Abstract
Complex I of the respiratory chain is composed of at least 45 subunits that assemble together at the mitochondrial inner membrane. Defects in human complex I result in energy generation disorders and are also implicated in Parkinson's disease and altered apoptotic signaling. The assembly of this complex is poorly understood and is complicated by its large size and its regulation by two genomes, with seven subunits encoded by mitochondrial DNA (mtDNA) and the remainder encoded by nuclear genes. Here we analyzed the assembly of a number of mtDNA- and nuclear-gene-encoded subunits into complex I. We found that mtDNA-encoded subunits first assemble into intermediate complexes and require significant chase times for their integration into the holoenzyme. In contrast, a set of newly imported nuclear-gene-encoded subunits integrate with preexisting complex I subunits to form intermediates and/or the fully assembly holoenzyme. One of the intermediate complexes represents a subassembly associated with the chaperone B17.2L. By using isolated patient mitochondria, we show that this subassembly is a productive intermediate in complex I assembly since import of the missing subunit restores complex I assembly. Our studies point to a mechanism of complex I biogenesis involving two complementary processes, (i) synthesis of mtDNA-encoded subunits to seed de novo assembly and (ii) exchange of preexisting subunits with newly imported ones to maintain complex I homeostasis. Subunit exchange may also act as an efficient mechanism to prevent the accumulation of oxidatively damaged subunits that would otherwise be detrimental to mitochondrial oxidative phosphorylation and have the potential to cause disease.
Complex I (NADH-ubiquinone oxidoreductase) is the major entry point of electrons into the electron transport chain and contributes to the establishment of a proton gradient that is required for the bulk of cellular ATP synthesis (30). Complex I is the largest and most complicated structure of the mitochondrial respiratory chain. We have yet to gain a clear understanding of how this enzyme functions in the cell due to the lack of detailed structural information, as well as significant evolutionary divergence between its human and lower cellular forms. In mammals, complex I contains 45 different subunits and forms a complex of ∼1 MDa (5). Seven complex I subunits are encoded by mitochondrial DNA (mtDNA), while the remainder are encoded by nuclear genes and then translated in the cytosol before being imported into the organelle via the protein import machineries (11, 40). Following import, the targeting signals are often, but not always, cleaved before the protein is folded and assembled. Assembled complex I is also known to associate with complex III and complex IV into supercomplexes or “respirasomes” (26). The role of these supercomplexes is not clear but may involve substrate channeling, as well as complex stability (1, 17, 27).
Defects in complex I activity are the most common diagnosis in patients with energy generation disorders (32). In most cases, these defects seem to correlate with a reduction in complex I activity and/or defects in its assembly (31). Complex I defects, and the associated generation of reactive oxygen species, have also been implicated in common neurodegenerative diseases such as Parkinson's disease, Alzheimer's disease, and multiple sclerosis, as well as aging and apoptosis (39). A clearer understanding of the complex I assembly pathway, including the roles of additional cofactors and chaperones, will provide insights into how dysfunction of this enzyme results in disease.
While the assembly process is poorly understood, a number of proteins involved in complex I biogenesis have been reported. The human ortholog of Neurospora crassa CIA30, termed NDUFAF1, is required for complex I assembly in a process that has yet to be clearly defined (38). In addition, roles for apoptosis-inducing factor, complex III, and complex IV in complex I biogenesis have been identified (1, 7, 27, 36). More recently, the protein B17.2L was found associated with an ∼800-kDa form of complex I present in mitochondria from patients lacking the subunit NDUFS4. A patient with a mutation in the B17.2L gene also exhibited reduced levels of complex I. Although the exact function of B17.2L is not known, its absence from fully assembled complex I suggests that it acts as a molecular chaperone/assembly factor (19).
Previous studies of mammalian complex I biogenesis have used patient cells containing assembly defects to generate models of complex I assembly (3, 34). Antonicka et al. (3) identified a number of complexes in different patient mitochondria, classifying them as assembly intermediates. These intermediates were then used to construct a unique assembly model for complex I. However, it was argued that these complexes may not be true assembly intermediates but rather misassembled or partly degraded complexes that are due to the complex I subunit mutations present in the patients studied (34, 37, 42). In a separate study, incorporation of subunits into new complex I assemblies was monitored in cells that had been depleted of complex I holoenzyme by pretreatment with doxycycline (35). Assembly intermediates different from those observed by Antonicka et al. (3) were identified, leading to the proposal that human complex I is assembled in an evolutionarily conserved, modular form (34, 37). In this study, we have addressed the biogenesis of complex I in mitochondria containing the preexisting holoenzyme, as well as in patient mitochondria containing complex I assembly defects. We analyzed the assembly profiles of newly synthesized mtDNA-encoded subunits and performed in vitro assays of the mitochondrial import and assembly of individual nuclear-gene-encoded subunits and provide evidence for two processes in complex I biogenesis—de novo complex I synthesis and regeneration of existing complexes by subunit exchange.
MATERIALS AND METHODS
Cloning procedures.
The cDNAs encoding human NDUFV3, NDUFS4, NDUFS6, NDUFS7, NDUFA9, NDUFA10, and NDUFS2 (accession numbers BC021217, BC005270, BC038664, BC111517, BC009311, BC003417, and BC008868, respectively) were obtained from the I.M.A.G.E Consortium (Medical Research Council, Cambridge, United Kingdom). The cDNAs encoding NDUFS1, NDUFV1, NDUFV2, NDUFB6, NDUFB8, and B17.2L (accession numbers NM_005006, NM_007103, NM_021074, NM_002493, NM_005004, and NM_174889) were amplified by PCR from a human placental cDNA library (BD Biosciences, Clontech) and cloned into pGEM-4Z (Promega). For expression of NDUFA9, the cDNA, lacking its initiation codon and mitochondrial targeting sequence, was cloned into pQE-30 (QIAGEN) at BamHI and SmaI restriction sites. All clones were verified by dideoxynucleotide sequencing.
Cell culture and mitochondrial isolation.
Primary skin fibroblasts grown from patient skin biopsy material were cultured in Dulbecco's modified Eagle's medium (DMEM; GIBCO-BRL) containing 10% (vol/vol) fetal calf serum (FCS) at 37°C under an atmosphere of 5% CO2 and 95% air, supplemented with 50 μg/ml uridine. Mitochondria were isolated by homogenization of cells in 20 mM HEPES (pH 7.6)-220 mM mannitol-70 mM sucrose-1 mM EDTA-0.5 mM phenylmethylsulfonyl fluoride. Cell homogenates were centrifuged at 700 × g at 4°C for 10 min to obtain a postnuclear supernatant. Mitochondria were pelleted by centrifugation at 10,000 × g at 4°C for 10 min.
mtDNA-encoded subunit labeling and chase assays.
Labeling of mtDNA-encoded subunits was performed essentially as previously described (6, 16). Control lymphoblasts were incubated with or without 50 μg/ml chloramphenicol (CAP) in DMEM containing 5% (vol/vol) FCS for 16 h before washing and addition of 0.1 mg/ml cycloheximide in methionine-free DMEM-5% (vol/vol) dialyzed FCS for 15 min at 37°C. Labeling was performed by addition of 20 μCi of [35S]methionine-cysteine (EXPRE35S35S Protein Labeling Mix; Perkin-Elmer Life Sciences) and incubation for 2 h, followed by addition of unlabeled methionine to a final concentration of 1 mM. After 15 min, the solution was removed and replaced with normal DMEM-5% (vol/vol) FCS. Cells were harvested at various chase times, and mitochondria were isolated as described above.
Mitochondrial import and assembly assays.
Generation of radiolabeled, nuclear-gene-encoded precursor proteins was performed by in vitro transcription, followed by translation with rabbit reticulocyte lysates (Promega) in the presence of [35S]methionine-cysteine protein labeling mix (Perkin-Elmer) as previously described (23). Translation products were incubated with freshly isolated mitochondria in 20 mM HEPES-KOH (pH 7.4)-250 mM sucrose-80 mM potassium acetate-5 mM magnesium acetate-10 mM sodium succinate-5 mM methionine at 37°C for various times, as indicated in the figure legends. Dissipation of membrane potential (Δψm) was carried out with 10 μM valinomycin (Sigma). Samples subjected to protease treatment were incubated on ice for 10 min with 100 μg/ml proteinase K (PK; Sigma) before treatment with 1 mM phenylmethylsulfonyl fluoride for 10 min. For Tris-Tricine sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (28), mitochondrial pellets (25 μg) were first precipitated with trichloroacetic acid according to Alconada et al. (2). For blue native PAGE (BN-PAGE), all mitochondrial pellets (50 μg protein) were resuspended in either 50 μl 1% (wt/vol) digitonin (Calbiochem), 1% (wt/vol) n-dodecyl-β-d-maltoside (DDM; Sigma), or 1% (wt/vol) Triton X-100 (Sigma) in 50 mM NaCl-10% (vol/vol) glycerol-20 mM Bis-Tris (pH 7.0) and subjected to 4 to 13% gradient BN-PAGE, with or without SDS-PAGE in the second dimension, according to McKenzie et al. (16). Subunit spots on two-dimensional PAGE were correlated based on migration patterns, assembly into appropriate complexes, and in some cases Western blot analysis. Thyroglobulin (669 kDa), ferritin (440 kDa), and bovine serum albumin (134 and 67 kDa) were used as markers for BN-PAGE.
Miscellaneous.
NDUFA9 polyclonal antibodies were raised in rabbits and used to detect complex I. Antibodies against the 70-kDa subunit and the core I subunit were used to detect complex II and complex III, respectively (Invitrogen). Radiolabeled proteins were detected by PhosphorImager analysis (Molecular Dynamics). Western blotting was performed by a semidry transfer method (9). Horseradish peroxidase-coupled secondary antibodies and ECL chemiluminescent substrate (Amersham) were used to detect immunoreactive proteins in blots. Images were captured with a ChemiGenius chemiluminescence detection system (SynGene).
RESULTS
Mitochondrially encoded subunits assemble into complex I via intermediates.
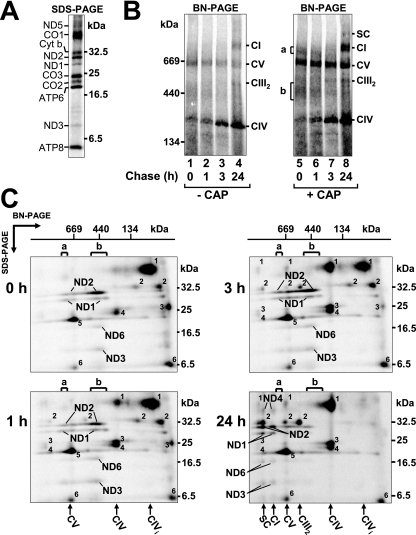
In order to analyze the assembly of mtDNA-encoded complex I subunits, cells were incubated with [35S]Met-Cys in the presence of cycloheximide to block cytosolic protein synthesis. This results in the preferential radiolabeling of the seven complex I subunits, as well as the other six subunits of complexes III, IV, and V. The specific labeling of mtDNA-encoded subunits can be seen through SDS-PAGE analysis (Fig. 1A). Cells were directly harvested or incubated for longer times following removal of radiolabel and cycloheximide (chase), before solubilization in 1% Triton X-100 and analysis by BN-PAGE (Fig. 1B). While some complexes (e.g., complex V [CV]) were found to contain radiolabeled subunits immediately after the labeling period (0 h of chase), assembly of subunits into complex I occurred later—between 3 and 24 h of chase (Fig. 1B, lanes 1 to 4). Neither the levels of steady-state complex I holoenzyme nor the labeling profiles of mtDNA-encoded subunits were altered over this chase period (data not shown). The assembly of mtDNA-encoded subunits was enhanced by first treating cells with CAP, which increases the pool of unassembled nuclear-gene-encoded subunits within mitochondria (6, 16). When this was performed (Fig. 1B, lanes 5 to 8), the complexes were more strongly radiolabeled yet assembly of the subunits into complex I still required a chase time of between 3 and 24 h. However, with the CAP pretreatment, additional broadly labeled complexes were visible from 0 to 3 h of chase (Fig. 1B, lanes 5 to 7, labeled a and b) before disappearing after 24 h of chase (Fig. 1B, lane 8).
FIG. 1.
Mitochondrially encoded subunits assemble into intermediate subcomplexes. mtDNA-encoded subunits were pulse-chase labeled in wild-type lymphoblasts, followed by mitochondrial isolation. (A) Labeling profile of mtDNA-encoded subunits on SDS-PAGE. (B) BN-PAGE analysis of mitochondria solubilized in Triton X-100. mtDNA-encoded subunits were labeled without (left side) or following (right) CAP pretreatment. The letters a and b indicate regions of potential assembly intermediates. CI, complex I; CIII2, complex III homodimer; CIV, complex IV; SC, CI/CIII2 supercomplex. (C) Two-dimensional PAGE analysis of mitochondria isolated from radiolabeled lymphoblasts after 0, 1, 3, and 24 h of chase. For the first-dimension BN-PAGE, mitochondria were solubilized in 1% Triton X-100 as in panel B but at a higher mitochondrial protein/detergent ratio. Regions a and b include complex I intermediate subcomplexes containing subunits ND1, ND2, ND3, and ND6. Subunits CO1 (1), cytochrome b (2), CO3 (3), CO2 (4), ATP6 (5), and ATP8 (6) are indicated. CIVi, complex IV intermediate.
We next addressed whether these broad complexes contain complex I subunits by performing SDS-PAGE in the second dimension (Fig. 1C). Mitochondria were again solubilized in 1% Triton X-100 for the first-dimension BN-PAGE, but in this case the mitochondrial protein-to-detergent ratio was higher than that used for the experiment shown in Fig. 1B, resulting in the presence of more complex I/complex III2 supercomplex (SC). In this analysis, spots representing mtDNA-encoded subunits of complexes I (ND1, ND2, ND3, ND4, and ND6), III, IV, and V are seen. At the 0- and 1-h chase times, radiolabeled complex I subunits ND1, ND2 ND3, and ND6 were preferentially found in a number of complexes in the 350- to 500-kDa range (Fig. 1C, labeled b) or in an ∼800-kDa complex (Fig. 1C, labeled a). At 3 h of chase, these subunits accumulated more in the ∼800-kDa species before being chased to the complex I holoenzyme (CI) and the supercomplex (SC) after 24 h (Fig. 1C). We conclude that mtDNA-encoded subunits assemble into the complex I holoenzyme through a number of intermediates and that significant maturation times are required, even in the presence of a matrix pool of unassembled nuclear-gene-encoded subunits.
Import and assembly of nuclear-gene-encoded subunits.
The biogenesis of individual nuclear-gene-encoded complex I subunits can be individually characterized by in vitro importing radiolabeled subunits into isolated mitochondria and addressing assembly by BN-PAGE. Since no other proteins are made during this time, any assembly represents an interaction between the imported radiolabeled subunit with preexisting subunits or complexes. The subunit NDUFV3 (10 kDa) contains an N-terminal, cleavable targeting signal and, as a flavoprotein subunit, is predicted to assemble into the tip of the matrix arm of complex I (10, 34). NDUFV3 was in vitro translated in rabbit reticulocyte lysate as a 35S-labeled precursor and incubated for various times with mitochondria that were freshly isolated from cultured human primary fibroblasts. In one sample, the mitochondrial membrane potential (Δψm) was dissipated with uncouplers prior to addition of 35S-labeled precursor, thereby blocking the import process. External PK was added to half of each sample to degrade the nonimported radiolabeled precursor. Samples of each were then subjected to SDS-PAGE, and radioactive protein was subsequently detected by phosphorimaging (Fig. 2A). As can be seen, the 35S-labeled NDUFV3 precursor binds to mitochondria and over time an additional faster-migrating species accumulates (Fig. 2A, lanes 1 to 3). Based on (i) its processing, (ii) protection from externally added PK (Fig. 2A, lanes 5 to 7), and (iii) the fact that it does not appear in mitochondria lacking a Δψm (lanes 4 and 8), we conclude that this species represents the imported mature form of NDUFV3.
FIG. 2.
Import and assembly of NDUFV3 into preexisting complex I. 35S-labeled precursor to NDUFV3 (pre-NDUFV3) was incubated for different times with mitochondria isolated from fibroblasts in the presence or absence of a membrane potential (Δψm). Samples were treated with or without PK and subjected to SDS-PAGE (A) or solubilized in DDM-containing buffer and then subjected to BN-PAGE (B). Radiolabeled proteins were detected by PhosphorImager analysis. CI, complex I; CIII2, complex III homodimer; CIII2/CIV, complex III/complex IV supercomplex; CI/CIII2, complex I/complex III2 supercomplex. As shown at the right side of panel B, the migration of complex I (CI) and its supercomplex with dimeric complex III (CI/CIII2) was identified by Western blot analysis with antibodies to complex I subunit NDUFA9 (α-CI) and the complex III core 1 subunit (α-CIII). (C) 35S-labeled NDUFV3 was incubated for 5 to 45 min with mitochondria isolated from control fibroblasts that had been pretreated with (lanes 4 to 6) or without (lanes 1 to 3) CAP for 24 h. All samples were treated with externally added PK before being solubilized in DDM-containing buffer and subjected to BN-PAGE, Western transfer, and PhosphorImager analysis (top). After the radiolabeled signals were acquired, complex I (CI) and the CI/CIII2 supercomplex were identified by Western blot analysis with antibodies to complex I subunit NDUFA9 (α-CI). The anti-70-kDa subunit antibody (α-CII) was used to detect complex II (CII) as a loading control (bottom).
In addition to SDS-PAGE, each sample was also solubilized in DDM and subjected to BN-PAGE (Fig. 2B). With this detergent, complex I is found in both its monomeric (∼980 kDa) and supercomplex (complex I/III2) forms (17), as demonstrated by immunoblot analysis with antibodies against complex I subunit NDUFA9 and the complex III core 1 subunit (Fig. 2B, right side). Imported 35S-labeled NDUFV3 readily assembles into both forms of complex I with the signal accumulating over time, consistent with its import shown in Fig. 2A. When Δψm was dissipated, the assembly of 35S-labeled NDUFV3 was dramatically reduced (Fig. 2B, compare lanes 4 and 8 versus lanes 3 and 7). These results indicate that 35S-labeled NDUFV3 assembles into complex I with the other 44 subunits that are already present in the isolated mitochondria. The possibility exists that NDUFV3 is the only complex I subunit in limiting amounts in mitochondria. However, if other individual subunits can also assemble into complex I following their import into isolated mitochondria, it would mean that either (i) newly imported subunits preferentially assemble into new complex I forms at the expense of preexisting subunits or (ii) the imported subunits are exchanged for previously assembled subunits via a dynamic process. We therefore imported 35S-labeled NDUFV3 into mitochondria isolated from cells that had been preincubated with CAP. In this case, complex I assembly intermediates containing mtDNA-encoded subunits are unlikely to be present while a pool of nuclear-gene-encoded subunits will also accumulate in mitochondria. Radiolabeled NDUFV3 is therefore less likely to assemble into complex I via intermediates and also needs to compete with the increased pool of free NDUFV3. As shown at the top of Fig. 2C, 35S-labeled NDUFV3 still rapidly assembles into complex I and its supercomplex in mitochondria from control and CAP-treated cells. Western blot analysis of these lanes (Fig. 2C, bottom parts) revealed that the steady-state levels of complex I were reduced in CAP-treated cells, relative to complex II (which lacks mtDNA-encoded subunits), consistent with the prevention of any new complex I assembly by CAP treatment.
We next tested the import and assembly of a variety of different nuclear-gene-encoded complex I subunits. Representative subunits with cleavable (NDUFV1, NDUFV2, NDUFV3, NDUFB8, NDUFS2, NDUFS4, NDUFS6, NDUFS7, NDUFA9, and NDUFA10) and noncleavable (NDUFS1) presequences, predicted transmembrane domains (NDUFA9, NDUFB8, and NDUFS7), and/or cofactor attachment (NDUFV1, NDUFV2, NDUFS1, NDUFS2, and NDUFS7) were chosen. The predicted subunit locations in holocomplex I (based on the models described in references 4, 10, and 25) are shown schematically in Fig. 3A. Radiolabeled precursors were incubated with isolated mitochondria in the presence or absence of Δψm, and all samples were treated with PK before BN-PAGE analysis. As shown in Fig. 3A, in addition to NDUFV3 (lanes 5 to 6), the newly imported subunits NDUFV1, NDUFV2 (lanes 1 to 4), NDUFS4, NDUFS6 (lanes 13 to 16), and NDUFA9 (lanes 19 to 20) clearly assembled into both monomeric complex I (CI) and its supercomplex form (with dimeric complex III; CI/CIII2) in a Δψm-dependent fashion. The relative positions of complex I, complex III, and the CI/CIII2 supercomplex are shown by Western blot analysis (Fig. 3A, lanes 23 to 24). Radiolabeled NDUFB8 (Fig. 3A, lane 7), NDUFS7 (lane 17), and NDUFA10 (lane 21) also assembled into complex I, but to a lesser degree. In addition to assembly into complex I, some subunits also appeared to assemble into smaller complexes. NDUFS1 (Fig. 3A, lane 9) and NDUFS2 (lane 11) assembled into additional complexes of ∼480 and ∼550 kDa. Subunits NDUFB8 (lanes 7) and NDUFA9 (lanes 19) also appeared to be present in an ∼800-kDa complex (Fig. 3A, asterisk) that resembles the subassembly of complex I that has been found to contain the assembly factor/molecular chaperone B17.2L (19). Thus, some subunits may also assemble into intermediate complexes. To verify that these precursors were imported into mitochondria, SDS-PAGE analysis was also performed (Fig. 3B). All proteins were efficiently radiolabeled (Fig. 3B, lane 4) and imported into mitochondria in a Δψm-dependent fashion. An exception was NDUFS4, which appeared to be processed in the absence of a Δψm (Fig. 3B, lane 3); however, this form is most likely not imported into the matrix since it could not assemble into complex I (Fig. 3A, lane 14). We conclude that following their import, a number of subunits can independently assemble with preexisting subunits into complex I.
FIG. 3.
Import and assembly of complex I subunits. 35S-labeled complex I subunits were individually incubated with isolated fibroblast mitochondria for 60 min in the presence or absence of a membrane potential (Δψm). (A) Samples were solubilized in DDM-containing buffer and subjected to BN-PAGE and PhosphorImager analysis. The approximate location of each subunit within complex I is indicated schematically above each pair of lanes. Complex I (CI), complex III (CIII2), and their supercomplex form (CI/CIII2) were identified by Western blot analysis with antibodies to complex I subunit NDUFA9 (α-CI) and the core I subunit of complex III (α-CIII; lanes 23 to 24). (B) SDS-PAGE analysis of imported radiolabeled complex I subunits. The precursor (p) and mature (m) forms of the subunits are identified. Samples were imported into mitochondria (mit.) in the presence or absence of a Δψm and treated with or without externally added PK. A sample of lysate (representing 20% of the added protein/import) is also shown (lane 4).
Analysis of B17.2L intermediates.
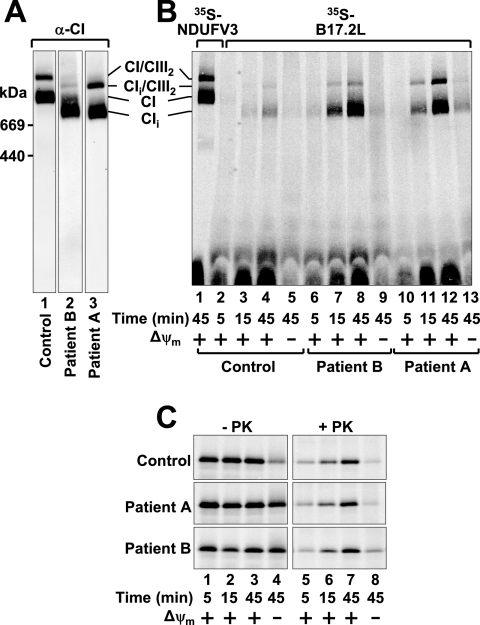
We previously identified a patient with lethal infantile mitochondrial disease lacking NDUFS6 (patient B) whose fibroblasts contained an ∼800-kDa complex I subassembly (15). This subassembly also resembles a complex reported in a patient lacking the subunit NDUFS4, although a different estimate of the size of the complex was found (19). We have also identified a patient with Leigh syndrome (patient A) harboring a 1-bp nonsense homozygous deletion in exon 3 of the NDUFS4 gene. This mutation prevents NDUFS4 subunit expression and enables us to directly compare the subassemblies between these patients. Western blot analysis indicates that patient A, lacking NDUFS4, contains a complex I subassembly of ∼800 kDa (CIi) that is indistinguishable in size from that seen in patient B, lacking NDUFS6 (Fig. 4A, lanes 2 to 3). The subassembly is also found associated with complex III in a supercomplex (CIi/CIII2, Fig. 4A, lanes 2 to 3) as previously reported (19). Given that the ∼800-kDa subassembly is significantly smaller than the ∼980-kDa holocomplex I (CI), these results suggest that a number of subunits are absent from this smaller complex. NDUFS4 and NDUFS6 are therefore crucial to the final steps in the assembly of complex I, possibly providing attachment points for the integration of other subunits.
FIG. 4.
The assembly factor B17.2L is associated with a complex I subassembly in patient and control mitochondria. (A) Mitochondria from control cells (lane 1) and patient cells lacking subunit NDUFS6 (patient B, lane 2) or NDUFS4 (patient A, lane 3) were solubilized in DDM-containing buffer and subjected to BN-PAGE before Western blot analysis with complex I antibodies (anti-NDUFA9). (B) 35S-labeled B17.2L was incubated with mitochondria isolated from control or patient cells for increasing times in the presence or absence of a Δψm. Half of each sample was treated with external PK before being split in two and subjected to BN-PAGE (protease-treated samples only) or (C) SDS-PAGE and phosphorimaging. As shown in lane 1 of panel B, 35S-labeled NDUFV3 was imported into control mitochondria to illustrate the fully assembled forms of complex I. CI/CIII2, complex I/complex III2 supercomplex; CIi/CIII2, complex I intermediate/complex III2 supercomplex; CIi, complex I intermediate; CI, complex I.
To further support the notion that the subassemblies are the same, we analyzed for the additional presence of B17.2L. In vitro import and assembly assays revealed that B17.2L associated with the ∼800-kDa subassembly (CIi) in both patients, as well as the supercomplex form (CIi/CIII2, Fig. 4B). Both complexes were indistinguishable in migration from the steady-state complex I subcomplexes (Fig. 4, compare panels A and B). 35S-labeled NDUFV3, which efficiently assembles into complex I, was used as a control and shows that the B17.2L-containing complex is clearly different from complex I (Fig. 4B, lane 1). Of note, B17.2L was also found in the subassembly in control mitochondria (Fig. 4B, lane 4). SDS-PAGE analysis of imported 35S-labeled B17.2L showed that it was efficiently imported into both patient and control mitochondria (Fig. 4C). The precursor also appears to contain a noncleavable presequence since the imported form is indistinguishable from the translation product (data not shown).
Import of missing subunits into patient mitochondria corrects complex I assembly.
It has been suggested that the subcomplexes observed in patient cells may represent not stalled complex I assembly intermediates but rather partially degraded complexes (37). We asked whether import of the missing subunit into patient mitochondria can drive assembly of complex I to its holoenzyme form. However, this would require that the other subunits absent from the subcomplex still be present within the isolated fibroblast mitochondria. We first imported 35S-labeled NDUFV3 into mitochondria from control or patient A fibroblasts (lacking NDUFS4) and compared its assembly profile against the complexes found under steady-state conditions by Western blot analysis (Fig. 5A). As expected, 35S-labeled NDUFV3 assembled into both monomeric (CI) and supercomplex (CI/CIII2) forms of complex I in control mitochondria; however, it did not assemble into patient mitochondria (Fig. 5B, lanes 1 to 3 versus lanes 4 to 6). NDUFV3 is predicted to associate with the tip of the matrix arm of complex I (10), and since no assembly was observed in the patient mitochondria, it follows that this tip is missing. We next imported 35S-labeled NDUFS4 into isolated mitochondria. As previously shown, NDUFS4 assembled into complex I in control mitochondria, forming the monomeric (CI) and supercomplex (CI/CIII2) forms (Fig. 5B, lanes 7 to 9). In patient mitochondria, imported 35S-labeled NDUFS4 also assembled into both complexes that were identical in size to those in control mitochondria (Fig. 5B, lanes 10 to 12). This indicates that assembly of complex I can be restored upon import of a single subunit. In addition, the assembly of 35S-labeled NDUFS4 into complex I was more efficient in the patient mitochondria than in the control mitochondria. This is consistent with the fact that in control mitochondria, imported 35S-labeled NDUFS4 competes with preexisting NDUFS4 for assembly into complex I.
FIG. 5.
Import of NDUFS4 into patient mitochondria restores complex I assembly. Mitochondria from control or NDUFS4-deficient patient A cells were incubated with 35S-labeled NDUFV3 (as a control) or 35S-labeled NDUFS4 for 5 to 45 min as indicated before BN-PAGE analysis. (A) Western blot analysis of complex I (probed with NDUFA9 antibodies) and (B) PhosphorImager analysis of import and assembly. Schematic models depicting complex I forms are shown. CI/CIII2, complex I/complex III2 supercomplex; CIi/CIII2, complex I intermediate/complex III2 supercomplex; CIi, complex I intermediate; CI, complex I. The asterisk denotes a nonspecific complex.
We tested whether restoration of complex I could also be seen by importing 35S-labeled NDUFS6 into mitochondria isolated from patient B fibroblasts (lacking endogenous NDUFS6). Some 35S-labeled NDUFV3 appeared to assemble into a complex of the same size as complex I (CI) (Fig. 6B, lanes 4 to 6), suggesting that complex I subunits can form higher-molecular-weight assemblies in the absence of NDUFS6. Closer examination of immunoreactive complex I assemblies by BN-PAGE revealed that, in addition to the ∼800-kDa subassembly (CIi) and its supercomplex form (CIi/CIII2), a small amount of complex I (CI, at ∼970 kDa; i.e., still lacking NDUFS6) was visible in patient B mitochondria (Fig. 6A). Import and assembly of 35S-labeled NDUFS6 was again more efficient in patient mitochondria that lack this subunit and resulted in the appearance of fully assembled complex I (CI) (Fig. 6B, lanes 10 to 12). Our findings indicate that those subunits absent from the subassembly are still present within mitochondria and introduction of either missing NDUFS4 or NDUFS6 promotes and stabilizes their assembly into mature complex I.
FIG. 6.
Import of NDUFS6 into patient mitochondria restores complex I assembly. Mitochondria from control or NDUFS6-deficient patient B cells were incubated with 35S-labeled NDUFV3 (as a control) or 35S-labeled NDUFS6 for 5 to 45 min as indicated before BN-PAGE analysis. (A) Western blot analysis of complex I (probed with NDUFA9 antibodies) and (B) PhosphorImager analysis of import and assembly. Schematic models depicting complex I forms are shown. CI/CIII2, complex I/complex III2 supercomplex; CIi/CIII2, complex I intermediate/complex III2 supercomplex; CIi, complex I intermediate; CI, complex I. The asterisk denotes a nonspecific complex.
Assembly of subunits in patient mitochondria.
A survey of assembly in mitochondria from both patients was conducted to identify subunits that can integrate into the ∼800-kDa complex I subassembly (CIi, Fig. 7 and 8), thereby providing additional information about the location of subunits within complex I. The steady-state levels and migration of complex I assemblies were detected by Western blot analysis (Fig. 7A and 8A, lanes 9 to 10 and 23 to 24). In patient A mitochondria (NDUFS4 mutation), there was almost no assembly of NDU subunit FV1, FV2, FV3, FS1, or FS6 (Fig. 7A, lanes 1 to 8 and 15 to 16), consistent with the finding that these subunits are located in the unassembled tip of the complex. Subunits NDUFS2, NDUFS7, NDUFA9, and NDUFA10 were able to integrate into the subassembly of complex I (CIi), suggesting that they must lie within this complex (Fig. 7A, lanes 11 to 12 and 17 to 22). To confirm that protein import was not impaired in patient mitochondria, representative subunits were also analyzed by SDS-PAGE (Fig. 7B). As can be seen, these subunits were imported in a Δψm-dependent manner in a fashion comparable to that of control mitochondria (see Fig. 3B). The survey of assembly in patient B (NDUFS6 mutation) showed that subunits NDUFS2, NDUFS7, NDUFA9, and NDUFA10 were also able to integrate into the CIi subassembly (Fig. 8A, lanes 11 to 12 and 17 to 22). Subunits NDUFV1, FV2, FV3, and FS1 weakly assembled into the residual complex I (CI) that forms in the absence of NDUFS6 (Fig. 8A, lanes 1 to 8). As for patient A, the import of representative subunits into patient B mitochondria appeared to be unaffected (Fig. 8B). In summary, our results suggest that NDUFS4 and NDUFS6 are required for the assembly and stabilization of a portion of complex I that contains a number of subunits including, NDUFV1, NDUFV2, and NDUFV3 (Fig. 9). Furthermore, we conclude that some subunits can assemble with preexisting subunits into complex I while others, such as NDUFS1, NDUFS2, NDUFS7, and the mtDNA-encoded subunits, assemble into complexes that represent earlier intermediates in complex I assembly.
FIG. 7.
Import and assembly of complex I subunits into patient mitochondria lacking NDUFS4. 35S-labeled complex I subunits were individually incubated for 60 min with mitochondria isolated from patient A fibroblasts. (A) Samples were treated with or without externally added PK before being solubilized in DDM-containing buffer and subjected to BN-PAGE and PhosphorImager analysis. The approximate location of each subunit is indicated schematically above each pair of lanes. The migration of the ∼800-kDa complex I intermediate (CIi), complex III (CIII2), and their supercomplex form (CIi/CIII2) were identified by Western blot analysis with antibodies to complex I subunit NDUFA9 (α-CI, lanes 9 and 23) and the core I subunit of complex III (α-CIII, lanes 10 and 24). (B) SDS-PAGE analysis of the import of a selection of radiolabeled complex I subunits into patient A mitochondria. The precursor (p) and mature (m) forms of the proteins are identified. Samples were imported into mitochondria (mit.) in the presence or absence of a Δψm and treated with or without externally added PK. A sample of lysate (representing 20% of added protein/import) is also shown (lane 4).
FIG. 8.
Import and assembly of complex I subunits into patient mitochondria lacking NDUFS6. 35S-labeled complex I subunits were individually incubated for 60 min with mitochondria isolated from patient B fibroblasts. (A) Samples were treated with or without externally added PK before being solubilized in DDM-containing buffer and subjected to BN-PAGE and PhosphorImager analysis. The approximate location of each subunit is indicated schematically above each pair of lanes. The migration of the ∼800-kDa complex I intermediate (CIi), complex III (CIII2), and their supercomplex form (CIi/CIII2) was identified by Western blot analysis with antibodies to complex I subunit NDUFA9 (α-CI, lanes 9 and 23) and the core I subunit of complex III (α-CIII, lanes 10 and 24). (B) SDS-PAGE analysis of the import of a selection of radiolabeled complex I subunits into patient B mitochondria. The precursor (p) and mature (m) forms of the proteins are identified. Samples were imported into mitochondria (mit.) in the presence or absence of a Δψm and treated with or without externally added PK. A sample of lysate (representing 20% of added protein/import) is also shown (lane 4).
FIG. 9.
Model depicting the assembly of complex I. De novo assembly occurs via the seeding of intermediate complexes through the synthesis of mitochondrially encoded (ND) subunits, followed by recruitment of other nuclear-gene-encoded subunits. ND1 is found in a separate, smaller complex before associating with other ND subunits. An intermediate containing the assembly factor B17.2L is present in the absence of NDUFS4 or NDUFS6. Their incorporation into the complex drives complete assembly and releases B17.2L. Complex I homeostasis can also occur by competition of newly imported subunits with preexisting ones for assembly into complex I. Complex I intermediates may be found in supercomplexes that include complex III.
DISCUSSION
In vitro import and assembly assays, combined with BN-PAGE, have been powerful tools for the analysis of mitochondrial membrane protein complexes, in particular, the protein import translocases (12, 13, 18, 22, 41). Here we have used a similar approach to address the assembly of respiratory complex I—an elaborate membrane protein complex containing subunits encoded by two separate genomes. A greater understanding of complex I assembly will not only provide insights into complex I function and subunit interactions but may also lead to new techniques for the diagnostic screening of patients with mitochondrial disorders.
Complex I biogenesis.
All proteins encoded by mtDNA are subunits of the respiratory chain complexes. Most are very hydrophobic and appear to be cotranslationally inserted into the inner mitochondrial membrane through the actions of a number of proteins involved in ribosome docking and membrane insertion (8, 20). Labeling and chase assays revealed that complex I subunits encoded by mtDNA required longer chase times for their assembly into the holoenzyme, compared to mtDNA-encoded subunits of the other respiratory complexes. This was also seen in other cell lines, including primary fibroblasts and lymphoblasts (data not shown). Even in the presence of an increased pool of nuclear-gene-encoded subunits following treatment with CAP, assembly of mtDNA subunits into holocomplex I was not accelerated, although larger, stable intermediates were observed. The presence of assembly intermediates containing mtDNA-encoded subunits is in concordance with other studies (3, 33, 35) and suggests that even in the presence of fully assembled endogenous complex I, mtDNA-encoded subunits assemble via intermediate modules. The modular model of assembly of mtDNA subunits would be consistent with the highly hydrophobic nature of these proteins and their location in the core of complex I.
In contrast, many nuclear-gene-encoded subunits are not hydrophobic and their assembly profile may differ from that of mtDNA-encoded subunits. Until now, the assembly of newly imported subunits in mitochondria containing endogenous levels of complex I has not been investigated. Initially, NDUFV3 was identified as a subunit that can assemble into both monomeric and supercomplex forms of complex I. Although assembly into the complex was kinetic, an assembly pathway composed of intermediates prior to formation of the holoenzyme was not observed. This indicates that NDUFV3 is incorporated at a late stage in complex I assembly, consistent with its location in the matrix tip of the complex and the fact that it is not present in the ∼800-kDa subassembly in patient cell lines. An assembly scan with subunits located in different regions within the complex was performed to identify other subunits that can assemble into complex I in vitro. This analysis also addressed whether there is a correlation between subunit characteristics (such as location, presence of cofactors, etc.) and their competency to assemble into complex I. This scan resulted in the identification of a host of different subunits that could assemble into complex I to various degrees; however, there did not appear to be any correlation between the assembly competency of a subunit and its location in the complex or its posttranslational modifications.
The assembly survey also revealed that a number of subunits, such as NDUFB8, NDUFS1, NDUFS2, NDUFS7, and NDUFA9, integrate into additional complexes. Investigations by Antonicka et al. (3) found an ∼480-kDa complex in patients containing at least NDUFA2, NDUFA9, NDUFS2, NDUFS3, and ND1. We also found that subunits NDUFS1 and NDUFS2 assembled into two complexes in the ∼480- and 550-kDa ranges. NDUFS7 was also found in a similar-sized complex (Fig. 7). These complexes also resemble those seen for mtDNA-encoded subunit ND1 (Fig. 1C). The bovine orthologs of these subunits (75 kDa [NDUFS1], 49 kDa [NDUFS2], PSST [NDUFS7], and ND1) are thought to integrate into a common assembly intermediate as part of the modular model of complex I assembly (37). This is also consistent with the recent crystal structure of the peripheral arm of complex I from Thermus thermophilus where subunits Nqo4, Nqo3, and Nqo6, which are orthologs of NDUFS2, NDUFS1, and NDUFS7, respectively, are found adjacent to one another (24). In addition to these subunits, NDUFV1, NDUFV2, NDUFA9, and NDUFA10 also appeared to partially assemble into these subcomplexes. NDUFV1 and NDUFV2 also form part of the T. thermophilus peripheral arm, consistent with their association in human mitochondrial subassemblies.
Subunits NDUFV3, NDUFS4, and NDUFS6 all assembled efficiently into complex I with an absence of detectable intermediates. Given that no single complex I subunit tested is limiting for assembly, it is unlikely that separate preformed intermediates are present in mitochondria which await the import of a single subunit to drive complete complex I assembly. This therefore indicates that individual, newly imported subunits can compete with their preexisting counterparts to assemble into complex I. In support of this, mitochondria isolated from cells pretreated with CAP did not hinder the assembly of newly imported NDUFV3. This is also consistent with the more efficient assembly of either newly imported NDUFS4 or NDUFS6 in patient mitochondria which lack the subunit compared to control mitochondria which contain preexisting forms.
B17.2L-associated subassemblies in patient mitochondria.
Ogilvie et al. (19) identified a molecular chaperone, termed B17.2L, that is associated with an ∼800-kDa subassembly of complex I in patients with mutations in the genes encoding subunit NDUFV1 or NDUFS4. Our investigations into the import and assembly of B17.2L showed that this subassembly is also present in control mitochondria that already contain fully assembled complex I. The ∼800-kDa intermediate is present at very low levels, and this is reflected by our inability to clearly detect the subassembly with antibodies against complex I. However, the signal we observed by using in vitro import and assembly assays is representative of only a small number of molecules imported into each mitochondrion and hence this is why it is possible to detect complexes that are present at such low levels. The presence of this subassembly in control mitochondria provides further evidence that the ∼800-kDa complex observed in patient mitochondria is not an artifact of destabilized complex I but in fact represents a stalled assembly intermediate of the holoenzyme.
Although the mutations in patients A and B are in different genes, the subassemblies in both patients comigrate on BN-PAGE. However, patient B also appears to have low levels of a complex I indistinguishable in size from the holoenzyme, which presumably only lacks the 13-kDa NDUFS6 subunit. The ∼800-kDa subassemblies appear to be associated with B17.2L, as observed by in vitro import and assembly assays (Fig. 4) and by immunoblot analysis (19). Furthermore, the assembly of B17.2L is much more efficient in patient mitochondria compared to the control, presumably because of the accumulation of the ∼800-kDa form in the patients. Interestingly, we did not observe any B17.2L in the small amount of ∼970-kDa complex I lacking NDUFS6 in patient B mitochondria, indicating that it must dissociate before complex I is completely assembled.
In addition to its association with the ∼800-kDa complex, B17.2L was also found in a high-molecular-weight complex that represents its association with complex III in the supercomplex (CIi/CIII2). Presumably, the interaction of B17.2L with this higher-molecular-weight complex is still via its association with the complex I intermediate. These results indicate that complex I does not need to be fully assembled before associating into supercomplexes. Indeed, complex III and, perhaps, complex IV may act as scaffolds to facilitate complex I assembly since loss of either has been found to lead to reduced complex I levels (1, 7).
Import and assembly of complex I subunits in patient mitochondria.
Reintroduction of either subunit missing from patient mitochondria (NDUFS4 in patient A and NDUFS6 in patient B) by in vitro import facilitated the completion of complex I assembly. This indicates that the patient's mitochondria must still harbor the subunits that are lacking from the unassembled portion of the complex and that these subunits are maintained in a form that allows for their productive assembly. We have been unable to detect whether these unassembled subunits form a separate complex, perhaps in association with novel assembly factors. Of note, NDUFV1 and NDUFV2 assembled into an ∼300-kDa complex in mitochondria lacking NDUFS4 (Fig. 7A), which was not observed in control mitochondria. In addition, given that reintroduction of the missing subunit into patient mitochondria could drive complete assembly of complex I, the ∼800-kDa subassembly can be viewed as a productive intermediate. This is also consistent with the association of B17.2L with the subassembly in patient mitochondria containing separate gene mutations.
A subunit survey in the patients provided further information as to the predicted locations of certain subunits and also aided in determining those subunits which are found in the ∼800-kDa subassembly and those which are not. Subunits NDUFS2, NDUFA9, and NDUFA10 assembled into the ∼800-kDa subassembly in the patients, while NDUFS6, NDUFS4, and NDUFV3 did not, suggesting that the latter are located in the missing tip of the complex.
The ability to follow the assembly of newly imported, nuclear-gene-encoded complex I subunits has opened a new perspective on the biogenesis of complex I. Since complex IV of the respiratory chain is assembled via intermediates that involve a number of chaperones (29), it is suspected that complex I is assembled in a similar fashion. Although it is likely that newly assembled complex I is formed via an assembly pathway, not every subunit that is imported into mitochondria must be incorporated into new assemblies; instead, a subunit may assemble with preexisting subunits that undergo dynamic transitions between intermediate and fully assembled complex I forms. Furthermore, these forms are most likely found in higher-molecular-weight supercomplexes. This is depicted schematically in Fig. 9. Such a mechanism would explain how multiple subunits can individually assemble into complex I in the absence of other subunit synthesis or import.
Complex I transitions between assembled and intermediate forms may enable a mechanism to aid in the turnover of subunits, some of which are prone to oxidative damage. Indeed, it was recently found that a number of complex I subunits had become oxidatively damaged in the brains of patients with Parkinson's disease (14). Given our findings, it is possible that the underlying basis of mitochondrial disease is due to decreased rates in mitochondrial protein import and/or assembly which slows the exchange of preexisting complex I subunits for newly imported ones. Decreased import or assembly may lead to increased oxidative damage and complex I deficiency that results in Bax-dependent apoptosis (21) and subsequent disease.
Acknowledgments
We thank Ann Frazier for critical reading of the manuscript.
This work was supported by grants from the Australian National Health and Medical Research Council, the Australian Research Council, the Ramaciotti Foundation, and the Muscular Dystrophy Association (United States). D.T. is supported by an NHMRC Senior Research Fellowship, M.M. is supported by an NHMRC Peter Doherty Fellowship, M.L. is supported by an Australian Postgraduate Award, and A.O. is supported by a grant-in-aid for Scientific Research from the Japan Society for the Promotion of Science (16591052).
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Acin-Perez, R., M. P. Bayona-Bafaluy, P. Fernandez-Silva, R. Moreno-Loshuertos, A. Perez-Martos, C. Bruno, C. T. Moraes, and J. A. Enriquez. 2004. Respiratory complex III is required to maintain complex I in mammalian mitochondria. Mol. Cell 13:805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alconada, A., F. Gartner, A. Honlinger, M. Kubrich, and N. Pfanner. 1995. Mitochondrial receptor complex from Neurospora crassa and Saccharomyces cerevisiae. Methods Enzymol. 260:263-286. [DOI] [PubMed] [Google Scholar]
- 3.Antonicka, H., I. Ogilvie, T. Taivassalo, R. P. Anitori, R. G. Haller, J. Vissing, N. G. Kennaway, and E. A. Shoubridge. 2003. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J. Biol. Chem. 278:43081-43088. [DOI] [PubMed] [Google Scholar]
- 4.Carroll, J., I. M. Fearnley, R. J. Shannon, J. Hirst, and J. E. Walker. 2003. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol. Cell. Proteomics 2:117-126. [DOI] [PubMed] [Google Scholar]
- 5.Carroll, J., I. M. Fearnley, J. M. Skehel, R. J. Shannon, J. Hirst, and J. E. Walker. 2006. Bovine complex I is a complex of forty-five different subunits. J. Biol. Chem. 281:32724-32727. [DOI] [PubMed] [Google Scholar]
- 6.Chomyn, A. 1996. In vivo labeling and analysis of human mitochondrial translation products. Methods Enzymol. 264:197-211. [DOI] [PubMed] [Google Scholar]
- 7.Diaz, F., H. Fukui, S. Garcia, and C. T. Moraes. 2006. Cytochrome c oxidase is required for the assembly/stability of respiratory complex I in mouse fibroblasts. Mol. Cell. Biol. 26:4872-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frazier, A. E., R. D. Taylor, D. U. Mick, B. Warscheid, N. Stoepel, H. E. Meyer, M. T. Ryan, B. Guiard, and P. Rehling. 2006. Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J. Cell Biol. 172:553-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harlow, E., and D. Lane. 1999. Using antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 10.Hirst, J., J. Carroll, I. M. Fearnley, R. J. Shannon, and J. E. Walker. 2003. The nuclear encoded subunits of complex I from bovine heart mitochondria. Biochim. Biophys. Acta 1604:135-150. [DOI] [PubMed] [Google Scholar]
- 11.Hoogenraad, N. J., L. A. Ward, and M. T. Ryan. 2002. Import and assembly of proteins into mitochondria of mammalian cells. Biochim. Biophys. Acta 1592:97-105. [DOI] [PubMed] [Google Scholar]
- 12.Humphries, A. D., I. C. Streimann, D. Stojanovski, A. J. Johnston, M. Yano, N. J. Hoogenraad, and M. T. Ryan. 2005. Dissection of the mitochondrial import and assembly pathway for human Tom40. J. Biol. Chem. 280:11535-11543. [DOI] [PubMed] [Google Scholar]
- 13.Johnston, A. J., J. Hoogenraad, D. A. Dougan, K. N. Truscott, M. Yano, M. Mori, N. J. Hoogenraad, and M. T. Ryan. 2002. Insertion and assembly of human tom7 into the preprotein translocase complex of the outer mitochondrial membrane. J. Biol. Chem. 277:42197-42204. [DOI] [PubMed] [Google Scholar]
- 14.Keeney, P. M., J. Xie, R. A. Capaldi, and J. P. Bennett, Jr. 2006. Parkinson's disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J. Neurosci. 26:5256-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirby, D. M., R. Salemi, C. Sugiana, A. Ohtake, L. Parry, K. M. Bell, E. P. Kirk, A. Boneh, R. W. Taylor, H. H. Dahl, M. T. Ryan, and D. R. Thorburn. 2004. NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. J. Clin. Investig. 114:837-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenzie, M., M. Lazarou, D. R. Thorburn, and M. T. Ryan. 2007. Analysis of mitochondrial subunit assembly into respiratory chain complexes using blue native polyacrylamide gel electrophoresis. Anal. Biochem. 364:2128-2137. [DOI] [PubMed] [Google Scholar]
- 17.McKenzie, M., M. Lazarou, D. R. Thorburn, and M. T. Ryan. 2006. Mitochondrial respiratory chain supercomplexes are destabilized in Barth syndrome patients. J. Mol. Biol. 361:462-469. [DOI] [PubMed] [Google Scholar]
- 18.Model, K., C. Meisinger, T. Prinz, N. Wiedemann, K. N. Truscott, N. Pfanner, and M. T. Ryan. 2001. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat. Struct. Biol. 8:361-370. [DOI] [PubMed] [Google Scholar]
- 19.Ogilvie, I., N. G. Kennaway, and E. A. Shoubridge. 2005. A molecular chaperone for mitochondrial complex I assembly is mutated in a progressive encephalopathy. J. Clin. Investig. 115:2784-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ott, M., M. Prestele, H. Bauerschmitt, S. Funes, N. Bonnefoy, and J. M. Herrmann. 2006. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 25:1603-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perier, C., K. Tieu, C. Guegan, C. Caspersen, V. Jackson-Lewis, V. Carelli, A. Martinuzzi, M. Hirano, S. Przedborski, and M. Vila. 2005. Complex I deficiency primes Bax-dependent neuronal apoptosis through mitochondrial oxidative damage. Proc. Natl. Acad. Sci. USA 102:19126-19131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rapaport, D., R. D. Taylor, M. Kaser, T. Langer, W. Neupert, and F. E. Nargang. 2001. Structural requirements of Tom40 for assembly into preexisting TOM complexes of mitochondria. Mol. Biol. Cell 12:1189-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan, M. T., W. Voos, and N. Pfanner. 2001. Assaying protein import into mitochondria. Methods Cell Biol. 65:189-215. [DOI] [PubMed] [Google Scholar]
- 24.Sazanov, L. A., and P. Hinchliffe. 2006. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311:1430-1436. [DOI] [PubMed] [Google Scholar]
- 25.Sazanov, L. A., S. Y. Peak-Chew, I. M. Fearnley, and J. E. Walker. 2000. Resolution of the membrane domain of bovine complex I into subcomplexes: implications for the structural organization of the enzyme. Biochemistry 39:7229-7235. [DOI] [PubMed] [Google Scholar]
- 26.Schagger, H. 2002. Respiratory chain supercomplexes of mitochondria and bacteria. Biochim. Biophys. Acta 1555:154-159. [DOI] [PubMed] [Google Scholar]
- 27.Schagger, H., R. de Coo, M. F. Bauer, S. Hofmann, C. Godinot, and U. Brandt. 2004. Significance of respirasomes for the assembly/stability of human respiratory chain complex I. J. Biol. Chem. 279:36349-36353. [DOI] [PubMed] [Google Scholar]
- 28.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 29.Shoubridge, E. A. 2001. Cytochrome c oxidase deficiency. Am. J. Med. Genet. 106:46-52. [DOI] [PubMed] [Google Scholar]
- 30.Smeitink, J. A., M. Zeviani, D. M. Turnbull, and H. T. Jacobs. 2006. Mitochondrial medicine: a metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab. 3:9-13. [DOI] [PubMed] [Google Scholar]
- 31.Thorburn, D. R., C. Sugiana, R. Salemi, D. M. Kirby, L. Worgan, A. Ohtake, and M. T. Ryan. 2004. Biochemical and molecular diagnosis of mitochondrial respiratory chain disorders. Biochim. Biophys. Acta 1659:121-128. [DOI] [PubMed] [Google Scholar]
- 32.Triepels, R. H., L. P. Van Den Heuvel, J. M. Trijbels, and J. A. Smeitink. 2001. Respiratory chain complex I deficiency. Am. J. Med. Genet. 106:37-45. [DOI] [PubMed] [Google Scholar]
- 33.Tuschen, G., U. Sackmann, U. Nehls, H. Haiker, G. Buse, and H. Weiss. 1990. Assembly of NADH: ubiquinone reductase (complex I) in Neurospora mitochondria. Independent pathways of nuclear-encoded and mitochondrially encoded subunits. J. Mol. Biol. 213:845-857. [DOI] [PubMed] [Google Scholar]
- 34.Ugalde, C., R. J. Janssen, L. P. van den Heuvel, J. A. Smeitink, and L. G. Nijtmans. 2004. Differences in assembly or stability of complex I and other mitochondrial OXPHOS complexes in inherited complex I deficiency. Hum. Mol. Genet. 13:659-667. [DOI] [PubMed] [Google Scholar]
- 35.Ugalde, C., R. Vogel, R. Huijbens, B. Van Den Heuvel, J. Smeitink, and L. Nijtmans. 2004. Human mitochondrial complex I assembles through the combination of evolutionary conserved modules: a framework to interpret complex I deficiencies. Hum. Mol. Genet. 13:2461-2472. [DOI] [PubMed] [Google Scholar]
- 36.Vahsen, N., C. Cande, J. J. Briere, P. Benit, N. Joza, N. Larochette, P. G. Mastroberardino, M. O. Pequignot, N. Casares, V. Lazar, O. Feraud, N. Debili, S. Wissing, S. Engelhardt, F. Madeo, M. Piacentini, J. M. Penninger, H. Schagger, P. Rustin, and G. Kroemer. 2004. AIF deficiency compromises oxidative phosphorylation. EMBO J. 23:4679-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel, R., L. Nijtmans, C. Ugalde, L. van den Heuvel, and J. Smeitink. 2004. Complex I assembly: a puzzling problem. Curr. Opin. Neurol. 17:179-186. [DOI] [PubMed] [Google Scholar]
- 38.Vogel, R. O., R. J. Janssen, C. Ugalde, M. Grovenstein, R. J. Huijbens, H. J. Visch, L. P. van den Heuvel, P. H. Willems, M. Zeviani, J. A. Smeitink, and L. G. Nijtmans. 2005. Human mitochondrial complex I assembly is mediated by NDUFAF1. FEBS J. 272:5317-5326. [DOI] [PubMed] [Google Scholar]
- 39.Wallace, D. C. 2005. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu. Rev. Genet. 39:359-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiedemann, N., A. E. Frazier, and N. Pfanner. 2004. The protein import machinery of mitochondria. J. Biol. Chem. 279:14473-14476. [DOI] [PubMed] [Google Scholar]
- 41.Wiedemann, N., V. Kozjak, A. Chacinska, B. Schonfisch, S. Rospert, M. T. Ryan, N. Pfanner, and C. Meisinger. 2003. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424:565-571. [DOI] [PubMed] [Google Scholar]
- 42.Yadava, N., T. Houchens, P. Potluri, and I. E. Scheffler. 2004. Development and characterization of a conditional mitochondrial complex I assembly system. J. Biol. Chem. 279:12406-12413. [DOI] [PubMed] [Google Scholar]