Abstract
Par 6 acts as a scaffold protein to facilitate atypical protein kinase C-mediated phosphorylation of cytoplasmic protein complexes, leading to epithelial and neuronal cell polarization. In addition to its location in the cytoplasm, Par 6 is localized to the nucleus. However, its organization and potential functions in the nucleus have not been examined. Using an affinity-purified Par 6 antibody and a chimera of Par 6 and green fluorescent protein, we show that Par 6 localizes precisely to nuclear speckles, but not to other nuclear structures, and displays characteristics of speckle proteins. We show that Par 6 colocalizes in the nucleus with Tax, a transcriptional activator of the human T-cell leukemia virus type 1 long terminal repeat, but multiple lines of evidence show that Par 6 is not directly involved in known functions of speckle proteins, including general transcription, splicing, or mRNA transport. Significantly, however, the structure of nuclear speckles is lost when Par 6 levels are reduced by Par 6-specific small interfering RNA. Therefore, we hypothesize that Par 6 in the nucleus acts as a scaffolding protein in nuclear speckle complexes, similar to its role in the cytoplasm.
Cytoplasmic protein complexes associated with the plasma membrane play important roles in the development of overall cell polarity, such as in epithelial and neuronal cells, the functional organization of different components of cell-cell adhesion complexes, the regulation of cell proliferation, and the orientation of the plane of cell division (1, 32, 64, 72). One of the protein complexes thought to play a major role in cell organization and function is the Par complex, comprised of Par 3, Par 6, and an atypical protein kinase C (aPKC) (33, 51, 54). Par proteins were first discovered in a genetic screen for mutations that cause defects in the partitioning of cell fate determinants in Caenorhabditis elegans (24), but they have since emerged as being critical in the establishment and maintenance of cell polarity in diverse species and cell types (70).
Par 6 acts as a scaffold protein in regulating diverse downstream signaling outputs from the Par complex. Through a Phox/Bem1 (PB1) domain near the N terminus, Par 6 binds to and inhibits aPKC. Activated Cdc42, a member of the Rho family GTPases, can bind to Par 6 through its CRIB/PDZ domain and activate aPKC, which initiates a signaling cascade leading to changes in the structural and functional organization of different cell types (20, 57, 76). In epithelial cells, for example, activated Par 6/aPKC interacts with Par 3, the Crumbs complex, and the Scribble complex to regulate the establishment of apical-basal polarity (26, 41, 70). In migrating astrocytes and keratinocytes, activated Par 6/aPKC at the leading edge orients microtubules required for polarized migration (17, 18, 34). Recently, Par 6/aPKC has also been shown to bind to dimerized ErbB2, resulting in a loss of polarity in MDCK cells and the formation of multiacini in MCF10A cells (3).
In addition to its localization in the cytoplasm, Par 6 has been reported to localize in the nucleus. Johansson et al. (29) noted that an antibody against Par 6α detected not only Par 6 staining at the tight junction, as has since been shown by others (19), but also Par 6 staining in the nucleus of MDCK cells; they also reported that overexpressed Par 6 localized to the nucleus. Though not specifically noted, the nuclear localization of Par 6 is apparent in other studies using other antibodies and constructs (19, 52). The authenticity and significance of Par 6 localization in the nucleus have not been studied. Nevertheless, that Par 6 may be localized at both the nucleus and plasma membrane is not without precedence. Such “dual-location” proteins have been described with increasing frequency (6, 9, 13, 43). For example, proteins that localize to the tight junction also function in the nucleus as a transcription factor that promotes proliferation (ZONAB) (5, 7) and as a scaffold for the RNA polyadenylation machinery (Symplekin) (27, 71). Here we show that Par 6 localizes to structures called nuclear speckles, has characteristics of other speckle proteins, and is required for the maintenance of nuclear speckle structure. Assays to assess roles of Par 6 in transcription, splicing, and mRNA transport are described, and Par 6 is shown to colocalize with nuclear structures formed by Tax, a transcriptional activator of the human T-cell leukemia virus type 1 (HTLV-1) long terminal repeat.
MATERIALS AND METHODS
Cell culture.
MDCK G II and HeLa cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum and antibiotics. Transient DNA transfections and small interfering RNA (siRNA) transfections were carried out using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's instructions. The medium was changed at 4 to 6 h posttransfection to avoid toxic side effects of the cationic transfection reagent. siRNA was purchased from Dharmacon. The duplexes used were human ParD6A duplexes no. 2 (5′CUACUUGGCUAUACGGAUGUU) and no. 4 (5′GAGUCGCAUUCGAGGAGAUUU) from the siGenome collection. siControl no. 1, also from Dharmacon, was used as the nontargeting control.
Immunofluorescence.
Cells were fixed in 2% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min, washed in PBS, and then extracted in CSK buffer [50 mM NaCl, 300 mM sucrose, 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.8), 3 mM MgCl2, 0.5% Triton X-100, 1 mM Pefabloc] for 5 min. Cells were blocked in either goat or donkey blocking buffer (0.2% bovine serum albumin, 50 mM NH4Cl2, 1% serum), depending on the host species of the secondary antibody to be used. In cases where extraction preceded fixation, the same procedures were used but in reverse order.
Antibodies for immunofluorescence were diluted in the appropriate blocking buffer (goat or donkey) as follows: Par 6, 1:75 (see below); SC-35, 1:300 (BD Biosciences Pharmingen); 9a9, 1:5 (a kind gift of P. J. Utz, Stanford University); Y12, 1:150 (Abcam); hemagglutinin (HA), 1:150 (Roche); T7, 1:500 (Novagen); bromodeoxyuridine (BrdU), 1:300 (Sigma); and TIA-1, 1:100 (Santa Cruz Biotechnology; sc-1751).
Fluorescein isothiocyanate-, Cy5-, and rhodamine RedX-labeled secondary antibodies were purchased from Jackson Immunoresearch and were used at a dilution of 1:200. Alexa 350-labeled secondary antibodies were purchased from Molecular Probes and were also used at a dilution of 1:200. Secondary antibodies were used in a variety of combinations, with equivalent results. Nuclei were labeled with Hoechst (Molecular Probes) diluted in PBS at 1:2,000.
Par 6 antibody production.
Glutathione S-transferase (GST)-Par 6 C terminus (a kind gift of R. G. Qiu and G. S. Martin, University of California, Berkeley) was produced in bacteria, purified with glutathione beads, and then used to immunize rabbits (Covance). For Western blotting applications, immunoglobulin G was purified from the serum by using a DEAE-Affi-Gel Blue column (Bio-Rad Laboratories). For the affinity-purified antibody used in immunofluorescence, the serum was first cleared of antibodies that recognized GST by using GST-conjugated glutathione beads and then passed over a column of GST-Par 6 C terminus attached to CNBr beads. Bound antibodies were eluted at low pH and neutralized with Tris.
SDS-PAGE and Western blotting.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out using standard protocols. Proteins were transferred to nitrocellulose, which was then blocked with a 5% solution of fat-free powdered milk in Tris-buffered saline (TBS) (10 mM Tris [pH 7.5], 150 mM NaCl). For Western blotting, primary antibodies were diluted in TBS plus 0.1% Tween 20 with 5% powdered milk as follows: Par 6, 1:10; HSP-90 (Santa Cruz Biotechnology, Inc.), 1:500; tandem affinity purification system (TAP), 1:200 (Santa Cruz Biotechnology, Inc.); green fluorescent protein (GFP), (Molecular Probes), 1:400. Fluorescently tagged secondary antibodies (Molecular Probes and Rockland) were diluted in the same solution. Washes were done with TBS plus 0.1% Tween 20. A final rinse in TBS was done before scanning on an Odyssey infrared imaging system (LI-COR Biosciences).
GAL4 transcriptional transactivation assay.
Human Par 6 was cloned into pM2 to create a fusion protein containing the GAL4 DNA binding domain (GAL4-DBD) (60). Empty vector was used as a control. For the positive control GAL4-LPP, pSM421 was used (56). Transcriptional transactivation activity was detected using pSM222, which contains five GAL4-DBD sites upstream of a minimal promoter and a luciferase reporter gene (W. Schaffner and D. Escher, Zurich, Switzerland). A β-galactosidase reporter plasmid was included in all transfections to control for transfection efficiency.
Subconfluent HeLa cells in 60-mm plates were transfected with 1.6 μg GAL4-DBD plasmid, 2 μg luciferase reporter plasmid, and 0.25 μg β-galactosidase plasmid. After 24 h, the cells were analyzed in triplicate for luciferase expression by using the dual-light luciferase and β-galactosidase reporter gene assay system (Applied Biosystems) according to the manufacturer's instructions.
Splicing assay.
pSV-mini-LCA 18 was previously described (38); 0.5 μg was cotransfected with 3 μg of GFP control vector, GFP-Par 6, or T7-ASF/SF2 into subconfluent HeLa cells in 60-mm plates. After 24 h, the cells were split onto 100-mm plates. RNA was harvested 24 h later using the RNeasy minikit (QIAGEN) according to the manufacturer's instructions. The QIAGEN OneStep reverse transcription-PCR (RT-PCR) kit was then used to amplify the CD45 minigene, using the forward primer 5′TGCAGGTCGACTCTAGAG and the reverse primer 5′CACATGTTGGCTTAGATGG. After 30 min at 50°C for RT and an initial PCR activation step of 15 min at 95°C, the PCR cycling was as follows: 30 seconds at 94°C, 30 seconds at 51°C, and 1 min at 72°C. Thirty-five cycles were completed before a final extension at 72°C.
Oligo(dT) pulldown assay.
Eight 150-mm plates of nearly confluent HeLa cells were separated into a cytosolic extract and nuclei by using the technique of Dignam et al. (16). The nuclei were then lysed in 5 ml of lysis and binding buffer (100 mM Tris [pH 8.0], 10 mM EDTA, 100 mM NaCl, 0.5% Triton X-100). Both lysates (cytosolic and nuclear) were then incubated for 12 min at room temperature with 100 mg of oligo(dT) cellulose (Monomer Sciences, Inc.) that had been preequilibrated in lysis and binding buffer. The cellulose was washed three times with 1 ml of lysis and binding buffer. Bound material was then eluted with 2.5 ml elution buffer (100 mM Tris [pH 7.5], 500 mM NaCl, 10 mM EDTA, 1% sodium dodecyl sulfate [SDS], 5 mM dithiothreitol). The supernatant and eluate were then concentrated in Vivaspin 15 protein concentrators (Sartorius AG) by centrifugation. The resulting concentrated proteins were then loaded onto a gel for analysis by SDS-PAGE and Western blotting.
Par 6 constructs.
The MDCK cells stably expressing GFP-Par 6 (a kind gift of A. Muesch, Cornell Medical College) and the GAL4 transactivation construct used human Par 6α. All other constructs used rat Par 6α (a kind gift of L. Braiterman, Johns Hopkins University).
Microscopy.
Unless otherwise indicated, all micrographs were taken using a Leica AOBS confocal microscope at the Stanford Beckman Center Cell Sciences Imaging Facility. Confocal stacks were merged into a single plane by using Volocity software (Improvision Ltd.). Images for GFP ratios were taken using a Zeiss Axioplan epifluorescence microscope equipped with a digital camera controlled by Axiovision software. All micrographs were adjusted for brightness and contrast using Photoshop 7.0. In cases where comparisons between the intensities of fluorescence of two samples were made, the micrographs were adjusted with identical intensity settings.
RESULTS
Par 6α is localized to the nucleus and the tight junction.
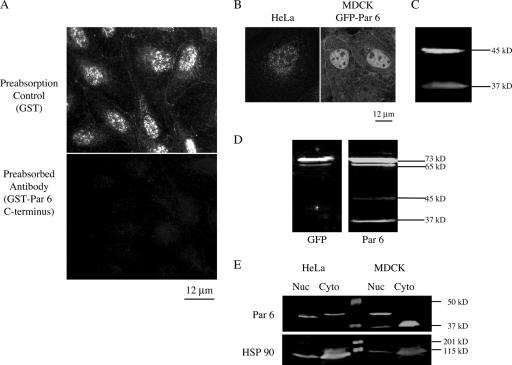
An affinity-purified polyclonal rabbit antibody raised against a GST fusion to the C terminus of human Par 6α (amino acids 266 to 347) detected Par 6 at cell-cell junctions, the cytoplasm, and the nuclei of confluent monolayers of MDCK cells (Fig. 1A). The epitope is specific for Par 6α alone. Preabsorption of the Par 6 antibody with GST did not affect either of these staining patterns (Fig. 1A, top panel). However, preabsorption of the antibody with the antigen (GST-Par 6 C terminus) abolished both staining patterns (Fig. 1A, bottom panel), indicating that the antibody staining is specific. This affinity-purified antibody also detected nuclear Par 6α in HeLa cells (Fig. 1B) and in MCF-10A, Jurkat, and Eph-4 cells (data not shown). In addition to detection of endogenous Par 6α in the nucleus, we found that exogenous GFP-Par 6 stably expressed in MDCK cells also clearly localized to both cell-cell junctions and nuclei (Fig. 1B). Therefore, the nuclear localization of Par 6α appears to a general characteristic and not cell type specific.
FIG. 1.
Par 6α is localized to the nucleus as well as at the tight junction. (A) MDCK cells stained with a polyclonal antibody to Par 6 preabsorbed with either GST or GST-Par 6 C terminus. Micrographs were taken using a Zeiss Axioplan epifluorescence microscope equipped with a digital camera controlled by Axiovision software. (B) Left, HeLa cell stained with Par 6 antibody; right, GFP signal from MDCK cells stably expressing GFP-Par 6. (C) Western blot of SDS lysate of MDCK cells. (D) Western blots of extracts from GFP-Par 6 stably transfected MDCK cells. Left, SDS extract immunoblotted with GFP antibody; right, immunoblot with Par 6 antibody. (E) Western blots of the fractionation of HeLa and MDCK cells into nuclear (Nuc) and cytosolic (Cyto) fractions. Nuclear extracts were prepared using the NE-PER kit (Promega) according to manufacturer's instructions.
Par 6α nuclear localization was confirmed biochemically. In Western blots of whole MDCK cell extract, the Par 6 antibody recognized two protein bands with apparent molecular masses of approximately 37 kDa and 45 kDa (Fig. 1C). The presence of the 45-kDa band is similar to other reports (29, 39), despite the fact that the predicted molecular mass of Par 6α based on its amino acid sequence is approximately 37 kDa. In extracts from GFP-Par 6 MDCK stable cells, the Par 6 antibody detected two bands with apparent molecular masses of 73 and 65 kDa (Fig. 1D, right panel). These molecular masses correspond to those predicted for the two endogenous proteins fused to GFP. The 73- and 65-kDa bands are also recognized by a GFP antibody (Fig. 1D, left panel), confirming that they are in fact the fusion proteins.
We tested but did not obtain evidence that the 37-kDa protein was a degradation product of the 45-kDa protein or that the 45-kDa protein was a phosphorylated from of the 37-kDa protein. Note that the 37-kDa protein recognized by our Par 6α antibody cofractionated in iodixanol gradients with other Par complex proteins (Par 3 and aPKC) and components of the tight junction from MDCK cells (73). Although the relationship between the 37-kDa and 45-kDa proteins is not understood at present, we considered both proteins to represent Par 6α, and hereafter they are referred to as “Par 6.”
Figure 1E shows extracts of MDCK and HeLa cells fractionated into a nuclear fraction and a combined cytoplasmic and membrane fraction (referred to as the cytosol fraction). The 45-kDa band is detected in both fractions from HeLa cells; increasing the brightness of the scanned image revealed a slight amount of the 37-kDa band in the nuclear fraction of HeLa cells (data not shown). In MDCK cells, only the 37-kDa band is detected in the cytosolic fraction, whereas both bands are found in the nuclear fraction. Thus, Par 6 is detected in the nuclear fractions of both types of cells.
Before quantifying the relative amounts of the 37-kDa and 45-kDa proteins in these cell fractions, we determined the degree of contamination of the nuclear fraction by cytoplasmic proteins. Western blots were reprobed with an anti-HSP 90 antibody (Fig. 1D, bottom panel) revealing that ∼20% of this cytosolic protein contaminated the nuclear fraction. Taking this contamination into account, we calculated that ∼40% of total Par 6 in HeLa cells (45-kDa band) is in the nucleus. In MDCK cells, ∼80% of the 45-kDa band is nuclear, while all of the 37-kDa band is cytoplasmic (the small amount visible in the MDCK nuclear fraction is within the contamination range).
Par 6 is a constitutive dual-location protein.
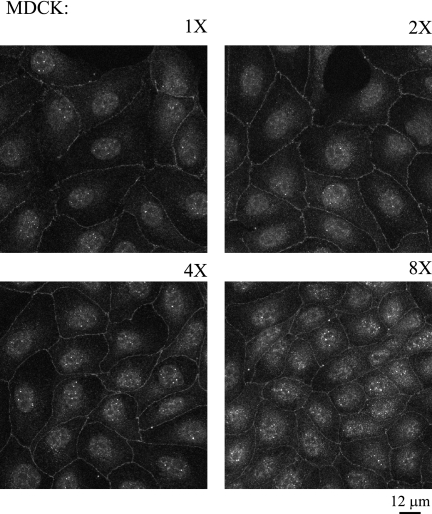
Factors controlling the localization of dual-location proteins are varied, and many are not understood. Since we found that Par 6 is localized to the nucleus and the tight junction, we tested whether, like that of another dual-location tight junction protein, ZONAB (7), Par 6 nuclear localization is dependent on cell density.
Figure 2 shows the localizations of Par 6 in MDCK cells after plating at different cell densities for 24 h. Each panel of Fig. 2 represents a twofold increase in plated cell density. After 24 h, the cells plated at the 1× density were in small colonies of approximately 20 cells, and the densities then increased to a confluent monolayer for the cells plated at 8×. Par 6 is constitutively present at the plasma membrane, cytoplasm, and nucleus at all cell densities. Par 6 is also present in the nuclei of single MDCK cells and during all stages of tight junction formation when contact-naïve single cells are induced to form E-cadherin-mediated contacts and initiate polarization following addition of calcium to the growth medium (data not shown). From the data shown in Fig. 1 and 2, we conclude that Par 6 is a dual-location protein that is constitutively localized to the nucleus.
FIG. 2.
Par 6 is a constitutive dual-location protein. MDCK cells were plated at increasing densities (1× to 8×) and stained with Par 6 antibody. Micrographs were taken using a Zeiss Axioplan microscope.
A putative NLS is partially responsible for Par 6 nuclear localization.
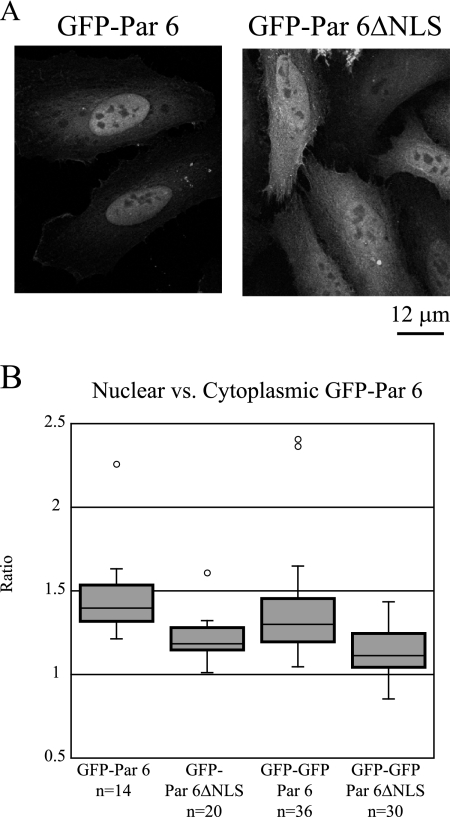
Johansson et al. (29) noted that Par 6 contains a putative nuclear localization signal (NLS) (112-RRKK-115). This sequence is conserved in human, canine, rat, mouse, and zebrafish Par 6. We substituted alanine residues across this putative NLS and expressed the mutated sequence in a construct consisting of GFP fused to the N terminus of rat Par 6 (GFP-Par 6ΔNLS). GFP-Par 6ΔNLS transiently expressed in HeLa cells localized in both the nucleus and the cytoplasm (Fig. 3A, right panel), but compared to the distribution of control GFP-Par 6 (Fig. 3A, left panel), the proportion of GFP-Par 6ΔNLS in the nucleus was reduced. Quantitation of the ratio of nuclear to cytoplasmic fluorescence intensities of control GFP-Par 6 and GFP-Par 6ΔNLS revealed that the median ratio for control GFP-Par 6 (n = 14) was 1.4, while the median ratio for GFP-Par 6ΔNLS (n = 20) was 1.18. Because the distributions were not normally distributed, the nonparametric Mann-Whitney U test was used to determine that this difference is significant (P < 0.001) (Fig. 3B).
FIG. 3.
The putative NLS of Par 6 is at least partially responsible for its nuclear localization. (A) HeLa cells were transfected with the indicated GFP constructs. (B) Quantitation of the ratio of nuclear to cytoplasmic GFP signal in HeLa cells transfected with the indicated constructs. Ratios were determined by measuring fluorescence intensity in equally sized areas within and immediately adjacent to the nucleus using Image J. Outliers are indicated by open circles.
The fusion of GFP to Par 6 yields a protein with a predicted molecular mass of approximately 65 kDa, which is close to the size range in which proteins can diffuse through nuclear pores (42). Therefore, we added a second GFP moiety to increase the molecular mass of the chimeric Par 6 protein to inhibit its diffusion into the nucleus. The distribution of the di-GFP chimera was similar to that of the mono-GFP chimera; median ratios of 1.3 for control GFP-GFP-Par 6 (n = 36) and 1.14 for GFP-GFP-Par 6ΔNLS (n = 30) were measured. The Mann-Whitney U test determined that these populations were significantly different from each other (P < 0.001) but not significantly different from their cognate single GFP constructs (GFP-Par 6 versus GFP-GFP-Par 6, P > 0.05; GFP-Par 6ΔNLS versus GFP-GFP-Par 6ΔNLS, P > 0.05) (Fig. 3B). Therefore, we conclude that diffusion is not responsible for the GFP signal seen in the nuclei of GFP-Par 6ΔNLS-transfected cells. The change in the ratio of nuclear to cytoplasmic GFP signal in the mutated constructs indicates that the RRKK sequence has at least some effect on nuclear localization. The fact that Par 6 still enters the nucleus despite the NLS mutation indicates the presence of another cryptic NLS or that Par 6 could bind to a protein that acts as a nuclear import chaperone. Further studies will be required to test these additional possibilities.
Par 6 does not traffic between the cytoplasm and the nucleus.
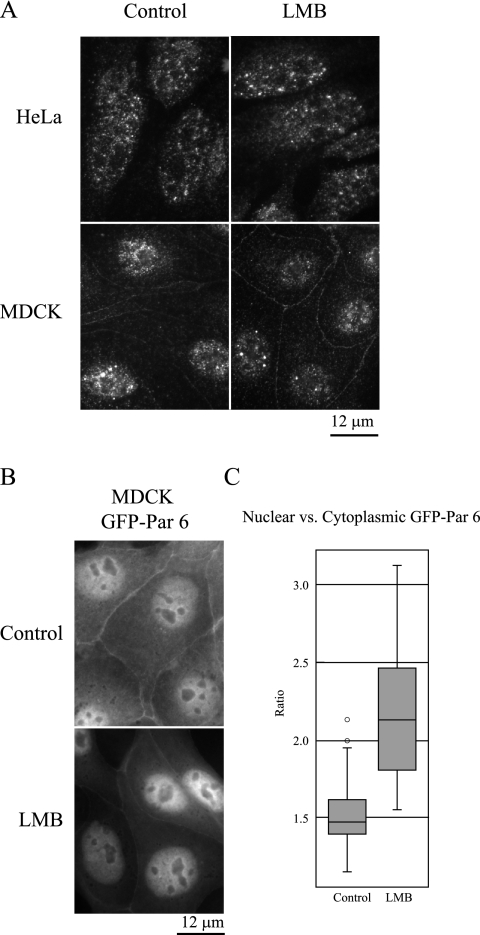
To investigate whether Par 6 shuttles between the cytoplasm and the nucleus or instead is stably localized to both of these compartments, we treated MDCK and HeLa cells with leptomycin B (LMB), a CRM1-specific nuclear export inhibitor. If Par 6 shuttles between cellular compartments, the expectation would be that inhibiting nuclear export would cause an accumulation of Par 6 in the nucleus. Figure 4A shows that in both cell types, nuclear and cytoplasmic Par 6 localization (and membrane-bound Par 6 localization in MDCK cells) is unchanged upon treatment with LMB, indicating that the protein is not exchanging between these compartments over the time course of the experiment. Figure 4B, however, shows that in cells stably expressing GFP-Par 6, LMB causes the overexpressed protein to accumulate in the nucleus. To quantify this accumulation, the ratio of the nuclear to cytoplasmic GFP signal was calculated as in Fig. 3, yielding median ratios of 1.473 for control cells and 2.130 for LMB-treated cells (Fig. 4C). A Mann-Whitney U test determined this difference to be significant (P < 0.001). Together these results indicate that endogenous Par 6 is stably localized in the nucleus but that the excess Par 6 that is transported into the nucleus is exported in a CRM1-dependent manner.
FIG. 4.
Par 6 does not dynamically traffic between the cytoplasm and the nucleus. (A) HeLa and MDCK cells were treated with either 70% methanol (carrier control) or 20 nM LMB for 4 h at 37°C. (B) MDCK cells stably expressing GFP-Par 6 were treated as for panel A. (C) Quantitation of the ratio of nuclear to cytoplasmic GFP signal in GFP-Par 6 MDCK cells. Ratios were determined as described for Fig. 3.
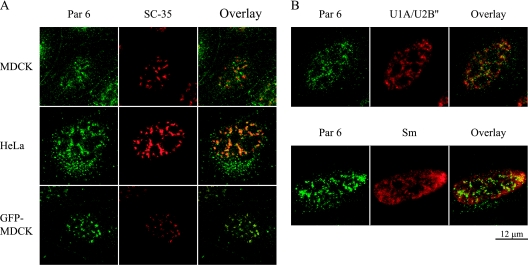
Par 6 is localized to discrete nuclear domains called nuclear speckles.
Close inspection of Fig. 1A and B shows that Par 6 staining in the nucleus is not homogenous. Discrete Par 6-positive structures are clearly labeled, but the nucleolus is excluded from staining. This resembles the staining of structures known as “nuclear speckles.” Nuclear speckles are structures identified by immunofluorescence staining of constituent proteins and are the equivalent of the interchromatin granule clusters seen by electron microscopy (37). Nuclear speckles are dynamic; are enriched in factors involved in pre-mRNA splicing, including small nuclear ribonucleoproteins (snRNPs) and the serine-arginine (SR) proteins; and are thought to be storage depots for splicing factors that participate in cotranscriptional splicing (37, 46, 65).
MDCK and HeLa cells costained with antibodies to Par 6 and SC-35, a marker protein of speckles, showed identical staining patterns for endogenous Par 6 and SC-35 in speckles (Fig. 5A). In MDCK cells stably expressing GFP-Par 6, the exogenous protein also colocalized with SC-35. However, due to a high diffuse background of GFP-Par6 in the nucleus, colocalization of GFP-Par 6 with SC-35 was most clearly detected after extraction prior to fixation (Fig. 5A).
FIG. 5.
Nuclear Par 6 is localized to discrete domains called nuclear speckles. (A) Top two rows, the indicated cell types were stained with Par 6 and SC-35 antibody to show colocalization. Bottom row, GFP-Par 6 MDCK cells were extracted prior to fixation and then stained with SC-35 antibody. (B) HeLa cells were stained with Par 6 antibody and either monoclonal antibody 9A9 (U1A/U2B‴) or Y12 (Smith antigen [Sm]) to show colocalization with speckle proteins.
We tested whether Par 6 colocalized with other speckle proteins by using (i) monoclonal antibody 9A9, which recognizes the splicing factors snRNP U1A/U2B‴, and (ii) monoclonal antibody Y12, which recognizes Smith antigen present in snRNPs U1, U2, U4, U5, and U6, which localize to speckles and more diffusely in the nucleoplasm (36, 66). Figure 5B shows that Par 6 colocalized with speckle staining of both snRNP U1A/U2B‴ and the Smith antigen. Therefore, we conclude that Par 6 is localized to nuclear speckles.
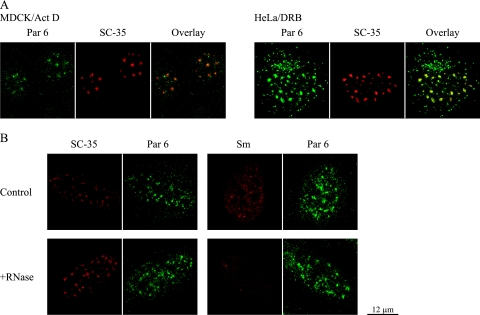
Par 6 has properties of an RNase-insensitive nuclear speckle protein.
The irregular shape of nuclear speckles is thought to reflect the constant addition and subtraction of splicing factors as they are recruited to and returned from sites of transcription throughout the nucleus. Inhibition of transcription causes speckles to adopt a more rounded-up appearance (45, 67, 68), presumably due to a stop in the flow of splicing factors to transcription sites (65). To test if Par 6 shares this characteristic with speckle proteins, transcription in HeLa and MDCK cells was inhibited with either actinomycin D or 5,6-dichloro-1-beta-d-ribobenzimidazole. Figure 6A shows that Par 6 adopted the same rounded-up structure as SC-35 in the presence of the transcription inhibitors (compare to untreated cells in Fig. 5A).
FIG. 6.
Par 6 behaves like an RNase-insensitive nuclear speckle protein. (A) MDCK and HeLa Cells Were treated with either 5 ng/ml actinomycin D (Act D) or 100 μM 5,6-dichloro-1-beta-d-ribobenzimidazole (DRB) in cell culture medium for 4 h at 37°C and then processed for immunofluorescence. (B) HeLa cells were fixed and extracted as described in Materials and Methods and then treated with either PBS with 5 mM MgCl2 alone (control) or plus 100 μg/ml RNase A (Sigma) for 2 h at room temperature. Coverslips were then stained with Par 6 and either SC-35 or Smith (Sm) antigen antibody.
There are two classes of protein interactions in speckles: one that is resistant to RNase treatment (e.g., the SR protein SC-35) and another that is RNase sensitive (e.g., snRNPs identified with the Smith antigen-recognizing antibody Y12) (67). To test which class Par 6 belongs to, HeLa cells were fixed, extracted, and then treated with RNase. Figure 6B shows examples of control and RNase-treated cells stained with Par 6 antibody and either SC-35 or Smith antigen antibody. Par 6 staining remained unchanged after RNase treatment, similar to that of SC-35. In contrast, Smith antigen staining becomes completely diffuse after RNase treatment. We conclude that Par 6 localization to nuclear speckles is RNA independent.
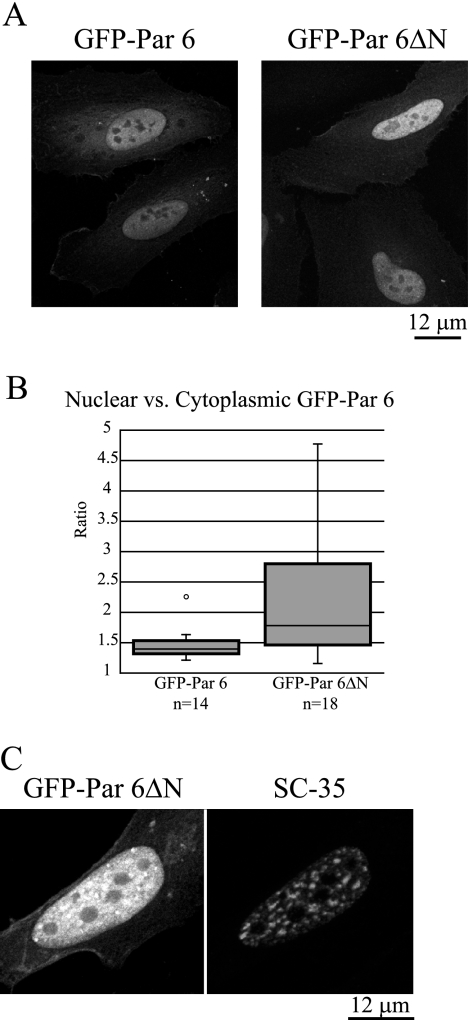
Par 6 nuclear localization is not dependent on aPKC.
In the literature to date, the protein complexes formed with Par 6 in the cytoplasm involve aPKC (26, 70), and therefore we determined if this interaction is necessary for the nuclear and/or speckle localization of Par 6. A GFP fusion to a truncated rat Par 6 protein lacking the N terminus (amino acids 1 to 96) (GFP-Par 6ΔN), which deletes the PB1 domain that binds to aPKC, is localized in the nucleus and specifically in speckles (Fig. 7A). Quantitation of the ratio of nuclear to cytoplasmic GFP-Par 6ΔN signal revealed a median ratio of 1.78 for GFP-Par 6ΔN (n = 18), while the median ratio of control GFP-Par 6 was 1.4 (Fig. 3A, left panel). The Mann-Whitney U statistic for this population compared to the control gives a P value of <0.02, indicating that there is a significant increase in the amount of nuclear Par 6ΔN compared to control Par 6. Figure 7B shows a magnified view of GFP-Par 6ΔN localization in a representative, transiently transfected HeLa cell. Through a diffuse haze of GFP signal in the nucleus, distinct puncta that correspond to nuclear speckles marked by SC-35 staining are visible. We could detect some aPKC in the nucleus with an antibody that recognizes both aPKCλ and aPKCζ, as reported previously (55), though not in nuclear speckles (data not shown). Taken together, these data indicate that the nuclear location and possible function of Par 6 are not as a scaffold for aPKC, in contrast to its function in the cytoplasm.
FIG. 7.
Par 6 nuclear localization is not dependent on aPKC. (A) HeLa cells were transfected with the indicated constructs. The GFP control is the same as for Fig. 3. (B) The ratio of nuclear to cytoplasmic GFP signal was measured as described for Fig. 3. The GFP-Par 6 population is the same as used for the analysis in Fig. 3. (C) GFP-Par 6ΔN can be seen in puncta in the nucleus. The puncta correspond to puncta observed with SC-35 staining.
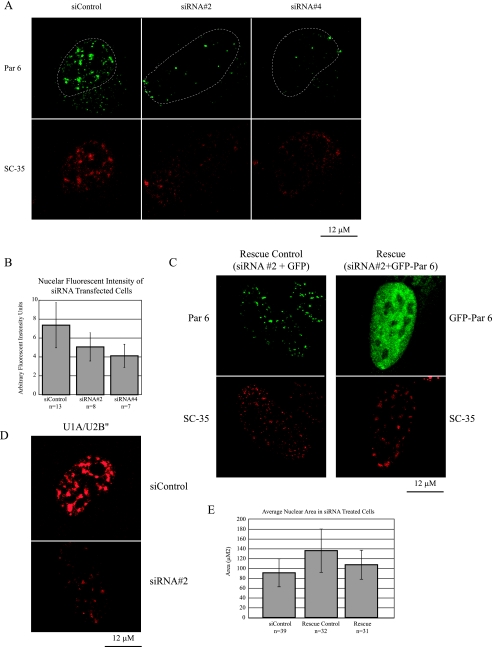
Par 6 knockdown results in dispersed speckles and enlarged nuclei.
To examine the consequences of Par 6 expression in the nucleus and on speckle organization, we knocked down Par 6 with specific siRNAs. The top panels of Fig. 8A show Par 6 immunostaining of HeLa cells transfected with a control siRNA duplex or one of two different duplexes directed against human Par 6 (no. 2 and no. 4); the cells were transfected two times over the course of 6 days. It is clear that Par 6 levels are reduced in the Par 6 siRNA-treated cells compared to the control cells. However, we were unable to completely knock down Par 6 using a number of different transfection protocols and siRNA duplexes. We sought to quantify the level of Par 6 knockdown following siRNA treatment. Using extracts from cells grown at the scale used for siRNA experiments, we were unable to obtain a sufficient signal for Western blotting or RT-PCR to determine knockdown efficiency. Instead, we quantified the fluorescent signals of individual cells. To track cells that had been transfected twice and thus maximally affected by siRNA treatment, we cotransfected cells with DsRed during the first transfection and GFP during the second transfection. Figure 8B shows that cells that had been transfected with either Par 6 siRNA construct showed a significant reduction in the total intensity of nuclear fluorescence. With siRNA no. 2 the average intensity was 31% lower than that of the control (5.063 arbitrary units versus 7.365 arbitrary units; t test, P < 0.03). With siRNA no. 4 the average intensity was 44% lower (4.119 arbitrary units versus 7.365 arbitrary units; t test, P < 0.001).
FIG. 8.
Par 6 knockdown results in dispersed speckles and enlarged nuclei. (A) HeLa cells were treated with the indicated siRNA constructs and stained with Par 6 and SC-35 antibodies. Dotted lines show the outline of the nucleus in the top row. (B) The nuclear fluorescence intensity for cells transfected with the different siRNA constructs was quantified from confocal images using Image J. (C) Cells were transfected with siRNA no. 2 and either GFP (rescue control) or GFP-Par 6 (rescue). Par 6 staining is shown for the control cell, while GFP signal is shown for the rescued cell. The bottom row shows staining for SC-35. (D) Cells were treated with either siControl or Par 6 siRNA no. 2. Panels show staining for U1A/U2B‴. (E) Quantitation of the nuclear area of cell subjected to the indicated treatments was done with Image J. Error bars in panels B and E represent standard deviations from the means.
Significantly, we found that staining for the speckle marker protein SC-35 was reduced in cells treated with Par 6 siRNAs but not in those treated with control siRNAs (Fig. 8A, bottom panels). Instead of the normally large, bright speckles, SC-35 staining was reduced to smaller, dimmer foci dispersed through the nucleus. When GFP-Par6 that had been engineered to have silent mutations rendering it resistant to siRNA duplex no. 2 was cotransfected with the second dose of siRNA to the cells, we saw a rescue of the SC-35 dispersal phenotype (Fig. 8C). Finally, to test whether this effect of Par 6 siRNA was on speckles in general or on SC-35 in particular, we also stained siRNA-treated cells for snRNPs U1A/U2B‴. Figure 8D shows that, as with SC-35, Par 6 siRNA treatment causes these snRNP antigens to change from large, bright puncta to a dimmer, more diffuse staining.
Inspection of Fig. 8 also shows that in addition to the dispersed speckle staining, the nuclei of Par 6 knockdown cells appeared larger than those of control cells. We quantified the nuclear areas for cells transfected with siRNA no. 2 and the control siRNA (Fig. 8E). The average nuclear area was 136 μm2 for Par 6 siRNA no. 2-treated cells, compared to 90.9 μm2 for control siRNA-treated cells; a t test determined that this difference is significant (P < 0.0001). In the rescued population, we observed that the enlarged-nucleus phenotype was partially reversed. The nuclear areas of these cells were significantly different from those of both control cells and nonrescued siRNA-treated cells (average nuclear area, 107 μm2; P < 0.02 in comparison to both control and nonrescued siRNA no. 2-treated cells).
Tests of possible nuclear functions of Par 6.
The data thus far show clearly that Par 6 is a dual-location protein localized to nuclear speckles. Furthermore, the siRNA experiments show that depletion of Par 6 from the nucleus leads to both a dissolution of nuclear speckles and an increase in nuclear size. To investigate nuclear functions of Par 6, we carried out assays to test its role in functions defined previously for other nuclear speckle proteins.
(i) Transcription.
Although transcription does not take place in nuclear speckles, splicing factors are recruited from speckles to sites of transcription by RNA polymerase II (14, 47). Furthermore, many dual-location proteins have been shown to have a role in transcription (7, 8, 10).
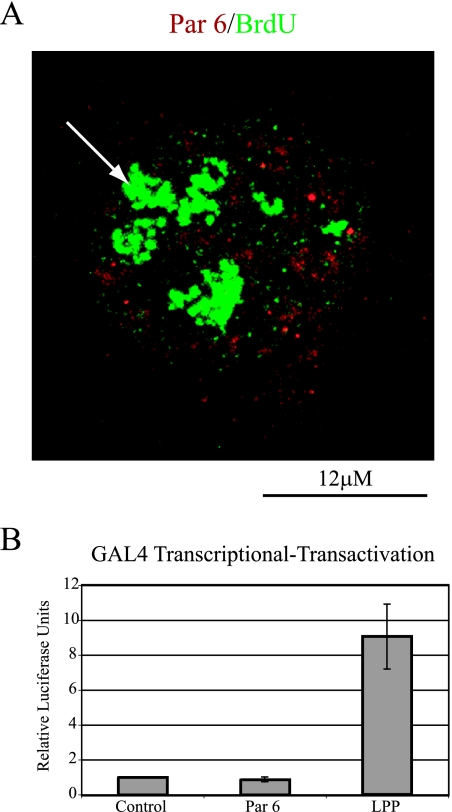
To test if Par 6 plays a role in transcription, two assays were performed. First, HeLa cells were pulse-labeled with 5′-fluorouridine (5′-FU), which becomes incorporated into newly synthesized RNA and can be detected with an antibody to BrdU. As shown in Fig. 9A, Par 6 staining concentrated in nuclear speckles did not colocalize with the 5′-FU sites of transcription. The larger densities of 5′-FU staining are nucleoli, which are visualized due to 5′-FU incorporation into rRNA. A mock incorporation control showed that the 5′-FU puncta representing nascent mRNA were specific (data not shown).
FIG. 9.
Assays for Par 6 involvement in transcription. (A) HeLa cells were pulsed with 2 mM 5′-FU for 6 min and then fixed and processed for immunofluorescence with Par 6 and BrdU antibodies. The arrow points to nucleolar incorporation of 5′-FU. (B) Quantitation of the amount of luciferase production in the GAL4 transcriptional-transactivation assay. The results represent four independent experiments. Error bars indicate standard deviations.
Next, we used a GAL4 transcriptional-transactivation assay to test for Par 6 involvement in transcription. A luciferase reporter construct was cotransfected into HeLa cells with a plasmid encoding either the GAL4-DBD alone or GAL4-DBD fused to Par 6. As a positive control we used GAL4-DBD fused to LPPΔNES; LPP is a dual-location protein that localizes to focal adhesions and the nucleus, and LPPΔNES has been shown to activate transcription in this assay (56). A β-galactosidase plasmid was included in all transfections to normalize luciferase expression to transfection efficiency. Figure 9B shows that GAL4-DBD-Par 6 did not increase luciferase production compared to the control, although it did localize to the nucleus (data not shown). The positive control LPPΔNES caused on average a ninefold increase in relative light units. The results of these two independent assays indicate that Par 6 does not play a direct role in general transcription.
(ii) Splicing.
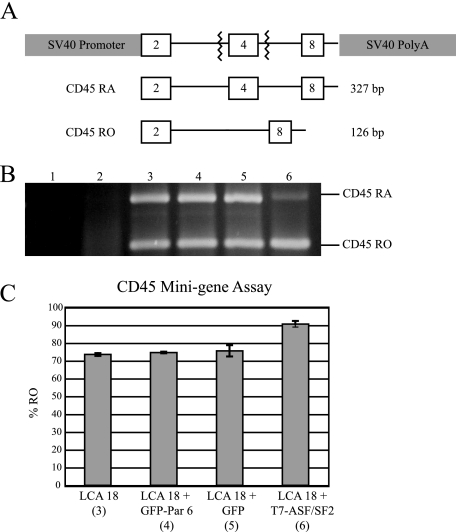
Due to its localization to nuclear speckles, in which many splicing factors reside, we tested whether Par 6 was involved in alternative splicing by using the CD45 minigene splicing assay. CD45 is a transmembrane tyrosine phosphatase expressed by cells of the lymphopoietic lineage and is alternatively spliced as T-cell memory develops (38). pSV-mini LCA-18 (LCA-18) is a CD45 minigene construct that recapitulates exon 4 alternative splicing in this gene (38, 69). Exons 2, 4, and 8 are cloned between a simian virus 40 promoter and a poly(A) site such that splice sites flank exon 4 (Fig. 10A). Note that overexpression of SR protein SC-35, SRp40, SRp75, or ASF/SF2 in cells expressing LCA-18 increases exon 4 splicing, resulting in changes the relative amounts of the RA and RO forms of the transcript (38).
FIG. 10.
Assay for Par 6 involvement in alternative splicing. (A) Schematic of pSV-mini LCA-18. (B) Representative gel of RT-PCR for exon 4 of pSV-mini LCA 18. (C) Quantitation of the percentage of exon 4 that is in the RO form. Numbers beneath the construct names indicate the corresponding gel lane. Error bars indicate standard deviations.
GFP-Par 6 was coexpressed with LCA-18 in HeLa cells, and RT-PCR was then used to detect the amounts of the RA and RO forms of the CD45 minigene expressed. Figure 10B shows representative RT-PCR results, and Fig. 9C shows the quantitation from three experiments. Controls for RNA contamination of RT-PCR components (lane 1) and nonspecific amplification of RA- or RO-sized bands from HeLa cells (lane 2) were negative. When LCA-18 alone was expressed in HeLa cells, approximately 72% of the transcripts were spliced to the RO form (lane 3). Lane 4 shows that when GFP-Par 6 was coexpressed with LCA-18, there was no significant change in the amount of the RO form. GFP vector alone also had no significant effect on exon 4 splicing (lane 5). As a positive control, T7-ASF/SF2 was coexpressed with LCA-18. As has been published previously, a significant shift to the RO form was observed (lane 6) (38). These results indicate that Par 6 does not play a role in general alternative splicing.
(iii) mRNA transport.
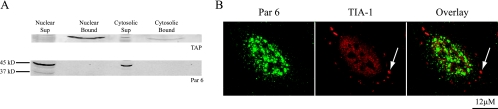
Many proteins localized to nuclear speckles are involved not only in splicing but also in transport of mRNAs out of the nucleus. For example, it is known that several of the SR proteins shuttle with mRNA out of the nucleus and play a role in determining when and where a message will be translated once it is in the cytoplasm (28, 61). To test whether Par 6 is complexed with mRNA and thus is a candidate for a transport factor, mRNA was precipitated from HeLa nuclear and cytosolic extracts with oligo(dT) cellulose. Bound proteins eluted from the oligo(dT) cellulose were concentrated 2.6 times over the supernatant to ensure that any bound Par 6 would be detected. Figure 11A shows a Western blot of the supernatants and bound proteins from each compartment and is representative of three experiments. Par 6 (both the 45-kDa band and a small amount of the 37-kDa band) was present in the supernatants but was not in the fraction bound to oligo(dT) cellulose. As a positive control, we blotted for TAP, a protein that mediates RNA transport out of the nucleus (75). The top panel of Fig. 11A shows that TAP was precipitated by the oligo(dT) cellulose from both the nuclear and cytosolic fractions. Some TAP is also detected in the cytoplasmic supernatant, presumably due to the fact that TAP is released from transcripts in the cytoplasm before recycling back to the nucleus.
FIG. 11.
Assays for Par 6 involvement in mRNA transport. (A) Representative Western blot of oligo(dT) pulldown to detect Par 6 bound to mRNA. Sup, supernatant; bound, Protein bound to oligo(dT). (B) HeLa cells were treated with 50 mM arsenite for 1 h at 37°C and then stained with Par 6 and TIA-1 antibodies. The arrow points to an example of a stress granule marked by TIA-1 staining.
As a second test for Par 6 involvement in mRNA transport, oxidative stress was applied to cells by using arsenite. This has been shown to induce the formation of cytoplasmic aggregates of mRNA and proteins called “stress granules” (30, 31, 44). Figure 11B shows that Par 6 did not colocalize with stress granules in HeLa cells (marked by TIA-1 staining) after arsenite treatment. The results of these two experiments suggest that Par 6 is not associated with general mature mRNA either in the nucleus or as it is transported out of the nucleus.
Par 6 colocalizes with HTLV-1 Tax protein.
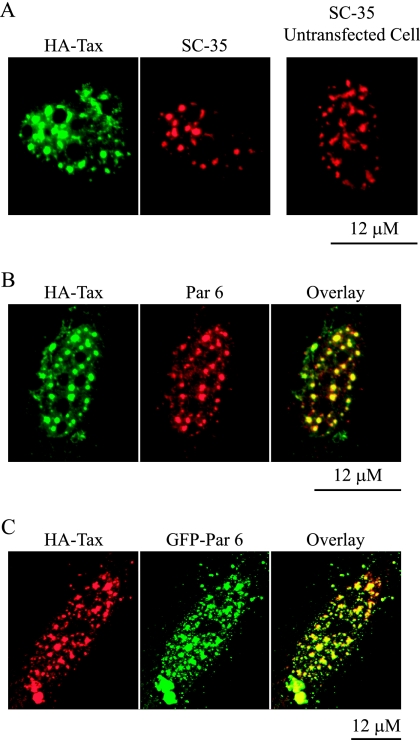
The results described above show that Par 6 is a constituent of nuclear speckles and that a reduction in its levels affected nucleus and nuclear speckle morphology, but it does not appear to play a general role in transcription, splicing, or mRNA transport. To further investigate the nuclear function of Par 6, we turned our attention to the fact that human Par 6 was first identified not as a protein involved in cell polarity but as an interaction partner for HTLV-1 Tax protein (58). In HTLV-1-infected cells or cells overexpressing Tax, Tax is localized to the nucleus, where it forms bodies called “Tax-speckled structures” (TSS) (11, 63). These structures contain several classes of proteins, including the transcription factor NF-κB (11), the nuclear corepressor SMRT (4), DNA damage response factors chk2 and 53BP1 (25), and, significantly, SC-35 and snRNPs recognized by the Smith antigen antibody that localize normally to speckles (11, 63).
The interaction between Par 6 and Tax was indicated by results from a yeast two-hybrid screen and coimmunoprecipitation (58). Given that Par 6 colocalizes with nuclear speckles and that many speckle proteins are localized in TSS, we tested whether endogenous Par 6 colocalizes with Tax in these structures. HA-tagged Tax was transiently expressed in HeLa cells and formed TSS that contained SC-35 (Fig. 12A, middle panel), as previously reported. Significantly, we found that the formation of TSS caused SC-35 to redistribute from irregularly shaped speckles to larger, rounder puncta (compare the transfected and untransfected cells in Fig. 12A). Costaining for endogenous Par 6 showed that this protein was also localized in the TSS (Fig. 12B).
FIG. 12.
Par 6 colocalizes with HTLV-1 Tax protein. (A) HeLa cells were transfected with HA-tagged Tax. Shown is a transfected cell stained with an antibody to the HA tag and SC-35. The right panel shows SC-35 staining in an untransfected cell. (B) Transfected cell stained with anti-HA and Par 6 antibodies. (C) Cell transfected with both HA-Tax and GFP-Par 6. The micrographs in panels A and B were taken with a Zeiss Axioplan microscope.
Coexpression of GFP-Par 6 and HA-Tax resulted in colocalization of both proteins to TSS (Fig. 12C). It is noteworthy that in these cells GFP-Par 6 was no longer diffuse in the nucleus, as we had found in cells not expressing Tax (compare to Fig. 1B and 3A). Instead, the GFP-Par 6 was completely localized to the TSS. This may indicate that the formation of TSS provides additional binding sites for overexpressed GFP-Par 6.
Together these results show that the previously identified interaction of Par 6 with Tax takes place in the nucleus and that Par 6 is recruited along with other speckle proteins to the TSS.
DISCUSSION
We showed that Par 6 is a constitutive dual-location protein that localizes to cell-cell contacts and nuclear speckles. Although some nuclear speckle proteins have been shown to play roles in transcription, alternative splicing, and mRNA transport, we could not find evidence for similar generalized roles for Par 6. Although we could not find evidence of a role for Par 6 as an mRNA binding protein, recent evidence has suggested that Par 6 interacts genetically and physically with the Drosophila homolog of fragile X mental retardation protein (dFMRP), as did the Par 6/aPKC target protein dLgl (77). FMRP is a cytoplasmic protein associated with mRNP complexes involved in the transport and translational control of its target mRNAs, and loss of FMRP is responsible for fragile X syndrome, the most common form of inherited mental retardation (62). It is possible that Par 6 interacts with only selected mRNAs, as FMRP does, and this would have been missed in our more general screen for binding bulk mRNAs.
Although we did not find evidence of direct involvement of Par 6 in several speckle protein functions, we found that it colocalized with the Tax protein of HTLV-1. HTLV-1 is the causative agent of not only adult T-cell leukemia but also the progressive neurological disease tropical spastic paraparesis. HTLV-1 lacks a classical oncogene but instead expresses Tax, which acts as a transcriptional activator of the HTLV long terminal repeat (21, 22). Tax expression causes the formation of TSS that contain a variety of nuclear speckle proteins in addition to transcription factors and components of the DNA repair machinery. It is possible that Tax evolved to advance virus production by acting as a scaffold for various nuclear processes, which it then hijacks for virus production. Thereby, Tax binding to Par 6 could facilitate aggregation of factors necessary for virus replication. Though Tax has a PDZ binding domain at its C terminus, it was shown in vitro that its interaction with Par 6 is not through this domain (58). Non-PDZ binding between Par 6 and Tax would allow Tax to bind to Par 6 without competing with other protein-protein interactions involving Par 6. We found that TSS were still able to form in Par 6 siRNA-treated cells, but our lack of a complete knockdown prevents us from concluding whether or not Par 6 is necessary for TSS formation.
We found that reduced levels of Par 6 caused the breakdown of nuclear speckles and loss of the speckle marker proteins SC-35 and U1A/U2B‴. These results indicate that Par 6 plays a role in the maintenance of speckle structure. Several other proteins regulate speckle morphology and function. A phenotype similar to that induced by reduced Par 6 expression was reported in cells overexpressing Clk/Sty or SRPK2 kinases that phosphorylate SR proteins (15, 23, 35, 74). Overexpression of the kinase DYRK1A also caused speckle disassembly (2). The speckle dispersal phenotype was also seen when PPI, a phosphatase that dephosphorylates SR proteins, was overexpressed (48). Overexpression of SR-cyclophilin, an RS domain-containing member of a large class of proteins called cyclophilins that function as peptidylprolyl-isomerase, caused speckle proteins to become diffuse within the nucleus. It was suggested that the peptidylprolyl-isomerase function of SR-cyclophilin could in some way contribute to the protein-protein interactions that hold speckles together (40).
The loss of certain proteins has also been found to affect speckle morphology. Syntenin 2 is a PDZ protein that is localized to nuclear speckles. Syntenin 2 targeting to nuclear speckles is mediated by interaction of its PDZ domains with nuclear PIP2. When syntenin 2 was knocked down using siRNA, speckles were disrupted (49). Speckles also become dispersed within the nucleus when the SR-related dual-location protein pinin is knocked down using siRNA (12). This phenotype is similar to that which we found in the Par 6 knockdown cells.
Speckles are thought to be the result of protein-protein interactions, as opposed to attachment to a preformed scaffold (59). We suggest that Par 6 plays a structural role in the nucleus to facilitate such protein-protein interactions. Par 6 contains two protein-protein interaction domains. The first, the PB1 domain, can cause proteins to homodimerize or heterodimerize with other PB1 domain-containing proteins (50). The second domain, the PDZ domain, can bind to other PDZ domains or to PDZ binding domains at the C termini of proteins (53). It is of note that syntenin 2, the speckle protein whose knockdown caused dissolution of speckles, also contains a PDZ domain. The constitutive localization of Par 6 to nuclear speckles and the resulting dissolution of the speckles when Par 6 levels were reduced with siRNA make this an attractive hypothesis.
Acknowledgments
Work in W.J.N.'s laboratory is supported by NIH GM35227. E.G.C. was supported in part by NIH NIGMS predoctoral training program GM07276.
We thank F. Bex of the University of Brussels for the HA-Tax construct, and we thank those who contributed reagents as described in Materials and Methods.
Footnotes
Published ahead of print on 9 April 2007.
REFERENCES
- 1.Aijaz, S., M. S. Balda, and K. Matter. 2006. Tight junctions: molecular architecture and function. Int. Rev. Cytol. 248:261-298. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez, M., X. Estivill, and S. de la Luna. 2003. DYRK1A accumulates in splicing speckles through a novel targeting signal and induces speckle disassembly. J. Cell Sci. 116:3099-3107. [DOI] [PubMed] [Google Scholar]
- 3.Aranda, V., T. Haire, M. E. Nolan, J. P. Calarco, A. Z. Rosenberg, J. P. Fawcett, T. Pawson, and S. K. Muthuswamy. 2006. Par6-aPKC uncouples ErbB2 induced disruption of polarized epithelial organization from proliferation control. Nat. Cell Biol. 8:1235-1245. [DOI] [PubMed] [Google Scholar]
- 4.Ariumi, Y., T. Ego, A. Kaida, M. Matsumoto, P. P. Pandolfi, and K. Shimotohno. 2003. Distinct nuclear body components, PML and SMRT, regulate the trans-acting function of HTLV-1 Tax oncoprotein. Oncogene 22:1611-1619. [DOI] [PubMed] [Google Scholar]
- 5.Balda, M. S., M. D. Garrett, and K. Matter. 2003. The ZO-1-associated Y-box factor ZONAB regulates epithelial cell proliferation and cell density. J. Cell Biol. 160:423-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balda, M. S., and K. Matter. 2003. Epithelial cell adhesion and the regulation of gene expression. Trends Cell Biol. 13:310-318. [DOI] [PubMed] [Google Scholar]
- 7.Balda, M. S., and K. Matter. 2000. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 19:2024-2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behrens, J., J. P. von Kries, M. Kuhl, L. Bruhn, D. Wedlich, R. Grosschedl, and W. Birchmeier. 1996. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382:638-642. [DOI] [PubMed] [Google Scholar]
- 9.Benmerah, A., M. Scott, V. Poupon, and S. Marullo. 2003. Nuclear functions for plasma membrane-associated proteins. Traffic 4:503-511. [DOI] [PubMed] [Google Scholar]
- 10.Betanzos, A., M. Huerta, E. Lopez-Bayghen, E. Azuara, J. Amerena, and L. Gonzalez-Mariscal. 2004. The tight junction protein ZO-2 associates with Jun, Fos and C/EBP transcription factors in epithelial cells. Exp. Cell Res. 292:51-66. [DOI] [PubMed] [Google Scholar]
- 11.Bex, F., A. McDowall, A. Burny, and R. Gaynor. 1997. The human T-cell leukemia virus type 1 transactivator protein Tax colocalizes in unique nuclear structures with NF-κB proteins. J. Virol. 71:3484-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu, Y., and P. Ouyang. 2006. Loss of Pnn expression attenuates expression levels of SR family splicing factors and modulates alternative pre-mRNA splicing in vivo. Biochem. Biophys. Res. Commun. 341:663-671. [DOI] [PubMed] [Google Scholar]
- 13.Cline, E. G., and W. J. Nelson. 2005. Dual location proteins; communication between cell adhesions and the nucleus, p. 47-69. In D. Wedlich (ed.), Cell migration in development and disease. Wiley-VCH, Weinheim, Germany.
- 14.Cmarko, D., P. J. Verschure, T. E. Martin, M. E. Dahmus, S. Krause, X. D. Fu, R. van Driel, and S. Fakan. 1999. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell 10:211-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colwill, K., T. Pawson, B. Andrews, J. Prasad, J. L. Manley, J. C. Bell, and P. I. Duncan. 1996. The Clk/Sty protein kinase phosphorylates SR splicing factors and regulates their intranuclear distribution. EMBO. J. 15:265-275. [PMC free article] [PubMed] [Google Scholar]
- 16.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Etienne-Manneville, S., and A. Hall. 2003. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature 421:753-756. [DOI] [PubMed] [Google Scholar]
- 18.Etienne-Manneville, S., and A. Hall. 2001. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell 106:489-498. [DOI] [PubMed] [Google Scholar]
- 19.Gao, L., and I. G. Macara. 2004. Isoforms of the polarity protein par6 have distinct functions. J. Biol. Chem. 279:41557-41562. [DOI] [PubMed] [Google Scholar]
- 20.Garrard, S. M., C. T. Capaldo, L. Gao, M. K. Rosen, I. G. Macara, and D. R. Tomchick. 2003. Structure of Cdc42 in a complex with the GTPase-binding domain of the cell polarity protein, Par6. EMBO. J. 22:1125-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gatza, M. L., J. C. Watt, and S. J. Marriott. 2003. Cellular transformation by the HTLV-I Tax protein, a jack-of-all-trades. Oncogene 22:5141-5149. [DOI] [PubMed] [Google Scholar]
- 22.Grassmann, R., M. Aboud, and K. T. Jeang. 2005. Molecular mechanisms of cellular transformation by HTLV-1 Tax. Oncogene 24:5976-5985. [DOI] [PubMed] [Google Scholar]
- 23.Gui, J. F., W. S. Lane, and X. D. Fu. 1994. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature 369:678-682. [DOI] [PubMed] [Google Scholar]
- 24.Guo, S., and K. J. Kemphues. 1996. Molecular genetics of asymmetric cleavage in the early Caenorhabditis elegans embryo. Curr. Opin. Genet. Dev. 6:408-415. [DOI] [PubMed] [Google Scholar]
- 25.Haoudi, A., R. C. Daniels, E. Wong, G. Kupfer, and O. J. Semmes. 2003. Human T-cell leukemia virus-I tax oncoprotein functionally targets a subnuclear complex involved in cellular DNA damage-response. J. Biol. Chem. 278:37736-37744. [DOI] [PubMed] [Google Scholar]
- 26.Henrique, D., and F. Schweisguth. 2003. Cell polarity: the ups and downs of the Par6/aPKC complex. Curr. Opin. Genet. Dev. 13:341-350. [DOI] [PubMed] [Google Scholar]
- 27.Hofmann, I., M. Schnolzer, I. Kaufmann, and W. W. Franke. 2002. Symplekin, a constitutive protein of karyo- and cytoplasmic particles involved in mRNA biogenesis in Xenopus laevis oocytes. Mol. Biol. Cell 13:1665-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang, Y., R. Gattoni, J. Stevenin, and J. A. Steitz. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11:837-843. [DOI] [PubMed] [Google Scholar]
- 29.Johansson, A., M. Driessens, and P. Aspenstrom. 2000. The mammalian homologue of the Caenorhabditis elegans polarity protein PAR-6 is a binding partner for the Rho GTPases Cdc42 and Rac1. J. Cell Sci. 113:3267-3275. [DOI] [PubMed] [Google Scholar]
- 30.Kedersha, N., and P. Anderson. 2002. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem. Soc. Trans. 30:963-969. [DOI] [PubMed] [Google Scholar]
- 31.Kim, S. H., W. K. Dong, I. J. Weiler, and W. T. Greenough. 2006. Fragile X mental retardation protein shifts between polyribosomes and stress granules after neuronal injury by arsenite stress or in vivo hippocampal electrode insertion. J. Neurosci. 26:2413-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein, T. J., and M. Mlodzik. 2005. Planar cell polarization: an emerging model points in the right direction. Annu. Rev. Cell Dev. Biol. 21:155-176. [DOI] [PubMed] [Google Scholar]
- 33.Knust, E., and O. Bossinger. 2002. Composition and formation of intercellular junctions in epithelial cells. Science 298:1955-1959. [DOI] [PubMed] [Google Scholar]
- 34.Kodama, A., I. Karakesisoglou, E. Wong, A. Vaezi, and E. Fuchs. 2003. ACF7: an essential integrator of microtubule dynamics. Cell 115:343-354. [DOI] [PubMed] [Google Scholar]
- 35.Kuroyanagi, N., H. Onogi, T. Wakabayashi, and M. Hagiwara. 1998. Novel SR-protein-specific kinase, SRPK2, disassembles nuclear speckles. Biochem. Biophys. Res. Commun. 242:357-364. [DOI] [PubMed] [Google Scholar]
- 36.Lamond, A. I., and M. Carmo-Fonseca. 1993. Localisation of splicing snRNPs in mammalian cells. Mol. Biol. Rep. 18:127-133. [DOI] [PubMed] [Google Scholar]
- 37.Lamond, A. I., and D. L. Spector. 2003. Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4:605-612. [DOI] [PubMed] [Google Scholar]
- 38.Lemaire, R., A. Winne, M. Sarkissian, and R. Lafyatis. 1999. SF2 and SRp55 regulation of CD45 exon 4 skipping during T cell activation. Eur. J. Immunol. 29:823-837. [DOI] [PubMed] [Google Scholar]
- 39.Lemmers, C., D. Michel, L. Lane-Guermonprez, M. H. Delgrossi, E. Medina, J. P. Arsanto, and A. Le Bivic. 2004. CRB3 binds directly to Par6 and regulates the morphogenesis of the tight junctions in mammalian epithelial cells. Mol. Biol. Cell 15:1324-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin, C. L., S. Leu, M. C. Lu, and P. Ouyang. 2004. Over-expression of SR-cyclophilin, an interaction partner of nuclear pinin, releases SR family splicing factors from nuclear speckles. Biochem. Biophys. Res. Commun. 321:638-647. [DOI] [PubMed] [Google Scholar]
- 41.Margolis, B., and J. P. Borg. 2005. Apicobasal polarity complexes. J. Cell Sci. 118:5157-5159. [DOI] [PubMed] [Google Scholar]
- 42.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 43.Matter, K., and M. S. Balda. 2003. Signalling to and from tight junctions. Nat. Rev. Mol. Cell Biol. 4:225-236. [DOI] [PubMed] [Google Scholar]
- 44.Mazroui, R., M. E. Huot, S. Tremblay, C. Filion, Y. Labelle, and E. W. Khandjian. 2002. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 11:3007-3017. [DOI] [PubMed] [Google Scholar]
- 45.Melcak, I., S. Cermanova, K. Jirsova, K. Koberna, J. Malinsky, and I. Raska. 2000. Nuclear pre-mRNA compartmentalization: trafficking of released transcripts to splicing factor reservoirs. Mol. Biol. Cell 11:497-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science 291:843-847. [DOI] [PubMed] [Google Scholar]
- 47.Misteli, T., J. F. Caceres, and D. L. Spector. 1997. The dynamics of a pre-mRNA splicing factor in living cells. Nature 387:523-527. [DOI] [PubMed] [Google Scholar]
- 48.Misteli, T., and D. L. Spector. 1996. Serine/threonine phosphatase 1 modulates the subnuclear distribution of pre-mRNA splicing factors. Mol. Biol Cell 7:1559-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mortier, E., G. Wuytens, I. Leenaerts, F. Hannes, M. Y. Heung, G. Degeest, G. David, and P. Zimmermann. 2005. Nuclear speckles and nucleoli targeting by PIP2-PDZ domain interactions. EMBO. J. 24:2556-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moscat, J., M. T. Diaz-Meco, A. Albert, and S. Campuzano. 2006. Cell signaling and function organized by PB1 domain interactions. Mol. Cell 23:631-640. [DOI] [PubMed] [Google Scholar]
- 51.Nelson, W. J. 2003. Adaptation of core mechanisms to generate cell polarity. Nature 422:766-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noda, Y., R. Takeya, S. Ohno, S. Naito, T. Ito, and H. Sumimoto. 2001. Human homologues of the Caenorhabditis elegans cell polarity protein PAR6 as an adaptor that links the small GTPases Rac and Cdc42 to atypical protein kinase C. Genes Cells 6:107-119. [DOI] [PubMed] [Google Scholar]
- 53.Nourry, C., S. G. Grant, and J. P. Borg. 2003. PDZ domain proteins: plug and play. Sci. STKE 2003:RE7. [DOI] [PubMed] [Google Scholar]
- 54.Ohno, S. 2001. Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr. Opin. Cell Biol. 13:641-648. [DOI] [PubMed] [Google Scholar]
- 55.Perander, M., G. Bjorkoy, and T. Johansen. 2001. Nuclear import and export signals enable rapid nucleocytoplasmic shuttling of the atypical protein kinase C lambda. J. Biol. Chem. 276:13015-13024. [DOI] [PubMed] [Google Scholar]
- 56.Petit, M. M., J. Fradelizi, R. M. Golsteyn, T. A. Ayoubi, B. Menichi, D. Louvard, W. J. Van de Ven, and E. Friederich. 2000. LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol. Biol. Cell 11:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qiu, R. G., A. Abo, and G. Steven Martin. 2000. A human homolog of the C. elegans polarity determinant Par-6 links Rac and Cdc42 to PKCzeta signaling and cell transformation. Curr. Biol. 10:697-707. [DOI] [PubMed] [Google Scholar]
- 58.Rousset, R., S. Fabre, C. Desbois, F. Bantignies, and P. Jalinot. 1998. The C-terminus of the HTLV-1 Tax oncoprotein mediates interaction with the PDZ domain of cellular proteins. Oncogene 16:643-654. [DOI] [PubMed] [Google Scholar]
- 59.Sacco-Bubulya, P., and D. L. Spector. 2002. Disassembly of interchromatin granule clusters alters the coordination of transcription and pre-mRNA splicing. J. Cell Biol. 156:425-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sadowski, I., B. Bell, P. Broad, and M. Hollis. 1992. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene 118:137-141. [DOI] [PubMed] [Google Scholar]
- 61.Sanford, J. R., N. K. Gray, K. Beckmann, and J. F. Caceres. 2004. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 18:755-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schaeffer, C., M. Beaulande, C. Ehresmann, B. Ehresmann, and H. Moine. 2003. The RNA binding protein FMRP: new connections and missing links. Biol. Cell 95:221-228. [DOI] [PubMed] [Google Scholar]
- 63.Semmes, O. J., and K. T. Jeang. 1996. Localization of human T-cell leukemia virus type 1 tax to subnuclear compartments that overlap with interchromatin speckles. J. Virol. 70:6347-6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sheng, M., and C. Sala. 2001. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 24:1-29. [DOI] [PubMed] [Google Scholar]
- 65.Singer, R. H., and M. R. Green. 1997. Compartmentalization of eukaryotic gene expression: causes and effects. Cell 91:291-294. [DOI] [PubMed] [Google Scholar]
- 66.Spector, D. L. 1993. Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 9:265-315. [DOI] [PubMed] [Google Scholar]
- 67.Spector, D. L., X. D. Fu, and T. Maniatis. 1991. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO. J. 10:3467-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spector, D. L., W. H. Schrier, and H. Busch. 1983. Immunoelectron microscopic localization of snRNPs. Biol. Cell 49:1-10. [DOI] [PubMed] [Google Scholar]
- 69.Streuli, M., and H. Saito. 1989. Regulation of tissue-specific alternative splicing: exon-specific cis-elements govern the splicing of leukocyte common antigen pre-mRNA. EMBO. J. 8:787-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suzuki, A., and S. Ohno. 2006. The PAR-aPKC system: lessons in polarity. J. Cell Sci. 119:979-987. [DOI] [PubMed] [Google Scholar]
- 71.Takagaki, Y., and J. L. Manley. 2000. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 20:1515-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Van Itallie, C. M., and J. M. Anderson. 2006. Claudins and epithelial paracellular transport. Annu. Rev. Physiol. 68:403-429. [DOI] [PubMed] [Google Scholar]
- 73.Vogelmann, R., and W. J. Nelson. 2005. Fractionation of the epithelial apical junctional complex: reassessment of protein distributions in different substructures. Mol. Biol. Cell 16:701-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang, H. Y., W. Lin, J. A. Dyck, J. M. Yeakley, Z. Songyang, L. C. Cantley, and X. D. Fu. 1998. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J. Cell Biol. 140:737-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weis, K. 2002. Nucleocytoplasmic transport: cargo trafficking across the border. Curr. Opin. Cell Biol. 14:328-335. [DOI] [PubMed] [Google Scholar]
- 76.Yamanaka, T., Y. Horikoshi, A. Suzuki, Y. Sugiyama, K. Kitamura, R. Maniwa, Y. Nagai, A. Yamashita, T. Hirose, H. Ishikawa, and S. Ohno. 2001. PAR-6 regulates aPKC activity in a novel way and mediates cell-cell contact-induced formation of the epithelial junctional complex. Genes Cells 6:721-731. [DOI] [PubMed] [Google Scholar]
- 77.Zarnescu, D. C., P. Jin, J. Betschinger, M. Nakamoto, Y. Wang, T. C. Dockendorff, Y. Feng, T. A. Jongens, J. C. Sisson, J. A. Knoblich, S. T. Warren, and K. Moses. 2005. Fragile X protein functions with lgl and the par complex in flies and mice. Dev. Cell 8:43-52. [DOI] [PubMed] [Google Scholar]