37°C in 20 mM Tris-EDTA, pH 5.5. Untreated wild-type LEI was incubated with DNA as a negative control. The AP10T mutant shows a DNase activity comparable to that of the wild type, while H368A has no endonuclease activity. No endonuclease activity could be detected for the 358stop mutant, but DNA was slowed down with incubation time, suggesting its binding to this protein.
We then asked whether the change in LEI activity might be associated with splicing of the protein (43). This posttranslational modification has already been described in prokaryotes and lower eukaryotes (6) and concerns proteins which are often related to DNA metabolism (11). This possibility was not assessed further since no consensus site for protein splicing was found in LEI.
We finally studied a possible conformational change as a source of the enzymatic switch in LEI activity. Concerning this possibility, it is worth noting that LEI, like all serpins, is a metastable protein. Its RCL (P5-P15 for LEI) is flexible and can assume different conformations (see Fig. S1 in the supplemental material). We have previously shown that cleavage of LEI (42 kDa) by elastase results in the appearance of a 35-kDa band in denaturing gels. This is a mandatory condition for the molecule to display endonuclease activity. Treatment of wild-type LEI with an inactivated elastase (heat treated or inhibited with phenylmethylsulfonyl fluoride) (13) did not cause its cleavage and transformation into L-DNase II (data not shown). This indicated that only active elastase can induce endonuclease activity.
In order to further investigate this hypothesis, we introduced a single base mutation into the hinge region of LEI (Fig. 1). This mutation resulted in the replacement of a small noncharged alanine with a bigger polar threonine (AP10T). This transforms the LEI interaction with its substrate from tight binding to competitive inhibition. This mutant lost its antiprotease activity (Fig. 2a, AP10T compared with WT). Nevertheless, after overnight treatment with elastase, the endonuclease activity was conserved (Fig. 2b, AP10T). It is worth noting that incubation of this mutant with elastase overnight was done to keep the experimental conditions identical to those of the wild-type molecule. In fact, the same activity was obtained with shorter incubation times (2 to 3 h).
Taken together, these experiments show that cleavage of the RCL of LEI is involved in the appearance of the endonuclease activity. They also suggest that endonuclease and antiprotease activities are not physically linked in the molecule.
Therefore, of the three hypotheses raised before, the conformational modification induced by cleavage of the RCL appears the most likely explanation for the change in LEI activity.
Endonuclease activity of L-DNase II.
The following three possibilities may then explain the emergence of the endonuclease activity generated by insertion of the RCL into A β-sheet: (i) the RCL establishes new interactions in A β-sheet, (ii) it creates a new interaction elsewhere in the molecule, or (iii) it uncovers a preexisting active site hidden underneath.
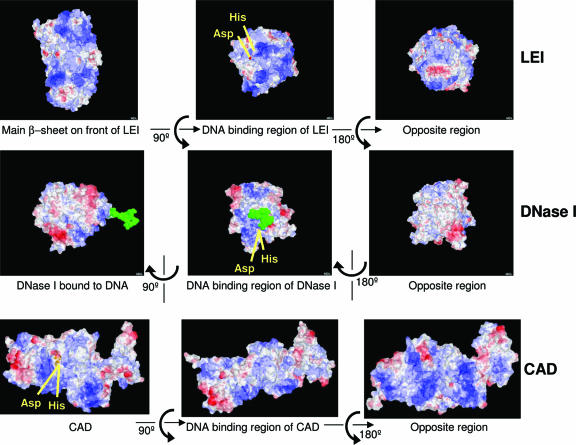
In order to distinguish among these possibilities, we analyzed the three-dimensional crystal structure of cleaved horse LEI. This was done by using Protein Explorer and structural data from the PDB (entry 1HLE) to evaluate the charge distribution of cleaved LEI. The molecule seems quite polarized (top panels of Fig. 3), with a higher number of positive charges in the RCL pole. The negative charge of DNA would better interact with this region, which is, in addition, exposed after cleavage of the RCL. Moreover, comparison of LEI with two well-characterized endonucleases, DNase I and CAD, shows a similar charge distribution (Fig. 3). Therefore, the presence of a preexisting endonuclease active site should be first investigated.
FIG. 3.
Comparison of charges of different endonucleases. The structures of LEI, DNase I, and CAD were analyzed with Protein Explorer (23). Perpendicular views of cleaved LEI (PDB number 1HLE) (top), DNase I (PDB number 3DNI) (middle), and CAD (PDB number 1V0D) (bottom) were charge colored (basic regions are blue, and acidic regions are red). DNA binding regions of the three molecules are more basic than the opposite side. That area would easily interact with acidic charges of DNA. Aspartate and histidine residues that seem important for endonuclease activity are yellow. DNA associated with DNase I is green. CAD seems to be polarized slightly differently, maybe because it is the only molecule that works as a dimer.
All apoptotic endonucleases show a DH doublet in the active site. Analysis of the three-dimensional structure of LEI shows the presence of two histidines (H287 and H368) in the more positively charged region. Of these histidines, we retained His368, which is conserved in all species, while His287 is absent in humans (Fig. 1). We therefore introduced a point mutation of His368 by replacing it with an alanine (H368A). We expressed it in bacteria, which produced the mutant protein, and measured its endonuclease activity (Fig. 2b, lowest part). As shown in Fig. 2, no endonuclease activity was detected in this mutant when it was exposed to DNA. This was not due to a lack of efficient cleavage of the mutant, as it was able to inhibit elastase. Therefore, this mutation did not affect the antiprotease activity of LEI (Fig. 2a, lowest part).
Nuclear translocation of L-DNase II is due to its binding to importin α.
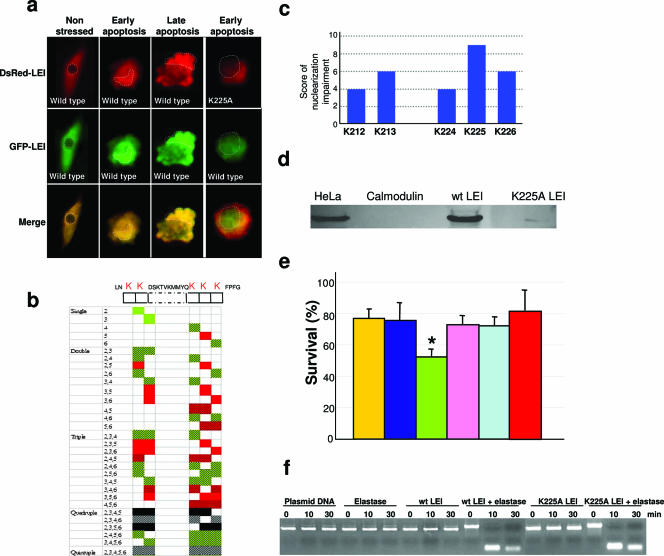
The change in the enzymatic activity of LEI induced during its transformation into L-DNase II is also followed by a change in its cellular localization (41). We therefore analyzed the structure of LEI by looking for a nuclear localization signal (NLS). We found a consensus bipartite NLS at positions 212 to 213 and 224 to 226 (Fig. 1). In order to verify that these lysines are really responsible for the nuclear import of L-DNase II, we systematically mutated them to alanine. This led to the generation of 31 constructs with one to five mutated lysines in different combinations. To monitor the subcellular localization of the protein, we performed the mutation on a DsRed2-LEI chimera. In order to exclude artifactual nuclear translocation due to a change in nuclear envelope permeability, we made a green fluorescent protein-wild-type LEI construct that we systematically cotransfected with the red chimeras. A mutant was considered to be impaired for nuclear translocation when cells expressing both plasmids presented a green nucleus and a yellow or red cytoplasm after induction of apoptosis with 40 μM EIPA (a Na+/H+ pump inhibitor) (Fig. 4a). The right part of Fig. 4a shows a default in the nuclear translocation of a DsRed2-LEI NLS mutant (the K225A LEI mutant). Analysis of the cotransfected cells allowed us to classify mutants as having a normal or altered nuclear translocation pattern during apoptosis. Relevant results are presented in tabular form in Fig. 4b. In these experiments, some red and/or green aggregates rendered the nuclear translocation of a few LEI mutants difficult to evaluate. There were also mutants that did not fluoresce sufficiently to produce a score. Nevertheless, by counting the mutants with impaired nuclear translocation for a given amino acid and using this table for support, we were able to score the importance of the different lysines of the NLS (Fig. 4c).
FIG. 4.
Site-directed mutagenesis of the consensus bipartite NLS. (a) Impairment of nuclear translocation is shown for the K225A NLS LEI mutant. Cotransfection was done with a green fluorescent protein-LEI plasmid to validate the nuclear translocation of wild-type LEI. The right part of the panel shows the different distributions of wild-type and NLS/K225A mutant LEI. Pictures were taken 16 h after induction of apoptosis with 40 μM EIPA. Nuclei or apoptotic bodies containing nuclear material are indicated by broken outlines. (b) The lysines of the putative L-DNase II NLS were changed to alanines, and it was cotransfected with a wild-type LEI construct into BHK cells (as described in Materials and Methods). Evaluation of the nuclearization of the constructs during apoptosis induced with 40 μM EIPA is summarized on the left. Each number on the left corresponds to the last number of the mutated lysine. Green squares represent wild-type behavior of the mutated protein, and red squares represent impaired translocation. Black and gray squares represent no or little fluorescent proteins, and hatched squares indicate the presence of fluorescent aggregates in cells. (c) Impairment of the nuclear translocation of a mutated lysine was scored 1. This allowed the assignment of a score to each mutant represented on the histogram. Lysine 225 is the most important for L-DNase II nuclearization. (d) The K225A LEI mutant is impaired in the ability to fix importin α compared to wild-type (wt) LEI. Calmodulin was used as a negative pull-down control, as it does not bind directly to importin α, and a brut extract of HeLa cells was used as a positive control for the presence and molecular mass of importin α. (e) Proapoptotic activity of wild-type LEI. The rate of survival of BHK cells overexpressing different LEI constructs shows increased cell death for wild-type LEI, compared to the mutated constructs. Vectors: empty (yellow), LacZ (dark blue), wild-type LEI (green), H368A/endonuclease active site LEI mutant (pink), K225A/NLS LEI mutant (light blue), and nontransfected cells (red). Data are means ± standards deviations. *, P < 0.001 (Student's test) versus H368A and K225A. (f) Endonuclease activity of the K225A NLS LEI mutant. Site-directed mutagenesis was performed on the middle lysine of the major cluster of lysines of the NLS. The K225A LEI mutant was then incubated with elastase overnight at 37°C to obtain the cleaved form of LEI. Incubation of this cleaved form with plasmid DNA for 10 to 30 min allowed us to measure the endonuclease activity of the K225A LEI mutant and compare it with that of wild-type (wt) LEI. K225A LEI showed endonuclease activity comparable to that displayed by wild-type LEI previously treated with elastase. NLS mutant or wild-type LEI was not able to cleave DNA if it had not been treated previously with elastase. Plasmid DNA showed no autodegradation or cleavage by elastase.
Proteins bearing a bipartite NLS are transported to the nucleus by carriers such as importin α (16). In order to verify the ability of the K225A LEI mutant to bind importin α, we pulled down importin α with recombinant LEI. To do this, we produced the His-tagged wild type and K225A mutant in bacteria, fixed them to an Ni2+ column, and added a protein extract from HeLa cells to each column. The extract was loaded onto an acrylamide gel, and Western blotting was done with an anti-importin α antibody (Fig. 4d). As shown in this Fig. 4, compared with wild-type LEI, the K225A mutant is impaired in the ability to bind importin α. His-tagged calmodulin was used as a negative pull-down control. HeLa whole-cell extract was used as a positive control for immunoblotting.
LEI must be transformed into L-DNase II to mediate apoptosis.
The apoptotic effect of L-DNase II probably depends on its endonuclease activity but also on its nuclear translocation. In order to explore this point, LEI/L-DNase II inactivated for endonuclease activity (H368A mutant) or impaired for nuclear translocation (K225A mutant) was transfected into BHK cells. After induction of apoptosis with HMA (a Na+/H+ pump inhibitor), cells transfected with wild-type LEI showed a significant decrease in their survival rate (Fig. 4e), in agreement with previous data (2). On the contrary, the NLS mutant (K225A) and the endonuclease active-site mutant (H368A) showed higher survival recovery compared to the wild type. It is worth noting that the K225A mutation does not modify endonuclease or antiprotease activity significantly (Fig. 4f and data not shown).
DISCUSSION
LEI belongs to the intracellular subgroup of ov-serpins (17). With the exception of ovalbumin, all members of this subgroup contain conserved residues in the hinge region that are essential for their inhibitory activity (Fig. 1). Modification of cellular behavior is the main function throughout this subgroup (8). Some regulate lysosomal proteases (squamous cell carcinoma antigen), monocyte/granulocyte proteinases (proteinase inhibitor 6), fibrinolysis (plasminogen activator inhibitor 2), or bone marrow differentiation (bomapine). Others are tumor suppressors (maspin) or are implicated in angiogenesis (10). Several serpins can inhibit apoptosis, as the viral serpin CrmA inhibits Fas- or tumor necrosis factor alpha-induced apoptosis (14). Overexpression of PAI-2 or proteinase inhibitor 9 protects cells from tumor necrosis factor alpha (15)- or granzyme B-induced apoptosis (7), respectively. All of these data and some of our results suggested that LEI, as a member of this serpin subgroup, could act as a molecular switch between life and death by apoptosis.
In a previous study, we have shown that a limited cleavage transforms LEI into L-DNase II (41). Three hypotheses were initially raised to explain the change in LEI activity. The transition may (i) result from proteolytic cleavage of the N- or C-terminal region, (ii) be linked to splicing of the protein (43), or (iii) involve a conformational change. Of these hypotheses, the first was not proven. Many experiments to find the released peptide were performed in our laboratory without success (not shown). The second hypothesis was also rejected since no consensus site for protein splicing was found in LEI (43). Proteins that undergo posttranslational splicing have junction sequences flanked by two cysteines, and these are only found in pig LEI (Fig. 1). Moreover, peptides between these cysteines were found in both LEI and L-DNase II by Edman degradation (41) (Fig. 1).
The third hypothesis was first investigated by introducing a single base mutation into the hinge region of LEI (Fig. 1). Previous studies showed that this replacement slows down the insertion of the RCL into β-sheet A (30) without changing protease specificity. This mutant LEI loses its antiprotease activity but keeps its endonuclease activity after elastase cleavage (Fig. 2). This indicates that endonuclease and antiprotease activities are not physically linked in the molecule and suggests that the insertion of the RCL into β-sheet A of LEI is involved in the establishment of endonuclease activity.
The endonuclease activity may be generated from new interactions in β-sheet A either locally or in other parts of the molecule or by uncovering a preexisting active site. Of these hypotheses, the last one was explored, since cleavage of the RCL uncovers a basic region and transforms LEI into a polarized molecule, a property common to many endonucleases. This region of the molecule that could potentially interact with DNA should then carry the DNase active site. Studies performed by our group did not reveal any consensus sequence for endonuclease activity (22). However, it is worth noting that the active site of endonucleases activated in apoptosis displays a DH doublet. Of these two amino acid residues, the histidine moiety is involved in the proton exchange during DNA cleavage (28). Two histidines (H287 and H368) were good candidates. We retained His368 because it is conserved in the species shown in Fig. 1, as well as in mice (UniProtKB/TrEMBL entry Q5SV42), rats (UniProtKB/TrEMBL entry Q4GG075), and zebra fish (UniProtKB/TrEMBL entry Q6DG30). This histidine is surrounded by a basic ring that may bind DNA. Mutation of this histidine completely abolishes the endonuclease activity and proapoptotic properties of L-DNase II, suggesting that this histidine is essential for endonuclease activity. It is interesting that the entire basic region is exposed by the RCL insertion in β-sheet A and this insertion also uncovers a bipartite nuclearization signal. This is not surprising since L-DNase II has to be translocated to the nucleus, where its new target is located. This feature is not due to the increase in nucleus permeability in late apoptosis (33), since in apoptosis induced by caspases, LEI remains in the cytoplasm (2).
Bipartite NLSs are formed by five lysines divided into two groups and are thought to interact with importin α. Here we showed that mutation of these lysines impairs nuclear translocation and binding to importin α. However, it should be stressed that the highest nuclearization impairment corresponds to the lysine at position 225, the central lysine of the major cluster. This result is in agreement with data in the literature concerning the relevance of the second lysine cluster of a bipartite NLS to importin α binding (16).
We have previously shown that, under certain apoptotic conditions, LEI is transformed into L-DNase II (2). In addition, several authors have demonstrated that overexpression of a DNase in a cell may induce apoptosis (18, 21, 25, 27, 31). This is also true for L-DNase II (2; Fig. 4). The apoptotic effect of L-DNase II depends on its endonuclease activity but also on its nuclear translocation because overexpression of the K225A mutant decreases the proapoptotic activity of L-DNase II as much as the endonuclease-inactive protein. This effect depends upon the inability of the K225A mutant to be nuclearized, since this mutation does not affect endonuclease activity.
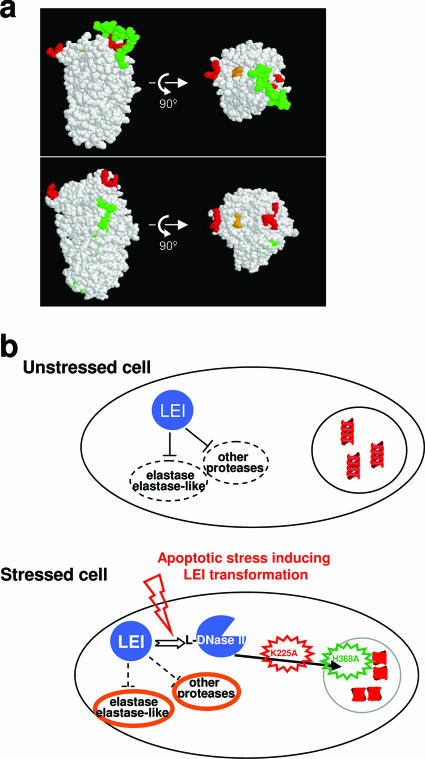
Taken together, these results indicate that the active endonuclease site of L-DNase II is located underneath the RCL. This site is flanked by a positively charged region that contains a bipartite NLS. Transformation of LEI into L-DNase II is then explained by the conformational modification induced by cleavage of the RCL, which uncovers the endonuclease active site. This conformational modification also unmasks a bipartite NLS that allows the nuclear translocation of L-DNase II by binding to importin α (Fig. 5a).
FIG. 5.
Molecular structure of LEI and its implication in cellular fate. (a) The structure of LEI was analyzed with Protein Explorer. Native LEI (upper part) shows the RCL (green) covering the major cluster of lysines of the bipartite NLS (red). The endonuclease active site is orange. This three-dimensional structure was obtained from the pig LEI sequence. The lower part of the panel shows cleaved horse LEI (PDB number 1HLE) with an unmasked NLS (red) after insertion of the RCL into β-sheet A of serpin. (b) In unstressed cells, LEI is localized in the cytoplasm and has an antiprotease activity that preferentially inhibits elastase. When the cell undergoes a stress that causes the transformation of LEI into L-DNase II, the antiprotease activity is released. L-DNase II enters the nucleus, cleaves the DNA, and finally leads to apoptosis. Different mutant forms of LEI have been constructed to understand the LEI/L-DNase II pathway. NLS mutant LEI (K225A, red) is not able to enter the nucleus, and endonuclease active-site mutant LEI (H368A, green) is able to enter the nucleus but is unable to degrade DNA. In both cases, apoptosis is reduced. LEI has to be cleaved into L-DNase II and translocated to the nucleus to induce apoptosis.
Current data concerning the dual activity of LEI/L-DNase II, including those obtained in the present study, are summarized in Fig. 5b. They indicate that LEI, as well as other members of the intracellular serpin subfamily, is highly implicated in cellular fate. Thus, LEI mediates caspase-independent apoptosis under some particular conditions, i.e., when serine proteases having LEI as a target are massively activated. This takes place during metabolic and oxidative stress, when cells are unable to activate caspases, due to terminal differentiation (as in retinal pigmented epithelium cells) (9) or to acquired mutations in cancer cells (4, 5). In this work, we show that the change in activity of LEI is mediated by the classical conformational modification mediating antiprotease activity of serpins. This is a novel example of the use of conformational modification of serpins to perform tasks different from protease inhibition.
The involvement of this serpin in the activation of a cell death program might turn out to be of interest in the development of therapeutic strategies against cancer cells, for instance. Malignant transformation is sometimes followed by loss of apoptotic effectors, leading to resistance to chemotherapy. Induction of apoptosis by stimuli leading to the transformation of LEI into L-DNase II may overcome this resistance. The LEI AP10T mutant described in this work could also be used to increase the efficiency of anticancer therapy. Indeed, this mutant is not very efficient at inhibiting its target protease and releases it quickly. The protease is no longer trapped in a covalent complex, as opposed to wild-type LEI. As a result, equilibrium will be displaced toward the production of L-DNase II, as the released target protease will be able to cleave more LEI molecules. This would trigger apoptosis in cancer cells. Moreover, a point mutation of the RCL might allow changing the protease specificity of the serpin, which may then be adapted to the protease expression of the target cell.
The existence of a protein that is able to induce apoptosis-like cell death by a single conformational modification should encourage research on other key molecules involved in cell fate decisions.
Supplementary Material
Acknowledgments
We thank Paolo Perani and Samia Zeggai for help with LEI wild-type and 358stop constructs and Sabine Chahory, Yves Courtois, Marie-Thérèse Dimanche-Boitrel, Patricia Lassiaz, Bernard Mignotte, Flore Renaud, Evelyne Segal, and Ivana Scovassi for critical reading of the manuscript. We acknowledge Slavica Kantric for editing the English manuscript.
This work was supported by Retina France, Fondation Raymonde et Guy Strittmatter, and Fondation Singer-Polignac.
Footnotes
Published ahead of print on 2 April 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Altairac, S., S. C. Wright, Y. Courtois, and A. Torriglia. 2003. L-DNase II activation by the 24 kDa apoptotic protease (AP24) in TNFα-induced apoptosis. Cell Death Differ. 10:1109-1111. [DOI] [PubMed] [Google Scholar]
- 2.Altairac, S., S. Zeggai, P. Perani, Y. Courtois, and A. Torriglia. 2003. Apoptosis induced by Na+/H+ antiport inhibition activates the LEI/L-DNase II pathway. Cell Death Differ. 10:548-557. [DOI] [PubMed] [Google Scholar]
- 3.Barry, M. A., and A. Eastman. 1993. Identification of deoxyribonuclease II as an endonuclease involved in apoptosis. Arch. Biochem. Biophys. 300:440-450. [DOI] [PubMed] [Google Scholar]
- 4.Belmokhtar, C. A., J. Hillion, C. Dudognon, S. Fiorentino, M. Flexor, M. Lanotte, and E. Segal-Bendirdjian. 2003. Apoptosome-independent pathway for apoptosis. Biochemical analysis of APAF-1 defects and biological outcomes. J. Biol. Chem. 278:29571-29580. [DOI] [PubMed] [Google Scholar]
- 5.Belmokhtar, C. A., A. Torriglia, M. F. Counis, Y. Courtois, A. Jacquemin-Sablon, and E. Segal-Bendirdjian. 2000. Nuclear translocation of a leukocyte elastase inhibitor/elastase complex during staurosporine-induced apoptosis: role in the generation of nuclear L-DNase II activity. Exp. Cell Res. 254:99-109. [DOI] [PubMed] [Google Scholar]
- 6.Bezerra, W. M., C. P. Carvalho, A. Moreira Rde, and T. B. Grangeiro. 2006. Establishment of a heterologous system for the expression of Canavalia brasiliensis lectin: a model for the study of protein splicing. Genet. Mol. Res. 5:216-223. [PubMed] [Google Scholar]
- 7.Bird, C. H., V. R. Sutton, J. Sun, C. E. Hirst, A. Novak, S. Kumar, J. A. Trapani, and P. I. Bird. 1998. Selective regulation of apoptosis: the cytotoxic lymphocyte serpin proteinase inhibitor 9 protects against granzyme B-mediated apoptosis without perturbing the Fas cell death pathway. Mol. Cell. Biol. 18:6387-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bird, P. I. 1999. Regulation of pro-apoptotic leucocyte granule serine proteinases by intracellular serpins. Immunol. Cell Biol. 77:47-57. [DOI] [PubMed] [Google Scholar]
- 9.Brossas, J. Y., R. Tanguy, F. Brignole-Baudouin, Y. Courtois, A. Torriglia, and J. Treton. 2004. L-DNase II associated with active process during ethanol induced cell death in ARPE-19. Mol. Vis. 10:65-73. [PubMed] [Google Scholar]
- 10.Cao, Y. 2001. Endogenous angiogenesis inhibitors and their therapeutic implications. Int. J. Biochem. Cell Biol. 33:357-369. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, A. A., Y. J. Chen, M. A. Lindorfer, and T. H. Stevens. 1993. Protein splicing of the yeast TFP1 intervening protein sequence: a model for self-excision. EMBO J. 12:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Counis, M. F., E. Chaudun, C. Arruti, L. Oliver, M. Sanwal, Y. Courtois, and A. Torriglia. 1998. Analysis of nuclear degradation during lens cell differentiation. Cell Death Differ. 5:251-261. [DOI] [PubMed] [Google Scholar]
- 13.El Ouriaghli, F., H. Fujiwara, J. J. Melenhorst, G. Sconocchia, N. Hensel, and A. J. Barrett. 2003. Neutrophil elastase enzymatically antagonizes the in vitro action of G-CSF: implications for the regulation of granulopoiesis. Blood 101:1752-1758. [DOI] [PubMed] [Google Scholar]
- 14.Enari, M., H. Hug, and S. Nagata. 1995. Involvement of an ICE-like protease in Fas-mediated apoptosis. Nature 375:78-81. [DOI] [PubMed] [Google Scholar]
- 15.Furlong, I. J., R. Ascaso, A. Lopez Rivas, and M. K. Collins. 1997. Intracellular acidification induces apoptosis by stimulating ICE-like protease activity. J. Cell Sci. 110(Pt. 5):653-661. [DOI] [PubMed] [Google Scholar]
- 16.Hodel, M. R., A. H. Corbett, and A. E. Hodel. 2001. Dissection of a nuclear localization signal. J. Biol. Chem. 276:1317-1325. [DOI] [PubMed] [Google Scholar]
- 17.Irving, J. A., R. N. Pike, A. M. Lesk, and J. C. Whisstock. 2000. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. 10:1845-1864. [DOI] [PubMed] [Google Scholar]
- 18.Krieser, R. J., and A. Eastman. 1998. The cloning and expression of human deoxyribonuclease II. A possible role in apoptosis. J. Biol. Chem. 273:30909-30914. [DOI] [PubMed] [Google Scholar]
- 19.Leist, M., and M. Jaattela. 2001. Four deaths and a funeral: from caspases to alternative mechanisms. Nat. Rev. Mol. Cell Biol. 2:589-598. [DOI] [PubMed] [Google Scholar]
- 20.Li, L. Y., X. Luo, and X. Wang. 2001. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412:95-99. [DOI] [PubMed] [Google Scholar]
- 21.Los, M., D. Neubuser, J. F. Coy, M. Mozoluk, A. Poustka, and K. Schulze-Osthoff. 2000. Functional characterization of DNase X, a novel endonuclease expressed in muscle cells. Biochemistry 39:7365-7373. [DOI] [PubMed] [Google Scholar]
- 22.Martin, E., M. F. Counis, P. Perani, Y. Courtois, and A. Torriglia. 2002. LEI/L-DNase II: les implications structurales d'un détournement de fonction. Med. Sci. 18:111-120. [Google Scholar]
- 23.Martz, E. 2002. Protein Explorer: easy yet powerful macromolecular visualization. Trends Biochem. Sci. 27:107-109. [DOI] [PubMed] [Google Scholar]
- 24.Montague, J. W., M. L. Gaido, C. Frye, and J. A. Cidlowski. 1994. A calcium-dependent nuclease from apoptotic rat thymocytes is homologous with cyclophilin. Recombinant cyclophilins A, B, and C have nuclease activity. J. Biol. Chem. 269:18877-18880. [PubMed] [Google Scholar]
- 25.Mukae, N., M. Enari, H. Sakahira, Y. Fukuda, J. Inazawa, H. Toh, and S. Nagata. 1998. Molecular cloning and characterization of human caspase-activated DNase. Proc. Natl. Acad. Sci. USA 95:9123-9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagata, S. 2005. DNA degradation in development and programmed cell death. Annu. Rev. Immunol. 23:853-875. [DOI] [PubMed] [Google Scholar]
- 27.Oliveri, M., A. Daga, C. Cantoni, C. Lunardi, R. Millo, and A. Puccetti. 2001. DNase I mediates internucleosomal DNA degradation in human cells undergoing drug-induced apoptosis. Eur. J. Immunol. 31:743-751. [DOI] [PubMed] [Google Scholar]
- 28.Pan, C. Q., J. S. Ulmer, A. Herzka, and R. A. Lazarus. 1998. Mutational analysis of human DNase I at the DNA binding interface: implications for DNA recognition, catalysis, and metal ion dependence. Protein Sci. 7:628-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peitsch, M. C., B. Polzar, H. Stephan, T. Crompton, H. R. MacDonald, H. G. Mannherz, and J. Tschopp. 1993. Characterization of the endogenous deoxyribonuclease involved in nuclear DNA degradation during apoptosis (programmed cell death). EMBO J. 12:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perani, P., S. Zeggai, A. Torriglia, and Y. Courtois. 2000. Mutations on the hinge region of leukocyte elastase inhibitor determine the loss of inhibitory function. Biochem. Biophys. Res. Commun. 274:841-844. [DOI] [PubMed] [Google Scholar]
- 31.Polzar, B., M. C. Peitsch, R. Loos, J. Tschopp, and H. G. Mannherz. 1993. Overexpression of deoxyribonuclease I (DNase I) transfected into COS-cells: its distribution during apoptotic cell death. Eur. J. Cell Biol. 62:397-405. [PubMed] [Google Scholar]
- 32.Potempa, J., A. Dubin, W. Watorek, and J. Travis. 1988. An elastase inhibitor from equine leukocyte cytosol belongs to the serpin superfamily. Further characterization and amino acid sequence of the reactive center. J. Biol. Chem. 263:7364-7369. [PubMed] [Google Scholar]
- 33.Roehrig, S., A. Tabbert, and E. Ferrando-May. 2003. In vitro measurement of nuclear permeability changes in apoptosis. Anal. Biochem. 318:244-253. [DOI] [PubMed] [Google Scholar]
- 34.Samejima, K., and W. C. Earnshaw. 2005. Trashing the genome: the role of nucleases during apoptosis. Nat. Rev. Mol. Cell Biol. 6:677-688. [DOI] [PubMed] [Google Scholar]
- 35.Shiokawa, D., H. Ohyama, T. Yamada, K. Takahashi, and S. Tanuma. 1994. Identification of an endonuclease responsible for apoptosis in rat thymocytes. Eur. J. Biochem. 226:23-30. [DOI] [PubMed] [Google Scholar]
- 36.Silverman, G. A., P. I. Bird, R. W. Carrell, F. C. Church, P. B. Coughlin, P. G. Gettins, J. A. Irving, D. A. Lomas, C. J. Luke, R. W. Moyer, P. A. Pemberton, E. Remold-O'Donnell, G. S. Salvesen, J. Travis, and J. C. Whisstock. 2001. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 276:33293-33296. [DOI] [PubMed] [Google Scholar]
- 37.Susin, S. A., H. K. Lorenzo, N. Zamzami, I. Marzo, B. E. Snow, G. M. Brothers, J. Mangion, E. Jacotot, P. Costantini, M. Loeffler, N. Larochette, D. R. Goodlett, R. Aebersold, D. P. Siderovski, J. M. Penninger, and G. Kroemer. 1999. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397:441-446. [DOI] [PubMed] [Google Scholar]
- 38.Torriglia, A., E. Chaudun, F. Chany-Fournier, Y. Courtois, and M. F. Counis. 2001. Involvement of L-DNase II in nuclear degeneration during chick retina development. Exp. Eye Res. 72:443-453. [DOI] [PubMed] [Google Scholar]
- 39.Torriglia, A., E. Chaudun, F. Chany-Fournier, J. C. Jeanny, Y. Courtois, and M. F. Counis. 1995. Involvement of DNase II in nuclear degeneration during lens cell differentiation. J. Biol. Chem. 270:28579-28585. [DOI] [PubMed] [Google Scholar]
- 40.Torriglia, A., C. Negri, E. Chaudun, E. Prosperi, Y. Courtois, M. F. Counis, and A. I. Scovassi. 1999. Differential involvement of DNases in HeLa cell apoptosis induced by etoposide and long term-culture. Cell Death Differ. 6:234-244. [DOI] [PubMed] [Google Scholar]
- 41.Torriglia, A., P. Perani, J. Y. Brossas, E. Chaudun, J. Treton, Y. Courtois, and M. F. Counis. 1998. L-DNase II, a molecule that links proteases and endonucleases in apoptosis, derives from the ubiquitous serpin leukocyte elastase inhibitor. Mol. Cell. Biol. 18:3612-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Gent, D., P. Sharp, K. Morgan, and N. Kalsheker. 2003. Serpins: structure, function and molecular evolution. Int. J. Biochem. Cell Biol. 35:1536-1547. [DOI] [PubMed] [Google Scholar]
- 43.Wallace, C. J. 1993. The curious case of protein splicing: mechanistic insights suggested by protein semisynthesis. Protein Sci. 2:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.