Abstract
DNA methyltransferase 1 (DNMT1) has been reported to interact with a wide variety of factors and to contain intrinsic transcriptional repressor activity. When a conservative point mutation was introduced at the key catalytic residue, mutant DNMT1 failed to rescue any of the phenotypes of Dnmt1-null embryonic stem (ES) cells, which indicated that the biological functions of DNMT1 are exerted through the methylation of DNA. ES cells that expressed the mutant protein did not survive differentiation. Intracisternal A-particle family retrotransposons were no longer methylated and were transcribed at high levels. The proper localization of DNMT1 depended on normal genomic methylation, and we discuss the implications of this finding for epigenetic dysregulation in cancer.
Patterns of methylated and unmethylated cytosines at CpG dinucleotides are propagated with a fidelity of >99%, and their stable inheritance for >80 cell generations has been documented (48). DNA methylation is required for the transcriptional silencing of transposons, imprinted genes, and genes on the inactive X chromosome. The methylation landscape of the human genome reflects this function: methylation is concentrated in transposons and repeat sequences and is rare in promoter regions, except for certain imprinted genes, certain cancer-testis antigen genes, and genes on the inactive X chromosome in females (49). DNA methylation is also required for normal chromosome structure and stability. Demethylation and chromosome rearrangements at juxtacentromeric heterochromatin are observed in patients with immunodeficiency, centromere instability, and facial anomaly syndrome (OMIM 242860), which is caused by mutations in the DNA methyltransferase 3B (DNMT3B) gene (63).
Cancer cells commonly exhibit aberrant genomic methylation patterns (3, 20). The epigenetic dysregulation is complex, and the causes are not known. Hypomethylation of repeat sequences often coexists with hypermethylation of a subset of promoters. Hypomethylation and hypermethylation can independently predict tumor malignancy (21). A loss of imprinting increased the incidence of tumorigenesis (31, 51), and aberrant methylation had different effects on the progressions of different cancers (64).
The mechanism of transcriptional repression of methylated DNA is not known. The chromatin-associated factors MeCP2, methyl-binding domain 1 (MBD1), MBD2, and Kaiso have been reported to repress methylated promoters, but methylated promoters are not reactivated in the absence of these factors, and the phenotypes of mice that lack these factors do not resemble those of mice that lack DNA methyltransferases (29, 45, 57, 66).
DNA methylation in mammals is catalyzed by three methyltransferases, DNMT1, DNMT3A, and DNMT3B (27). Biochemical and genetic data support a model in which all three enzymes have de novo and maintenance activities and cooperate to establish and maintain genomic methylation patterns. Of the methyltransferases, DNMT1 has the highest expression level in vivo and the highest specific activity in vitro, and mutations in Dnmt1 have the most severe phenotypes. Mouse embryos homozygous for null alleles of Dnmt1 have developmental delays and do not survive past day 8.5 of development (38). Mouse Dnmt1−/− embryonic stem (ES) cells have severely demethylated genomes and undergo apoptosis when induced to differentiate (35, 38), but the mechanism is not known. Several studies have documented defects in mismatch repair or microsatellite instability in Dnmt1−/− cells (28, 33, 60). Mice bearing a hypomorphic allele of Dnmt1 exhibit chromosome instability and a high incidence of T-cell lymphomas (18, 25). Maintenance of genomic methylation patterns in HCT116 colorectal cancer cells was reported to be largely independent of DNMT1 (46), but it was recently shown that the DNMT1 disruption in that study caused only a minor reduction in DNMT1 activity (19) and that the complete disruption of DNMT1 in HCT116 cells leads to mitotic defects and cell death (9). Together, these results demonstrate a requirement for DNMT1 during development and in the survival of differentiated cells.
DNMT1 contains an amino-terminal domain of 1,120 residues and a methyltransferase domain of 500 residues (27). The amino-terminal domain includes a nuclear localization signal and a replication focus-targeting domain as well as a cysteine-rich Zn2+-binding domain and two bromo-adjacent homology domains of unknown function (Fig. 1A). DNA binding and allosteric control of the methyltransferase domain have been reported for regions of the amino-terminal domain (2, 22). In addition, some domains of DNMT1 have been reported to repress transcription and to associate with histone deacetylase 1 and histone deacetylase 2; methyl-binding proteins MeCP2, MBD2, and MBD3; retinoblastoma protein; PCNA; DMAP1; heterochromatin protein HP1β; histone methyltransferase SUV39H1; the PML-retinoic acid receptor fusion oncoprotein; and Polycomb group protein EZH2 (10, 24, 58). However, the biological functions of these interactions have not been demonstrated, and the region of DNMT1 that interacts with DMAP1 has been shown to be entirely dispensable for DNMT1 function in vivo (16).
FIG. 1.
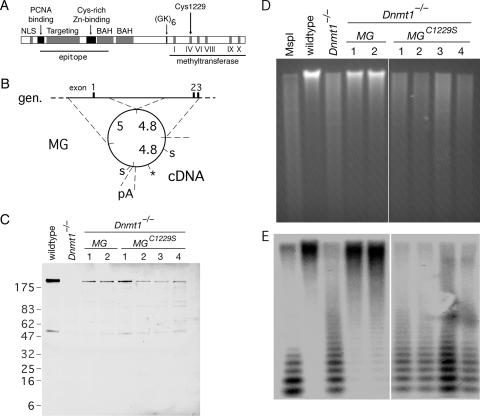
A point mutation in DNMT1 abolishes methyltransferase activity. (A) Domains of DNMT1 include the nuclear localization signal (NLS), PCNA binding domain, targeting to replication foci, cysteine-rich Zn2+-binding domain, bromo-adjacent homology domain (BAH), a glycine- and lysine-rich segment (GK), and the methyltransferase domain with diagnostic motifs. A large segment of the amino terminus served as an epitope for the anti-DNMT1 (pATH52) antibody (4). A conservative cysteine→serine mutation at residue 1229 (C1229S) was introduced into a murine Dnmt1 minigene. (B) Schematic of the Dnmt1 minigene (MG) described previously by Tucker et al. (56). The minigene is comprised of two genomic (gen.) fragments that contain a regulatory sequence and the first three exons, cDNA that encodes the remainder of the protein, and a polyadenylation sequence (pA) from the PGK locus. The lengths of these fragments (in kb) are shown. To generate the C1229S mutation (marked by the asterisk), the 4.4-kb fragment between SalI sites (“S”) was subcloned into a cloning vector, subjected to site-directed mutagenesis, and subcloned back into the minigene. (C) The Dnmt1 minigene or the Dnmt1C1229S minigene was electroporated into Dnmt1−/− ES cells to establish stable cell lines in which the minigene construct was the only source of DNMT1. Cell lysates from wild-type cells, Dnmt1−/− cells, two clones of Dnmt1−/− MG, and four clones of Dnmt1−/− MGC1229S were resolved on a 10 to 20% gradient gel and immunoblotted with anti-DNMT1 (pATH52) antibody. Some nonspecific bands are apparent. (D) Ethidium-stained gel of HpaII-digested genomic DNA from the panel of cell lines shown in C. HpaII is sensitive to CpG methylation, but isoschizomer MspI is not. (E) Southern blot of gel in D with a probe to the minor satellite (centromeric) repeats.
We have investigated the functions of DNMT1 by making a conservative point mutation that eliminates methyltransferase activity. We report that the essential functions of DNMT1 require its methyltransferase activity. We found that the localization of DNMT1 depended on genomic methylation levels, which has implications for the dysregulation of methylation patterns in cancer.
MATERIALS AND METHODS
Construction of cell lines.
The wild-type Dnmt1 minigene MT80 was a kind gift of R. Jaenisch. The C1229S mutation was introduced by site-directed mutagenesis with QuikChange (Stratagene) to generate MTCS. Either MT80 or MTCS was electroporated with PGK-PURO into Dnmt1c/c cells to establish stable cell lines. Genomic integration was not targeted; however, the minigene contains 9.8 kb of genomic sequence (Fig. 1B). Clones were selected in 2 μg/ml puromycin, expanded, and screened by PCR. Candidate clones were then screened by immunoblot for expression of full-length DNMT1 with anti-DNMT1 (pATH52) antibody. Southern blot analysis was used to estimate the minigene copy number. All six clones had the normal complement of 40 chromosomes.
Cell culture and sample preparation.
Mouse ES cells were cultured on gelatinized tissue culture dishes in Dulbecco's modified Eagle's medium (catalog no. 11965; Gibco) supplemented with modified Eagle's medium nonessential amino acids (catalog no. 11140; Gibco), 2 mM l-glutamine, 100 IU/ml penicillin, 100 μg/ml streptomycin, 0.12 mM β-mercaptoethanol, leukemia-inhibitory factor (LIF) (from conditioned medium of LIF-secreting cells), and 15% of either fetal bovine serum (HyClone) or serum replacement for ES cells (catalog no. 10828; Gibco). Genomic DNA was isolated by using a DNeasy kit (QIAGEN). Total RNA was isolated with TRIzol reagent (Invitrogen) and briefly treated with RNase-free DNase (Roche) in the presence of RNase inhibitor (RNasin; Promega). For immunoblots, cells were lysed in ice-cold phosphate-buffered saline with 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), and protease inhibitors. Lysates were clarified for 5 min at 14 krpm, and protein concentrations were measured with the BCA assay (Pierce). Ponceau S stain confirmed the transfer of proteins to the membrane.
Differentiation and competitive growth assay.
Differentiation was induced in three ways. First, for low-density plating, cells were plated at 500 cells/cm2 in ES cell medium without LIF. Second, comparable results (not shown) were obtained when cells were plated at 104 cells/cm2, cultured in medium without LIF, and treated with 10−7 M all-trans-retinoic acid for 3 days starting the day after plating. Third, for the competitive growth assay (35), a 1:1 mixture of the two cell types indicated was cultured in several dishes for 2 days in ES cell medium. Next, on “day 0,” embryoid bodies were generated from some dishes, other dishes were maintained in ES cell medium, and a small aliquot of cells was reserved for DNA isolation. Embryoid bodies were generated from confluent dishes by an 80-s treatment with trypsin-EDTA followed by growth in suspension in petri dishes in ES cell medium without LIF and containing only 10% fetal bovine serum. Medium was changed by allowing the embryoid bodies to sediment in conical tubes. The experiment was repeated on independent occasions. For the Southern blots, genomic DNA was digested with EcoRI and EcoRV and probed with a 600-bp PCR-amplified fragment that contained the genomic sequence from 4.1 to 3.5 kb upstream of the Cys1229 codon. Each cell line had been probed individually so that it could be identified in the patterns generated by cell mixtures.
Antibodies.
Primary antibodies with dilutions for immunofluorescence (IF) and immunoblot (IB) were anti-DNMT1 (pATH52) (4) (1:500 for IF and 1:2,000 for IB), anti-histone macroH2A1 (catalog no. 07-219; Upstate) (1:100 for IF and 1:500 for IB), and fluorescein isothiocyanate-conjugated anti-bromodeoxyuridine (BrdU) (Becton Dickinson) (1:10 for IF). Secondary antibodies were obtained from Jackson Immunoresearch.
Immunofluorescence.
All solutions were based on phosphate-buffered saline (pH 7.2) with 1.5 mM MgCl2 and 0.9 mM CaCl2. Cells were harvested, fixed in 2% formaldehyde at 4°C, centrifuged onto glass slides, permeabilized in 0.2% Triton X-100, and blocked in 5% goat serum-0.2% fish scale gelatin-0.2% Tween 20 (all from Sigma). Antibody incubations in blocking solution at room temperature were followed by several washes in buffer. DNA was stained with 10 ng/ml Hoechst 33258, and slides were mounted in n-propyl gallate. Costaining of DNMT1 and active replication sites was performed essentially as described previously (36). Briefly, cells were pulsed with 10 μM BrdU for 15 min prior to harvesting; following DNMT1 immunofluorescence as described above, the antibodies were fixed in place with formaldehyde, and DNA was then denatured with HCl and probed with fluorescein isothiocyanate-anti-BrdU.
ChIP.
Chromatin immunoprecipitation (ChIP) was repeated for at least three independent biological replicates, starting with cell culture. The protocol was based on a method described previously by Takahashi (53), with modifications. For cross-linking, 1 × 108 cells were harvested and treated with 0.4% formaldehyde for 10 min at room temperature. Chromatin was sheared by sonication such that the DNA fragment size was 200 to 1,000 bp, and some aliquots were reserved as “input.” Protein A-Sepharose beads (catalog no. 17-1279-01; Amersham) were washed and blocked in 1 ml/sample of phosphate-buffered saline with 5 mg/ml bovine serum albumin and 2 μg/ml denatured salmon sperm DNA. For each sample, chromatin from 1 × 107 cells was precleared with blocked beads and then incubated overnight with or without 5 μl per immunoprecipitation (IP) of anti-histone macroH2A1 antibody. The next day, a fresh aliquot of blocked beads was incubated for 2 h with the chromatin-antibody mix plus 1 μg additional salmon sperm DNA. The beads were washed twice in a solution containing 20 mM Tris (pH 8), 150 mM NaCl, 2 mM EDTA, 1% Triton, and 0.1% SDS; once in a solution containing 20 mM Tris (pH 8), 500 mM NaCl, 2 mM EDTA, 1% Triton, and 0.1% SDS; twice in a solution containing 10 mM Tris (pH 8), 1 mM EDTA, 1% NP-40, 1% sodium deoxycholate, and 250 mM LiCl; and once in a solution containing 10 mM Tris (pH 8) and 1 mM EDTA.
Quantitative real-time PCR.
We used an ABI 7700 apparatus (Columbia University Cancer Center) with SYBR Green PCR Master Mix (catalog no. 4309155; Applied Biosystems). Primer pairs were designed from previously published sequences (14, 42, 65) and tested to ensure linear amplification and a single product. Many primer sets that were designed for the intracisternal A-particle (IAP) long terminal repeat (LTR) did not satisfy these criteria, possibly owing to the heterogeneity of IAP LTRs (14). A single PCR product represented amplification from many individual IAP elements with the same LTR sequence. The primers for GAPDH were specific to the active gene on chromosome 6 and did not have homology to any of the GAPDH pseudogenes. For PCR, input DNA was diluted 65-fold more than immunoprecipitated DNA so that amplification was comparable. The threshold cycle (CT) method was used to determine the relative DNA concentration in input and IP samples for each primer set. The enrichment in the IP was calculated by subtracting input CT from IP CT (ΔCT) and then transforming by 2−x and calculating for the dilution factors. This normalization, along with the confirmed linear amplification of the PCR, allowed the comparison of multicopy IAP sequences with single-copy ACTB and GAPDH. For analysis of data from different primer sets, the two-tailed Student's t test was used. For analysis of wild-type versus Dnmt1−/− cells, the ΔCT values for the cell lines were compared for each experiment (by subtraction), and the averages and standard deviations from three experiments were calculated. The two-tailed t test for paired samples was used.
Primer sequences (5′ to 3′) used were as follows:, GACATCCTGTGTTCTAAGTGG and CCAGAGCCAGAGAGCAAAAGC (IAP LTR), GTGTCCATTGACACATGTTCTGG and GTGGGTTACGTCCATCTGACG (IAP internal 8), GCAGCCACATCTAATGATTGGG and CATTAGCCACATAGGCACCTATGC (IAP internal 4), CACCAGGTAAGTGACCTGTTAC and GGTCAGGATACCTCTCTTGCTC (ACTB), and GCTCATGGTATGTAGGCAGTGG and GATGGCATGGACTGTGGTCTAG (GAPDH).
RESULTS
Conservative point mutation in DNMT1.
To separate the methyltransferase activity from other putative functions of DNMT1, we introduced a Dnmt1 minigene with a point mutation at the key catalytic cysteine residue into Dnmt1−/− ES cells. The cysteine residue is conserved in all DNA methyltransferases, and its mutation to serine abolishes catalytic activity but does not affect DNA binding (32, 41, 61, 62). We introduced the Cys1229→Ser (C1229S) mutation into a functional minigene of mouse Dnmt1 composed of ∼5 kb of cDNA and ∼10 kb of genomic sequence that contains the regulatory sequence and the first three exons (56). A schematic of the minigene is shown in Fig. 1B, and a complete description of sequence and functionality was provided previously by Tucker et al. (56).
The wild type or C1229S minigene was electroporated into Dnmt1−/− mouse ES cells to establish stable cell lines. Clones were expanded and assayed for the expression of full-length DNMT1 by IB (Fig. 1C). IB following separation on a 3 to 8% gradient gel provided further evidence that the proteins were full length (data not shown). Two clones that expressed full-length DNMT1 (designated Dnmt1−/− MG, where MG indicates minigene) and four clones that expressed full-length DNMT1C1229S (designated Dnmt1−/− MGC1229S) were selected for further analysis. The minigene construct was the only source of DNMT1 in these cells. The level of DNMT1C1229S in the Dnmt1−/− MGC1229S clones was comparable to the levels of DNMT1 in the Dnmt1−/− MG clones (Fig. 1C and data not shown), which permitted a controlled comparison of Dnmt1−/− MG to Dnmt1−/− MGC1229S. Southern blot analysis of the clones indicated that the minigene copy number per clone was between 1 and 3 and correlated with the protein level (see Fig. S1 in the supplemental material).
To assess genome-wide methylation levels in the panel of cell lines, genomic DNA was isolated and digested with the methylation-sensitive enzyme HpaII. Electrophoresis and ethidium bromide staining revealed that DNA from wild-type cells was resistant to HpaII digestion but that DNA from Dnmt1−/− cells was not, which confirmed that the Dnmt1−/− genome was extensively demethylated (Fig. 1D). The Dnmt1 minigene restored genomic methylation, but Dnmt1C1229S did not (Fig. 1D), which indicated that the minigene was functional and that DNMT1C1229S did not have methyltransferase activity. For comparison, the pattern that would correspond to fully demethylated DNA was obtained by digestion with MspI, an isoschizomer of HpaII that is not sensitive to methylation at CpG sites.
To examine the methylation of a specific compartment of the genome, the digested DNA shown in Fig. 1D was transferred onto nitrocellulose and hybridized with a probe to minor satellite DNA, which consists of 120-bp centromeric repeats that constitute 0.2 to 0.4% of the mouse genome (13). We found that minor satellite DNA was methylated in wild-type and Dnmt1−/− MG cells and demethylated in Dnmt1−/− and Dnmt1−/− MGC1229S cells (Fig. 1E).
The Dnmt1−/− ES cells used in this study, known as Dnmt1c/c, are homozygous for a mutation in the methyltransferase domain of DNMT1 and are viable in the undifferentiated state (35). No mRNA transcript or full-length protein was detected in Dnmt1c/c cells, and genomic DNA was extensively demethylated (35). However, reverse transcription-PCR indicated some transcription upstream of the deletion (6). The interpretation of the data from this study required a precise characterization of the Dnmt1−/− cells used in these experiments. If the full-length or truncated DNMT1 protein were present even at low levels in Dnmt1−/− cells, similar phenotypes in Dnmt1−/− and Dnmt1−/− MGC1229S cells could be trivially explained as being due to the presence of the DNMT1 protein in both cell types. However, if no DNMT1 protein was detected in Dnmt1−/− cells, similar phenotypes in Dnmt1−/− and Dnmt1−/− MGC1229S cells could be interpreted to mean that the noncatalytic DNMT1 did not rescue any phenotypes of the null alleles and thus that the biological functions of DNMT1 require its methyltransferase activity. We therefore performed a detailed analysis of the Dnmt1c/c cells used in the current study. Immunoblot analysis with anti-DNMT1 (pATH52) antibody following resolution in a 10 to 20% gradient gel did not show any specific bands in Dnmt1c/c cells (Fig. 1C). The pATH52 antibody (4) was raised against a large segment of the DNMT1 amino terminus (Fig. 1A) and should recognize peptides that may result from truncated transcripts of the Dnmt1c/c allele. Northern analysis with a full-length cDNA probe revealed little or no transcript in Dnmt1c/c cells (see Fig. S2 in the supplemental material). Reverse transcription-PCR of Dnmt1c/c RNA with four primer pairs that spanned the deletion yielded specific fragments, although they were very weak compared to those from wild-type cells. The fragments were amplified by additional PCR and sequenced; the results are shown in Fig. S2 in the supplemental material. Based on these results and the phenotypes of Dnmt1c/c cells, we conclude that Dnmt1c/c is functionally a null allele; there may be trace-level transcription at the locus, but no DNMT1 protein was detectable. The Northern and immunoblot analyses also confirmed that no truncated transcript or truncated DNMT1 protein was detectable in the Dnmt1−/− MG or Dnmt1−/− MGC1229S cell lines.
Demethylated cells do not survive differentiation.
Several phenotypes of Dnmt1−/− cells were examined in the panel of cell lines to determine whether the phenotypes were caused by low genomic methylation or the absence of other functions of DNMT1. Dnmt1−/− ES cells are viable as stem cells but undergo apoptosis when induced to differentiate; the “lethal differentiation” phenotype has been characterized previously (35, 38), but the mechanism is not understood. To investigate this phenotype, we induced ES cells to differentiate by three methods.
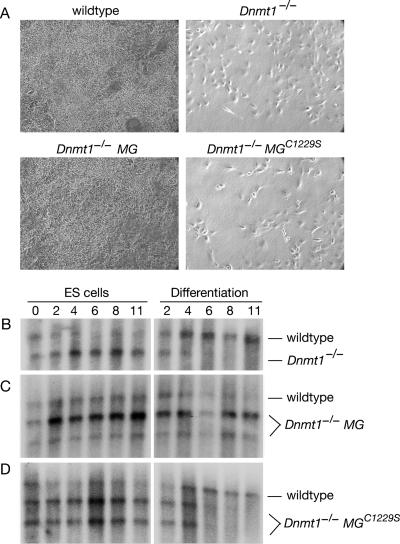
Cells were plated at low density and cultured for 7 days under established conditions (52). Wild-type and Dnmt1−/− MG cells grew robustly under these conditions, but Dnmt1−/− and Dnmt1−/− MGC1229S cells did not (Fig. 2A). Similar results were obtained with an alternate protocol that included treatment with retinoic acid (data not shown). These results indicated that the lethal differentiation phenotype was caused by low genomic methylation.
FIG. 2.
Lethal differentiation of demethylated ES cells. (A) ES cells were plated at low density to induce differentiation and were photographed after 7 days of culture. Methylated cells grew robustly, whereas demethylated cells did not. Both clones of Dnmt1−/− MG and all four clones of Dnmt1−/− MGC1229S showed the same result (not shown). (B to D) Competitive growth assay. Mixtures (1:1) of wild-type and Dnmt1−/− (B), Dnmt1−/− MG (C), or Dnmt1−/− MGC1229S (D) cells were cultured for 2 days and then split in half (“day 0”) for culture as ES cells and as embryoid bodies (differentiation). Genomic DNA was extracted from samples on days 0, 2, 4, 6, 8, and 11. Southern blot analysis with a probe to the Dnmt1 genomic sequence yielded a diagnostic band for the wild-type or Dnmt1−/− background and an additional band for the minigene.
The phenotype was also investigated by the induction of differentiation in embryoid bodies. Using a competitive growth assay (35), we determined the relative growth rates of two cell types during their coculture as ES cells and as differentiating cells in embryoid bodies. The relative amount of each cell type at various time points was determined by Southern blotting. When wild-type and Dnmt1−/− cells were cocultured, Dnmt1−/− cells were not detected in embryoid bodies after day 6, but they survived when cultured as ES cells (Fig. 2B). The minigene rescued the phenotype, as Dnmt1−/− MG cells competed well with wild-type cells in embryoid bodies (Fig. 2C); in contrast, Dnmt1−/− MGC1229S cells did not (Fig. 2D). These results are consistent with the results from low-density differentiation and indicate that the lethal differentiation phenotype is caused by low genomic methylation and not the absence of other functions of DNMT1. We note that the wild-type cells were outcompeted by cells in the Dnmt1−/− background during coculture as ES cells (Fig. 2). This may be due to the demethylation and overexpression of growth-promoting imprinted genes in Dnmt1−/− cells (30, 37); genomic imprints are not restored by the wild-type Dnmt1 minigene without germ line passage (55).
Transcriptional activation of IAP retrotransposons in demethylated cells.
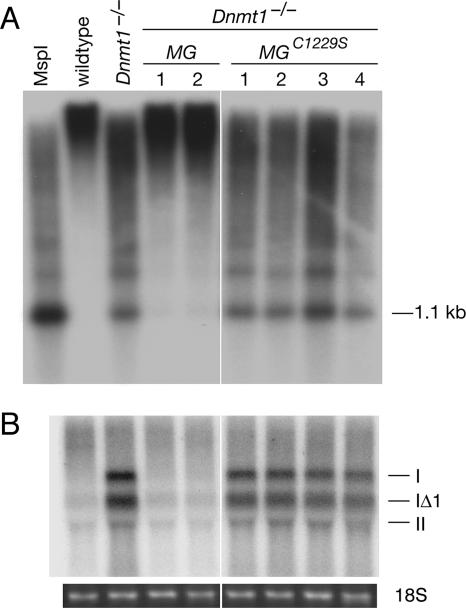
The silencing of retrotransposons is an established function of DNA methylation (5, 59). We investigated the methylation status and transcription of the IAP family of retrotransposons in the panel of cell lines. The haploid mouse genome contains approximately 1,000 IAP elements (34). IAP elements are demethylated and transcribed in Dnmt1-null embryos and in demethylated ES cells (54, 59). We found extensive demethylation of IAP retrotransposons in Dnmt1−/− and Dnmt1−/− MGC1229S cells (Fig. 3A). The demethylation was not complete (compare to MspI digestion), which was consistent with results for the minor satellite (Fig. 1E) and previous findings (54). The Dnmt1 minigene restored methylation at the IAPs to nearly wild-type levels (Fig. 3A).
FIG. 3.
Transcriptional activation of IAP retrotransposons in demethylated cells. (A) Southern blot of HpaII-digested genomic DNA with a probe to the LTR of the IAP family of retrotransposons. A separate aliquot of wild-type DNA was digested with MspI. (B) Northern blot of IAP elements. The order of samples corresponds to A but without the leftmost lane. The transcripts from three subtypes of IAP elements (I, IΔI, and II) are indicated. Ethidium staining of the gel confirmed equal loading (18S rRNA is shown).
Northern blot analysis revealed high levels of IAP transcripts in Dnmt1−/− and Dnmt1−/− MGC1229S cells but not in wild-type or Dnmt1−/− MG cells (Fig. 3B). The banding pattern represents mRNA from three subtypes (I, IΔI, and II) of IAP elements (34). These results demonstrate that the transcriptional activation of IAPs is caused by the demethylation of the genome and not by the absence of other putative functions of DNMT1, which was reported by several studies to repress transcription (23, 47, 50).
Methylation-dependent association of histone macroH2A at retrotransposons.
We hypothesized that the repression of methylated IAP elements could involve the histone variant macroH2A. Histone macroH2A, which is composed of an H2A-like domain and a nonhistone transcriptional repressor domain (1, 17, 44), is localized at the inactive X chromosome (15) and imprinted loci (12). It was recently shown that macroH2A is mislocalized from euchromatin to foci of pericentric heterochromatin in Dnmt1−/− ES cells (40), which we also observed (Fig. 4A). We found that the Dnmt1 minigene restored proper localization of macroH2A but that the Dnmt1C1229S minigene did not (Fig. 4A), which indicated that the localization of histone macroH2A depends on genomic methylation. Immunoblot analysis of whole-cell extracts demonstrated that the levels of macroH2A were comparable in all cell lines (Fig. 4B).
FIG. 4.
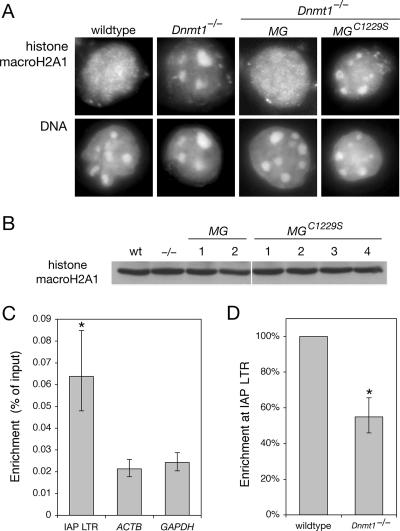
Methylation-dependent localization of histone macroH2A1. (A) Immunofluorescence with anti-histone macroH2A1 antibody. The regions of intense Hoechst DNA staining correspond to pericentric heterochromatin. Both clones of Dnmt1−/− MG and all four clones of Dnmt1−/− MGC1229S showed the same result (not shown). (B) IB of cell lysates with anti-histone macroH2A1 antibody. The lanes correspond to those shown in Fig. 1C. wt, wild type. (C) Association of histone macroH2A1 with the IAP LTR as determined by ChIP. The enrichment at the LTR is highly significant compared to the signal at ACTB and GAPDH (P = 0.001 and P = 0.002, respectively; the asterisk indicates statistical significance). Error bars represent standard deviations of the means. Quantitative real-time PCR was performed on samples from at least three independent biological replicates. (D) Methylation-dependent association of histone macroH2A1 with the IAP LTR. ChIP was performed with wild-type and Dnmt1−/− cells. The enrichment in Dnmt1−/− cells was significantly lower than that in wild-type cells (P = 0.028; the asterisk indicates statistical significance). The error bar represents the standard deviation of the mean.
To investigate whether macroH2A is associated with IAP elements, we performed ChIPs with anti-macroH2A1 antibody and analyzed the immunoprecipitated DNA by quantitative real-time PCR. We found that histone macroH2A was enriched at IAP LTRs in wild-type cells (Fig. 4C). The enrichment at the IAP LTR was highly significant compared to those at two transcribed loci, ACTB (β-actin) and GAPDH, with P values of 0.001 and 0.002, respectively. The results for the IAP represent many individual IAP elements with LTR sequences matched by the primers; it was essential to normalize the ChIP data for the difference in copy number between IAP elements and ACTB or GAPDH (see Materials and Methods). The signal at ACTB and GAPDH most likely represents the background level in the assay, because macroH2A is generally depleted at active genes (8). CpG dinucleotides are located throughout IAP elements, and macroH2A was associated with internal IAP regions but only at ∼70% of the level at the LTR (data not shown).
We found that the association of histone macroH2A at the IAP LTR depends on DNA methylation (Fig. 4D). macroH2A enrichment at the LTR in Dnmt1−/− cells was significantly lower than that in wild-type cells (P = 0.028). In contrast, no difference between wild-type and Dnmt1−/− cells was observed at internal IAP regions (see Fig. S3 in the supplemental material). The distinction between the LTR and internal regions is consistent with the finding that repression by macroH2A occurs specifically at the level of transcription initiation and not elongation (17). The change in macroH2A enrichment at the IAP LTR in methylated and demethylated cells may be greater than what we measured, because the IAPs are not fully demethylated in Dnmt1c/c cells, full background corrections cannot be estimated, and fixation of chromatin may have reduced the number of epitopes available for ChIP.
Together, our results indicate that histone macroH2A associates with the IAP LTR in a methylation-dependent manner. The change in macroH2A association with the IAP LTR parallels the change in IAP transcription and methylation (Fig. 3 and 4). We propose that macroH2A is involved in the transcriptional repression of methylated DNA.
The DNMT1C1229S protein mislocalizes to pericentric heterochromatin.
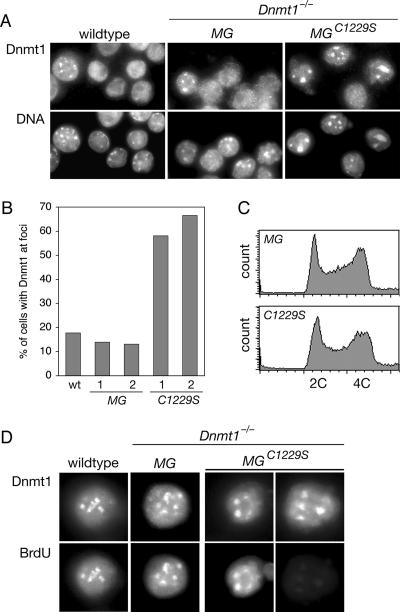
DNMT1 normally localizes at DNA replication foci during S phase, and the foci coincide with the pericentric heterochromatin in 15 to 20% of cells in asynchronous culture (36). When we performed anti-DNMT1 immunofluorescence in the panel of cell lines, we observed DNMT1 foci at pericentric heterochromatin in a large fraction of Dnmt1−/− MGC1229S cells compared to wild-type and Dnmt1−/− MG cells (Fig. 5A). Approximately 60% of Dnmt1−/− MGC1229S cells had DNMT1 foci at pericentric heterochromatin, in contrast to the expected ∼15% of wild-type and Dnmt1−/− MG cells (Fig. 5B).
FIG. 5.
DNMT1C1229S is mislocalized to pericentric heterochromatin. (A) Immunofluorescence with anti-DNMT1 antibody. Representative fields indicate the higher fraction of Dnmt1−/− MGC1229S cells with DNMT1 foci. The regions of intense Hoechst DNA staining correspond to pericentric heterochromatin. Both clones of Dnmt1−/− MG and all four clones of Dnmt1−/− MGC1229S showed the same result (not shown). (B) The percentage of cells with DNMT1 foci was determined in wild-type (wt) cells and in cells from two clones each of Dnmt1−/− MG (“MG”) and Dnmt1−/− MGC1229S (“C1229S”). Approximately 300 to 400 cells were counted for each sample. (C) Cell cycle distribution of asynchronous culture as determined by flow cytometry. Cells were fixed in ethanol and treated with RNase A, and DNA was stained with propidium iodide. The wild type, Dnmt1−/−, both clones of Dnmt1−/− MG (“MG”), and all four clones of Dnmt1−/− MGC1229S (“C1229S”) showed the same distribution (not shown). (D) Costaining of DNMT1 and BrdU after a 15-min pulse with BrdU. All wild-type and Dnmt1−/− MG cells with DNMT1 foci were BrdU positive. In Dnmt1−/− MGC1229S cells, some cells with DNMT1 foci were BrdU positive, and some were BrdU negative.
We determined that the large fraction of cells with DNMT1C1229S foci was due specifically to the mislocalization of DNMT1C1229S. First, the cell cycle distribution of each cell line was assessed by flow cytometry. The distributions were comparable in all cell lines, including wild-type, Dnmt1−/−, Dnmt1−/− MG, and Dnmt1−/− MGC1229S cells (Fig. 5C and data not shown). Thus, the large fraction of cells with DNMT1C1229S foci could not be explained by an abnormally high fraction of cells in S phase. Second, cells were pulsed with BrdU to mark sites of active DNA replication and then costained with antibodies to BrdU and DNMT1. In wild-type and Dnmt1−/− MG cells, all of the cells with DNMT1 foci stained positive for BrdU. In contrast, some Dnmt1−/− MGC1229S cells with DNMT1C1229S foci were BrdU positive, and others were BrdU negative (Fig. 5D), which indicated that the localization of DNMT1C1229S to pericentric heterochromatin did not require the coincident replication of that compartment of the genome. Together, these data indicate that DNMT1C1229S is mislocalized to the pericentric heterochromatin and that the proper localization of DNMT1 depends on normal patterns of genomic methylation.
DISCUSSION
Functions of the amino-terminal domain of DNMT1.
We separated the methyltransferase activity of DNMT1 from its other putative functions by introducing a conservative point mutation into the key catalytic residue. This genetic approach was necessary because demethylating drugs such as 5-aza-2′-deoxycytidine and zebularine cause the degradation of the DNMT1 protein (11, 26). All of the examined phenotypes of Dnmt1−/− were also observed in Dnmt1−/− MGC1229S cells, which demonstrates that low genomic methylation is responsible for these phenotypes. The demethylation of IAP elements was sufficient for their transcriptional activation, even in the presence of DNMT1C1229S. The lethal differentiation phenotype and the mislocalization of histone macroH2A were observed in both Dnmt1−/− and Dnmt1−/− MGC1229S cells. Together, these data indicate that the essential functions of DNMT1 are exerted through its methyltransferase activity and that the reported interactions of DNMT1 with other factors do not constitute a major source of phenotype in cells null for Dnmt1. These findings do not exclude the possibility that the amino terminus of DNMT1 acts as a transcriptional repressor under some conditions. However, such additional functions would be secondary to and dependent on the methyltransferase activity. The in vivo functional significance of any repressor activity of DNMT1 remains to be established.
Transcriptional repression of methylated DNA.
Many factors have been proposed to be global repressors of methylated DNA, but significant changes in gene expression have not been detected in the corresponding mutant mice. MeCP2 encodes a putative methyl-CpG binding protein, and its mutation causes Rett syndrome in humans. Mice with mutations in MeCP2 exhibited Rett-like symptoms but no detectable changes in gene expression and no reactivation of methylated promoters (57). Mice with homozygous mutations in MBD2 are healthy and fertile, but females have minor defects in maternal behavior. No activation of imprinted genes or retroviruses (including IAP elements) was observed in MBD2−/− mice (29). Mice with homozygous mutations in MBD1 are healthy and fertile, with minor defects in adult neurogenesis. No changes in gene expression were observed in MBD1−/− mice except for minor IAP transcription in adult neural stem cells (66). Mice with homozygous mutations in Kaiso are healthy and fertile, with no observed changes in gene expression (45).
In light of the studies that have indicated that there is an overlap between DNA methylation and histone macroH2A, we hypothesized that the repression of methylated DNA involves macroH2A. We have shown that macroH2A associates with IAP elements when they are methylated and silenced and that this association is reduced when IAPs are demethylated and actively transcribed. However, mice with homozygous mutations in histone macroH2A1 were recently shown to be viable and fertile, with no obvious phenotypes or defects in X inactivation in the liver (7). Currently, the mechanism by which DNA methylation represses transcription remains essentially unknown.
Mechanisms that might destabilize genomic methylation patterns.
By generating cells that expressed only DNMT1C1229S, we were able to observe DNMT1 localization in the context of a demethylated genome. DNMT1C1229S is mislocalized to pericentric heterochromatin in demethylated cells, and we interpret this result to mean that the proper localization of DNMT1 requires normal genomic methylation patterns. Pericentric heterochromatin is still partially methylated in Dnmt1−/− cells due to the presence of DNMT3B (43), and this methylation might recruit DNMT1C1229S. Similarly, the mislocalization of histone macroH2A to pericentric heterochromatin in Dnmt1−/− and Dnmt1−/− MGC1229S cells might be explained by residual methylation in that compartment. Consistent with this model, an MBD fused to green fluorescent protein was located at pericentric heterochromatin in wild-type and Dnmt1−/− mouse ES cells but not in Dnmt1−/− Dnmt3a−/− Dnmt3b−/− ES cells (54).
The recruitment of DNMT1 to methylated compartments may promote the maintenance of methylated sequences, and the exclusion of DNMT1 from unmethylated compartments may reduce spurious de novo activity. Notably, the de novo activity of DNMT1 (assessed in Dnmt3a−/− Dnmt3b−/− cells) required preexisting methylation (39). Other factors that may contribute to the fidelity of DNA methylation patterns include the 5- to 30-fold preference of DNMT1 for hemimethylated substrates, the concentration of DNMT1 at replication foci, the partial redundancy and/or cooperation of DNMT1 with DNMT3a and DNMT3b, histone modifications, and DNA sequence. None of these factors alone can account for the >99% fidelity of inheritance of methylation patterns.
Abnormal DNA methylation patterns in cancer have not been explained by changes in DNA methyltransferase expression. An extensive analysis of ovarian tumors found no correlation between DNMT1 expression and locus-specific hypermethylation or global hypomethylation (21). The coexistence of hypermethylation and hypomethylation in many tumors underscores the complexity of the epigenetic dysregulation. Our results suggest that altered genomic methylation patterns in tumor cells may partly be caused or propagated by the mislocalization of DNA methyltransferases. An understanding of the mechanisms that underlie epigenetic dysregulation in cancer will be critical for the design of effective biomarkers and therapeutics.
Supplementary Material
Acknowledgments
We are grateful to R. Jaenisch for the DNMT1 minigene and to C.-S. Lin for reagents. We thank members of the Bestor laboratory for discussions and assistance; J. Pehrson for helpful discussions; A. Brodsky, G. Adelmant, and K. Tucker for advice; and M. Goll, A. O'Donnell, S. Ooi, and C. Schaefer for comments on the manuscript.
This work was supported by a fellowship from the Damon Runyon Cancer Research Foundation to M.D. and by grants from the NIH to T.H.B.
Footnotes
Published ahead of print on 19 March 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Angelov, D., A. Molla, P. Y. Perche, F. Hans, J. Cote, S. Khochbin, P. Bouvet, and S. Dimitrov. 2003. The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol. Cell 11:1033-1041. [DOI] [PubMed] [Google Scholar]
- 2.Bacolla, A., S. Pradhan, J. E. Larson, R. J. Roberts, and R. D. Wells. 2001. Recombinant human DNA (cytosine-5) methyltransferase. III. Allosteric control, reaction order, and influence of plasmid topology and triplet repeat length on methylation of the fragile X CGG·CCG sequence. J. Biol. Chem. 276:18605-18613. [DOI] [PubMed] [Google Scholar]
- 3.Baylin, S., and T. H. Bestor. 2002. Altered methylation patterns in cancer cell genomes: cause or consequence? Cancer Cell 1:299-305. [DOI] [PubMed] [Google Scholar]
- 4.Bestor, T. H. 1992. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 11:2611-2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bestor, T. H. 2003. Cytosine methylation mediates sexual conflict. Trends Genet. 19:185-190. [DOI] [PubMed] [Google Scholar]
- 6.Chan, M. F., R. van Amerongen, T. Nijjar, E. Cuppen, P. A. Jones, and P. W. Laird. 2001. Reduced rates of gene loss, gene silencing, and gene mutation in Dnmt1-deficient embryonic stem cells. Mol. Cell. Biol. 21:7587-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Changolkar, L. N., C. Costanzi, N. A. Leu, D. Chen, K. J. McLaughlin, and J. R. Pehrson. 2007. Developmental changes in histone macroH2A1-mediated gene regulation. Mol. Cell. Biol. 27:2758-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Changolkar, L. N., and J. R. Pehrson. 2006. macroH2A1 histone variants are depleted on active genes but concentrated on the inactive X chromosome. Mol. Cell. Biol. 26:4410-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, T., S. Hevi, F. Gay, N. Tsujimoto, T. He, B. Zhang, Y. Ueda, and E. Li. 2007. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 39:391-396. [DOI] [PubMed] [Google Scholar]
- 10.Chen, T., and E. Li. 2006. Establishment and maintenance of DNA methylation patterns in mammals. Curr. Top. Microbiol. Immunol. 301:179-201. [DOI] [PubMed] [Google Scholar]
- 11.Cheng, J. C., C. B. Yoo, D. J. Weisenberger, J. Chuang, C. Wozniak, G. Liang, V. E. Marquez, S. Greer, T. F. Orntoft, T. Thykjaer, and P. A. Jones. 2004. Preferential response of cancer cells to zebularine. Cancer Cell 6:151-158. [DOI] [PubMed] [Google Scholar]
- 12.Choo, J. H., J. D. Kim, J. H. Chung, L. Stubbs, and J. Kim. 2006. Allele-specific deposition of macroH2A1 in imprinting control regions. Hum. Mol. Genet. 15:717-724. [DOI] [PubMed] [Google Scholar]
- 13.Choo, K. H. 1997. The centromere. Oxford University Press, Oxford, United Kingdom.
- 14.Christy, R. J., A. R. Brown, B. B. Gourlie, and R. C. Huang. 1985. Nucleotide sequences of murine intracisternal A-particle gene LTRs have extensive variability within the R region. Nucleic Acids Res. 13:289-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costanzi, C., and J. R. Pehrson. 1998. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393:599-601. [DOI] [PubMed] [Google Scholar]
- 16.Ding, F., and J. R. Chaillet. 2002. In vivo stabilization of the Dnmt1 (cytosine-5)-methyltransferase protein. Proc. Natl. Acad. Sci. USA 99:14861-14866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doyen, C. M., W. An, D. Angelov, V. Bondarenko, F. Mietton, V. M. Studitsky, A. Hamiche, R. G. Roeder, P. Bouvet, and S. Dimitrov. 2006. Mechanism of polymerase II transcription repression by the histone variant macroH2A. Mol. Cell. Biol. 26:1156-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eden, A., F. Gaudet, A. Waghmare, and R. Jaenisch. 2003. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 300:455. [DOI] [PubMed] [Google Scholar]
- 19.Egger, G., S. Jeong, S. G. Escobar, C. C. Cortez, T. W. Li, Y. Saito, C. B. Yoo, P. A. Jones, and G. Liang. 2006. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc. Natl. Acad. Sci. USA 103:14080-14085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ehrlich, M. 2002. DNA methylation in cancer: too much, but also too little. Oncogene 21:5400-5413. [DOI] [PubMed] [Google Scholar]
- 21.Ehrlich, M., C. B. Woods, M. C. Yu, L. Dubeau, F. Yang, M. Campan, D. J. Weisenberger, T. Long, B. Youn, E. S. Fiala, and P. W. Laird. 2006. Quantitative analysis of associations between DNA hypermethylation, hypomethylation, and DNMT RNA levels in ovarian tumors. Oncogene 25:2636-2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fatemi, M., A. Hermann, S. Pradhan, and A. Jeltsch. 2001. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J. Mol. Biol. 309:1189-1199. [DOI] [PubMed] [Google Scholar]
- 23.Fuks, F., W. A. Burgers, A. Brehm, L. Hughes-Davies, and T. Kouzarides. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 24:88-91. [DOI] [PubMed] [Google Scholar]
- 24.Fuks, F., P. J. Hurd, R. Deplus, and T. Kouzarides. 2003. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31:2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gaudet, F., J. G. Hodgson, A. Eden, L. Jackson-Grusby, J. Dausman, J. W. Gray, H. Leonhardt, and R. Jaenisch. 2003. Induction of tumors in mice by genomic hypomethylation. Science 300:489-492. [DOI] [PubMed] [Google Scholar]
- 26.Ghoshal, K., J. Datta, S. Majumder, S. Bai, H. Kutay, T. Motiwala, and S. T. Jacob. 2005. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell. Biol. 25:4727-4741. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Goll, M. G., and T. H. Bestor. 2005. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74:481-514. [DOI] [PubMed] [Google Scholar]
- 28.Guo, G., W. Wang, and A. Bradley. 2004. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature 429:891-895. [DOI] [PubMed] [Google Scholar]
- 29.Hendrich, B., J. Guy, B. Ramsahoye, V. A. Wilson, and A. Bird. 2001. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 15:710-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez, L., S. Kozlov, G. Piras, and C. L. Stewart. 2003. Paternal and maternal genomes confer opposite effects on proliferation, cell-cycle length, senescence, and tumor formation. Proc. Natl. Acad. Sci. USA 100:13344-13349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holm, T. M., L. Jackson-Grusby, T. Brambrink, Y. Yamada, W. M. Rideout III, and R. Jaenisch. 2005. Global loss of imprinting leads to widespread tumorigenesis in adult mice. Cancer Cell 8:275-285. [DOI] [PubMed] [Google Scholar]
- 32.Hsieh, C. L. 1999. In vivo activity of murine de novo methyltransferases, Dnmt3a and Dnmt3b. Mol. Cell. Biol. 19:8211-8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, M., B. N. Trinh, T. I. Long, S. Oghamian, and P. W. Laird. 2004. Dnmt1 deficiency leads to enhanced microsatellite instability in mouse embryonic stem cells. Nucleic Acids Res. 32:5742-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuff, E. L., and K. K. Lueders. 1988. The intracisternal A-particle gene family: structure and functional aspects. Adv. Cancer Res. 51:183-276. [DOI] [PubMed] [Google Scholar]
- 35.Lei, H., S. P. Oh, M. Okano, R. Juttermann, K. A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122:3195-3205. [DOI] [PubMed] [Google Scholar]
- 36.Leonhardt, H., A. W. Page, H. U. Weier, and T. H. Bestor. 1992. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell 71:865-873. [DOI] [PubMed] [Google Scholar]
- 37.Li, E., C. Beard, and R. Jaenisch. 1993. Role for DNA methylation in genomic imprinting. Nature 366:362-365. [DOI] [PubMed] [Google Scholar]
- 38.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69:915-926. [DOI] [PubMed] [Google Scholar]
- 39.Lorincz, M. C., D. Schübeler, S. R. Hutchinson, D. R. Dickerson, and M. Groudine. 2002. DNA methylation density influences the stability of an epigenetic imprint and Dnmt3a/b-independent de novo methylation. Mol. Cell. Biol. 22:7572-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma, Y., S. B. Jacobs, L. Jackson-Grusby, M. A. Mastrangelo, J. A. Torres-Betancourt, R. Jaenisch, and T. P. Rasmussen. 2005. DNA CpG hypomethylation induces heterochromatin reorganization involving the histone variant macroH2A. J. Cell Sci. 118:1607-1616. [DOI] [PubMed] [Google Scholar]
- 41.Mi, S., and R. J. Roberts. 1993. The DNA binding affinity of HhaI methylase is increased by a single amino acid substitution in the catalytic center. Nucleic Acids Res. 21:2459-2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mietz, J. A., Z. Grossman, K. K. Lueders, and E. L. Kuff. 1987. Nucleotide sequence of a complete mouse intracisternal A-particle genome: relationship to known aspects of particle assembly and function. J. Virol. 61:3020-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 44.Pehrson, J. R., and V. A. Fried. 1992. MacroH2A, a core histone containing a large nonhistone region. Science 257:1398-1400. [DOI] [PubMed] [Google Scholar]
- 45.Prokhortchouk, A., O. Sansom, J. Selfridge, I. M. Caballero, S. Salozhin, D. Aithozhina, L. Cerchietti, F. G. Meng, L. H. Augenlicht, J. M. Mariadason, B. Hendrich, A. Melnick, E. Prokhortchouk, A. Clarke, and A. Bird. 2006. Kaiso-deficient mice show resistance to intestinal cancer. Mol. Cell. Biol. 26:199-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rhee, I., K. W. Jair, R. W. Yen, C. Lengauer, J. G. Herman, K. W. Kinzler, B. Vogelstein, S. B. Baylin, and K. E. Schuebel. 2000. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature 404:1003-1007. [DOI] [PubMed] [Google Scholar]
- 47.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 48.Rollins, R. A., M. Damelin, and T. H. Bestor. 2006. Inheritance of genomic methylation patterns, p. 141-153. In M. L. DePamphilis (ed.), DNA replication and human disease. CSHL Press, New York, NY.
- 49.Rollins, R. A., F. Haghighi, J. R. Edwards, R. Das, M. Q. Zhang, J. Ju, and T. H. Bestor. 2006. Large-scale structure of genomic methylation patterns. Genome Res. 16:157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 51.Sakatani, T., A. Kaneda, C. A. Iacobuzio-Donahue, M. G. Carter, S. de Boom Witzel, H. Okano, M. S. Ko, R. Ohlsson, D. L. Longo, and A. P. Feinberg. 2005. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science 307:1976-1978. [DOI] [PubMed] [Google Scholar]
- 52.Smith, A. 1991. Culture and differentiation of embryonic stem cells. J. Tiss. Cult. Methods 13:89-94. [Google Scholar]
- 53.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 54.Tsumura, A., T. Hayakawa, Y. Kumaki, S. Takebayashi, M. Sakaue, C. Matsuoka, K. Shimotohno, F. Ishikawa, E. Li, H. R. Ueda, J. Nakayama, and M. Okano. 2006. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11:805-814. [DOI] [PubMed] [Google Scholar]
- 55.Tucker, K. L., C. Beard, J. Dausmann, L. Jackson-Grusby, P. W. Laird, H. Lei, E. Li, and R. Jaenisch. 1996. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes Dev. 10:1008-1020. [DOI] [PubMed] [Google Scholar]
- 56.Tucker, K. L., D. Talbot, M. A. Lee, H. Leonhardt, and R. Jaenisch. 1996. Complementation of methylation deficiency in embryonic stem cells by a DNA methyltransferase minigene. Proc. Natl. Acad. Sci. USA 93:12920-12925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tudor, M., S. Akbarian, R. Z. Chen, and R. Jaenisch. 2002. Transcriptional profiling of a mouse model for Rett syndrome reveals subtle transcriptional changes in the brain. Proc. Natl. Acad. Sci. USA 99:15536-15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vire, E., C. Brenner, R. Deplus, L. Blanchon, M. Fraga, C. Didelot, L. Morey, A. Van Eynde, D. Bernard, J. M. Vanderwinden, M. Bollen, M. Esteller, L. Di Croce, Y. de Launoit, and F. Fuks. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439:871-874. [DOI] [PubMed] [Google Scholar]
- 59.Walsh, C. P., J. R. Chaillet, and T. H. Bestor. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20:116-117. [DOI] [PubMed] [Google Scholar]
- 60.Wang, K. Y., and C. K. J. Shen. 2004. DNA methyltransferase Dnmt1 and mismatch repair. Oncogene 23:7898-7902. [DOI] [PubMed] [Google Scholar]
- 61.Wilke, K., E. Rauhut, M. Noyer-Weidner, R. Lauster, B. Pawlek, B. Behrens, and T. A. Trautner. 1988. Sequential order of target-recognizing domains in multispecific DNA-methyltransferases. EMBO J. 7:2601-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wyszynski, M. W., S. Gabbara, E. A. Kubareva, E. A. Romanova, T. S. Oretskaya, E. S. Gromova, Z. A. Shabarova, and A. S. Bhagwat. 1993. The cysteine conserved among DNA cytosine methylases is required for methyl transfer, but not for specific DNA binding. Nucleic Acids Res. 21:295-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu, G.-L., T. H. Bestor, D. Bourc'his, C.-L. Hsieh, N. Tommerup, M. Bugge, M. Hulten, X. Qu, J. J. Russo, and E. Viegas-Péquignot. 1999. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402:187-191. [DOI] [PubMed] [Google Scholar]
- 64.Yamada, Y., L. Jackson-Grusby, H. Linhart, A. Meissner, A. Eden, H. Lin, and R. Jaenisch. 2005. Opposing effects of DNA hypomethylation on intestinal and liver carcinogenesis. Proc. Natl. Acad. Sci. USA 102:13580-13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ymer, S., W. Q. Tucker, H. D. Campbell, and I. G. Young. 1986. Nucleotide sequence of the intracisternal A-particle genome inserted 5′ to the interleukin-3 gene of the leukemia cell line WEHI-3B. Nucleic Acids Res. 14:5901-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhao, X., T. Ueba, B. R. Christie, B. Barkho, M. J. McConnell, K. Nakashima, E. S. Lein, B. D. Eadie, A. R. Willhoite, A. R. Muotri, R. G. Summers, J. Chun, K. F. Lee, and F. H. Gage. 2003. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc. Natl. Acad. Sci. USA 100:6777-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.