Abstract
Human cytomegalovirus (HCMV) can bind, fuse, and initiate gene expression in a diverse range of vertebrate cell types. This broad cellular tropism suggests that multiple receptors and/or universally distributed receptors mediate HCMV entry. Our laboratory has recently discovered that certain β1 and β3 integrin heterodimers are critical mediators of HCMV entry into permissive fibroblasts (A. L. Feire, H. Koss, and T. Compton, Proc. Natl. Acad. Sci. USA 101:15470-15475, 2004). It has also been reported that epidermal growth factor receptor (EGFR) is necessary for HCMV-mediated signaling and entry (X. Wang, S. M. Huong, M. L. Chiu, N. Raab-Traub, and E. E. Huang, Nature 424:456-461, 2003). Integrins are known to signal synergistically with growth factor receptors, and this coordination was recently reported for EGFR and β3 integrins in the context of HCMV entry (X. Wang, D. Y. Huang, S. M. Huong, and E. S. Huang, Nat. Med. 11:515-521, 2005). However, EGFR-negative cell lines, such as hematopoietic cells, are known to be infected by HCMV. Therefore, we wished to confirm a role for EGFR in HCMV entry and then examine any interaction between β1 integrins and EGFR during the entry process. Surprisingly, we were unable to detect any role for EGFR in the process of HCMV entry into fibroblast, epithelial, or endothelial cell lines. Additionally, HCMV did not activate the EGFR kinase in fibroblast cell lines. We first examined HCMV entry into two EGFR-positive or -negative cell lines but observed no increase in entry when EGFR was expressed to high levels. Physically blocking EGFR with a neutralizing antibody in fibroblast, epithelial, or endothelial cell lines or blocking EGFR kinase signaling with a chemical inhibitor in fibroblast cells did not inhibit virus entry. Lastly, we were unable to detect phosphorylation of EGFR in fibroblasts cells in response to HCMV stimulation. Our findings demonstrate that EGFR does not play a significant role in HCMV entry or signaling. These results suggest that specific integrin heterodimers either act alone as the primary entry receptors or interact in conjunction with an additional receptor(s), other than EGFR, to facilitate virus entry.
Human cytomegalovirus (HCMV), a member of the Herpesviridae family of viruses, is an opportunistic pathogen that causes significant disease in immunocompromised individuals, including AIDS patients, organ transplant recipients, and neonates (2, 21). Like all herpesviruses, HCMV maintains a lifelong infection of its host, and persistent infection is associated with proliferative diseases such as atherosclerosis (1, 11, 18) and gastrointestinal cancer (10, 23). HCMV has broad tissue tropism and can infect a variety of vertebrate cell types, including fibroblasts, monocytes/macrophages, smooth muscle cells, vascular endothelial cells, astrocytes, epithelial cells, trophoblasts, stromal cells, and hepatocytes (2, 19, 21). Consequently, HCMV can manifest disease in almost every organ system and tissue type (2, 21).
In vitro, HCMV entry is promiscuous as it can bind, penetrate, and initiate infection in nearly all of the vertebrate cell lines tested (20). HCMV initially tethers itself to cell surface heparan sulfate proteoglycans (HSPGs) mediated via two viral envelope glycoproteins, gM and gB (5, 14). HSPGs are ubiquitously expressed and absolutely required for HCMV entry; however, HSPGs alone are not sufficient to mediate HCMV entry (5). Virion interaction with HSPGs is easily dissociable with soluble heparin, but the virus quickly transitions to a second, more stable interaction with one or more different receptors that ultimately leads to pH-neutral fusion of the virion, presumably at the cell surface (5). The ability of HCMV to enter a wide variety of cell types suggests that HCMV utilizes multiple receptors and/or a widely distributed cell surface receptor for entry.
Recently, cellular integrins (α2β1, α6β1, and αvβ3) were identified as entry receptors for HCMV that may constitute the second, more stable attachment receptor (7, 28). Neutralizing antibodies to the α2, α6, αV, β1, and β3 integrin subunits inhibit HCMV entry in a dose-dependent manner without blocking virion attachment (7). Additionally, β1 integrin-null fibroblasts showed markedly reduced HCMV entry compared to the same cell line with restored β1 expression (7). It was also discovered that HCMV gB contains an integrin recognition motif, the disintegrin-like domain, which is highly conserved throughout the beta and gamma herpesviruses (7). Peptides encompassing this disintegrin-like domain of gB are able to specifically block HCMV entry, supporting a role for this region in the entry process (7). HCMV also induces the phosphorylation and activation of β1 (7) and β3 (28) integrins and focal adhesion kinase and causes distinct morphological changes in the cell due to actin rearrangements (7, 28).
Epidermal growth factor (EGF) receptor (EGFR) was also recently implicated as a receptor for HCMV (28, 29). Many of the downstream consequences of EGFR activation are consistent with HCMV infection, including activation of mitogen-activated protein kinases through Ras and Raf, the phosphatidylinositol-3-OH kinase-Akt pathway, and phospholipase C-γ, which leads to intracellular mobilization of Ca2+, as well as activation of protein kinase C (29). Furthermore, Wang et al. report that HCMV entry correlated with EGFR expression and was blocked by an EGFR neutralizing antibody (29). Although EGFR is a broadly expressed cellular receptor, it is not found on all HCMV-permissive cell types, including hematopoietic cells, such as monocytes/macrophages, dendritic cells, and neutrophils (22). It is possible that other cellular receptors, including other growth factor receptors, may substitute for EGFR on these cell types, although this has not been demonstrated. The primary goal of this study was to confirm a role for EGFR in HCMV entry, specifically into permissive fibroblast cells, as well as epithelial and endothelial cell lines. A secondary goal of this study was to determine if β1 integrins and EGFR are able to coordinately mediate early HCMV entry events. Surprisingly, we could find no role for EGFR in HCMV entry or signaling pathways.
High levels of EGFR expression by a human breast cancer cell line failed to enhance HCMV infection compared to an EGFR-negative human breast cancer cell line, indicating that EGFR alone is not able to mediate HCMV entry. Physically blocking EGFR on fibroblast, epithelial, or endothelial cells with a neutralizing antibody failed to inhibit HCMV infection to any degree, suggesting that EGFR is not needed in a structural capacity to mediate HCMV entry. Additionally, blocking EGFR kinase activation in fibroblasts with a chemical inhibitor failed to block HCMV entry, suggesting that HCMV also does not require signaling through the EGFR kinase. Last, we were unable to detect EGFR phosphorylation in fibroblasts upon stimulation with HCMV, even at high multiplicities of infection (MOIs), indicating that the EGFR kinase, generally a good indicator of EGFR activation, does not become stimulated by HCMV. Together, these data indicate that EGFR is not required for HCMV infection, in either a structural or a signaling role; however, this does not rule out the existence of another receptor(s), in addition to cellular integrins, in the HCMV entry process.
MATERIALS AND METHODS
Cell lines and viruses.
Normal human dermal fibroblasts (NHDFs; Clonetics, San Diego, CA) and human breast cancer cells, MDA-MB-453 (MB453; HTB-131) and MDA-MB-468 (MB468; HTB-132), obtained from the American Type Culture Collection (ATCC; Manassas, VA) were grown at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; Omega Scientific, Inc., Tarzana, CA), 1% l-glutamine (Invitrogen), and 1% penicillin-streptomycin-amphotericin B (pen-strep-ampho B; BioWhittaker, Walkersville, MD). Human embryonic lung (HEL) fibroblasts (ATCC; CCL-137) were grown at 37°C in 5% CO2 in minimum essential alpha medium (Invitrogen) supplemented with 10% FBS, 1% l-glutamine, 1% pen-strep-ampho B, 1 mM sodium pyruvate (Invitrogen), 0.1 mM nonessential amino acids (Invitrogen), and 1.5 g/liter sodium bicarbonate (Invitrogen). Spontaneously arising retinal pigment epithelial (ARPE-19; CRL-2302) cells were purchased from ATCC and grown at 37°C in 5% CO2 in DMEM-F12 (1:1) (BioWhittaker) medium supplemented with 10% FBS, 1% l-glutamine, and 1% pen-strep. Human umbilical vein endothelial cells (HUVEC) and myometrial uterine microvascular endothelial cells (UtMVEC) were purchased from Clonetics and grown at 37°C in 5% CO2 in endothelial cell basal medium (Clonetics) supplemented with 3 mg/ml bovine brain extract, 10 ng/ml human EGF, 1 μg/ml hydrocortisone, 10% FBS, and 1% gentamicin sulfate and ampho B (GA-1000). HCMV strain AD169 was propagated in NHDFs as previously described (4), and HCMV strain Towne was propagated in HEL fibroblasts. Both were cushioned over 20% sorbitol, resuspended in Tris-buffered saline (TBS), repelleted, and resuspended in TBS, and titers were determined by immediate-early (IE1-72 and IE2-86) gene expression detection via indirect immunofluorescence as previously described (3). HCMV AD169-GFP (green fluorescent protein) indicator virus encoding an IE2-GFP fusion product was kindly provided by Deborah H. Spector (University of California, San Diego) (24) and propagated in NHDF cells (4). Clinical strain VR1814 was a generous gift from Giuseppe Gerna (IRCCS Policlinico San Matteo) (9) and was propagated in HUVEC, pelleted through a 20% sorbitol cushion, and resuspended in TBS. Strain AD169 repaired at the UL131 locus, BADrUL131, was a kind gift from Thomas Shenk (Princeton University) (27) and was propagated in ARPE-19 cells, pelleted through a 20% sorbitol cushion, and resuspended in TBS.
Reagents and antibodies.
Recombinant human EGF was purchased from Invitrogen. The EGFR kinase inhibitor tyrphostin AG1478 was purchased from Calbiochem (La Jolla, CA). Heparin was purchased from Sigma-Aldrich (St. Louis, MO). Restore Western blot stripping buffer was purchased from Pierce Biotechnology (Rockford, IL) and used according to the manufacturer's instructions. Monoclonal antibody 1203, which recognizes the immediate-early (IE1-72 and IE2-86) gene products of HCMV, was purchased from the Rumbaugh-Goodwin Institute for Cancer Research, Inc. (Plantation, FL). A rabbit polyclonal antibody that recognizes the immediate-early (IE1-72 and IE2-86) gene products was kindly provided by Jay A. Nelson (Oregon Health Sciences University) (16). Anti-EGFR (1005) and phosphospecific anti-EGFR (Tyr-1173) polyclonal antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti-EGFR neutralizing monoclonal antibody and P-Tyr-conjugated agarose were purchased from Upstate Biotechnology (Lake Placid, NY). Alexa Fluor 594 goat anti-mouse secondary antibody was purchased from Molecular Probes (Eugene, OR). A goat anti-rabbit horseradish peroxidase-conjugated secondary antibody was purchased from Pierce Biotechnology.
SDS-PAGE and Western immunoblotting.
Cells were harvested in phosphate-buffered saline (PBS) containing 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS; Sigma; St. Louis, MO) for EGFR and IE gene product detection or in RIPA cell lysis buffer (50 mM Tris, 150 mM NaCl, 1 mM Na3VO4, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1% Triton X-100, 0.25% deoxycholate) for phospho-EGFR (P-EGFR) detection. Protein concentration was determined with a Bio-Rad (Hercules, CA) protein assay kit with bovine immunoglobulin G as the standard. Equivalent amounts of total cellular protein were boiled in sodium dodecyl sulfate (SDS) gel loading buffer and subjected to SDS-8% polyacrylamide gel electrophoresis (PAGE), followed by Western immunoblotting with anti-EGFR (1005), anti-IE, or anti-P-EGFR (Tyr-1173).
Time course of HCMV infection.
Subconfluent NHDF, MB468, and MB453 cells were grown in six-well plates and infected at an MOI of 1 PFU/cell at 37°C for 2 h. Virus that had not penetrated cells was inactivated with low-pH citrate buffer (40 mM citric acid, 10 mM KCl, 135 mM NaCl [pH 3.0]) (12). The cells were incubated for 8, 24, 48, or 72 h postinfection at 37°C in DMEM supplemented with 2% FBS, 1% glutamine, and 1% pen-strep-ampho B. Cells were harvested, subjected to SDS-PAGE, and then subjected to Western immunoblotting for IE gene expression as described above.
Virus entry assays.
Subconfluent NHDF or HEL cells were grown in 12-well plates for AD169-GFP infection and on glass coverslips in 12-well plates for Towne infection. Cells were washed twice with PBS and then treated with 0, 0.1, 1.0, or 10 μg/ml EGFR neutralizing antibody or 0, 5, 50, or 500 nM AG1478 diluted in serum-free DMEM at 37°C for 30 min. Medium was aspirated from the cells, HCMV (AD169-GFP or Towne) was added to the cells at an MOI of 0.5 PFU/cell, and the mixture was incubated at 37°C for 2 h. For the EGFR kinase inhibitor entry assay, AG1478 was also added to the virus at the same concentrations used in the pretreatment. Virus that had not penetrated cells was inactivated with low-pH citrate buffer. The cells were incubated for 24 h in DMEM supplemented with 2% FBS, 1% glutamine, and 1% pen-strep-ampho B. Infected cells were detected via flow cytometry for IE2-GFP (AD169) or indirect immunofluorescence assay for IE gene expression (Towne). For detection of GFP by flow cytometry, cells were released with trypsin, DMEM supplemented with 10% FBS was added to inactivate the trypsin, and the cells were then pelleted and resuspended in PBS. Propidium iodide (Molecular Probes) was added, and samples were analyzed on a FACScan flow cytometer (Becton Dickinson, Mountain View, CA) with a standard filter set. Samples were gated for propidium iodide exclusion (live cells) and assayed for GFP expression. The data were analyzed with FlowJo (version 6.1; Tree Star, Inc., Ashland, OR). Indirect immunofluorescence analysis was performed as previously described (5), with mouse anti-IE monoclonal antibody 1203, followed by detection with Alexa Fluor 594 goat anti-mouse secondary antibody. Experiments were performed in duplicate with a minimum of 500 cells scored per coverslip.
Positive controls were performed by incubating NHDF or HEL cells with the EGFR neutralizing antibody or AG1478 for 30 min, washing them twice in PBS, and then stimulating them with 100 ng/ml EGF for 10 min. Cells were harvested and analyzed by SDS-PAGE and Western immunoblotting for P-EGFR (Tyr-1173) as described above.
For the VR1814 entry assay, EGFR neutralizing antibody was incubated with ARPE-19 cells, UtMVEC, or HUVEC at 0 or 10 μg/ml for 1 h at 37°C. For the BADrUL131 entry assay, EGFR neutralizing antibody was incubated with ARPE-19 cells, UtMVEC, or HUVEC at 0, 0.01, 0.1, 1.0, or 10 μg/ml for 1 h at 37°C. Antibody was removed, and cells were infected with VR1814 or BADrUL131 at an MOI of 1.0 PFU/ml for 4 or 2 h, respectively. BADrUL131 was also incubated with soluble heparin prior to addition to cells. Cells were washed three times in complete medium and incubated for 24 h at 37°C. Cells infected with VR1814 were then fixed in 4% paraformaldehyde, and immunofluorescence analysis was performed to visualize IE antigens. Percent GFP in cells infected with BADrUL131 was determined by flow cytometry.
EGFR activation assay.
Subconfluent NHDF or HEL cells were grown in six-well plates, washed twice in PBS, serum starved for 24 h, and then stimulated with HCMV (AD169 or Towne) at an MOI of 5.0 PFU/ml for 0, 5, 10, 30, or 60 min. Cells were washed with PBS, harvested in RIPA buffer, and analyzed by SDS-PAGE and Western immunoblotting for P-EGFR (Tyr-1173) as described above.
Immunoprecipitation assay.
Subconfluent NHDF or HEL cells were grown in 100-mm plates, washed twice in PBS, serum starved for 24 h, and then stimulated with HCMV (AD169 or Towne) at an MOI of 10 PFU/ml for 10 min. Cells were washed with PBS, lysed in RIPA buffer, and centrifuged at 13,000 rpm at 4°C for 10 min. Cell supernatant was incubated with 40 μl of P-Tyr antibody conjugated to agarose at 4°C while rotating overnight. Samples were washed three times in RIPA buffer and then three times in RIPA buffer without detergent and analyzed by SDS-PAGE and Western immunoblotting for EGFR (1005) as described above.
RESULTS
EGFR expression does not correlate with HCMV infection.
It was previously reported that an EGFR-negative human breast cancer cell line (MB453) was resistant to HCMV infection, whereas an EGFR-positive human breast cancer cell line (MB468) was permissive for viral gene expression (29). We initially performed immunoblot analysis to confirm EGFR expression levels in these cell lines. As reported in Fig. 1A, MB468 cells greatly overexpress EGFR. MB453 cells also appear to express some EGFR, although at significantly reduced levels compared to MB468 cells. HCMV-permissive NHDF cells also express EGFR. When HCMV viral gene expression was evaluated in the variably EGFR-expressing cell lines, we were unable to detect major IE proteins at 8, 24, 48, or 72 h postinfection with HCMV strain AD169 (Fig. 1B). Similar results were obtained with the Towne strain of HCMV at 72 h postinfection (Fig. 1C); however, in both cases infection of NHDF cells resulted in high levels of IE1 gene expression (Fig. 1B and C). These results were confirmed by flow cytometric analysis of MB468 and MB453 cells infected with an HCMV IE2-GFP reporter in which no GFP was present in either cell line at 24 h postinfection (data not shown).
FIG. 1.
EGFR expression and HCMV infection in EGFR-positive and EGFR-negative cell lines. (A) Immunoblot detection of EGFR expression in NHDF and mammary breast cancer cell lines MB468 (EGFR positive) and MB453 (EGFR negative). (B) HCMV AD169 was unable to infect EGFR-negative or -positive mammary breast cancer cell lines as detected by immunoblotting. The IE1 viral protein was detected in NHDF, MB468, and MB453 cells infected with HCMV strain AD169 at an MOI of 10 PFU/ml. (C) HCMV Towne was unable to infect EGFR-negative or -positive mammary breast cancer cell lines as detected by immunoblotting. The IE1 viral protein was detected in NHDF, MB468, and MB453 cells infected with HCMV strain Towne at an MOI of 5.0 PFU/ml. h.p.i., hours postinfection. M, mock infection.
An EGFR neutralizing antibody does not inhibit HCMV entry into fibroblasts.
EGF binds EGFR in a specific ligand-binding pocket, leading to receptor dimerization and activation of EGFR (25). Activation can be inhibited by a neutralizing antibody, which binds the extracellular domain of EGFR and blocks EGF access to the ligand-binding pocket to prevent EGFR activation; importantly, the antibody itself does not activate the intrinsic tyrosine kinase of EGFR (13). It was reported by Wang et al. that an EGFR neutralizing antibody (Upstate Biotechnology) at 1 μg/ml inhibited HCMV attachment by 65% and infection by 97% (28). Therefore, we hypothesized that this same monoclonal antibody would inhibit HCMV (AD169 and Towne strains) entry into fibroblasts. However, as reported in Fig. 2A and B, pretreatment of NHDF or HEL cells with the EGFR neutralizing antibody, even at concentrations of up to 10 μg/ml, failed to inhibit HCMV infection as measured by flow cytometric detection of IE2-GFP gene expression at 24 h postinfection. The neutralizing antibody did, however, inhibit EGF-mediated EGFR phosphorylation (Tyr-1173) at concentrations as low as 0.1 μg/ml (Fig. 2C).
FIG. 2.
HCMV infection in EGFR neutralizing antibody-treated fibroblast cells. (A) Increasing concentrations of an EGFR neutralizing antibody were unable to inhibit HCMV AD169 entry into fibroblast cells. IE2-GFP gene expression was detected by flow cytometry in NHDF and HEL fibroblasts treated with EGFR neutralizing antibody, followed by infection with HCMV AD169-GFP at an MOI of 0.5 PFU/ml. (B) Increasing concentrations of an EGFR neutralizing antibody were unable to inhibit HCMV Towne entry into fibroblast cells. IE1 and IE2 gene expression was detected by immunofluorescence in NHDF and HEL cells treated with an EGFR neutralizing antibody, followed by infection with HCMV Towne at an MOI of 0.5 PFU/ml. (C) Positive control verifying that EGFR phosphorylation is inhibited in fibroblast cells treated with an EGFR neutralizing antibody (Ab.). NHDF and HEL cells were treated with EGFR neutralizing antibody, followed by stimulation with EGF. P-EGFR (1173) was detected by immunoblotting. Blot was stripped and reprobed for total EGFR.
AG1478, an EGFR kinase inhibitor, does not inhibit HCMV entry into fibroblasts.
EGFR stimulation results in the activation of its intrinsic tyrosine kinase, which subsequently autophosphorylates the receptor and leads to many downstream signaling events (30). A potent EGFR-specific kinase inhibitor, tyrphostin AG1478 (50% inhibitory concentration = 3 nM), efficiently blocks EGFR tyrosine kinase activation (8). It was previously reported that AG1478 completely inhibits HCMV (strain Towne) gene expression in MB468 cells (29). Therefore, we hypothesized that AG1478 would be able to inhibit HCMV IE gene expression in human fibroblasts. However, even at high concentrations of AG1478 (500 nM), no inhibition of HCMV (AD169 and Towne strains) IE2-GFP gene expression, as measured by flow cytometry 24 h postinfection, was observed in NHDF or HEL fibroblast cells (Fig. 3A and B). In a parallel experiment, EGF-mediated EGFR phosphorylation (Tyr-1173) was inhibited by AG1478 treatment (Fig. 3C).
FIG. 3.
HCMV infection of AG1478-treated fibroblast cells. (A) Increasing concentrations of AG1478 were unable to inhibit HCMV AD169 entry into fibroblast cells. IE2-GFP gene expression was detected by flow cytometry in NHDF and HEL cells treated with AG1478, followed by infection with HCMV AD169-GFP at an MOI of 0.5 PFU/ml. (B) Increasing concentrations of AG1478 were unable to inhibit HCMV Towne entry into fibroblast cells. IE1 and IE2 gene expression was detected by immunofluorescence in NHDF and HEL cells treated with AG1478, followed by infection with HCMV Towne at an MOI of 0.5 PFU/ml. (C) Positive control verifying that EGFR phosphorylation is inhibited in fibroblast cells treated with AG1478. NHDF and HEL cells were treated with increasing concentrations of AG1478, followed by stimulation with EGF. P-EGFR (1173) was detected by immunoblotting. The blot was stripped and reprobed for total EGFR.
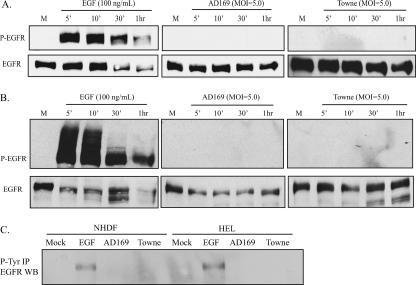
HCMV does not induce EGFR phosphorylation in fibroblasts.
EGFR is rapidly autophosyphorylated at specific tyrosine residues upon activation (30), and it has been reported that EGFR becomes phosphorylated within minutes of infection with HCMV (strain Towne) (29). We hypothesized that HCMV would be able to stimulate EGFR phosphorylation in fibroblast cells. To test this hypothesis, NHDF and HEL cells were stimulated with HCMV (AD169 and Towne strains) at various time intervals between 5 min and 1 h. As demonstrated in Fig. 4A and B, neither HCMV strain was able to stimulate EGFR phosphorylation (Tyr-1173) in NHDF or HEL cells at an MOI of 5 PFU/ml, although EGF (100 ng/ml) activates EGFR in these cell lines within minutes of stimulation. In addition, HCMV strain AD169 at an MOI of 10 PFU/ml or HCMV strain Towne at an MOI of 10 or 100 PFU/ml was not able to stimulate EGFR phosphorylation (Tyr-1173) at 10 min poststimulation (data not shown).
FIG. 4.
EGF and HCMV stimulation of fibroblast cells. (A) HCMV is unable to stimulate EGFR phosphorylation in NHDF cells. NHDF cells were stimulated with EGF, HCMV AD169, or HCMV Towne. P-EGFR (Tyr-1173) was detected by immunoblotting. The blot was stripped and reprobed for EGFR. (B) HCMV is unable to stimulate EGFR phosphorylation in HEL cells. HEL cells were stimulated with EGF, HCMV AD169, or HCMV Towne. P-EGFR (Tyr-1173) was detected by immunoblotting. The blot was stripped and reprobed for EGFR. (C) HCMV is unable to stimulate EGFR phosphorylation in NHDF or HEL cells. Cells were stimulated with EGF, HCMV AD169, or HCMV Towne. All phospho-tyrosine proteins were immunoprecipitated (IP), followed by detection of EGFR by Western blotting (WB). M, mock infection.
NHDF or HEL fibroblasts were also stimulated at an MOI of 10 PFU/ml for 10 min with HCMV strain AD169 or Towne, followed by immunoprecipitation of all phospho-tyrosine proteins and EGFR detection by Western blotting. As demonstrated in Fig. 4C, EGFR does not become phosphorylated on any tyrosine residues following stimulation with HCMV. Additionally, EGFR was immunoprecipitated, followed by detection of all phosphorylated tyrosine residues by Western blotting, again indicating that no such phosphorylation occurs following HCMV stimulation (data not shown).
An EGFR neutralizing antibody does not inhibit VR1814 and BADrUL131 strain entry into epithelial and endothelial cell lines.
To test if differences between laboratory-adapted and clinical strains of HCMV would allow the utilization of EGFR in the entry process, ARPE-19 cells, UtMVEC, and HUVEC were incubated with an EGFR neutralizing antibody, followed by infection with clinical endotropic VR1814 strain or BADrUL131, an AD169 strain with the UL131 locus repaired, conferring the ability to enter epithelial or endothelial cells (27). As reported in Fig. 5A, EGFR neutralizing antibody at 0 and 10 μg/ml did not inhibit VR1814 virus entry into the various cell lines. Additionally, BADrUL131 entry was also not inhibited following pretreatment with EGFR neutralizing antibody at concentrations up to 10 μg/ml but was inhibited following pretreatment with soluble heparin (Fig. 5B).
FIG. 5.
HCMV VR1418 and BADrUL131 infection in EGFR neutralizing antibody-treated epithelial and endothelial cells. (A) An EGFR neutralizing antibody (Ab) at a concentration of 10 μg/ml was unable to inhibit HCMV VR1418 entry into epithelial and endothelial cells. IE2 gene expression was detected by immunofluorescence in ARPE-19 cells, UtMVEC, and HUVEC treated with EGFR neutralizing antibody, followed by infection with VR1814 at an MOI of 1.0 PFU/ml for 4 h. (B) Increasing concentrations of an EGFR neutralizing antibody were unable to inhibit HCMV BADrUL131 entry into epithelial and endothelial cells. GFP was detected by flow cytometry in ARPE-19 cells, UtMVEC, and HUVEC treated with an EGFR neutralizing antibody, followed by infection with HCMV BADrUL131 at an MOI of 1.0 PFU/ml for 2 h.
DISCUSSION
The promiscuous nature of HCMV entry, presumably due to its ability to utilize multiple and/or ubiquitously expressed receptors, has made identification of essential cellular receptors difficult. Although many entry receptors for HCMV have been proposed, few have fully satisfied the criteria of true virus receptors. Heparan sulfate proteoglycans (HSPGs) are well established, at least in cell culture systems, to act as the initial attachment receptors for HCMV (5). Supporting the role of HSPGs in HCMV entry, virions are able to bind HSPGs and soluble heparin completely blocks HCMV entry (5). More recently, two laboratories have verified the role of cellular integrins as entry receptors for HCMV (7, 28). Evidence supporting this includes HCMV-mediated activation of β1 and β3 integrins and downstream integrin-specific signaling pathways (7, 28). Additionally, neutralizing antibodies to specific integrin heterodimers are able to inhibit HCMV entry (7, 28). Moreover, a cell line lacking β1 integrin expression supports minimal HCMV infection, while infection returns to wild-type levels when β1 is reintroduced into cells (7). HCMV glycoprotein B also contains a highly conserved integrin-binding domain known as the disintegrin-like domain (7).
Recent reports have also indicated that EGFR can serve as an entry and signaling receptor for HCMV (29). We sought to determine what, if any, interaction EGFR has with β1 integrins. Surprisingly, we could not confirm a role for EGFR in either HCMV entry or signaling events. The presence of EGFR did not confer permissiveness to HCMV in human breast cancer cell lines, contradicting previously reported results (29). It is important to note that these breast cancer cell lines are not from matched backgrounds and instead were isolated from two different patients. Differences other than EGFR expression levels, such as FGFR expression, are also known to exist (17). Regardless, expression of more than 1 × 106 EGFR molecules/cell (15) failed to allow HCMV to infect MB468 cells and initiate IE gene expression. We next wanted to focus on permissive fibroblasts, a more relevant cell type for HCMV infection.
We found that physically blocking EGFR with a potent monoclonal EGFR neutralizing antibody failed to inhibit HCMV infection of NHDF and HEL fibroblast cells, even at a high concentration (10 μg/ml). Although the neutralizing antibody efficiently blocks EGF-mediated EGFR stimulation, it was possible that HCMV interacted with EGFR at a site different from the endogenous EGF ligand. However, the same neutralizing antibody was previously reported to inhibit HCMV glycoprotein B interaction with EGFR, HCMV attachment, and HCMV entry, even at a low (1 μg/ml) concentration (28, 29). Additionally, Söderberg et al., when investigating the contribution of CD13 to HCMV entry, used an EGFR polyclonal antibody to treat HEL fibroblasts but also failed to inhibit HCMV entry (26), again indicating a limited role for EGFR in the HCMV entry process.
It remained possible, however, that EGFR was not needed in a structural role to physically mediate HCMV entry, but rather its kinase signaling capabilities were required. However, we found that a potent (50% inhibitory concentration = 3 nM) EGFR kinase inhibitor, AG1478 (8), was unable to block HCMV gene expression in either NHDF or HEL fibroblast cells. This again was surprising given previous reports that this inhibitor blocks all stages of HCMV gene expression in MB468 breast cancer and HEL fibroblast cells (28, 29). Our results suggest that the EGFR tyrosine kinase signaling activity is not necessary to mediate HCMV entry or to induce or maintain HCMV IE gene expression.
Additionally, we found that HCMV virions, even at high MOIs, are not capable of activating the EGFR tyrosine kinase, as measured by receptor autophosphorylation. Although Wang et al. have demonstrated HCMV-mediated EGFR phosphorylation within minutes of stimulation (29), we were not able to reproduce these results, even when using a comparable MOI. Additionally, Fairley et al. have reported that HCMV actually promotes a loss of the EGFR receptor along with its phosphorylation and signaling. Receptor down-regulation, however, does not occur with UV-inactivated virus, suggesting that virus binding itself is not necessary, but instead, this viral function has been mapped to an HCMV early gene (6).
It remained possible that differences between or within laboratory-adapted and clinical HCMV isolates caused our results to conflict with previously published reports. Therefore, clinical endotropic strain VR1814 and BADrUL131, an AD169 strain with a repair at the UL131 locus restoring its ability to enter both epithelial and endothelial cells (27), were tested for the ability to enter epithelial and endothelial cell lines following treatment with an EGFR neutralizing antibody. However, no difference was found in IE gene expression at 24 h postinfection, indicating that clinical strains also do not require EGFR to mediate virus entry into epithelial or endothelial cells.
Together, our data indicate that EGFR does not play a significant role in the entry or signaling of HCMV. EGFR does not appear to be needed in a structural capacity to mediate HCMV entry, its kinase activity does not enhance HCMV gene expression, and the EGFR tyrosine kinase itself is not activated by HCMV stimulation. No obvious explanation has been found for why the results presented herein differ from previously published reports. To our knowledge, the preparation and purity of the virus stocks used for both studies are comparable. Although clinical isolates of HCMV were tested, it remains a formal possibility that variability between or within HCMV strains results in the ability to activate, signal through, and/or enter cells via EGFR. It is also possible that other tyrosine kinases, including other growth factor receptors, play a role in the HCMV signaling and entry cascade, perhaps in coordination with cellular integrins. In fact, Charles Cobbs and colleagues (University of Alabama—Birmingham) have also found that HCMV does not activate EGFR but indicate the possible presence of another, as yet unidentified, tyrosine kinase that may be involved (personal communication). Furthermore, it is very likely that other entry receptors exist for HCMV which also may act synchronously with cellular integrins to mediate the virus-cell fusion event.
Acknowledgments
We thank Deborah H. Spector for HCMV AD169-GFP, Giuseppe Gerna for HCMV VR1814, Thomas Shenk for HCMV BADrUL131, and Jay A. Nelson for IE antibody.
This work was supported by NIH grant R01 AI034998-10A1 (to T.C.).
Footnotes
Published ahead of print on 11 April 2007.
REFERENCES
- 1.Adam, E., J. L. Melnick, J. L. Probtsfield, B. L. Petrie, J. Burek, K. R. Bailey, C. H. McCollum, and M. E. DeBakey. 1987. High levels of cytomegalovirus antibody in patients requiring vascular surgery for atherosclerosis. Lancet 2:291-293. [DOI] [PubMed] [Google Scholar]
- 2.Alford, C. A., and W. J. Britt. 1993. Cytomegalovirus, p. 227-255. In B. Roizman, R. J. Whitley, and C. Lopez (ed.), The human herpesviruses. Raven Press, New York, NY.
- 3.Compton, T. 1993. An immortalized human fibroblast cell line is permissive for human cytomegalovirus infection. J. Virol. 67:3644-3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Compton, T., R. R. Nepomuceno, and D. M. Nowlin. 1992. Human cytomegalovirus penetrates host cells by pH-independent fusion at the cell surface. Virology 191:387-395. [DOI] [PubMed] [Google Scholar]
- 5.Compton, T., D. M. Nowlin, and N. R. Cooper. 1993. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology 193:834-841. [DOI] [PubMed] [Google Scholar]
- 6.Fairley, J. A., J. Baillie, M. Bain, and J. H. Sinclair. 2002. Human cytomegalovirus infection inhibits epidermal growth factor (EGF) signalling by targeting EGF receptors. J. Gen. Virol. 83:2803-2810. [DOI] [PubMed] [Google Scholar]
- 7.Feire, A. L., H. Koss, and T. Compton. 2004. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc. Natl. Acad. Sci. USA 101:15470-15475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fry, D. W., A. J. Kraker, A. McMichael, L. A. Ambroso, J. M. Nelson, W. R. Leopold, R. W. Connors, and A. J. Bridges. 1994. A specific inhibitor of the epidermal growth factor receptor tyrosine kinase. Science 265:1093-1095. [DOI] [PubMed] [Google Scholar]
- 9.Grazia Revello, M., F. Baldanti, E. Percivalle, A. Sarasini, L. De-Giuli, E. Genini, D. Lilleri, N. Labo, and G. Gerna. 2001. In vitro selection of human cytomegalovirus variants unable to transfer virus and virus products from infected cells to polymorphonuclear leukocytes and to grow in endothelial cells. J. Gen. Virol. 82:1429-1438. [DOI] [PubMed] [Google Scholar]
- 10.Harkins, L., A. L. Volk, M. Samanta, I. Mikolaenko, W. J. Britt, K. I. Bland, and C. S. Cobbs. 2002. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet 360:1557-1563. [DOI] [PubMed] [Google Scholar]
- 11.Hendrix, M. G., M. M. Salimans, C. P. van Boven, and C. A. Bruggeman. 1990. High prevalence of latently present cytomegalovirus in arterial walls of patients suffering from grade III atherosclerosis. Am. J. Pathol. 136:23-28. [PMC free article] [PubMed] [Google Scholar]
- 12.Highlander, S. L., S. L. Sutherland, P. J. Gage, D. C. Johnson, M. Levine, and J. C. Glorioso. 1987. Neutralizing monoclonal antibodies specific for herpes simplex virus glycoprotein D inhibit virus penetration. J. Virol. 61:3356-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson, G. R., B. Kannan, M. Shoyab, and K. Stromberg. 1993. Amphiregulin induces tyrosine phosphorylation of the epidermal growth factor receptor and p185erbB2. Evidence that amphiregulin acts exclusively through the epidermal growth factor receptor at the surface of human epithelial cells. J. Biol. Chem. 268:2924-2931. [PubMed] [Google Scholar]
- 14.Kari, B., and R. Gehrz. 1992. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J. Virol. 66:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kraus, M. H., N. C. Popescu, S. C. Amsbaugh, and C. R. King. 1987. Overexpression of the EGF receptor-related proto-oncogene erbB-2 in human mammary tumor cell lines by different molecular mechanisms. EMBO J. 6:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margolis, M. J., S. Pajovic, E. L. Wong, M. Wade, R. Jupp, J. A. Nelson, and J. C. Azizkhan. 1995. Interaction of the 72-kilodalton human cytomegalovirus IE1 gene product with E2F1 coincides with E2F-dependent activation of dihydrofolate reductase transcription. J. Virol. 69:7759-7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McLeskey, S. W., I. Y. Ding, M. E. Lippman, and F. G. Kern. 1994. MDA-MB-134 breast carcinoma cells overexpress fibroblast growth factor (FGF) receptors and are growth-inhibited by FGF ligands. Cancer Res. 54:523-530. [PubMed] [Google Scholar]
- 18.Melnick, J. L., B. L. Petrie, G. R. Dreesman, J. Burek, C. H. McCollum, and M. E. DeBakey. 1983. Cytomegalovirus antigen within human arterial smooth muscle cells. Lancet 2:644-647. [DOI] [PubMed] [Google Scholar]
- 19.Myerson, D., R. C. Hackman, J. A. Nelson, D. C. Ward, and J. K. McDougall. 1984. Widespread presence of histologically occult cytomegalovirus. Hum. Pathol. 15:430-439. [DOI] [PubMed] [Google Scholar]
- 20.Nowlin, D. M., N. R. Cooper, and T. Compton. 1991. Expression of a human cytomegalovirus receptor correlates with infectibility of cells. J. Virol. 65:3114-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pass, R. F. 2001. Cytomegalovirus, p. 2675-2706. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4 ed., vol. 2. Raven Press, New York, NY. [Google Scholar]
- 22.Real, F. X., W. J. Rettig, P. G. Chesa, M. R. Melamed, L. J. Old, and J. Mendelsohn. 1986. Expression of epidermal growth factor receptor in human cultured cells and tissues: relationship to cell lineage and stage of differentiation. Cancer Res. 46:4726-4731. [PubMed] [Google Scholar]
- 23.Rich, J. D., J. M. Crawford, S. N. Kazanjian, and P. H. Kazanjian. 1992. Discrete gastrointestinal mass lesions caused by cytomegalovirus in patients with AIDS: report of three cases and review. Clin. Infect. Dis. 15:609-614. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez, V., C. L. Clark, J. Y. Yen, R. Dwarakanath, and D. H. Spector. 2002. Viable human cytomegalovirus recombinant virus with an internal deletion of the IE2 86 gene affects late stages of viral replication. J. Virol. 76:2973-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlessinger, J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103:211-225. [DOI] [PubMed] [Google Scholar]
- 26.Söderberg, C., T. D. Giugni, J. A. Zaia, S. Larsson, J. M. Wahlberg, and E. Moller. 1993. CD13 (human aminopeptidase N) mediates human cytomegalovirus infection. J. Virol. 67:6576-6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, D., and T. Shenk. 2005. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J. Virol. 79:10330-10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, X., D. Y. Huang, S. M. Huong, and E. S. Huang. 2005. Integrin αvβ3 is a coreceptor for human cytomegalovirus. Nat. Med. 11:515-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, X., S. M. Huong, M. L. Chiu, N. Raab-Traub, and E. S. Huang. 2003. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature 424:456-461. [DOI] [PubMed] [Google Scholar]
- 30.Yarden, Y., and M. X. Sliwkowski. 2001. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell. Biol. 2:127-137. [DOI] [PubMed] [Google Scholar]