Abstract
Fra-1, a transcription factor that is phylogenetically and functionally related to the proto-oncoprotein c-Fos, controls many essential cell functions. It is expressed in many cell types, albeit with differing kinetics and abundances. In cells reentering the cell cycle, Fra-1 expression is transiently stimulated albeit later than that of c-Fos and for a longer time. Moreover, Fra-1 overexpression is found in cancer cells displaying high Erk1/2 activity and has been linked to tumorigenesis. One crucial point of regulation of Fra-1 levels is controlled protein degradation, the mechanism of which remains poorly characterized. Here, we have combined genetic, pharmacological, and signaling studies to investigate this process in nontransformed cells and to elucidate how it is altered in cancer cells. We report that the intrinsic instability of Fra-1 depends on a single destabilizer contained within the C-terminal 30 to 40 amino acids. Two serines therein, S252 and S265, are phosphorylated by kinases of the Erk1/2 pathway, which compromises protein destruction upon both normal physiological induction and tumorigenic constitutive activation of this cascade. Our data also indicate that Fra-1, like c-Fos, belongs to a small group of proteins that may, under certain circumstances, undergo ubiquitin-independent degradation by the proteasome. Our work reveals both similitudes and differences between Fra-1 and c-Fos degradation mechanisms. In particular, the presence of a single destabilizer within Fra-1, instead of two that are differentially regulated in c-Fos, explains the much faster turnover of the latter when cells traverse the G0/G1-to-S-phase transition. Finally, our study offers further insights into the signaling-regulated expression of the other Fos family proteins.
The transcriptional regulator AP-1 is actually a large family of dimeric protein complexes that bind to DNA motifs (AP-1/TRE) found in a wide array of target genes (16). It is involved in the regulation of many biological processes at the cellular level, such as proliferation, differentiation, apoptosis, and response to stresses, up to the whole organism, where it is involved in organogenesis, immune responses, and control of cognitive functions, among others (28, 29, 33, 43, 59, 67). AP-1 is also implicated in a variety of pathological situations, notably tumorigenesis, and certain components can be oncogenes and/or tumor suppressors depending on the cell context (20, 33, 44, 65, 68).
Within the variety of transcription factors that form AP-1, the best known ones are the members of the Fos family, namely, c-Fos, FosB, Fra-1, and Fra-2, and those of the Jun family, namely, c-Jun, JunB, and JunD (16, 43). They all share two adjacent, highly conserved domains: the basic DNA-binding domain and the leucine zipper (LZ), which mediates dimerization. Together, these domains are known as the bZip region. Fos proteins must heterodimerize with other AP-1 components to acquire transcriptional competence. In contrast, Jun proteins can also function as homodimers, even though heterodimerization with partners like Fos proteins is favored. Importantly, AP-1 can act as either a positive or negative regulator of transcription depending on its composition, the target gene, the cell context, and the signals received from the environment (16).
c-Fos, the prototype of the family, and Fra-1 are the most studied Fos proteins. They are expressed constitutively in a limited number of tissues (33). However, the c-fos and fra-1 genes are best characterized as immediate early genes, as they are rapidly induced, i.e., within 15 min and 2 h, respectively, in many cell types by a variety of extracellular stimuli. They also show differences in protein expression. For example, upon mitogen stimulation of quiescent cells, c-Fos accumulates transiently, disappearing within a few hours, whereas Fra-1 appears later and persists well beyond the G1 phase of the cell cycle. Importantly, variations in abundances are not limited to c-Fos and Fra-1 in cells reentering the cell cycle but also concern other AP-1 family proteins, which are responsible for continuous and dynamic changes in AP-1 dimer composition (35, 36, 38). It is noteworthy that c-Fos participates in the transcriptional activation of fra-1 (8, 68) together with other components that have yet to be fully elucidated.
The expression of c-Fos and Fra-1 is altered in many tumors (44, 68). c-Fos is oncogenic in several in vitro and in vivo settings (20, 33), while Fra-1 is not transforming on its own (20, 33). However, it is associated with tumor progression, where it can contribute to cell survival (66), proliferation (7), and invasiveness (7). To avoid the deleterious effects of improper expression, both the c-fos and fra-1 genes are regulated at multiple transcriptional and posttranscriptional levels. In particular, their transcription is activated by various mitogen-activated protein kinase (MAPK) pathways, and their protein products are targeted by the Erk1/2 (48, 68) and Erk5 (62) cascades. Physiological regulation of the c-Fos protein by the Erk1/2 pathway has been more extensively studied than that of Fra-1. Both Erk1/2 and their effector kinases Rsk1 and Rsk2 phosphorylate c-Fos (14, 15, 26, 45, 46, 50, 51, 60), which alters both its degradation rate (15, 23, 50, 51) and its transcriptional activity (45, 46, 50). Similarly, phosphorylation catalyzed by the Erk5 cascade stabilizes c-Fos and enhances its ability to activate reporter genes (62). Importantly also, the pattern of AP-1 proteins, including Fra-1, is perturbed in Ras-, Raf-, and Mek1-transformed cells, where Erk1/2 plays a key role in the expression and the phosphorylation of several of them (18, 42, 64).
As for many key cell regulators, the tight control of Fos protein degradation rates is essential to ensure the correct timing and level of expression. Thus far, only the mechanisms of c-Fos breakdown have been studied in detail. Most of the studies have been performed in two different experimental contexts where the protein is massively nuclear. One is constitutive expression during asynchronous growth, and the other is transient induction during the G0-to-G1-phase transition following mitogen stimulation of quiescent cells. In both situations, c-Fos is unstable, with a half-life in the hour range, and the bulk of the protein is degraded by the proteasome (1, 9, 23, 55) independently of prior ubiquitylation (9), although a fraction of c-Fos can undergo ubiquitylation in vivo in certain circumstances (9). This is unusual, as most substrates require polyubiquitylation to be addressed to and/or processed by the proteasome (22). In line with this observation and the fact that c-Fos is not detectably ubiquitylated in serum-stimulated cells (9), Sasaki et al. recently reported the lack of ubiquitylation of unstable, nuclear c-Fos induced by tetradecanoyl phorbol acetate (56). However, they also elegantly showed that c-Fos, when retained in the cytoplasm upon activation of the STAT3 pathway in the presence of the inactive Erk5 kinase pathway, is subjected to ubiquitylation-dependent and proteasome-dependent degradation (56). These data raise the possibility that alternative pathways may contribute to c-Fos degradation in different subcellular compartments. They may also explain why c-Fos was originally found to be more unstable in the cytoplasm than in the nucleus (53).
Another peculiarity of c-Fos is that its breakdown is controlled by differentially regulated autonomous destabilizers located at its two extremities (9, 23). A C-terminal element is functional in c-Fos during both asynchronous growth and the G0-to-G1-phase transition, whereas an N-terminal destabilizer is active only in G0/G1 cells (9, 23). Moreover, the cytoplasmic degradation of c-Fos described previously by Sasaki et al. depends on a single destabilizer colocalizing with the N-terminal one active in G0/G1 cells (56). Further work will establish whether the c-Fos N-terminal region contains a single or two distinct destabilizing elements. Importantly, the activity of the C-terminal destabilizer is reduced upon the phosphorylation of two C-terminal serines by Erk1/2 and Rsk1/2 (9, 23). As a consequence, this domain is less active in G0/G1 cells, where the Erk1/2 pathway is strongly activated, than in asynchronously growing cells, where Erk1/2 is at best weakly active (48). c-Fos turnover is, however, maintained in G0/G1 phase by the functional activation of the N-terminal destabilizer via an undefined mechanism (9, 23). Finally, the expression of Ras, Mos, and Raf oncogenes, which activate the Erk1/2 pathway (48), also inhibits c-Fos C-terminal destabilizer activity in proliferating transformed cells.
There are a number of reports suggesting that Fra-1 is also an unstable protein that is stabilized upon Erk1/2 MAPK pathway activation. Thus, in the case of high Erk1/2 pathway activity resulting from either physiological stimulation by mitogens (26, 49) or oncogenic activation of upstream signaling effectors in thyroid (13), colon (66), and breast (7) cancer cells, Fra-1 accumulates to high levels and shows a characteristic diffuse and retarded electrophoretic mobility due to phosphorylation at multiple, unmapped sites. By contrast, Fra-1 shows reduced phosphorylation and destabilization when Erk1/2 activity is reduced, as is the case in control nontransformed thyroid cells (13), in cells treated with a pharmacological inhibitor (66), or upon mitogen withdrawal (26, 49). Finally, pharmacological inhibition of the proteasome in colon tumor cells with a high content of stabilized Fra-1 led to a further overaccumulation of the latter protein. This suggested that the turnover of at least a fraction of Fra-1 is controlled by proteasomal degradation (66).
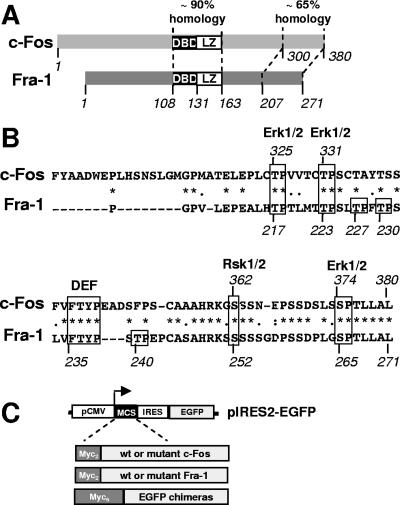
Several lines of evidence suggest that c-Fos and Fra-1 may be degraded by, at least partially, similar mechanisms. In addition to the bZip domain, the two proteins show a second region of high homology at the C terminus, where one of the two destabilizers of c-Fos is located (9, 23). Several serines and threonines phosphorylated by Erk1/2 and Rsk1/2 in c-Fos are conserved in Fra-1 (see Fig. 1A and below) (45, 46, 49, 50, 69), where only Thr231 of rat Fra-1 has been studied in depth (69). Finally, the activation of the Erk1/2 and Erk5 pathways stabilizes both c-Fos and Fra-1 (62). Nevertheless, the two proteins show differences in their turnovers. For example, in Swiss 3T3 mouse embryo fibroblasts (MEFs) undergoing a G0-to-G1-phase transition, Fra-1 has a half-life of more than 5 h, while that of c-Fos is 45 min (26). To elucidate the molecular basis of Fra-1 degradation and that of its physiological and pathological Erk1/2 pathway-dependent stabilization, we have undertaken here genetic and signaling studies of MEFs and several cancer cell lines.
FIG. 1.
Comparison of Fra-1 and c-Fos and analysis of mutants. (A) Structures of c-Fos and Fra-1. The bZIP domain is central. It is the most conserved region between c-Fos and Fra-1. The second region of high homology is the C-terminal domain of both proteins. DBD, DNA-binding domain. (B) Comparison of c-Fos and Fra-1 C-terminal domains. The C-terminal 80 amino acids of human c-Fos were compared with the homologous region in human Fra-1 using the Clustal W program. Identical amino acids are indicated with *, conserved ones are indicated with •, and semiconserved ones are indicated with •. The Erk1/2 and Rsk1/2 target sites and the DEF domain of c-Fos are boxed together with their conserved equivalents in Fra-1. The other putative Erk1/2 target sites of Fra-1 are also boxed. (C) Structure of bicistronic expression plasmids and principle of the immunoblotting assay. pIRES2-EGFP is a CMV promoter-based eukaryotic expression vector. Wild-type (wt) and mutant Fra-1 proteins and EGFP chimeras are cloned in a multicloning site (MCS) linker situated upstream of an encephalomyocarditis virus internal ribosome entry site (IRES)-EGFP expression cassette. Asynchronous cell cultures were transfected in parallel with pIRES2-EGFP-based plasmids. Total cell extracts were prepared 16 to 24 h later, which is a time that is sufficient to reach equilibrium in protein accumulation. They were subsequently analyzed by immunoblotting using specific antibodies against EGFP and the protein to be analyzed (most often the 9E10 anti-Myc tag monoclonal antibody). Note that for EGFP chimeras, a Myc6 tag entails electrophoretic retardation (approximately 10 kDa) that allows easy discrimination with EGFP used as an internal reference. Protein decay can, in the first approximation, be considered to be exponential, which implies that the relative steady-state levels of different proteins synthetized at the same rate are (nearly) proportional to their half-lives. The comparison of protein-to-be-analyzed/EGFP ratios between samples, which can be determined by densitometry scanning of luminograms or direct chemiluminescence signal quantification with a camera-based system, therefore gives the relative stabilities of compared proteins. Importantly, using EGFP as an internal standard and comparing such ratios intrinsically corrects for variations in protein synthesis rates, whether those are due to slight differences in transfection efficiencies or to variations in CMV promoter activity resulting from an alteration of intracellular signaling (see Results).
MATERIALS AND METHODS
Plasmids, cloning, and mutagenesis.
Mutagenesis and clonings were carried out using standard PCR-based techniques with a mutagenesis kit from Stratagene. All mutants were entirely sequenced. All details on expression plasmids are available upon request. Table 1 summarizes the names of the Fra-1 variants and chimeras, the mutations, and the name of the expression vectors. The Myc2K/R-Fra-1K/R cDNA was synthetized by Genecust. The parental human Fra-1 cDNA plasmid is a gift from H. Iba. The pIRES2-EGFP plasmid was obtained from Clontech. The serum-inducible PM302 vector, which contains a c-Fos insert, was described previously by Acquaviva et al. (1). The vectors for mouse Mos (pcDNA3-HA-Moswt; vector CD294) and kinase-dead MosKD (pcDNA3-HA-MosKD; vector CD295) (39) were kind gifts from S. Leibovitch. The pECE-HA/p45mapkkDD vector (CD511) (11), a kind gift from G. Pages, expresses a hemagglutinin-tagged version of the hamster MEK1 kinases in which S218 and S222 have been mutated in D. All plasmids were purified using a Nucleobond kit.
TABLE 1.
Correspondences between names of Fra-1 mutants and chimeras, mutations, and protein structures and plasmid names
| Protein | Mutation(s) | Plasmid |
|---|---|---|
| pIRES2-based expression vectors for Fra-1 mutants | ||
| Fra-1 | None | PM1155 |
| Fra-1-S252A | S252A | PM1167 |
| Fra-1-S265A | S265A | PM1157 |
| Fra-1-2S/A | S252A, S265A | PM1156 |
| Fra-1-S252D | S252D | PM1178 |
| Fra-1-S265D | S265D | PM1179 |
| Fra-1-2S/D | S252D, S265D | PM1166 |
| Fra-1-4T/A | T217A, T223A, T227A, T230A | PM1192 |
| Fra-1-4T/A-2S/D | T217A, T223A, T227A, T230A, S252D, S265D | PM1193 |
| Fra-1-4T/D | T217D, T223D, T227D, T230D | PM1194 |
| Fra-1-2T/A | T217A, T223A | PM1195 |
| Fra-1-2T/A-2S/D | T217A, T223A, S252D, S265D | PM1196 |
| Fra-1-T240A | T240A | PM1212 |
| Fra-1-T240D | T240D | PM1213 |
| Fra-1-T240A-2S/A | T240A, S252A, S265A | PM1214 |
| Fra-1-T240D-2S/A | T240D, S252A, S265A | PM1215 |
| Fra-1-T240A-2S/D | T240A, S252D, S265D | PM1216 |
| Fra-1-T240D-2S/D | T240D, S252D, S265D | PM1217 |
| Fra-1/1-261 | Δ262-271 | PM1158 |
| Fra-1/1-251 | Δ252-271 | PM1159 |
| Fra-1/1-221 | Δ222-271 | PM1168 |
| Myc6-EGFP | Myc6-EGFP | PM1172 |
| Myc2K/R-Fra-1K/R | Lysineless mutant of Myc2-tagged Fra-1 | PM1248 |
| Myc2K/R-Fra-1K/R/1-261 | Lysineless mutant of Myc2-tagged Fra-1; Δ262-271 | PM1251 |
| Myc2K/R-Fra-1K/R-2S/A | Lysineless mutant of Myc2-tagged Fra-1; S252A, S265A | PM1252 |
| Myc2K/R-Fra-1K/R-2S/D | Lysineless mutant of Myc2-tagged Fra-1; S252D, S265D | PM1253 |
| pIRES2-based expression vectors for EGFP/Fra-1 chimeras | ||
| Myc6-EGFP/231-271-2S/A | Myc6-EGFP-Fra-1 amino acids 231-271; S252A, S265A | PM1236 |
| Myc6-EGFP/221-271 | Myc6-EGFP-Fra-1 amino acids 221-271 | PM1177 |
| Myc6-EGFP/231-271 | Myc6-EGFP-Fra-1 amino acids 231-271 | PM1176 |
| Myc6-EGFP/241-271 | Myc6-EGFP-Fra-1 amino acids 241-271 | PM1175 |
| Myc6-EGFP/251-271 | Myc6-EGFP-Fra-1 amino acids 251-271 | PM1174 |
| Myc6-EGFP/261-271 | Myc6-EGFP-Fra-1 amino acids 261-271 | PM1173 |
| Myc6-EGFP/231-271-2S/D | Myc6-EGFP-Fra-1 amino acids 231-271; S252D, S265D | PM1237 |
| PM302-based expression vectors for Fra-1 mutants | ||
| Fra-1 | None | PM1189 |
| Fra-1-2S/A | S252A, S265A | PM1190 |
| Fra-1-2S/D | S252D, S265D | PM1180 |
Cell culture and transfections.
All cells were grown in Dulbecco's modified Eagle medium containing 10% fetal calf serum. HeLa, BALB/c 3T3, MCF7, LS174T, and HCT116 cells are available from the American Type Culture Collection. Transient transfections were carried out under standardized conditions using the Jet PEI transfection reagent (Ozyme). Usually, 2 μg of plasmid was used per 0.5 × 106 cells. Stable transfectants of BALB/c fibroblasts were obtained after cotransfection of PM302-based Fra-1 expression plasmids and the neomycin gene-expressing pCDNA3 vector (Clontech). Transfected cells were selected in the presence of 1 mg/ml G418 and pooled for analysis. For serum synchronization experiments, BALB/c 3T3 cells were serum starved for 36 h to arrest them in G0 phase and stimulated with fresh medium containing 20% fetal calf serum. UO126 (Cell Signaling) was usually added at a concentration of 40 μM. To eliminate the possibility of UO126 exhaustion during the experiments, efficient inhibition of the Erk1/2 pathway was always verified in immunoblotting experiments with anti-phospho-Erk1/2 antibodies. MG132 (BiolMol) and epoxomycin (Affiniti) were used at concentrations of 5 and 1 μM, respectively.
Immunoblotting analysis and antibodies.
Immunoblotting analyses were carried out as previously described after the electrotransfer of sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis-fractionated proteins onto polyvinylidene difluoride membranes and using the Western Lightening chemiluminescence kit from Perkin-Elmer Life Sciences (10). Luminograms of appropriate exposures on Kodak X-OmatAR films are presented in the figures. When necessary, bioluminescent signals were quantified using the camera-based GeneGnome system from Syngene BioImaging and the GeneTool analysis program provided by the supplier. Home-prepared 9E10 monoclonal antibody was used to detect Myc-tagged proteins. All other antibodies were rabbit antisera. The anti-enhanced green fluorescent protein (EGFP) antiserum was obtained from Roche, and that against GAPDH (glyceraldehyde-3-phoshpate dehydrogenase) was homemade. Erk1/2 was detected using the anti-p42/44 MAPK antibody and phospho-Erk1/2 with the anti-phospho-p42/44 MAPK (Thr202/Tyr204) antibody from Cell Signaling. Anti-Fra-1 antibodies (sc183) were from Santa Cruz Biotechnology. The anti-phosphoserine 252 and 265 antibodies were obtained after immunization of rabbits with the CSAHRKSpSSSSGD and SSDPLGpSPTLLAL peptides coupled to keyhole limpet hemocyanin, respectively. They were produced and affinity chromatography purified by Eurogentec (Belgium). The purification procedure involved two successive steps using phosphorylated and nonphosphorylated forms of the above-described peptides. Specific blocking by phosphorylated peptides, but not by the nonphosphorylated ones, was verified both in enzyme-linked immunosorbent assays using the phosphorylated peptides as targets and by immunoblotting using extracts from HeLa cells cotransfected with plasmids for wild-type Fra-1 and Mos. The horseradish peroxidase-conjugated anti-mouse and anti-rabbit immunoglobulin secondary antibodies were obtained from Sigma.
Pulse-chase experiments.
Twenty-four hours after transient transfection of pIRES2-EGFP-based vectors for Fra-1, Fra-1-2S/A, and Fra-1-2S/D, 107 cells were radiolabeled in the presence of 150 μCi of a [35S]methionine-cysteine mixture (NEG-772; Perkin-Elmer Life Sciences) per ml of culture medium for 1 h. Chases and cell lysis were conducted as described previously (9, 23). Immunoprecipitations were carried out with an anti-Fra-1 antibody (sc183; Santa Cruz Biotechnology) using 20 μl of magnetic protein G Dynabeads (Dynal). The immunocomplexes were directly resuspended in the electrophoresis loading buffer and fractionated through sodium dodecyl sulfate-containing 12% polyacrylamide gels. The quantification of signals on dried gels was performed using the 445SI PhosphorImager 5 device from Molecular Dynamics and the ImageQuant program.
Sequence alignments.
Sequence alignments were performed using the Clustal W (1.83) program available from EMBL-EBI (www.ebi.ac.uk/).
RESULTS
c-Fos and Fra-1 C-terminal domains.
Our study of Fra-1 (271 amino acids) was prompted by its structural similarity with c-Fos (380 amino acids) (Fig. 1A) and, in particular, the homology revealed by the alignment of their C-terminal domains (Fig. 1B). In c-Fos, serines 374 and 362 are phosphorylated by Erk1/2 and Rsk1/2, respectively. This facilitates the docking of Erk1/2 to the upstream DEF (docking site for erk, FXFP) domain FTYP and the subsequent phosphorylation of threonines 325 and 331 (49, 50). Phosphorylation of S362 and S374 leads to c-Fos stabilization (15, 23, 50, 51), and that of T325 and T331 increases transactivation by c-Fos (45, 46, 50). Serines and threonines are found at the equivalent positions in Fra-1 (S265, S252, T223, and T217), as is the DEF domain. Three additional threonine-proline motifs (T227, T230, and T240) lie in the vicinity of the Fra-1 DEF motif and therefore represent other potential sites for phosphorylation by Erks (34, 48).
Erk1/2 pathway-driven phosphorylations of serines 252 and 265, but not those of threonines 217, 223, 227, 230, and 240, stabilize Fra-1 in asynchronous cells.
We first addressed the phosphorylation and stabilization of Fra-1 by the Erk1/2 pathway in exponentially growing cells to determine if the two events were linked directly or indirectly. A series of nonphosphorylatable and phosphomimetic mutants were generated in human Fra-1 at positions corresponding to known Erk1/2 and Rsk1/2 target sites in c-Fos (Fig. 1B and 2A and Table 1). The stability of these mutants was evaluated in HeLa cells that display low, albeit somewhat variable, Erk1/2 activity. We developed an assay system that is simpler than pulse-chase analysis to measure the relative stabilities of the numerous wild-type, mutant, and chimeric proteins in our study (Fig. 1C). This assay involves parallel transfections of asynchronous cells with expression vectors derived from pIRES2-EGFP (Fig. 1C) that give rise to both the proteins under study and EGFP from the same bicistronic mRNA. The steady-state levels of the proteins were analyzed using immunoblotting. EGFP, which is highly stable in our system (23), serves as an internal standard for normalization and allows the estimation relative protein stabilities, if needed (see the legend of Fig. 1C). This approach was validated (not shown) by verifying that differences in the accumulation of c-Fos mutants relative to that of the wild-type protein correlated with differences in half-lives measured by pulse-chase in a previous work (23).
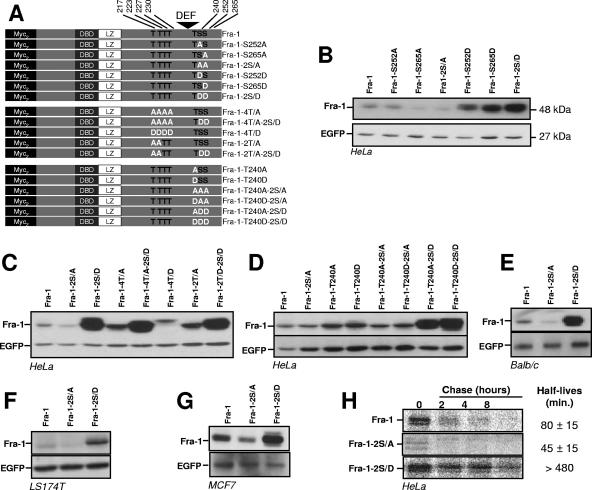
FIG. 2.
Relative stabilities of C-terminal-domain serine and threonine mutants of Fra-1. pIRES2-EGFP-based expression plasmids of the various Fra-1 mutants were transfected in asynchronously growing cells. Immunoblotting experiments were carried out 24 h later using total cell extracts. Fra-1 proteins and EGFP were visualized using the 9E10 anti-Myc tag monoclonal antibody and an anti-EGFP antiserum, respectively. (A) Structure of Fra-1 mutants. DBD, DNA-binding domain. (B) Analysis of serine 252 and serine 265 mutants in HeLa cells. (C) Analysis of threonine 217, 223, 227, and 230 mutants in HeLa cells. (D) Analysis of threonine 240 mutants in HeLa cells. (E) Analysis of Fra-1 serine mutants in BALB/c 3T3 fibroblasts. (F) Analysis of Fra-1 serine mutants in LS174T cells. (G) Analysis of Fra-1 serine mutants in MCF7 cells. (H) Pulse-chase analysis of Fra-1, Fra-1-2S/A, and Fra-1-2S/D in HeLa cells. HeLa cells were transfected to express Fra-1, Fra-1-2S/A, and Fra-1-2S/D, and pulse-chase analyses were carried out as described in Materials and Methods. All data presented are representative of at least three independent experiments.
In Fra-1, individual phosphomimetic mutants of S252 (Fra-1-S252D) and S265 (Fra-1-S265D) were both stabilized (Fig. 2B), with Fra-1-S265D appearing to be more stable than Fra-1-S252D. Notably, the double S252D-S265D phosphomimetic mutant (Fra-1-2S/D) showed a cumulative effect (Fig. 2B). In contrast, the nonphosphorylatable Fra-1-S265A mutant and the double S252A-S265A mutant (Fra-1-2S/A) were slightly less stable than Fra-1, likely reflecting a low level of phosphorylation-dependent stabilization of wild-type Fra-1 by weak basal Erk1/2 pathway activity in our HeLa cells. Strikingly, Fra-1-S252A was not destabilized, confirming the observation with the S-to-D mutants that the phosphorylation of S252 has a weaker effect than that of S265 on Fra-1 turnover (Fig. 2B).
Since phosphorylation occurs preferentially at sites that are N terminal to DEF motifs after the docking of Erk1/2 (21, 50), we analyzed mutants in threonines 217, 223, 227, and 230 alone or in combination with mutants in S252 and S265 (Fig. 2C). Mutation of T217 and T223 (Fra-1-2T/D), which are conserved between Fra-1 and c-Fos, or of all four threonines (Fra-1-4T/D) into alanines curiously led to a slight stabilization. This increase (on average less than 50%) was modest relative to that found with Fra-1-2S/D, either alone or together with the four T-to-A mutants (Fra-1-4T/A-2S/D). Similarly, no increase in stability was seen upon the mutation of the four threonines into aspartic acids (Fra-1-4T/D). Taken together, these data indicate that the phosphorylation of these threonines plays no crucial role in the stabilization of Fra-1 turnover. Instead, it may slightly attenuate the stabilizing effect of S252 and S265 phosphorylation. Given this minor effect, these threonines were not investigated further.
We also studied the potential role of T240, located downstream of the DEF motif (Fig. 2D). Neither its mutation in D nor its mutation in A affected Fra-1 accumulation, either alone (Fra-1-T240D and Fra-1-T240A, respectively) or with S252 and S265 mutated to D (Fra-1-T240D-2S/D and Fra-1-T240A-2S/D) or A (Fra-1-T240D-2S/A and Fra-1-T240A-2S/A). Importantly, all stabilized constructs, like wild-type Fra-1, localized predominantly within the nucleus in indirect immunofluorescence assays (not shown), indicating that stabilization was not a direct consequence of gross intracellular protein redistribution. Moreover, expression of wild-type Fra-1, Fra-1-2S/A, and Fra-1-2S/D in three other cell lines with low Erk1/2 pathway activity, namely, BALB/c 3T3 mouse embryo fibroblasts, LS174T human colon carcinoma cells, and MCF7 human breast cancer cells, gave results similar to those described above (Fig. 2E to G). This strongly suggests that the regulation of Fra-1 stability by modification of serines 252 and 265 is not cell type specific.
Pulse-chase experiments (Fig. 2H) were then used to estimate the half-lives of wild-type Fra-1, Fra-1-2S/A, and Fra-1-2S/D in asynchronous HeLa cells. Those of Fra-1 and Fra-1-2SA were 80 ± 15 and 45 ± 15 min, respectively. This matched the twofold difference usually seen in steady-state-level assays such as the one presented in Fig. 2B and further validated that approach. Consistently, with the 10- to 20-fold higher accumulation of Fra-1-2S/D compared with Fra-1 in parallel transfection experiments, we were unable to determine the half-life of Fra-1-2S/D, as only an approximately 30% decay could be monitored in an 8-h chase.
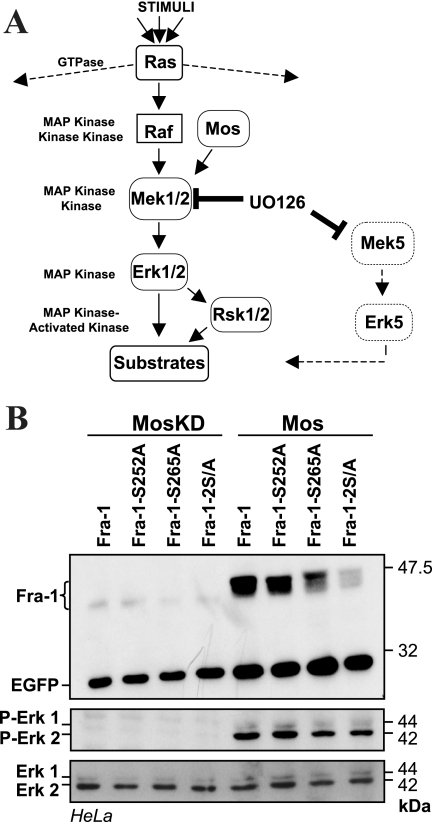
Next, we demonstrated that the activation of the Erk1/2 pathway leads to the stabilization of Fra-1 via the phosphorylation of S252 and S265. The Erk1/2 and the Erk5 cascades are blocked by U0126, a pharmacological inhibitor of the Erk1/2-activating kinases Mek1 and Mek2 as well as of the Erk5 activator Mek5 (see references 34 and 48) (Fig. 3A). In contrast, the Erk1/2 cascade is activated selectively upon ectopic expression of the Mek1/2 kinase Mos or of an activated form of the Erk1/2-activating kinase Mek1 (MEK1DD). HeLa cells were transfected with expression vectors for single or double alanine mutants of S252 and S265 either alone or together with one for Mos or a mutant Mos lacking kinase activity (MosKD). The activation of Erk1/2 by Mos, but not by MosKD, was shown by immunoblotting using antibodies specific to residues phosphorylated by Mek1/2 (34, 48) (Fig. 3B). Mos led to increased levels of EGFP, presumably by stimulating the promoter activity of the pIRES2-EGFP-derived expression vectors. More importantly, Mos induced much stronger expression of wild-type Fra-1 than Fra-1-2S/A, indicating stabilization of the former protein but not of the latter. Interestingly, the single S mutants showed intermediate degrees of stabilization (Fig. 3B), with Fra-1-S252A being higher than Fra-1-S265A. This is consistent with the increased accumulation of Fra-1-S265D relative to that of Fra-1-S252D in Fig. 2B. Notably, all proteins, including Fra-1-2S/A, showed retarded electrophoretic mobility in the presence of Mos. This is most likely due to the phosphorylation of the DEF domain-proximal threonines T217, T223, T227, and T230, as suggested previously by other researchers (see above), since Fra-1-4T/D (Fig. 2C) and another Fra-1-4T/D-2S/A (not shown) were also subjected to electrophoretic retardation. These phosphorylations were not further investigated because of the lack of an effect on Fra-1 stability in our setting. Identical results were obtained in BALB/c MEFs and upon cotransfection of wild-type and mutant Fra-1 with expression vectors for MEK1DD and Erk1 in HeLa cells (not shown).
FIG. 3.
S252 and S265 phosphorylation-dependent stabilization of Fra-1 upon Erk1/2 pathway activation. (A) Erk1/2 pathway. Erk1 and Erk2 MAPKs (MAP kinase) (Erk1/2) are activated upon phosphorylation by the Mek1 and Mek2 MAPK kinases (Mek1/2). The UO126 drug can reversibly inhibit the latter as well as Mek5. Mek1/2 is itself activated upon phosphorylation by kinases such as Raf and Mos. The activation of Raf is controlled by Ras small GTPases, which also control other pathways. Erk1/2 phosphorylates its substrates at S/T-P motifs. It activates the Rsk1 and Rsk2 MAPK-activated kinases. Raf and Mos specifically control the Erk1/2 pathway. (B) Effect of Mos on the S252 and S265 mutants of Fra-1. HeLa cells were transfected to express the indicated proteins (Fig. 2A) in the presence or in the absence of vectors for the wild type (Mos) or a kinase-dead mutant (MosKD) of Mos. Note that EGFP levels were higher in the presence of Mos due to the stimulation of the CMV promoter. Taking this point into account, chemiluminescent quantification indicated the absence of Fra-1-2S/A stabilization. The level and the activation of Erk1/2 were assayed using anti-Erk1/2 and anti-phospho-Erk1/2 antisera. The data presented are representative of three independent experiments.
To facilitate further studies, we developed specific antisera against Fra-1 peptides phosphorylated on S252 and S265. In HeLa cells cotransfected with expression vectors for Mos and either wild-type or serine-mutated Fra-1, these antisera specifically detected the corresponding phosphorylated protein in immunoblots (Fig. 4A). This indicates that these serines are actual targets of the Erk1/2 pathway and that the phosphorylation of one serine was not dependent on that of the other.
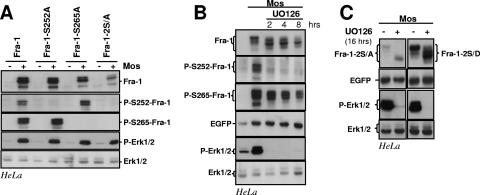
FIG. 4.
Erk1/2 pathway-induced phosphorylations of S252 and S265 and Fra-1 stabilization. (A) Erk1/2 pathway-induced phosphorylations of Fra-1 S252 and S265. HeLa cells were transfected to express the indicated proteins (Fig. 2A) in the presence or in the absence of Mos. Total cell extracts were probed with the various rabbit antisera designated for the detection of total Fra-1 (Fra-1), S252-phosphoylated Fra-1 (P-S252-Fra-1), and S265-phosphorylated Fra-1 (P-S265-Fra-1) as well as total Erk1/2 and phosphorylated Erk1/2 (P-Erk1/2). The differences in Fra-1-2S/A abundances in the presence and in the absence of Mos result largely from differences in the transcriptional activity of the expression vector as shown in Fig. 3. (B) UO126 chase of HeLa cells coexpressing Fra-1 and Mos. The UO126 chase was started 16 h after cotransfection of asynchronous HeLa cells transfected with Fra-1 and Mos expression vectors. Immunoblotting experiments were conducted with extracts from cells taken at various time points. (C) UO126 chase in HeLa cells coexpressing Mos with either Fra-1-2S/A or Fra-1-2S/D. The experiment was carried out as in B. + corresponds to 8 h in the presence of UO126. The data presented are representative of three independent experiments.
We used UO126 to block the activation of the Erk1/2 cascade by Mos in HeLa cells coexpressing it and Fra-1. Fra-1 returned to its faster-migrating form in sodium dodecyl sulfate-polyacrylamide gels within 2 h of inhibitor addition, and its levels decreased by two- to threefold at most after 8 h of inhibition (Fig. 4B). Thus, in the presence of UO126, Fra-1 had not recovered its original half-life of less than 1 h in its unphosphorylated state (Fig. 2H). Immunoblots showed that the dephosphorylation of S252 was fast in the presence of UO126, while that of S265 was slower (Fig. 4B). This explained why Fra-1 was not fully destabilized. This finding also raised the possibility that basal levels of Erk1/2 activity may be necessary for the efficient dephosphorylation of Fra-1-S265, since UO126 treatment diminished Erk1/2 activity below that found in nontreated HeLa cells (Fig. 4B). However, we cannot rule out that the phosphatase specific for P-S265 may have been rate limiting in this overexpression experiment. Notably, the slowed electrophoretic mobility of both Fra-1-2S/A and Fra1-2S/D induced by the coexpression of Mos is reversed by the addition of UO126 (Fig. 4C). This indicates that the slowed migration of Fra-1 essentially reflects its modification at multiple sites that are not S252 and S265.
Taken together, these data identify S252 and S265 as being targets of the Erk1/2 pathway and implicate their phosphorylation, and not that of other putative Erk1/2 phosphorylation sites, as the principal regulator of Fra-1 stability.
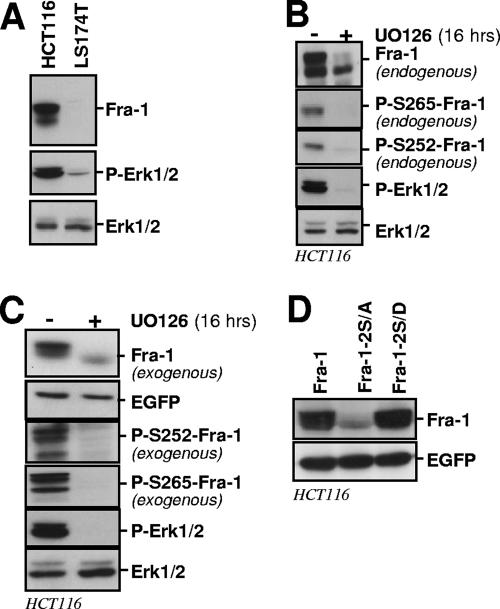
Phosphorylation of serines 252 and 265 in high-Erk1/2-activity-displaying HCT116 cancer cells is responsible for stabilization of Fra-1.
HCT116 colon cancer cells display 10-fold-higher Erk1/2 activity than LS174T, as estimated from immunoblotting quantification of phospho-Erk1/2 in the two cell types (Fig. 5A). This correlates with a high level of Fra-1 showing reduced electrophoretic mobility, reflecting hyperphosphorylation (66). We used several strategies to address whether this high Fra-1 accumulation is contributed by S252 and S265 phosphorylation-dependent protein stabilization. In a first step, we used immunoblotting to show that endogenous Fra-1 is phosphorylated on S252 and S265 in HCT116 cells (Fig. 5B). Next, the addition of UO126 to HCT116 cells led to the dephosphorylation of S252 and S265, which correlated with an 8- to 10-fold reduction in protein levels in a 16-h chase (Fig. 5B). Next, before resorting to the analysis of Fra-1 mutants, we tested whether exogenous Fra-1 behaves similarly to the endogenous protein. This was the case: when overexpressed by the transfection of a bicistronic expression vector, the level of exogenous wild-type Fra-1 decreased by approximately 10-fold upon U0126 treatment, which correlated with faster electrophoretic mobility and the absence of phosphorylation of S252 and S265 (Fig. 5C). Finally, transfected Fra-1 accumulated to a level comparable to that of Fra-1-2S/D in parallel transfection experiments, in contrast to the Fra-1-2S/A level, which was much lower (Fig. 5D). Notably, the latter data contrasted with those obtained using LS174T cells, where Erk1/2 activity was low and where Fra-1 and Fra-1-2S/A showed only a modest difference in their level of accumulation, whereas Fra-1-2S/D accumulated to a high level (Fig. 2F).
FIG. 5.
Stabilization of Fra-1 in HCT116 colon cancer cells. (A) Relative Erk1/2 activities in HCT116 and LS174T cells. Cell extract immunoblots were probed with antibodies specific for the indicated proteins. (B) Phosphorylation of endogenous Fra-1 on S252 and S265. Extracts from nontreated cells and cells treated with UO126 for 16 h were analyzed by immunoblotting for visualization of the indicated proteins. (C) Destabilization of overexpressed ectopic Fra-1 in HCT116 cells upon Erk1/2 pathway inactivation. HCT116 cells were transiently transfected in the presence or in the absence of UO126 for 16 h to inhibit the phosphorylation of Fra-1. Levels of ectopic total Fra-1 were assayed with the 9E10 antibody, and S252-phosphorylated Fra-1 and S265-phosphorylated Fra-1 were assayed with the relevant anti-phosphoserine antiserum. Transfected Fra-1 was expressed well over the level of endogenous Fra-1, which avoided interference with endogenous Fra-1 when probing with the latter two antisera. The efficiency of UO126 was demonstrated by assaying the levels of phosphorylated and nonphosphorylated Erk1/2. (D) Relative stabilities of Fra-1, Fra-1-2S/A, and Fra-1-2S/D. Transfection experiments were performed as described in the legend of Fig. 2.
Thus, while they do not exclude that other mechanisms also contribute to the high accumulation of Fra-1 in HCT116 cells, these data, when taken together, strongly suggest an important role of the Erk1/2 pathway in Fra-1 stabilization via the phosphorylation of S252 and S265.
Stability of Fra-1 during the G0/G1-to-S-phase transition is due to Erk pathway-dependent phosphorylation of serines 252 and 265.
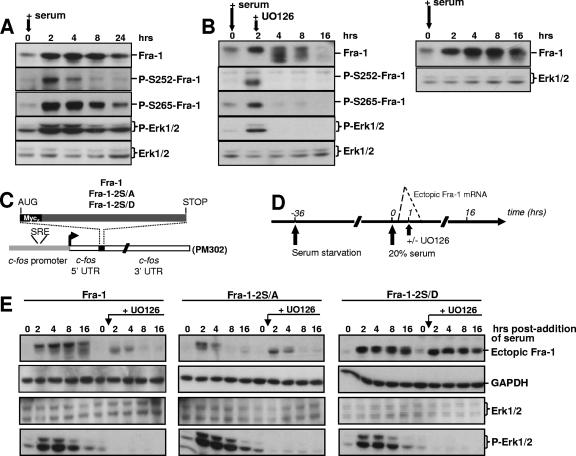
Fra-1 levels are low in most nondividing, nontransformed cells. They increase within 2 h upon mitogenic stimulation and return to an intermediate level by 16 to 24 h poststimulation, depending on the cell type. During the G0/G1-to-S-phase transition, Fra-1 is phosphorylated and relatively stable but becomes destabilized if mitogens are removed (see above). Moreover, mitogenic stimulation leads to the strong activation of the Erk1/2 cascade within minutes, which then decreases to a moderate level that persists for hours. Therefore, we investigated whether the Erk1/2 pathway-dependent phosphorylation of S252 and S265 regulates the stability of endogenous Fra-1 in the G0/G1-to-S-phase transition.
First, we assessed S252 and S265 phosphorylation during a G0/G1-to-S-phase transition in quiescent BALB/c MEFs stimulated for growth by serum. The typical kinetics of endogenous Fra-1 accumulation are presented in Fig. 6A. The phosphorylation of S252 and S265 was detectable 2 h after serum addition. Thereafter, the decrease in phospho-S265 levels paralleled those of total Fra-1 and of active Erk1/2, while phospho-S252 decayed more rapidly. The addition of UO126 2 h after stimulation by serum led to (i) the rapid dephosphorylation of Fra-1, as visualized by its faster electrophoretic migration at the 4- and 8-h points, (ii) the loss of phospho-S252 and phospho-S265 signals as early as 4 h poststimulation, and (iii) the faster decay of Fra-1 starting at 8 h (Fig. 6B, left).
FIG. 6.
Wild-type and mutant Fra-1 stability during a G0/G1-to-S-phase transition. (A) Phosphorylation of Fra-1 in serum-stimulated cells. BALB/c 3T3 fibroblasts were brought in G0 phase by serum deprivation for 36 h. They were then stimulated for growth by the readdition of culture medium containing 20% serum. Immunoblotting experiments were conducted with extracts from cells stimulated for various periods of time with the indicated antibodies. GAPDH (not shown) and Erk1/2 were used as an invariant internal standards. (B) Fate and phosphorylation of Fra-1 in serum-stimulated cells treated with UO126. In the left panel, cells were treated as described above (A), except that UO126 was added 2 h after stimulation with serum. The right panel corresponds to control cells treated in parallel with no UO126 addition and allows the visualization of the faster Fra-1 decay in the presence of UO126. (C) Structure of transient expression vectors. Fra-1, Fra-1-2S/A, and Fra-1-2S/D open reading frames were cloned in the PM302 vector after the removal of its original c-Fos insert (1). They were stably transfected in BALB/c 3T3 fibroblasts. UTR, untranslated region; SRE, serum-responsive element. (D) Design of the synchronization experiment. Deprivation of and stimulation by serum of the various cells stably transfected with the plasmids described in C were performed as described in A. When required, UO126 was added 1 h poststimulation, which gives sufficient time for Fra-1 accumulation. Ectopic mRNA levels peaked by 45 to 60 min and were back to the basal level by 90 to 120 min after serum addition (1). (E) Immunoblotting assays. Immunoblotting experiments were carried out as described above (A) on the cells stably transfected with the various PM302-based vectors. The data presented are representative of at least three independent experiments.
Although not proof per se, the above-described observations are compatible with the idea of Erk-dependent stabilization of Fra-1 through the phosphorylation of S252 and S265. To address this point formally, we next resorted to an expression system permitting the pulsed expression of mRNAs coding for wild-type Fra-1, nonphosphorylatable Fra-1-2S/A, and phosphomimetic Fra-1-2S/D at the beginning of the G0-to-G1-phase transition. The relevant open reading frames were cloned in a vector faithfully reproducing the transient accumulation of c-fos mRNA at the beginning of the G0-to-G1-phase transition (1), i.e., with mRNA expression peaking by 45 to 60 min and being back to the basal level 90 to 120 min after stimulation owing to (i) a minimal c-fos promoter carrying the serum-responsive element responsible for mitogen-induced transcriptional switching on and off and (ii) the natural 3′ c-fos untranslated region harboring the main mRNA destabilizer (Fig. 6C). Thus, ectopic proteins are no longer synthetized to significant levels after 90 to 120 min, which means that the analysis of their steady-state levels by immunoblotting at later time points reflects their stability. Stably transfected BALB/c 3T3 MEFs were arrested in G0 phase by serum deprivation and then stimulated by the readdition of serum in the presence or in the absence of UO126 (Fig. 6D). Importantly, as c-Fos promoter activation necessitates an active Erk1/2 pathway (25, 30), the drug was added 1 h after stimulation, i.e., a time sufficient to reach maximal levels of ectopic proteins. Levels of the latter, as well as Erk1/2 levels and activity, were monitored over time by immunoblotting. The data presented in Fig. 6E show that (i) in the absence of UO126, the wild-type Fra-1 level remained nearly constant, except for a slight decrease by 16 h that correlated with a reduction in Erk1/2 activity and partial dephosphorylation visualized by faster electrophoretic mobility, (ii) Fra-1 was dephosphorylated and destabilized in the presence of UO126, (iii) Fra-1-2S/A expression was transient whether UO126 was present or not, (iv) the similar kinetics of expression of Fra-1, in the presence of UO126, and Fra-1-2S/A, in the presence and in the absence of UO126, indicated that the addition of UO126 had no incidence on our transient expression system, at least under the conditions used, and (v) Fra-1-2S/D was stable whatever the experimental conditions. Altogether, the data indicate that Fra-1 is an intrinsically unstable protein. However, during the period extending from G0 to S phase, its degradation is antagonized by Erk1/2 pathway-dependent phosphorylation of S252 and/or S265. The data also indicate a half-life for nonphosphorylated Fra-1 in the 1-h range, which is consistent with the value obtained in pulse-chase experiments with cells showing low Erk1/2 activity (Fig. 2G).
Delineation of the Fra-1 destabilizer.
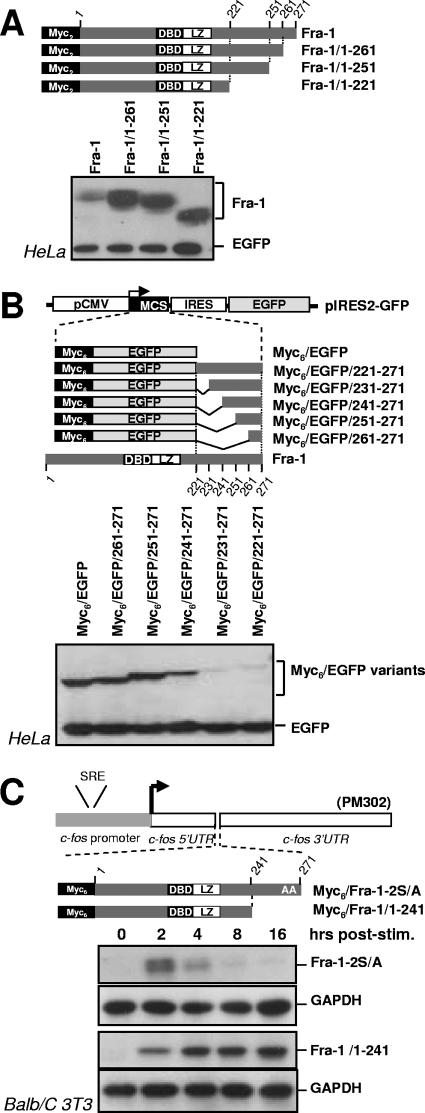
We next delineated the regions in Fra-1 responsible for its instability. The stability of Fra-1 C-terminal truncation mutants was compared to that of the wild-type protein in asynchronous HeLa cells. The removal of 10 amino acids led to approximately a 12- to 15-fold stabilization, as deduced from densitomer scanning analysis of the luminogram shown in Fig. 7A. Further deletions did not enhance this effect. We then tested whether the fusion of Fra-1 C-terminal fragments to the C terminus of EGFP would destabilize this normally very stable protein. The addition of an N-terminal Myc6 tag (78 amino acids) allowed us to differentiate between the EGFP-Fra-1 chimeras and EGFP that served as the control for expression. A 2.2-fold destabilizing effect was already detectable with the last 30 amino acids of Fra-1, as deduced from densitomer scanning of the luminogram shown Fig. 7B, and the fusion of the last 40 or 50 amino acids further destabilized EGFP with similar efficiencies (12-fold) (Fig. 7B). Taken together, the above-described data indicate that a unique structural determinant contained within the last 30 to 40 amino acids is sufficient to destabilize Fra-1 in asynchronous cells.
FIG. 7.
Delineation of the Fra-1 destabilizer. (A) Analysis of C-terminal-truncation Fra-1 mutants. Various C-terminal-truncation mutants were cloned in the pIRES2-EGFP expression vectors, and their relative accumulation in transiently transfected HeLa cells was assayed as described in the legend of Fig. 2. DBD, DNA-binding domain. (B) Analysis of EGFP-Fra-1 chimera in asynchronous HeLa cells. The various chimeras were cloned in the pIRES2-EGFP vector for transfection analysis in HeLa cells as described above (A). Immunodetections of Fra-1 proteins and EGFP were performed together with an appropriate combination of anti-Myc tag and anti-EGFP antisera. MCS, multiple cloning site; IRES, internal ribosome entry site. (C) Analysis of Fra-1 mutants during the G0/G1-to-S-phase transition. BALB/c 3T3 fibroblasts, stably transfected to express the indicated proteins from PM302-based plasmids (Fig. 6C), were serum synchronized and analyzed as described in the legend of Fig. 6. Fra-1 was immunodetected with anti-Myc monoclonal antibodies, and GAPDH was used as an internal invariant control. The data presented are representative of at least three independent experiments. UTR, untranslated region.
As c-Fos contains two destabilizers that are active in G0/G1 phase, we considered the possibility that S252 and S265 phosphorylation inhibits not only the C-terminal destabilizer in serum-stimulated cells but also another one located towards the N terminus. To test this, we compared the stability of Fra-1 mutants lacking the last 30 (Fig. 7C), 40 (not shown), and 50 (not shown) amino acids to that Fra-1-2S/A, which is unstable. All three deletion mutants were stable for at least 16 h after serum addition. As they contain neither the inhibiting phosphorylation sites nor the C-terminal destabilizer, this confirmed the lack of an N-terminal destabilizer whose action could be antagonized by the phosphorylation of serines 252 and 265 during the G0/G1-S period.
Proteasomal degradation of Fra-1.
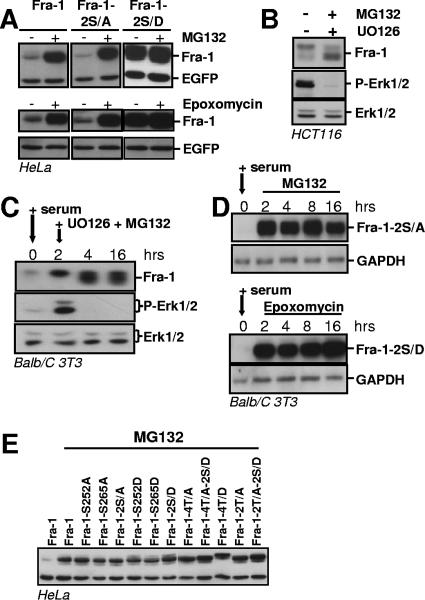
We then asked to what extent Fra-1 is degraded by the proteasome. Asynchronous HeLa cells were first transfected to express wild-type Fra-1, Fra-1-2S/A, and Fra-1-2S/D, respectively, and then treated with the proteasome inhibitors MG132 or epoxomycin for 8 h before immunoblotting analysis (Fig. 8A). Fra-1 and Fra-1-2S/A levels increased more than 20- to 30-fold, which is consistent with their half-lives in the 1- to 1.5-h range and indicates that most, if not all, Fra-1 undergoes proteasomal degradation. In contrast, there was no detectable change in Fra-1-2S/D abundance, in keeping with our observation that the half-life of phosphorylated Fra-1 exceeds 8 h (Fig. 2G). This also ruled out a significant contribution by another major proteolytic system to Fra-1 turnover under these experimental conditions. Consistently, Fra-1 decay induced by UO126 treatment in HCT116 cells (Fig. 5B) was blocked by MG132 (Fig. 8B). We also verified in two steps that the stabilization of phosphorylated Fra-1 in the G0/G1-to-S-phase transition was due to the antagonism of proteasomal degradation and not of another proteolytic pathway. First, serum-stimulated BALB/c 3T3 fibroblasts were treated with both UO126 and MG132 2 h after the addition of serum. Endogenous Fra-1 did not decay during the experiments, even though its faster electrophoretic migration indicated that it was hypophosphorylated (Fig. 8C), which contrasted with its disappearance in the absence of MG132 (compare with the 16-h time point in Fig. 6B, left). Second, quiescent BALB/c 3T3 cells stably transfected with the serum-inducible nonphosphorylatable Fra-1-2S/A mutant were serum stimulated and treated with either MG132 or epoxomycin 1 h later. Under these conditions, Fra-1-2S/A was stable for 16 h (Fig. 8D), which contrasted with its disappearance in the experiments presented in Fig. 6E. Finally, we verified that the instability of the nonphosphorylatable Fra-1 mutants was reversed in the presence of MG132. Indeed, all these mutants accumulated to a level comparable to that of the most stable phosphomimetic mutants processed similarly in parallel experiments (Fig. 8E). This confirmed that the bulk of Fra-1 undergoes proteasomal degradation under the experimental conditions used.
FIG. 8.
Proteasomal degradation of Fra-1. (A) Proteasome-dependent degradation of Fra-1 in asynchronous cells. HeLa cells were transfected with pIRES2-EGFP expression vectors to express the indicated proteins. Twenty-four hours later, they were treated with MG132 or epoxomycin for 8 h. Immunoblotting experiments were carried out as described in the legend of Fig. 2. (B) Inhibition of endogenous Fra-1 decay in HCT116 cells treated with UO126. HCT116 cells were treated with UO126, as described in the legend of Fig. 5B, in the presence of MG132 before immunoblotting analysis. (C) Inhibition of endogenous Fra-1 decay by MG132 in serum-stimulated fibroblasts treated with UO126. BALB/c 3T3 cells were serum stimulated as described in the legend of Fig. 6B. UO126 and MG132 were added 2 h poststimulation. (D) Proteasomal degradation of Fra-1-2S/A during the G0-to-G1-phase transition. BALB/c cells stably transfected to express Fra-1-2S/A from a PM302-based vector (Fig. 6C) were serum deprived for 36 h and then stimulated by the addition of 20% serum. MG132 or epoxomycin was added 1 h later. Immunoblotting assays were performed as described in the legend of Fig. 6 by using GAPDH as an internal control. The data presented are representative of at least three independent experiments. (E) Stabilization of unstable Fra-1 mutants by MG132. HeLa cells were transfected with pIRES2-derived expression vectors coding for the indicated proteins for 24 h and treated for another 8 h in the presence of MG132 before immunoblotting analysis. Normalization of luminograms with EGFP in three independent experiments indicates comparable levels of protein accumulation in the presence of MG132.
Degradation of the bulk of Fra-1 does not depend on its prior ubiquitylation.
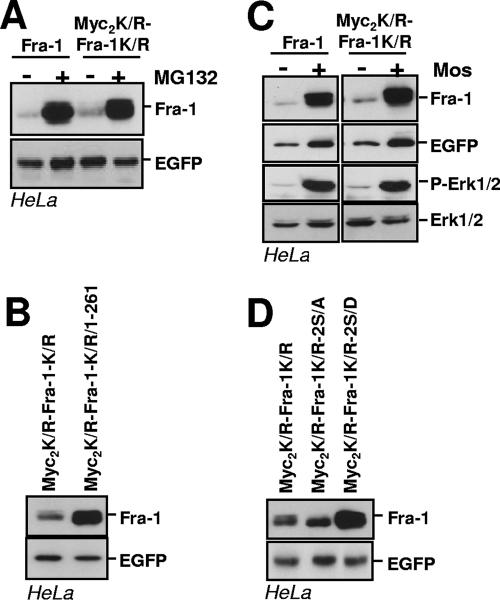
Ubiquitin is conjugated to proteins at the ɛ-amino group of lysines and, in rare cases, the N terminus (22). We previously showed that a nonubitiquitylable c-Fos mutant with all lysines turned into arginines and with the N terminus blocked by a Myc tag undergoes properly regulated degradation by the proteasome, which thereby demonstrated that prior ubiquitylation was not necessary for c-Fos destruction (9). We therefore tested whether a similar mutant of Fra-1 (Myc2K/R-Fra-1K/R) would undergo proteasomal proteolysis and show regulated degradation like the wild-type protein. In parallel transient transfection assays in HeLa cells, Fra-1 and Myc2K/R-Fra-1K/R accumulated to the same level (Fig. 9A). Moreover, the degradation of both proteins was dependent on the proteasome, as shown by the stabilization by MG132 (Fig. 9A), and on the presence of a C-terminal destabilizer, since mutants lacking the C-terminal 10 amino acids were stabilized (Fra-1/1-261 and Myc2K/R-Fra-1K/R/1-261 in Fig. 7A and 9B, respectively). Finally, the degradation of Fra-1 and Myc2K/R-Fra-1K/R was similarly antagonized by Mos-driven activation of the Erk1/2 pathway (Fig. 9C). Mutation of S252 and S265 to A led to a slight destabilization, while mutating these two S's to D's strongly stabilized both proteins (Fig. 2B and 9D). Taken together, these data indicate that the ubiquitylation of Fra-1 is not required for regulated proteasomal degradation.
FIG. 9.
Prior ubiquitylation is not necessary for proteasomal degradation of Fra-1. (A) Proteasomal degradation of Myc2K/R-Fra-1K/R. HeLa cells were transiently transfected to express Fra-1 and Myc2K/R-Fra-1K/R from pIRES2-EGFP-based vectors. MG132 was added 24 h later for a period of 8 h before immunoblotting analysis. (B) Relative stabilities of Myc2K/R-Fra-1K/R and Myc2K/R-Fra-1K/R/1-261. Experiments were carried out in HeLa cells with pIRES2-EGFP-based vectors for the indicated proteins as described above (A). (C) Stabilization of Myc2K/R-Fra-1K/R upon Mos expression. HeLa cells were transfected with pIRES2-EGFP-based vectors for the indicated proteins in the presence of the Mos expression vector. Immunoblotting experiments were conducted 16 h later. (D) Relative stabilities of S252 and S265 mutants of MycK/R-Fra-1K/R. S252 and S265 of Myc2K/R-Fra-1K/R were mutated either in A or in D, and the resulting pIRES2-EGFP-based plasmids were transfected in parallel with that for Myc2K/R-Fra-1K/R. Immunoblotting analysis of the various Fra-1 proteins was performed 16 h later.
The C-terminal 40 amino acids are sufficient for Erk1/2 pathway-dependent stabilization of Fra-1.
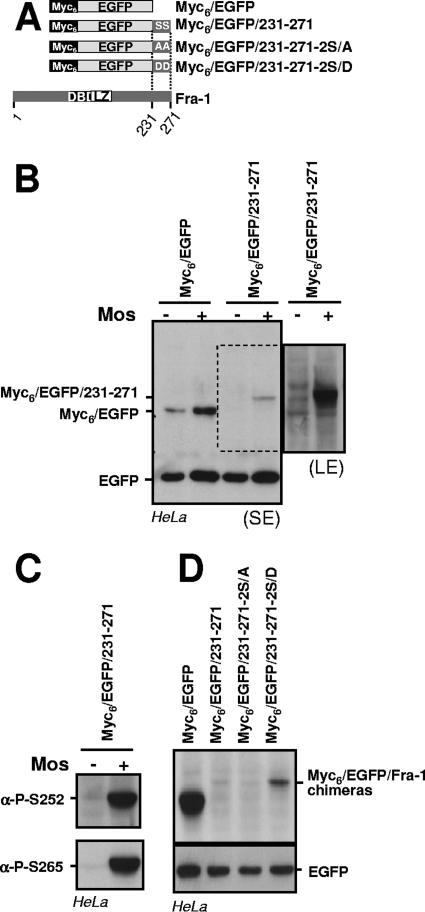
To further evaluate how S252 and S265 phosphorylation inhibits the Fra-1 destabilizer, we determined the effect of Mos expression on the stability of EGFP fused to the C-terminal 40 amino acids of Fra-1 (Fig. 10A). In keeping with the above-mentioned data, Mos entailed a higher accumulation of Myc6-EGFP due to the transcriptional activation of the cytomegalovirus (CMV) promoter of pIRES2-EGFP. However, the difference in accumulations of Myc6-EGFP in the absence and in the presence of Mos was smaller than that of Myc6-EGFP/231-271 transfected under the same conditions (long-exposure luminogram), which pointed to an increase of the stability of Myc6-EGFP/231-271 in the presence of Mos (Fig. 10B). This correlated with the phosphorylation of the fusion protein, as detected by immunoblotting with the anti-S252- and anti-S265-phosphorylated Fra-1 antisera (Fig. 10C). Although not excluding the influence of other domains of the protein for more efficient phosphorylation, this indicated that the Fra-1 destabilizer contains sufficient information for being recognized and antagonized by Erk1/2 pathway kinases. The observation that Myc6-EGFP/231-271 is less stable than Myc6-EGFP under these conditions supports the idea that phosphorylation reduces the destabilizing activity without abolishing it. This was already suggested by the fact that a small fraction of Fra-1-2S/D had disappeared at the 8-h time point in the pulse-chase experiments presented in Fig. 2G. Consistently, a mutant protein where S252 and S265 were mutated to D (Myc6-EGFP/231-271-2S/D) accumulated to a level (i) higher than that of Myc6-EGFP/231-271 or of a mutant where S252 and S265 were mutated to A (Myc6-EGFP/231-271-2S/A) but (ii) lower than that of Myc6-EGFP in parallel transfection experiments (Fig. 10D).
FIG. 10.
Phosphorylation-driven antagonization of the Fra-1 destabilizer. (A) Structure of EGFP-Fra-1 chimeras. DBD, DNA-binding domain. (B) Mos-driven inhibition of Fra-1 destabilizer activity. Asynchronous HeLa cells were transfected to express the indicated proteins from a pIRES2-EGFP-based plasmid in the presence or in the absence of the Mos expression vector. The immunoblotting analysis was carried out 24 h later with anti-EGFP antibodies. A short-exposure luminogram (SE) is presented in the left panel. Because of the signal saturation for EGFP on this luminogram, transcriptional activation of pIRES2-EGFP is barely appreciable but is similar to that shown in Fig. 3B. For better visualization of Myc6-EGFP/231-271, a longer-exposure luminogram (LE) of the relevant part of the left panel is presented in the right panel. (C) Phosphorylation of Myc6-EGFP/231-271 in the presence of Mos. The protein extracts from cells transfected with a pIRES2-EGFP-based vector for Myc6-EGFP/231-271 in the presence or in the absence of Mos were probed with anti-phospho-S252 and anti-phospho-S265 antisera. (D) Comparison of EGFP chimeras made with wild-type and mutated Fra-1 destabilizer. Asynchronous HeLa cells were transfected with pIRES2-based plasmids for the indicated proteins, and immunoblotting analysis was carried out 24 h later. All data presented are representative of at least three independent experiments.
DISCUSSION
We report here that under the conditions studied, the degradation of the bulk of the intrinsically unstable Fra-1 protein occurs in a proteasome-dependent but ubiquitin-independent manner and depends on a single destabilizer residing within the C-terminal 30 to 40 amino acids. Fra-1 destruction is antagonized upon the phosphorylation of two serines of the latter domain that are targets of Erk1/2 pathway kinases. Consistently, Fra-1 stabilization was observed under physiological and oncogenic conditions of high Erk1/2 activity.
Fra-1 stabilization upon phosphorylation by kinases of the Erk1/2 pathway.
We have combined several approaches to demonstrate that S252 and S265 are actual targets of the Erk1/2 pathway. The approaches include the analysis of site-specific mutants of Fra-1, cell signaling studies, and the use of both a pharmacological inhibitor and phosphoserine-specific antisera. The phosphorylation of one serine is not strictly dependent on that of the other, but whether one event can modulate the other cannot be excluded at this stage. The decreased electrophoretic mobility of Fra-1-2S/A upon the activation of the Erk1/2 pathway is consistent with previous suggestions by others that multiple sites are phosphorylated in Fra-1 (49, 69). This also indicates that the latter phosphorylations do not necessarily depend on prior S252 and/or S265 modifications. However, we cannot rule out that they might have an effect, as is the case with c-Fos (49, 50), under more moderate activation levels of Erk1/2. Whatever the case, these additional phosphorylations do not affect Fra-1 turnover control, a situation similar to that of c-Fos, where a mutation of T325 and T331 neither accelerates nor slows down the rate of degradation (45, 46, 50).
An important question is which kinases are responsible for S252 and S265 phosphorylation. By analogy with c-Fos and given our data, it is reasonable to assume that S265, which resides in an SP motif, is a target for Erk1 and/or Erk2. Our data do not rule out its phosphorylation on endogenous Fra-1 by Erk5 (62) in serum-induced BALB/c 3T3 cells, since this pathway is also inhibited by UO126. In contrast, S252 is not followed by a proline but instead is preceded by an arginine at position −3, a motif recognized by several kinases. The most likely candidates are members of the Rsk and Msk families, both of which are activated by Erk1/2 and phosphorylate numerous transcriptional regulatory proteins (19, 24). Accordingly, c-Fos is phosphorylated by Rsk in vitro and in vivo, a modification that regulates its activity and turnover (14, 15, 50). Phosphorylation of neither Fra-1 by Rsk's nor Fos family proteins by Msks has been described. Future studies will determine the contributions of these two kinase families in vivo.
The addition of UO126 to HeLa cells expressing phosphorylated Fra-1 due to the presence of Mos entailed a dephosphorylation of S265 that was slower than that of S252 (Fig. 4B), suggesting that the two phosphoserines are targeted by different still-to-be-identified phosphatases. Although we do not exclude the possibility that the phosphatase(s) specific for S265 may have been rate limiting under our conditions of Fra-1 overexpression, an interesting scenario is that basal Erk1/2 pathway activity may be required for the dephosphorylation of S265. The possibility that a kinase responsible for the stabilization of a protein is also necessary for the destabilization of the same protein may appear contradictory. However, it is well documented that both the nature and the intensity of the processes under the control of Erk1/2 depend on not only the timing but also the level of their activation (41, 48). Characterizing the relative contributions of phosphorylation and dephosphorylation events will help elucidate how Fra-1 turnover is regulated.
Several lines of evidence show that the principal mechanism responsible for Fra-1 stabilization in HCT116 cells is Erk1/2 pathway-dependent phosphorylation of S252 and S265 (Fig. 5). As high Fra-1 levels are observed in a variety of tumors also displaying high MAPK activity (see above), it will be interesting to determine whether the phosphorylation of S252 and S265 is also a major stabilization factor in these situations. Should this be the case, it is important to underline that other mechanisms might also contribute to Fra-1 overaccumulation, especially because the Erk1/2 pathway is already known to positively regulate fra-1 gene transcription (68), including in tumor cells (13, 66).
Proteasome-dependent, ubiquitylation-independent degradation of Fra-1.
Vial and Marshall previously reported that the proteasome is responsible for the residual degradation of Fra-1 in HCT116 cells (66). Our work, even though it does not exclude possible minor contributions by other proteolytic systems, establishes that Fra-1, when unstable, is essentially degraded by the proteasome (Fig. 8C). The vast majority of substrates subjected to proteasomal hydrolysis are thought to depend on prior ubiquitylation for being addressed and/or recognized by the proteasome (22). We show that the degradation, as well as the regulation of degradation, of the nonubiquitylatable Myc2K/R-Fra-1K/R mutant is undistinguishable from that of wild-type Fra-1. Thus, Fra-1 belongs to the small group of proteins that can undergo ubiquitylation-independent proteasomal destruction. The hamster E36-ts20 cell line is thermosensitive for ubiquitylation activity due to a mutation in the first enzyme (E1) of the ubiquitin cycle (37). It is notable that Fra-1 is not stabilized upon a shift of these cells cultured asynchronously at the nonpermissive temperature (not shown). Even though caution is required for the interpretation of data from this experiment due to the usually leaky phenotype of E1-thermosensitive cell lines (54), these data are consistent with our turnover analysis of Myc2K/R-Fra-1K/R. Another point of consistency is our previous observation that the activity of the C-terminal c-Fos destabilizer, closely related to that of Fra-1, is independent of an active ubiquitin cycle (9). It remains possible that a small fraction of Fra-1 underwent ubiquitylation-dependent degradation in our experiments, as approaches such as those that we have used for studying c-Fos and Fra-1 measure the behavior of the bulk of a protein only. It is also possible that Fra-1 may undergo ubiquitin-dependent degradation under other conditions, as it is now clear that some proteins can be addressed to the proteasome via several mechanisms. This is well illustrated for the p53 oncosuppressor (4) and c-Fos, whose cytoplasmic degradation can involve the UBR1 E3 ubiquitin ligase (56).
Although the ubiquitylation of Fra-1 itself was not necessary for proteasomal degradation in our experiments, one possibility to consider is the polyubiquitin chains present in trans on a protein partner. Such an interactor cannot be an LZ dimerization partner since (i) an LZ-deficient mutant of Fra-1 was as unstable as the wild-type protein (not shown) and (ii) EGFP is destabilized by a 40-amino-acid C-terminal fragment of Fra-1 that does not contain the LZ.
Very interestingly, Hoffman et al. showed previously that transfected Fra-1 is also ubiquitylatable in an in vivo ubiquitylation assay, like that initially described by Treier et al. (63), and that Fra-1 ubiquitylation is stimulated by the activation of the Erk1/2 pathway, i.e., when the protein is stabilized (31). This poses the question regarding the actual role of this modification, as ubiquitylation is also involved in the control of a variety of protein functions independent of proteasomal proteolysis (27, 57). The regulation of Fra-1 transcriptional activity is certainly to be considered a priority, as it is stimulatable by the Erk1/2 pathway (69). Ubiquitylation is, however, not an absolute prerequisite, as the lysineless Myc2K/R-Fra-1K/R mutant was transcriptionally active in a transient luciferase transcription assay performed using HeLa cells (not shown).
Fra-1 destabilizer and degradation of other Fos proteins.
We show here that Fra-1 breakdown depends on a unique C-terminal destabilizer. This helps us to understand why c-Fos expression is transient in mitogen-stimulated cells, whereas that of Fra-1 is much longer lasting. The molecular explanations are multiple. An obvious first explanation is the differential transcriptional regulations of the two genes. Thus, c-fos mRNA induction in G0/G1 phase is rapid and transient, whereas that of Fra-1 occurs later and is longer before returning to a basal level (2, 17, 68). A second mechanism is protein stability control. Using pulsed ectopic mRNA expression for wild-type and mutant Fra-1 proteins in serum-stimulated fibroblasts (Fig. 6), we show that the Erk1/2 pathway kinases are sufficiently active for at least 16 h to phosphorylate S252 and S265 and thereby prevent Fra-1 degradation during this period of time. By contrast, c-Fos protein instability is preserved by the activation of an N-terminal destabilizer even though the activity of its C-terminal destabilizer is also reduced (23).
The Fra-1 destabilizer is contained within the last 30 to 40 amino acids at the C terminus. This region contains striking similarities with the equivalent domains in the other Fos proteins (Fig. 11). It is therefore plausible that its counterparts in Fra-2 and FosB also show destabilizing activities, as these two proteins are also unstable. Work is currently under way to investigate this point and to determine whether (i) the destabilizers of the four Fos proteins are equally efficient or not and (ii) those of Fra-2 and FosB are also regulated by the Erk1/2 pathway-driven phosphorylation of the serine counterparts of S252 and S265 of human Fra-1. Because the N-terminal regions of c-Fos, Fra-2, and FosB share several homologous segments that are not found in Fra-1, it will also be important to address whether Fra-2 and FosB carry another N-terminally-located destabilizer as c-Fos. In turn, the dissimilarity between the N-terminal regions of c-Fos and Fra-1 raises the question of whether UBR1-dependent, ubiquitin-dependent degradation of c-Fos (56) may also occur on Fra-1. Preliminary studies using RNA interference to knock down UBR1 showed no increase in Fra-1 levels in asynchronous HeLa cells (not shown). However, to truly determine if Fra-1 behaves like c-Fos with respect to UBR1-mediated degradation, we must establish conditions where Fra-1 is quantitatively retained within the cytoplasm.
FIG. 11.
Alignments of Fos protein C termini. The domains of the four human Fos proteins corresponding to the C-terminal 40 amino acids of Fra-1 were aligned using the Clustal W multiple sequence alignment program. *:, and • correspond to identical, conserved, and semiconserved amino acids, respectively. The residues corresponding to Fra-1-S252 and Fra-1-S265 are boxed.
The C-terminal half of c-Fos shows a disorganized structure (12). Because (i) c-Fos and Fra-1 primary structures are closely related in the C-terminal region and (ii) no particular motif or structure could be identified in a bioinformatic analysis (not shown), it is very likely that the Fra-1 C-terminal domain is unstructured as well. It is therefore interesting that ubiquitylation-independent proteasomal degradation is thought to preferentially concern poorly structured polypeptides possibly exposing hydrophobic segments (32). Several possibilities could be put forward to explain why the lack of structure would favor efficient Fra-1 proteasomal degradation and how the latter could be inhibited upon the phosphorylation of S252 and S265. Adapter proteins for the recruitment and delivery of ubiquitylated proteins to the proteasome have already been described (22). A first possibility would therefore be the existence of comparable adaptors, maybe of the chaperone type, recognizing loosely structured and/or hydrophobic peptide motifs. Another possibility would consist of the direct recognition by the proteasome, as proposed for most proteins processed independently of prior ubiquitylation. In fact, several types of proteasomal complexes do coexist at any time in any eukaryotic cell (52), which poses the question of which one(s) is responsible for Fra-1 destruction. Thus, a large fraction of the proteolytic core of the system, called the 20S proteasome, is found free in vivo (61), whereas the rest associates with different regulatory complexes (52) displaying different biochemical properties. Although it has been considered to be a latent protease for a long time, there is accumulating evidence that the 20S proteasome contributes to cell protein hydrolysis (3, 5, 6, 32, 47, 52, 58). It is a four-ring cylinder-shaped multisubunit complex with its proteolytic sites hidden in a central cavity (52), and two orifices, one at each tip, are gated by flexible interdigitated extensions of the subunits forming the outer two rings. Various poorly structured protein substrates have now been shown to be capable of perturbing the organization of these extensions to gain access to the proteolytic chamber (32, 52). It is therefore possible that Fra-1 could do the same via its unstructured C terminus. The best-known proteasomal regulators are the 19S regulatory (or PA700) and 11S (or PA28α/β and PA28γ) activator complexes, which can attach alone, in duplicate, or in combination at the extremities of the 20S particle (52). The 19S complex recognizes ubiquitylated as well as nonubiquitylated substrates (52). In contrast, PA28 activators, which influence the proteolytic activities of the 20S proteasome, are not capable of recognizing ubiquitylated proteins, although they may show some ubiquitylation-independent substrate selectivity (40). These observations thus leave open the possibility that the 19S and/or the PA28 regulators contribute to Fra-1 degradation. Biochemical studies involving various proteasomal complexes are under way to identify which proteasomal complex(es) is responsible for Fra-1 hydrolysis as well as whether recognition occurs directly via its C terminus or indirectly through a peptidic adaptor.
Acknowledgments
M.P.'s laboratory is an “Equipe Labelisée” supported by the Ligue Nationale contre le Cancer. This work has also been supported by grants from the CNRS, the ARC, and the ACI program of the French Ministry for Research. J.B. was supported by a fellowship for the Ligue Nationale contre le Cancer.
We are grateful to G. Bossis for critical reading of the manuscript.
Footnotes
Published ahead of print on 19 March 2007.
REFERENCES
- 1.Acquaviva, C., F. Brockly, P. Ferrara, G. Bossis, C. Salvat, I. Jariel-Encontre, and M. Piechaczyk. 2001. Identification of a C-terminal tripeptide motif involved in the control of rapid proteasomal degradation of c-Fos proto-oncoprotein during the G(0)-to-S phase transition. Oncogene 20:7563-7572. [DOI] [PubMed] [Google Scholar]
- 2.Adiseshaiah, P., S. Peddakama, Q. Zhang, D. V. Kalvakolanu, and S. P. Reddy. 2005. Mitogen regulated induction of FRA-1 proto-oncogene is controlled by the transcription factors binding to both serum and TPA response elements. Oncogene 24:4193-4205. [DOI] [PubMed] [Google Scholar]
- 3.Asher, G., Z. Bercovich, P. Tsvetkov, Y. Shaul, and C. Kahana. 2005. 20S proteasomal degradation of ornithine decarboxylase is regulated by NQO1. Mol. Cell 17:645-655. [DOI] [PubMed] [Google Scholar]
- 4.Asher, G., and Y. Shaul. 2005. p53 proteasomal degradation: poly-ubiquitination is not the whole story. Cell Cycle 4:1015-1018. [DOI] [PubMed] [Google Scholar]
- 5.Asher, G., P. Tsvetkov, C. Kahana, and Y. Shaul. 2005. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 19:316-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baugh, J. M., and E. V. Pilipenko. 2004. 20S proteasome differentially alters translation of different mRNAs via the cleavage of eIF4F and eIF3. Mol. Cell 16:575-586. [DOI] [PubMed] [Google Scholar]
- 7.Belguise, K., N. Kersual, F. Galtier, and D. Chalbos. 2005. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene 24:1434-1444. [DOI] [PubMed] [Google Scholar]
- 8.Bergers, G., P. Graninger, S. Braselmann, C. Wrighton, and M. Busslinger. 1995. Transcriptional activation of the fra-1 gene by AP-1 is mediated by regulatory sequences in the first intron. Mol. Cell. Biol. 15:3748-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bossis, G., P. Ferrara, C. Acquaviva, I. Jariel-Encontre, and M. Piechaczyk. 2003. c-Fos proto-oncoprotein is degraded by the proteasome independently of its own ubiquitinylation in vivo. Mol. Cell. Biol. 23:7425-7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bossis, G., C. E. Malnou, R. Farras, E. Andermarcher, R. Hipskind, M. Rodriguez, D. Schmidt, S. Muller, I. Jariel-Encontre, and M. Piechaczyk. 2005. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Mol. Cell. Biol. 25:6964-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brunet, A., G. Pages, and J. Pouyssegur. 1994. Constitutively active mutants of MAP kinase kinase (MEK1) induce growth factor-relaxation and oncogenicity when expressed in fibroblasts. Oncogene 9:3379-3387. [PubMed] [Google Scholar]
- 12.Campbell, K. M., A. R. Terrell, P. J. Laybourn, and K. J. Lumb. 2000. Intrinsic structural disorder of the C-terminal activation domain from the bZIP transcription factor Fos. Biochemistry 39:2708-2713. [DOI] [PubMed] [Google Scholar]
- 13.Casalino, L., D. De Cesare, and P. Verde. 2003. Accumulation of Fra-1 in ras-transformed cells depends on both transcriptional autoregulation and MEK-dependent posttranslational stabilization. Mol. Cell. Biol. 23:4401-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, R. H., C. Abate, and J. Blenis. 1993. Phosphorylation of the c-Fos transrepression domain by mitogen-activated protein kinase and 90-kDa ribosomal S6 kinase. Proc. Natl. Acad. Sci. USA 90:10952-10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, R. H., P. C. Juo, T. Curran, and J. Blenis. 1996. Phosphorylation of c-Fos at the C-terminus enhances its transforming activity. Oncogene 12:1493-1502. [PubMed] [Google Scholar]
- 16.Chinenov, Y., and T. K. Kerppola. 2001. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 20:2438-2452. [DOI] [PubMed] [Google Scholar]
- 17.Cohen, D. R., and T. Curran. 1988. fra-1: a serum-inducible, cellular immediate-early gene that encodes a fos-related antigen. Mol. Cell. Biol. 8:2063-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook, S. J., N. Aziz, and M. McMahon. 1999. The repertoire of Fos and Jun proteins expressed during the G1 phase of the cell cycle is determined by the duration of mitogen-activated protein kinase activation. Mol. Cell. Biol. 19:330-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deak, M., A. D. Clifton, L. M. Lucocq, and D. R. Alessi. 1998. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 17:4426-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eferl, R., and E. F. Wagner. 2003. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3:859-868. [DOI] [PubMed] [Google Scholar]
- 21.Fantz, D. A., D. Jacobs, D. Glossip, and K. Kornfeld. 2001. Docking sites on substrate proteins direct extracellular signal-regulated kinase to phosphorylate specific residues. J. Biol. Chem. 276:27256-27265. [DOI] [PubMed] [Google Scholar]
- 22.Farras, R., G. Bossis, E. Andermarcher, I. Jariel-Encontre, and M. Piechaczyk. 2005. Mechanisms of delivery of ubiquitylated proteins to the proteasome: new target for anti-cancer therapy? Crit. Rev. Oncol. Hematol. 54:31-51. [DOI] [PubMed] [Google Scholar]
- 23.Ferrara, P., E. Andermarcher, G. Bossis, C. Acquaviva, F. Brockly, I. Jariel-Encontre, and M. Piechaczyk. 2003. The structural determinants responsible for c-Fos protein proteasomal degradation differ according to the conditions of expression. Oncogene 22:1461-1474. [DOI] [PubMed] [Google Scholar]
- 24.Frodin, M., and S. Gammeltoft. 1999. Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol. Cell. Endocrinol. 151:65-77. [DOI] [PubMed] [Google Scholar]
- 25.Gille, H., M. Kortenjann, O. Thomae, C. Moomaw, C. Slaughter, M. H. Cobb, and P. E. Shaw. 1995. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 14:951-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruda, M. C., K. Kovary, R. Metz, and R. Bravo. 1994. Regulation of Fra-1 and Fra-2 phosphorylation differs during the cell cycle of fibroblasts and phosphorylation in vitro by MAP kinase affects DNA binding activity. Oncogene 9:2537-2547. [PubMed] [Google Scholar]
- 27.Haglund, K., and I. Dikic. 2005. Ubiquitylation and cell signaling. EMBO J. 24:3353-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herdegen, T., and V. Waetzig. 2001. AP-1 proteins in the adult brain: facts and fiction about effectors of neuroprotection and neurodegeneration. Oncogene 20:2424-2437. [DOI] [PubMed] [Google Scholar]
- 29.Hess, J., P. Angel, and M. Schorpp-Kistner. 2004. AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 117:5965-5973. [DOI] [PubMed] [Google Scholar]
- 30.Hipskind, R. A., M. Baccarini, and A. Nordheim. 1994. Transient activation of RAF-1, MEK, and ERK2 coincides kinetically with ternary complex factor phosphorylation and immediate-early gene promoter activity in vivo. Mol. Cell. Biol. 14:6219-6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoffmann, E., A. Thiefes, D. Buhrow, O. Dittrich-Breiholz, H. Schneider, K. Resch, and M. Kracht. 2005. MEK1-dependent delayed expression of Fos-related antigen-1 counteracts c-Fos and p65 NF-kappaB-mediated interleukin-8 transcription in response to cytokines or growth factors. J. Biol. Chem. 280:9706-9718. [DOI] [PubMed] [Google Scholar]
- 32.Hoyt, M. A., and P. Coffino. 2004. Ubiquitin-free routes into the proteasome. Cell. Mol. Life Sci. 61:1596-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jochum, W., E. Passegue, and E. F. Wagner. 2001. AP-1 in mouse development and tumorigenesis. Oncogene 20:2401-2412. [DOI] [PubMed] [Google Scholar]
- 34.Kolch, W. 2005. Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6:827-837. [DOI] [PubMed] [Google Scholar]
- 35.Kovary, K., and R. Bravo. 1992. Existence of different Fos/Jun complexes during the G0-to-G1 transition and during exponential growth in mouse fibroblasts: differential role of Fos proteins. Mol. Cell. Biol. 12:5015-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovary, K., and R. Bravo. 1991. Expression of different Jun and Fos proteins during the G0-to-G1 transition in mouse fibroblasts: in vitro and in vivo associations. Mol. Cell. Biol. 11:2451-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulka, R. G., B. Raboy, R. Schuster, H. A. Parag, G. Diamond, A. Ciechanover, and M. Marcus. 1988. A Chinese hamster cell cycle mutant arrested at G2 phase has a temperature-sensitive ubiquitin-activating enzyme, E1. J. Biol. Chem. 263:15726-15731. [PubMed] [Google Scholar]
- 38.Lallemand, D., G. Spyrou, M. Yaniv, and C. M. Pfarr. 1997. Variations in Jun and Fos protein expression and AP-1 activity in cycling, resting and stimulated fibroblasts. Oncogene 14:819-830. [DOI] [PubMed] [Google Scholar]
- 39.Lenormand, J. L., B. Benayoun, M. Guillier, M. Vandromme, M. P. Leibovitch, and S. A. Leibovitch. 1997. Mos activates myogenic differentiation by promoting heterodimerization of MyoD and E12 proteins. Mol. Cell. Biol. 17:584-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, X., D. M. Lonard, S. Y. Jung, A. Malovannaya, Q. Feng, J. Qin, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2006. The SRC-3/AIB1 coactivator is degraded in a ubiquitin- and ATP-independent manner by the REGgamma proteasome. Cell 124:381-392. [DOI] [PubMed] [Google Scholar]
- 41.Mackeigan, J. P., L. O. Murphy, C. A. Dimitri, and J. Blenis. 2005. Graded mitogen-activated protein kinase activity precedes switch-like c-Fos induction in mammalian cells. Mol. Cell. Biol. 25:4676-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mechta, F., D. Lallemand, C. M. Pfarr, and M. Yaniv. 1997. Transformation by ras modifies AP1 composition and activity. Oncogene 14:837-847. [DOI] [PubMed] [Google Scholar]
- 43.Mechta-Grigoriou, F., D. Gerald, and M. Yaniv. 2001. The mammalian Jun proteins: redundancy and specificity. Oncogene 20:2378-2389. [DOI] [PubMed] [Google Scholar]
- 44.Milde-Langosch, K. 2005. The Fos family of transcription factors and their role in tumourigenesis. Eur. J. Cancer 41:2449-2461. [DOI] [PubMed] [Google Scholar]
- 45.Monje, P., J. Hernandez-Losa, R. J. Lyons, M. D. Castellone, and J. S. Gutkind. 2005. Regulation of the transcriptional activity of c-Fos by ERK. A novel role for the prolyl isomerase PIN1. J. Biol. Chem. 280:35081-35084. [DOI] [PubMed] [Google Scholar]
- 46.Monje, P., M. J. Marinissen, and J. S. Gutkind. 2003. Phosphorylation of the carboxyl-terminal transactivation domain of c-Fos by extracellular signal-regulated kinase mediates the transcriptional activation of AP-1 and cellular transformation induced by platelet-derived growth factor. Mol. Cell. Biol. 23:7030-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moorthy, A. K., O. V. Savinova, J. Q. Ho, V. Y. Wang, D. Vu, and G. Ghosh. 2006. The 20S proteasome processes NF-kappaB1 p105 into p50 in a translation-independent manner. EMBO J. 25:1945-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy, L. O., and J. Blenis. 2006. MAPK signal specificity: the right place at the right time. Trends Biochem. Sci. 31:268-275. [DOI] [PubMed] [Google Scholar]
- 49.Murphy, L. O., J. P. MacKeigan, and J. Blenis. 2004. A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Mol. Cell. Biol. 24:144-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy, L. O., S. Smith, R. H. Chen, D. C. Fingar, and J. Blenis. 2002. Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 4:556-564. [DOI] [PubMed] [Google Scholar]
- 51.Okazaki, K., and N. Sagata. 1995. The Mos/MAP kinase pathway stabilizes c-Fos by phosphorylation and augments its transforming activity in NIH 3T3 cells. EMBO J. 14:5048-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rechsteiner, M., and C. P. Hill. 2005. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 15:27-33. [DOI] [PubMed] [Google Scholar]
- 53.Roux, P., J. M. Blanchard, A. Fernandez, N. Lamb, P. Jeanteur, and M. Piechaczyk. 1990. Nuclear localization of c-Fos, but not v-Fos proteins, is controlled by extracellular signals. Cell 63:341-351. [DOI] [PubMed] [Google Scholar]
- 54.Salvat, C., C. Acquaviva, M. Scheffner, I. Robbins, M. Piechaczyk, and I. Jariel-Encontre. 2000. Molecular characterization of the thermosensitive E1 ubiquitin-activating enzyme cell mutant A31N-ts20. Requirements upon different levels of E1 for the ubiquitination/degradation of the various protein substrates in vivo. Eur. J. Biochem. 267:3712-3722. [DOI] [PubMed] [Google Scholar]
- 55.Salvat, C., I. Jariel-Encontre, C. Acquaviva, S. Omura, and M. Piechaczyk. 1998. Differential directing of c-Fos and c-Jun proteins to the proteasome in serum-stimulated mouse embryo fibroblasts. Oncogene 17:327-337. [DOI] [PubMed] [Google Scholar]
- 56.Sasaki, T., H. Kojima, R. Kishimoto, A. Ikeda, H. Kunimoto, and K. Nakajima. 2006. Spatiotemporal regulation of c-Fos by ERK5 and the E3 ubiquitin ligase UBR1, and its biological role. Mol. Cell 24:63-75. [DOI] [PubMed] [Google Scholar]
- 57.Schnell, J. D., and L. Hicke. 2003. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 278:35857-35860. [DOI] [PubMed] [Google Scholar]
- 58.Sdek, P., H. Ying, D. L. Chang, W. Qiu, H. Zheng, R. Touitou, M. J. Allday, and Z. X. Xiao. 2005. MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol. Cell 20:699-708. [DOI] [PubMed] [Google Scholar]
- 59.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4:E131-E136. [DOI] [PubMed] [Google Scholar]
- 60.Swanson, K. D., L. K. Taylor, L. Haung, A. L. Burlingame, and G. E. Landreth. 1999. Transcription factor phosphorylation by pp90(rsk2). Identification of Fos kinase and NGFI-B kinase I as pp90(rsk2). J. Biol. Chem. 274:3385-3395. [DOI] [PubMed] [Google Scholar]
- 61.Tanahashi, N., Y. Murakami, Y. Minami, N. Shimbara, K. B. Hendil, and K. Tanaka. 2000. Hybrid proteasomes. Induction by interferon-gamma and contribution to ATP-dependent proteolysis. J. Biol. Chem. 275:14336-14345. [DOI] [PubMed] [Google Scholar]
- 62.Terasawa, K., K. Okazaki, and E. Nishida. 2003. Regulation of c-Fos and Fra-1 by the MEK5-ERK5 pathway. Genes Cells 8:263-273. [DOI] [PubMed] [Google Scholar]
- 63.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 64.Treinies, I., H. F. Paterson, S. Hooper, R. Wilson, and C. J. Marshall. 1999. Activated MEK stimulates expression of AP-1 components independently of phosphatidylinositol 3-kinase (PI3-kinase) but requires a PI3-kinase signal to stimulate DNA synthesis. Mol. Cell. Biol. 19:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Dam, H., and M. Castellazzi. 2001. Distinct roles of Jun:Fos and Jun:ATF dimers in oncogenesis. Oncogene 20:2453-2464. [DOI] [PubMed] [Google Scholar]
- 66.Vial, E., and C. J. Marshall. 2003. Elevated ERK-MAP kinase activity protects the FOS family member FRA-1 against proteasomal degradation in colon carcinoma cells. J. Cell Sci. 116:4957-4963. [DOI] [PubMed] [Google Scholar]
- 67.Wagner, E. F., and R. Eferl. 2005. Fos/AP-1 proteins in bone and the immune system. Immunol. Rev. 208:126-140. [DOI] [PubMed] [Google Scholar]
- 68.Young, M. R., and N. H. Colburn. 2006. Fra-1 a target for cancer prevention or intervention. Gene 379:1-11. [DOI] [PubMed] [Google Scholar]
- 69.Young, M. R., R. Nair, N. Bucheimer, P. Tulsian, N. Brown, C. Chapp, T. C. Hsu, and N. H. Colburn. 2002. Transactivation of Fra-1 and consequent activation of AP-1 occur extracellular signal-regulated kinase dependently. Mol. Cell. Biol. 22:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]