Abstract
The three-dimensional (3D) organization of the chromosomal fiber in the human interphase nucleus is an important but poorly understood aspect of gene regulation. Here we quantitatively analyze and compare the 3D structures of two types of genomic domains as defined by the human transcriptome map. While ridges are gene dense and show high expression levels, antiridges, on the other hand, are gene poor and carry genes that are expressed at low levels. We show that ridges are in general less condensed, more irregularly shaped, and located more closely to the nuclear center than antiridges. Six human cell lines that display different gene expression patterns and karyotypes share these structural parameters of chromatin. This shows that the chromatin structures of these two types of genomic domains are largely independent of tissue-specific variations in gene expression and differentiation state. Moreover, we show that there is remarkably little intermingling of chromatin from different parts of the same chromosome in a chromosome territory, neither from adjacent nor from distant parts. This suggests that the chromosomal fiber has a compact structure that sterically suppresses intermingling. Together, our results reveal novel general aspects of 3D chromosome architecture that are related to genome structure and function.
The cell nucleus harbors a number of precisely coordinated processes, including transcription, DNA repair, and replication. To exert these interdependent functions, the genome is assembled into a chromatin fiber, which is highly folded inside the interphase cell nucleus. Chromatin folding seems to be tightly related to genome function, including transcription and replication (18, 65). However, basic principles of three-dimensional (3D) chromatin and chromosome structures are still elusive. Studies on in situ chromatin and chromosome structure predominantly employ light microscopy. Due to limited spatial resolution, light microscopy does not allow one to follow individual chromatin or chromosomal fibers in 3D space in the nucleus. Therefore, 3D chromatin structure has been studied in several indirect ways, e.g., by measuring (i) the relative positions of specific loci in the interphase nucleus (2, 18, 22, 45), (ii) the average chromatin concentration per unit volume of the nucleus (18), and (iii) the relationship between genomic and physical distances between fixed points on the genome (29, 36, 38, 48, 63). The last two types of analyses directly relate to chromatin condensation. Results show, for instance, that active genes are generally positioned more towards the nuclear interior and that their chromatin state is more open (18, 46, 51). It has also been shown that transcription activation can change the positioning of genes (64). However, there are also many examples where active genes are present in the periphery of the eukaryotic nucleus, while inactive genes are positioned in the nuclear interior (8, 26, 31, 39). Also, evidence has been presented that transcription can take place in condensed chromatin (19, 31). Evidently, the transcriptional status of an individual gene is not related in a simple way to the local 3D organization of chromatin. Nearby genes and regulatory elements in the 1D genome codetermine the 3D chromatin organization and therefore the functional states of individual genes.
Genes are distributed far from randomly in the 1D eukaryotic genome. Genes coding for proteins with similar functions are occasionally clustered, such as genes coding for beta-globin (51), Hox proteins (25), core histones, and olfactory receptors (for a review see reference 30). These and other gene clusters may become decondensed upon transcriptional activation (10, 58, 60). Another, more frequent type of genomic clustering that is related to gene expression is shown by the human transcriptome map (HTM) (7, 56). The HTM reveals that many highly expressed human genes are clustered in about 40 genomic domains, each spanning 5 to 15 Mb, called regions of increased gene expression (ridges). Ridges are gene rich and contain both housekeeping genes and highly expressed genes that are active only in certain tissues (33, 56). Furthermore, the HTM shows clusters of similar size that are enriched in genes that have low expression (antiridges). In contrast to ridges, antiridges are relatively gene poor.
In this study we systematically and quantitatively analyze the 3D chromatin and chromosome architectures in cultured human cells on the basis of the HTM, with special focus on transcriptionally highly active ridges and their much less active counterparts, the antiridges (56). Ridges and antiridges are general features in the human genome, observed in all human cell types and tissues studied so far (56). Starting from the 1D organization of the human genome, as displayed by the HTM, we aimed at identifying general principles of human chromatin and chromosome structure, i.e., features that are the same in all cells in a homogeneous cell population and that are independent of cell type. We quantitatively determined various structural parameters of chromatin (including volume, shape, nuclear position, and spatial overlap) of ridges and antiridges on chromosomes 1, 3, and 11 in G1 phase cells of six human cell lines, using fluorescent in situ hybridization (FISH). We identified chromatin-folding-related properties that stand out against a considerable cell-to-cell variation in 3D chromatin structure. These properties are conserved in the six different human cell lines and therefore seem to be independent of cell-type-specific gene expression.
MATERIALS AND METHODS
Cell culture.
Human female primary fibroblast (04-147), neuroblastoma (SH-EP2), human embryonic kidney 293, K562 lymphoblastoid, and HeLa cells were cultured in Dulbecco modified Eagle medium containing 10% fetal calf serum, 20 mM glutamine, 60 μg/ml penicillin, and 100 μg/ml streptomycin. Human colon carcinoma (HCT 116) cells were cultivated in McCoy's 5A containing 10% fetal calf serum. Primary fibroblasts were used up to passage 25 to avoid effects related to senescence. In addition, primary fibroblasts were stained for senescence-associated β-galactosidase activity at different passages (20). Up to passage 30, no senescence-associated β-galactosidase activity was detected. To prepare metaphase spreads, human primary fibroblasts were blocked in metaphase with 0.1 μg/ml Colcemid for 4 h prior to a hypotonic treatment step with 0.075 mM KCl.
RNA isolation for microarray analysis.
Cells were spun down and snap frozen in liquid nitrogen. Total RNA was isolated using TRIzol (Invitrogen, Carlsbad, CA) and then purified with RNeasy (QIAGEN, Valencia, CA) according to the manufacturer's protocol. Sample integrity was checked on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Four micrograms of total RNA was used for cRNA synthesis and fragmented. Sample quality was checked on a Bioanalyzer prior to and after fragmentation. Labeling was performed with the One-Cycle cDNA synthesis kit (Affymetrix). Ten micrograms of labeled cRNA was hybridized to Affymetrix Human Genome U133 Plus 2.0 arrays according to the manufacturer's protocol (Affymetrix, Santa Clara, CA) at the Microarray Department of the University of Amsterdam (Amsterdam, The Netherlands). Arrays were scanned with a GeneChip scanner 3000 (Affymetrix).
Microarray data analysis and generation of transcription profiles.
Expression data (.cel files) were normalized using the MAS5 algorithm. Affymetrix probe sets were linked to transcriptional units (TUs), which have been described previously (56), in the following manner. All consensus and exemplar sequences used for Affymetrix U133 Plus 2.0 probe set generation were mapped to the human genome sequence HG15 (2003 to 2004) using the BLAST algorithm (http://genome.ucsc.edu/). We then checked for overlap of every mapped sequence with the exons of TUs. If the TU and Affymetrix sequence orientations were in agreement or if the TU could not be assigned an orientation (1,273 TUs), then the probe set associated with the Affymetrix sequence was linked to the TU. All probe sets linked to more than three TUs were removed from the data set. This resulted in 16,841 (of 20,382) TUs being linked to one or more probe sets. TUs not coupled to any probe set were treated as missing values. For each TU the average signal for all linked probe sets was calculated. Transcriptome maps were generated by calculating a moving median over 49 TUs (MM49). In short, the median is calculated over the 24 TUs situated upstream and downstream of the TU in question and the TU itself. This is done separately for each TU on every chromosome, hence giving a moving median.
DNA probe preparation and FISH.
Human centromeric alphoid DNA probes were obtained from plasmids pAL1 and pBR11 (1). Bacterial artificial chromosomes (BACs) were selected from libraries available in the RP11 collection (Sanger Institute) with an average distance of 400 kb between their centers (see material at http://staff.science.uva.nl/∼roeld/). This resulted in a homogeneously spaced set of probes with an average gap size of approximately 220 kb. At the light microscopy level this homogenous BAC distribution leads to a contiguous and even labeling of the domains investigated. BAC DNA was isolated using the QIAGEN REAL Prep 96 kit (QIAGEN, Venlo, The Netherlands). The identity of BACs was confirmed by end sequencing. For the preparation of FISH domain probes, all BACs of a given region were pooled and labeled with the same fluorophore. All BACs of a chromosomal domain were pooled prior to degenerate oligonucleotide primer-PCR (DOP-PCR) amplification (52). In short, during DOP-PCR an unspecific annealing was achieved in the first cycles through the application of an annealing temperature of 30°C. Subsequently, the annealing temperature was increased to 62°C for the specific thermal cycles. Four different DOP-PCR primers were used, each containing a different highly abundant annealing sequence (23): 6MW, CCGACTCGAGNNNNNNATGTGG; DOP1, CCGACTCGAGNNNNNNCTAGAA; DOP2, CCGACTCGAGNNNNNNTAGGAG; and DOP3, CCGACTCGAGNNNNNNTTCTAG.
PCR products of four independent PCRs, using the different DOP-PCR primers, were pooled prior to nick translation in order to achieve a maximum DNA-probe coverage and to prevent any PCR-based sequence bias. As an additional control for the efficiency of PCR-based DNA labeling, the chromosome 1 ridge and antiridge domains were directly labeled by pooling 15 individual BACs without prior PCR amplification, and hybridization results were compared with those for PCR-amplified BAC domain probes (see Fig. S1C and D at http://staff.science.uva.nl/∼roeld/). Measurements of 3D parameters confirmed that DOP-PCR-based probe amplification results in the same labeling and hybridization efficiency as does direct labeling of the domains.
Nick translation was used to label the probes with digoxigenin, biotin, or fluorescein isothiocyanate (FITC) (Roche Molecular Biochemicals, Basel, Switzerland). All probes were tested for hybridization specificity and domain coverage on metaphase spreads (see Fig. S1A and B at http://staff.science.uva.nl/∼roeld/).
FISH on interphase nuclei was carried out as described elsewhere, with slight modifications (15, 49). In short, cells in interphase were incubated with a 30-min pulse of 25 μM bromodeoxyuridine (BrdU) (Sigma-Aldrich, MO) to label replicating DNA prior to fixation in 4% (wt/vol) formaldehyde, in order to detect S-phase cells. Denaturation was carried out at 78°C in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 50% formamide. Hybridization was allowed to proceed overnight at 37°C. Posthybridization washes were carried out with 2× SSC-50% formamide at 45°C. All incubations for probe detection were done at room temperature in 4× SSC containing 5% (wt/vol) nonfat dried milk. FITC- and Cy5-conjugated antibodies (Jackson Immunoresearch Laboratories, Inc., West Grove, PA) and Cy3-conjugated streptavidin (Vector Laboratories, Burlingame, CA) were used to visualize the hybridization signals. BrdU was covisualized using a primary anti-BrdU antibody from Roche (Roche Molecular Biochemicals, Basel, Switzerland) and an FITC-labeled secondary antibody. Slides were mounted in Vectashield containing DAPI (4′,6′-diamidino-2-phenylindole) (Vector Laboratories, Burlingame, CA) to reduce photobleaching and outline the cell nucleus.
Confocal laser scanning microscopy.
All experiments were performed in duplicate. For each experiment 40 to 60 nuclei were 3D imaged. S-phase cells, as detected by BrdU labeling, as well as cells in G2, as determined by the size of the nuclei and a double signal per allele, were excluded. Twelve-bit images were recorded using an LSM 510 confocal laser scanning microscope (Carl Zeiss, Göttingen, Germany) equipped with a 63×/1.4 NA Apochromat objective. We used an Ar ion laser at 364 nm, an Ar laser at 488 nm, an He/Ne laser at 543 nm, and an He/Ne laser at 633 nm to excite DAPI, FITC, Cy3, and Cy5 fluorochromes, respectively. Fluorescence was detected with a 385- to 470-nm band-pass filter (DAPI), a 505- to 530-nm band-pass filter (FITC), a 560- to 615-nm band-pass filter (Cy3), and a 650-nm long-pass filter (Cy5). Images were scanned as 512 by 512 by 90 to 512 by 512 by 140 voxel images with a sampling rate of 50 by 50 by 100 nm (x, y, z).
Image processing and data evaluation.
All images were deconvolved using Huygens Professional 2.7 software (Scientific Volume Imaging BV, Hilversum, The Netherlands), using measured point spread functions. Chromatic shifts were measured using Tetraspeck fluorescent beads (Molecular Probes-Invitrogen, Eugene, OR) and corrected by the analysis software. Image analysis was carried out with Argos software (homepages.cwi.nl/∼wimc/argos). To identify and quantitatively analyze FISH-labeled areas, deconvolved 3D images were treated with a band-pass filter and subsequently segmented using a range of thresholds (see Fig. S5 at http://staff.science.uva.nl/∼roeld/). Normalization of deconvolved images was carried out beforehand to scale the voxel values such that the minimum and maximum voxel intensity values were equal to the minimum and maximum values of 16-bit images (65,536 gray levels). Upper and lower limits of the thresholding range were tested, and a fixed threshold was finally chosen in a useful range and kept constant during the course of all measurements in order to make data sets comparable. Collections of connected voxels were regarded as domains. All measurements of volume, shape, and nuclear position were subsequently carried out on these domains (see Fig. 2). For testing of statistical significance, a one-way nonparametric analysis of variance with the Kruskal-Wallis test was performed, using GraphPad Prism version 4.00 (GraphPad Software, California) (see Fig. S4 and S5 at http://staff.science.uva.nl/∼roeld/).
FIG. 2.
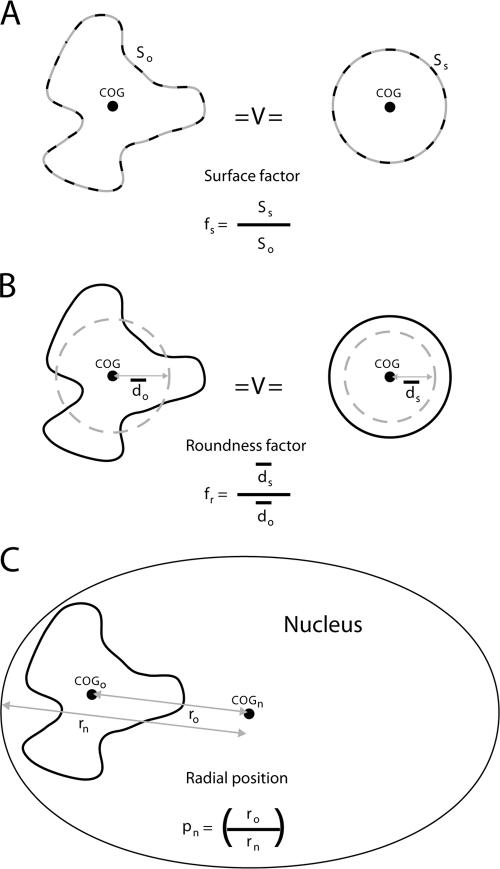
Surface factor, roundness factor, and radial position. Two parameters were determined as a measure of the shapes of ridges and antiridges: the surface factor fs and the roundness factor fr. (A) The surface factor (fs = Ss/So) is the surface So of a given chromosomal domain/object normalized to the surface of a sphere (Ss) with an equal volume. (B) The roundness factor (fr) is the average distance ds of any point in a sphere to the center of gravity (COG) divided by the same parameter do in a chromosomal domain with identical volume. The value fs is a measure of the irregularity of the domain surface, whereas fr is a measure of the overall shape of a domain. (C) The radial nuclear position pn (pn = ro/rn) of a chromosomal domain was calculated as the distance between the center of gravity of a domain and the center of the nucleus (ro) divided by the length of a line from the nuclear center to the nuclear envelope through the center of gravity of the domain (rn).
Microarray accession numbers.
Microarray data were deposited in MIAME-compliant format in GEO (http://www.ncbi.nlm.nih.gov/geo/) at NCBI under accession no. GSE6890 and GSM153780.
RESULTS
Experimental approach.
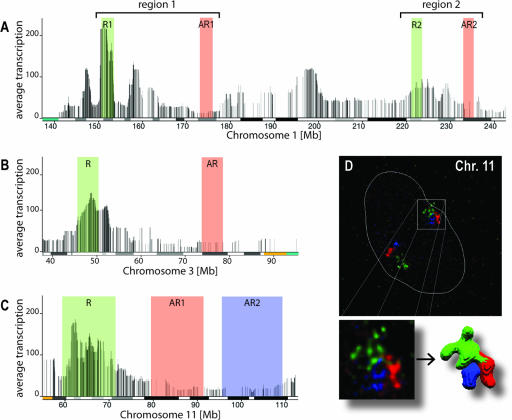
The aim of this study was to establish relationships between the 1D organization of the human genome as described by the HTM and the 3D structure of chromatin and chromosomes in order to identify principles of 3D chromatin organization. We investigated in detail the 3D structures of several ridges and antiridges on chromosomes 1, 3, and 11, since these are pronounced examples of such HTM-defined gene clusters (56) (Fig. 1A to C). Multicolor FISH in combination with 3D confocal imaging was used to quantitatively determine structural parameters of these ridges and antiridges in primary human female fibroblasts (04-147) and in five established human cell lines with diverse karyotypes. To minimize cell cycle effects on chromosome structure, we analyzed only cells in G1. Using sets of BACs, the four ridges and five antiridges marked in Fig. 1 were FISH labeled homogeneously over their entire lengths, spanning 3 to 12 Mb.
FIG. 1.
Investigated ridges and antiridges on chromosomes 1, 3, and 11. The HTMs of parts of chromosomes 1, 3, and 11 illustrate the clustering of highly expressed genes in ridges (R, green) and genes expressed at low levels in antiridges (AR, green and blue). Each vertical line in the map represents a gene, and the height of the line indicates the median gene activity averaged over a moving window of 49 genes. Below the HTM the Giemsa banding pattern is indicated, marking G-negative bands (white bars), G-positive bands (black bars), intermediate levels of Giemsa staining (gray bars), and pericentromeric repeats (colored) (see also Table S1 at http://staff.science.uva.nl/∼roeld/). (D) Representative 3D FISH images of the investigated ridge and two antiridges on chromosome 11 in human primary fibroblasts (04-147), shown as a maximum-intensity projection and after volume rendering, using the same color coding as in the HTM. The dotted white lines mark the contour of the nucleus.
After 3D confocal imaging, image deconvolution, and image segmentation, the relative volumes, shapes, and radial nuclear positions of ridges and antiridges were determined. In addition, 3D volume rendering allowed visual inspection of the nuclear positions of the domains and their 3D sizes and shapes (Fig. 1D). Two parameters were determined to quantitatively analyze the shapes of ridges and antiridges: the surface factor fs and the roundness factor fr (Fig. 2A and B; see figure legend for definitions of fs and fr). The value fs is a measure of the irregularity of the domain surface, whereas fr is a measure of the overall shape of a domain. To analyze the position of a chromosomal domain in the interphase nucleus, the radial nuclear position pn was determined (Fig. 2C; see figure legend for definition of pn). All pn values are squared to account for the higher probability of a domain to be located more towards the nuclear periphery if distributed randomly inside the nucleus.
To establish how these 3D structural parameters relate to differentiation state, gene expression pattern, and karyotype, we measured ridges and antiridges on chromosomes 1 and 11 (for their genomic locations, see Fig. 1A and C) in human primary fibroblasts and five human cell lines that represent different differentiation states and karyotypes: (i) neuroblastoma cells (SH-EP cells, containing extra material from the long arm of chromosome 1 (q10) (14, 34), (ii) cervical carcinoma cells (HeLa cells, containing three normal chromosomes 1 and a chromosome 1 derivative, resulting in tetraploidy for chromosome 1, and two normal copies of chromosome 11 and one derivative of chromosome 11 containing a complex intrachromosomal rearrangement) (35), (iii) human colon carcinoma cells (HCT 116 cells, near diploid), (iv) human embryonic kidney cells (293 cells, a hypotriploid human cell line with two normal copies of chromosome 1 and one derivative), and (v) lymphoblastoid cells (K562 cells, triploid for chromosomes 1 and 11 (11). For each experiment, about 50 3D images were obtained, allowing a statistical analysis of the data and a quantitative assessment of cell-to-cell variation. (For a summary of the measured values for the six cell lines, see the first two columns of Fig. 4.)
FIG. 4.
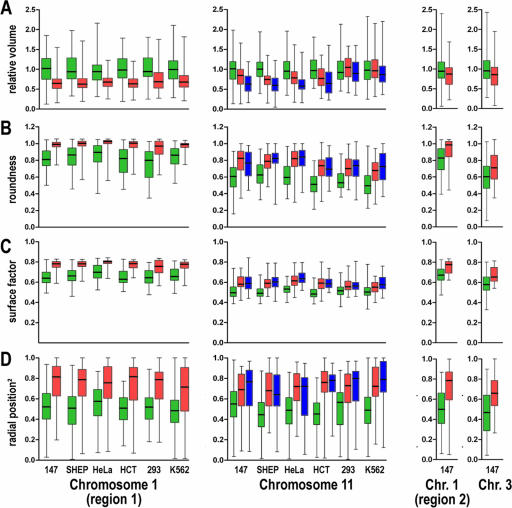
Structural parameters of ridges and antiridges in 04-147, SH-EP, HeLa, HCT, 293, and K562 cells. Box plots show the four quartiles of the distribution of various structural parameters measured for ridges (green) and antiridges (red and blue) on the q arms of chromosomes 1 and 11 and the p arm of chromosome 3. The boxes extend from the 25th to the 75th percentiles, with a horizontal line marking the median. The whiskers above and below the boxes show the highest and the lowest values determined in the course of our experiments. Each distribution contains information for approximately 50 cells. Results are shown on two chromosomes for primary fibroblasts and five different established cell lines. The third column summarizes results acquired with human primary fibroblasts only. (A) Relative volume per Mb of ridges and antiridges, showing that ridges have an about 1.4-times-larger volume per Mb than antiridges. (B) Roundness of ridges and antiridges, showing that antiridges are more sphere-like than ridges. (C) Surface factors of ridges and antiridges, showing that the surfaces of antiridges are less convoluted than those of ridges. (D) Squared radial positions of ridges and antiridges, showing that ridges are located more towards the nuclear center and antiridges more towards the nuclear periphery. All measurements show a considerable cell-to-cell variation, implying a high flexibility in 3D chromatin structure. The six cell types are indicated below the x axis. Chr., chromosome.
Expression profiles of six human cell lines.
The HTM averages expression levels of over 133 pooled SAGE libraries from a range of different human tissues and cell lines, as depicted for chromosomes 1, 3, and 11 in Fig. 1 (7). Figure 3 shows the transcription profiles of chromosomes 1 (R1 and AR1) and 11 of the six human cell lines used in the present study, based on mRNA gene expression values as measured with Affymetrix U133 Plus 2.0 microarrays. Like the HTM, the profiles for the individual cell lines show at each gene position the median gene expression level of a moving window of 49 genes. As expected, the profiles display the same ridges and antiridges as the HTM. At the same time, the profiles show considerable differences in gene expression for the six cell lines, reflecting their different differentiation states. Note that the representation in Fig. 3 underestimates these differences, due to the moving median in the 49-gene-wide moving window. Figure S2 at http://staff.science.uva.nl/∼roeld/ shows the differences for the chromosome 1a ridge and antiridge in detail.
FIG. 3.
Transcription profiles of relevant areas on chromosomes 1 (region 1) and 11 of the six human cell lines investigated. Each vertical line represents a gene. Its height depicts the median expression of that gene and the 24 genes at the left and at the right of the gene (median of a moving window of 49 genes). The ridge and antiridge areas investigated are color coded in green (ridges) and red and blue (antiridges). The transcription profiles illustrate that the large differences in transcriptional activity between ridges and antiridges are conserved in all cell types. At the same time, the expression levels of individual genes vary significantly. (See also Fig. S2 at http://staff.science.uva.nl/∼roeld/.)
Relative volumes of ridges and antiridges.
Volumes of ridges and antiridges were expressed as volume per Mb. Absolute volumes cannot be extracted from these measurements, due to the lack of objective image segmentation criteria. This problem can largely be circumvented by determining the ratio of volumes of antiridges to those of ridges, using the same image segmentation parameters for both. This ratio is considerably less dependent on segmentation parameters than absolute volumes. The volumes of ridges and antiridges differ significantly (Fig. 4A; see Fig. S3 and S4 at http://staff.science.uva.nl/∼roeld/). The volume per Mb of antiridges on chromosomes 1 and 11 is on average 1.4 times smaller than that of ridges, showing that the chromatin of antiridges is more condensed. For all cell lines, the volume of these chromosomal domains showed a considerable cell-to-cell variation. The differences in volumes of ridges and antiridges, as well as the cell-to-cell variation, are similar for all six investigated cell lines. The only exceptions are the ridge and antiridges on chromosome 11 in 293 and K562 cells, where the volumes of the two types of domains do not differ significantly. It may be that this is related to the spherical shape of the nuclei of these two cell types, in contrast to the flat ellipsoidal nuclei of the other cell lines, since earlier work has shown that nuclear shape may affect large-scale chromosome organization (2). If true, this may suggests that the chromatin of antiridges is more easily compressed by forces exerted by the cytoskeleton upon cell flattening than the chromatin of ridges.
Shapes of ridges and antiridges.
Ridge and antiridge types of genomic domains not only differ in chromatin condensation but also have different 3D shapes. Figure 4B and C show that the shape factors fs and fr systematically and significantly differ for ridges and antiridges in all cell types analyzed. The differences in fs and fr show that the ridges have a more irregular shape and surface than the antiridges, which are more spherical and have a less convoluted surface. Interestingly, similar differences in shape have been observed for the active and inactive X-chromosome territory (21). Like the volumes, the shape values show a substantial cell-to-cell variation for all cell lines.
Nuclear positions of ridges and antiridges.
Ridges and antiridges of chromosomes 1 and 11 occupy different radial positions (Fig. 4D). The two ridges are predominantly located more towards the nuclear interior than the three antiridges, which on average are closer to the nuclear envelope. Again, this aspect of chromosome structure is cell type independent and therefore apparently independent of cell-type-specific gene expression and karyotype. Radial position shows a considerable cell-to-cell variation, similar to what is observed for the parameters volume and shape. The different positions of ridges and antiridges suggest that chromosomes have a radial orientation in G1 nuclei, with their transcriptionally highly active ridges more interior and the less active antiridges more peripheral.
Structures and positions of ridges and antiridges elsewhere in the genome.
In the above section we analyzed two ridges and three antiridges on chromosomes 1 and 11 in six different cell types. The results show that ridges on average are more decondensed, irregularly shaped, and located closer to the nuclear center than antiridges. To be able to generalize these observations, we investigated four further domains in human primary fibroblasts: an additional ridge and an antiridge on the long arm of chromosome 1 (region 2) and a ridge and an antiridge on chromosome 3p (Fig. 4, column 3). Experiments were carried out with human primary fibroblasts only, because structural parameters did not differ significantly between various cell types (see above). We found that these genomic domains are organized in the same way as the ridges and antiridges on chromosomes 1 and 11 described above. Again, the relative volume of ridges significantly exceeded that of antiridges (P < 0.05) (Fig. 4A). Also, the surface and roundness factors were smaller for ridges than for antiridges (P < 0.001), confirming that antiridges have a more spherical shape (Fig. 4B and C). With respect to nuclear position, the antiridges show a clear preference for the nuclear periphery, unlike ridges (P < 0.001) (Fig. 4D). Evidently, the proximity of the (probably heterochromatic) centromere to the antiridge on chromosome 3p and the ridges on chromosomes 1q and 11q does not affect the radial position (for locations of centromeres, see Fig. 1). Our results indicate that ridges and antiridges in the human genome have similar large-scale chromatin structures (i.e., at the level observable by light microscopy) and that they share structural parameters irrespective of their position on the linear chromosome.
Is cell-to-cell variation due to different substages of G1?
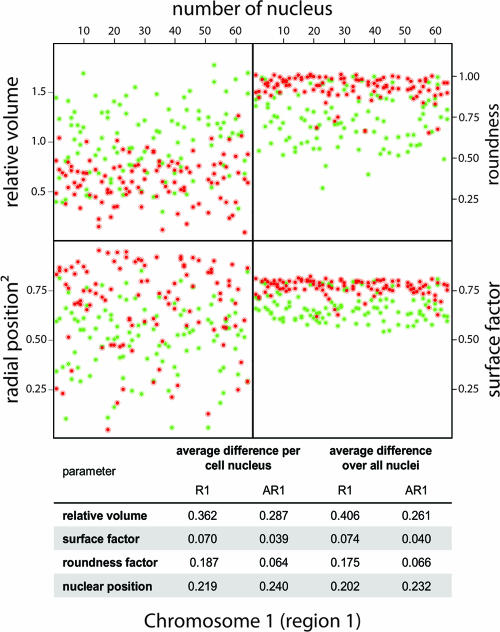
We observe a considerable cell-to-cell variation for all parameters in all cell types. This may be due to different substages of G1 (41). To explore this possibility, we reanalyzed our data for human primary fibroblasts on chromosome 1 region 1 (R1 and AR1) (Fig. 5). If the observed cell-to-cell variation is due to cells being in different substages of G1, one would predict that the variation of the parameters measured for different ridges (and for different antiridges), measured in the same cell (and therefore being in the same G1 substage), will be significantly smaller than the variation observed for the complete cell population. Therefore, we (i) determined the differences in 3D parameters (volume, shape factors, and radial position) between the two ridges (and the three antiridges) in every individual cell, (ii) evaluated the distribution of these values measured in 65 individual nuclei, and (iii) compared this distribution to the average distribution of the parameters over the whole population. The last was determined by calculating the differences between, for instance, the nuclear position value of a certain ridge and all position values acquired for ridges in every other nucleus of the population. The table in Fig. 5 summarizes the average difference we found within individual cells and compares it to the average difference over all nuclei. This result shows that the intracellular variation is similar to the variation we observe for the whole population.
FIG. 5.
Structural parameters of ridges and antiridges on homologous chromosomes in identical nuclei. Values for relative volume, roundness, surface factor, and nuclear position of the chromosome 1 ridge R1 and antiridge AR1 in human primary fibroblasts are shown per cell nucleus. Green spots indicate ridges, while red spots indicate antiridges. In total, 65 nuclei were evaluated (number of nucleus). The graphical representation and the table show that the variation between relative volume, roundness, surface factor, and nuclear position in identical nuclei (average difference per nucleus) is similar to the cell-to-cell variation observed for the whole population.
The methodology used, i.e., microscopy, image segmentation, and the FISH procedure, may contribute to the observed variation. To estimate the contribution of errors in 3D measurements, we determined the distance between the centers of gravity of genomic sites labeled with two identical FISH probes, each tagged with a different fluorophore. Theoretically, this should result in a measured distance of zero. The actual measured values are 7 ± 9 nm in x, 40 ± 11 nm in y, and 22 ± 12 nm in z. Evidently, this cannot account for the observed cell-to-cell variation. Image segmentation also is unlikely to cause the observed cell-to-cell variation. Segmentation parameters have been kept constant in all our measurements (see Fig. S5 at http://staff.science.uva.nl/∼roeld/), which therefore may result in a systematic rather than a random error. The FISH procedure is another potential source of error. However, under the sample preparation conditions we used, others have shown that structural perturbations of nuclear structure that can be measured by light microscopy are small (49, 55). FISH labeling results in a reproducible increase in chromatin and nuclear volume (18). Taken together, it is likely that the cell-to-cell variation that we observe represents real biological variation, i.e., that the 3D folding of the chromatin fiber itself varies considerably between otherwise identical cells. This suggests that chromatin structure has a high plasticity and that the chromatin fiber can fold in different ways without impairing genome function. This is in agreement with results from the Trask group, who presented evidence that the chromatin fiber follows a random walk (62).
Interphase chromosome architecture.
The different radial nuclear positions that we found for ridges and antiridges suggest that chromosomes in G1 interphase nuclei have a radial orientation. To analyze the overall structure and orientation of chromosomes more closely, we subdivided the 79-Mb q arm of chromosome 11 into nine domains, three of them representing the ridge and the two antiridges analyzed above, another representing an antiridge located near the q-arm telomere, another representing a domain containing the centromeric area, and four domains between ridges and antiridges (Fig. 6A). In a similar manner, we analyzed four domains on chromosome 1: the ridge R1 and the antiridge AR1 investigated above, the centromeric area (alpha satellite repeats), and an area I between the ridge and the antiridge (Fig. 7A). We determined the average radial position of each domain in human primary fibroblasts. The results show that not only antiridges but also chromosomal domains connecting two antiridges tend to be close to the nuclear envelope. Similarly, chromosomal domains adjacent to ridges have a high probability to be located closer to the nuclear center, like ridges themselves. The radial nuclear localization of domains with intermediate gene expression levels and gene densities seems to be determined by the flanking ridge or antiridge domains. In conclusion, chromosome arms tend to have a radial arrangement in the G1 nucleus, with the less transcriptionally active antiridges closer to the nuclear envelope and the ridges more inside the nucleus. Others have found that the radial positions of entire chromosome territories are influenced by their overall transcriptional activity (2, 5). It is likely that the radial positions of complete chromosomes are determined by the sum of the positions of their subchromosomal domains.
FIG. 6.
Radial position and absence of intermingling between subchromosomal domains on chromosome 11. (A) The 11q arm was subdivided into nine domains as indicated by horizontal bars in the chromosome 11q HTM: the centromere (C), the ridge (R), three antiridges (AR1 to AR3), and four areas of intermediate transcriptional activity (I to IV). The horizontal bars in the HTM mark the extents of the nine domains. The vertical positions of the bars relate to the squared radial nuclear position (y axis on the right). The vertical lines above and below the bars correspond to the second and third quartile of a box plot. The white line in the image marks the edge of the nucleus. (B to D) Three simultaneously FISH-labeled domains are indicated and color coded on the chromosome 11 transcriptome map. The volumes of the subchromosomal domains and the overlap between them were determined. The 3D-rendered image represent a typical triad of FISH-labeled domains and their respective overlap (n = 40 to 50). The surface areas of the colored circles correspond to the relative average domain volumes and display the measured overlap between the domains. Our measurements reveal remarkably little intermingling between subchromosomal domains. Note that AR1 and domain III in panel C share 22% of the FISH probes on the linear DNA and serve as a positive control for measurements of overlap.
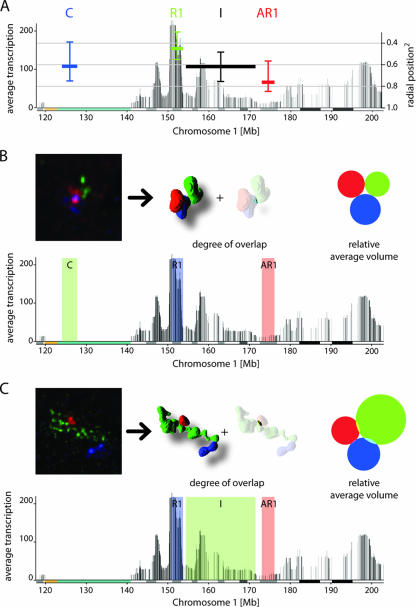
FIG. 7.
Radial position and absence of intermingling between subchromosomal domains on chromosome 1. (A) Four domains on the HTM of chromosome 1 were investigated: centromeric repeats (C), ridge R1, antiridge AR1, and an area of intermediate transcriptional activity (I). The horizontal bars in the HTM mark the extents of the four domains. The vertical positions of the bars relate to the squared radial nuclear position (y axis on the right). The vertical lines above and below the bars correspond to the second and third quartiles of a box plot. Nuclear position measurements showed a radial orientation of the chromosome arm, with the ridge being oriented more to the nuclear interior (pn2 ∼ 0.5) and the antiridge more peripheral (pn2 ∼ 0.8). (B and C) Three simultaneously FISH labeled domains on the chromosome 1 transcriptome map. 3D-rendered images represent a typical example of FISH-labeled domains and their respective overlap. The surface areas of the colored circles relate to the average domain volumes and display the overlap of the domains in 40 to 50 nuclei.
Analysis of the nine domains of chromosome 11q and the four domains of 1q, using three-color FISH labeling and analyzing various combinations of three domains, allowed us to systematically measure the degree of overlap between any pair of subchromosomal domains within a chromosome territory (Fig. 6B to D; see Fig. S6 at http://staff.science.uva.nl/∼roeld/). Strikingly, we never found significant overlap between any two domains, except in the trivial case when we used overlapping sets of FISH probes, such as, for instance, for domain III and antiridges 1 and 2 on chromosome 11 (Fig. 6C). Since the degree of overlap depends on the thresholding in image processing, we evaluated one set of images at different threshold levels (see Fig. S7 at http://staff.science.uva.nl/∼roeld/). We found that even at very low threshold levels, the degree of overlap was remarkably small, i.e., on the order of a few percent. These results indicate that the bulk of chromatin from different parts of the same chromosome arm does not intermingle as the chromosome folds into its chromosome territory in the G1 nucleus, suggesting that the chromosomal fiber has a compact structure that does not allow merging. This property mirrors that of complete chromosomes, since different chromosome territories also show little overlap, and the overlap they show is mainly restricted to the periphery of the territories (4, 16).
DISCUSSION
The aim of this study was to identify principles of chromatin organization, defined as those features that are the same in different cell types, i.e., independent of differentiation state and cell-type-specific expression patterns. To achieve this, we systematically and quantitatively investigated the relationship between the 1D arrangement of the protein-coding genes in the human genome, as established by the HTM, and the 3D organization of the chromatin fiber in the G1 interphase nuclei of six cultured human cell lines, including primary fibroblasts. Our analyses primarily concentrated on gene-dense clusters in the human genome that are enriched in highly transcribed genes (ridges) and on gene-sparse clusters that contain mainly genes that are expressed at low levels (antiridges).
Structural information from approximately 100 measurements per chromosomal domain and cell line (i.e., about 50 nuclei) was acquired, allowing a thorough statistical analysis of our data sets. Our results reveal three key aspects of the structure of human chromosomes in G1 nuclei. First, chromosome/chromatin structure shows a remarkably large cell-to-cell variation. This indicates that chromosomes are highly flexible structures. Second, despite the large cell-to-cell variation, ridges and antiridges are shown to have different chromatin structures that are conserved in all six cell lines studied. These structural parameters seem to be independent of gene expression pattern, differentiation state, and karyotype. Conserved features are as follows: (i) the chromatin of antiridges is on average 1.4 times more compact than that of ridges; (ii) antiridges have a more spherical shape than ridges; and (iii) antiridges are closer to the nuclear envelope, whereas ridges are found more in the nuclear interior (Fig. 4). Since these structural properties significantly differ for ridges and antiridges (P < 0.01; see Fig. S3 and S4 at http://staff.science.uva.nl/∼roeld/) and are similar for all six cell lines and therefore independent of differentiation state, they may represent general principles of chromatin structure in human cells. Third, the results show a remarkable lack of intrachromosomal intermingling between different subchromosomal regions on the q arms of chromosome 1 and chromosome 11 (Fig. 6 and 7). This suggests that the chromosomal fiber in interphase is relatively compact, sterically hindering intermingling.
Cell-to-cell variation of chromatin structure.
A remarkably large cell-to-cell variation is observed for all parameters analyzed here. One may argue that this is due to changes in chromatin structure during progress of the G1 phase of the cell cycle (41). However, our measurements show that the variation between the structural properties of the two loci in the same nucleus, which therefore are in the same G1 cell cycle substage, is similar to that of the structural parameters averaged over the whole cell population (Fig. 5). Other studies, concentrating on specific genomic loci and on complete chromosomes, also showed a considerable cell-to-cell variation in chromatin structure. For instance, the looping out of specific loci from chromosome territories displayed a large variability (36, 58). Also, the radial positions of gene-rich and gene-poor chromosome territories have been shown to differ from cell to cell, while the average values display a significant correlation with gene density (3). Evidently, chromatin folding has a high plasticity, apparently not affecting overall cell properties. It is possible that part of this plasticity is related to cell-to-cell variation in gene expression or epigenetic state at the moment of fixation. Fluctuations of gene expression over time have been observed in various biological systems (32, 40, 42). Obviously, the HTM does not take such variation into account, since the methods used to assemble the HTM average over large numbers of cells. An important potential explanation of the observed variation is the constrained diffusion of chromatin in interphase (12). On a time scale of minutes, a fluorescently labeled locus may move in an interphase nucleus over distances of a several tenths of a micrometer or more (13, 27). This may therefore be one of the main causes of the cell-to-cell variation.
Structural parameters related to the HTM.
The observed differences in structural properties of ridges and antiridges reflect differences in chromatin folding between these genomic domains with different transcriptional activity and gene density. The volume per Mb chromatin of the antiridges, which are gene sparse and expressed at low levels, is on average 1.4 times smaller than that of the gene-dense and highly expressed ridges (Fig. 4A). This difference in compaction is probably not due to transcription per se. Recently, the Bickmore group developed an elegant way to biochemically fractionate chromatin on the basis of differences in compactness (29). Interestingly, fractions enriched in open chromatin were derived predominantly from ridges in the HTM, indicating that the differences between ridges and other chromatin domains (e.g., antiridges and heterochromatin) persist during cell breakage and chromatin isolation. Apparently, relatively strong interactions of chromatin with, probably, protein complexes establish the open structure of ridges, regardless of ongoing transcription.
Differences in compaction of chromatin folding have also been studied extensively in transgenic systems that undergo condensation and decondensation in relation to changes in gene activity or epigenetic transitions (18, 53, 54). In these studies, chromatin compaction of specific mammalian loci in their transcriptionally active (or epigenetically permissive) and inactive (epigenetically repressive) states has been compared. Upon activation, over 10-fold decompaction has been observed (53). However, it has been questioned to what extent these observations may be generalized (51).
In several studies chromatin compaction and nuclear positioning have been related to Giemsa banding patterns of metaphase chromosomes, in particular G-light (R) and G-dark (G) bands, and to replication timing (24, 44, 45, 47, 61, 62, 65). Correlations have been found between banding patterns and nuclear architecture, as well as replication timing. However, the relationship between banding and human genome structure and function, including the HTM, is far from perfect. For instance, an open chromatin fiber conformation has been reported for a subset of R bands (29, 62), and several G bands have been shown to have a low gene density and belong to the mid-replicating fraction of DNA, which is found predominantly as a small rim at the nuclear periphery (45). Moreover, recent publications provide evidence for early-replicating DNA segments in G bands and find gene-poor areas and late-replicating DNA in R bands (29, 61, 65). Also, early replication does not itself specify gene expression: there is clear evidence that transcription and replication occur in spatially distinct nuclear zones during S phase (9, 59). Evidently, neither Giemsa banding nor replication timing is consistently related to transcription levels of protein-coding genes and/or gene density. In contrast, ridges and antiridges in the HTM are based on actual transcription levels and explicitly relate to gene density. As expected, ridges and antiridges show only a limited correlation with the chromosomal banding patterns, as listed in the UCSC cytogenetic map (see Table S1A and B at http://staff.science.uva.nl/∼roeld/) (24). A clear example is provided by ridge R1 and antiridge AR1 of chromosome 1. Although they position in the same class of Giemsa bands (see Table S1A at http://staff.science.uva.nl/∼roeld/ and banding pattern in Fig. 1), these domains show striking structural and functional differences. From this, one can conclude that ridges and antiridges in the HTM reflect functional properties of the human genome considerably better than Giemsa banding.
Ridges and antiridges have different shapes (Fig. 4B and C). Ridges are less spherical and have a more convoluted surface than antiridges. Little is known about the relationship between the shape of chromatin domains and transcriptional activity. One of the few cases that have been described is the comparison between the active and inactive X chromosomes of human cells. These display a similar difference in shape as antiridges and ridges; i.e., the inactive X chromosome is less asymmetric than the active one (21). Another example of shape measurements on chromatin domains is the analysis of the length of artificially and naturally occurring gene clusters under different conditions of gene activity. Similarly, lacO tandem arrays in transcriptionally active domains were more elongated than those in inactive areas (53). Evidently, lacO arrays do not require active transcription for maintaining that elongated state. Similar results, referring to changes in shape and looping, have been observed for the MHCII locus after interferon induction (37, 58). The chromatin fiber of the activated domain looped out from its chromosome territory (58). Similar observations have been made for the ridge on the 11p15.5 locus of chromosome 11 (29). Taken together, the results suggest that high transcriptional activity results in a more irregular 3D domain structure, possibly due to local and maybe transient hyperdecondensation.
The chromosomal fiber in interphase does not intermingle with itself in interphase.
Our results show that different parts of the same chromosome arm show remarkably little intermingling. A similar low degree of overlap has been shown by others for gene-rich and gene-poor domains (48), for the p and q arms of different chromosomes (17), and for complete chromosome territories (2, 4). Recently, Branco and Pombo (4) found that intermingling between different chromosome territories can be as high as 20% of the nuclear volume, with the majority of overlap at the borders of the territories. This is more than we observed for intrachromosomal intermingling. If this is true, then intermingling within a chromosome territory is more restricted than intermingling between different chromosomes. Alternatively, this difference may be due to differences in sample preparation, microscopy, and image processing and analysis. Late-replicating chromosomal subdomains have also been shown not to mix (57). For instance, it has been demonstrated that pulse-labeled replication foci persist during subsequent cell cycle stages after labeling (50, 66) These observations indicate a compact structure of the chromosomal fiber. Our data suggest that this chromosomal fiber is thicker, i.e., more decondensed, and more irregularly shaped in gene-rich domains (ridges) and more compact and thinner in gene-poor areas (antiridges) (Fig. 8).
FIG. 8.
Chromosomal fiber model of the interphase chromosome. Human interphase chromosomes consist of a compact chromosomal fiber (indicated by a dashed line surrounding the chromatin fiber). This chromosomal fiber is thicker in ridges (green) and thinner in antiridges (red). Consequently, in agreement with volume and shape measurements, the chromatin fiber in such a superordinate chromosomal fiber is more decondensed and flexible in ridges than in antiridges. In the human interphase nucleus the chromosomal fiber is arranged so that antiridges are more peripheral, while ridges are more internal.
What causes the differences between the 3D chromatin structures of ridges and antiridges? It may be that the difference in transcriptional activity causes the difference in structure between the gene-rich and highly expressed ridges and the gene-poor antiridges that are expressed at low levels. This, however, is unlikely, because differences in chromatin compactness between ridges and antiridges persist during biochemical fractionation (29). More likely, differences in gene density, length of intergenic sequences, or specific, not-yet-identified sequence elements are responsible. An interesting possibility is that specific sets of proteins and/or noncoding RNAs are bound to functionally different types of genomic domains, imposing different types of chromatin folding. Whatever the molecular mechanism, it seems that specific subnuclear microenvironments are formed that promote high transcription rates (ridges) or allow only low to moderate transcriptional activities (antiridges). Conceptually, this is similar to, for instance, pericentromeric heterochromatic domains in mouse cells, which constitute microenvironments that suppress transcription (6). Currently it is unknown what molecular systems define the formation of such microenvironments. A thorough biochemical analysis, exploiting biochemical chromatin fractionation methods, such as described by Gilbert and coworkers (29), might be suitable to address this issue. Potential players are posttranslationally modified histones, relative amounts of canonical and noncanonical (i.e., variant) histones, nonhistone protein composition, and local DNA methylation. In a first high-resolution genome-wide mapping approach, Roh and coworkers (43) found that transcriptionally active chromatin domains coincide with histone H3 acetylation islands. The genomic distribution of these acetylation islands correlates remarkably well with that of ridges. A critical experiment to test the hypothesis that ridges and antiridges represent microenvironments of intrinsic high and low gene expression, respectively, is to insert a reporter gene with a weak promoter at different positions in the genome. The above hypothesis predicts that the reporter gene will have higher activity if inserted into ridges than into antiridges. Recent experiments by the Versteeg group show this to be the case (28), supporting the idea that ridges and antiridges represent different subnuclear environments imposing different rates of expression on their constituent genes.
Acknowledgments
We thank the Sanger Institute and Eric Schoenmakers (University Nijmegen) for providing BACs. We thank Peter van Sluis (AMC) for his assistance with RNA quality control and Richard Volckmann for his assistance with the Affymetrix data. We acknowledge the Centre of Advanced Microscopy of the University of Amsterdam for technical support and Scientific Volume Imaging BV (Hilversum, The Netherlands) for assistance with 3D deconvolution.
This work was supported by the European Commission as part of the 3DGENOME program, contract LSHG-CT-2003-503441.
Footnotes
Published ahead of print on 9 April 2007.
REFERENCES
- 1.Archidiacono, N., R. Antonacci, R. Marzella, P. Finelli, A. Lonoce, and M. Rocchi. 1995. Comparative mapping of human alphoid sequences in great apes using fluorescence in situ hybridization. Genomics 25:477-484. [DOI] [PubMed] [Google Scholar]
- 2.Bolzer, A., G. Kreth, I. Solovei, D. Koehler, K. Saracoglu, C. Fauth, S. Muller, R. Eils, C. Cremer, M. R. Speicher, and T. Cremer. 2005. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 3:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle, S., S. Gilchrist, J. M. Bridger, N. L. Mahy, J. A. Ellis, and W. A. Bickmore. 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211-219. [DOI] [PubMed] [Google Scholar]
- 4.Branco, M. R., and A. Pombo. 2006. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 4:e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bridger, J. M., S. Boyle, I. R. Kill, and W. A. Bickmore. 2000. Re-modelling of nuclear architecture in quiescent and senescent human fibroblasts. Curr. Biol. 10:149-152. [DOI] [PubMed] [Google Scholar]
- 6.Brown, K. E., S. S. Guest, S. T. Smale, K. Hahm, M. Merkenschlager, and A. G. Fisher. 1997. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91:845-854. [DOI] [PubMed] [Google Scholar]
- 7.Caron, H., B. van Schaik, M. van der Mee, F. Baas, G. Riggins, P. van Sluis, M. C. Hermus, R. van Asperen, K. Boon, P. A. Voute, S. Heisterkamp, A. van Kampen, and R. Versteeg. 2001. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science 291:1289-1292. [DOI] [PubMed] [Google Scholar]
- 8.Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus, and P. A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117:427-439. [DOI] [PubMed] [Google Scholar]
- 9.Chakalova, L., E. Debrand, J. A. Mitchell, C. S. Osborne, and P. Fraser. 2005. Replication and transcription: shaping the landscape of the genome. Nat. Rev. Genet. 6:669-677. [DOI] [PubMed] [Google Scholar]
- 10.Chambeyron, S., N. R. Da Silva, K. A. Lawson, and W. A. Bickmore. 2005. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development 132:2215-2223. [DOI] [PubMed] [Google Scholar]
- 11.Chen, T. R. 1985. Modal karyotype of human leukemia cell line, K562 (ATCC CCL 243). Cancer Genet. Cytogenet. 17:55-60. [DOI] [PubMed] [Google Scholar]
- 12.Chubb, J. R., and W. A. Bickmore. 2003. Considering nuclear compartmentalization in the light of nuclear dynamics. Cell 112:403-406. [DOI] [PubMed] [Google Scholar]
- 13.Chubb, J. R., S. Boyle, P. Perry, and W. A. Bickmore. 2002. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12:439-445. [DOI] [PubMed] [Google Scholar]
- 14.Cohen, N., D. R. Betts, G. Rechavi, N. Amariglio, and L. Trakhtenbrot. 2003. Clonal expansion and not cell interconversion is the basis for the neuroblast and nonneuronal types of the SK-N-SH neuroblastoma cell line. Cancer Genet. Cytogenet. 143:80-84. [DOI] [PubMed] [Google Scholar]
- 15.Cremer, M., J. vonHase, T. Volm, A. Brero, G. Kreth, J. Walter, C. Fischer, I. Solovei, C. Cremer, and T. Cremer. 2001. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 9:541-567. [DOI] [PubMed] [Google Scholar]
- 16.Cremer, T., and C. Cremer. 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2:292-301. [DOI] [PubMed] [Google Scholar]
- 17.Dietzel, S., A. Jauch, D. Kienle, G. Qu, H. Holtgreve-Grez, R. Eils, C. Munkel, M. Bittner, P. S. Meltzer, J. M. Trent, and T. Cremer. 1998. Separate and variably shaped chromosome arm domains are disclosed by chromosome arm painting in human cell nuclei. Chromosome Res. 6:25-33. [DOI] [PubMed] [Google Scholar]
- 18.Dietzel, S., K. Zolghadr, C. Hepperger, and A. S. Belmont. 2004. Differential large-scale chromatin compaction and intranuclear positioning of transcribed versus non-transcribed transgene arrays containing beta-globin regulatory sequences. J. Cell Sci. 117:4603-4614. [DOI] [PubMed] [Google Scholar]
- 19.Dimitri, P., N. Corradini, F. Rossi, and F. Verni. 2005. The paradox of functional heterochromatin. Bioessays 27:29-41. [DOI] [PubMed] [Google Scholar]
- 20.Dimri, G. P., X. Lee, G. Basile, M. Acosta, G. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, M. Peacocke, and J. Campisi. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. USA 92:9363-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eils, R., S. Dietzel, E. Bertin, E. Schrock, M. R. Speicher, T. Ried, M. Robert-Nicoud, C. Cremer, and T. Cremer. 1996. Three-dimensional reconstruction of painted human interphase chromosomes: active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J. Cell Biol. 135:1427-1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferreira, J., G. Paolella, C. Ramos, and A. I. Lamond. 1997. Spatial organization of large-scale chromatin domains in the nucleus: a magnified view of single chromosome territories. J. Cell Biol. 139:1597-1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiegler, H., P. Carr, E. J. Douglas, D. C. Burford, S. Hunt, C. E. Scott, J. Smith, D. Vetrie, P. Gorman, I. P. Tomlinson, and N. P. Carter. 2003. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer 36:361-374. [DOI] [PubMed] [Google Scholar]
- 24.Furey, T. S., and D. Haussler. 2003. Integration of the cytogenetic map with the draft human genome sequence. Hum. Mol. Genet. 12:1037-1044. [DOI] [PubMed] [Google Scholar]
- 25.Garcia-Fernandez, J. 2005. The genesis and evolution of homeobox gene clusters. Nat. Rev. Genet. 6:881-892. [DOI] [PubMed] [Google Scholar]
- 26.Gartenberg, M. R., F. R. Neumann, T. Laroche, M. Blaszczyk, and S. M. Gasser. 2004. Sir-mediated repression can occur independently of chromosomal and subnuclear contexts. Cell 119:955-967. [DOI] [PubMed] [Google Scholar]
- 27.Gasser, S. M. 2002. Visualizing chromatin dynamics in interphase nuclei. Science 296:1412-1416. [DOI] [PubMed] [Google Scholar]
- 28.Gierman, H. J., M. H. G. Indemans, J. Koster, S. Goetze, J. Seppen, D. Geerts, R. van Driel, and R. Versteeg. Domain-wide regulation of gene expression in the human genome. Genome Res., in press. [DOI] [PMC free article] [PubMed]
- 29.Gilbert, N., S. Boyle, H. Fiegler, K. Woodfine, N. P. Carter, and W. A. Bickmore. 2004. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118:555-566. [DOI] [PubMed] [Google Scholar]
- 30.Horton, R., L. Wilming, V. Rand, R. C. Lovering, E. A. Bruford, V. K. Khodiyar, M. J. Lush, S. Povey, C. C. Talbot, Jr., M. W. Wright, H. M. Wain, J. Trowsdale, A. Ziegler, and S. Beck. 2004. Gene map of the extended human MHC. Nat. Rev. Genet. 5:889-899. [DOI] [PubMed] [Google Scholar]
- 31.Janicki, S. M., T. Tsukamoto, S. E. Salghetti, W. P. Tansey, R. Sachidanandam, K. V. Prasanth, T. Ried, Y. Shav-Tal, E. Bertrand, R. H. Singer, and D. L. Spector. 2004. From silencing to gene expression: real-time analysis in single cells. Cell 116:683-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaern, M., T. C. Elston, W. J. Blake, and J. J. Collins. 2005. Stochasticity in gene expression: from theories to phenotypes. Nat. Rev. Genet. 6:451-464. [DOI] [PubMed] [Google Scholar]
- 33.Lercher, M. J., A. O. Urrutia, and L. D. Hurst. 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 31:180-183. [DOI] [PubMed] [Google Scholar]
- 34.Lutz, W., M. Stohr, J. Schurmann, A. Wenzel, A. Lohr, and M. Schwab. 1996. Conditional expression of N-myc in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene 13:803-812. [PubMed] [Google Scholar]
- 35.Macville, M., E. Schrock, H. Padilla-Nash, C. Keck, B. M. Ghadimi, D. Zimonjic, N. Popescu, and T. Ried. 1999. Comprehensive and definitive molecular cytogenetic characterization of HeLa cells by spectral karyotyping. Cancer Res. 59:141-150. [PubMed] [Google Scholar]
- 36.Mahy, N. L., P. E. Perry, and W. A. Bickmore. 2002. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159:753-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller, W. G., D. Rieder, G. Kreth, C. Cremer, Z. Trajanoski, and J. G. McNally. 2004. Generic features of tertiary chromatin structure as detected in natural chromosomes. Mol. Cell. Biol. 24:9359-9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munkel, C., R. Eils, S. Dietzel, D. Zink, C. Mehring, G. Wedemann, T. Cremer, and J. Langowski. 1999. Compartmentalization of interphase chromosomes observed in simulation and experiment. J. Mol. Biol. 285:1053-1065. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen, J. A., L. D. Hudson, and R. C. Armstrong. 2002. Nuclear organization in differentiating oligodendrocytes. J. Cell Sci. 115:4071-4079. [DOI] [PubMed] [Google Scholar]
- 40.Osborne, C. S., L. Chakalova, K. E. Brown, D. Carter, A. Horton, E. Debrand, B. Goyenechea, J. A. Mitchell, S. Lopes, W. Reik, and P. Fraser. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 36:1065-1071. [DOI] [PubMed] [Google Scholar]
- 41.Pardee, A. B. 1989. G1 events and regulation of cell proliferation. Science 246:603-608. [DOI] [PubMed] [Google Scholar]
- 42.Raj, A., C. S. Peskin, D. Tranchina, D. Y. Vargas, and S. Tyagi. 2006. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4:e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roh, T. Y., S. Cuddapah, and K. Zhao. 2005. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 19:542-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saccone, S., C. Federico, and G. Bernardi. 2002. Localization of the gene-richest and the gene-poorest isochores in the interphase nuclei of mammals and birds. Gene 300:169-178. [DOI] [PubMed] [Google Scholar]
- 45.Sadoni, N., S. Langer, C. Fauth, G. Bernardi, T. Cremer, B. M. Turner, and D. Zink. 1999. Nuclear organization of mammalian genomes: polar chromosome territories build up functionally distinct higher order compartments. J. Cell Biol. 146:1211-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheuermann, M. O., J. Tajbakhsh, A. Kurz, K. Saracoglu, R. Eils, and P. Lichter. 2004. Topology of genes and nontranscribed sequences in human interphase nuclei. Exp. Cell Res. 301:266-279. [DOI] [PubMed] [Google Scholar]
- 47.Schubeler, D., D. Scalzo, C. Kooperberg, B. van Steensel, J. Delrow, and M. Groudine. 2002. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat. Genet. 32:438-442. [DOI] [PubMed] [Google Scholar]
- 48.Shopland, L. S., C. R. Lynch, K. A. Peterson, K. Thornton, N. Kepper, J. Hase, S. Stein, S. Vincent, K. R. Molloy, G. Kreth, C. Cremer, C. J. Bult, and T. P. O'Brien. 2006. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J. Cell Biol. 174:27-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Solovei, I., A. Cavallo, L. Schermelleh, F. Jaunin, C. Scasselati, D. Cmarko, C. Cremer, S. Fakan, and T. Cremer. 2002. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH). Exp. Cell Res. 276:10-23. [DOI] [PubMed] [Google Scholar]
- 50.Sparvoli, E., M. Levi, and E. Rossi. 1994. Replicon clusters may form structurally stable complexes of chromatin and chromosomes. J. Cell Sci. 107:3097-3103. [DOI] [PubMed] [Google Scholar]
- 51.Sproul, D., N. Gilbert, and W. A. Bickmore. 2005. The role of chromatin structure in regulating the expression of clustered genes. Nat. Rev. Genet. 6:775-781. [DOI] [PubMed] [Google Scholar]
- 52.Telenius, H., A. H. Pelmear, A. Tunnacliffe, N. P. Carter, A. Behmel, M. A. Ferguson-Smith, M. Nordenskjold, R. Pfragner, and B. A. Ponder. 1992. Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosomes Cancer. 4:257-263. [DOI] [PubMed] [Google Scholar]
- 53.Tumbar, T., G. Sudlow, and A. S. Belmont. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verschure, P. J., I. van der Kraan, W. de Leeuw, J. van der Vlag, A. E. Carpenter, A. S. Belmont, and R. van Driel. 2005. In vivo HP1 targeting causes large-scale chromatin condensation and enhanced histone lysine methylation. Mol. Cell. Biol. 25:4552-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verschure, P. J., I. van der Kraan, E. M. Manders, and R. van Driel. 1999. Spatial relationship between transcription sites and chromosome territories. J. Cell Biol. 147:13-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Versteeg, R., B. D. van Schaik, M. F. van Batenburg, M. Roos, R. Monajemi, H. Caron, H. J. Bussemaker, and A. H. van Kampen. 2003. The human transcriptome map reveals extremes in gene density, intron length, GC content, and repeat pattern for domains of highly and weakly expressed genes. Genome Res. 13:1998-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Visser, A. E., and J. A. Aten. 1999. Chromosomes as well as chromosomal subdomains constitute distinct units in interphase nuclei. J. Cell Sci. 112:3353-3360. [DOI] [PubMed] [Google Scholar]
- 58.Volpi, E. V., E. Chevret, T. Jones, R. Vatcheva, J. Williamson, S. Beck, R. D. Campbell, M. Goldsworthy, S. H. Powis, J. Ragoussis, J. Trowsdale, and D. Sheer. 2000. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J. Cell Sci. 113:1565-1576. [DOI] [PubMed] [Google Scholar]
- 59.Wei, X., J. Samarabandu, R. S. Devdhar, A. J. Siegel, R. Acharya, and R. Berezney. 1998. Segregation of transcription and replication sites into higher order domains. Science 281:1502-1506. [DOI] [PubMed] [Google Scholar]
- 60.Williams, R. R., S. Broad, D. Sheer, and J. Ragoussis. 2002. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp. Cell Res. 272:163-175. [DOI] [PubMed] [Google Scholar]
- 61.Woodfine, K., H. Fiegler, D. M. Beare, J. E. Collins, O. T. McCann, B. D. Young, S. Debernardi, R. Mott, I. Dunham, and N. P. Carter. 2004. Replication timing of the human genome. Hum. Mol. Genet. 13:191-202. [DOI] [PubMed] [Google Scholar]
- 62.Yokota, H., M. J. Singer, G. J. van den Engh, and B. J. Trask. 1997. Regional differences in the compaction of chromatin in human G0/G1 interphase nuclei. Chromosome Res. 5:157-166. [DOI] [PubMed] [Google Scholar]
- 63.Yokota, H., G. van den Engh, J. E. Hearst, R. K. Sachs, and B. J. Trask. 1995. Evidence for the organization of chromatin in megabase pair-sized loops arranged along a random walk path in the human G0/G1 interphase nucleus. J. Cell Biol. 130:1239-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zink, D., M. D. Amaral, A. Englmann, S. Lang, L. A. Clarke, C. Rudolph, F. Alt, K. Luther, C. Braz, N. Sadoni, J. Rosenecker, and D. Schindelhauer. 2004. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J. Cell Biol. 166:815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zink, D., H. Bornfleth, A. Visser, C. Cremer, and T. Cremer. 1999. Organization of early and late replicating DNA in human chromosome territories. Exp. Cell Res. 247:176-188. [DOI] [PubMed] [Google Scholar]
- 66.Zink, D., T. Cremer, R. Saffrich, R. Fischer, M. F. Trendelenburg, W. Ansorge, and E. H. Stelzer. 1998. Structure and dynamics of human interphase chromosome territories in vivo. Hum. Genet. 102:241-251. [DOI] [PubMed] [Google Scholar]