Abstract
The microphthalmia transcription factor (Mitf) is critical for the survival and differentiation of a variety of cell types. While on the transcript level it has been noted that melanocytes and cardiomyocytes express specific Mitf isoforms, mast cells express several isoforms, mainly Mitf-H and Mitf-MC, whose function has not been thoroughly investigated. We found that in mast cells the expression of the specific Mitf isoforms is dependent on physiological stimuli that cause a major shifting of promoter usage and internal splicing. For example, activation of the c-kit signaling pathway almost totally abolished one of the main splice isoforms. Since cardiomyocytes express only the Mitf-H isoform, they were an ideal system to determine this isoform's physiological role. We identified that the expression of myosin light-chain 1a (MLC-1a) is regulated by Mitf-H. Interestingly, the transactivation of MLC-1a by Mitf-H in cardiomyocytes is decreased by overexpression of the splice form with exon 6a. In conclusion, we found that there is physiological switching of Mitf isoforms and that the promoter context and the cell context have a combined influence on gene expression programs.
The microphthalmia transcription factor (Mitf) is a basic helix-loop-helix leucine zipper (bHLH-Zip) DNA-binding protein (11). Its gene resides at the mi locus in mice (12), and mutations of this gene (21) result in deafness, bone loss, small eyes, and poorly pigmented eyes and skin. In humans, mutation in this gene causes Waardenburg syndrome type II (35). Mitf regulates transcription of the genes that encode tyrosinase and pink-eyed Pmel 17 (silver) (1), c-kit (13), p75 receptor of nerve growth factor (15), granzyme B (14), tryptophan hydroxylase (14), Bcl-2 (17), and mast cell proteases, such as mMCP6 (23).
Mitf regulates gene transcription by binding to E-box elements in the 5′-flanking regions of Mitf-responsive genes (23). Like other DNA-binding proteins, the transcriptional activation of Mitf is influenced in a complex manner by different intracellular proteins. For example, in vitro studies have indicated that Mitf can form heterodimers with the four related family members TFEB, TFEC, TFE3, and USF2 (10, 28). In our efforts to explore the role played by this transcription factor in the growth, differentiation, and maturation of mast cells, we recently identified two inhibitors of Mitf transcriptional activity, Hint (32) and protein inhibitor of activated STAT3 (PIAS3) (16).
Mitf is regulated at both the transcriptional and posttranscriptional levels. Mitf is transcribed from several alternative promoters, giving rise to various splice isoforms with potentially distinct biological functions (Fig. 1A) (reviewed by Steingrimsson et al. [33]). While exon 1m is melanocyte specific, and exon 1h is extensively used in the heart (11), other exons (such as 1a) are more widely expressed. These exons are widely spaced in the genomic DNA and are expressed from different promoters (M, A, and H). The regulation of Mitf expression has only been studied in depth in melanocytes, and very little is known about the regulation of its expression in other cells. The expression of Mitf isoforms has been investigated in several mast cell lines (29, 36). Expression of Mitf-MC and Mitf-H was detected in the C57 mast cell line and in bone marrow-derived mast cells (BMMC) (36), while spleen-derived mast cells expressed Mitf-E and Mitf-M (29), and peritoneal mast cells expressed mainly Mitf-M. However, the regulation of Mitf isoforms by physiological stimuli has never been explored. Also, the biological function of the different Mitf isoforms is as yet unclear, although it has been shown that Mitf-M has much greater activation potential of the tyrosinase promoter than Mitf-MC in NIH 3T3 cells (36).
FIG. 1.
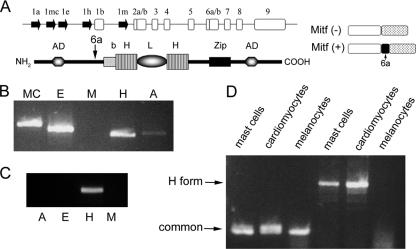
Expression of various Mitf isoforms. (A) Genomic organization of the mi locus and the structure of Mitf protein. The genomic organization of the Mitf gene is depicted (top). Filled arrows represent alternative promoters. Empty boxes represent common exons. The structure of Mitf protein is depicted below: b, basic domain; HLH, helix-loop-helix; Zip, leucine zipper; AD, activation domain. The location of the alternatively spliced exon 6a is indicated by an arrow. The structures of Mitf-(−) and Mitf-(+) are also depicted (right). (B) Expression of Mitf isoforms in bone marrow-derived mast cells. Variable 5′ primers from exons 1mc, 1e, 1m, 1h, and 1a and from the common exon 2 were amplified by PCR. (C) Expression of Mitf isoforms in normal heart of mice. Variable 5′ primers from exons 1e, 1m, 1h, and 1a and from the common exon 2 were amplified by PCR. (D) Expression of Mitf and Mitf-H in mast cells, melanocytes, and primary cardiomyocytes. Mitf was amplifed by PCR using primers from the common exon 5 to exon 7 (common; lanes 1 to 3 from the left), and Mitf-H was amplified by PCR using different primers for exon 1h to exon 1b (H form; lanes 4 to 6).
Differential splicing of the 18-bp exon 6a leads to two types of all of the Mitf isoforms (Fig. 1A). The positive (+) isoform, containing the 6-amino-acid sequence ACIFPT, and the negative (−) isoform, without these amino acids, have been found to accumulate to similar levels (8, 34). The Mitf-(−) isoform exhibited a slightly lower binding affinity to a CACGTG probe as a homodimer but normal binding affinity as a heterodimer (10). Transfection of Mitf-(+) into HEK293 cells inhibited cell proliferation compared to Mitf-(−). This inhibitory effect seemed to be dependent on the presence of an intact N terminus and to be unrelated to Mitf transcriptional activity (3).
Since it is known that only the Mitf-H isoform is expressed in the heart, this seemed an ideal system to explore the importance of cell context on the activity of the different Mitf isoforms by comparison to mast cells in which a variety of isoforms are expressed.
Our results indicate tissue-specific differences in the expression and regulation of Mitf isoforms, depending on the physiological stimuli. Furthermore, we found that the transcriptional activation potential of the different Mitf isoforms depends on the cell type and the DNA target. In addition, we have demonstrated that Mitf is expressed in cardiomyocytes and regulates the expression of myosin light-chain 1a (MLC-1a).
MATERIALS AND METHODS
Cell culture.
RBL-2H3, NIH 3T3, and H9C2 cells were cultured in growth medium as previously described (16). Primary cardiac myocytes were cultured as previously described (9). Femoral bone marrow cells derived from mice were cultured in interleukin-3 (IL-3)-containing medium for 3 weeks to generate BMMC, as previously described (31).
BMMC were either triggered with 1 μg/ml anti-2,4-dinitrophenol (DNP) immunoglobulin E (IgE) monoclonal antibody alone overnight (SPE-7; Sigma-Aldrich Corp., St. Louis, MO) or first sensitized with IgE overnight and then challenged with 300 ng/ml DNP for 2 h. IgE antibody was ultracentrifuged before use to remove aggregates.
Mice.
All mouse lines were held and propagated in a specific-pathogen-free environment. Mitfsp/sp and Mitfsp/ce mice were kindly provided by Lynne Lamoreux from the College of Veterinary Medicine, Texas A&M University, and bred to produce sp/sp and ce/ce mice for experiments. The Mitf encoded by the mutated mouse allele (ce/ce) lacks the Zip domain of Mitf because of a stop codon between the HLH and Zip domains (34). VGA-9-tg/+ mice were kindly provided by H. Arnheiter (NIH, Bethesda, MD) and were propagated by heterozygous crossing. Mice carrying the tg/tg mutation have an insertion of approximately 50 copies of a transgene integrated upstream of the Mitf-M promoter and are unable to express Mitf (11). Normal littermates of ce/ce mice were distinguished by coat color. Normal littermates of tg/tg mice were determined by genomic DNA extraction from tails and subsequent PCR analysis.
PCR amplification.
Total RNA was prepared using TRI reagent (Sigma, Saint Louis, MO). For PCR analysis, RNA was reverse transcribed using Expand reverse transcriptase (Roche Applied Science, Mannheim, Germany). Primers were used to amplify the region between alternative exon 1 and the constant exon 2 or 5 of Mitf. The resulting PCR products were electrophoresed on an agarose gel. The primers used for Mitf amplification were as follows: exon 1a, 5′-AAGTCGGGGAGGAGTTTCAT-3′; exon 1e, 5′-TCACAGAGGTTAGTAGGTGGATGGG-3′; exon 1h, 5′-GGCGCTTAGATTTGAGATGC-3′; exon 1m, 5′-GAGGACTAAGTGGTCTGCGG-3′; exon 2, 5′-TGTGGTACTTGGTGGGGTTT-3′; and exon 5, 5′-GGACAGGAGTTGCTGATGGT-3′. The presence of exon 6a was analyzed by PCR amplification and subsequent scanning and densitometry on ImageMaster VDS-CL using TINA 2.10 software. The primers used were exon 5 (5′-ACCATCAGCAACTCCTGTCC-3′) and exon 7 (5′-TAGCTCCTTAATGCGGTCGT-3′).
Real-time quantitative PCR.
Candidate Mitf-responsive genes were measured using real-time quantitative PCR. Total RNA was extracted from hearts of wild-type, ce/ce, and tg/tg mice. mRNA levels of various genes were quantified by SYBR green incorporation or by Taqman probe (SYBR green PCR master mix and Taqman master mix; Applied Biosystems, Foster City, CA) on the ABI Prism 7000 sequence detection system (Applied Biosystems).
The primers used for gene amplification for real-time PCR were as follows: β-actin sense, 5′-CCTGATCCACATCTGCTGGAA-3′; β-actin antisense, 5′-ATTGCCGACAGGATG CAGAA-3′; MLC-1a 574F, 5′-GAGGTGGAGCAGCTGTTGTCT-3′; and MLC-1a 714R, 5′-CGCTGTGGATCTCTTGC TTTC-3′. The primers used for amplification of Mitf isoforms are described in Table 1.
TABLE 1.
Primers used for amplification of Mitf isoforms
| Exon | Primer
|
|
|---|---|---|
| Forward | Reverse | |
| 1h | GATGGAGGCGCTTAGATTTGA | CATGAGTTGCTGGCGTAGCA |
| 1e | CCACAGGCTGCTCTTCTGTGT | TGCATCTGCTCACGCATGA |
| 1mc | TGGCAGCTTTGAGGATGGA | TGGAGGCCCCAGAATGC |
Plasmid construction.
pcDNA vectors containing wild-type Mitf and the ce/ce mutation were kindly provided by David Fisher, Boston, MA. These were amplified by PCR and then cut and ligated into pGEX-4T-3 vector (Amersham Biosciences, Uppsala, Sweden) using NotI and EcoRI (New England Biolabs, Beverly, MA).
The MLC-1a promoter was amplified using PCR of genomic mouse DNA, and the fragment from −638 to +58 was excised using PstI and SacI (New England Biolabs, Inc.) and ligated into a pSP72 vector (Promega Co., Madison, WI) containing a luciferase reporter gene. Mutations of MLC-1a promoter were generated using the QuickChange mutagenesis kit (Stratagene, LaJolla, CA) with the following primers: E3 mutant sense, 5′-GATAATACTGAGATGTGAGTTGCACCCGGCTGGTGTC-3′; E3 mutant antisense, 5′-GACACCAGCCGGGTGCAACTCACATCTCAGTATTATC-3′; E2 mutant sense, 5′-GTCAGCTGCACCCGGGTGTTGTCTCTTCCTTTTATAG-3′; and E2 mutant antisense, 5′-CTATAAAAGGAAGAGACAACACCCGGGTGCAGCTGAC-3′.
N termini of Mitf-H and Mitf-MC were amplified by PCR from cDNA derived from BMMC and cloned into a pcDNA-Mitf-M expression vector. The mMCP-6 promoter reporter vector was kindly provided by Y. Kitamura (Osaka, Japan), and the tyrosinase reporter was kindly provided by David Fisher.
Transient cotransfection and luciferase assay.
NIH 3T3 cells (2 × 105) were cotransfected using Transfast reagent (Promega Co.) with 0.1 μg of wild-type or E23-mutated MLC-1a-pSPLuc reporter and 0.1 μg of pcDNA-Mitf wild-type or ce/ce mutant Mitf. NIH 3T3 and H9C2 cells were similarly cotransfected with 0.1 μg of various Mitf isoform expression vectors and either mMCP-6, tyrosinase, or MLC-1a reporter. The cells were incubated in 24-well plates for 48 h. The cells were lysed and assayed for luciferase activity. The luciferase activity was normalized to the total protein concentration.
Nucleofection of RBL cells.
RBL cells (5 × 106) were used in luciferase assay experiments. Cells were transfected using Nucleofector Technology solution R and program T-20 (Amaxa Biosystems, Amaxa GmbH, Cologne, Germany). Cells were transfected with 1 μg of different luciferase reporter genes of MITF (mMCP-6 and tyrosinase) and 1 μg of the different Mitf splicing variants (M, MC, and H). The cells were incubated in plates for 24 h, lysed, and assayed for luciferase activity. The luciferase activity was normalized to the total protein concentration. The ratio was expressed as the relative luciferase activity.
EMSA.
For the electrophoretic mobility shift assay (EMSA), cold, mutated, and hot probes were synthesized by PCR amplification of the following primers: E23F, 5′-GGATTGGTGGCTCAAACCTGG-3′; E23R, 5′-AGAGACACCAGCCGGGTGC-3′; E1F, 5′-TCTCTTCCTTTTATAGTCAGCAGC-3′; E1R, 5′-TAGGAGCCCCCACAGAGG-3′; E4F, 5′-CCTGCGGTTGGGAGAGTGG-3′; and E4R, 5′-AGCCCTGAAGCAACAGCTGG-3′.
E23 was used as a probe by labeling with [32P]dCTP. Cold E23, including mutations in E23, E1, and E4 were used for competition with 5,000 cpm of radiolabeled E23 on binding to 10 μg of glutathione S-transferase (GST)-Mitf or GST-ce/ce mutant Mitf. NIH 3T3 and H9C2 nuclear extracts were prepared as previously described (38). Binding and running were performed as previously described (16).
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed as follows. H9C2 cells were treated with formaldehyde for protein-DNA cross-linking, and chromatin was extracted and then sonicated to give an average size of 300 to 3,000 bp. Chromatin was incubated with either affinity-purified anti-Mitf antibody or preimmune IgG serum from the same rabbit. Immunoprecipitation was performed overnight at 4°C with rotation. After washing and elution, DNA was purified by phenol-chloroform extraction and resuspended in 20 μl of Tris (10 mM)-EDTA (1 mM) buffer. DNA solution (2 μl) was used as a template for 27 cycles of PCR amplification with the following primers: forward, 5′-GGAGGACGAACTGGTGACAA-3′; and reverse, 5′-GCCTTCCCCTGCTTTTATTC-3′.
Prediction of Mitf binding sites.
Twenty-one previously published Mitf binding sites were used to build a nucleotide distribution matrix, which was processed by MatInd (30). The resulting output was used by MatInspector to scan sequences for Mitf binding sites, with a core similarity threshold of 0.80 and matrix similarity threshold of 0.85.
RESULTS
Characterization of Mitf splicing isoforms in mast cells and in cardiomyocytes.
Several alternate first exons of Mitf have been described in mice (Fig. 1A). In order to characterize Mitf subtypes in mast cells and in the heart, PCR was performed on cDNA from mast cells, cardiomyocytes, and melanocytes. In BMMC, expression of Mitf-H, Mitf-MC, and Mitf-E was detected, while expression of Mitf-A was barely detected and Mitf-M was not expressed at all (Fig. 1B). In contrast, spleen-derived mast cells express mainly Mitf-E and Mitf-M and peritoneal mast cells express Mitf-M (29). In the heart, however, expression of the Mitf-H isoform alone was detected (Fig. 1C). Expression of the Mitf-H isoform was also detected in mast cells but not in melanocytes (Fig. 1D).
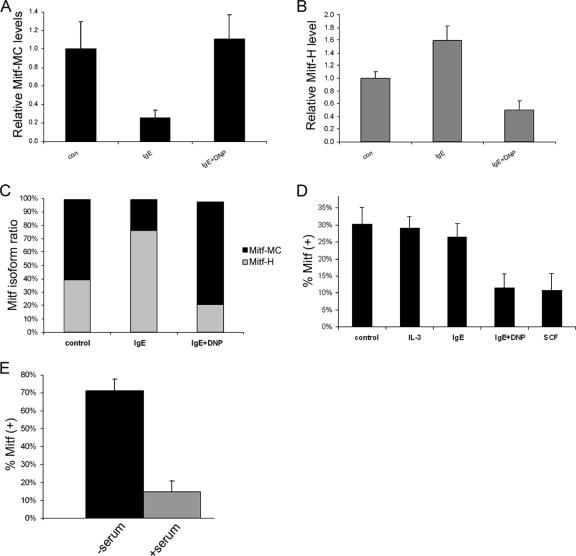
Analysis of the different Mitf isoforms expressed in BMMC was performed using real-time PCR-specific 5′ primers and 3′ primers from the common exon 1b. The level of the E isoform was very low in both resting and activated BMMC (less than 3% of the total Mitf). In resting mast cells (control), both the MC and H isoforms were expressed (Fig. 2A and B). Mitf-MC isoform expression was repressed by IgE alone, while activation with IgE and antigen (IgE plus DNP) increased its expression (Fig. 2A). In contrast, IgE alone increased the expression of the Mitf-H isoform, while activation with IgE plus DNP decreased this isoform's expression (Fig. 2B). Overall, IgE alone caused a shift to the Mitf-H isoform, while IgE plus DNP caused a shift to the Mitf-MC isoform (Fig. 2C).
FIG. 2.
Influence of physiological stimuli on Mitf isoform expression. (A and B) Real-time PCR analysis of Mitf-MC (A) and Mitf-H (B) expression in BMMC. Cells were activated with either IgE alone or IgE followed by DNP (IgE + DNP). con, control. Results represent mean ± standard error (n = 3). (C) Results of real-time PCR analysis of Mitf-H and Mitf-MC isoforms showing the relative amounts of the two isoforms expressed as a percentage of the total Mitf. (D) Analysis of the expression of exon 6a in BMMC. Cells were activated with either IL-3, IgE alone, IgE followed by DNP (IgE + DNP), or SCF. Exon 5 to exon 7 were amplified by PCR (25 cycles) and separated on 8% acrylamide gel, and the ratio between the positive and negative isoforms was determined by densitometry. Results are expressed as percentage of Mitf-(+) out of total Mitf. Results represent the mean ± standard error (n = 3). (E) Analysis of the expression of exon 6a in H9C2 cardiomyoctes. Cells were grown initially in Dulbecco's modified Eagle's medium (DMEM) supplemented with fetal calf serum (FCS). The medium was then replaced with either serum-free DMEM or fresh DMEM with FCS for 6 h. Results are expressed as percentage of Mitf-(+) out of total Mitf. Results represent the mean ± standard error (n = 5).
Resting mast cells express both the Mitf-(−) and Mitf-(+) splicing isoforms (Fig. 2D). Activation of BMMC with IgE alone had no effect on the ratio between the Mitf-(+) and Mitf-(−) isoforms. However, aggregation of the FcɛRI by either IgE followed by DNP (IgE plus DNP) for 4 h or activation of mast cells by stem cell factor (SCF), the ligand of c-kit receptor, significantly reduced the expression of the Mitf-(+) isoform.
H9C2 cardiomyoctes also express both the Mitf-(+) and Mitf-(−) isoforms (Fig. 2E). Cells grown in growth medium express the Mitf-(−) isoform, but when these cells were cultured in serum-free medium for 6 h, the Mitf-(+) isoform became dominant. We were unable to detect a similar shifting of Mitf isoforms in melanocytes, where the Mitf-(+) isoform was dominant under all conditions tested, constituting about two-thirds of all transcripts (data not shown).
Melanocytes and cardiomyocytes express specific Mitf isoforms (M and H, respectively), while mast cells express a variety of isoforms, which are switched according to different physiologic stimuli, such as the aggregation of FcɛRI. Switching between Mitf-(+) and Mitf-(−) isoforms was detected in both mast cells and cardiomyocytes but not in melanocytes.
The identification of a cardiac-specific target for Mitf, MLC-1a.
Since we have demonstrated that the Mitf-H isoform is expressed in cardiomyocytes (37), and it seems to be the only isoform expressed, we were interested in studying the impact of different isoforms on the expression of cardiomyocyte-specific target genes in contrast to mast cell-specific targets in both cell types. However, since no cardiac-specific targets of Mitf have been described so far, we set out to find such a target.
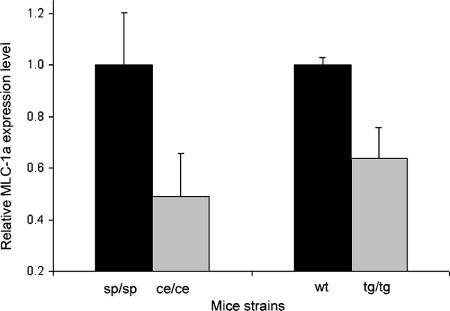
Since several bHLH-Zip transcription factors were shown to regulate the expression of the various myosin chains (4, 20, 27), the mRNA levels of the different myosin heavy and light chains in the hearts of wild-type and Mitf-mutated mice were determined using real-time PCR. No significant differences were apparent for either myosin heavy chains or for the regulatory myosin light chains. However, the level of MLC-1a, which is specifically expressed in the heart and is known to be an important regulator of force generation (6), was significantly decreased in both tg/tg and ce/ce Mitf-mutated mice (P < 0.01, n = 14, and P < 0.01, n = 13, respectively) (Fig. 3).
FIG. 3.
Real-time PCR analysis of MLC-1a expression levels in hearts of tg/tg and ce/ce mutant Mitf mice and their wild-type littermates. mRNA was extracted from hearts of wild-type (wt) and Mitf-mutated mice. mRNA quantitation was determined by SYBR green incorporation. Expression levels were normalized to that of the β-actin housekeeping gene. Results represent the mean ± standard error (n = 14 for tg/tg and n = 13 for ce/ce mice).
Mitf binds CANNTG E-box elements in the promoters of its target genes (10). Four E-box elements have previously been described in the MLC-1a promoter between −638 and the transcriptional start site (Fig. 4A). Prediction of Mitf binding to the MLC-1a promoter was performed. A nucleotide distribution matrix was constructed from 21 previously described Mitf binding sites using MatInd (30). The MatInspector program was used to analyze the MLC-1a promoter, resulting in the prediction of E3 as an Mitf binding site.
FIG. 4.
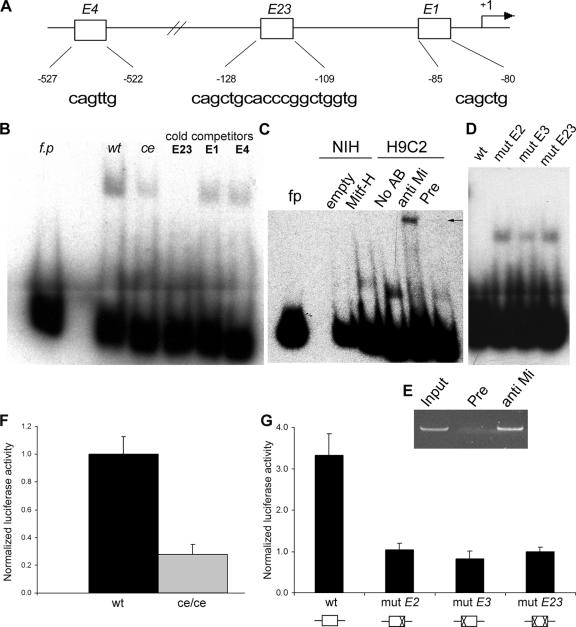
Mitf regulates MLC-1a expression through binding to E-box elements in MLC-1a promoter. (A) Four E boxes have been described in the proximal promoter region. E2 and E3 were included in one PCR fragment (E23). The sequence and the position of each of the four E boxes are depicted below. (B) EMSA results for MLC-1a promoter fragments are shown. E23 was used as a radiolabeled probe. fp, free probe; wt, wild-type Mitf; ce, ce/ce Mitf. E23, E1, and E4 were used as cold competitors with labeled E23. One representative experiment out of four is shown. (C) Binding of nuclear extracts of NIH 3T3 cells either overexpressing Mitf-H or transfected with an empty vector (empty). Supershifting of H9C2 nuclear extracts with either polyclonal anti-Mitf antibody directed against the common C terminus (anti Mi), preimmune serum (Pre), or without any sera (No AB) was performed. An arrow indicates the supershifted band. (D) E boxes E2 and E3 in the E23 element were point mutated to gTGtTG and gAGtTG, respectively, and were used for cold competition. Either wild-type E23 oligonucleotide (wt) or E23-mutated cold oligonucleotides (depicted as mut E2, mut E3, and mut E23), as indicated above each lane, competed with the normal E23 probe. (E) Chromatin immunoprecipitation assay in cardiomyocytes. Chromatin was immunoprecipitated with an anti-Mitf antibody directed against the C terminus of Mitf (anti Mi) or preimmune rabbit IgG (Pre) and then PCR amplified using primers for the MLC-1a promoter. (F) Transient transfection of NIH 3T3 cells with the MLC-1a promoter construct. Wild-type Mitf-H or ce/ce mutant plasmid constructs were cotransfected with the MLC-1a promoter reporter construct. The luciferase activity was normalized to that of total protein and divided by the value obtained for the wild type. The results shown represent the mean ± standard error (n = 4). (G) MLC-1a mutations in the E23 element. Either the wild-type MLC-1a promoter (wt) or MLC-1a E23 mutants (mut E2, mut E3, and mut E23) were cotransfected with Mitf expression vector. Luciferase activity was normalized as described above and divided by the value obtained for the E23 mutant promoter. A schematic representation of the different mutations is depicted below. The results shown represent the mean ± standard error (n = 3).
In order to demonstrate direct binding of Mitf to the normal and mutated MLC-1a promoter, we used an EMSA. Short PCR fragments were synthesized and either labeled with radioactive dCTP or used as cold competitors. Two of the putative E boxes are very close to one another and were therefore included in the same PCR fragment (E23).
The E-box element E23, located at −128, was able to bind wild-type Mitf, and competition with cold E23 completely abolished this binding (Fig. 4B). Furthermore, E23 very weakly bound the ce/ce mutant Mitf (Fig. 4B). E1 and E4 competed weakly with E23. Using NIH 3T3 nuclear extracts, we were able to detect a band in cells overexpressing Mitf-H but not in cells that were transfected with an empty vector. To further verify Mitf binding to the MLC-1a promoter, supershift experiments were carried out on H9C2 nuclear extracts using polyclonal anti-Mitf antibody directed against the common C terminus. The E23-DNA complex was supershifted by anti-Mitf antibody but not by the preimmune serum of the same rabbit (Fig. 4C). We next used cold competitors with mutations in either E-box E2 (E2), E3 (E3), or both (E23). The mutated E3 oligonucleotide only partially competed with the normal E23 probe, while E2- and E23-mutated olingonucleotides were unable to compete (Fig. 4D), indicating that the double-E-box E23 is indeed necessary for the binding of Mitf.
Using the ChIP assay, endogenous Mitf from cardiomyocytes was detected at the MLC-1a promoter in vivo (Fig. 4E).
The transcriptional activity of Mitf on MLC-1a promoter was then determined by construction of a pSP72 reporter plasmid which harbored the −638 to +57 fragment from MLC-1a promoter upstream of the luciferase reporter gene. NIH 3T3 fibroblasts were cotransfected with either full-length wild-type or ce/ce mutant Mitf expression vectors and the MLC-1a promoter reporter construct. Wild-type Mitf was able to activate the MLC-1a promoter approximately four times more efficiently than ce/ce Mitf (Fig. 4F).
E-boxes E2 and E3 in the E23 element were point mutated to gTGtTG and gAGtTG, respectively. NIH 3T3 fibroblasts were cotransfected with the three mutant MLC-1a promoters or wild-type promoter with Mitf expression vector. Mitf was able to activate the wild-type MLC-1a promoter roughly fourfold more than the mutated MLC-1a promoters (Fig. 4G).
Thus, real-time PCR, EMSA, ChIP, and transient transfection assay results indicate that Mitf regulates MLC-1a expression.
Activation of Mitf target genes by different Mitf isoforms in mast cells.
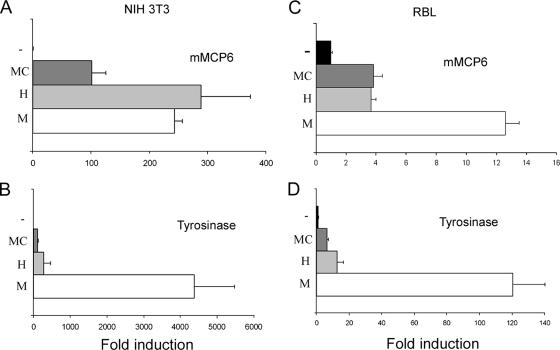
The Mitf isoforms were cotransfected into NIH 3T3 cells with luciferase reporter vectors of the mast cell protease mMCP-6 and the melanocyte-specific tyrosinase (Fig. 5A and B). The MC isoform had the weakest transactivation potential for both promoters, while the H isoform caused two- to threefold-stronger transactivation. In addition, in contrast to the observation that M isoform transactivation of the mMCP-6 promoter was similar to that of the H isoform, M isoform activation of the tyrosinase promoter was over 10-fold greater than its activation by both the MC and the H isoforms. We observed slightly different results in mast cells. Tyrosinase activation in mast cells was similar to that in NIH 3T3 cells (Fig. 5C and D). However, whereas the mMCP-6 promoter in RBL cells was similarly activated by the MC and H isoforms, activation by the M isoform was threefold greater. The activation of the tyrosinase promoter by the various Mitf isoforms in mast cells showed the same pattern as in NIH 3T3 cells, whereas the mMCP-6 activation pattern was dependent on the cell type. These results also indicate that while Mitf-M and Mitf-MC transactivation potential is not influenced by cell type, as previously described (36), Mitf-H activity is sensitive to cell type, suggesting a new role for the differential splicing of Mitf. Thus, Mitf transactivation potential is dependent on cell type, DNA targets, and the isoform of Mitf, with Mitf-H being sensitive to the cell type.
FIG. 5.
Transactivation of the mMCP-6 and tyrosinase promoters by different Mitf isoforms. (A and B) Mitf-M, Mitf-H, and Mitf-MC expression vectors or empty pcDNA vector (−) was cotransfected with reporters of mMCP-6 (A) and tyrosinase (B) into NIH 3T3 cells. Luciferase activity was normalized as described above and divided by the value obtained for the empty vector. The results shown represent the mean ± standard error (n = 4). (C and D) Expression vectors of Mitf-M, Mitf-H, and Mitf-MC or empty pcDNA vector (−) were cotransfected with reporters of mMCP-6 (C) and tyrosinase (D) into RBL cells. Luciferase activity was normalized as described above and divided by the value obtained for the empty vector. The results shown represent the mean ± standard error (n = 4).
Regulation of the expression of MLC-1a by two Mitf spliced isoforms in cardiomyocytes.
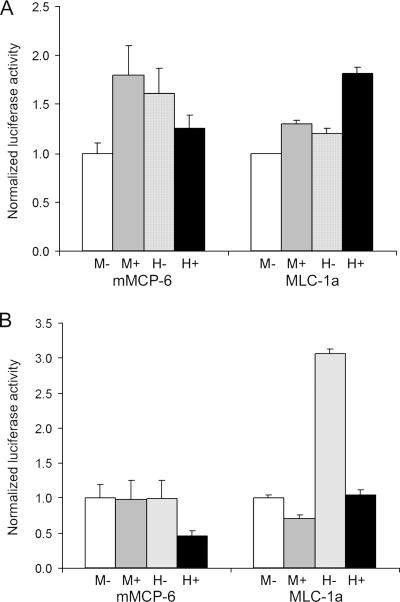
Mitf-M and Mitf-H spliced forms, with or without exon 6a, were cotransfected with either mMCP-6 or MLC-1a reporter vectors. In NIH 3T3 cells, levels of activation of both reporters by the two spliced forms were about the same (Fig. 6A), except for slightly lower activation of mMCP-6 by Mitf-M-(−). However, in H9C2 cardiomyocytes (Fig. 6B), Mitf-H-(−) had much greater activation potential for both mMCP-6 and MLC-1a promoters than Mitf-H-(+). Activation of MLC-1a promoter by Mitf-H-(−) was roughly threefold greater than that by all other Mitf isoforms. These results indicate that Mitf-H is more sensitive to internal alternate splicing than Mitf-M. Thus, it would seem that the fine-tuning of Mitf-H activity is more complex than that of Mitf-M.
FIG. 6.
Transactivation of mMCP-6 and MLC-1a promoters by different Mitf isoforms. Mitf-M-(−), Mitf-M-(+), Mitf-H-(−), and Mitf-H-(+) expression vectors were cotransfected with mMCP-6 and MLC-1a reporters into NIH 3T3 cells (A) and H9C2 cardiomyocytes (B). Luciferase activity was normalized to the result for Mitf-M-(−). The results shown represent the mean ± standard error (n = 4).
These results indicate that both cell type and 5′ and internal splicing influence the transcriptional activation potential of Mitf-H on different target genes.
DISCUSSION
Mitf has an essential role in the development and function of several cell types, including melanocytes and mast cells. Mitf is also abundantly expressed in cardiomyocytes. Multiple Mitf products, both those with different N termini and those resulting from differential internal splicing, have been described, but the relevance of this diversity to biological activity has yet to be determined.
We have studied the regulation of Mitf isoforms in several cell types. Melanocytes and cardiomyocytes express specific Mitf isoforms (M and H, respectively), but mast cells can express a variety of isoforms (MC, E, H, A, and M), and isoform expression is determined by the cell type and is dependent on physiological stimuli. Mast cells can also down-regulate the expression of exon 6a as a result of activation with either IgE plus DNP or SCF. Similarly, up-regulation of exon 6a was detected in cardiomyocytes in response to serum deprivation but was not detected in melanocytes.
Although the splicing variants both including and excluding the 18-bp exon 6a (Fig. 1A) have been extensively described in the literature, the role of exon 6a splicing still has to be investigated. The splicing of exon 6a does not affect either sumoylation of Mitf (18) or its phosphorylation by extracellular signal-regulated kinase (ERK) (19). Furthermore, while binding of Mitf without exon 6a to DNA as a homodimer is only slightly decreased, there is no effect on its binding as a heterodimer (10). An indication of the physiological role of these splicing products was recently provided (3). Mitf-(+) was shown to have a strong inhibitory effect on cell proliferation by transfection of Mitf-M into 293HEK cells. This activity seems not to be related to Mitf transcriptional activity, and deletion of the N terminus resulted in loss of the Mitf-(+) inhibitory effect. Depletion of Mitf-(+), therefore, might account in part for the increased proliferation of mast cells exposed to SCF (reviewed by Bischoff and Sellge [2]).
Our results indicate that while the M isoform had a high and constant transcriptional activity in all cell types used in our study, particularly for the tyrosinase promoter, and while the activity of the MC is consistently lower, the activity of the H isoform is dependent on cell type. Furthermore, activity of the H isoform is dependent on the presence or absence of the alternative exon 6a in cardiomyocytes, but not in NIH 3T3 cells, indicating the involvement of cell-specific regulators in the function of Mitf isoforms. Thus, we describe here for the first time a role for the alternative exon 6a in the regulation of Mitf's transcriptional activity. This role seems to be cell context sensitive, indicating that a protein-protein interaction might be required to exert this fine-tuning effect.
The M isoform includes only 11 amino acids upstream of the common exon 2, while all other isoforms include at least the 83 amino acids of exon 1b and a variable N terminus of 20 to 40 amino acids. These alternate N termini could enable Mitf to become sensitive to cell-specific inhibitors and activators and to the presence or absence of the alternatively spliced exon 6a.
Sequence analysis of exon 1b revealed a glutamine-rich domain containing 20 glutamine residues, most of which are absent from the M isoform, but are present in all other Mitf isoforms, including the E isoform, which lacks a part of exon 1b. This glutamine-rich domain is highly conserved between species (Fig. 7) and is predicted to assume a coiled-coil structure. Glutamine-rich domains can act as transcriptional activators (7) or could participate either in cis or in trans transcriptional inhibition (5). Thus, Mitf gains greater complexity in the regulation of its various target genes in different cells, enabling it to fine-tune its response to physiological stimuli. The presence or absence of the glutamine-rich domain together with the myriad of splicing options of Mitf (8) could not only enable great variety in the activation of the different Mitf targets by Mitf but also could permit Mitf to function as a transcriptional repressor of related transcription factors. The transcription factor Sp3, for example, carries an inhibitor domain that not only inhibits the Sp3 glutamine-rich domain mediated transactivation but also represses the activity of its family member Sp1 (5). Whether all MITF transcripts are translated in vivo remains to be determined. However, the shortness of the different N termini presents a problem in designing efficient specific antibodies and hinders answering this question.
FIG. 7.
Sequence alignment of the putative glutamine-rich domain of Mitf. The sequences of the putative glutamine-rich domain from different species were aligned. Glutamine residues appear with gray boxes. Highly conserved residues are represented by asterisks.
We have identified a cardiac cell-specific target of Mitf by studying mice carrying mutations in Mitf. These mice exhibit down-regulation of MLC-1a, a cardiac cell-specific myosin light chain. Mutations in two E-box elements in MLC-1a promoter, E2 and E3, resulted in the loss of Mitf's ability both to bind this promoter and to transactivate it. Either an extended form of an E-box is necessary for efficient binding of Mitf to MLC-1a promoter, or the synergistic binding of two Mitf molecules is needed for transactivation. E2, as has previously been described (4), is not a classical CANNTG E-box. Two previous studies (23, 24) have identified Mitf binding and activation of elements that do not conform to the CANNTG paradigm. These accumulated data imply the importance of nucleotide sequences other than the classical E-box for transcriptional regulation by Mitf. Expression of MLC-1a causes a pronounced positive inotropic effect (39) and is associated with increased Ca2+ sensitivity (6). Moreover, MLC-1a expression is augmented following myocardial infarction (25), in dilated cardiomyopathy (22), and under hemodynamic overload conditions (26). Therefore, MLC-1a up-regulation seems to be an important compensating mechanism. Very little is known about the regulation of MLC-1a expression, but our results indicate that Mitf is one of its regulators.
Our results clearly demonstrate that the various Mitf isoforms are regulated by physiological stimuli and that this enables refined control of target gene expression through a cell context and promoter context combinatory effect. Further investigation is needed in order to fully decipher the role played by Mitf and its various isoforms in the physiology of the heart, mast cells, and possibly other cell types.
Acknowledgments
This work was supported by the United States Binational Science Foundation (96-111 to E. Razin), the Israeli Academy of Science (E. Razin), the German-Israel Foundation for Scientific Research and Development (grant I-726-10.2 to E. Razin), and in part by the Dean's Fund of the Faculty of Medicine of Hebrew University.
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Bentley, N. J., T. Eisen, and C. R. Goding. 1994. Melanocyte-specific expression of the human tyrosinase promoter: activation by the microphthalmia gene product and role of the initiator. Mol. Cell. Biol. 14:7996-8006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bischoff, S. C., and G. Sellge. 2002. Mast cell hyperplasia: role of cytokines. Int. Arch. Allergy Immunol. 127:118-122. [DOI] [PubMed] [Google Scholar]
- 3.Bismuth, K., D. Maric, and H. Arnheiter. 2005. MITF and cell proliferation: the role of alternative splice forms. Pigment Cell Res. 18:349-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catala, F., R. Wanner, P. Barton, A. Cohen, W. Wright, and M. Buckingham. 1995. A skeletal muscle-specific enhancer regulated by factors binding to E and CArG boxes is present in the promoter of the mouse myosin light-chain 1A gene. Mol. Cell. Biol. 15:4585-4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dennig, J., M. Beato, and G. Suske. 1996. An inhibitor domain in Sp3 regulates its glutamine-rich activation domains. EMBO J. 15:5659-5667. [PMC free article] [PubMed] [Google Scholar]
- 6.Diffee, G. M., and D. F. Nagle. 2003. Regional differences in effects of exercise training on contractile and biochemical properties of rat cardiac myocytes. J. Appl. Physiol. 95:35-42. [DOI] [PubMed] [Google Scholar]
- 7.Gill, G., E. Pascal, Z. H. Tseng, and R. Tjian. 1994. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc. Natl. Acad. Sci. USA 91:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hallsson, J. H., J. Favor, C. Hodgkinson, T. Glaser, M. L. Lamoreux, R. Magnusdottir, G. J. Gunnarsson, H. O. Sweet, N. G. Copeland, N. A. Jenkins, and E. Steingrimsson. 2000. Genomic, transcriptional and mutational analysis of the mouse microphthalmia locus. Genetics 155:291-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han, B., R. Fixler, R. Beeri, Y. Wang, U. Bachrach, and Y. Hasin. 2003. The opposing effects of endothelin-1 and C-type natriuretic peptide on apoptosis of neonatal rat cardiac myocytes. Eur. J. Pharmacol. 474:15-20. [DOI] [PubMed] [Google Scholar]
- 10.Hemesath, T. J., E. Steingrimsson, G. McGill, M. J. Hansen, J. Vaught, C. A. Hodgkinson, H. Arnheiter, N. G. Copeland, N. A. Jenkins, and D. E. Fisher. 1994. Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8:2770-2780. [DOI] [PubMed] [Google Scholar]
- 11.Hodgkinson, C. A., K. J. Moore, A. Nakayama, E. Steingrimsson, N. G. Copeland, N. A. Jenkins, and H. Arnheiter. 1993. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell 74:395-404. [DOI] [PubMed] [Google Scholar]
- 12.Hughes, M. J., J. B. Lingrel, J. M. Krakowsky, and K. P. Anderson. 1993. A helix-loop-helix transcription factor-like gene is located at the mi locus. J. Biol. Chem. 268:20687-20690. [PubMed] [Google Scholar]
- 13.Isozaki, K., T. Tsujimura, S. Nomura, E. Morii, U. Koshimizu, Y. Nishimune, and Y. Kitamura. 1994. Cell type-specific deficiency of c-kit gene expression in mutant mice of mi/mi genotype. Am. J. Pathol. 145:827-836. [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, A., E. Morii, K. Maeyama, T. Jippo, D. K. Kim, Y. M. Lee, H. Ogihara, K. Hashimoto, Y. Kitamura, and H. Nojima. 1998. Systematic method to obtain novel genes that are regulated by mi transcription factor: impaired expression of granzyme B and tryptophan hydroxylase in mi/mi cultured mast cells. Blood 91:3210-3221. [PubMed] [Google Scholar]
- 15.Jippo, T., E. Morii, T. Tsujino, T. Tsujimura, Y.-M. Lee, D.-K. Kim, H. Matsuda, H.-M. Kim, and Y. Kitamura. 1997. Involvement of transcription factor encoded by the mouse mi locus (MITF) in expression of p75 receptor of nerve growth factor in cultured mast cells of mice. Blood 90:2601-2608. [PubMed] [Google Scholar]
- 16.Levy, C., H. Nechushtan, and E. Razin. 2002. A new role for the STAT3 inhibitor, PIAS3: a repressor of microphthalmia transcription factor. J. Biol. Chem. 277:1962-1966. [DOI] [PubMed] [Google Scholar]
- 17.McGill, G. G., M. Horstmann, H. R. Widlund, J. Du, G. Motyckova, E. K. Nishimura, Y. L. Lin, S. Ramaswamy, W. Avery, H. F. Ding, S. A. Jordan, I. J. Jackson, S. J. Korsmeyer, T. R. Golub, and D. E. Fisher. 2002. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell 109:707-718. [DOI] [PubMed] [Google Scholar]
- 18.Miller, A. J., C. Levy, I. J. Davis, E. Razin, and D. E. Fisher. 2005. Sumoylation of MITF and its related family members TFE3 and TFEB. J. Biol. Chem. 280:146-155. [DOI] [PubMed] [Google Scholar]
- 19.Molina, D. M., S. Grewal, and L. Bardwell. 2005. Characterization of an ERK-binding domain in microphthalmia-associated transcription factor and differential inhibition of ERK2-mediated substrate phosphorylation. J. Biol. Chem. 280:42051-42060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Molkentin, J. D., R. S. Brogan, S. M. Jobe, and B. E. Markham. 1993. Expression of the alpha-myosin heavy chain gene in the heart is regulated in part by an E-box-dependent mechanism. J. Biol. Chem. 268:2602-2609. [PubMed] [Google Scholar]
- 21.Moore, K. J. 1995. Insight into the microphthalmia gene. Trends Genet. 11:442-450. [DOI] [PubMed] [Google Scholar]
- 22.Morano, I., K. Hadicke, H. Haase, M. Bohm, E. Erdmann, and M. C. Schaub. 1997. Changes in essential myosin light chain isoform expression provide a molecular basis for isometric force regulation in the failing human heart. J. Mol. Cell Cardiol. 29:1177-1187. [DOI] [PubMed] [Google Scholar]
- 23.Morii, E., T. Tsujimura, T. Jippo, K. Hashimoto, K. Takebayashi, K. Tsujino, S. Nomura, M. Yamamoto, and Y. Kitamura. 1996. Regulation of mouse mast cell protease 6 gene expression by transcription factor encoded by the mi locus. Blood 88:2488-2494. [PubMed] [Google Scholar]
- 24.Murakami, M., T. Ikeda, K. Ogawa, and M. Funaba. 2003. Transcriptional activation of mouse mast cell protease-9 by microphthalmia-associated transcription factor. Biochem. Biophys. Res. Commun. 311:4-10. [DOI] [PubMed] [Google Scholar]
- 25.Nakao, K., H. Yasue, K. Fujimoto, M. Jougasaki, H. Yamamoto, Y. Hitoshi, K. Takatsu, and E. Miyamoto. 1992. Increased expression and regional differences of atrial myosin light chain 1 in human ventricles with old myocardial infarction. Analyses using two monoclonal antibodies. Circulation 86:1727-1737. [DOI] [PubMed] [Google Scholar]
- 26.Nakao, K., H. Yasue, K. Fujimoto, K. Okumura, H. Yamamoto, Y. Hitoshi, T. Murohara, K. Takatsu, and E. Miyamoto. 1992. Increased expression of atrial myosin light chain 1 in the overloaded human left ventricle: possible expression of fetal type myocytes. Int. J. Cardiol. 36:315-328. [DOI] [PubMed] [Google Scholar]
- 27.Navankasattusas, S., M. Sawadogo, M. van Bilsen, C. V. Dang, and K. R. Chien. 1994. The basic helix-loop-helix protein upstream stimulating factor regulates the cardiac ventricular myosin light-chain 2 gene via independent cis regulatory elements. Mol. Cell. Biol. 14:7331-7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nechushtan, H., Z. Zhang, and E. Razin. 1997. Microphthalmia (mi) in murine mast cells: regulation of its stimuli-mediated expression on the translational level. Blood 89:2999-3008. [PubMed] [Google Scholar]
- 29.Oboki, K., E. Morii, T. R. Kataoka, T. Jippo, and Y. Kitamura. 2002. Isoforms of mi transcription factor preferentially expressed in cultured mast cells of mice. Biochem. Biophys. Res. Commun. 290:1250-1254. [DOI] [PubMed] [Google Scholar]
- 30.Quandt, K., K. Frech, H. Karas, E. Wingender, and T. Werner. 1995. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23:4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Razin, E., C. Cardon-Cardo, and R. A. Good. 1981. Growth of a pure population of mouse mast cells in vitro using conditioned medium derived from Con A-stimulated splenocytes. Proc. Natl. Acad. Sci. USA 78:2559-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Razin, E., Z. C. Zhang, H. Nechushtan, S. Frenkel, Y. N. Lee, R. Arudchandran, and J. Rivera. 1999. Suppression of microphthalmia transcriptional activity by its association with protein kinase C-interacting protein 1 in mast cells. J. Biol. Chem. 274:34272-34276. [DOI] [PubMed] [Google Scholar]
- 33.Steingrimsson, E., N. G. Copeland, and N. A. Jenkins. 2004. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 38:365-411. [DOI] [PubMed] [Google Scholar]
- 34.Steingrimsson, E., K. J. Moore, M. L. Lamoreux, A. R. Ferre-D'Amare, S. K. Burley, D. C. Zimring, L. C. Skow, C. A. Hodgkinson, H. Arnheiter, N. G. Copeland et al. 1994. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat. Genet. 8:256-263. [DOI] [PubMed] [Google Scholar]
- 35.Tachibana, M., K. Takeda, Y. Nobukuni, K. Urabe, J. E. Long, K. A. Meyers, S. A. Aaronson, and T. Miki. 1996. Ectopic expression of MITF, a gene for Waardenburg syndrome type 2, converts fibroblasts to cells with melanocyte characteristics. Nat. Genet. 14:50-54. [DOI] [PubMed] [Google Scholar]
- 36.Takemoto, C. M., Y. J. Yoon, and D. E. Fisher. 2002. The identification and functional characterization of a novel mast cell isoform of the microphthalmia-associated transcription factor. J. Biol. Chem. 277:30244-30252. [DOI] [PubMed] [Google Scholar]
- 37.Tshori, S., D. Gilon, R. Beeri, H. Nechushtan, D. Kaluzhny, E. Pikarsky, and E. Razin. 2006. Transcription factor MITF regulates cardiac growth and hypertrophy. J. Clin. Investig. 116:2673-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wadman, I. A., H. Osada, G. G. Grutz, A. D. Agulnick, H. Westphal, A. Forster, and T. H. Rabbitts. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zacharzowsky, U. B., G. Wolff, M. Kott, H. Haase, H. Bartsch, A. K. Nuessler, L. G. Baltas, L. Karawajew, and I. Morano. 2002. Analysis of the energetic state of heart cells after adenovirus-mediated expression of hALC-1. J. Cell Biochem. 86:422-431. [DOI] [PubMed] [Google Scholar]