Abstract
Nucleosomes fulfill the apparently conflicting roles of compacting DNA within eukaryotic genomes while permitting access to regulatory factors. Central to this is their ability to stably associate with DNA while retaining the ability to undergo rearrangements that increase access to the underlying DNA. Here, we have studied different aspects of nucleosome dynamics including nucleosome sliding, histone dimer exchange, and DNA wrapping within nucleosomes. We find that alterations to histone proteins, especially the histone tails and vicinity of the histone H3 αN helix, can affect these processes differently, suggesting that they are mechanistically distinct. This raises the possibility that modifications to histone proteins may provide a means of fine-tuning specific aspects of the dynamic properties of nucleosomes to the context in which they are located.
The organization of DNA into chromatin creates the functional template for any process requiring access to the genetic information such as transcription, DNA replication, recombination, and repair. The basic repeating unit of chromatin is the nucleosome core particle, which consists of 147 bp of DNA wrapped almost twice around an octamer of histone H2A, H2B, H3, and H4 proteins (12, 32). Chromatin packaging tends to restrict access to the underlying DNA, meaning that regulation of chromatin structure is a key feature of many genetic processes.
Eukaryotes have adopted an assortment of different strategies by which they can manipulate chromatin structure. One means involves the posttranslational modification of histone proteins by, for example, acetylation, methylation, and phosphorylation. These are often correlated with particular chromatin states, and the levels of many of these modifications are observed to change during the course of gene regulation (38). To date, the majority of the best-characterized modifications occur in the N-terminal extensions of the histone proteins. These N-terminal tails extend beyond the globular core of the nucleosome and do not take up distinct conformations in high-resolution crystal structures (12, 32). Modification of residues within the tail domains has been shown to affect chromatin structure by generating epitopes for the recruitment of external chromatin binding proteins. These include the chromodomains of HP1 and PRC1, which interact specifically with histones methylated at histone H3 lysine 9 and 27, respectively (4, 14, 27), and bromodomain-containing proteins which bind acetylated histone tails (26, 42).
An alternative means by which histone tails can influence chromatin structure is through direct alteration of the dynamic properties of nucleosomes. Chromatin is not a static entity but can undergo an assortment of dynamic alterations. Arrays of nucleosomes spontaneously condense to form chromatin fibers (5, 9, 57), the positions of nucleosomes on DNA fragments can change as a result of thermally driven nucleosome sliding (16, 37), and the outer turns of DNA are prone to transient dissociation from the histone octamer via a process referred to as site exposure (30, 31, 47). Previous studies have implicated histone tails and/or their modification as a means of directly altering the properties of nucleosomes (1, 50, 55, 56).
In addition to modifications of histones in the N- and C-terminal extensions, a number of modifications have been identified within the globular core of the histone octamer (21). As even conservative single-amino acid changes to nucleosome structure can cause significant changes in nucleosome mobility (19, 41), the idea that modifications within the globular core of nucleosomes may modulate their dynamic properties is an attractive one (11).
Here, we investigate the role of individual histone tails on nucleosome mobility. We find that histone tails have nonredundant effects on the rate of thermal nucleosome repositioning. Deletion of the entire H3 tail was found to have additional effects on DNA wrapping and histone dimer exchange. Mutations of residues located at the base of the H3 tail and the adjoining H3 αN helix were also found to have effects on nucleosome sliding, H2A/H2B dimer stability, and the trajectory of linker DNA. Introducing mutations that mimic modifications detected within this region also affected these dynamic processes. These observations add to the range of mechanisms by which chromatin modifications may influence access to the underlying genetic information.
MATERIALS AND METHODS
Octamer preparation.
Recombinant Xenopus laevis histone proteins were expressed and purified as previously described (33). Site-directed mutagenesis was carried out using a Stratagene Quickchange Kit.
Nucleosome assembly.
Nucleosomes were assembled by mixing equimolar amounts of histone octamer and DNA in high salt concentrations and performing stepwise dialysis into low salt concentrations as previously described (20). For mutants that did not form octamers during dialysis into high salt concentrations, stoichiometric quantities of tetramer and dimer were added to DNA. For these mutants assembly of nucleosomes with fluorescently labeled dimers demonstrated that the nucleosome remained intact without significant levels of dimer loss during thermal incubation (data not shown). DNA was generated by preparative PCR using fluorescently labeled primers from Eurogentec (Belgium). The PCR fragments were purified by ion exchange chromatography on a 1.8-ml SOURCE 15Q (Pharmacia) column. Radiolabeled DNA was prepared using T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (Molecular Bioproducts) according to the manufacturer's specifications.
Nucleosome repositioning reactions.
Thermal remodeling reactions were performed by incubating nucleosomes in a thin-walled 200-μl PCR tube (ABgene, United Kingdom) in 50 mM NaCl-50 mM Tris, pH 7.5, at 47°C in a PCR machine with a heated lid for the amount of time indicated in the figures. At the end of the reaction, sucrose was added to 5% (wt/vol), and the reaction mixtures were placed on ice. The samples were run on preprepared 0.2× Tris-borate-EDTA-5% native polyacrylamide gels for 3.5 h at 300 V at 4°C with pump recirculation. Gels were scanned in a Fuji Phosphoimager FLA-5100, and the bands corresponding to nucleosomes at the +70 and +22 locations were quantified with Aida software (Fujifilm). The initial rate of shifting was calculated through nonlinear fitting of the data to a hyperbolic equation within the Solver add-in for Excel over 1,000 iterative cycles.
Site-directed nucleosome mapping.
Site-directed nucleosome mapping was performed as described previously (15). Briefly, nucleosomes with site-specifically attached mapping reagent were incubated with ammonium ferrous sulfate, ascorbic acid, and hydrogen peroxide to cleave DNA. DNA was isolated by phenol-chloroform extraction and run on 8% polyacrylamide denaturing gels.
DNA FRET measurement.
Fluorescent resonance energy transfer (FRET) measurements were performed in a Cary Eclipse fluorimeter (Varian, Australia). Polarizers were used to compensate for anisotropy. The photomultiplier tube voltage was set to high, and the slowest scan speed setting was selected (0.8 nm/s). All measurements were taken at room temperature. For the donor-plus-acceptor scan, the excitation was set to 515 nm, and emission was scanned from 550 nm to 700 nm, whereas for the acceptor-only scan the excitation wavelength was set to 610 nm, and emission was scanned from 630 nm to 700 nm.
Nucleosomes were assembled onto a 181-bp fragment derived from the 601.3 nucleosome positioning sequence such that 24 and 11 bp of DNA of linker DNA were present at the Pst1 and BsaAI ends of the nucleosome, respectively. The ends of the DNA were labeled with Cy5 and Cy3 dyes, respectively. Following nucleosome assembly by salt dialysis, samples were spun for 1 min at 14,000 rpm on a benchtop centrifuge to remove any precipitate. Five microliters of 10× reaction buffer (500 mM NaCl, 500 mM Tris, pH 7.5, 10 mM MgCl2) was added to 10 μl of nucleosome (final concentration, 320 nM), and the volume was made up to 50 μl with water, with the exception of the experiment shown in Fig. 3 (as described in the legend). Readings were taken at 70, 170, 270, 370, 470, and 2,020 mM NaCl by the addition of 1.3, 1.37, 1.44, 1.52, and 43.1 μl of 4 M NaCl. FRET efficiency was measured using the Cary Eclipse software by calculating the ratio of the donor-plus-acceptor signal to the acceptor-only signal over the region of 640 to 690 nm, across which the response should ideally be flat.
FIG. 3.
Deletion of the H3 tail affects DNA end-to-end FRET. (A) Nucleosomes were assembled onto DNA containing Cy3 on one end and Cy5 on the other. Once assembled, the two arms of DNA were brought into close enough proximity to generate a FRET signal. (B) gH3 nucleosomes are not able to constrain their arms of linker DNA, as indicated by the greatly reduced FRET signal. This does not occur if the H4 tail is deleted instead. The measurements were made in 50 mM Tris (pH 7.5)-1 mM MgCl2 with 60, 160, 260, 360, 460, and 2,010 mM NaCl. WT, wild type.
Dimer exchange.
Thermal dimer exchange was performed as described previously (8). Briefly, this exchange was performed as for nucleosome repositioning, except with a twofold molar excess of H3/H4 tetrasomes assembled onto 147 bp of DNA (54).
RESULTS
Histone tails have opposing contributions to intrinsic nucleosome mobility.
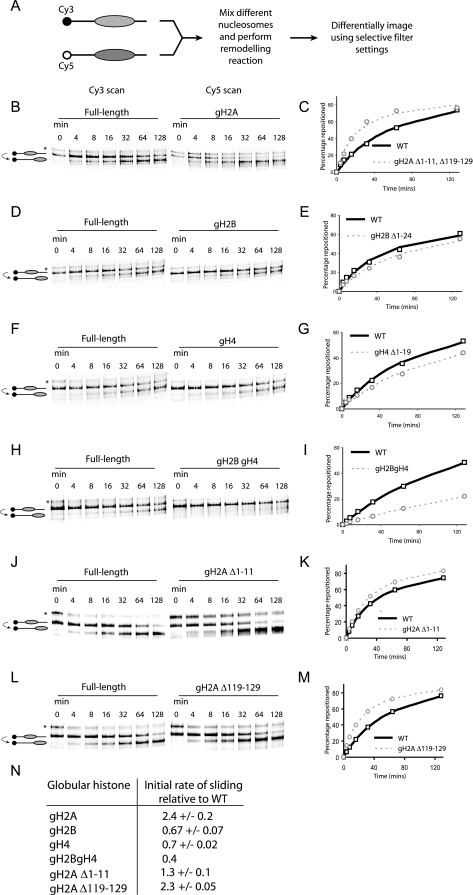
As a first step toward investigating the effect of histone tails in nucleosome mobility, N-terminally truncated histones were generated by site-directed mutagenesis. In order to assess the contribution of individual histone tails on nucleosome mobility, we used a DNA sequence derived from the mouse mammary tumor virus (MMTV) promoter to direct the assembly of positioned nucleosomes (16). Following chromatin assembly by salt dialysis, nucleosomes were initially deposited predominantly at a location 54 bp from the upstream end and 18 bp from the downstream end of the DNA fragment used (16). Upon thermal incubation the majority of these nucleosomes equilibrated to a location 48 bp upstream, which could be monitored as an increase in the mobility of these nucleosomes during native gel electrophoresis (16). In order to allow a direct comparison to be made between two samples, we modified the assay so that two different histone octamers were assembled onto DNA fragments labeled with spectrally separated fluorescent dyes. These were mixed and thermally incubated in the same tube, and following gel electrophoresis, the behavior of the two types of nucleosomes could be distinguished using selective filter sets (Fig. 1A). We found that this approach greatly improved the reliability and sensitivity with which it is possible to compare nucleosome mobility (data not shown), allowing us to routinely make comparisons between altered nucleosomes and the intact Xenopus histone octamer as a reference.
FIG. 1.
Effect of histone tail deletions on thermal nucleosome sliding. (A) Two picomoles of truncated and full-length octamers was assembled onto differentially labeled fluorescent DNA, mixed, and incubated at 47°C for the specified amount of time. Nucleosomes at different translational positions were separated on a native polyacrylamide gel, and the individual types of nucleosomes were visualized using the appropriate excitation and emission settings as illustrated. The majority of nucleosomes assembled at a position +70 relative to the MMTV transcriptional start site; following thermal incubation these equilibrated to a more favorable location at +22 (16). However, a proportion of nucleosomes (indicated by an asterisk) was deposited at a less strongly favored location at +47 (53). (B to M) Individual tail deletions have distinct effects on the rate of nucleosome repositioning. The graphs show the fraction of nucleosomes repositioned as a function of time. The data points are the average of three independent repeats, with the exception of panel F, and the curve describes the line of best fit used to calculate the initial rates. (N) Initial rate of repositioning for globular nucleosomes relative to a full-length control. WT, wild type.
Truncation of the N- and C-terminal extensions of H2A increased the rate at which nucleosomes spontaneously repositioned on this DNA fragment (Fig. 1B). By measuring the proportion of nucleosomes at the starting and finishing locations, it was possible to plot the rate at which nucleosomes were repositioned (Fig. 1C). These data could be fitted to a hyperbolic curve, providing a means of calculating differences in the initial rate of nucleosome sliding. In the case of gH2A octamers, sliding was observed to occur 2.4-fold faster than the intact octamer (Fig. 1N). The ability of globular H2A (gH2A) nucleosomes to slide faster thermally was also seen on the completely distinct Lytechinus variegatus 5S promoter DNA sequence (data not shown), suggesting that the increase in mobility is likely to be generally applicable to other DNA fragments. In order to distinguish the effects of the H2A N- and C-terminal truncations, these were analyzed separately. Truncation of the C terminus alone caused a 2.3-fold increase in mobility, whereas N-terminal truncation had only a 1.3-fold effect (Fig. 1J to N). In addition, truncation of the H2A N terminus caused an increase in the proportion of nucleosomes deposited at the alternative +22 location (Fig. 1J, asterisk).
Surprisingly, deleting the H2B N-terminal tail had the opposite effect, and the initial rate of sliding of gH2B-containing nucleosomes was slower that that of the control nucleosome (Fig. 1D and E). This was also seen with deletion of the H4 N-terminal tail (Fig. 1F and G). Although the effects are modest, deleting both the H2B and H4 N-terminal tails in combination had a cumulative effect, with the gH2B/gH4-containing nucleosome sliding at a rate 2.5-fold slower than the full-length control (Fig. 1H and I). This suggests that these histone tails make a positive contribution to nucleosome mobility.
Deletion of the entire H3 tail causes an alteration to the wrapping of nucleosomal DNA.
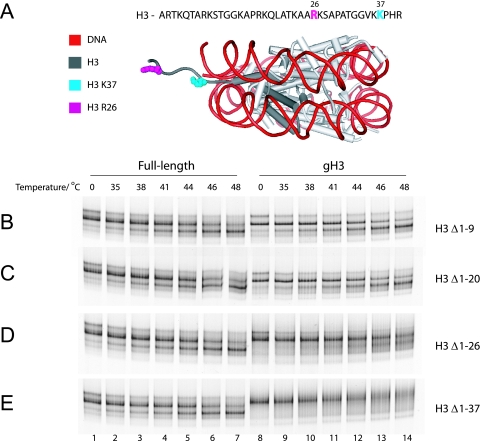
Deletion of the first 20 amino acids was observed to have no effect on nucleosome mobility (Fig. 2B and C). However, when the H3 tail was progressively deleted up to the published H3 trypsin cleavage site at amino acid 26 (3, 7) and finally up to residue 37, an interesting change in the behavior of nucleosomes was observed (Fig. 2A to E). Instead of a shift in mobility to a discrete, faster-migrating species, these nucleosomes became diffusely distributed about the starting location following thermal incubation (Fig. 2D and E, lanes 12 to 14). This suggested that the nucleosomes were not sliding to a new location, as was the case with intact histones, but were undergoing some other form of transition in structure.
FIG. 2.
Thermal repositioning of H3 truncated nucleosomes. (A) Cartoon illustration of the nucleosome core particle and the relative positions of residues R26, historically considered the last tail residue, and K37, which is the last residue. (B to E) Two picomoles of nucleosomes with either full-length or the indicated truncated H3 was incubated for 1 h at the temperatures indicated. Deletion of the entire H3 tail results in the nucleosomes' becoming diffusely distributed about the start position rather than moving to a new location.
To investigate this further we utilized a FRET-based assay to monitor the wrapping of DNA around nucleosomes (44). This involves the labeling of DNA fragments with Cy3 and Cy5 dyes at either end. The spectral overlap between these fluorophores allows them to undergo FRET when they are in close proximity. When DNA fragments of close to mononucleosome length are assembled into nucleosomes, the ends of the DNA fragments are brought into close proximity and can undergo FRET (Fig. 3A). In order to increase the homogeneity of the template and to reduce the chances of nucleosome sliding during the course of our experiments, we used a DNA fragment derived from the 601.3 strong nucleosome positioning sequence to direct the assembly of nucleosomes (54) (see Materials and Methods). Site-directed mapping and mobility shift assays indicated that the proportion of nucleosomes assembled at alternative locations was always less than 5% (data not shown). The amount of free DNA present in reconstitutions was always less than 8%, with an average of 2% that did not change significantly for any of the different mutations. As the changes in FRET efficiency we observed were of the order of 30%, the contribution of nucleosome movement or dissociation to this would be negligible.
As expected, increasing the salt concentration decreases FRET between DNA ends until at 2 M NaCl the nucleosome is completely disrupted and histones no longer bind DNA (Fig. 3B). Deletion of the H3 tail resulted in a markedly lower apparent FRET efficiency over a broad range of salt concentrations, indicating that the DNA ends move apart. In addition to being observed on a 181-bp fragment, this effect was also observable on fragments of different lengths (data not shown). The effect also appears to be specific for the H3 tail as deletion of the H4 tail had only a minimal effect on FRET (Fig. 3B).
Deletion of the H3 tail alters histone-DNA contacts within the nucleosome core.
One mechanism through which deletion of the H3 tail could cause DNA ends to move further apart involves increasing the frequency with which DNA unwraps from the outer turns of the nucleosome. There is good evidence that DNA can transiently dissociate from the edge of nucleosomes for periods of approximately 30 ms around 4 times per s (30) (Fig. 3A). If an increase in the frequency or duration of site exposure is the explanation for the reduced FRET we observed, then it would be anticipated that histone-DNA contacts nearer the edge of the nucleosome would also be reduced.
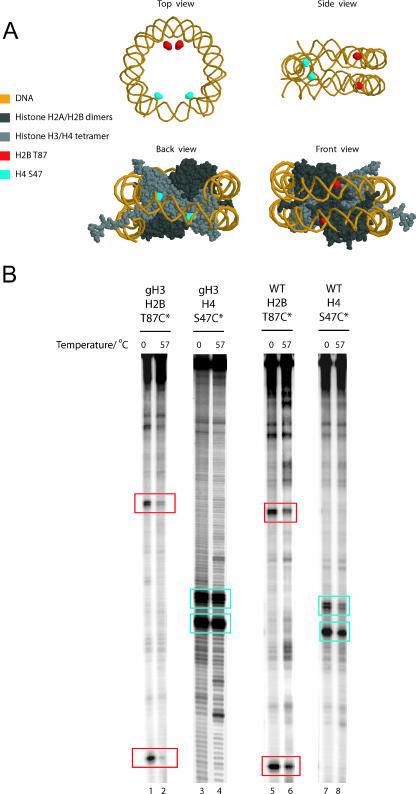
In order to test this, we attached reporter compounds capable of cleaving DNA to engineered cysteine residues at locations near the center and the edge of the nucleosome. Attachment of this mapping reagent to intact histone octamers results in the cleavage of DNA predominantly at two locations that have a spacing determined by the dyad symmetry of the nucleosome (Fig. 4A and B). Following thermal incubation, the proportion of DNA cleaved was reduced more significantly at the edge of the gH3 nucleosomes (96% reduction in mapping signal) compared to that close to the dyad (22% reduction in mapping signal) (Fig. 4B, compare lanes 1 and 2 with 3 and 4). This change was fivefold greater for gH3 octamers than for intact octamers (Fig. 4B, compare lanes 1 and 2 with 5 and 6). This supports the idea that there is a reduction in the stability with which DNA interacts with this region of the histone octamer following H3 truncation.
FIG. 4.
Deletion of the H3 tail affects histone-DNA contacts in the outer gyre. (A) Cysteine-EDTA nucleosome mapping reagent was conjugated to nucleosomes via cysteine residues introduced either at the dyad (H4 S47C) or the outer DNA gyre (H2B T87C). (B) Nucleosomes were incubated for 1 h at the indicated temperatures, followed by initiation of the Fenton reaction which causes site-specific DNA cleavage. The absence of the H3 tail results in the preferential loss of histone-DNA contacts at H2B T87C (cleavage sites in red box). Compare lanes 1 and 2 with lanes 5 and 6. In contrast, the DNA contacts in the region of the nucleosome dyad (H4 S47C contacts in blue box) are relatively unaffected.
Deletion of the H3 tail destabilizes H2A/H2B dimers within nucleosomes.
Site exposure provides a means of transiently removing DNA from the edge of nucleosomes. In addition to making contacts with the base of the H3 N-terminal tail, the outer turns of DNA interact significantly with histone H2A/H2B dimers. As association with DNA is required to maintain octamer structure at physiological salt concentrations, it is possible that removal of DNA in this region could destabilize histone dimers. Indeed, we previously observed that this was indeed the case when DNA was removed from the edge of nucleosomes as a consequence of ATP-dependent chromatin remodeling (8). In order to investigate whether the association of histone dimers was reduced following removal of the histone H3 tail, we incubated nucleosomes bearing fluorescently labeled dimers with a tetrasome acceptor for increasing lengths of time at 47°C (Fig. 5A). Under these conditions, up to 6% of dimers dissociate from intact nucleosomes after an 80-min incubation (Fig. 5B, lane 4). However, when gH3 nucleosomes were incubated at an identical temperature, 19% of H2A/H2B dimers were observed to transfer to the tetrasome acceptor (Fig. 5B, lane 8). This confirms that deletion of the H3 tail results in destabilization of the histone dimers within nucleosomes.
FIG. 5.
Measuring histone dimer stability with a dimer exchange assay. (A) Histone octamers containing Cy5-labeled H2B were assembled onto a 219-bp DNA fragment and incubated at 47°C with histone H3-H4 tetramers assembled on a 147-bp DNA fragment. Any histone dimers that dissociate from the donor nucleosome are likely to associate with the tetrasome which acts as a “sink.” (B) Deletion of the H3 tail results in increased transfer of histone dimers to the acceptor chromatin. WT, wild type.
Residues in the H3 αN helix affect different aspects of nucleosome dynamics.
The observation that truncations extending to the base of the H3 tail had the most significant effects on nucleosome dynamics indicates that this region plays an important role. This could be because amino acids in this region directly contact linker DNA (2) or because this region of the H3 tail affects the conformation of a nearby region of the histone octamer, which in turn regulates DNA association. For example, alterations to the base of the H3 tail may affect the conformation of the H3 αN helix from which the tail protrudes. To investigate this further, amino acids in the region at the base of the H3 N-terminal tail and the adjoining region that extends into the αN helix were systematically altered by site-directed mutagenesis to alanine residues. The effect of these mutations on the thermal mobility of nucleosomes was assessed, and the results are summarized in Table 1 as the average of at least three independent experiments. Individual mutations at the base of the H3 tail from K23 to V35 had very little effect on sliding (data not shown). However, for residues from K36 to S57, mutations frequently resulted in increased nucleosome sliding, although to various degrees (Table 1; Fig. 6A).
TABLE 1.
Effects of alanine mutagenesis within the H3 αN helix on nucleosome dynamicsa
| Mutation | Octamer formation | Initial nucleosome sliding rate relative to WT | Initial DNA end-to-end FRET (%)c | H2A/H2B exchange relative to WT at 60 min (fold)d |
|---|---|---|---|---|
| WT | + | 1 | 100 ± 6 | 1 |
| gH3 (Δ1-37) | + | −b | 35 ± 15 | 3.1 ± 0.3 |
| H3 V35A | + | 0.9 ± 0.1 | ND | ND |
| H3 K36A | + | 1.3 ± 0.2 | ND | ND |
| H3 K37A | + | 1.5 ± 0.4 | ND | ND |
| H3 H39A | + | 3.1 ± 0.1 | 76 ± 9 | 1.0 ± 0.3 |
| H3 R40A | + | 3.5 ± 0.6 | 71 ± 3 | 2.3 ± 0.2 |
| H3 Y41A | + | 1.1 ± 0.3 | 72 ± 7 | 0.3 ± 0.2 |
| H3 R42A | + | 2.7 ± 0.2 | 54 ± 3 | 0.9 ± 0.1 |
| H3 P43A | + | 1.4 ± 0.01 | 91 ± 3 | 1.0 ± 0.2 |
| H3 G44A | + | 3.6 ± 0.2 | 68 ± 9 | 2.2 ± 0.6 |
| H3 T45A | + | 1.7 ± 0.1 | 52 ± 5 | 0.8 ± 0.1 |
| H3 V46A | + | 2.0 ± 0.3 | 89 ± 5 | 1.0 ± 0.1 |
| H3 L48A | − | 2.5 ± 0.4 | 86 ± 4 | 1.7 ± 0.3 |
| H3 R49A | + | 2.5 ± 0.2 | 67 ± 3 | 1.4 ± 0.2 |
| H3 E50A | + | 1.5 ± 0.5 | 98 ± 6 | 1.8 ± 0.3 |
| H3 I51A | − | 9.2 ± 1.0 | 81 ± 13 | 3.4 ± 0.7 |
| H3 R52A | + | 1.7 ± 0.1 | 78 ± 3 | 0.9 ± 0.2 |
| H3 R53A | + | 1.2 ± 0.02 | 80 ± 4 | 1.3 ± 0.2 |
| H3 Y54A | + | 1.3 ± 0.1 | 96 ± 3 | 1.7 ± 0.2 |
| H3 Q55A | − | 5.4 ± 0.4 | 69 ± 11 | 3.4 ± 0.5 |
| H3 K56A | + | 1.6 ± 0.3 | 95 ± 4 | 0.7 ± 0.3 |
| H3 S57A | − | 1.9 ± 0.2 | 102 ± 5 | 2.3 ± 0.3 |
| H4 R45H | + | 4.5 ± 0.7 | 100 ± 4 | 0.5 ± 0.2 |
WT, wild type; ND, not determined. All data are expressed as means ± standard deviations of the means.
−, not measurable; nucleosome unstable.
Values represent the initial FRET signal relative to wild-type signal as a percentage. FRET was measured as described in the legend of Fig. 3.
Values represent changes of H2A/H2B exchange for mutant octamer relative to a wild-type octamer after 60 min.
FIG. 6.
Point mutations in the H3 αN helix affect nucleosome dynamics. (A) Wild-type and H3 I51A nucleosomes were incubated for the indicated amount of time at 47°C. H3 I51A nucleosomes slide approximately ninefold faster than wild type. (B) H3 Q55A nucleosomes show decreased dimer stability. After 60 min at 47°C, H3 Q55A nucleosomes have lost almost threefold more dimers than wild type. (C) Specific point mutations within the αN helix reduce DNA end-to-end FRET (R49A), indicating that DNA wrapping is severely altered in these nucleosomes. WT, wild-type.
We also tested the effects of selected mutations on other aspects of nucleosome dynamics. In order to interpret these data, we classified these amino acids as interacting with four regions of the nucleosome—the DNA entry gyre, the DNA central gyre, the H2A C-terminal extension, and the H4 α1 helix—based on which face of the αN helix they interact with (Fig. 7). Residues whose side chains interacted with the DNA entry gyre affected DNA end-to-end FRET when mutated (Table 1; Fig. 6C), consistent with a role in constraining the “site exposure” mode of DNA unpeeling from the entry of the nucleosome (30). Amino acids contacting the central DNA gyre had a generally lesser effect, although they did lead to a limited increase in breathing of DNA ends. In contrast, with the regions making DNA contacts, residues that made contact with the H2A C-terminal extension exhibited very high dimer exchange and, indeed, did not even form octamers in the standard refolding method (Table 1; Fig. 6B). This is consistent with the idea that interactions of the H2A C-terminal extension with the H3 αN helix play an important role in stabilizing the nucleosome (19, 32, 58). Interestingly, the mutations I51A and Q55A that displayed significantly increased histone dimer exchange were also able to reposition nucleosomes up to ninefold faster than wild type (Table 1; Fig. 7).
FIG. 7.
Interactions of the H3 αN helix with DNA, H2A, and H4 help to explain the effects of mutations in this region on nucleosome dynamics. (A) Overview of where the H3 αN helix sits within the nucleosome and an enlargement illustrating some of the regions it interacts with. These include the DNA entry gyre, the central DNA gyre, the H2A C-terminal extension, or the H4 α1 helix. Residues oriented on the same surface of the helix make similar contacts so the residues within the helix can be assigned into these four interaction surfaces. In panel B the residues within each horizontal bar interact with a common adjacent region of the nucleosome. This helical net annotation provides a means of relating how the different interaction surfaces affect the different aspects of nucleosome dynamics we have studied. Residues H3 R40 to P43 do not form part of the αN helix but are shown for completeness. S57 is joined with a dotted line to K56 to indicate that it is in an unusual conformation. The four interaction surfaces are displayed linearly; however, due to its circular nature, residues pointing toward the DNA central gyre are also close to the H4 α1 helix. (C) Data from Table 1 are superimposed onto the helical net to uncover patterns in how residues affect the dynamic properties of nucleosomes. This analysis reveals that DNA end FRET is most influenced by mutation to residues pointing toward the entry DNA gyre, while dimer exchange is affected by residues interacting with the H2A C-terminal extension as well as the central DNA gyre. Nucleosome sliding is strikingly affected by residues contacting H2A and also by residues interacting with DNA.
The histone H3 N-terminal tail and αN helix contain several sites that are subject to posttranslational modification (61). In order to investigate whether these might affect the dynamic properties of nucleosomes, site-directed mutagenesis was carried out to introduce side chains that mimic these modifications. These included mutation to methionine to mimic lysine and arginine methylation, mutation to glutamine to mimic lysine acetylation, and mutation to glutamic acid to mimic serine phosphorylation. Although these mutations have different structures from the modifications they were selected to mimic, they provide an insight as to the type of effects the bona fide modifications are likely to have. The data presented in Table 2 illustrate that many of these modification mimics affected DNA end-to-end FRET (the FRET signal generated when the two arms of the DNA are brought into close proximity) and nucleosome mobility. H3 K56Q increased nucleosome thermal mobility almost twofold, and R52M reduced DNA end-to-end FRET to the levels of mutations such as R40A and R49A, which are key residues identified in this screen. This suggests that posttranslational modifications at these sites can directly influence the dynamic properties of nucleosomes.
TABLE 2.
Effects of mutations mimicking posttranslational modifications
| Position | Modification | Mutation | Initial nucleosome sliding rate relative to wild type | Initial DNA end-to-end FRET (%)a |
|---|---|---|---|---|
| K36 | Methylation | K36M | 1.4 ± 0.2 | 89 ± 6 |
| K36 | Acetylation | K36Q | 1.3 ± 0.1 | ND |
| K37 | Methylation | K37M | 1.6 ± 0.1 | 81 ± 2 |
| K37 | Acetylation | K37Q | 1.4 ± 0.01 | ND |
| P38 | Proline isomerization | P38A | 1.1 ± 0.1 | 95 ± 1 |
| R52 | Methylation | R52M | 1.7 ± 0.06 | 73 ± 3 |
| R53 | Methylation | R53M | 1.4 ± 0.1 | 94 ± 1 |
| K56 | Acetylation | K56Q | 1.8 ± 0.2 | 82 ± 2 |
| S57 | Phosphorylation | S57E | 3.0 ± 0.4 | 105 ± 11 |
DISCUSSION
Here, we report that different histone tails have distinct effects on nucleosome mobility. Particularly noteworthy is the observation that truncation of the H2A tails promotes nucleosome mobility, whereas deletion of the H4 and H2B histone tails reduces it. This indicates that the H2B and H4 tails act to assist the process of nucleosome sliding. Our observation that the H2B tail acts to increase nucleosome mobility may relate to a role for this region in transcriptional repression of a large subset of the Saccharomyces cerevisiae genome (45). Contrasting effects of H2A and H2B truncations on the structure and function of chromatin have been reported previously (46, 59). On a genome-wide scale truncations of H3, H4, and H2B have effects on different subsets of genes (45, 51), while truncation of the H2A N terminus, like many other alterations that act to increase nucleosome mobility, confers a Sin phenotype (19, 24, 41). The observation that histone tails can play opposing roles in nucleosome behavior is worth consideration in light of previous studies where all tails have been removed simultaneously by trypsin cleavage (6, 10, 40, 48). Although the mechanism by which the tails exert their effects is not clear from our study, it is worth noting that basic amino acids in the histone tails are capable of making contacts with DNA (2, 13, 23, 28).
Previously, it has been observed that that deletion of the H2B tail alters the locations to which nucleosomes are initially deposited (22). We would have expected to observe a similar change in the positions to which nucleosomes are initially deposited. However, as we have not observed this on DNA fragments derived from MMTV or sea urchin 5S DNA (data not shown), we think that this likely reflects a species-specific difference between Drosophila and Xenopus histones.
Deletion of the complete H3 tail had the most striking effect on nucleosome structure, causing a qualitative defect in DNA wrapping in contrast to the quantitative effects on nucleosome sliding that resulted from the other truncations. With respect to this, it is interesting that in S. cerevisiae, deletion of the tails of histone H3 or H2A leads to derepression of subtelomeric genes to the levels seen due to nucleosome depletion (34, 51, 59). These observations are consistent with the interpretation that the H3 tail together with the structured histone octamer core plays a role in rendering nucleosomal DNA less accessible. It is also notable that H3 deletion has been observed to cause changes in the number of supercoils constrained by chromatin both in vivo and in vitro (29, 52) and that the changes to DNA wrapping that we have detected would be expected to affect the number of supercoils constrained by nucleosomes.
One explanation for the effect of H3 tail deletion is the perturbation of the adjacent H3 αN helix, which makes important contacts with DNA at the edge of the nucleosome. To study this, we performed scanning mutagenesis along this region and monitored three different aspects of nucleosome dynamics: thermal mobility, DNA end-to-end FRET, and histone dimer stability. Alterations to residues involved in making contacts with the outer DNA gyre were observed to have larger effects on DNA end FRET than contacts with the inner gyre. This is consistent with a key role for DNA contacts with the outer gyre in regulating the rate of DNA breathing. Reductions in DNA end-to-end FRET did not in general correlate with increased histone dimer exchange. This suggests that increased DNA breathing is not the primary limiting factor for histone dimer exchange. Instead, protein-protein interactions, especially those with the H2A C-terminal extension and H4 α1 helix, have the largest effects on nucleosome stability. Many of these mutations affecting dimer exchange also had significant effects on nucleosome mobility. For example, H3 I51A is the point mutation with the strongest effect on in vitro nucleosome mobility identified to date, shifting approximately twofold faster than the Sin mutation H4 R45H. These observations are consistent with previous work implicating interactions between H3 and the H2A C-terminal extension in nucleosome mobility (19).
Although the mechanisms that result in these alterations to nucleosome structure remain unclear, it has been tempting to speculate that they are mechanistically linked. For example, the transient release of the outer turns of DNA might provide an opportunity for the capture of DNA loops on the surface of the histone octamer. Nucleosomes bearing this additional DNA are intermediates in one of the pathways that have been proposed to explain nucleosome redistribution (17, 18, 39). The removal of DNA from the edges of nucleosomes has also been mentioned as a possible means by which histone dimers might dissociate from histone octamers. However, there are also clear examples where one aspect of nucleosome dynamics is altered without affecting other properties. For example, we have observed that it is possible to increase nucleosome mobility without increasing histone dimer exchange or DNA unwrapping. Similarly, changes in DNA wrapping do not always result in increased histone dimer exchange or nucleosome mobility. This suggests that the mechanisms underlying these processes are not as closely related as might have been thought previously. It should be noted that we have studied only a selected subset of characterized dynamic fluctuations in nucleosome structure here. An assortment of additional assays has been used to monitor nucleosome dynamics, and it is as yet unclear whether these will be related to any of the properties we have measured here. For example, the processes we have studied might be anticipated to impede chromatin fiber formation.
The sensitivity of nucleosomes to alterations within the H3 αN region is consistent with strong phenotypes associated with this region of the nucleosome. For example, a recent study found that three of the four mutations we found to prevent octamer formation, H3-L48A, H3-I51A, and H3-Q55A, are lethal in yeast (36). In addition, mutation of the amino acids in the region that includes the H3 αN helix confers detectable phenotypes at a high frequency (25, 36). This suggests that the function of the nucleosome is especially sensitive to alterations in this region. Nevertheless, we have not been able to identify a direct correlation between any of the biochemical properties we have studied and a specific phenotype in yeast. For example, there is not a good correlation between the severity of the Spt phenotype and dimer stability. However, such phenotypes are likely to be influenced by both the inherent dynamic properties of nucleosomes and the presence of interaction surfaces important for recruitment of nucleosome binding factors. This complicates the identification of such correlations.
Widespread acetylation of H3 at K56 has been found to occur during S phase in budding yeast (35, 43, 49, 60). Given the proximity of this region to DNA, several researchers have proposed that this modification may act to directly affect histone-DNA contacts (35, 43, 60). Indeed, the K56Q mutation designed to mimic H3 K56 acetylation has been observed to reduce the resistance of native yeast chromatin to micrococcal nuclease digestion and affect the linking number of endogenous plasmid DNA (35). Our finding that this mutation alters nucleosome mobility and the wrapping of DNA around nucleosomes adds further support to the hypothesis that this modification can directly alter chromatin structure. Other modifications within this region of the nucleosome may act similarly as we have found that several mutations designed to mimic modifications had similar effects. Although we have focused on this region of the nucleosome, there are many other locations distributed through the globular core of the histone octamer that have been characterized as being subject to posttranslational modification (21), and further modifications may remain to be identified. Many of these have the potential to affect various aspects of nucleosome dynamics.
It has previously been proposed that histone modifications, especially those within the globular domain of the histone octamer, may act to alter nucleosome mobility (11). Here we provide experimental support for this hypothesis with the finding that mutations designed to mimic histone modifications within the histone octamer have effects on the rates of nucleosome repositioning. We also reveal additional complexity to the number of potential ways in which the dynamic properties of nucleosomes can be manipulated. The observation that different dynamic properties can be altered separately raises the possibility that nucleosomes can be fine-tuned for the context in which they function. In concert with the ability of many modifications to serve as marks for recruitment of chromatin binding proteins, this extends the range of different means by which histone modifications can act to influence gene regulation.
Acknowledgments
We thank Jon Widom for the 601.3 sequence and Chris Stockdale and members of the division of Gene Regulation for helpful discussions.
H.F. is supported by the Wellcome Trust. J.S. is supported by the Wellcome Trust, Todd Foundation Awards for Excellence, William Georgetti Scholarship, and ORSAS and is a Fellow of the New Zealand Federation of Graduate Women.
Footnotes
Published ahead of print on 26 March 2007.
REFERENCES
- 1.Anderson, J. D., P. T. Lowary, and J. Widom. 2001. Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 307:977-985. [DOI] [PubMed] [Google Scholar]
- 2.Angelov, D., J. M. Vitolo, V. Mutskov, S. Dimitrov, and J. J. Hayes. 2001. Preferential interaction of the core histone tail domains with linker DNA. Proc. Natl. Acad. Sci. USA 98:6599-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausio, J., F. Dong, and K. E. van Holde. 1989. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone tails in the stabilization of the nucleosome. J. Mol. Biol. 206:451-463. [DOI] [PubMed] [Google Scholar]
- 4.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 5.Bednar, J., R. A. Horowitz, S. A. Grigoryev, L. M. Carruthers, J. C. Hansen, A. J. Koster, and C. L. Woodcock. 1998. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA 95:14173-14178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertin, A., A. Leforestier, D. Durand, and F. Livolant. 2004. Role of histone tails in the conformation and interactions of nucleosome core particles. Biochemistry 43:4773-4780. [DOI] [PubMed] [Google Scholar]
- 7.Bohm, L., and C. Crane-Robinson. 1984. Proteases as structural probes for chromatin: the domain structure of histones. Biosci. Rep. 4:365-386. [DOI] [PubMed] [Google Scholar]
- 8.Bruno, M., A. Flaus, C. Stockdale, C. Rencurel, H. Ferreira, and T. Owen-Hughes. 2003. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol. Cell 12:1599-1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carruthers, L. M., and J. C. Hansen. 2000. The Core Histone N-termini function independently of linker histones during chromatin condensation. J. Biol. Chem. 275:37285-37290. [DOI] [PubMed] [Google Scholar]
- 10.Chirinos, M., F. Hernandez, and E. Palacian. 1998. Repressive effect on oligonucleosome transcription of the core histone tail domains. Biochemistry 37:7251-7259. [DOI] [PubMed] [Google Scholar]
- 11.Cosgrove, M. S., J. D. Boeke, and C. Wolberger. 2004. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 11:1037-1043. [DOI] [PubMed] [Google Scholar]
- 12.Davey, C. A., D. F. Sargent, K. Luger, A. W. Maeder, and T. J. Richmond. 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 319:1097-1113. [DOI] [PubMed] [Google Scholar]
- 13.Ebralidse, K. K., S. A. Grachev, and A. D. Mirzabekov. 1988. A highly basic histone H4 domain bound to the sharply bent region of nucleosomal DNA. Nature 331:365-367. [DOI] [PubMed] [Google Scholar]
- 14.Fischle, W., Y. Wang, S. A. Jacobs, Y. Kim, C. D. Allis, and S. Khorasanizadeh. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17:1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flaus, A., K. Luger, S. Tan, and T. J. Richmond. 1996. Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc. Natl. Acad. Sci. USA 93:1370-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Flaus, A., and T. Owen-Hughes. 2003. Dynamic properties of nucleosomes during thermal and ATP-driven mobilization. Mol. Cell. Biol. 23:7767-7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flaus, A., and T. Owen-Hughes. 2001. Mechanisms for ATP-dependent chromatin remodelling. Curr. Opin. Genet. Dev. 11:148-154. [DOI] [PubMed] [Google Scholar]
- 18.Flaus, A., and T. Owen-Hughes. 2004. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 14:165-173. [DOI] [PubMed] [Google Scholar]
- 19.Flaus, A., C. Rencurel, H. Ferreira, N. Wiechens, and T. Owen-Hughes. 2004. Sin mutations alter inherent nucleosome mobility. EMBO J. 23:343-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flaus, A., and T. J. Richmond. 1999. Base-pair resolution mapping of nucleosome positions using site-directed hydroxy radicals. Methods Enzymol. 304:251-263. [DOI] [PubMed] [Google Scholar]
- 21.Freitas, M. A., A. R. Sklenar, and M. R. Parthun. 2004. Application of mass spectrometry to the identification and quantification of histone post-translational modifications. J. Cell Biochem. 92:691-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamiche, A., J. G. Kang, C. Dennis, H. Xiao, and C. Wu. 2001. Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc. Natl. Acad. Sci. USA 98:14316-14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill, C. S., and J. O. Thomas. 1990. Core histone-DNA interactions in sea urchin sperm chromatin. The N-terminal tail of H2B interacts with linker DNA. Eur. J. Biochem. 187:145-153. [DOI] [PubMed] [Google Scholar]
- 24.Hirschhorn, J. N., A. L. Bortvin, S. L. Ricupero-Hovasse, and F. Winston. 1995. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol. 15:1999-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyland, E. M., M. S. Cosgrove, H. Molina, D. Wang, A. Pandey, R. J. Cottee, and J. D. Boeke. 2005. Insights into the role of histone H3 and histone H4 core modifiable residues in Saccharomyces cerevisiae. Mol. Cell. Biol. 25:10060-10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 27.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 28.Lee, K. M., and J. J. Hayes. 1997. The N-terminal tail of histone H2A binds to two distinct sites within the nucleosome core. Proc. Natl. Acad. Sci. USA 94:8959-8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenfant, F., R. K. Mann, B. Thomsen, X. Ling, and M. Grunstein. 1996. All four core histone N termini contain sequences required for the repression of basal transcription in yeast. EMBO J. 15:3974-3985. [PMC free article] [PubMed] [Google Scholar]
- 30.Li, G., M. Levitus, C. Bustamante, and J. Widom. 2005. Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 12:46-53. [DOI] [PubMed] [Google Scholar]
- 31.Li, G., and J. Widom. 2004. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 11:763-769. [DOI] [PubMed] [Google Scholar]
- 32.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 33.Luger, K., T. J. Rechsteiner, and T. J. Richmond. 1999. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 304:3-19. [DOI] [PubMed] [Google Scholar]
- 34.Martin, A. M., D. J. Pouchnik, J. L. Walker, and J. J. Wyrick. 2004. Redundant roles for histone H3 N-terminal lysine residues in subtelomeric gene repression in Saccharomyces cerevisiae. Genetics 167:1123-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masumoto, H., D. Hawke, R. Kobayashi, and A. Verreault. 2005. A role for cell-cycle-regulated histone H3 lysine 56 acetylation in the DNA damage response. Nature 436:294-298. [DOI] [PubMed] [Google Scholar]
- 36.Matsubara, K., N. Sano, T. Umehara, and M. Horikoshi. 2007. Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells 12:13-33. [DOI] [PubMed] [Google Scholar]
- 37.Meersseman, G., S. Pennings, and E. M. Bradbury. 1992. Mobile nucleosomes—a general behavior. EMBO J. 11:2951-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mellor, J. 2006. Dynamic nucleosomes and gene transcription. Trends Genet. 22:320-329. [DOI] [PubMed] [Google Scholar]
- 39.Mihardja, S., A. J. Spakowitz, Y. Zhang, and C. Bustamante. 2006. Effect of force on mononucleosomal dynamics. Proc. Natl. Acad. Sci. USA 103:15871-15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morales, V., and H. Richard-Foy. 2000. Role of histone N-terminal tails and their acetylation in nucleosome dynamics. Mol. Cell. Biol. 20:7230-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muthurajan, U. M., Y. Bao, L. J. Forsberg, R. S. Edayathumangalam, P. N. Dyer, C. L. White, and K. Luger. 2004. Crystal structures of histone Sin mutant nucleosomes reveal altered protein-DNA interactions. EMBO J. 23:260-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Owen, D. J., P. Ornaghi, J. C. Yang, N. Lowe, P. R. Evans, P. Ballario, D. Neuhaus, P. Filetici, and A. A. Travers. 2000. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J. 19:6141-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozdemir, A., S. Spicuglia, E. Lasonder, M. Vermeulen, C. Campsteijn, H. G. Stunnenberg, and C. Logie. 2005. Characterization of lysine 56 of histone H3 as an acetylation site in Saccharomyces cerevisiae. J. Biol. Chem. 280:25949-25952. [DOI] [PubMed] [Google Scholar]
- 44.Park, Y. J., P. N. Dyer, D. J. Tremethick, and K. Luger. 2004. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J. Biol. Chem. 279:24274-24282. [DOI] [PubMed] [Google Scholar]
- 45.Parra, M. A., D. Kerr, D. Fahy, D. J. Pouchnik, and J. J. Wyrick. 2006. Deciphering the roles of the histone H2B N-terminal domain in genome-wide transcription. Mol. Cell. Biol. 26:3842-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Placek, B. J., and L. M. Gloss. 2002. The N-terminal tails of the H2A-H2B histones affect dimer structure and stability. Biochemistry 41:14960-14968. [DOI] [PubMed] [Google Scholar]
- 47.Polach, K. J., and J. Widom. 1995. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 254:130-149. [DOI] [PubMed] [Google Scholar]
- 48.Protacio, R. U., G. Li, P. T. Lowary, and J. Widom. 2000. Effects of histone tail domains on the rate of transcriptional elongation through a nucleosome. Mol. Cell. Biol. 20:8866-8878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Recht, J., T. Tsubota, J. C. Tanny, R. L. Diaz, J. M. Berger, X. Zhang, B. A. Garcia, J. Shabanowitz, A. L. Burlingame, D. F. Hunt, P. D. Kaufman, and C. D. Allis. 2006. Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc. Natl. Acad. Sci. USA 103:6988-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ren, Q., and M. A. Gorovsky. 2001. Histone H2A.Z acetylation modulates an essential charge patch. Mol. Cell 7:1329-1335. [DOI] [PubMed] [Google Scholar]
- 51.Sabet, N., F. Tong, J. P. Madigan, S. Volo, M. M. Smith, and R. H. Morse. 2003. Global and specific transcriptional repression by the histone H3 amino terminus in yeast. Proc. Natl. Acad. Sci. USA 100:4084-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sivolob, A., F. De Lucia, M. Alilat, and A. Prunell. 2000. Nucleosome dynamics. VI. Histone tail regulation of tetrasome chiral transition. A relaxation study of tetrasomes on DNA minicircles. J. Mol. Biol. 295:55-69. [DOI] [PubMed] [Google Scholar]
- 53.Stockdale, C., A. Flaus, H. Ferreira, and T. Owen-Hughes. 2006. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J. Biol. Chem. 281:16279-16288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thastrom, A., P. T. Lowary, H. R. Widlund, H. Cao, M. Kubista, and J. Widom. 1999. Sequence motifs and free energies of selected natural and non-natural nucleosome positioning DNA sequences. J. Mol. Biol. 288:213-229. [DOI] [PubMed] [Google Scholar]
- 55.Tse, C., and J. C. Hansen. 1997. Hybrid trypsinized nucleosomal arrays: identification of multiple functional roles of the H2A/H2B and H3/H4 N-termini in chromatin fiber compaction. Biochemistry 36:11381-11388. [DOI] [PubMed] [Google Scholar]
- 56.Tse, C., T. Sera, A. P. Wolffe, and J. C. Hansen. 1998. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18:4629-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Holde, K., and J. Zlatanova. 1995. Chromatin higher order structure: Chasing a mirage? J. Biol. Chem. 270:8373-8376. [DOI] [PubMed] [Google Scholar]
- 58.Wood, C. M., J. M. Nicholson, S. J. Lambert, L. Chantalat, C. D. Reynolds, and J. P. Baldwin. 2005. High-resolution structure of the native histone octamer. Acta Crystallogr. F 61:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wyatt, H. R., H. Liaw, G. R. Green, and A. J. Lustig. 2003. Multiple roles for Saccharomyces cerevisiae histone H2A in telomere position effect, Spt phenotypes and double-strand-break repair. Genetics 164:47-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu, F., K. Zhang, and M. Grunstein. 2005. Acetylation in histone H3 globular domain regulates gene expression in yeast. Cell 121:375-385. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, L., E. E. Eugeni, M. R. Parthun, and M. A. Freitas. 2003. Identification of novel histone post-translational modifications by peptide mass fingerprinting. Chromosoma 112:77-86. [DOI] [PubMed] [Google Scholar]