Abstract
The inducible expression of antimicrobial peptide genes in Drosophila melanogaster is regulated by the conserved Toll and peptidoglycan recognition protein LC/immune deficiency (PGRP-LC/IMD) signaling pathways. It has been proposed that the two pathways have independent functions and mediate the specificity of innate immune responses towards different microorganisms. Scattered evidence also suggests that some antimicrobial target genes can be activated by both Toll and IMD, albeit to different extents. This dual activation can be mediated by independent stimulation or by cross-regulation of the two pathways. We show in this report that the Toll and IMD pathways can interact synergistically, demonstrating that cross-regulation occurs. The presence of Spätzle (the Toll ligand) and gram-negative peptidoglycan (the PGRP-LC ligand) together caused synergistic activation of representative target genes of the two pathways, including Drosomycin, Diptericin, and AttacinA. Constitutive activation of Toll and PGRP-LC/IMD could mimic the synergistic stimulation. RNA interference assays and promoter analyses demonstrate that cooperation of different NF-κB-related transcription factors mediates the synergy. These results illustrate how specific ligand binding by separate upstream pattern recognition receptors can be translated into a broad-spectrum host response, a hallmark of innate immunity.
Upon infection, insects mount a rapid antimicrobial response that consists of many components, including antimicrobial peptides, hemocytes, and phenoloxidase-based melanization (3, 19, 23, 38, 49). This insect antimicrobial response is equivalent to the innate immune response in mammals (14). Even though insects do not possess B and T lymphocytes, the insect innate immune system can recognize different classes of microorganisms and respond accordingly (25, 26).
A critical aspect of the Drosophila melanogaster innate immune response is the inducible expression of a spectrum of antimicrobial peptides which function synergistically to fight microbial infections (3, 19, 23, 38, 49). The inducible expression of antimicrobial peptide genes is regulated by the Toll and immune deficiency (IMD) pathways. In the absence of these two pathways, the antimicrobial peptide genes cannot be induced and the flies become more susceptible to many microbes, even those that are normally nonpathogenic (12, 51, 53).
The IMD pathway regulates the response to gram-negative bacterial infection. Gram-negative bacterium-derived diaminopimelic acid (DAP)-type peptidoglycan is recognized by upstream receptors called peptidoglycan recognition proteins LC (PGRP-LC) and LE (4, 6, 12, 21, 26, 29, 44, 47, 48). The recognition leads to activation of the adaptor protein IMD, a homologue of the mammalian tumor necrosis factor receptor-interacting protein RIP (5, 10). Other downstream components of the IMD pathway include the Drosophila homologues of the kinases TAK1, IKK, and JNK (12, 30, 35, 42, 43, 52). These regulatory proteins converge onto the NF-κB-related transcription factor Relish. The signal-induced proteolytic cleavage of Relish allows nuclear translocation of the active N-terminal portion, which interacts with other proteins to modulate the expression of antimicrobial peptide genes such as Diptericin and AttacinA (15, 22, 45).
The Toll pathway regulates the response to gram-positive bacterial and fungal infections. Gram-positive bacterium-derived Lys-type peptidoglycan is recognized by upstream receptors PGRP-SA and -SD, as well as gram-negative binding protein 1 (GNBP-1) (2, 9, 11, 33, 37). The fungal component that stimulates the Toll pathway is not well defined. Nonetheless, both infections cause activation of protease cascades and ultimately cleavage of the host protein Spätzle (20, 27, 28). Cleaved Spätzle serves as the ligand for Toll and induces the expression of a subset of antimicrobial peptide genes, such as Drosomycin and Immune induced molecule 1 (IM1) (18, 54). The cytoplasmic components of the Toll signaling pathway include MyD88, Pelle, Cactus, DIF, and Dorsal (17, 32, 39, 46, 50). These components are homologous to mammalian MyD88, IRAK, IκB, and NF-κB, which are used in Toll-like receptor, interleukin-1 receptor, and tumor necrosis factor receptor signaling (14). Therefore, evolutionarily conserved pathways are employed in Drosophila and mammals to regulate innate immunity.
The signaling molecules of the Toll and IMD pathways are clearly distinct. Some antimicrobial peptide genes preferentially respond to one of the two pathways. Therefore, it has been generally accepted that the two pathways serve independent functions and provide specificity in Drosophila innate immunity (3, 19, 38, 49). Meanwhile, a number of reports demonstrate that double mutants of the two pathways have increased susceptibility to microbial challenge and that some antimicrobial peptide genes are regulated, albeit to different extents, by both pathways (7, 15, 16, 24, 40). Antimicrobial peptide genes can be grouped based on the complex response to the two signaling pathways. Induction of Diptericin and Drosocin is highly defective in IMD pathway mutants; induction of Drosomycin and IM1 is highly defective in Toll pathway mutants; induction of AttacinA, CecropinA, and Defensin is defective to different degrees in either Toll or IMD pathway mutants; and induction of Metchnikowin is not affected in either Toll or IMD mutants but is defective in double mutants. A mechanism that can explain these results is independent activation of overlapping target genes by the two pathways. For instance, some target promoters may contain binding sites for both DIF and Relish: thus, they can be activated independently by both pathways and the response depends on the affinity and number of κB sites. This mechanism fits nicely with all of the available genetic and molecular analyses. Another mechanism is cross-regulation of the two pathways, such as cross-modification by a kinase, better formation of a common adaptor complex, or cooperation of transcription factors. All of these possible mechanisms do not need to be mutually exclusive and can be employed at the same time in vivo. The mechanism of pathway interaction so far has received little support from experimental evidence. We show in this report that the two pathways can synergistically activate the expression of antimicrobial peptide genes. When both pathways are simultaneously stimulated at lower levels, the immunity genes are already induced efficiently. We also demonstrate that the cooperation is mediated through an interaction of the NF-κB-related transcription factors in the two pathways. This synergistic interaction of two immune regulatory pathways illustrates how specific ligand binding by separate upstream pattern recognition receptors can be translated into broad-spectrum host response, a hallmark of innate immunity.
MATERIALS AND METHODS
Molecular cloning and dsRNA synthesis.
Site-directed mutagenesis of κB sites on the Drosomycin promoter was performed in pBluescript Clone. The Stratagene QuickChange mutagenesis kit was used on a double-stranded DNA (dsDNA) template. The first GGG residues in all of the κB sites were changed to ATT.
Constructs for dsRNA synthesis were made by inserting an EcoRI-PstI dorsal cDNA fragment (nucleotides [nt] 555 to 1737 [ATG is +1]) and an XhoI-PstI Relish cDNA fragment (nt 591 to 2334) into the pCCM113 vector, which contains T7 promoter sequence on both ends. Three different Dif cDNA fragments (nt 135 to 725, 720 to 1580, and 1575 to 1990) were used for this series of experiments. The experiments shown in Fig. 5 used the Dif fragment (nt 720 to 1580). The pCCM113 clones were used as templates for PCR using Pfu DNA polymerase (Stratagene) and T7 primer. The PCR products were purified from agarose gel with QIAEX II (QIAGEN) and used as templates for in vitro transcription, using the MEGAscript transcription kit (Ambion) and T7 polymerase. The transcription products were denatured at 95°C for 5 min, renatured by gradual cooling down to 24°C, and then purified with NucAway spin columns (Ambion).
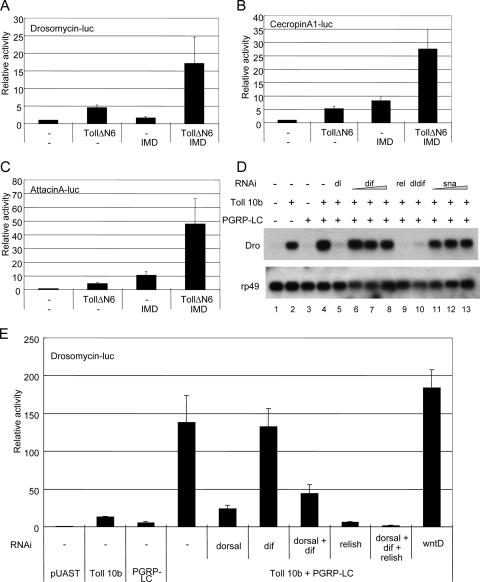
FIG. 5.
Requirement of NF-κB-related proteins for synergistic activation. S2 cells were transiently transfected with different combinations of the expression vector for TollΔN6 and IMD, along with the luciferase (luc) reporter gene of Drosomycin (A), CecropinA (B), or AttacinA (C). Luciferase activity was analyzed and plotted as described in the legend to Fig. 2. (D) S2 cells were transiently transfected with the expression vector for Toll10b and PGRP-LC, together with purified dsRNA. For dorsal and relish, 3 μg of dsRNA was used. For Dif and snail, 3, 6, or 9 μg of dsRNA was used. The expression of the endogenous Drosomycin gene was analyzed by Northern blot hybridization after total RNA was extracted from the S2 cells. The autoradiographs are shown. (E) S2 cells were transiently transfected with the expression vector for Toll10b and PGRP-LC, together with 3 μg of dsRNA for dorsal, Dif, relish, wntD, or the indicated combination. All samples also included the Drosomycin-luciferase reporter gene, and luciferase activity was analyzed and plotted. The results together show that in the S2 cells, Dorsal and Relish are both required for the synergistic activation of Drosomycin, while DIF does not play a role.
Cell culture and RNA expression analyses.
Drosophila Schneider-2 (S2) cells were maintained at 25°C in Schneider's Drosophila medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone), 100 U/ml penicillin, and 100 μg/ml streptomycin (Invitrogen). For stimulation experiments, 2 × 106 cells were cultured in six-well plates for 50 h and purified SpätzleC106 protein (54) and Escherichia coli O111:B4 peptidoglycan (InvivoGen) were added to the culture cells. When ecdysone was included in the experiment, 20-hydroxyecdysone (20-HE [Sigma]) was added at a concentration of 1 μM 24 h before adding other ligands. Cells were harvested 6 h or 20 h after stimulation. The RNA preparation and Northern blot analysis were performed as described previously (13).
Quantitative reverse transcription-PCR (RT-PCR) was carried out with the iScript cDNA synthesis kit (Bio-Rad) for cDNA synthesis and the iQ SYBR green Supermix (Bio-Rad) and MyiQ single-color real-time PCR detection system (Bio-Rad) for real-time PCR. The following gene-specific primers were used: Drosomycin, 5′-TACTTGTTCGCCCTCTTCG-3′ and 5′-GTATCTTCCGGACAGGCAGT-3′; Diptericin, 5′-CCGCAGTACCCACTCAATCT-3′ and 5′-ACTGCAAAGCCAAAACCATC-3′; AttacinA, 5′-AGGTTCCTTAACCTCCAATC-3′ and 5′-CATGACCAGCATTGTTGTAG-3′; and Ribosomal protein 49, 5′-AAGCTAGCCCAACCTGCTTC-3′ and 5′-GTGCGCTTCTTCACGATCT-3′.
Transfection assay.
Transient transfection was performed as described in the protocol for the Lipofectin reagent (Invitrogen). Approximately 2 × 106 cells in each well of six-well plates were transfected with 0.5 μg of the Actin5C-Gal4 plasmid, 0.05 μg of pUAST plasmids (Toll, PGRP-LC, and IMD), 0.5 μg of luciferase reporter plasmids, and 1 μg of copia-lacZ plasmids. The AttacinA, AttacinD, CecropinA1, Defensin, and Drosomycin promoter-luciferase reporters contained 1.0 kb, 2.0 kb, 0.8 kb, 2.7 kb, and 2.9 kb of upstream sequences, respectively. The Drosomycin promoter deletion mutants contained various amounts of promoter sequence as indicated in Fig. 6A, and the κB site mutants were constructed within the 0.43-kb promoter. The transfected cells were grown for 70 h and then harvested. For stimulation, the transfected cells were treated with E. coli or peptidoglycans for 24 h before harvest. The bacteria used were E. coli O55:B5, the cells of which were cultured overnight in 2× YT broth (QBIOgene), heat treated at 65°C for 15 min, and then added to S2 cells at a 1/100 final dilution. Gram-negative peptidoglycan [PGN(−)] was from E. coli O111:B4 (Invivogen), and gram-positive peptidoglycan [PGN(+)] was from Staphylococcus aureus (Fluka), with a final concentration of 10 μg/ml. The harvested cells were lysed in 0.1 M potassium phosphate buffer (pH 7.0) by repeated freeze-thaw. Luciferase and β-galactosidase activities were measured using d(−)-luciferin (Roche) and o-nitrophenyl-β-d-galactopyranoside (Sigma) as substrates, respectively.
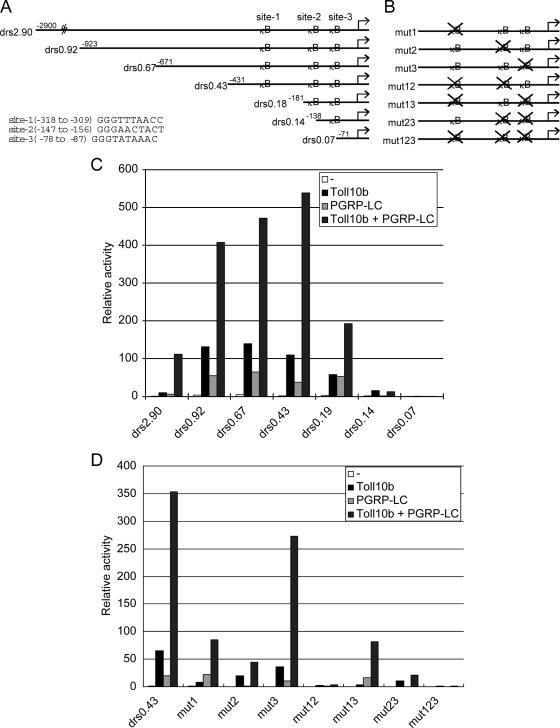
FIG. 6.
Synergistic activation of Drosomycin promoter involves two κB sites. (A) Schematic representation of the promoter and the deletion mutants analyzed for Drosomycin. The transcription start site is +1, and the end point of the upstream sequence is indicated. The three κB sites on the Drosomcyin promoter are as indicated, and the exact location and sequence are shown in the lower left corner. (B) Schematic representation and nomenclature of the six κB site point mutants of the 0.43-kb Drosomycin promoter-luciferase reporter. (C) Luciferase activity of the deletion mutants of the Drosomycin promoter driven by Toll10b and PGRP-LC in transiently transfected S2 cells. The results shown are the average of three independent experiments. (D) Luciferase activity of the κB point mutants of the 0.43-kb Drosomycin promoter driven by Toll10b and PGRP-LC in transiently transfected S2 cells. The results shown are the average of three independent experiments. The results indicate that κB sites 1 and 2 are involved in the response to Toll and PGRP-LC stimulation.
For RNA interference (RNAi) experiments, 3 μg of synthesized and column-purified dsRNA was added with the plasmids and the Lipofectin reagent for transient transfection.
Transgenic flies and natural infection.
Male transgenic flies carrying the gain-of-function mutant construct TollD (Toll755/781Y) on the 2nd chromosome and PGRP-LC on the 3rd chromosome under the regulation by UAS promoter were crossed with female flies carrying Yp1-, nanos-, Hsp70-, arm-, 132-, 127-, 362-, Actin5C-, rhomboid-, e33C-, or daughterless-Gal4 transgenes. Adult flies were counted with the help of balancer marker chromosomes, and viability was designated based on the number of expected flies obtained. For the crosses that produced viable offspring, RNA was extracted from adult females of the next generation containing both Gal4 and UAS transgenes. The RNA was subjected to Northern blot analysis.
For natural infection, Canton-S females of age 2 to 4 days after eclosion were dipped into a solution containing 1 × 106 spores/ml of Beauveria bassiana (ATCC 9453), and then starved at 29°C for 2 h in empty vials. The flies were then transferred into new vials which contain 2 cm by 3 cm of 3MM chromatography paper (Whatman) soaked with 500 μl of 5% sucrose-Pseudomonas entomophila culture. The Pseudomonas entomophila culture used in these experiments was in the logarithmic growth phase and was concentrated by centrifugation and added to the sucrose solution to make the final concentration of 1 at an optical density of 600 nm. Flies were fed with this sucrose-bacterium mixture at 29°C for 24 h and then collected for RNA preparation.
RESULTS
Spätzle and gram-negative peptidoglycan stimulate antimicrobial peptide genes cooperatively.
Even though the Toll and IMD pathways have separate components and preferential target genes, some target genes seem to be regulated by both pathways (7, 16). Thus, we hypothesized that these two pathways could interact at some levels. We used Drosophila S2 cells to test our hypothesis because S2 cells respond to immune stimulants and allow more precise manipulation. The method of injecting microbes into whole flies may stimulate more than one pattern recognition receptor, and injury itself can cause some level of gene induction or prime the innate immune response (1, 26, 31, 36).
In our experiments, a truncated Spätzle protein that is constitutively active was used to stimulate the Toll pathway and PGN(−) was used to stimulate the IMD pathway (26, 54). Spätzle and PGN(−) are ligands of the receptors Toll and PGRP-LC, respectively. The expression of endogenous antimicrobial peptide genes was used to monitor the stimulations (Fig. 1A). The Drosomycin gene is a widely used readout for the Toll pathway, and the Diptericin, AttacinA, and CecropinA genes are common readouts for the IMD pathway. The results showed that direct stimulation for 20 h by the two ligands at the same time induced the highest expression levels of four different antimicrobial peptide genes (Fig. 1A, lane 4). In comparison, adding Spätzle alone did not show detectable induction of these genes, while adding PGN(−) alone induced only low-level expression of AttacinA and CecropinA (Fig. 1A, lanes 1 to 3).
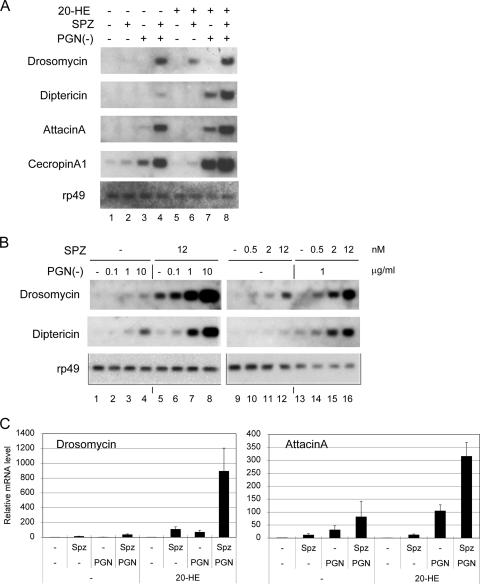
FIG. 1.
Synergistic induction of antimicrobial peptide genes by Spätzle and peptidoglycan. (A) Total RNA was isolated from S2 cells treated with different combinations of 1 μM 20-hydroxyecdysone (20-HE), 2.4 nM SpätzleC106 (SPZ), and 10 mg/ml peptidoglycan from Escherichia coli [(PGN(−)] for 20 h as indicated. The RNA samples were analyzed by Northern blotting using the indicated gene probes for hybridization. The autoradiographs are shown. (B) Dose-response study using various amounts of SpätzleC106 and PGN(−) as indicated at the top of the panel. S2 cells were treated with the ligands, and RNA was isolated and analyzed by Northern blotting. The autoradiographs are shown. The results show that all of the antimicrobial peptide genes tested can be activated synergistically by the two ligands in the S2 cells. (C) Total RNA was isolated from S2 cells 6 h after stimulation. The RNA samples were analyzed by RT-PCR. The results are the average of three independent experiments, and standard deviations are shown as error bars.
Previous experiments show that S2 cells are more responsive to stimulation after treatment with ecdysone, the master regulatory hormone of metamorphosis (8). After ecdysone (20-HE) treatment, the cells became responsive to Spätzle by expressing detectable level of Drosomycin (lane 6) and to PGN(−) by expressing higher levels of Diptericin, AttacinA, and CecropinA (lane 7). These results recapitulated the specificity of the response to the two ligands in whole flies. More importantly, under the same condition, stimulation of the cells with the two ligands together induced even higher expression of all four genes (lane 8). Therefore, the cooperation of the two pathways also takes place when the cells are primed to respond after ecdysone treatment.
We then analyzed the dose response of S2 cells to this cooperative stimulation. In this series of experiments, no ecdysone was included. Spätzle was used up to 12 nM and PGN(−) was used up to 10 μg/ml; these ranges of concentration are similar to those used in previous reports (21, 26, 54). The two representative target genes of the Toll and IMD pathways, Drosomycin and Diptericin, were assayed (Fig. 1B). As expected, PGN(−) alone stimulated S2 cells to express a detectable level of Diptericin and, to a lesser extent, of Drosomycin (lanes 1 to 4). Meanwhile, Spätzle alone stimulated Drosomycin better than Diptericin (lanes 9 to 12). When different concentrations of Spätzle and PGN(−) were added together to the S2 cells, we observed a synergistic activation of both Drosomycin and Diptericin. At the highest concentrations used, the two target genes were expressed approximately 10-fold higher than the additive signal induced by the individual ligands (compare lane 8 to lanes 4 and 5). Therefore, there is cooperation between these two ligands at various concentrations to stimulate the innate immune response.
Antimicrobial peptide genes are induced with very different kinetics. Diptericin and Attacin mRNA reach peak levels in about 6 h, while Drosomycin mRNA reaches its peak level in about 24 h. The different kinetics may represent complex regulation, both positive and negative, by multiple pathways on target promoters. We tested whether the synergy also occurs at an earlier time. The same conditions were used as in Fig. 1A, and RNA samples isolated from the S2 cells were assayed by quantitative real-time PCR. As shown in Fig. 1C, synergistic activation of Drosomycin and AttacinA was detected at 6 h poststimulation. The expression of other genes was also assayed, and a lower degree of synergy was observed (data not shown).
Synergy is not mediated through direct stimulation of Toll by bacterial compounds.
A logical explanation of the results presented above is that Spätzle and PGN(−) activate the Toll and IMD pathways, respectively, and the two pathways cooperate to cause better activation of antimicrobial response. However, it is also possible that PGN(−) acts on Toll directly or that PGN(−) interacts with Spätzle and the complex binds to Toll. These situations will be similar to mammalian Toll-like receptors, which can bind to microbial compounds or to microbial compound/host protein complexes (14). Therefore, we examined further how the observed cooperation occurred.
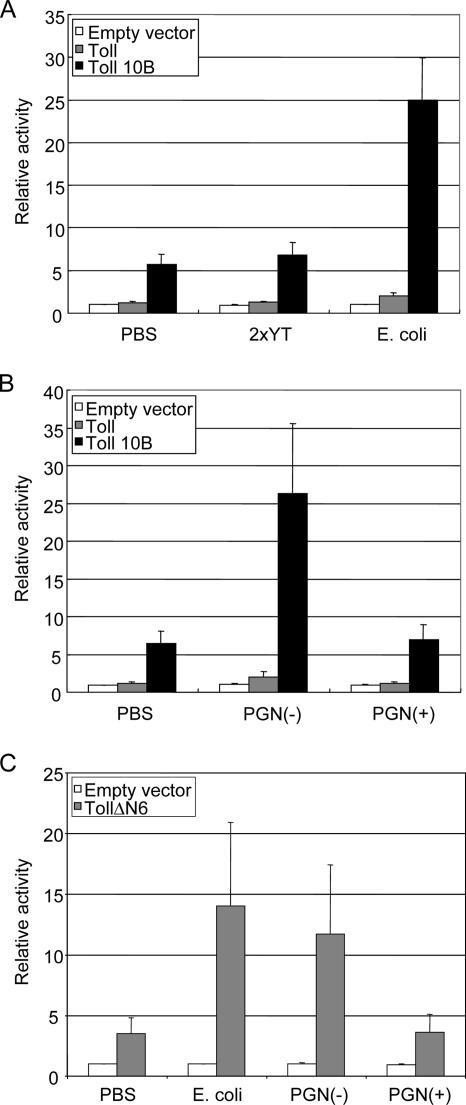
Toll10b is a Toll point mutant, and the mutation renders the receptor constitutively active in the absence of Spätzle (18). In these transfection experiments, we used an approximately 10-fold-smaller amount (50 ng) of the plasmids compared to that used (500 ng) in our previous report (34). This amount of Toll10b plasmid caused a fivefold increase in reporter expression, while Toll plasmid had no effect. The addition of the gram-negative bacterium E. coli further increased the response, from 6- to 25-fold, of Toll10b-transfected cells (Fig. 2A). More importantly, the bacteria did not stimulate this reporter in parental cells or Toll-transfected cells. These results are consistent with faster kinetics of gene induction in Toll10b mutant flies after septic injury (24). We further show that similar synergistic interaction with Toll10b was observed using PGN from gram-negative bacteria but not PGN from gram-positive bacteria (Fig. 2B). These results together demonstrate that PGN(−) synergistically stimulates the reporter gene when the Toll pathway is already partially activated. However, the Toll10b protein still contains the whole extracellular domain. Thus, we transfected another constitutively active Toll construct, TollΔN6, which contains only the transmembrane and intracellular domains (18). The TollΔN6-transfected cells were also responsive to E. coli and PGN(−) stimulation (Fig. 2C). This result further supports the idea that PGN(−) exerts the synergistic effect not by binding to the extracellular domain of Toll but by activating another pathway.
FIG. 2.
Constitutively active Toll synergizes with PGN(−) to stimulate Drosomycin. S2 cells were transiently transfected with the expression vector for wild-type Toll, gain-of-function mutant Toll10b, or gain-of-function deletion mutant TollΔN6, as indicated. All samples were cotransfected with the Drosomycin-luciferase reporter gene. The samples were then treated with gram-negative bacterium E. coli or with 2× YT growth medium as a control (A), and the luciferase activity in the extract was assayed. All of the samples were normalized with β-galactosidase activity from the cotransfected lacZ plasmid. The luciferase activity of the S2 cells transfected with empty vector and treated with PBS was set to 1; all other samples were plotted as relative activity to this control. The results were the average of three independent experiments, and the standard deviation is shown as error bars. In panel B, similar transfection experiments were performed using peptidoglycan from E. coli [PGN(−)] or S. aureus [PGN(+)], and the luciferase activities were analyzed. In panel C, similar experiments were performed, except the TollΔN6 construct was used. This construct has the whole extracellular domain deleted and is constitutively active as Toll10b. The results show that the constitutively active Toll pathway, but not the extracellular domain, can synergize with PGN(−) to stimulate Drosomycin.
Constitutive activation of both Toll and IMD pathways can cause synergy.
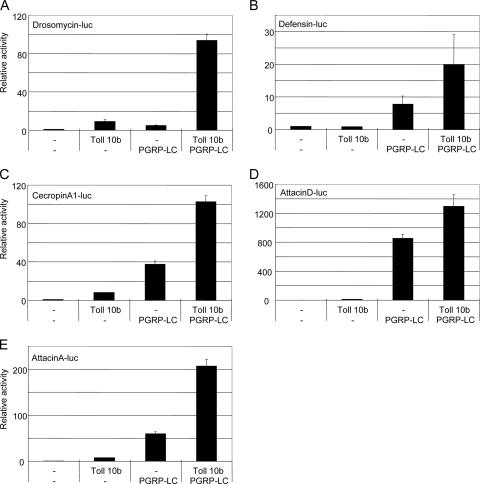
Previous reports have demonstrated that DAP-type PGNs from gram-negative bacteria and some gram-positive bacteria such as bacilli stimulate the IMD pathway. These DAP-type PGNs activate the IMD pathway by binding to PGRP-LC and -LE (4, 6, 12, 21, 26, 29, 44, 47, 48). Therefore, we speculated that the PGN(−) we used caused stimulation of the IMD pathway receptor to synergize with the activated Toll. Thus, we quantitatively analyzed the cooperative activation of target genes in S2 cells by transfecting plasmids encoding Toll10b and PGRP-LC. It has been demonstrated that overexpression of PGRP-LC or -LE is sufficient to activate the IMD pathway constitutively (12, 48). Under our transfection conditions, the Drosomycin-luciferase reporter was modestly activated by Toll10b or PGRP-LC. In contrast, the cotransfection of both Toll10b and PRGP-LC highly activated this reporter (Fig. 3A), demonstrating a synergistic effect of the two activated receptors. We similarly tested four other representative promoters, the Defensin, CecropinA1, AttacinA, and AttacinD gene promoters (Fig. 3B to E). The same amounts of Toll10b- and PGRP-LC-encoding plasmids (50 ng each) were used in the assay. Consistent with previous reports, CecropinA1, AttacinA, and AttacinD exhibited strong responses to PGRP-LC stimulation. The Defensin promoter had a relatively weak response. Nonetheless, the presence of both Toll10b and PGRP-LC always led to synergistic responses: that is, responses higher than the additive effect of the two receptors when transfected alone. Although the levels of synergy differ among target genes, the results clearly indicate that Toll and PGRP-LC synergistically activate the promoters of a battery of antimicrobial peptide genes.
FIG. 3.
Synergistic activation of antimicrobial peptide gene reporters by Toll10b and PGRP-LC. S2 cells were transiently transfected with the expression vector for Toll10b and/or PGRP-LC, along with the luciferase reporter gene as indicated in each panel (A to E). Analysis of luciferase activity, normalization of the results, and presentation of the graphs are the same as described in the legend to Fig. 2.
Synergistic interaction of Toll and IMD pathways in whole flies.
We used various means to examine whether the synergy also occurs in whole animals. Septic injury experiments using combinations of gram-positive (S. aureus and M. luteus) and gram-negative (E. coli, Enterbacter cloacae, and Erwinia carotovora) bacteria did not result in further increase of antimicrobial peptide gene expression (data not shown). We also injected various amounts of gram-positive and gram-negative peptidoglycans together, but again no further increase of gene expression was observed. We surmised that the complexity of whole bacteria used and the act of injury might have primed the animals so that the response was already optimized and thus no synergy could be detected.
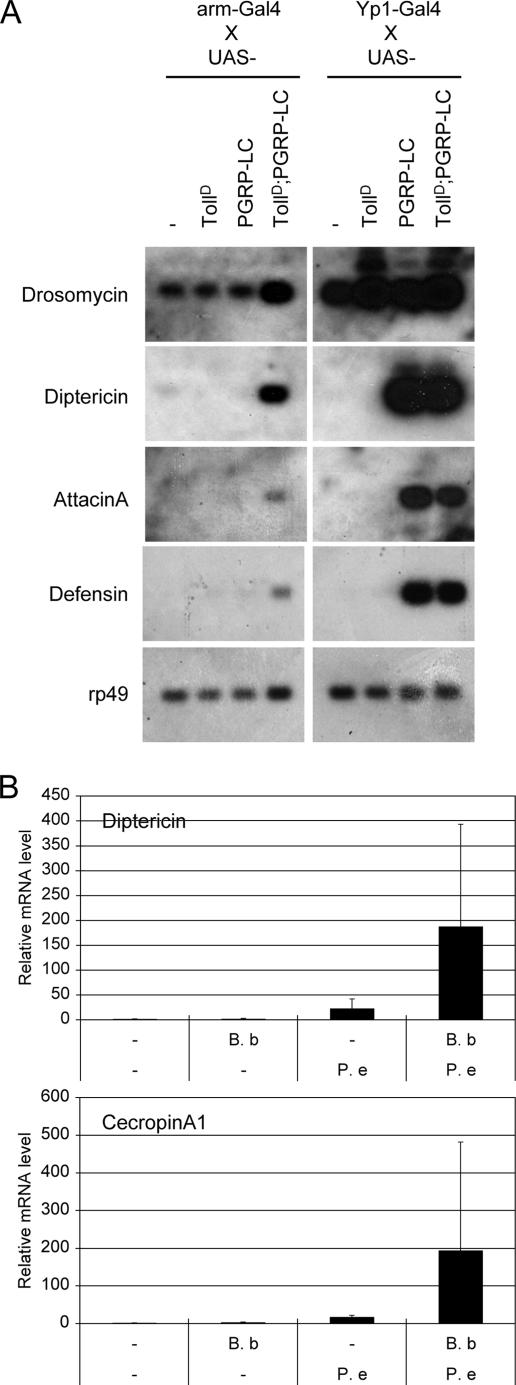
To better control the activation of the two pathways, we turned to the Gal4-UAS transgenic expression system. We crossed Gal4 strains with UAS-TollD (a mutant similar to Toll10b) (18) and UAS-PGRP-LC. A number of Gal4 lines we tested affected viability of the flies. The offspring from Yp1-, nanos-, Hsp70-, armadillo-, and 132-Gal4 crosses showed normal viability, while 127-, 362-, Actin5C-, and rhomboid-Gal4 caused lethality when both TollD and PRGP-LC were expressed (data not shown). Thus, we examined the expression of antimicrobial peptide genes using the Gal4 crosses that produced viable adults. The Yp1-Gal4 driver has been widely used to express transgenic constructs in adult female fat bodies (18, 50). The coexpression of TollD and PGRP-LC using this driver did not result in synergy (Fig. 4A, right panel). We surmised that the expression level driven by Yp1-Gal4 might be too high so that each of the pathways was already activated optimally, as supported by the robust expression of Drosomycin in TollD and Diptericin in PGRP-LC lines. Meanwhile, there was a significant increase in Drosomycin expression when both receptors were expressed under the control of arm-Gal4. This driver is supposed to cause a lower level of expression compared to Yp1-Gal4. The Diptericin expression also showed a clear increase, while modest synergy was observed for AttacinA and Defensin (Fig. 4A, left panel). As controls, the use of arm-Gal4 crosses with either TollD or PGRP-LC led to no stimulation of these antimicrobial target genes. Overall, the results are consistent with the idea that when both pathways are stimulated at lower levels, synergy can occur, but when stimulated at a higher level each pathway can induce the target genes optimally and independently.
FIG. 4.
Interaction of activated Toll and PGRP-LC in whole flies. (A) The gain-of-function mutant TollD (see Materials and Methods) and PGRP-LC were expressed in transgenic flies using the Gal4-UAS system. The Gal4 drivers used are indicated. Expression of Drosomycin, Diptericin, AttacinA, and Defensin in the transgenic adult females expressing TollD and PGRP-LC was examined. Northern blot analysis was carried out using RNA from the female flies that resulted from armadillo- or Yp1-Gal4 crosses with the indicated UAS lines. The autoradiographs are shown. There is an increase in gene expression when both TollD and PGRP-LC are expressed using the armadillo-Gal4 driver. (B) Double natural infection of adult flies with fungi and gram-negative bacteria. Adult female flies were soaked with fungal spores of Beauveria bassiana (B.b) and then fed with entomopathogenic gram-negative bacterium Pseudomonas entomophila (P.e) for 24 h. The gene expression was analyzed by quantitative RT-PCR. The results are the average of five independent experiments, and standard deviations are shown as error bars.
We then tested whether synergy can occur when both pathways are stimulated during immune response in whole animals. Previous reports show that natural infection by the fungus B. bassiana activates antimicrobial peptide gene expression through the Toll pathway (25). Meanwhile oral feeding of the gram-negative bacterium P. entomophila causes activation of antimicrobial response through the IMD pathway (53). We first titrated down the amount of microbes so that there is minimal expression of target genes after natural infection of either microbe. Then we combined the two infection protocols and after 24 h assayed for the expression of antimicrobial peptide genes. We performed this experiment five times, and the average result shows that there was detectable, albeit variable, synergistic activation of Diptericin and CecropinA (Fig. 4B). Other antimicrobial peptide genes assayed showed only marginal expression. The natural infection experiments are difficult to control, and we used a smaller amount of microbes, which thus may contribute to the highly variable results. Overall, the results suggest that at least under some circumstances there is synergistic activation of antimicrobial peptide genes through stimulation of Toll and IMD pathways during the innate immune response.
Synergy depends on NF-κB-related proteins.
To investigate the underlying mechanism that leads to the synergistic response, we first performed a transfection assay using plasmids encoding TollΔN6 and IMD. The expression of these two proteins in S2 cells also synergistically activated the Drosomycin, AttacinA, and CecropinA reporters (Fig. 5A to C). Because TollΔN6 contains no extracelluar domain and IMD is a cytoplasmic adaptor, the synergy observed between the Toll and IMD pathways should occur at an intracellular step.
Because of the varieties of intracellular signaling molecules involved in the two pathways, various mechanisms can be envisaged, such as cross regulation by a kinase, better formation of a common adaptor complex, or cooperation of transcription factors. Our previous report using stably transfected S2 cells showed that coexpression of DIF and Relish, as well as Dorsal and Relish, activated Drosomycin more efficiently (13). Dorsal and DIF act in the Toll pathway, and Relish acts in the IMD pathway. If interaction of NF-κB-related transcription factors mediates the synergy, a prediction will be that the two receptors stimulate the respective pathway independently down to the transcription factors. Then the transcription factors in both pathways will be critical for this response. If cross talk occurs upstream of the transcription factors, then only one of the two transcription factors will be involved. Thus, rather than testing the many signaling components in the two pathways, we used RNAi to directly knock down the expression of Dorsal, DIF, and Relish and examined whether the activation of the target gene was affected (Fig. 5D and E). We first showed that cotransfection of Toll10b and PGRP-LC can efficiently activate the expression of endogenous Drosomycin in the S2 cells (Fig. 5D, lanes 1 to 4). When the transfection experiment also included dsRNA for dorsal or relish, we observed that each caused almost total loss of expression of Drosomycin induced by the two pathways (Fig. 5D, lanes 5 and 9). To our surprise, we did not observe an involvement of DIF using the RNAi approach; the amount of dsRNA used was sufficient to knock down DIF expressed through transient transfection (data not shown). DIF is the key transcription factor in the Toll-mediated immune response in adult flies, while DIF and Dorsal have redundant functions in larval immune response (32, 39). In our S2 cells, the endogenous DIF may not be expressed or functional: thus, Dorsal becomes the key factor in Toll signaling.
Using the transiently transfected Drosomycin reporter as a more quantitative assay, we observed similar results. Transfected dorsal or relish dsRNA abolished the reporter activity induced by the two pathways, but Dif RNAi had no effect (Fig. 5E). All of the results demonstrate that Dorsal and Relish, downstream transcription factors of the Toll and IMD pathways, respectively, are critical for the synergistic activation in the S2 cells. Thus, the interaction of the two pathways likely occurs at the level of utilization of the NF-κB-related proteins in both pathways.
Synergistic activation through κB sites on a target promoter.
To gain further insight into how the transcription factors regulate the response, we examined the Drosomycin promoter in detail because it is a representative target and shows a good response to activation by both pathways. The activity of a series of deletion mutants (Fig. 6A and C) was analyzed by transient transfection assay. The deletion of upstream sequences down to 0.43 kb increased the overall activity compared to that of the parental 2.9-kb construct (Fig. 6C), suggesting the presence of negative regulatory elements between the 2.9- and 0.43-kb regions. Further deletion caused a steady decline of promoter activity. The biggest drop of activity was caused by deleting the region between the 0.43- and 0.14-kb regions, demonstrating that the Toll and IMD pathway response elements are mainly located within this 300-bp region upstream of the transcription start site.
Within the 0.43-kb 5′-flanking sequence, there are three identifiable κB sites (Fig. 6A). We made specific mutations of each of these three κB sites by changing the most conserved 5′-GGG sequence to ATT (Fig. 6A and B) and analyzed them in the context of the 0.43-kb promoter construct. Mutating the three sites individually caused different effects, with site 1 and site 2 having severe effects while site 3 had a minor effect (Fig. 6D). Moreover, mutating sites 1 and 2 together (mut12) or simultaneously mutating all three sites (mut123) abolished the activity. These results are consistent with the deletion analysis, which reveals that the region between 0.43 and 0.14 kb, where sites 1 and 2 are located, is responsible for most of the activity. Thus, κB sites 1 and 2 are critical elements that mediate the Toll and IMD stimulation, while site 3 has only an auxiliary role.
Careful analysis of the results reveals that the κB sites 1 and 2 serve different functions. Mut1 showed a clear reduction of the response to Toll10b stimulation, suggesting that DIF or Dorsal binds to this site after stimulation of the Toll pathway. The response to PGRP-LC, however, was not affected by mutating site 1. Moreover, the synergistic response of mut1 was still apparent, although the overall level was reduced. mut2, on the other hand, showed no response to PGRP-LC stimulation, while the response to Toll10b was partially reduced. Thus, site 2 is critical for the response to the IMD pathway and contributes to the response to the Toll pathway. Most importantly, without site 2, the synergistic activation of the promoter by the two pathways is much reduced. Thus, Relish likely binds to site 2 to mediate the IMD response. Overall the mutational analysis supports the idea that different NF-κB-related proteins bind to two κB sites on the Drosomycin promoter and provide the optimal condition for a synergistic response to the two pathways.
DISCUSSION
In this study, we have shown that cooperation of the Toll and IMD pathways can occur. One favorable condition is when both pathways are activated at lower levels. This condition perhaps resembles the invasion of microbes when flies suffer injury in the natural environment. The cooperation of the NF-κB proteins will be particularly helpful when both signaling pathways are stimulated by a low level of infection with multiple microbes, which is probably the most common scenario when injury occurs in nature. A hallmark of innate immunity is broad recognition and protection. Thus, our results reveal that the innate immune system can combine two different pathogen recognition signals to activate a wider spectrum of target genes by cooperation of signaling pathways.
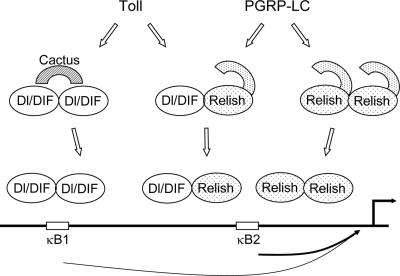
As shown in Fig. 7, we propose a model to illustrate how Dorsal, DIF, and Relish interact with each other and the κB sites to mediate the synergistic response to Toll and PGRP-LC. In our S2 cell experiments, Dorsal but not Dif is essential for Drosomycin activation. In adult flies, DIF but not Dorsal is critical (32, 39). In larvae, DIF and Dorsal have redundant functions. Thus, we have presented DIF and Dorsal as having equal functions in this model. Because the presence of both κB sites 1 and 2 gave the best overall response, binding of homodimers or heterodimers of the transcription factors to these two sites can account for the cooperative activation.
FIG. 7.
Possible synergistic mechanisms involving the three NF-κB-related proteins downstream of the Toll and IMD pathways. We have presented DIF and Dorsal (Dl) having equal functions in this model. The Toll pathway activates DIF and Dorsal, and the IMD pathway activates Relish (Rel). Our results so far do not indicate interaction of these two pathways upstream of the transcription factors. After signal stimulation, homodimers of the three factors can bind to site 1 and site 2 independently. After binding, Relish homodimer can interact with DIF or Dorsal homodimer to activate transcription synergistically. Alternatively, signal stimulation may enhance the formation or function of Dorsal/Relish and DIF/Relish heterodimers. Relish homodimer or heterodimer binds to site 2 preferentially. This binding to site 2 and interaction with site 1 provide the optimal condition for the synergistic response to the two pathways.
Site 1 matches the Dorsal binding consensus (GGGA/TA/TT/AA/T/CCT/G/C), while site 2 matches the Relish binding consensus (GGGAA/T/CNC/TC/AC/T) (41) (Fig. 6A). The DIF binding selection showed ambiguous sequence except for the first three G's; thus, DIF may bind to a broad array of κB sites (41). It is likely that Dorsal or DIF homodimers can bind to sites 1 and 2. Thus, mut1 and mut2 (Fig. 6D) both showed a reduced response to Toll. Meanwhile, Relish can only bind to site 2. Mutating site 2, therefore, abolished the response to PGRP-LC. For cooperation to occur, the receptors may activate the respective homodimers, which then bind to sites 1 and 2 independently. DIF or Dorsal homodimers on site 1 then cooperate with Relish homodimer on site 2 to activate the transcription complex efficiently.
Alternatively, site 2 can bind Relish heterodimers (Dorsal/Relish or DIF/Relish). DIF/Relish heterodimer has been shown to have higher binding specificity in vitro (GGGAA/TTCC/AC) (41), which is still similar to site 2. Using electrophoretic mobility shift assays, we previously showed that site 2 can bind Relish homodimer and heterodimers (13). Moreover, DIF and Relish, as well as Dorsal and Relish, in stably transfected S2 cells synergistically activate Drosomycin (13). In these extracts, heterodimer formation was detected at a low level. The binding of heterodimer to site 2 in vivo is further supported by the finding that mut2 had the most reduced synergy (Fig. 6D) and that mut13 still has some synergistic response. Therefore, it is possible that heterodimers of Relish on site 2 alone account for some response to both pathways and can cooperate with Dorsal or DIF on site 1. Finally, we want to emphasize that the utilizations of various dimers of the three NF-κB-related proteins can all happen in vivo and need not be mutually exclusive.
IMD pathway mutants have preferential loss of induction of genes including Diptericin. Toll pathway mutants on the other hand have preferential loss of induction of Drosomycin. While these genetic analyses clearly suggest that the two pathways can function independently, the mutant animals represent extreme cases such that one of the two pathways is totally inactivated. For example, in dorsal Dif double mutants, the Relish homodimer is the only combination that exists. In such a situation, the solo Relish binding site (site 2) on the Drosomycin promoter may not be sufficient for the activation. The Diptericin and Attacin promoters may have a sufficient number of binding sites for Relish so that the response to IMD pathway remains sufficient. In a relish mutant, DIF or Dorsal homodimers either cannot bind to the Diptericin promoter or cannot activate without the help of Relish. In wild-type flies, both pathways are working at the same time and all NF-κB dimer combinations are available. These dimer combinations may all interact with the target promoters. Promoters of antimicrobial peptide genes contain clusters of κB sites (41). The occupation of these κB sites may determine the overall expression levels and the responsiveness to the Toll and IMD pathways.
Acknowledgments
We thank J. Royet for the PGRP-LC transgenic flies and B. Lemaitre for bacterial and fungal strains.
This work was supported by a grant from NIH (GM53269). Core resources supported by Diabetes Endocrinology Research Center grant DK32520 were also used.
Footnotes
Published ahead of print on 16 April 2007.
REFERENCES
- 1.Bettencourt, R., H. Asha, C. Dearolf, and Y. T. Ip. 2004. Hemolymph-dependent and -independent responses in Drosophila immune tissue. J. Cell. Biochem. 92:849-863. [DOI] [PubMed] [Google Scholar]
- 2.Bischoff, V., C. Vignal, I. G. Boneca, T. Michel, J. A. Hoffmann, and J. Royet. 2004. Function of the drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat. Immunol. 5:1175-1180. [DOI] [PubMed] [Google Scholar]
- 3.Brennan, C. A., and K. V. Anderson. 2004. Drosophila: the genetics of innate immune recognition and response. Annu. Rev. Immunol. 22:457-483. [DOI] [PubMed] [Google Scholar]
- 4.Chang, C. I., K. Ihara, Y. Chelliah, D. Mengin-Lecreulx, S. Wakatsuki, and J. Deisenhofer. 2005. Structure of the ectodomain of Drosophila peptidoglycan-recognition protein LCa suggests a molecular mechanism for pattern recognition. Proc. Natl. Acad. Sci. USA 102:10279-10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choe, K. M., H. Lee, and K. V. Anderson. 2005. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc. Natl. Acad. Sci. USA 102:1122-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choe, K. M., T. Werner, S. Stoven, D. Hultmark, and K. V. Anderson. 2002. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296:359-362. [DOI] [PubMed] [Google Scholar]
- 7.De Gregorio, E., P. T. Spellman, P. Tzou, G. M. Rubin, and B. Lemaitre. 2002. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21:2568-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimarcq, J. L., J. L. Imler, R. Lanot, R. A. Ezekowitz, J. A. Hoffmann, C. A. Janeway, and M. Lagueux. 1997. Treatment of l(2)mbn Drosophila tumorous blood cells with the steroid hormone ecdysone amplifies the inducibility of antimicrobial peptide gene expression. Insect Biochem. Mol. Biol. 27:877-886. [DOI] [PubMed] [Google Scholar]
- 9.Filipe, S. R., A. Tomasz, and P. Ligoxygakis. 2005. Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep. 6:327-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgel, P., S. Naitza, C. Kappler, D. Ferrandon, D. Zachary, C. Swimmer, C. Kopczynski, G. Duyk, J. M. Reichhart, and J. A. Hoffmann. 2001. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev. Cell 1:503-514. [DOI] [PubMed] [Google Scholar]
- 11.Gobert, V., M. Gottar, A. A. Matskevich, S. Rutschmann, J. Royet, M. Belvin, J. A. Hoffmann, and D. Ferrandon. 2003. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science 302:2126-2130. [DOI] [PubMed] [Google Scholar]
- 12.Gottar, M., V. Gobert, T. Michel, M. Belvin, G. Duyk, J. A. Hoffmann, D. Ferrandon, and J. Royet. 2002. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416:640-644. [DOI] [PubMed] [Google Scholar]
- 13.Han, Z. S., and Y. T. Ip. 1999. Interaction and specificity of Rel-related proteins in regulating Drosophila immunity gene expression. J. Biol. Chem. 274:21355-21361. [DOI] [PubMed] [Google Scholar]
- 14.Hargreaves, D. C., and R. Medzhitov. 2005. Innate sensors of microbial infection. J. Clin. Immunol. 25:503-510. [DOI] [PubMed] [Google Scholar]
- 15.Hedengren, M., B. Asling, M. S. Dushay, I. Ando, S. Ekengren, M. Wihlborg, and D. Hultmark. 1999. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol. Cell 4:827-837. [DOI] [PubMed] [Google Scholar]
- 16.Hedengren-Olcott, M., M. C. Olcott, D. T. Mooney, S. Ekengren, B. L. Geller, and B. J. Taylor. 2004. Differential activation of the NF-kappaB-like factors Relish and Dif in Drosophila melanogaster by fungi and Gram-positive bacteria. J. Biol. Chem. 279:21121-21127. [DOI] [PubMed] [Google Scholar]
- 17.Horng, T., and R. Medzhitov. 2001. Drosophila MyD88 is an adapter in the Toll signaling pathway. Proc. Natl. Acad. Sci. USA 98:12654-12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu, X., Y. Yagi, T. Tanji, S. Zhou, and Y. T. Ip. 2004. Multimerization and interaction of Toll and Spatzle in Drosophila. Proc. Natl. Acad. Sci. USA 101:9369-9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hultmark, D. 2003. Drosophila immunity: paths and patterns. Curr. Opin. Immunol. 15:12-19. [DOI] [PubMed] [Google Scholar]
- 20.Jang, I. H., N. Chosa, S. H. Kim, H. J. Nam, B. Lemaitre, M. Ochiai, Z. Kambris, S. Brun, C. Hashimoto, M. Ashida, P. T. Brey, and W. J. Lee. 2006. A spatzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev. Cell 10:45-55. [DOI] [PubMed] [Google Scholar]
- 21.Kaneko, T., W. E. Goldman, P. Mellroth, H. Steiner, K. Fukase, S. Kusumoto, W. Harley, A. Fox, D. Golenbock, and N. Silverman. 2004. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20:637-649. [DOI] [PubMed] [Google Scholar]
- 22.Kim, T., J. Yoon, H. Cho, W. B. Lee, J. Kim, Y. H. Song, S. N. Kim, J. H. Yoon, J. Kim-Ha, and Y. J. Kim. 2005. Downregulation of lipopolysaccharide response in Drosophila by negative crosstalk between the AP1 and NF-kappaB signaling modules. Nat. Immunol. 6:211-218. [DOI] [PubMed] [Google Scholar]
- 23.Lemaitre, B. 2004. The road to Toll. Nat. Rev. Immunol. 4:521-527. [DOI] [PubMed] [Google Scholar]
- 24.Lemaitre, B., E. Nicolas, L. Michaut, J. M. Reichhart, and J. A. Hoffmann. 1996. The dorsoventral regulatory gene cassette spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86:973-983. [DOI] [PubMed] [Google Scholar]
- 25.Lemaitre, B., J. M. Reichhart, and J. A. Hoffmann. 1997. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 94:14614-14619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leulier, F., C. Parquet, S. Pili-Floury, J. H. Ryu, M. Caroff, W. J. Lee, D. Mengin-Lecreulx, and B. Lemaitre. 2003. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 4:478-484. [DOI] [PubMed] [Google Scholar]
- 27.Levashina, E. A., E. Langley, C. Green, D. Gubb, M. Ashburner, J. A. Hoffmann, and J. M. Reichhart. 1999. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science 285:1917-1919. [DOI] [PubMed] [Google Scholar]
- 28.Ligoxygakis, P., N. Pelte, J. A. Hoffmann, and J.-M. Reichhart. 2002. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science 297:114-116. [DOI] [PubMed] [Google Scholar]
- 29.Lim, J. H., M. S. Kim, H. E. Kim, T. Yano, Y. Oshima, K. Aggarwal, W. E. Goldman, N. Silverman, S. Kurata, and B. H. Oh. 2006. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem. 281:8286-8295. [Epub 20 January 2006.] [DOI] [PubMed] [Google Scholar]
- 30.Lu, Y., L. P. Wu, and K. V. Anderson. 2001. The antibacterial arm of the drosophila innate immune response requires an IkappaB kinase. Genes Dev. 15:104-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markus, R., E. Kurucz, F. Rus, and I. Ando. 2005. Sterile wounding is a minimal and sufficient trigger for a cellular immune response in Drosophila melanogaster. Immunol. Lett. 101:108-111. [DOI] [PubMed] [Google Scholar]
- 32.Meng, X., B. S. Khanuja, and Y. T. Ip. 1999. Toll receptor-mediated Drosophila immune response requires Dif, an NF-kappaB factor. Genes Dev. 13:792-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michel, T., J.-M. Reichhart, J. A. Hoffmann, and J. Royet. 2001. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414:756-759. [DOI] [PubMed] [Google Scholar]
- 34.Ooi, J. Y., Y. Yagi, X. Hu, and Y. T. Ip. 2002. The Drosophila Toll-9 activates a constitutive antimicrobial defense. EMBO Rep. 3:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park, J. M., H. Brady, M. G. Ruocco, H. Sun, D. Williams, S. J. Lee, T. Kato, Jr., N. Richards, K. Chan, F. Mercurio, M. Karin, and S. A. Wasserman. 2004. Targeting of TAK1 by the NF-kappa B protein Relish regulates the JNK-mediated immune response in Drosophila. Genes Dev. 18:584-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paterson, H. M., T. J. Murphy, E. J. Purcell, O. Shelley, S. J. Kriynovich, E. Lien, J. A. Mannick, and J. A. Lederer. 2003. Injury primes the innate immune system for enhanced Toll-like receptor reactivity. J. Immunol. 171:1473-1483. [DOI] [PubMed] [Google Scholar]
- 37.Pili-Floury, S., F. Leulier, K. Takahashi, K. Saigo, E. Samain, R. Ueda, and B. Lemaitre. 2004. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J. Biol. Chem. 279:12848-12853. [DOI] [PubMed] [Google Scholar]
- 38.Royet, J., J. M. Reichhart, and J. A. Hoffmann. 2005. Sensing and signaling during infection in Drosophila. Curr. Opin. Immunol. 17:11-17. [DOI] [PubMed] [Google Scholar]
- 39.Rutschmann, S., A. C. Jung, C. Hetru, J. M. Reichhart, J. A. Hoffmann, and D. Ferrandon. 2000. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity 12:569-580. [DOI] [PubMed] [Google Scholar]
- 40.Rutschmann, S., A. Kilinc, and D. Ferrandon. 2002. Cutting edge: the toll pathway is required for resistance to gram-positive bacterial infections in Drosophila. J. Immunol. 168:1542-1546. [DOI] [PubMed] [Google Scholar]
- 41.Senger, K., G. W. Armstrong, W. J. Rowell, J. M. Kwan, M. Markstein, and M. Levine. 2004. Immunity regulatory DNAs share common organizational features in Drosophila. Mol. Cell 13:19-32. [DOI] [PubMed] [Google Scholar]
- 42.Silverman, N., R. Zhou, S. Stoven, N. Pandey, D. Hultmark, and T. Maniatis. 2000. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 14:2461-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sluss, H. K., Z. Han, T. Barrett, R. J. Davis, and Y. T. Ip. 1996. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila. Genes Dev. 10:2745-2758. [DOI] [PubMed] [Google Scholar]
- 44.Stenbak, C. R., J. H. Ryu, F. Leulier, S. Pili-Floury, C. Parquet, M. Herve, C. Chaput, I. G. Boneca, W. J. Lee, B. Lemaitre, and D. Mengin-Lecreulx. 2004. Peptidoglycan molecular requirements allowing detection by the Drosophila immune deficiency pathway. J. Immunol. 173:7339-7348. [DOI] [PubMed] [Google Scholar]
- 45.Stoven, S., N. Silverman, A. Junell, M. Hedengren-Olcott, D. Erturk, Y. Engstrom, T. Maniatis, and D. Hultmark. 2003. Caspase-mediated processing of the Drosophila NF-kappa B factor Relish. Proc. Natl. Acad. Sci. USA 100:5991-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun, H., P. Towb, D. N. Chiem, B. A. Foster, and S. A. Wasserman. 2004. Regulated assembly of the Toll signaling complex drives Drosophila dorsoventral patterning. EMBO J. 23:100-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swaminathan, C. P., P. H. Brown, A. Roychowdhury, Q. Wang, R. Guan, N. Silverman, W. E. Goldman, G. J. Boons, and R. A. Mariuzza. 2006. Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs). Proc. Natl. Acad. Sci. USA 103:684-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takehana, A., T. Yano, S. Mita, A. Kotani, Y. Oshima, and S. Kurata. 2004. Peptidoglycan recognition protein (PGRP)-LE and PGRP-LC act synergistically in Drosophila immunity. EMBO J. 23:4690-4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanji, T., and Y. T. Ip. 2005. Regulators of the Toll and Imd pathways in the Drosophila innate immune response. Trends Immunol. 26:193-198. [DOI] [PubMed] [Google Scholar]
- 50.Tauzig-Delamasure, S., H. Bilak, M. Capovilla, J. A. Hoffmann, and J.-L. Imler. 2002. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat. Immunol. 3:91-97. [DOI] [PubMed] [Google Scholar]
- 51.Tzou, P., J. M. Reichhart, and B. Lemaitre. 2002. Constitutive expression of a single antimicrobial peptide can restore wild-type resistance to infection in immunodeficient Drosophila mutants. Proc. Natl. Acad. Sci. USA 99:2152-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vidal, S., R. S. Khush, F. Leulier, P. Tzou, M. Nakamura, and B. Lemaitre. 2001. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-kappaB-dependent innate immune responses. Genes Dev. 15:1900-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vodovar, N., M. Vinals, P. Liehl, A. Basset, J. Degrouard, P. Spellman, F. Boccard, and B. Lemaitre. 2005. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc. Natl. Acad. Sci. USA 102:11414-11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber, A. N., S. Tauszig-Delamasure, J. A. Hoffmann, E. Lelievre, H. Gascan, K. P. Ray, M. A. Morse, J. L. Imler, and N. J. Gay. 2003. Binding of the Drosophila cytokine Spatzle to Toll is direct and establishes signaling. Nat. Immunol. 4:794-800. [DOI] [PubMed] [Google Scholar]