Abstract
Hsmar1, one of the two subfamilies of mariner transposons in humans, is an ancient element that entered the primate genome lineage ∼50 million years ago. Although Hsmar1 elements are inactive due to mutational damage, one particular copy of the transposase gene has apparently been under selection. This transposase coding region is part of the SETMAR gene, in which a histone methylatransferase SET domain is fused to an Hsmar1 transposase domain. A phylogenetic approach was taken to reconstruct the ancestral Hsmar1 transposase gene, which we named Hsmar1-Ra. The Hsmar1-Ra transposase efficiently mobilizes Hsmar1 transposons by a cut-and-paste mechanism in human cells and zebra fish embryos. Hsmar1-Ra can also mobilize short inverted-repeat transposable elements (MITEs) related to Hsmar1 (MiHsmar1), thereby establishing a functional relationship between an Hsmar1 transposase source and these MITEs. MiHsmar1 excision is 2 orders of magnitude more efficient than that of long elements, thus providing an explanation for their high copy numbers. We show that the SETMAR protein binds and introduces single-strand nicks into Hsmar1 inverted-repeat sequences in vitro. Pathway choices for DNA break repair were found to be characteristically different in response to transposon cleavage mediated by Hsmar1-Ra and SETMAR in vivo. Whereas nonhomologous end joining plays a dominant role in repairing excision sites generated by the Hsmar1-Ra transposase, DNA repair following cleavage by SETMAR predominantly follows a homology-dependent pathway. The novel transposon system can be a useful tool for genome manipulations in vertebrates and for investigations into the transpositional dynamics and the contributions of these elements to primate genome evolution.
mariner elements make up a diverse family of eukaryotic DNA transposons present in a wide variety of genomes, including that of humans (references 6, 41, and 43 and references therein). These transposons contain a single gene encoding the transposase flanked by short, <30-bp terminal inverted-repeat (IR) sequences. mariner elements mobilize through a cut-and-paste mechanism catalyzed by the transposase, which belongs to a large family of recombinase proteins, including retroviral/retrotransposon integrases and transposases, characterized by the DDE/D signature in the catalytic domain of the proteins (40). Transposition results in the accumulation of hundreds or thousands of transposon copies over evolutionary time. However, most mariner copies appear to be dead remnants of once-active transposons inactivated by mutations (14). To date, only three mariner elements out of the hundreds of sequences that have been described have proven to be active. Two of these, Mos1 and Famar1, are natural elements isolated from the genomes of Drosophila mauritiana (20) and the earwig Forficula auricularia (3), respectively. The active Himar1 element is a majority-rule consensus of cloned genomic copies obtained from the horn fly Haematobia irritans (24). Mos1 and Himar1 have been used as molecular tools for genome manipulations in diverse species (reviewed in reference 40). However, the utility of these invertebrate mariner transposons in mammalian genetics is hindered by their limited activity in mammalian cells (12).
mariner elements are represented by two subfamilies in the human genome: Hsmar1 (44) and Hsmar2 (43). The first Hsmar1 element entered the primate genome lineage approximately 50 million years ago, and transposition was ongoing until at least 37 million years ago, producing about 200 Hsmar1 copies (44) (Fig. 1A). However, none of the present copies encodes a functional transposase protein, due to mutational inactivation. The Hsmar1 transposon copies are accompanied by about 4,500 copies of solo IRs (containing a single IR) and about 2,500 copies of an Hsmar1-related paired-IR element, MiHsmar1 (15, 44) (Fig. 1A). Such miniature IR transposable elements (MITEs) are thought to have been generated by internal deletions of longer transposons; they make up the predominant fraction of DNA elements in flowering plants and are often found in animal genomes (9). Even though the classical MITEs are longer than the MiHsmar1 elements, the common characteristics that link all of these elements are the following: significantly smaller size than the corresponding precursor element, significantly higher copy number than the corresponding precursor element, and lack of transposase-coding capacity. It is widely believed that MITEs can be mobilized only by transposases supplied in trans (9), but only one such instance has been documented at the molecular level (7). Although mechanisms of preferential transposition due to small size (25, 44) and transposition linked to the cellular process of DNA replication (10, 17) have been suggested, the mechanisms of MITE mobilization and amplification are incompletely understood.
FIG. 1.
Structure of Hsmar1 transposons in the human genome and transpositional activities of consensus and reconstructed ancestral transposases. (A) Summary of the structures and copy numbers of Hsmar1-derived sequences. The arrows represent IRs. SETMAR is a chimeric protein made up of a histone methyltransferase (SET) domain and a particularly conserved transposase (Tpase) domain. (B) The inset summarizes the principles of a transposition assay in tissue culture. The transposon containing a selectable antibiotic resistance gene (neo) is transfected either with or without a transposase- expressing helper plasmid. Transfected cells are placed under antibiotic selection. The dramatic increase in the number of resistant cell colonies in the presence of transposase is the result of transposition of the element from the plasmid vector into chromosomes. The diagram shows the transpositional activity of Hsmar1-Ra in human HeLa cells. The pHsmar1-neo donor plasmid was cotransfected with pCMV-Hsmar1 consensus, pCMV-SB (expressing the Sleeping Beauty transposase), and pCMV-Hsmar1-Ra. The efficiency of transgene integration was determined by counting G418-resistant colonies. The numbers on the y axis represent mean values obtained from at least three independent experiments. (C) Alignment of mariner transposase amino acid sequences of the cecropia subfamily in the region of the four predicted amino acid substitutions (asterisks). The alignment revealed that the changes predicted in silico correspond to the majority-rule consensus amino acid sequence at these positions. (D) In vitro DNA-binding activity of the Hsmar1-Ra transposase. A radioactively labeled DNA fragment of 83 bp containing the 5′ IR of the Hsmar1 transposon (depicted below) was incubated with the purified MBP-Hsmar1-Ra protein, and the DNA-protein complexes were visualized by EMSA autoradiography. Compared to free, unbound probe (lane 1), increasing concentrations of MBP-Hsmar1-Ra (lanes 2 to 4) produced DNA-protein complexes. Complex assembly was challenged with cold (unlabeled) probe as a specific competitor (lanes 5 to 7), or increasing amounts of pBluescript were used as nonspecific competitor DNA (lanes 8 to 10).
Despite their parasitic nature, there is increasing evidence that transposable elements are a powerful force in gene evolution. Indeed, about 50 human genes are derived from transposable elements (15), among them genes that are responsible for immunoglobulin gene recombination in all vertebrates (23). One of these “domesticated” transposase-derived genes is SETMAR, a fusion gene containing an N-terminal SET domain fused in frame to an Hsmar1 transposase (44). The SETMAR gene has apparently been under selection; the transposase open reading frame is conserved and shows only 2.4% divergence from a consensus Hsmar1 transposase gene sequence (versus 8% average divergence between Hsmar1 transposase genes) (44). SETMAR is broadly expressed in humans (30), suggesting a housekeeping function. The SET domain can be found in histone methyltransferases that regulate gene expression by chromatin modifications (21). The SETMAR protein has been shown to have histone H3 methyltransferase activity in vitro and has been proposed to play a role in DNA double-strand break (DSB) repair (30). Both the transposase domain of SETMAR and the full-length SETMAR protein were shown to bind to Hsmar1 IR sequences (5, 32), and it was recently demonstrated that SETMAR can perform transposition reactions using precleaved transposon substrates in vitro (32).
We used a computational approach to reconstruct the ancient sequence of an originally active Hsmar1 transposase gene, which was named Hsmar1-Ra (for Hsmar1-reconstructed ancestral). The reconstructed transposase and the Hsmar1 IRs constitute the first active vertebrate mariner transposon system. We found that the Hsmar1-Ra transposase efficiently mobilizes Hsmar1 transposons in mammalian cultured cells and in the zebra fish embryo with a cut-and-paste mechanism. Hsmar1-Ra can also excise MiHsmar1 elements, providing evidence for a functional relationship between these elements and an Hsmar1 transposase source. We also report that SETMAR is capable of introducing nicks at the 5′ ends of Hsmar1 transposon DNA in vitro. In line with these observations, we show that SETMAR triggers homology-dependent DSB repair events at the Hsmar1 transposon ends in human and rodent cells in vivo.
MATERIALS AND METHODS
Phylogenetic analysis and ancestral-sequence reconstruction.
We collected Hsmar1-related elements from the human genome by performing a TBLASTN (2) search for potential homologues of the Hsmar1 consensus sequence (44) in the translated chromosomal sequences of Homo sapiens at an E-value cutoff of 10−5. Only hits with the following criteria were retained for further analysis: (i) alignment covering at least 90% of the query sequence, (ii) less than 10% gaps in the alignment, and (iii) sequence identity of more than 30%. The resulting 51 sequences, along with those of the cecropia transposase subfamily (Bombyx mori mar2 [Bmmar2], D88671; Heterorhabditis bacteriophora mar1 [Hbmar1], U68392; Dugesia tigrina mar1 [Dtmar1], X71979; Hyalophora cecropia mar1 [Hcmar1] [31]; and Cecropius sp. mar1 [26]) were aligned by T-Coffee (37). The phylogeny of these sequences was reconstructed using the maximum likelihood method implemented in PhyML 2.4 with a JTT amino acid substitution model. We allowed for site-specific substitution rate heterogeneity by using a discrete gamma model with eight rate categories and the gamma shape parameter estimated from the data. The resulting phylogenetic tree was rooted using the invertebrate mariner elements as outgroups and was visualized by TreeView (38) (see Fig. S1 in the supplemental material). Protein sequences at ancestral nodes were inferred by PAML 3.14b (51) using maximum likelihood marginal reconstruction. The evaluations of the reconstructed ancestral amino acid states at the node connecting the cecropia mariner transposons with the branch leading to the human or chimpanzee sequences (see Fig. S1 in the supplemental material) predicted the following three amino acid substitutions in the consensus transposase protein sequence with posterior probabilities of ≥0.9: C53R, L201V, and A219C. The probabilities for the P167S substitution predicted from the human and the chimpanzee trees were 0.9 and 0.7, respectively. Other Hsmar1-like sequences for alignments were Hydra littoralis mar1 (AAB61385), acrobat ant mar 29.3 (42), and Bos taurus mar1 (Btmar1) (6). The Ornithorhynchus anatinus mar1 (Oamar1) sequence found in public databases is available upon request.
Plasmids.
The Klenow-filled EcoRI/BamHI fragment of pRc/CMV (Invitrogen), containing the simian virus 40 promoter-neo-poly(A) sequences, was cloned into the SmaI site of pUC19 to gain pNeo. Hsmar1 transposon sequences were assembled from 40- to 60-nucleotide-long oligonucleotides. Briefly, for the assembly of the left IR and the 5′ untranslated sequences of the consensus Hsmar1 transposon, 6 pmol (each) of the primers L1 to L8 was used in a PCR in a total volume of 50 μl with Pwo polymerase (Roche) with the following PCR program: 94°C for 1 min; 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 30 s; and 72°C for 2 min. The 3′ untranslated sequences and the right IR of the Hsmar1 transposon were assembled from the oligonucleotides R1 and R2 using PCR conditions similar to those described above. The PCR for the left end of the transposon was diluted 200-fold, and 1 μl of the dilution was used in a 100-μl PCR mixture with 15 pmol (each) of the primers L1 and L8. The PCR program was 94°C for 1 min; 25 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 30 s; and 72°C for 2 min. The fragments were then cloned into the EcoRI/KpnI (5′ IR and untranslated region [UTR]) and XbaI/HindIII (3′ UTR and IR) sites of pNeo, resulting in pHsmar1-neo. pHsmar1-zeo was obtained by cloning the Klenow-filled XbaI-BspHI fragment of pUT-SVZeo (Invitrogen) into the blunted KpnI/XbaI site of pHsmar1-neo.
The Hsmar1-like transposase coding sequence was obtained from chimpanzee genomic DNA with an open reading frame-trapping strategy as described previously (36) and cloned into the NotI/SpeI sites of pBS (Stratagene) to serve as the template for a series of PCR mutagenesis reactions performed with the QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene). The final transposase gene was cloned into the NheI/BamHI site of pEGFP-C2 (Clontech) to form pCMV-Hsmar1-Ra. The same fragment cloned into the KpnI/XbaI sites of pHsmar1-neo resulted in pHsmar1-Full.
The primers Amp-MITEFw and Amp-MITERev were used to create a cloning site within the beta lactamase (bla) gene in a zeo-containing pUC19-derived plasmid. The obtained fragment served as the donor backbone for all transposons tested in excision experiments with the bla gene. The consensus MiHsmar1 sequence (44) was assembled by annealing 0.5 nmol of each of the Hsmar1 MITEFw and Hsmar1 MITERev oligonucleotides in 100 μl 1× New England Biolabs buffer 2, followed by a fill-in reaction with Klenow using 20 μl of the annealed primers. The MITE and the PCR-amplified transposons from pHsmar1-neo and pHsmar1-Full (primer, Hsmar1IR) were inserted in the bla gene, deriving pAmpMITE, pAmp-neo, and pAmpFull, respectively. To obtain the longer IRs found in MiHsmar1 elements, pAmp-neo and pAmpFull were subjected to PCR mutagenesis with the primers HsmarLonger1 and -2, using a QuikChange Multi Site-Directed Mutagenesis Kit (Stratagene), yielding pAmp-neoL and pAmpFullL, respectively. The SETMAR gene was PCR amplified from the IMAGp956A05160Q24 cDNA clone (RZPD) and ligated into the BamHI/XbaI sites of pcDNA3.1/Zeo (Invitrogen), resulting in pCMV-SETMAR. All primer sequences are available in Table S1 in the supplemental material.
Cell culture transfection and plasmid rescue.
Cells were cultured, transfected, and selected as described previously (36) with some modifications: 2 μl JET-PEI/-RGD (Qbiogene) was used to transfect 100 ng pCMV-Hsmar1-Ra or pCMV-SB (16) with 500 ng of the transposon donor plasmids. For plasmid rescue, 10 μg of genomic DNA prepared from pooled zeocin (Zeo)-resistant HeLa colonies was digested with NheI, SpeI, and AvrII; precipitated and ligated under dilute conditions; and electroporated into DH10B cells, which were then selected with 50 μg/ml Zeo.
Luciferase reporter assays.
Reporter assays were performed as described previously (50); 150 ng of all reporter constructs (1) and p5′-UTR/Luc were cotransfected into HeLa cells with 100 ng of pCMV-βgal (Clontech) as an internal control for transfection efficiency. Two days posttransfection, luciferase activity was measured in a Lumat LB 9507 luminometer with a 10-second integration period. Luciferase units were normalized with β-galactosidase readouts for transfection efficiency and with the Bradford assay for total protein.
Transposon excision assays.
Hsmar1-Ra mRNA was transcribed from pSK/BG3′u-2a (a gift from Perry Hackett) with the mMessage mMachine kit (Ambion). One-cell-stage zebra fish embryos were injected with a mixture of 50 ng/μl mRNA and 20 ng/μl pHsmar1-neo. After 9 h, pools of 30 embryos were digested with 200 μg/ml proteinase K in 0.5 ml extraction buffer (10 mM Tris-HCl, pH 8.2, 10 mM EDTA, 0.2 M NaCl, 0.5% sodium dodecyl sulfate), and plasmid DNA was purified with a QIAprep Spin Kit (QIAgen). Plasmid purification from cultured cells and the conditions for the nested PCRs have been described previously (18).
For excision from the bla gene, HeLa cells were transfected with 100 ng of pCMV-Hsmar1-Ra and 500 ng of the various transposon donor plasmids. Plasmid DNA was purified from cells 3 days posttransfection by a standard miniprep protocol. After precipitation, the DNA pellet was dissolved in 10 μl water and transformed into Escherichia coli. Ten microliters of the 1-ml bacterial suspension in SOC medium (Super Optimal broth with catabolite repression) was plated on Zeo plates, with the rest on ampicillin (Amp)/Zeo plates. The numbers of the double-resistant colonies were normalized with the total amount of plasmid DNA recovered as follows: (number of Ampr/Zeor foci × 100)/number of Zeor foci.
Maltose-binding protein (MBP)-Hsmar1-Ra and MBP-SETMAR purification.
The SETMAR coding region from the IMAGp956A05160Q24 cDNA clone (RZPD) was cloned into the BamHI/XbaI site of pMAL-c2x (New England Biolabs); in the case of pHsmar1-Ra, the XbaI/HindIII sites were used for cloning. The purification of the proteins was carried out as described previously (32) with minor modifications. The plasmids were transfected into E. coli BL21-CodonPlus-RIL competent cells (Stratagene). The next day, the culture was diluted 100 times to 80 ml and grown to an optical density at 600 nm of 0.5, and protein expression was induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 30°C for 3 h. After being harvested by centrifugation, the pellet was resuspended in 3 ml of HSG buffer (32) and sonicated with a Bandelin Sonopuls sonicator for 3 min (cycle, 50%; 10% power). After centrifugation, 400 μl of the soluble crude extract was mixed with 100 μl of amilose resin (New England Biolabs) and rotated for 30 min at 4°C. The resin was washed seven times with 0.5 ml of HSG buffer containing 0.5 M NaCl and once with 0.5 ml HSG buffer. The proteins were eluted with 80 μl of HSG buffer containing 10 mM maltose.
Electrophoretic mobility shift assay (EMSA).
The 83-bp-long probe comprising the 5′ IR of Hsmar1 was obtained by annealing the primers Hsmar1/Apo1 and Hsmar1/AflII, followed by a Klenow fill-in reaction using [α-P32]dATP (Perkin Elmer); 0.24 pmol of MBP-Hsmar1-Ra or 2.5 pmol of MBP-SETMAR was incubated with 0.3 pmol of probe in a 20-μl binding reaction mixture as described previously (32) for 3 h at room temperature. The binding reaction products were separated on 6% Tris-acetic acid-polyacrylamide gels containing 1% glycerol, dried, exposed to a phosphoscreen for several hours, and visualized by a STORM phosphorimager (Amersham).
Cleavage site determination by linker-mediated PCR.
A 281-bp-long DNA fragment containing the left IR and the complete 5′ untranslated sequences of the Hsmar1 transposon was PCR amplified with the pUC6 and L8 primers using pHsmar1-Full as a template; 0.5 pmol of this DNA fragment was incubated with 1 pmol MBP-Hsmar1-Ra or 25 pmol MBP-SETMAR in a 30-μl reaction mixture containing 20 mM HEPES (pH 7.5), 100 mM NaCl, 250 μg/ml bovine serum albumin, 2 mM dithiothreitol, 10% glycerol, 5% dimethyl sulfoxide, and 6 mM MgCl2 overnight at 37°C, and 0.5 pmol of the probe was digested with EcoRI as a control. The proteins were heat inactivated at 70°C for 10 min and digested with 100 μg of proteinase K in a 100-μl reaction volume for 1 h. After heat inactivation at 95°C for 30 min, the DNA was precipitated with isopropanol in the presence of glycogene and dissolved in 10 μl water. The DNA solution was subsequently mixed with 10 pmol single-stranded linker oligonucleotide (Link1/2), heated to 95°C for 3 min, and chilled on ice before it was completed with buffer, T4 RNA ligase (New England Biolabs), and water to a 15-μl final volume. After ligation overnight at room temperature, the reaction mixture was heat inactivated at 65°C for 15 min and purified with a QIAquick PCR purification Kit (QIAgen). The eluate was split and used for templates for two rounds of PCR; 1 μl of eluate was used for the PCR when the ligation was performed on the EcoRI-digested probe. The primers used in the first round of PCR to detect cleavage of the lower strand were pUC6 and Link3; L8 and Link3 were used to detect upper-strand cleavage sites. The PCR program was 94°C for 30 s and 30 cycles of 94°C for 30s, 55°C for 30 s, and 72°C for 15 s. One microliter (each) of 100-fold-diluted PCR mixtures was used in the nested PCRs, using Link4 and pUC2 primers for the lower strand and Link4 and HsmarRev3 for determining the upper-strand cleavage sites. The PCR program for the nested PCR was 94°C for 30 s and 30 cycles of 94°C for 30 s, 62°C for 30 s, and 72°C for 15 s. The detected PCR products were gel isolated, cloned using the pGEM-T Vector System (Promega), and sequenced.
Nucleotide sequence accession number.
The sequence of the reconstructed ancestral Hsmar1-Ra element can be found under GenBank accession number EF517118.
RESULTS
The consensus Hsmar1 transposase is not active.
We previously reconstructed two functional transposable elements from vertebrate genomes: Sleeping Beauty (SB) from fish (16) and Frog Prince (FP) from amphibians (36). Both gene reconstructions were based on the hypothesis that a consensus sequence established from several cloned inactive copies represents an active gene. Encouraged by our former success, we set out to reconstruct an active Hsmar1 transposable element from the human genome.
We engineered the consensus sequence of the transposase gene (44) by site-directed mutagenesis of 21 codons of an Hsmar1 ortholog obtained from the chimpanzee genome. Transposition activity was assessed in human HeLa cells, using a two-component transposition system similar to those established for SB and FP (16, 36). The assay (Fig. 1B) is based on cotransfection of a donor plasmid carrying a nonautonomous transposon marked with a neomycin resistance gene (neo), with or without the helper plasmid expressing the transposase. Both random plasmid integrations and transposition events into chromosomes give rise to G418-resistant colonies. Following antibiotic selection, transposition efficiency is calculated from the numbers of G418-resistant cell colonies in the presence versus absence of the transposase. There was no indication of Hsmar1 transposition in these experiments (Fig. 1B), suggesting that the consensus of the Hsmar1 transposase gene represents an inactive sequence.
Molecular reconstruction of an ancestral Hsmar1 transposase gene.
The approach to gene sequence prediction based on consensus does not incorporate phylogenetic information. For example, an inactivating mutation in a transposable element may become overrepresented if that particular mutant was preferentially amplified over the active sequence. With the aim of reconstructing the ancient, active Hsmar1 transposase gene that colonized the genome lineage of primates, we applied a statistically rigorous approach based on maximum likelihood that has been successfully used to reconstruct ancestral gene sequences (48).
First, human Hsmar1 transposase-like amino acid sequences were obtained from the human genome by TBLASTN similarity searches. To infer the sequence of the last common ancestor of primate and invertebrate mariner transposases, a likely candidate for an active transposase protein capable of colonizing new hosts, invertebrate mariner transposase sequences of the cecropia subfamily were also included in the phylogenetic analysis (see Materials and Methods). The evaluations of the reconstructed ancestral amino acid states at the node connecting the invertebrate mariner elements with the branch leading to the human sequences (see Fig. S1 in the supplemental material) revealed the following four amino acid substitutions in the known consensus transposase protein sequence with posterior probabilities greater than or equal to 0.9: C53R, P167S, L201V, and A219C. Independent inferences from human nucleotide sequences (as identified by BLASTN) or from chimpanzee amino acid sequences also identified these amino acids as the most likely ancestral states at these sites. Furthermore, inspection of these positions in an alignment of cecropia-type transposase sequences revealed that the predicted substitutions represent conserved amino acids within the subfamily (Fig. 1C), suggesting that these residues may be important for transposase activity.
Next, the putative ancestral Hsmar1 transposase gene was engineered by incorporating the four predicted amino acid substitutions into the framework of the consensus Hsmar1 transposase, and the resulting protein was tested for DNA binding and transposition activities. A fusion protein consisting of the MBP and the reconstructed ancestral Hsmar1 transposase was expressed and purified from E. coli, and its ability to bind Hsmar1 IR sequences was tested in an EMSA. The experiment showed binding of the transposase to a transposon IR probe in a sequence-specific manner (Fig. 1D). The colony-forming assay was used to address the transpositional activity of the reconstructed ancestral transposase in tissue culture cells. Upon cotransfection of the neo-marked Hsmar1 transposon with a vector expressing the modified transposase, a 23-fold increase in the number of antibiotic-resistant colonies was observed (Fig. 1B), suggesting that the resurrected protein efficiently catalyzes transgene integration from the donor plasmids into human chromosomes. Thus, it is likely that the inferred sequence, which was named Hsmar1-Ra, represents or is very similar to the sequence of the ancient mariner element that colonized the genome lineage of primates.
We next sought to establish molecular evidence of Hsmar1 transposition. In the first step of the cut-and-paste transposition of Tc1/mariner elements, the transposase mediates excision of the transposon from its donor site with staggered cuts. The resulting DSB is sensed by host DNA repair mechanisms that seal the broken DNA, predominantly by nonhomologous end joining (NHEJ) (18, 52). NHEJ repair of the transposon excision sites generates a characteristic transposon footprint (a small, transposon-specific, 2- to 4-bp sequence signature), which can be identified with PCR (Fig. 2A), using primers flanking the transposon (18). PCR readily generated a product consistent with transposon excision and subsequent footprint formation in transfections with the Hsmar1-Ra transposase (Fig. 2B, lane 1), but not with the SB transposase (Fig. 2B, lane 2), consistent with specific Hsmar1-Ra-dependent excision events followed by DNA repair. Sequencing of the PCR products revealed that Hsmar1-Ra generates TTA or TAA triplets at the excision site, corresponding to the 5′- and 3′-terminal nucleotides of Hsmar1 transposons (Fig. 2C). This is consistent with footprint formation by the Himar1 (24, 27) and Mos1 (4) mariner transposons, which predominantly generate 3-bp footprints. Similar to footprints generated by SB in mammalian somatic cells (18), a minor fraction of Hsmar1 excision sites contained noncanonical footprints with small 1- to 2-bp deletions (data not shown).
FIG. 2.
Excision and integration of Hsmar1. (A) Transposon excision assay in cultured cells. The two functional components of the transposon system (a transposon and a transposase source) are introduced into cells. The transposase catalyzes transposon excision; Tc1/mariner elements leave behind a 2- to 4-nucleotide-long 3′ overhang at the DSBs. DNA repair rejoins the ends of the broken DNA, and the resulting sequences can be recovered by PCR using primers flanking the excision site. (B) Agarose gels showing PCR products of excision assays obtained from HeLa cells and zebra fish embryos. The appearance of the 260-bp bands in the presence of pCMV-Hsmar1-Ra (lane 1) or Hsmar1-Ra mRNA (lane 3) indicates transposon excision from the pHsmar1-neo donor plasmids, followed by excision site repair. Transfection of pCMV-SB (lane 2) and injection of SB mRNA (lane 4) served as controls. M, 250-bp size marker. (C) Hsmar1 transposon footprints identified from cloned excision PCR products from HeLa cells and zebra fish embryos. The sequence of the donor plasmid is schematically shown above. Flanking TA dinucleotides are in boldface, and footprints are in uppercase letters. (D) Three human chromosomal sequences that served as target sites for Hsmar1 transposons. Duplicated TA target dinucleotides are in boldface. (E) Distribution of endogenous genomic Hsmar1 copies (gray columns) and 47 de novo integrants (black columns) in human chromosomes. Mapping of transposon insertions was done using BLAST on the human genome sequence database at NCBI. The graph shows the genomic distribution of transposon insertions with respect to human genes, nongene regions, and repetitive sequences.
Tc1/mariner elements are able to transpose in organisms other than their natural hosts (40). This phenomenon makes these elements excellent tools for genome manipulation in model organisms. We tested Hsmar1 transposition in zebra fish embryos. Hsmar1-Ra transposase mRNA synthesized in vitro was coinjected with the donor plasmid into fertilized zebra fish oocytes, and the excision PCR assay was performed on DNA isolated from the embryos. The majority of PCR products obtained in the presence of the Hsmar1-Ra transposase mRNA (Fig. 2B, lane 3) contained the canonical TTA/TAA footprints (Fig. 2C), indicative of precise Hsmar1 excision followed by NHEJ repair.
Tc1/mariner elements insert into TA target dinucleotides, which become duplicated and flank the integrated transposons (40). In order to obtain formal molecular proof for cut-and-paste Hsmar1 transposition, 47 integration events were isolated from HeLa cells. All of the transposon insertions occurred at TA dinucleotides (Fig. 2D) scattered on 16 human chromosomes. We found that the chromosomal distributions of the endogenous genomic copies and the de novo integrants overlap and have a bias toward larger chromosomes (Fig. 2E). Forty-four percent of the hits were identified in introns of genes (Fig. 2E), indicating a fairly random genomic distribution similar to that found with SB in human cells (49).
In sum, the results described above indicate that we successfully reactivated the first vertebrate mariner transposon from the human genome. The new transposon system shows high-efficiency transposition into various loci in the human genome and retains its activity in zebra fish embryos.
Hsmar1 can function as an autonomous transposon.
Virtually all Tc1/mariner elements contain UTRs upstream of the initiation codon of the transposase gene, suggesting that they might have functions associated with transposition activity. Indeed, some of these 5′ UTRs harbor eukaryotic promoter motifs (28). However, previous studies did not reveal an internal promoter in the Tc1 element but showed that the elements are transcribed by read-through transcription from Caenorhabditis elegans genes (46).
To test whether the Hsmar1 element has promoter activity, the 180-bp-long 5′ UTR of the transposon was fused to a luciferase reporter gene (Fig. 3A). This plasmid and control reporter constructs were transfected into HeLa cells, and the expression of the reporter gene was followed by enzymatic luciferase assays. The 5′-UTR sequences of Hsmar1 showed pronounced promoter activity (Fig. 3A), indicating that the expression of the transposase gene (and thus transposition) might not depend on external promoters.
FIG. 3.
Hsmar1 can function as an autonomous transposon. A schematic representation of the luciferase reporter construct in p5′ UTR/Luc compared to the structure of an Hsmar1 transposon is depicted. (A) Reporter gene expression in HeLa cells from the plasmids indicated below. The error bars indicate standard deviations. (B) Transposon excision assay as shown in Fig. 2. HeLa cells were transfected with the pHsmar1-neo donor plasmid, together with the plasmids indicated. The presence of the characteristic PCR band indicates that the complete Hsmar1 element (Hsmar1-Full) can autonomously catalyze its own excision from the donor plasmid. M, 250-bp size marker.
To test such a possibility, the Hsmar1-Ra transposase gene was cloned between the transposon IRs, resulting in a transposon whose structure mimics that of the hypothetical ancestor of the full-length element. This element was transfected into HeLa cells, and excision of the complete transposon was tested by the excision PCR assay. The autonomous element produced PCR products comigrating with those produced by the two-component system (Fig. 3B), indicating excision of the element from its donor plasmid. These results suggest that the full-length Hsmar1 element can function as an autonomous transposon.
Excision of MiHsmar1.
The human genome contains about 2,500 copies of an Hsmar1-related MITE (Fig. 1A). The MiHsmar1 elements have a consensus sequence of 80 bp containing 37-bp IRs, the first 30 bp of which are identical to the IRs of Hsmar1 (44). The copy number of MiHsmar1 is at least an order of magnitude higher than that of the full-sized Hsmar1 elements in the human genome (44). One possible explanation that might account for their abundance is their small size, which could predispose them for efficient transposition (25, 44).
To test this hypothesis, transposon donor plasmids were constructed containing a Zeo resistance gene and either the consensus sequence of MiHsmar1 or the long versions of the transposon (Hsmar1-neo or the autonomous element) inserted into the same position in the coding region of the β-lactamase (bla) gene. In addition, long transposons with IRs identical to those of MiHsmar1 were created. The insertions disrupt the bla reading frame, which can be restored only if the transposons are excised by the transposase and NHEJ repairs the plasmid producing the canonical footprints (Fig. 4).
FIG. 4.
Excision of Hsmar1 and MiHsmar1 transposons. (A) Outline of the excision assay. A transposon (Tpson) insertion disrupts and inactivates the Amp resistance gene (Amp). Transposon excision, followed by DNA repair, can restore Amp resistance, which can be selected for in E. coli. (B) Coding triplets of the original, the modified, and the repaired excision sites are listed. Transposon footprints and amino acid sequences are uppercase. Various transposon donor plasmids (indicated on the x axis) were transfected into HeLa cells either with pCMV-Hsmar1-Ra (experiments 1 to 3, numbered below the graph), alone (experiments 4 and 5), or with pCMV-SB (experiment 6) as a control. Indicator plasmids contain the following transposons in the Amp resistance gene: pAmpMITE, the consensus MiHsmar1 sequence; pAmpneo, the neo-tagged Hsmar1 element; pAmpFull, the autonomous Hsmar1 transposon; pAmpneoL, the neo-tagged Hsmar1 element with IR sequences that match those of MiHsmar1; and pAmpFullL, the autonomous Hsmar1 transposon with IR sequences that match those of MiHsmar1. The Ampr/Zeor colonies represent excision events followed by canonical footprint formation at the excision site. The normalized numbers of the double-resistant colonies (shown above the columns) were obtained by dividing the numbers of Ampr/Zeor colonies with the corresponding Zeor colony numbers.
The donor plasmids harboring transposons of different lengths were transfected into HeLa cells, together with vectors expressing either the Hsmar1-Ra or the SB (control) transposase; the reporter containing the autonomous Hsmar1 element was transfected alone. Low-molecular-weight DNA purified from the cells was transformed into E. coli. Excision events were scored by counting Ampr/Zeor colonies; selection with Zeo alone served to control overall plasmid recovery. MiHsmar1 elements were excised from the donor plasmids 2 orders of magnitude more efficiently than any of the longer versions of the transposon (Fig. 4, experiments 1, 2, and 4). IR length did not influence the activities of the long versions of the transposons in the excision assays (Fig. 4, experiments 3 and 5) and in transposon integration assays (data not shown). Therefore, our data indicate that reduced size is responsible for the elevated excision frequency of MiHsmar1.
The human SETMAR protein binds Hsmar1 IRs and retains 5′ cleavage activity of its transposase domain.
The transposase domain of the SETMAR protein was recently shown to specifically bind to Hsmar1 IRs (5) and to catalyze transposition reactions in vitro (32). Since the protein used in those experiments lacked the SET domain, we addressed the question of whether the full-length physiological form of SETMAR can also exhibit transposase-related activities.
To test such a possibility, SETMAR was expressed in E. coli and purified as an N-terminal fusion with the MBP (MBP-SETMAR). EMSAs showed binding of MBP-SETMAR to the 5′ IR of Hsmar1 (Fig. 5, compare unbound probe in lane 1 to complexes formed at increasing transposase concentrations in lanes 2 to 4) in a sequence-specific manner, since binding was competed with cold specific DNA (Fig. 5, lanes 5 to 7) but not with excess nonspecific DNA (Fig. 5, lanes 8 to 10).
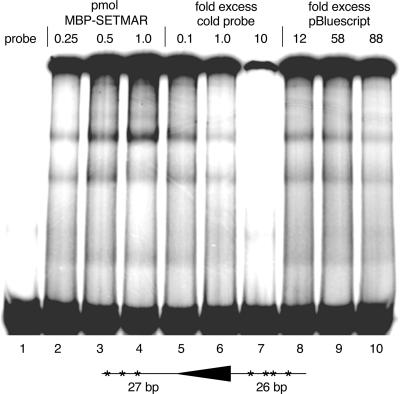
FIG. 5.
In vitro DNA binding activity of the SETMAR protein. A radioactively labeled DNA fragment of 83 bp containing the 5′ IR of Hsmar1 (depicted below) was incubated with purified MBP-SETMAR protein, and the DNA-protein complexes were visualized by EMSA. Compared to free, unbound probe (lane 1), increasing concentrations of MBP-SETMAR (lanes 2 to 4) produced DNA-protein complexes. Substantial amounts of the SETMAR protein formed aggregates at all concentrations used and were unable to enter the gel during electrophoresis. Complex assembly was challenged with cold (unlabeled) probe as a specific competitor (lanes 5 to 7), or increasing amounts of pBluescript were used as nonspecific competitor DNA (lanes 8 to 10).
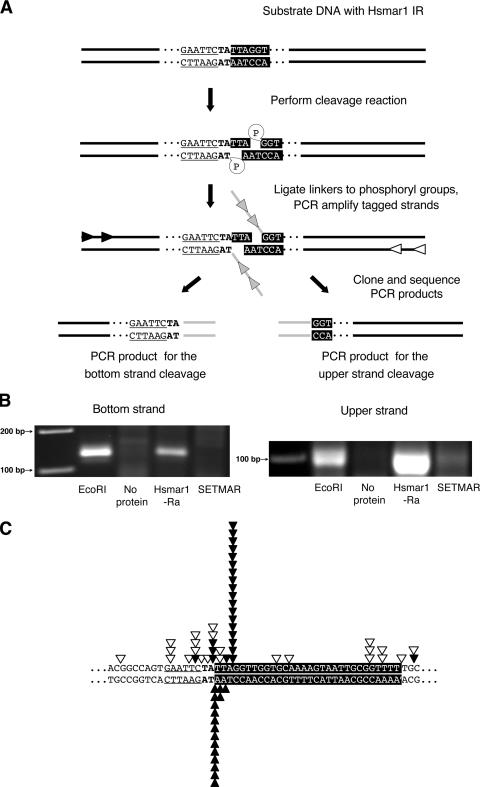
Despite the high sequence similarity of SETMAR to the Hsmar1-Ra transposase, SETMAR contains several amino acid substitutions that potentially compromise its catalytic functions. For example, the third D (Asp) of the catalytically essential DDD triad in the transposase domain of SETMAR is replaced by an N (Asn). Even the conservative D-to-E (Glu) mutation in this position abolished the catalytic activity of the Mos1 transposase (33). In order to test possible catalytic activity of SETMAR, an in vitro DNA cleavage assay was first applied. The assay is based on incubation of recombinant, purified protein with a double-stranded DNA substrate containing IR sequences of Hsmar1, followed by ligation of the 3′ end of a single-stranded oligonucleotide linker to phosphorylated 5′ ends of DNA exposed as a result of endonuclease activity and PCR using substrate- and linker-specific primers (Fig. 6A).
FIG. 6.
In vitro cleavage site analysis with ligation-mediated PCR. (A) Schematic overview of the assay. A 281-bp-long DNA fragment containing the 5′ IR of Hsmar1 was used as the substrate for the reaction. The 5′ end of the IR is shown on a black background, the TA nucleotides bordering the element are in boldface, and the neighboring EcoRI site is underlined. Cleavage of one or both strands of the substrate DNA by the purified proteins results in phosphoryl-terminated DNA ends, which can later be ligated to 3′-hydroxyl-terminated single-stranded oligonucleotide linkers (gray lines) with T4 RNA ligase. The linkers tag the cleavage sites and enable amplification of the cleavage products by nested PCR. (B) Agarose gels with PCR products obtained with linker-specific primers (gray arrows in panel A) and primers specific for the upper or bottom strand of the substrate (black or white arrows in panel A, respectively). PCRs performed on EcoRI-digested DNA served as positive controls. (C) Distribution of cleavage positions identified after cloning and sequencing of the PCR products. The black triangles indicate cleavage sites introduced by MBP-Hsmar1-Ra, and the white triangles mark positions where MBP-SETMAR nicked the upper strand of the substrate DNA.
Purified MBP-SETMAR or MBP-Hsmar1-Ra protein was incubated with a double-stranded DNA fragment containing the 5′ IR of Hsmar1. The substrate also contained an EcoRI recognition site, directly adjacent to the 5′ end of the Hsmar1 IR (Fig. 6A). Digestion of the probe with EcoRI served as a control for the assay. MBP-Hsmar1-Ra cleavage products were identified for both strands of the Hsmar1 transposon ends (Fig. 6B). The most prominent cleavage site on the upper strand was 3 nucleotides inside the transposon DNA, whereas the lower strand was predominantly cut at the end of the transposon (Fig. 6C). Therefore, the cleavage pattern of Hsmar1-Ra fully corresponds to the 3-bp transposon footprints that are generated after transposon excision. Cleavage products by MBP-SETMAR could only be identified for the upper strand (Fig. 6B), suggesting that the SETMAR protein has very weak 3′ nicking activity, if any. These findings are in good agreement with the results of Liu et al. who found inefficient 3′ cleavage activity with the transposase domain of SETMAR (32). Multiple SETMAR nicking sites were identified, none of them corresponding to the major 5′ cleavage site by Hsmar1-Ra (Fig. 6C). Altogether, the data indicate that SETMAR is a defective transposase in vitro and that it retains only a fraction of the biochemical activities of a transposase, namely, 5′ cleavage at transposon IR sequences.
In vivo catalytic activity of SETMAR was addressed by codelivery of an Hsmar1 transposon plasmid and a SETMAR expression plasmid into mammalian cultured cells, recovery of extrachromosomal plasmids from the transfected cells, and PCR amplification using primers flanking the transposable element in the donor plasmid (Fig. 2A). In contrast to the strong, dominant footprint products generated by the Hsmar1-Ra transposase (Fig. 7A, lane 1), SETMAR generated only weak, smeary products (Fig. 7A, lane 2), which were cloned and sequenced. As shown in Fig. 7B, 10 out of the 12 recovered products contained sequences from either one or both IRs. The majority (9/12) of the excision products contained deletions of pUC19 sequences flanking the transposon in the donor plasmid (Fig. 7B). The DNA ends were almost exclusively rejoined at 2- to 9-nucleotide-long microhomologies, shared either between the left and right transposon sequences or between transposon and vector backbone sequences flanking the element (Fig. 7B). No canonical footprints were identified at the excision sites.
FIG. 7.
In vivo cleavage activity of SETMAR. (A) The agarose gel shows PCR products of excision assays on plasmid DNA from HeLa cells transfected with pHsmar1-neo and the helper plasmids indicated. The faint, smeary products around the expected size in lane 2 indicate loss of Hsmar1 transposons from the donor plasmids catalyzed by the SETMAR protein. (B) The overall structures of footprints from HeLa and CHO-K1 cells. The schematic view of the donor site is depicted above. Δ represents deletions, and the sequences in the white boxes are microhomologies at the junctions.
These results are consistent with SETMAR-mediated nicking of Hsmar1 elements in vivo. However, in contrast to the DNA lesions generated by the Hsmar1-Ra transposase, which are predominantly repaired by NHEJ, the characteristics of transposon footprints by SETMAR could be best explained by the involvement of a homology-dependent repair (HDR) pathway.
DISCUSSION
Here, we report the resurrection of an ancient, extinct transposon in the human genome. The apparent inactivity of the confident majority-rule consensus Hsmar1 transposase sequence (Fig. 1B) implies that inactive Hsmar1 copies were efficiently mobilized in the past by a transposase source in trans; thus, nonautonomous elements contributed more effectively to the spread of Hsmar1 copies. One plausible mechanism that could explain such a phenomenon is that transposase-producing Hsmar1 copies suffered mutations within their IRs that compromised their ability to move. We undertook an in silico phylogenetic approach to infer the sequence of an archaic mariner transposase that colonized an ancestral primate genome ∼50 million years ago (44). The reconstructed transposase gene and its specific IRs make up the first functional vertebrate mariner transposon system.
Molecular characterization of transposition events demonstrated that Hsmar1 follows precise cut-and-paste transposition in its natural (human) host and in the zebra fish embryo (Fig. 2). Transpositional activity of Hsmar1 in HeLa cells is an order of magnitude higher than that of the invertebrate mariner elements Himar1 and Mos1 and about as efficient as FP and SB, two Tc1-like elements of vertebrate origin (data not shown). These observations suggest that the new Hsmar1 transposition system can be developed as a useful genetic tool.
We provide experimental evidence that the Hsmar1-Ra transposase can excise MiHsmar1 elements (Fig. 4). Almost all MITEs previously identified from different genomes are inactive, and thus, their mechanisms of transposition and accumulation in eukaryotic genomes have been poorly understood. Although there are strong indications that MITEs are mobilized in trans by a corresponding transposase (e.g., mPing/Pong and Stowaway/Osmar mobilization in the rice genome [11, 22], Tourist/PIF interaction in maize [53], or in vitro interaction between the Arabidopsis elements Emigrant and Lemi1 [35]), this has been experimentally demonstrated for MITE mobilization only by the impala transposase in Fusarium (7). The new Hsmar1-Ra transposon system provides a unique opportunity to investigate the origin and transpositional dynamics of these elements and their contribution to primate genome evolution.
MITEs can accumulate to copy numbers far exceeding those of transposase-encoding DNA transposons in different genomes (9). The suggestion that MITEs might be preferentially mobilized due to their small size has been speculative. Here, we show that MiHsmar1 elements can be excised 2 orders of magnitude more efficiently than their longer transposon versions (Fig. 4). This phenomenon could have contributed to the prevalence of Hsmar1-related MITEs in primate genomes.
The Human Genome Project identified about 50 human genes derived from transposable elements (15). However, to date, there is no evidence for current transpositional activity of any of these “domesticated” genes in humans. The only exceptions are the Rag genes, whose physiological function is to generate the immunoglobulin repertoire by a transposition-like process called V(D)J recombination (23).
We provided experimental evidence that SETMAR, the product of a domesticated gene in the human genome derived from an Hsmar1 transposase, retains its capacity to cleave Hsmar1 transposon DNA in vitro (Fig. 6). However, whereas the Hsmar1-Ra transposase efficiently cleaved both strands of DNA at transposon ends, thereby generating DSBs, the SETMAR protein exhibited only 5′ cleavage activity, generating single-strand nicks (Fig. 6). Inefficient 3′ cleavage by the transposase domain of SETMAR has been recently described (32). We showed that DNA damage inflicted by Hsmar1-Ra and SETMAR is processed differently by cells. Whereas transposon excision sites generated by the Hsmar1-Ra transposase predominantly contained the canonical 3-bp footprints (Fig. 2C), SETMAR activity in vivo is associated with extended stretches of transposon sequences, deletions of flanking DNA, and microhomologies at the junctions at the excision sites (Fig. 7B). The structure of these noncanonical footprints can be best explained by an interrupted synthesis-dependent strand-annealing (SDSA) pathway of HDR, completed by an end-joining process generating microhomologies (18). SDSA has been shown to play a role in the repair of transposon excision sites and to be responsible for generating internally deleted versions of diverse transposable elements in animals and plants (8, 13, 39, 45), including the Mos1 mariner element (34) and SB (18). Pathway choice in DSB repair can be influenced by the structure of the gap, the availability of repair factors, and the cell cycle phase (13, 18, 47). Our observations for the lack of detectable 3′ cleavage activity of SETMAR suggests that the differential utilization of repair pathways by Hsmar1-Ra and SETMAR can be explained by the different structures of the cleavage sites: DSBs for Hsmar1-Ra and single-strand nicks for SETMAR (Fig. 6). Single-strand nicks have been shown to be potent triggers of HDR in mammalian cells (29). For example, repair of DSBs generated by the RAG recombinase in V(D)J recombination is tightly linked to NHEJ (19). However, nick-only RAG mutants have been shown to stimulate robust homologous recombination, and RAG-mediated nicking has been proposed to contribute to gene duplication events and chromosomal rearrangements (29). Interestingly, some of the repair products obtained after SETMAR cleavage (products 1, 2, 4, 6, and 8 to 11 in Fig. 7B) resemble the structure of the Hsmar1-related solo IRs and MITEs present in the human genome. Thus, interrupted SDSA repair events following Hsmar1 transposon excision catalyzed by an Hsmar1 or SETMAR transposase source could have played a role in the emergence and proliferation of the MITEs and solo IRs.
The emergence of the SETMAR gene and the invasion of the ancient primate genome by the Hsmar1 transposons took place within an overlapping evolutionary time window, between 40 and 58 million years ago (5). Thus, it may be that the SETMAR protein played a role in regulating Hsmar1 transposition. The 5′ UTR of the Hsmar1 transposon has significant promoter activity, sufficient to drive transposase expression (Fig. 3). Through its ability to bind to Hsmar1 transposon IR sequences (Fig. 5), and to catalyze specific histone modifications (30), SETMAR could induce local chromatin changes at the Hsmar1 transposase gene promoter, thereby regulating transposase expression.
SETMAR has likely been under selection in human cells for a function other than its residual nicking activity, but this function remains enigmatic. Cordaux et al. (5) have found that selection has been preserving the IR-binding activity of the SETMAR transposase (5). Thus, the function of the SETMAR protein is likely associated with its ability to specifically recognize numerous genomic binding sites represented by the Hsmar1 IRs. It is tempting to speculate that some of these binding sites are conserved because targeted chromatin modifications by SETMAR at these genomic locations are required for normal cellular functions. Ongoing work will have to clarify the past and present functions of SETMAR, making use of the active Hsmar1 transposon as an experimental system.
Supplementary Material
Acknowledgments
The pSK/BG3′u-2a plasmid was a kind gift of P. Hackett, and luciferase reporter constructs were received from J. Altschmied.
This work was supported by EU grant QLG2-CT-2000-00821 and grant IV 21/3-2 from the Deutsche Forschungsgemeinschaft. B.P. is a Long-Term Fellow of the Human Frontier Science Program Organization.
Footnotes
Published ahead of print on 2 April 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Altschmied, J., and J. Duschl. 1997. Set of optimized luciferase reporter gene plasmids compatible with widely used CAT vectors. BioTechniques 23:436-438. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry, E. G., D. J. Witherspoon, and D. J. Lampe. 2004. A bacterial genetic screen identifies functional coding sequences of the insect mariner transposable element Famar1 amplified from the genome of the earwig, Forficula auricularia. Genetics 166:823-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryan, G., D. Garza, and D. Hartl. 1990. Insertion and excision of the transposable element mariner in Drosophila. Genetics 125:103-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordaux, R., S. Udit, M. A. Batzer, and C. Feschotte. 2006. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc. Natl. Acad. Sci. USA 103:8101-8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demattei, M. V., C. Auge-Gouillou, N. Pollet, M. H. Hamelin, M. Meunier-Rotival, and Y. Bigot. 2000. Features of the mammal mar1 transposons in the human, sheep, cow, and mouse genomes and implications for their evolution. Mamm. Genome 11:1111-1116. [DOI] [PubMed] [Google Scholar]
- 7.Dufresne, M., A. Hua-Van, H. A. El Wahab, S. Ben M'Barek, C. Vasnier, L. Teysset, G. H. Kema, and M. J. Daboussi. 2007. Transposition of a fungal miniature inverted-repeat transposable element through the action of a Tc1-like transposase. Genetics 175:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Engels, W. R., D. M. Johnson-Schlitz, W. B. Eggleston, and J. Sved. 1990. High-frequency P element loss in Drosophila is homolog dependent. Cell 62:515-525. [DOI] [PubMed] [Google Scholar]
- 9.Feschotte, C., N. Jiang, and S. R. Wessler. 2002. Plant transposable elements: where genetics meets genomics. Nat. Rev. Genet. 3:329-341. [DOI] [PubMed] [Google Scholar]
- 10.Feschotte, C., and C. Mouches. 2000. Evidence that a family of miniature inverted-repeat transposable elements (MITEs) from the Arabidopsis thaliana genome has arisen from a pogo-like DNA transposon. Mol. Biol. Evol. 17:730-737. [DOI] [PubMed] [Google Scholar]
- 11.Feschotte, C., M. T. Osterlund, R. Peeler, and S. R. Wessler. 2005. DNA-binding specificity of rice mariner-like transposases and interactions with Stowaway MITEs. Nucleic Acids Res. 33:2153-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer, S. E., E. Wienholds, and R. H. Plasterk. 2001. Regulated transposition of a fish transposon in the mouse germ line. Proc. Natl. Acad. Sci. USA 98:6759-6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gloor, G. B., J. Moretti, J. Mouyal, and K. J. Keeler. 2000. Distinct P-element excision products in somatic and germline cells of Drosophila melanogaster. Genetics 155:1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartl, D. L., A. R. Lohe, and E. R. Lozovskaya. 1997. Modern thoughts on an ancyent marinere: function, evolution, regulation. Annu. Rev. Genet. 31:337-358. [DOI] [PubMed] [Google Scholar]
- 15.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 16.Ivics, Z., P. B. Hackett, R. H. Plasterk, and Z. Izsvak. 1997. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91:501-510. [DOI] [PubMed] [Google Scholar]
- 17.Izsvak, Z., Z. Ivics, N. Shimoda, D. Mohn, H. Okamoto, and P. B. Hackett. 1999. Short inverted-repeat transposable elements in teleost fish and implications for a mechanism of their amplification. J. Mol. Evol. 48:13-21. [DOI] [PubMed] [Google Scholar]
- 18.Izsvák, Z., E. E. Stuwe, D. Fiedler, A. Katzer, P. A. Jeggo, and Z. Ivics. 2004. Healing the wounds inflicted by Sleeping Beauty transposition by double-strand break repair in mammalian somatic cells. Mol. Cell 13:279-290. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, S. P., and P. A. Jeggo. 1995. DNA double-strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem. Sci. 20:412-415. [DOI] [PubMed] [Google Scholar]
- 20.Jacobson, J. W., M. M. Medhora, and D. L. Hartl. 1986. Molecular structure of a somatically unstable transposable element in Drosophila. Proc. Natl. Acad. Sci. USA 83:8684-8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jenuwein, T., G. Laible, R. Dorn, and G. Reuter. 1998. SET domain proteins modulate chromatin domains in eu- and heterochromatin. Cell Mol. Life Sci. 54:80-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, N., Z. Bao, X. Zhang, H. Hirochika, S. R. Eddy, S. R. McCouch, and S. R. Wessler. 2003. An active DNA transposon family in rice. Nature 421:163-167. [DOI] [PubMed] [Google Scholar]
- 23.Jones, J. M., and M. Gellert. 2004. The taming of a transposon: V(D)J recombination and the immune system. Immunol. Rev. 200:233-248. [DOI] [PubMed] [Google Scholar]
- 24.Lampe, D. J., M. E. Churchill, and H. M. Robertson. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15:5470-5479. [PMC free article] [PubMed] [Google Scholar]
- 25.Lampe, D. J., T. E. Grant, and H. M. Robertson. 1998. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149:179-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lampe, D. J., K. K. Walden, and H. M. Robertson. 2001. Loss of transposase-DNA interaction may underlie the divergence of mariner family transposable elements and the ability of more than one mariner to occupy the same genome. Mol. Biol. Evol. 18:954-961. [DOI] [PubMed] [Google Scholar]
- 27.Lampe, D. J., K. K. Walden, J. M. Sherwood, and H. M. Roberstson. 2000. Genetic engineering of insects with mariner transposons, p. 237-248. In A. M. Handler and A. A. James (ed.), Insect transgenesis: methods and applications. CRC Press, Boca Raton, FL.
- 28.Leaver, M. J. 2001. A family of Tc1-like transposons from the genomes of fishes and frogs: evidence for horizontal transmission. Gene 271:203-214. [DOI] [PubMed] [Google Scholar]
- 29.Lee, G. S., M. B. Neiditch, S. S. Salus, and D. B. Roth. 2004. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell 117:171-184. [DOI] [PubMed] [Google Scholar]
- 30.Lee, S. H., M. Oshige, S. T. Durant, K. K. Rasila, E. A. Williamson, H. Ramsey, L. Kwan, J. A. Nickoloff, and R. Hromas. 2005. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc. Natl. Acad. Sci. USA 102:18075-18080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lidholm, D. A., G. H. Gudmundsson, and H. G. Boman. 1991. A highly repetitive, mariner-like element in the genome of Hyalophora cecropia. J. Biol. Chem. 266:11518-11521. [PubMed] [Google Scholar]
- 32.Liu, D., J. Bischerour, A. Siddique, N. Buisine, Y. Bigot, and R. Chalmers. 2007. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol. Cell. Biol. 27:1125-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lohe, A. R., D. De Aguiar, and D. L. Hartl. 1997. Mutations in the mariner transposase: the D,D(35)E consensus sequence is nonfunctional. Proc. Natl. Acad. Sci. USA 94:1293-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohe, A. R., C. Timmons, I. Beerman, E. R. Lozovskaya, and D. L. Hartl. 2000. Self-inflicted wounds, template-directed gap repair and a recombination hotspot. Effects of the mariner transposase. Genetics 154:647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loot, C., N. Santiago, A. Sanz, and J. M. Casacuberta. 2006. The proteins encoded by the pogo-like Lemi1 element bind the TIRs and subterminal repeated motifs of the Arabidopsis Emigrant MITE: consequences for the transposition mechanism of MITEs. Nucleic Acids Res. 34:5238-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miskey, C., Z. Izsvak, R. H. Plasterk, and Z. Ivics. 2003. The Frog Prince: a reconstructed transposon from Rana pipiens with high transpositional activity in vertebrate cells. Nucleic Acids Res. 31:6873-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 38.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 39.Plasterk, R. H. 1991. The origin of footprints of the Tc1 transposon of Caenorhabditis elegans. EMBO J. 10:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plasterk, R. H., Z. Izsvak, and Z. Ivics. 1999. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 15:326-332. [DOI] [PubMed] [Google Scholar]
- 41.Robertson, H. M. 1995. The Tc1-mariner superfamily of transposons in animals. J. Insect Physiol. 41:99-105. [Google Scholar]
- 42.Robertson, H. M., and E. G. MacLeod. 1993. Five major subfamilies of mariner transposable elements in insects, including the Mediterranean fruit fly, and related arthropods. Insect Mol. Biol. 2:125-139. [DOI] [PubMed] [Google Scholar]
- 43.Robertson, H. M., and R. Martos. 1997. Molecular evolution of the second ancient human mariner transposon, Hsmar2, illustrates patterns of neutral evolution in the human genome lineage. Gene 205:219-228. [DOI] [PubMed] [Google Scholar]
- 44.Robertson, H. M., and K. L. Zumpano. 1997. Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene 205:203-217. [DOI] [PubMed] [Google Scholar]
- 45.Rubin, E., and A. A. Levy. 1997. Abortive gap repair: underlying mechanism for Ds element formation. Mol. Cell. Biol. 17:6294-6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sijen, T., and R. H. Plasterk. 2003. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426:310-314. [DOI] [PubMed] [Google Scholar]
- 47.Takata, M., M. S. Sasaki, E. Sonoda, C. Morrison, M. Hashimoto, H. Utsumi, Y. Yamaguchi-Iwai, A. Shinohara, and S. Takeda. 1998. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17:5497-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thornton, J. W. 2004. Resurrecting ancient genes: experimental analysis of extinct molecules. Nat. Rev. Genet. 5:366-375. [DOI] [PubMed] [Google Scholar]
- 49.Vigdal, T. J., C. D. Kaufman, Z. Izsvak, D. F. Voytas, and Z. Ivics. 2002. Common physical properties of DNA affecting target site selection of Sleeping Beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 323:441-452. [DOI] [PubMed] [Google Scholar]
- 50.Walisko, O., Z. Izsvak, K. Szabo, C. D. Kaufman, S. Herold, and Z. Ivics. 2006. Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1. Proc. Natl. Acad. Sci. USA 103:4062-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 52.Yant, S. R., and M. A. Kay. 2003. Nonhomologous-end-joining factors regulate DNA repair fidelity during Sleeping Beauty element transposition in mammalian cells. Mol. Cell. Biol. 23:8505-8518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, X., C. Feschotte, Q. Zhang, N. Jiang, W. B. Eggleston, and S. R. Wessler. 2001. P instability factor: an active maize transposon system associated with the amplification of Tourist-like MITEs and a new superfamily of transposases. Proc. Natl. Acad. Sci. USA 98:12572-12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.