Abstract
Smad7 plays an essential role in the negative-feedback regulation of transforming growth factor β (TGF-β) signaling by inhibiting TGF-β signaling at the receptor level. It can interfere with binding to type I receptors and thus activation of receptor-regulated Smads or recruit the E3 ubiquitin ligase Smurf to receptors and thus target them for degradation. Here, we report that Smad7 is predominantly localized in the nucleus of Hep3B cells. The targeted expression of Smad7 in the nucleus conferred superior inhibitory activity on TGF-β signaling, as determined by reporter assay in mammalian cells and by its effect on zebrafish embryogenesis. Furthermore, Smad7 repressed Smad3/4-, Smad2/4-, and Smad1/4-enhanced reporter gene expression, indicating that Smad7 can function independently of type I receptors. An oligonucleotide precipitation assay revealed that Smad7 can specifically bind to the Smad-responsive element via its MH2 domain, and DNA-binding activity was further confirmed in vivo with the promoter of PAI-1, a TGF-β target gene, by chromatin immunoprecipitation. Finally, we provide evidence that Smad7 disrupts the formation of the TGF-β-induced functional Smad-DNA complex. Our findings suggest that Smad7 inhibits TGF-β signaling in the nucleus by a novel mechanism.
The roles of transforming growth factor β (TGF-β) and related growth factors in tissue development and homeostasis have been well documented (15, 20, 31, 55). By modulating the expression of various target genes, these factors regulate cell proliferation, migration, differentiation, and death, and dysregulation of their signaling led to a variety of human disorders, including cancer, tissue fibrosis, and bone-related diseases (8, 11, 12, 32, 33, 36, 37, 52). TGF-β transduces its signaling from the membrane to the nucleus through serine/threonine kinase receptors and their downstream effectors, Smad proteins. Of the three subtypes of Smads, receptor-regulated Smads (R-Smads) are directly phosphorylated by type I receptors and form oligomeric complexes with common mediator Smads. The Smad complexes then accumulate in the nucleus and regulate target gene transcription through cooperation with other DNA-binding factors and/or transcription factors (34, 47). The third class of Smads comprises the inhibitory Smads (I-Smads), Smad6 and Smad7. Unlike signaling mediator Smad proteins, I-Smads antagonize TGF-β signaling.
Although the receptor/Smad-mediated TGF-β signaling pathway is relatively simple, it is stringently regulated. One of the negative-feedback regulations of TGF-β signaling is via upregulation of I-Smad proteins (1, 22, 51). Smad7 is a direct gene target of Smad3/4 (9, 39). Smad7 also acts as a mediator for cross talk between TGF-β and other signaling pathways, as its expression is induced by gamma interferon/STAT, tumor necrosis factor alpha/NF-κB, and epidermal growth factor (1, 4, 53).
Although Smad7 has been suggested to be important for TGF-β-induced apoptosis by interacting with β-catenin (14), I-Smad proteins have been mainly suggested to exert their negative effects on TGF-β/BMP signaling by multiple mechanisms. First, it has been reported that I-Smads inhibit signaling through stable binding to activated type I receptors and competition with R-Smads for receptor activation (16, 19, 22, 40, 49). Second, Smad7 can recruit the E3 ubiquitin ligases Smurf1 and Smurf2 to the type I receptors, leading to the degradation of the type I receptors (13, 24), or recruit protein phosphatase 1 to inactivate TβRI (46). Smad6 can also mediate Smurf1 to induce ubiquitination and degradation of BMP type I receptors and Smad1/5 (38). Third, it has been shown that Smad6 forms a complex with phosphorylated Smad1 and therefore disrupts the formation of the functional R-Smad-common mediator Smad complex (17). Finally, it has been reported that Smad6 can function in the nucleus to inhibit BMP signaling. It was shown to act as an intracellular (probably in the nucleus) antagonist of TGF-β family responses in Xenopus (41, 42). Smad6 acts as a transcriptional repressor by interacting with Hoxc-8 (3) or binds to DNA and recruits transcriptional corepressor histone deacetylases or CtBP to inhibit the transcription of target genes (2, 28).
Although a nuclear function of Smad6 has been suggested, whether Smad7 could function in the nucleus to interfere with TGF-β signaling has not been explored. In the present study, we provide evidence that Smad7 can inhibit TGF-β signaling in the nucleus. Forced expression of Smad7 in the nucleus potently repressed the transcriptional activity of TGF-β, and the inhibitory effect of Smad7 could be TGF-β type I receptor (TβRI) independent, as demonstrated in TβRI-deficient R1B/L17 cells. Furthermore, Smad7 was able to bind DNA in vivo and in vitro and disrupted the formation of functional Smad-DNA complexes. Our results provide a novel mechanism underlying Smad7 inhibition of TGF-β signaling.
MATERIALS AND METHODS
Cells and plasmids.
HEK293T, COS1, COS7, and HeLa cells were maintained in Dulbecco's modified Eagle's medium (GIBCO) supplemented with 10% fetal bovine serum (HyClone) in 5% CO2 at 37°C in a humidified atmosphere. Hep3B and R1B/L17 cells were grown in minimum essential medium (GIBCO) supplemented with 10% fetal bovine serum. Deletion mutants of Smad7, NLS-Smad7, which contain a nuclear localization signal (NLS) sequence (KKKRK) of simian virus 40 (SV40) large T antigen at the N terminus, were constructed in pcDNA3.1(+) (Invitrogen) by a PCR-based approach, and the sequences were confirmed by DNA sequencing. Smad7(ΔPY) mutants, which lack the PY motif (PPPPY) in the middle region, were constructed by PCR, and the sequence was confirmed by DNA sequencing. For in vitro synthesis of mRNA, Smad7, NLS-Smad7, Smad7(1-408), and Smad7-MH2 were subcloned into pCS2.
Reporter assay and immunoblotting.
Reporter assays and immunoblotting were performed as described previously (7).
DNA oligonucleotide precipitation.
Biotinylated activin response element (ARE) oligonucleotide (for the sequence, see Fig. 7B) was synthesized by Sangon (Shanghai, China). Oligonucleotide precipitation was carried out as described previously (44). Briefly, 750 μl of whole-cell lysates from HEK293T cells was incubated with 10 μg biotinylated oligonucleotide and streptavidin beads (Pierce) at 4°C overnight in lysis buffer containing 100 mM KCl, 10 mM HEPES (pH 7.9), 10% glycerol, 1 mM dithiothreitol, 5 mM MgCl2, 0.5% NP-40, 10 mM NaF, 20 mM beta-glycerophosphate, and proteinase inhibitors (Roche). After being extensively washed with lysis buffer, DNA-bound protein was then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by immunoblotting using appropriate antibodies.
FIG. 7.
Smad7 binds to DNA via its MH2 domain. (A) Smad7 binds to the ARE oligonucleotide via its MH2 domain. HEK293T cells were transfected with expression plasmids as indicated and treated with 100 pM TGF-β1 for 2 h. The cell lysates were harvested for oligonucleotide precipitation assay with the biotin-labeled 51-bp ARE oligonucleotide. DNA-bound proteins (top) and total protein levels (middle and bottom) were analyzed by anti-Flag or anti-Myc immunoblotting. FL, Full length. (B) Smad7 binds to ARE as confirmed by EMSA. HEK293T cells were transfected with Flag-Smad7 and treated with 100 pM TGF-β1 for 2 h. Nuclear extracts were used to perform gel mobility shift assays by using the ARE oligonucleotide as a probe in the presence of anti-Smad7 antibody or preimmune antiserum as indicated. The asterisk above lane 5 indicates that the probe was incubated with anti-Smad7 antibody in the absence of any nuclear extract. (C) Sequence of the ARE of the Mix.2 promoter. The SBE and FoxH1 binding sites are underlined. The dots indicate the mutated nucleotides in SBE. (D) The Smad7 MH2 domain specifically binds to the ARE, but not to its mutant (mut) sequence. GST fused to N-terminal Smad7 MH2 was expressed and purified from Escherichia coli. The oligonucleotide precipitation assay was carried out as described for panel A. (E) The binding of GST-Smad7 protein to DNA is specific. GST-Smad7 protein was purified from E. coli. EMSA was carried out using ARE, PAI-1 sequence, and their mutants. (F) Smad7 associates with the PAI-1 promoter in Hep3B cells under physiological conditions. A ChIP assay was performed with anti-Smad7 antibody. PCR amplification of the distal PAI-1 promoter (−744/−522) was performed to detect Smad7-bound DNA (anti-Smad7 ChIP). DNA input and Smad7 expression are shown. β-Actin acted as a negative control. TGF-β1-induced PAI-1 mRNA expression was detected by RT-PCR in Hep3B cells (right). Total RNA was isolated from Hep3B cells treated with TGF-β1 (100 pM) for 3 h. GAPDH mRNA expression served as an RT-PCR control. IgG, immunoglobulin G.
Immunofluorescence.
Cells were grown on coverslips, fixed with 3% paraformaldehyde, and blocked with 10% bovine serum albumin in phosphate-buffered saline (pH 7.0). The cells were then incubated with primary antibodies, followed by fluorescein isothiocyanate (FITC)-conjugated secondary antibodies. The nuclei were counterstained with DAPI (4′,6′-diamidino-2-phenylindole). Images were obtained with a confocal Olmypus fluoview 500 microscope.
RNA interference (RNAi).
pSUPER.retro (OligoEngine) was used for the expression of small interfering RNA (siRNA). The target sequence of human Smad7 is 5′-AGGTCACCACCATCCCCAC-3′ (46). A nonspecific siRNA expression vector, pNS, containing the sequence 5′-AGCGGACTAAGTCCATTGC-3′, was constructed as a negative control. Oligonucleotides were synthesized (Bioasin, Shanghai, China) and inserted into the pSUPER.retro vector in the SalI and KpnI sites.
Apoptosis assay.
Cells were harvested and then stained with 5 μg/ml annexin V-FITC and 5 μg/ml propidium iodide in the dark for 15 min at room temperature. Samples were analyzed by flow cytometry with a FACScan instrument (Becton-Dickinson, San Jose, CA). Those cells with negative propidium iodide staining and positive annexin V staining were considered the ones actively undergoing apoptosis, and the total number of these cells compared to the total number of analyzed cells was recorded.
In vitro synthesis of mRNA and microinjection of zebrafish embryos.
Capped mRNAs were in vitro synthesized with the Cap-Scribe Kit (Roche). The synthesized mRNA was purified using an RNAeasy Mini Kit (QIAGEN) and dissolved in nuclease-free water. Appropriate amounts of synthetic mRNA, as indicated in the figure legends, were injected into one-cell embryos using a gas-driven microinjector (Sutter Instruments). The injection dose was an estimated amount received by a single embryo. For mRNA injection experiments, the control embryos were injected with green fluorescent protein mRNA.
Reverse transcription-PCR (RT-PCR).
Total RNA was prepared from Hep3B cells using Trizol reagent (Roche) and treated with DNase (Takara). Two micrograms of RNA was reverse transcribed at 42°C for 45 min in a 20-μl reaction mixture using the Reverse Transcription System (Promega). Expression levels of plasminogen activator inhibitor 1 (PAI-1) were detected by semiquantitative PCR with the following primers: 5′-GTGGTCTGTGTCACCGTATC-3′ (forward) and 5′-GTAGTTGAATCCGAGCTGCC-3′ (reverse). The primers for the glyceraldehyde 3′-phosphate dehydrogenase (GAPDH) gene were 5′-GAAGGTGAAGGTCGGAGTC-3′ (forward) and 5′-GAAGATGGTGATGGGATTTC-3′ (reverse).
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were carried out essentially as described previously (45). Hep3B cells were treated with 200 pM TGF-β1 for 3 h. The cell lysates were subjected to anti-Smad7 immunoprecipitation. Smad7-precipitated genomic-DNA pellets were subjected to PCR. The primers used to amplify the human PAI-1 promoter harboring the Smad-binding elements (SBE) were 5′-CCTCCAACCTCAGCCAGACAAG-3′ (forward) and 5′-CCCAGCCCAACAGCCACA-3′ (reverse) (26). β-Actin was used as a negative control. The primers were 5′-AGCCATGTACGTTGCTATCCAG-3′ (forward) and 5′-CTTCTCCTTAATGTCACGCACG-3′ (reverse).
Elecrophoretic mobility shift assay (EMSA).
After HEK293T cells were transfected with the indicated plasmids for 48 h, nuclear extracts were prepared by suspending the cells in a hypotonic buffer containing 10 mM HEPES (pH 7.9), 10 mM KCl, 0.1 mM EDTA, and 0.1% NP-40 with proteinase inhibitors on ice for 10 min. Then, the nuclear fraction was recovered by centrifugation and incubated at 4°C for 1 h with a hypertonic buffer containing 20 mM HEPES (pH 7.9), 400 mM NaCl, 0.5 mM EDTA, 0.5 mM EGTA, 1.2 mM MgCl2, 12.5% glycerol, and 0.2% NP-40 with proteinase inhibitors. After centrifugation, the supernatant was recovered as nuclear extract. The ARE and PAI-1 oligonucleotide probes were synthesized by Sangon (Shanghai, China) and labeled with [32P]ATP with T4 polynucleotide kinase (Takara). The PAI-1 oligonucleotides were 5′-TCGAGAGCCAGACAAAAAGCCAGACATTTAGCCAGACAC-3′ and its complementary sequence. The mutant sequence was 5′-TCGAGAGCTACATAAAAAGCTACATATTTAGCTACATAC-3′ and its complementary sequence (10). DNA-binding assays were performed essentially as described previously (21). DNA-protein complexes were resolved on 6% (40:1) polyacrylamide gels containing 1% glycerol.
RESULTS
Smad7 is predominantly located in the nuclei of Hep3B cells.
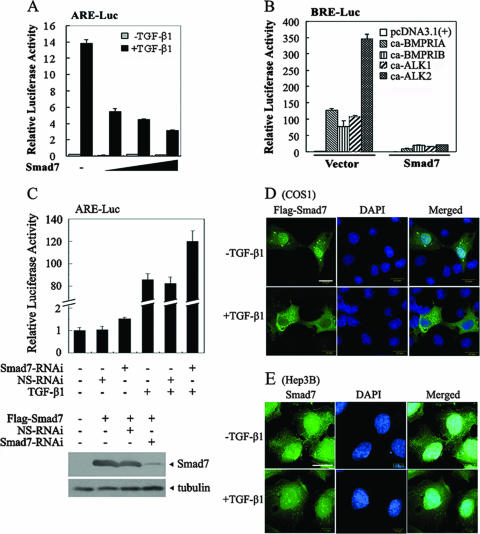
It has been reported that Smad7 negatively regulates TGF-β signaling by interacting with type I receptors and acting as a pseudosubstrate or by recruiting the ubiquitin E3 ligase Smurf and targeting receptors for degradation (13, 19, 24, 40). As Smad7 was shown to be exported from the nucleus upon TGF-β treatment in COS1 cells (23), it is generally thought that Smad7 exerts its inhibitory effect in the cytoplasm. However, a potential nuclear function of Smad7 has also been suggested in various cells under different conditions (16, 50, 57). To further delineate the mechanism whereby Smad7 inhibits TGF-β signaling, the effect of Smad7 on the transcriptional activity of TGF-β and the subcellular localization of Smad7 were investigated. Human heptoma Hep3B cells have been widely used to study the functions of TGF-β in transcription regulation and apoptosis. Hep3B cells were transfected with Smad7 and the TGF-β-responsive reporter ARE-luciferase, which expresses luciferase under the control of the ARE derived from the Mix.2 promoter (21). As shown in Fig. 1A, overexpression of Smad7 efficiently repressed TGF-β-induced expression of ARE-luciferase in Hep3B cells in a dose-dependent manner. Furthermore, Smad7 inhibited the expression of the BMP-responsive reporter BRE-Luc (18) induced by the constitutively active forms of BMPRIA, BMPRIB, ALK1, and ALK2 (Fig. 1B). To examine the function of endogenous Smad7, a vector-based short hairpin RNA was generated to interfere with Smad7 expression. Down-regulation of endogenous Smad7 by RNA interference not only led to a higher basal expression of the ARE-luciferase reporter, but also increased TGF-β1-induced expression of the reporter in Hep3B cells, whereas a nonspecific short hairpin RNA had no effect (Fig. 1C). Together, these data are consistent with the previous results showing that Smad7 functions as a general antagonist of the TGF-β superfamily.
FIG. 1.
Smad7 is localized in the nucleus in Hep3B cells regardless of TGF-β1 treatment. (A) Smad7 inhibits the TGF-β-induced expression of the ARE-luciferase reporter in a dose-dependent manner. ARE-luciferase (0.5 μg), Renilla luciferase (20 ng) and Flag-Smad7 (0.1 μg, 0.5 μg and 1 μg, respectively) were cotransfected into Hep3B cells. Twenty-four hours posttransfection, cells were either treated with TGF-β1 (50 pM) for 20 h or left untreated and then harvested for luciferase assay. The experiment was performed in triplicate, and the data represent the means and standard deviations of three independent experiments after normalization to Renilla luciferase activity. (B) Smad7 represses expression of the BRE-luciferase reporter induced by various constitutively active forms of type I receptors. Hep3B cells were cotransfected with BRE-luciferase (0.5 μg), and constitutively active (ca) type I receptor (0.3 μg) with or without Flag-Smad7 (0.1 μg). The reporter assay was performed similarly to that in panel A. (C) Knockdown of endogenous Smad7 results in enhanced expression of the ARE-luciferase reporter. Hep3B cells were cotransfected with ARE-luciferase and Smad7-specific siRNA (Smad7-RNAi) (0.2 μg) or a nonspecific siRNA (NS-RNAi). The reporter assay was performed similarly to that in panel A. RNAi efficiency was examined in HEK293T cells. Flag-Smad7 (2 μg) was cotransfected with Smad7-RNAi (4 μg) or NS-RNAi (4 μg). At 40 h posttransfection, the cells were harvested for anti-Flag immunoblotting. (D) TGF-β1 induces translocation of Smad7 from the nucleus to the cytoplasm in COS1 cells. COS1 cells were transfected with Flag-Smad7. After 40 h of transfection, cells were treated with 100 pM TGF-β1 or left untreated. Flag-Smad7 subcelluar localization was detected by indirect immunofluorescence using anti-Flag antibody followed by FITC-conjugated secondary antibody and visualized under a fluorescence microscope. (E) TGF-β1 does not alter the nuclear localization of Smad7 in Hep3B cells. Subcellular localization of endogenous Smad7 was detected by indirect immunofluorescence with anti-Smad7 antibody and FITC-conjugated secondary antibody and visualized under a fluorescence microscope. Nuclei were stained with DAPI (blue). Scale bar, 20 μm.
Next, we examined the subcellular localization of Smad7 in the presence or absence of TGF-β in several cell types by immunofluorescence. In agreement with the previous reports (23), Smad7 was predominantly distributed in the nucleus, and TGF-β treatment led to translocation of Smad7 to the cytoplasm in COS1 cells (Fig. 1D). However, endogenous Smad7 protein mostly remained in the nucleus in Hep3B cells regardless of TGF-β1 treatment (Fig. 1E), and similar results were obtained in HeLa cells (data not shown). We have examined the subcellular localization of Smad7 in other cell lines and found that Smad7 localization and responses to TGF-β stimulation vary depending on the cell types. Smad7 mainly resides in the cytoplasm in HepG2 and HaCat cells, which is in agreement with previous studies (14, 16). However, Smad7 mostly accumulated in the nucleus in Mv1Lu mutant L17 cells and in human normal lung epithelial HPL-1 cells, regardless of TGF-β stimulation (data not shown). These data together suggest that the subcellular localization of Smad7 varies in different cell types and that Smad7 may function as an antagonist of TGF-β signaling in the nucleus.
Smad7 inhibits TGF-β signaling in the nucleus.
The above-mentioned results suggest that Smad7 might function in the nucleus to interfere with TGF-β signaling. First, we attempted to map the domains responsible for the nuclear localization by generating a series of truncated forms of Smad7 (Fig. 2A). Smad7(180-426) and Smad7-MH2 resembled wild-type Smad7 [Smad7(WT)] and resided in the nucleus, and other deletion mutants, including Smad7(90-426) and C-terminally deleted mutants, were mainly found in the cytoplasm of both HeLa and Hep3B cells (Fig. 2B), indicating that the first 90 residues and the MH2 domain (amino acids 260 to 426) are required for nuclear localization.
FIG. 2.
Subcellular localization of Smad7 mutants. (A) Schematic diagram of the structure of Smad7 and its truncation mutants. Smad7(WT) represents wild-type Samd7. NLS-Smad7 contains an N-terminally fused NLS from the SV40 large T antigen. The Smad7 MH2 domain is located at the C terminus from amino acids 260 to 426. (B) The MH2 domain is important for Smad7 nuclear localization. Hep3B and HeLa cells were transfected with wild type or mutant Flag-tagged Smad7. After 40 h, their subcellular localizations were determined by indirect immunofluorescence with anti-Flag antibody and FITC-conjugated secondary antibody. Scale bars, 20 μm. (C) Nuclear localization of NLS-Smad7. Hep3B cells transfected with Smad7(WT) or NLS-Smad7 were treated with TGF-β1 for 45 min. Then, their subcellular localization was determined by indirect immunofluorescence with anti-Flag antibody and FITC-conjugated secondary antibody. Scale bar, 20 μm.
To directly test if Smad7 can function in the nucleus, we generated a Smad7 variant that contained the NLS sequence (KKKRK) of the SV40 large T antigen at the N terminus (NLS-Smad7). Immunofluorescence revealed that NLS-Smad7 was exclusively in the nucleus in Hep3B cells, and the nuclear localization was not apparently affected by TGF-β treatment (Fig. 2C).
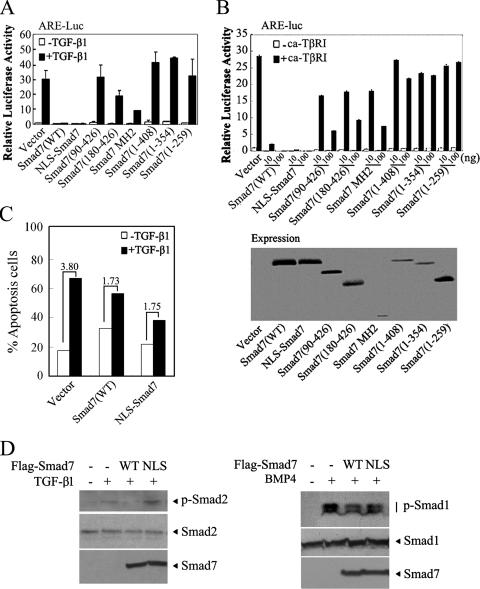
We next examined the abilities of these Smad7 mutants to inhibit TGF-β signaling in Hep3B cells. As shown in Fig. 3A, Smad7(WT), Smad7(180-426), and Smad7-MH2, all of which reside in the nucleus, retained the ability to inhibit the TGF-β1-stimulated expression of ARE-luciferase, although Smad7(180-427) was less effective. In contrast, the mutants that are mainly localized in the cytoplasm lost this inhibitory ability. The inhibitory effects of most mutants on the constitutively active form of TβRI (ca-TβRI) was very similar to their effects on TGF-β1, except for Smad7(90-426), which attenuated ARE-luciferase expression enhanced by ca-TβRI but not by TGF-β1 (Fig. 3B). The inhibitory function of Smad7(90-426) on ca-TβRI was probably due to its ability to interact with TβRI (data not shown). Similar results were obtained when CAGA-Luc, another TGF-β/Smad3-responsive reporter (10), was used to examine the activities of these Smad7 variants (data not shown).
FIG. 3.
The nuclear localization of Smad7 is correlated with its ability to repress the transcriptional activity of TGF-β. (A) Effects of Smad7 mutants on the TGF-β1-induced expression of ARE-luciferase. Smad7 and its variants (0.1 μg) were cotransfected with an ARE-luciferase construct into Hep3B cells. Twenty-four hours posttransfection, cells were treated with TGF-β1 (50 pM) for 20 h or left untreated and then harvested for luciferase assay. (B) Effects of Smad7 mutants on the expression of ARE-luciferase induced by ca-TβRI. ARE-luciferase was cotransfected with Smad7 and ca-TβRI into Hep3B cells as indicated. The cells were harvested for luciferase assay at 48 h posttransfection. The reporter assay was performed in triplicate, and the data represent the means and standard deviations of three independent experiments after normalization to Renilla luciferase activity. The expression levels of Smad7 variants are shown by immunoblotting (right). (C) NLS-Smad7 interferes with TGF-β-induced Hep3B apoptosis. After transfection with various constructs as indicated, the cells were treated with or without 200 pM TGF-β1 for 40 h and then subjected to apoptosis analysis. The degree of TGF-β-induced apoptosis is indicated. (D) NLS-Smad7 has no effect on ligand-induced R-Smad phosphorylation. Hep3B cells transfected with Flag-tagged Smad7(WT) or NLS-Smad7 were treated with 200 pM TGF-β1 or with 1 nM BMP4 for 45 min. The cells were subsequently harvested for immunoblotting with anti-phospho-Smad2 or anti-phospho-Smad1 (upper blots). Total protein expression was confirmed by immunoblotting with anti-Smad2 and anti-Flag antibodies (middle and lower blots).
Furthermore, NLS-Smad7 retained the ability to inhibit the expression of ARE-luciferase stimulated by TGF-β1 (Fig. 3A) and, even more effectively than Smad7(WT), to block ca-TβRI activity at low concentrations (Fig. 3B). To further investigate the significance of Smad7 in the nucleus, interference with TGF-β-induced apoptosis by NLS-Smad7 was studied. As shown in Fig. 3C, like Smad7(WT), NLS-Smad7 interfered with TGF-β-promoted apoptosis of Hep3B cells.
As NLS-Smad7 resides in the nucleus, it presumably loses its ability to influence R-Smad phosphorylation. To test this hypothesis, Hep3B cells were transfected with Smad7(WT) or NLS-Smad7 and treated with TGF-β1 or BMP4, and phosphorylation of Smad2 or Smad1 was examined, respectively. As shown in Fig. 3D, Smad7(WT) effectively attenuated ligand-induced R-Smad phosphorylation.
Expression of the Smad7 variants located in the nucleus in zebrafish embryos results in abnormal embryo development.
In order to confirm the nuclear functions of Smad7 in vivo, three variants of Smad7, NLS-Smad7, Smad7-MH2, and Smad7(1-408), were chosen, and their mRNAs were injected into zebrafish embryos at the one-cell stage. Injection of 30 pmol Smad7(WT) mRNA caused embryo abnormalities, such as cyclopic embryos, lack of axial structures, and reduced trunk mesoderm (Fig. 4A), phenotypes resembling those of embryos with defective Nodal signaling, as suggested in previous studies (43). When 15 pmol mRNA was injected, Smad7(WT) caused gastrulation defects in 53% of injected embryos, and NLS-Smad7 induced gastrulation defects in 93% of injected embryos (Fig. 4B and C), consistent with the above-mentioned reporter assay results showing that NLS-Smad7 is a more potent antagonist of TGF-β signaling than Smad7(WT). Furthermore, in agreement with the reporter assay data, expression of the MH2 domain interfered with embryo development, while Smad7(1-408) generated no apparent abnormal phenotype. These in vivo results further support the role of Smad7 in the nucleus.
FIG. 4.
Nuclear localization of Smad7 is important for its inhibitory activity in zebrafish embryos. (A) Embryos were injected with 30 pmol of mRNA encoding wild-type Smad7 at the single-cell stage. The control embryos were injected with the same amount of green fluorescent protein mRNA. The embryo phenotype was observed at 24 h postfertilization. The embryos are shown in lateral views, with anterior to the left. (B) Experiments were performed as in panel A, except that 15 pmol mRNA was injected. The embryo phenotype was observed at 24 h postfertilization. (C) Summary of embryo phenotype at 24 h postfertilization from panel B. The gastrulation defects were defined as cyclopic embryos, lack of axial structures, and reduced trunk mesoderm.
A Smurf-binding-deficient Smad7 mutant is localized in the nucleus and retains inhibitory activity.
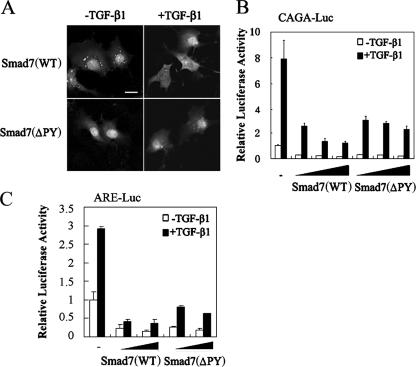
It has been shown that the the E3 ubiquitin ligase Smurf binds to Smad7 through a PPXY sequence (PY motif) in the linker region of Smad7 and induces the nuclear export of Smad7 (13, 24). It was proposed that Smad7 interferes with TGF-β signaling by recruiting Smurf proteins to TβRI and inducing receptor degradation. To further examine whether the nuclear export of Smad7 is essential for its inhibitory function and to exclude the involvement of Smurf in Smad7-mediated inhibition, a Smad7(ΔPY) mutant with deletion of the PY motif was constructed. Consistent with early reports (13, 24), Smad7(ΔPY) could not interact with Smurf1 and stayed in the nucleus with or without Smurf coexpression in COS7 cells (data not shown). Although Smad7(WT) translocated to the cytoplasm upon TGF-β stimulation in COS7 cells (Fig. 5A, top), Smad7(ΔPY) resided in the nucleus regardless of TGF-β treatment (Fig. 5A, bottom). Furthermore, Smad7(ΔPY) suppressed the TGF-β-induced expression of both CAGA-luciferase and ARE-luciferase in a dose-dependent manner, although it might have been slightly less effective than Smad7(WT) (Fig. 5B and C). These data further confirm the inhibitory function of Smad7 in the nucleus.
FIG. 5.
Smurf-binding-deficient Smad7(ΔPY) is localized in the nucleus and retains the ability to inhibit TGF-β signaling. (A) TGF-β induces the nuclear export of Smad7(WT) but not Smad7(ΔPY) in COS7 cells. Cells were transfected with Flag-Smad7(WT) and Flag-Smad7(ΔPY). At 40 h posttransfection, cells were treated with 100 pM TGF-β1 for 45 min or left untreated. Smad7 subcellular localization was detected by indirect immunofluorescence using anti-Flag antibody, followed by FITC-conjugated secondary antibody, and visualized under a fluorescence microscope. Scale bar, 20 μm. (B) Smad7(ΔPY) suppressed CAGA-luciferase expression induced by TGF-β1. COS7 cells were transfected with CAGA-Luc (0.5 μg), Smad7(WT), and Smad7(ΔPY) (0.1, 0.5, and 1 μg, respectively) as indicated. At 24 h posttransfection, the cells were treated with TGF-β1 (50 pM) for 20 h or left untreated and then harvested for luciferase assay. The experiment was performed in triplicate, and the data represent the means and standard deviations of three independent experiments after normalization to Renilla luciferase activity. (C) Smad7(ΔPY) suppressed ARE-luciferase expression induced by TGF-β1. COS7 cells were transfected with ARE-Luc (0.5 μg) and with Smad7(WT) and Smad7(ΔPY) (0.1 and 0.5 μg, respectively) as indicated. The reporter assay was performed similarly to that in panel B.
Smad7 inhibits TGF-β/BMP signaling independently of type I receptors.
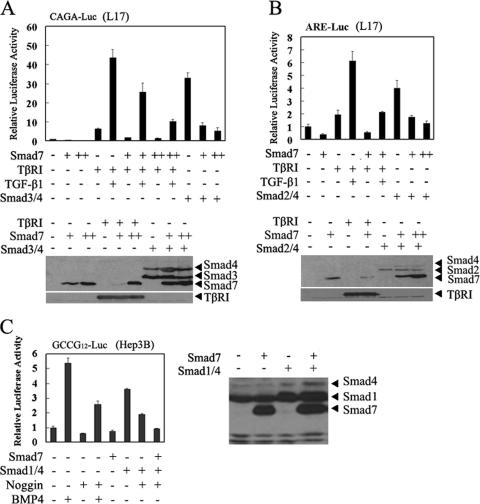
Several lines of evidence have demonstrated that Smad7 negatively regulates TGF-β signaling at the type I receptor level. To provide further evidence to support the notion that Smad7 can function in the nucleus, we took advantage of TβRI-defective R1B/L17 cells, which were derived from mink lung epithelial MvlLu cells (5). Consistent with previous data, introduction of TβRI restored the responsiveness of R1B/L17 cells to TGF-β, and Smad7 attenuated TGF-β-induced expression of CAGA-luciferase in a dose-dependent manner (Fig. 6A). As expected, overexpression of Smad3 and Smad4 stimulated CAGA-luciferase expression independently of TGF-β and TβRI. Importantly, the Smad3/4-enhanced expression of CAGA-luciferase was blocked by Smad7 in a dose-dependent manner (Fig. 6A), and this inhibition was specific, as Smad7 had no effect on the expression of Smad3 and Smad4 (Fig. 6A, bottom). Similar results were obtained with the Smad2-responsive reporter ARE-luciferase (Fig. 6B). Smad7 also inhibited the expression of the BMP-responsive reporter GCCG12-luciferase (27) induced by Smad1 and Smad4 in Hep3B cells (Fig. 6C). These results strongly indicate that Smad7 could repress the transcriptional activity of signaling Smads without the involvement of type I receptors.
FIG. 6.
Smad7 inhibits the transcriptional activities of Smad3/4, Smad2/4, and Smad1/4. (A) Smad7 inhibits the expression of CAGA-luciferase induced by overexpression of Smad3 and Smad4 in R1B/L17 cells. TβRI-deficient R1B/L17 cells were transfected with CAGA-luciferase (0.5 μg), TβRI (0.1 μg), Smad3 and Smad4 (0.1 μg), and Smad7 (0.1 μg and 0.2 μg) as indicated. At 24 h posttransfection, cells were treated with TGF-β1 (50 pM) for 20 h or left untreated and then harvested for luciferase assay and immunobloting. Protein expression is shown below. (B) Smad7 inhibits the expression of ARE-luciferase induced by overexpression of Smad2 and Smad4 in R1B/L17 cells. TβRI-deficient R1B/L17 cells were transfected with ARE-luciferase (0.5 μg), TβRI (0.1 μg), Smad2 and Smad4 (0.2 μg and 0.1 μg, respectively), and Smad7 (0.1 μg and 0.3 μg) as indicated. At 24 h posttransfection, cells were treated with TGF-β1 (50 pM) for 20 h or left untreated and then harvested for luciferase assay. Protein expression is shown below. (C) Smad7 represses the expression of the BMP-responsive reporter GCCG12-luciferase induced by overexpression of Smad1 and Smad4 in Hep3B cells. Smad7 (0.3 μg) as indicated and GCCG12-luciferase (0.4 μg) were coexpressed with and without Smad1 (0.2 μg) and Smad4 (0.1 μg) in Hep3B cells. At 24 h posttransfection, cells were treated with BMP4 (1.6 nM) or noggin (2 nM) for another 20 h or left untreated and then harvested for luciferase assay. Protein expression is shown below. All reporter assays were performed in triplicate, and the data represent the means and standard deviations of three independent experiments after normalization to Renilla luciferase activity.
The Smad7 MH2 domain can directly bind to DNA.
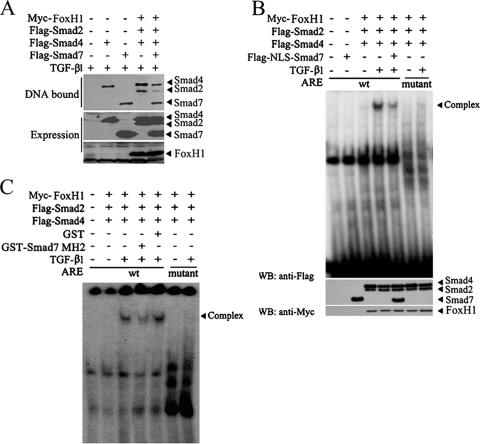
To explore the mechanism whereby Smad7 inhibits TGF-β signaling in the nucleus, we tested whether Smad7 possesses the capability to bind to DNA. An oligonucleotide precipitation assay was performed with a 51-bp biotinylated ARE DNA fragment derived from the Mix.2 promoter (21). The ARE fragment was incubated with the cell lysates from HEK293T cells transfected with Smad2, Smad4, and FoxH1, together with wild-type or mutant Smad7 constructs, and DNA-bound proteins were examined by immunoblotting. As shown in Fig. 7A, the ARE fragment could pull down Smad4, as well as Smad2, when FoxH1 was coexpressed, consistent with the previous observation that these proteins can bind to the DNA fragment (6, 29). Importantly, the ARE was also able to precipitate both Smad7(WT) and the MH2 domain, but not Smad7(1-259). To confirm this, the nuclear fraction was extracted from HEK293T cells transiently expressing Flag-Smad7 and used for EMSA. Flag-Smad7 specifically interacted with the ARE fragment, and the anti-Smad7 antibody supershifted the DNA-protein complex band to a slowly migrating one, whereas preimmune antiserum had no effect (Fig. 7B, lane 3 versus lane 4). A lack of apparent binding of anti-Smad7 to the ARE fragment further confirmed the specificity of Smad7-DNA interaction (lane 5). Notably, TGF-β had no effect on the Smad7-ARE complex (lanes 8 and 9). That the Smad7 MH2 domain is responsible for DNA binding was confirmed by recombinant glutathione S-transferase (GST)-Smad7-MH2, which specifically bound to the wild-type ARE sequence but not to the ARE mutant with two SBE mutated (Fig. 7C and D). EMSA with purified GST-Smad7 further confirmed that Smad7 can directly interact with DNA in a sequence-specific manner, not only with the ARE sequence but also with the DNA derived from the promoter of PAI-1 (10) (Fig. 7E). These results together indicate that the binding of Smad7 to DNA is sequence specific and that its binding sequences are possibly similar to those of other Smads.
The above-mentioned in vitro Smad7-DNA-binding results prompted us to investigate whether Smad7 can bind to the natural promoter of the TGF-β target genes in vivo. PAI-1 is a TGF-β immediately-early responsive gene in diverse cell types, including Hep3B cells (25, 54), and its expression was induced by TGF-β1 treatment (Fig. 7F, right). A ChIP assay showed that endogenous Smad7 could pull down the PAI-1 promoter DNA in Hep3B cells (Fig. 7F, left), indicating that Smad7 binds to the promoter of PAI-1 under physiological conditions.
Smad7 competes with the Smad2-Smad4-FoxH1 complex to bind to DNA.
Formation of the Smad-DNA complexes is essential for transcriptional activation of TGF-β target genes (6, 29, 56; reviewed in reference 35). As Smad7 is able to bind to a specific DNA sequence, we next tested whether Smad7 directly competes with the DNA binding of signaling Smad proteins and inhibits the formation of the functional Smad-DNA complex. Oligonucleotide precipitation was performed with the biotinylated ARE sequence and the cell lysates from the HEK293T cells transfected with the constructs indicated in Fig. 8A. As mentioned above, the ARE oligonucleotide could interact with Smad2 and Smad4 in the presence of FoxH1, as well as with Smad7. Coexpression of Smad7 apparently reduced the amount of DNA-binding Smad2/4 (Fig. 8A), suggesting that Smad7 directly impedes the formation of the Smad2/4-DNA complex.
FIG. 8.
Smad7 interferes with the DNA-binding activities of Smad2 and Smad4. (A) Smad7 decreases the DNA-binding activities of Smad2 and Smad4. Flag-Smad4, Flag-Smad2, and Myc-FoxH1 were cotransfected with Smad7 into HEK293T cells. At 48 h posttransfection, the cells were treated with 100 pM TGF-β1 for 2 h. Then, cell lysates were harvested for an oligonucleotide precipitation assay with the biotin-labeled ARE oligonucleotide. DNA-bound proteins (top) and total protein levels (middle and bottom) were analyzed by immunoblotting using anti-Flag or anti-Myc antibodies. (B) Smad7 attenuates TGF-β-induced formation of the Smad-DNA complex containing Smad2, Smad4, and FoxH1. HEK293T cells were transfected with expression plasmids as indicated. At 48 h posttransfection, the cells were treated with 100 pM TGF-β1 for 2 h. EMSA was performed with nuclear extracts (top). Protein expression was analyzed by immunoblotting (Western blotting [WB], middle and bottom). (C) Recombinant GST-Smad7 MH2 protein reduced TGF-β-induced formation of the functional Smad-DNA complex. HEK293T cells were transfected with expression plasmids as indicated and harvested after 48 h. EMSA was performed with whole-cell extracts. GST-Smad7 MH2 or GST protein (0.5 μg) was used.
To further confirm the effect of Smad7 on DNA binding of Smad2/4, we forced Smad7 expression in the nucleus with the NLS and examined its effect by EMSA. When Smad2, Smad4, and FoxH1 were coexpressed in HEK293T cells, TGF-β induced the formation of the Smad2/4-DNA complex with the wild-type ARE sequences, but not with the ARE mutant (Fig. 8B). Expression of NLS-Smad7 remarkably decreased the amount of Smad2/4 binding to DNA. To examine the inhibitory effect of Smad7 on the functional Smad-DNA complex formation, we generated recombinant GST-Smad7-MH2 in bacteria. As shown in Fig. 8C, GST-Smad7-MH2 effectively disrupted the Smad2/4-DNA complex, while GST had no effect. Taken together, these results strongly support the idea that Smad7 inhibits TGF-β/Smad signaling in the nucleus by directly binding to DNA and disrupting the formation of signaling Smad-DNA complex.
DISCUSSION
TGF-β signaling is tightly regulated in both positive and negative manners. One of the important negative-feedback regulations is via induction of I-Smad expression (1, 22, 51). Smad6 and Smad7 were initially found to stably associate with the active type I receptors and to act as pseudosubstrates to interfere with R-Smad activation (19, 22, 40). In addition, Smad7 can also function as an adaptor protein to recruit the ubiquitin E3 ligases Smurf1 and Smurf2 to target type I receptors for degradation (13, 24). However, accumulating evidence suggests there are other mechanisms for I-Smads to inhibit TGF-β signaling. For example, Smad6 can act as a transcriptional corepressor and recruit CtBP and histone deacetylases to inhibit the transcription of genes in the nucleus (2, 28). In our study, we demonstrated for the first time that Smad7 inhibits TGF-β signaling in the nucleus. Our immunofluorescence analysis revealed that endogenous Smad7 remains in the nucleus regardless of TGF-β treatment in Hep3B cells. Ectopic expression of Smad7 in the nucleus via its conjugation with the NLS of SV40 large T antigen rendered a stronger inhibitory activity in reporter transcription assays and in vivo embryonic assays. Moreover, Smad7(ΔPY), which loses the ability to bind to Smurf and stays in the nucleus, also exerts an inhibitory effect on TGF-β-induced CAGA-luciferase reporter expression. We also showed that Smad7 can interfere with Smad3/4- and Smad1/4-stimulated reporter expression in TβRI-deficient R1B/L17 and Hep3B cells, respectively. Finally, Smad7 was able to directly bind to the ARE sequence via its MH2 domain and interfered with the formation of the functional Smad-DNA complex.
Subcellular localization of Smad7.
The subcellular localization of Smad7 may vary in different cell types. Exogenously expressed Smad7 has been shown to be located in the nucleus in COS1, COS7, and Mv1Lu cells, while it was predominantly found in the cytoplasm in HepG2 cells and TβRI-deficient R mutant MvlLu cells (16, 23). Although the Smurf-mediated nuclear export of Smad7 has been demonstrated by several studies (16, 24, 50), the effect of TGF-β or constitutively active TβRI(T204D) on Smad7 distribution has been controversial. Smad7 has been reported to be transported from the nucleus into the cytoplasm in MvlLu cells upon TGF-β stimulation and in COS1 cells when coexpressed with TβRI(T204D) (23), although TβRI(T204D)-induced nuclear export was not observed by Hanyu et al. (16). In this study, we also observed that TGF-β treatment induces nuclear export to the cytoplasm of Smad7 in COS1 and COS7 cells. Intriguingly, our data revealed that in Hep3B cells, endogenous Smad7 predominantly remains in the nucleus and that this localization is not substantially altered by TGF-β1 treatment, and similar results were also obtained in HeLa cells with ectopically expressed Smad7. The difference in Smad7 localization in various cells might be due to the presence of Smad7-interacting proteins in only certain types of cells. It is unclear whether cell culture conditions play a role in influencing the subcellular localization of Smad7, as suggested by Zhu et al. (57). In addition to Smurfs, the adaptor protein Axin has also been implicated in the induction of Smad7 nuclear export (30).
To further define the regions important for its nuclear localization, we generated a series of deletion mutants of Smad7. In accordance with the previous report by Itoh et al. (23), we found that the MH2 domain is located in the nucleus, and its integrity is important for Smad7 nuclear localization, as deletion of the last 18 amino acids [Smad7(1-408)] led to cytoplasmic distribution. We also found that Smad7(90-426) is mainly located in the cytoplasm and Smad7(180-426) is in the nucleus, suggesting that the region comprising amino acids 90 to 180 may contain a nuclear export signal or function to interfere with nuclear import activity. These results are different from the conclusion of Smad6/7 chimera studies that this region may be critical for the nuclear localization of Smad7 (16).
Function of Smad7 in the nucleus.
Whereas both Smad6 and Smad7 are generally thought to inhibit TGF-β signaling at the receptor level, several studies have reported that Smad6 can inhibit BMP signaling in the nucleus (2, 3, 28). Here, we showed that Smad7 could also function as an antagonist of TGF-β signaling in the nucleus. NLS-mediated forced expression of Smad7 conferred a potent inhibitory activity of Smad7 on zebrafish embryos and on the reporter expression induced by TGF-β or the active TβRI without influencing the phosphorylation of R-Smads. Furthermore, Smad7 could directly bind the DNA fragment derived from the ARE in vitro and the natural PAI-1 promoter in vivo. This binding is specific, as mutations in the minimal SBE abolished Smad7 association. Although the DNA-binding activities of Smads have been shown to be mediated by the MH1 domain (35, 48), we showed here that, unlike in other Smad proteins, the Smad7-MH2 domain is responsible for DNA binding. It is noteworthy that the N-terminal domain of Smad7 does not interact with DNA, consistent with the fact that the amino acid sequence in the N-terminal part of I-Smads is divergent from the MH1 domains of other Smad proteins. We further provided evidence that Smad7 could compete with functional Smad complexes to bind DNA, which is different from the earlier reports demonstrating that Smad6 functions in the nucleus to repress transcription by recruiting transcriptional corepressors, such as histone deacetylase, CtBP, and Hoxc-8 (2, 3, 28). Furthermore, this competition mechanism may be ubiquitous in the regulation of TGF-β superfamily signaling by Smad7, since it could act as a general inhibitor to repress the Smad-mediated expression of both TGF-β and BMP reporters. Our results thus suggest a novel mechanism for Smad7 to antagonize TGF-β signaling.
In summary, our findings extend our understanding of the molecular mechanisms underlying the negative regulation of TGF-β signaling by Smad7. In addition to interfering with R-Smad activation and targeting receptor for degradation in the cytoplasm, Smad7 can exert its inhibitory effect in the nucleus. Depending on the cell type or state, the modes of Smad7 action may work together or independently.
Acknowledgments
We are grateful to A. Moustakas for Smad7 antibody and to M. Kawabata for GCCG12-luciferase reporter.
This work was supported by grants from the National Natural Science Foundation of China to Y.-G.C. (30125021 and 30430360) and to X.-H.F. (30428002) and from the 973 Program (2004CB720002 and 2006CB943401) to Y.-G.C. Y.-G.C. is a Chueng Kong Scholar.
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Afrakhte, M., A. Moren, S. Jossan, S. Itoh, K. Sampath, B. Westermark, C. H. Heldin, N. E. Heldin, and P. ten Dijke. 1998. Induction of inhibitory Smad6 and Smad7 mRNA by TGF-β family members. Biochem. Biophys. Res. Commun. 249:505-511. [DOI] [PubMed] [Google Scholar]
- 2.Bai, S., and X. Cao. 2002. A nuclear antagonistic mechanism of inhibitory Smads in transforming growth factor-beta signaling. J. Biol. Chem. 277:4176-4182. [DOI] [PubMed] [Google Scholar]
- 3.Bai, S., X. Shi, X. Yang, and X. Cao. 2000. Smad6 as a transcriptional corepressor. J. Biol. Chem. 275:8267-8270. [DOI] [PubMed] [Google Scholar]
- 4.Bitzer, M., G. von Gersdorff, D. Liang, A. Dominguez-Rosales, A. A. Beg, M. Rojkind, and E. P. Bottinger. 2000. A mechanism of suppression of TGF-beta/SMAD signaling by NF-kappa B/RelA. Genes Dev. 14:187-197. [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, F. T., and J. Massague. 1989. Transforming growth factor-beta inhibition of epithelial cell proliferation linked to the expression of a 53-kDa membrane receptor. J. Biol. Chem. 264:2272-2278. [PubMed] [Google Scholar]
- 6.Chen, X., E. Weisberg, V. Fridmacher, M. Watanabe, G. Naco, and M. Whitman. 1997. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature 389:85-89. [DOI] [PubMed] [Google Scholar]
- 7.Chen, Y. G., F. Liu, and J. Massague. 1997. Mechanism of TGFβ receptor inhibition by FKBP12. EMBO J. 16:3866-3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datto, M., and X. F. Wang. 2000. The Smads: transcriptional regulation and mouse models. Cytokine Growth Factor Rev. 11:37-48. [DOI] [PubMed] [Google Scholar]
- 9.Denissova, N. G., C. Pouponnot, J. Long, D. He, and F. Liu. 2000. Transforming growth factor beta-inducible independent binding of SMAD to the Smad7 promoter. Proc. Natl. Acad. Sci. USA 97:6397-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennler, S., S. Itoh, D. Vivien, P. ten Dijke, S. Huet, and J. M. Gauthier. 1998. Direct binding of Smad3 and Smad4 to critical TGF beta-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 17:3091-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derynck, R., R. J. Akhurst, and A. Balmain. 2001. TGF-β signaling in tumor suppression and cancer progression. Nat. Genet. 29:117-129. [DOI] [PubMed] [Google Scholar]
- 12.Derynck, R., and Y. E. Zhang. 2003. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 425:577-584. [DOI] [PubMed] [Google Scholar]
- 13.Ebisawa, T., M. Fukuchi, G. Murakami, T. Chiba, K. Tanaka, T. Imamura, and K. Miyazono. 2001. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J. Biol. Chem. 276:12477-12480. [DOI] [PubMed] [Google Scholar]
- 14.Edlund, S., S. Y. Lee, S. Grimsby, S. Zhang, P. Aspenstrom, C. H. Heldin, and M. Landstrom. 2005. Interaction between Smad7 and beta-catenin: importance for transforming growth factor beta-induced apoptosis. Mol. Cell. Biol. 25:1475-1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, X. H., and R. Derynck. 2005. Specificity and versatility in TGF-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 21:659-693. [DOI] [PubMed] [Google Scholar]
- 16.Hanyu, A., Y. Ishidou, T. Ebisawa, T. Shimanuki, T. Imamura, and K. Miyazono. 2001. The N domain of Smad7 is essential for specific inhibition of transforming growth factor-beta signaling. J. Cell Biol. 155:1017-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hata, A., G. Lagna, J. Massague, and A. Hemmati-Brivanlou. 1998. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 12:186-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hata, A., J. Seoane, G. Lagna, E. Montalvo, A. Hemmati-Brivanlou, and J. Massague. 2000. OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell 100:229-240. [DOI] [PubMed] [Google Scholar]
- 19.Hayashi, H., S. Abdollah, Y. Qiu, J. Cai, Y. Y. Xu, B. W. Grinnell, M. A. Richardson, J. N. Topper, M. A. Gimbrone, Jr., J. L. Wrana, and D. Falb. 1997. The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell 89:1165-1173. [DOI] [PubMed] [Google Scholar]
- 20.Heldin, C. H., K. Miyazono, and P. ten Dijke. 1997. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 390:465-471. [DOI] [PubMed] [Google Scholar]
- 21.Huang, H. C., L. C. Murtaugh, P. D. Vize, and M. Whitman. 1995. Identification of a potential regulator of early transcriptional responses to mesoderm inducers in the frog embryo. EMBO J. 14:5965-5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imamura, T., M. Takase, A. Nishihara, E. Oeda, J. Hanai, M. Kawabata, and K. Miyazono. 1997. Smad6 inhibits signalling by the TGF-β superfamily. Nature 389:622-626. [DOI] [PubMed] [Google Scholar]
- 23.Itoh, S., M. Landstrom, A. Hermansson, F. Itoh, C. H. Heldin, N. E. Heldin, and P. ten Dijke. 1998. Transforming growth factor beta1 induces nuclear export of inhibitory Smad7. J. Biol. Chem. 273:29195-29201. [DOI] [PubMed] [Google Scholar]
- 24.Kavsak, P., R. K. Rasmussen, C. G. Causing, S. Bonni, H. Zhu, G. H. Thomsen, and J. L. Wrana. 2000. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol. Cell 6:1365-1375. [DOI] [PubMed] [Google Scholar]
- 25.Keeton, M. R., S. A. Curriden, A. J. van Zonneveld, and D. J. Loskutoff. 1991. Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforming growth factor beta. J. Biol. Chem. 266:23048-23052. [PubMed] [Google Scholar]
- 26.Kurisaki, K., A. Kurisaki, U. Valcourt, A. A. Terentiev, K. Pardali, P. Ten Dijke, C. H. Heldin, J. Ericsson, and A. Moustakas. 2003. Nuclear factor YY1 inhibits transforming growth factor beta- and bone morphogenetic protein-induced cell differentiation. Mol. Cell. Biol. 23:4494-4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kusanagi, K., H. Inoue, Y. Ishidou, H. K. Mishima, M. Kawabata, and K. Miyazono. 2000. Characterization of a bone morphogenetic protein-responsive Smad-binding element. Mol. Biol. Cell 11:555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, X., Y. Y. Liang, B. Sun, M. Liang, Y. Shi, F. C. Brunicardi, and X. H. Feng. 2003. Smad6 recruits transcription corepressor CtBP to repress bone morphogenetic protein-induced transcription. Mol. Cell. Biol. 23:9081-9093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, F., C. Pouponnot, and J. Massague. 1997. Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional complexes. Genes Dev. 11:3157-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, W., H. Rui, J. Wang, S. Lin, Y. He, M. Chen, Q. Li, Z. Ye, S. Zhang, S. C. Chan, Y. G. Chen, J. Han, and S. C. Lin. 2006. Axin is a scaffold protein in TGF-β signaling that promotes degradation of Smad7 by Arkadia. EMBO J. 25:1646-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massague, J. 1998. TGF-β signal transduction. Annu. Rev. Biochem. 67:753-791. [DOI] [PubMed] [Google Scholar]
- 32.Massague, J., S. W. Blain, and R. S. Lo. 2000. TGFβ signaling in growth control, cancer, and heritable disorders. Cell 103:295-309. [DOI] [PubMed] [Google Scholar]
- 33.Massague, J., and Y. G. Chen. 2000. Controlling TGF-β signaling. Genes Dev. 14:627-644. [PubMed] [Google Scholar]
- 34.Massague, J., J. Seoane, and D. Wotton. 2005. Smad transcription factors. Genes Dev. 19:2783-2810. [DOI] [PubMed] [Google Scholar]
- 35.Massague, J., and D. Wotton. 2000. Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 19:1745-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyazono, K., H. Suzuki, and T. Imamura. 2003. Regulation of TGF-β signaling and its roles in progression of tumors. Cancer Sci. 94:230-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moustakas, A., and C. H. Heldin. 2002. From mono- to oligo-Smads: the heart of the matter in TGF-β signal transduction. Genes Dev. 16:1867-1871. [DOI] [PubMed] [Google Scholar]
- 38.Murakami, G., T. Watabe, K. Takaoka, K. Miyazono, and T. Imamura. 2003. Cooperative inhibition of bone morphogenetic protein signaling by Smurf1 and inhibitory Smads. Mol. Biol. Cell 14:2809-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nagarajan, R. P., J. Zhang, W. Li, and Y. Chen. 1999. Regulation of Smad7 promoter by direct association with Smad3 and Smad4. J. Biol. Chem. 274:33412-33418. [DOI] [PubMed] [Google Scholar]
- 40.Nakao, A., M. Afrakhte, A. Moren, T. Nakayama, J. L. Christian, R. Heuchel, S. Itoh, M. Kawabata, N. E. Heldin, C. H. Heldin, and P. ten Dijke. 1997. Identification of Smad7, a TGF-β-inducible antagonist of TGF-β signalling. Nature 389:631-635. [DOI] [PubMed] [Google Scholar]
- 41.Nakayama, T., L. K. Berg, and J. L. Christian. 2001. Dissection of inhibitory Smad proteins: both N- and C-terminal domains are necessary for full activities of Xenopus Smad6 and Smad7. Mech. Dev. 100:251-262. [DOI] [PubMed] [Google Scholar]
- 42.Nakayama, T., H. Gardner, L. K. Berg, and J. L. Christian. 1998. Smad6 functions as an intracellular antagonist of some TGF-β family members during Xenopus embryogenesis. Genes Cells 3:387-394. [DOI] [PubMed] [Google Scholar]
- 43.Pogoda, H. M., and D. Meyer. 2002. Zebrafish Smad7 is regulated by Smad3 and BMP signals. Dev. Dyn. 224:334-349. [DOI] [PubMed] [Google Scholar]
- 44.Seoane, J., C. Pouponnot, P. Staller, M. Schader, M. Eilers, and J. Massague. 2001. TGFβ influences Myc, Miz-1 and Smad to control the CDK inhibitor p15INK4b. Nat. Cell Biol. 3:400-408. [DOI] [PubMed] [Google Scholar]
- 45.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 46.Shi, W., C. Sun, B. He, W. Xiong, X. Shi, D. Yao, and X. Cao. 2004. GADD34-PP1c recruited by Smad7 dephosphorylates TGFβ type I receptor. J. Cell Biol. 164:291-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi, Y., and J. Massague. 2003. Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113:685-700. [DOI] [PubMed] [Google Scholar]
- 48.Shi, Y., Y. F. Wang, L. Jayaraman, H. Yang, J. Massague, and N. P. Pavletich. 1998. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell 94:585-594. [DOI] [PubMed] [Google Scholar]
- 49.Souchelnytskyi, S., T. Nakayama, A. Nakao, A. Moren, C. H. Heldin, J. L. Christian, and P. ten Dijke. 1998. Physical and functional interaction of murine and Xenopus Smad7 with bone morphogenetic protein receptors and transforming growth factor-beta receptors. J. Biol. Chem. 273:25364-25370. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki, C., G. Murakami, M. Fukuchi, T. Shimanuki, Y. Shikauchi, T. Imamura, and K. Miyazono. 2002. Smurf1 regulates the inhibitory activity of Smad7 by targeting Smad7 to the plasma membrane. J. Biol. Chem. 277:39919-39925. [DOI] [PubMed] [Google Scholar]
- 51.Takase, M., T. Imamura, T. K. Sampath, K. Takeda, H. Ichijo, K. Miyazono, and M. Kawabata. 1998. Induction of Smad6 mRNA by bone morphogenetic proteins. Biochem. Biophys. Res. Commun. 244:26-29. [DOI] [PubMed] [Google Scholar]
- 52.ten Dijke, P., and C. S. Hill. 2004. New insights into TGF-beta-Smad signalling. Trends Biochem. Sci. 29:265-273. [DOI] [PubMed] [Google Scholar]
- 53.Ulloa, L., J. Doody, and J. Massague. 1999. Inhibition of transforming growth factor-beta/SMAD signalling by the interferon-gamma/STAT pathway. Nature 397:710-713. [DOI] [PubMed] [Google Scholar]
- 54.Westerhausen, D. R., Jr., W. E. Hopkins, and J. J. Billadello. 1991. Multiple transforming growth factor-beta-inducible elements regulate expression of the plasminogen activator inhibitor type-1 gene in Hep G2 cells. J. Biol. Chem. 266:1092-1100. [PubMed] [Google Scholar]
- 55.Whitman, M. 1998. Smads and early developmental signaling by the TGFβ superfamily. Genes Dev. 12:2445-2462. [DOI] [PubMed] [Google Scholar]
- 56.Yingling, J. M., M. B. Datto, C. Wong, J. P. Frederick, N. T. Liberati, and X. F. Wang. 1997. Tumor suppressor Smad4 is a transforming growth factor beta-inducible DNA binding protein. Mol. Cell. Biol. 17:7019-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, H. J., J. Iaria, and A. M. Sizeland. 1999. Smad7 differentially regulates transforming growth factor beta-mediated signaling pathways. J. Biol. Chem. 274:32258-32264. [DOI] [PubMed] [Google Scholar]