Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) 7a protein, which is not expressed by other known coronaviruses, can induce apoptosis in various cell lines. In this study, we show that the overexpression of Bcl-XL, a prosurvival member of the Bcl-2 family, blocks 7a-induced apoptosis, suggesting that the mechanism for apoptosis induction by 7a is at the level of or upstream from the Bcl-2 family. Coimmunoprecipitation experiments showed that 7a interacts with Bcl-XL and other prosurvival proteins (Bcl-2, Bcl-w, Mcl-1, and A1) but not with the proapoptotic proteins (Bax, Bak, Bad, and Bid). A good correlation between the abilities of 7a deletion mutants to induce apoptosis and to interact with Bcl-XL was observed, suggesting that 7a triggers apoptosis by interfering directly with the prosurvival function of Bcl-XL. Interestingly, amino acids 224 and 225 within the C-terminal transmembrane domain of Bcl-XL are essential for the interaction with the 7a protein, although the BH3 domain of Bcl-XL also contributes to this interaction. In addition, fractionation experiments showed that 7a colocalized with Bcl-XL at the endoplasmic reticulum as well as the mitochondria, suggesting that they may form complexes in different membranous compartments.
Many virus genomes encode gene products that can modulate apoptosis, also called programmed cell death, and the regulation of apoptosis in the infected host is an important determinant in the struggle between virus and host for survival. In the case of the severe acute respiratory syndrome coronavirus (SARS-CoV), one of the most common abnormalities in infected patients is lymphopenia, which is caused by the depletion of T lymphocytes by apoptosis (4, 5, 22). In addition, apoptosis was observed in various infected tissues, such as lung, liver, and thyroid, obtained during autopsy studies on SARS casualties (3, 32). Interestingly, necrosis, another form of cell death, was also observed in different tissues obtained from SARS-CoV-infected patients (6, 9, 17). Taken together, these observations suggest that the modulation of cell death during SARS-CoV infection is important for viral replication and/or pathogenesis.
The family of Bcl-2-related proteins constitutes one of the biologically important gene products in the process of apoptosis. Several members of this family are prosurvival, as their overexpression inhibits apoptosis induced by many different stimuli, while other members are proapoptotic, as their overexpression promotes apoptosis (2, 34). Besides playing important roles in regulating apoptosis and maintaining homeostasis in healthy cells, members of the Bcl-2 family are also implicated in viral infections. Indeed, some viruses encode proteins that are functional homologues of members of the Bcl-2 family or are capable of interacting with members of the Bcl-2 family (8, 12, 23, 33). Interestingly, it was recently demonstrated that the prosurvival Bcl-2 protein inhibits the caspase-dependent apoptosis induced by SARS-CoV infection without affecting viral replication kinetics (1). In addition, it has been shown that the SARS-CoV envelope protein induces apoptosis in Jurkat T cells in the absence of growth factors and that this event could be inhibited by the overexpression of Bcl-XL, another prosurvival member of the Bcl-2 family (37).
The SARS-CoV 7a protein, which shows no significant sequence homology to viral proteins of other known coronaviruses, can induce caspase-dependent apoptosis in cell lines derived from different organs, including the lung, kidney, and liver (16, 28). In this study, we show that the overexpression of Bcl-XL blocks the induction of apoptosis by 7a, suggesting that 7a may be acting at the level of or upstream from the Bcl-2 family. A detailed analysis of the properties of 7a with respect to its ability to interact with different members of the Bcl-2 family and its cellular localization suggests that 7a induces apoptosis by interfering directly with the function of the prosurvival proteins like Bcl-XL, Bcl-w, Mcl-1, and A1.
MATERIALS AND METHODS
Materials.
All reagents used in this study were purchased from Sigma (St. Louis, MO) unless otherwise stated. All cell lines were purchased from the American Type Culture Collection (Manassas, VA) and cultured at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium containing 1 g/liter glucose, 2 mM liter glutamine, 1.5 g/liter sodium bicarbonate, 0.1 mM nonessential amino acids, 0.1 mg/ml streptomycin, 100 U penicillin, and 5% fetal bovine serum (HyClone).
Construction of plasmids.
Expression plasmids for 7a and Bcl-XL deletion and substitution mutants were generated by PCR using titanium Taq DNA polymerase (Clontech Laboratories Inc., Palo Alto, CA). All sequences were confirmed by sequencing performed by the core facilities at the Institute of Molecular and Cell Biology, Singapore. All primers used in this study were purchased from Research Biolabs, Singapore.
Generation of stable cell lines.
The Bcl-XL open reading frame (ORF) was cloned into the pCep4 vector (Invitrogen, Carlsbad, CA) and transfected into 293T cells by electroporation as previously described (30). Cells stably expressing Bcl-XL were obtained after selection in 0.2 mg/ml of hygromycin B (Roche Molecular Biochemicals, Indianapolis, IN). Control cells were stably transfected with an empty vector.
Transient transfections, CaspACE fluorometric assay, and Western blot analysis.
Transient transfections of 293T and Vero E6 cells were performed using Lipofectamine reagent (Invitrogen), according to the manufacturer's protocol. Approximately 16 h after transfection, the activation of caspase-3 was quantified by using a CaspACE fluorometric assay system from Promega Corporation (Madison, WI) as previously described (15, 28).
Western blot analysis was performed as previously described (31), and some of the primary antibodies (anti-myc monoclonal [Santa Cruz Biotechnology, Santa Cruz, CA], anti-SARS-CoV M rabbit polyclonal [Abgent, San Diego, CA], anti-actin and anti-tubulin monoclonal [Sigma], anti-pyruvate dehydrogenase [PDH] E2 subunit [Molecular Probes, OR], anti-Bcl-XL monoclonal [Transduction Laboratories], anti-BAD rabbit polyclonal [Cell Signaling Technology Inc. Beverly, MA], and anti-calreticulin and anti-alpha-COP I rabbit polyclonal [Affinity BioReagents, CO] antibodies) were purchased, while an anti-7a monoclonal antibody was generated for this study. Briefly, a bacterially expressed glutathione S-transferase (GST)-7a fusion protein was used to immunize BALB/c mice as previously described (10). The spleen was excised from a mouse that showed strong antibody response, and hybridoma fusion was performed to generate anti-7a monoclonal antibodies as previously described (20). All procedures involving the use of laboratory animals were performed by trained personnel in accordance with the regulations and guidelines of the National Advisory Committee for Laboratory Animal Research (NACLAR), Singapore. The other antibodies against SARS-CoV proteins (anti-N and anti-3a mouse polyclonal antibodies) have been described previously (11, 31).
Coimmunoprecipitation experiments.
For the coimmunoprecipitation experiments, each 6-cm dish of cells was resuspended in 200 μl of immunoprecipitation (IP) buffer (50 mM Tris [pH 8], 150 mM NaCl, 0.5% NP-40, 0.5% deoxycholic acid, 0.005% sodium dodecyl sulfate [SDS], and 1 mM phenylmethylsulfonyl fluoride) and subjected to freeze-thawing six times. One hundred microliters of the lysates was diluted with 50 μl of IP buffer, 5 μl of rabbit anti-myc polyclonal antibody (Santa Cruz Biotechnology) was added, and the mixture was subjected to end-over-end mixing at room temperature for 1 h. Protein A-agarose beads (Roche) were then added, and the mixing was continued for at least 4 h at 4°C. Beads were washed four times with cold IP buffer, and then 15 μl of Laemmli's SDS buffer was added and the samples were boiled at 100°C for 5 min to release the immunocomplexes. Samples were separated by SDS-polyacrylamide gel electrophoresis and subjected to Western blot analysis.
Coimmunoprecipitation experiments with SARS-CoV-infected cells were performed in a similar manner. Infection of Vero E6 cells with an isolate of SARS-CoV (strain HKU 39849) was carried out in a physical-containment level 4 laboratory as previously described (14). Lysates obtained from SARS-CoV-infected Vero E6 cells were subjected to Western blot analysis, to determine the expression levels of different viral proteins, or to IP with either rabbit anti-Bcl-XL polyclonal antibody (Santa Cruz Biotechnology) or an irrelevant antibody (anti-BID rabbit polyclonal antibody; Santa Cruz Biotechnology) and protein A-agarose beads. Western blot analyses were then performed to detect the amounts of 7a or Bcl-XL in the immunocomplexes captured on the protein A-agarose beads.
Fractionation experiments.
Intact mitochondria were isolated from 293T cells expressing 7a by using a mitochondrial isolation kit for cultured cells from Pierce (Rockford, IL). The reagent-based method was used according to the manufacturer's protocol, and this method gave two fractions. The first fraction contained nonmitochondrial proteins, while the second fraction contained only mitochondrial proteins. Both fractions were made up to equal volumes (500 μl, ∼8 × 106 cells) and subjected to Western blot analysis as described above. The same protocol was used to fractionate SARS-CoV-infected Vero E6 cells that were harvested at 0, 16, or 24 h postinfection.
Indirect immunofluorescence experiments.
Vero E6 cells were transfected with cDNA for expressing 7a as described above. After 16 h, the cells were incubated with 50 nM of MitroTracker red (Molecular Probes, Inc., Eugene, OR) for 15 min at 37°C. Then, the cells were processed for antibody staining as previously described (31).
RESULTS
Overexpression of Bcl-XL inhibits the induction of apoptosis by the SARS-CoV 7a protein.
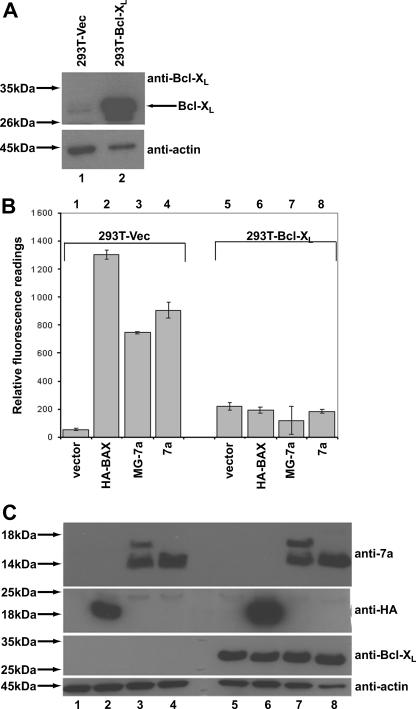
In order to determine if the SARS-CoV 7a protein acts upstream or downstream of the Bcl-2 family to induce apoptosis, 293T cells stably expressing Bcl-XL were generated. As shown in Fig. 1A, there is high expression of Bcl-XL in the chosen stable clone (293T-Bcl-XL) in comparison to the endogenous Bcl-XL level in the vector control cells (293T-Vec). These stable cell lines were then transiently transfected with cDNA to express HA-BAX (positive control) or 7a, and the induction of apoptosis was determined as previously described (28). The expression of HA-BAX and 7a caused apoptosis in 293T-Vec cells (Fig. 1B, columns 1 to 4) as determined by a fluorometric assay for measuring caspase-3 activation. However, in the 293T-Bcl-XL cells, the expression of HA-BAX and 7a did not induce significant levels of apoptosis (Fig. 1B, columns 5 to 8). Thus, the overexpression of Bcl-XL inhibited the 7a-induced apoptosis, suggesting that the regulation of cellular apoptosis by 7a occurs at or upstream of the Bcl-2 family in the apoptotic pathway.
FIG. 1.
Effects of Bcl-XL on the induction of apoptosis by the SARS-CoV 7a protein. (A) Western blot analysis was performed to determine the expression level of Bcl-XL in the stable cell line 293T-Bcl-XL compared to the expression level in the vector control (293T-Vec) cells. Equal amounts of cells were used in each lane, as verified by the levels of endogenous actin. Molecular mass markers are shown on the left. (B) A CaspACE fluorometric assay system from Promega Corporation (Madison, WI) was used to measure the activation of caspase-3 protease activity, which is a hallmark of apoptosis, in cells that were transfected with vector only, a classical apoptosis inducer (HA-BAX), 7a with two additional amino acids at its N terminus (MG-7a) or full-length 7a. The cells used were 293T-Vec or 293T-Bcl-XL. All experiments were performed in duplicate, and the average values with standard deviations are plotted. (C) A corresponding Western blot analysis was performed to determine the expression levels of 7a, HA-BAX, and Bcl-XL, using anti-7a, anti-HA, and anti-Bcl-XL antibodies, respectively. The amounts of total cell lysates loaded were verified by measuring the levels of endogenous actin (anti-actin). Molecular mass markers are shown on the left.
Previously, we constructed a 7a cDNA expression plasmid (MG-7a), with six additional base pairs (ATGGGA) which code for two additional amino acids (methionine and glycine) before the ATG codon of the 7a ORF, in order to achieve a Kozak consensus ribosome binding site for efficient translation initiation (10). Here, a new 7a cDNA expression plasmid without the Kozak sequence was constructed. As shown in Fig. 1C, the cleavage of the signal peptide of the 7a protein is more efficient than that of the protein expressed using the old cDNA expression plasmid (compare lanes 3 and 4 or lanes 7 and 8). However, there is no difference in the degree of apoptosis induced by the transfection of either construct, which is consistent with our previous finding that the cleavage of the signal peptide of 7a is not important for the induction of apoptosis (28). The 7a construct without the Kozak sequence was used for the subsequent studies.
SARS-CoV 7a interacts with Bcl-XL but not with proapoptotic members of the Bcl-2 family.
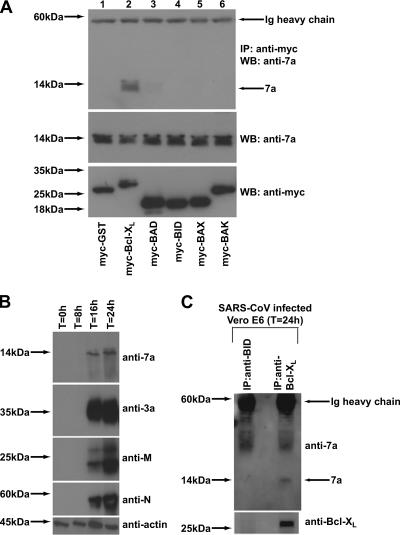
To further understand how the SARS-CoV 7a protein modulates the function of the Bcl-2 family of proteins, we performed coimmunoprecipitation experiments to determine if 7a can interact with members of this family of proteins. As shown in Fig. 2A (top panel), 7a was specifically coimmunoprecipitated by myc-Bcl-XL (lane 2) but not by the negative control, myc-GST (lane 1), or the proapoptotic proteins (myc-BAD, myc-BID, myc-BAX, or myc-BAK) tested (lanes 3 to 6). These results indicate that 7a may induce apoptosis by interfering with the prosurvival function of Bcl-XL.
FIG. 2.
Interactions of the SARS-CoV 7a protein with members of the Bcl-2 family. (A) 293T cells were transfected with cDNA constructs expressing full-length 7a and myc-tagged GST (negative control; myc-GST) or myc-tagged members of the Bcl-2 family (myc-Bcl-XL, myc-BAD, myc-BID, myc-BAX, or myc-BAK). The cells were harvested at ∼16 h posttransfection, lysed, and subjected to IP with myc-polyclonal antibody and protein A beads. The amount of 7a that coimmunoprecipitated with the myc-tagged proteins (IP: anti-myc) was determined by Western blot analysis with an anti-7a mouse monoclonal antibody (WB: anti-7a) (top). The amounts of 7a (WB: anti-7a) and myc-tagged proteins (WB: anti-myc) in the lysates before IP were determined by subjecting aliquots of the lysates to Western blot analysis (middle and bottom). (B) Lysates were obtained from SARS-CoV-infected Vero E6 cells harvested at different time points postinfection and Western blot analyses were performed to determine the expression of SARS-CoV proteins, using anti-7a mouse monoclonal antibody, anti-3a mouse polyclonal antibody, anti-M rabbit polyclonal antibody, and anti-N mouse polyclonal antibody. Antiactin monoclonal antibody was used to verify that equal amounts of lysates were used in each lane. (C) Cell lysates obtained from Vero E6 cells infected with SARS-CoV (at 24 h postinfection) were subjected to IP using anti-BID or anti-Bcl-XL rabbit polyclonal antibodies and protein A beads. The immunocomplexes were subjected to Western blot analysis with an anti-7a mouse monoclonal antibody. The membrane was then stripped and reprobed with an anti-Bcl-XL mouse monoclonal antibody. Ig, immunoglobulin. Molecular mass markers are shown on the left.
As 293T cells do not support SARS-CoV replication, we determined the temporal expression of 7a in infected Vero E6, an African green monkey kidney cell line. The expression of 7a and other SARS-CoV proteins (3a, M, and N) was detected in cells harvested at 16 h and 24 h postinfection (Fig. 2B). Coimmunoprecipitation experiments were then performed using lysates obtained at 24 h postinfection. As shown in Fig. 2C, 7a was coimmunoprecipitated by an anti-Bcl-XL antibody and not by an anti-BID antibody, suggesting that the 7a and endogenous Bcl-XL proteins formed complexes during SARS-CoV infection. This experiment was performed three times, and the data presented in Fig. 2C are a representative set.
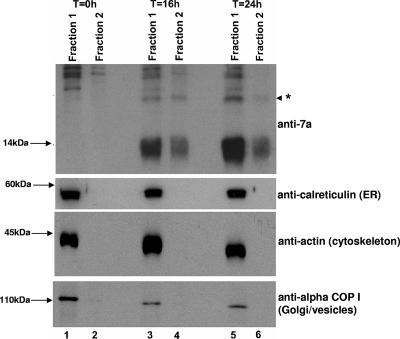
FIG. 7.
Cellular distribution of the 7a protein in SARS-CoV-infected Vero E6 cells as determined by fractionation experiments. SARS-CoV-infected Vero E6 cells harvested at different time points postinfection were fractionated into two fractions, one that contained mitochondria (fraction 2) and another that contained all other cellular compartments (fraction 1). Western blot analyses were performed to determine the amounts of 7a and cellular proteins in these two fractions. The protein marked with an asterisk represents an oligomeric form of 7a (lanes 3 to 6). Molecular mass markers are shown on the left.
The transmembrane domain of the SARS-CoV 7a protein is important for its interaction with Bcl-XL and for the induction of apoptosis.
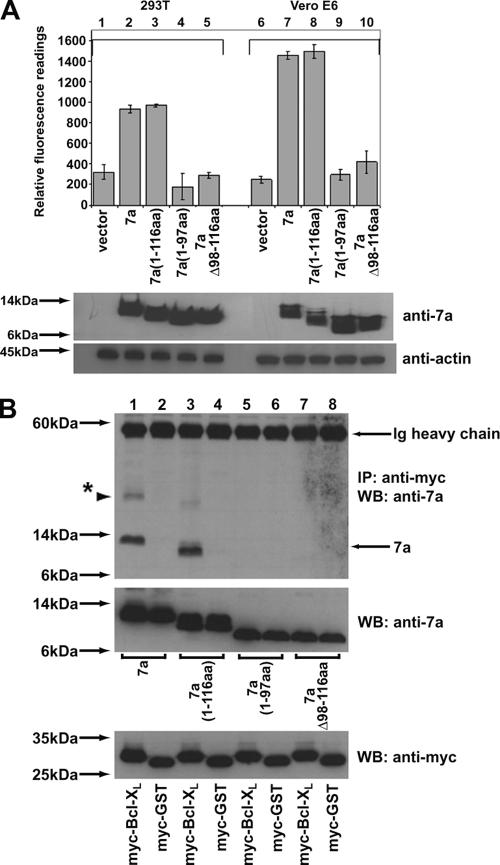
Next, we constructed a series of 7a mutants and compared their abilities to induce apoptosis. The C-terminal portion of the 7a protein contains a transmembrane domain (98 to 116 amino acids [aa]) and a short cytoplasmic tail (117 to 122 aa) (10). While the transmembrane domain is important for the insertion of 7a into intracellular membranes, the cytoplasmic tail contains an endoplasmic reticulum (ER) retrieval motif that mediates the transport of 7a between the Golgi and the ER. As demonstrated by the activation of caspase 3 (Fig. 3A), the 7a(1-116 aa) deletion mutant (columns 3 and 8) induced as much apoptosis as the full-length 7a, indicating that the cytoplasmic tail of 7a is not essential for apoptosis induction. However, the 7a(1-97aa) and 7aΔ98-116aa deletion mutants (columns 4, 5, 9, and 10) did not induce significant levels of apoptosis in either 293T or Vero E6 cells, implying that the transmembrane domain of 7a is essential for apoptosis induction (Fig. 3A). The same mutants were tested in coimmunoprecipitation experiments for their abilities to bind Bcl-XL. The results showed that the 7a(1-116aa) mutant (Fig. 3B, lane 3), but not the 7a(1-97aa) and 7aΔ98-116aa mutants (Fig. 3B, lanes 5 and 7, respectively), could bind Bcl-XL, indicating that the transmembrane domain of 7a is also important for its interaction with Bcl-XL.
FIG. 3.
Comparison of the abilities of SARS-CoV 7a deletion mutants to induce apoptosis and to interact with Bcl-XL. (A) A CaspACE fluorometric assay system from Promega Corporation (Madison, WI) was used to measure the activation of caspase-3 protease activity in cells that were transfected: vector only (vector), full-length 7a (7a), or 7a deletion mutants [7a(1-116aa), 7a(1-97aa) and 7aΔ98-116aa]. The cells used were 293T or Vero E6. All experiments were performed in duplicate and the average values with standard deviations are plotted. Western blot analyses were performed to determine the expression levels of 7a (anti-7a) and endogenous actin (anti-actin) in each sample. (B) 293T cells were transfected with cDNA constructs expressing full-length 7a or 7a deletion mutants [7a(1-116aa), 7a(1-97aa), and 7aΔ98-116aa] and myc-tagged full-length Bcl-XL (myc-Bcl-XL) or myc-tagged GST (negative control; myc-GST). Coimmunoprecipitation experiments were performed as described in the legend for Fig. 2A, and the amounts of 7a that coimmunoprecipitated with the myc-tagged proteins (IP: anti-myc) were determined by using an anti-7a mouse monoclonal antibody (WB: anti-7a; top). Ig, immunoglobulin. The amounts of 7a (WB: anti-7a) and myc-tagged (WB: anti-myc) proteins in the lysates before IP were determined by subjecting aliquots of the lysates to Western blot analysis (middle and bottom). The protein marked with an asterisk represents the oligomeric form of 7a. Molecular mass markers are shown on the left.
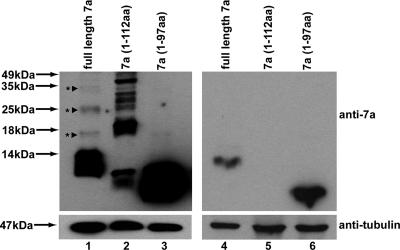
In this experiment, we observed that the 7a protein has a higher-molecular-mass form that migrates much more slowly than the monomeric 7a protein (Fig. 3B, top panel, lane 1). In order to determine the nature of this higher-molecular-mass form of 7a, a new deletion mutant that lacks the last 4 aa of the transmembrane domain [7a(1-112aa)] was constructed and analyzed together with full-length 7a and the 7a(1-97aa) mutant. The Western blot analysis was performed using two sets of lysates; one set contained 24 μg of total protein (Fig. 4, lanes 1 to 3), so that the dimeric and/or oligomeric forms of 7a are more clearly illustrated, while the second set contained 6 μg of total protein, so that the migrations of the 7a proteins are clearer (Fig. 4, lanes 4 to 6). As shown in Fig. 4 (lanes 1 and 2), higher-molecular-mass forms of full-length 7a and the 7a(1-112aa) mutant are detected, suggesting that they could be dimers and/or oligomers which are resistant to boiling, 20 mM dithiothreitol, and 1% SDS (which are found in the Laemmli SDS loading buffer). For example, the full-length 7a protein that migrated at ∼25 kDa (Fig. 4, lane 1) would match the size of the dimeric form of 7a, since the monomeric form of 7a is predicted to be 12.5 kDa. Nevertheless, the majority of the full-length 7a protein migrated at ∼12.5 kDa (Fig. 4, lanes 1 and 4), suggesting that a greater portion of full-length 7a exists as monomers. While the 7a(1-112aa) deletion mutant showed a more pronounced tendency to form oligomers (Fig. 4, lane 2), the 7a(1-97aa) deletion mutant, which lacks the transmembrane domain, showed little tendency to form dimers or oligomers (Fig. 4, lanes 3 and 6). Overall, these results suggested that the higher-molecular-mass forms of 7a arose from the transient unfolding of the transmembrane helix, which resulted in unfavorable exposure of hydrophobic residues and self-aggregation of 7a. The deletion of the last 4 amino acids of the transmembrane helix [i.e., the 7a(1-112aa) mutant] led to further destabilization of the helix, and hence, more aggregation is observed.
FIG. 4.
Comparison of the extent of protein aggregation of SARS-CoV 7a full-length protein and deletion mutants. 293T cells were transfected with cDNA constructs expressing full-length 7a or 7a deletion mutants [7a(1-112aa) and 7a(1-97aa)]. Lanes 1 to 3 were loaded with larger amounts of lysates, so that the dimeric and/or oligomeric forms of 7a are more clearly illustrated, while lanes 4 to 6 were loaded with smaller amounts (a quarter of the amounts of the corresponding samples in lanes 1 to 3) so that the migrations of the 7a proteins are clearer. Western blot analyses were performed to detect 7a (anti-7a). The proteins marked with asterisks represent dimeric or oligomeric forms of 7a. Equal amounts of the cell lysates were loaded, as verified by the level of endogenous tubulin (anti-tubulin). Molecular mass markers are shown on the left.
Amino acids 224 and 225 within the C-terminal transmembrane domain of Bcl-XL are essential for interacting with the SARS-CoV 7a protein.
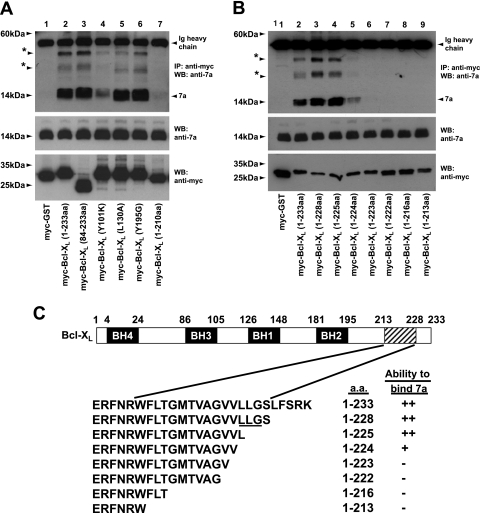
In order to determine which domain(s) in Bcl-XL is involved in the interaction with 7a, coimmunoprecipitation experiments were performed to examine several well-characterized mutants of Bcl-XL for their abilities to interact with 7a. The results showed that the N-terminally deleted mutant, Bcl-XL(84-233aa), retained the ability to bind 7a (Fig. 5A, lane 3), suggesting that the BH4 domain of Bcl-XL is not required for the interaction with 7a. In addition, three substitution mutants were examined, and in each of these mutants, a critical residue in one of the BH1 to BH3 domains was substituted. As deduced from an analysis of the three-dimensional structure of Bcl-XL, these residues are involved in hydrophobic interactions with the BH3 domains of death agonists but are not involved in intramolecular interactions (21). As shown in Fig. 5A, Bcl-XL(Y101K), in which a critical tyrosine residue in the BH3 domain was replaced, showed much-reduced binding to 7a (lane 4). However, Bcl-XL(L130A) and Bcl-XL(Y195G), where critical residues in the BH1 and BH2 domains, respectively, were mutated, bound 7a efficiently (Fig. 5A, lanes 5 and 6). Lastly, the C-terminally deleted mutant, Bcl-XL(1-210aa), which lacks the transmembrane at the C terminus, did not bind 7a significantly (Fig. 5A, lane 7). Hence, it appears that the transmembrane domain of Bcl-XL is essential for interacting with 7a, while its BH3 domain enhances the strength of the interaction.
FIG. 5.
Comparison of the abilities of Bcl-XL mutants to interact with the SARS-CoV 7a protein. (A and B) 293T cells were transfected with cDNA constructs expressing full-length 7a and myc-tagged GST (negative control; myc-GST), myc-tagged full-length Bcl-XL [myc-Bcl-XL(1-233aa)], or myc-tagged mutants of Bcl-XL. For panel A, the mutations created are a deletion at the N terminus [myc-Bcl-XL(84-233aa)]; single amino acid substitutions in the BH3 [myc-Bcl-XL(Y101K)], BH1 [myc-Bcl-XL(L130A)], and BH2 [myc-Bcl-XL(Y195G)] domains; and a deletion at the C terminus [myc-Bcl-XL(1-210aa)]. For panel B, the mutants used are C-terminally deleted mutants of Bcl-XL [myc-Bcl-XL(1-228aa), myc-Bcl-XL(1-225aa), myc-Bcl-XL(1-224aa), myc-Bcl-XL(1-223aa), myc-Bcl-XL(1-222aa), myc-Bcl-XL(1-216aa), and myc-Bcl-XL(1-213aa)]. IP experiments were performed as described in the legend for Fig. 2A, and the amounts of 7a that coimmunoprecipitated with the myc-tagged proteins (IP: anti-myc) were determined by using an anti-7a mouse monoclonal antibody (WB: anti-7a). The proteins marked with asterisks represent oligomeric forms of 7a. The amounts of 7a (WB: anti-7a) and myc-tagged (WB: anti-myc) proteins in the lysates before IP were determined by subjecting aliquots of the lysates to Western blot analysis (middle and bottom). Ig, immunoglobulin. Molecular mass markers are shown on the left. (C) A schematic diagram showing the construction of Bcl-XL deletion mutants and a summary of their abilities to bind 7a. ++, strong interaction; +, weak interaction; −, no interaction. LLG corresponds to aa 225 to 227 of Bel-XL.
Next, a series of C-terminally deleted mutants of Bcl-XL was constructed to further delineate the essential residues for interaction with 7a. As shown in Fig. 5B, the transmembrane domain of Bcl-XL (aa 213 to 228) is important for the interaction between 7a and Bcl-XL, as 7a bound to Bcl-XL(1-228aa) (lane 3) but not to Bcl-XL(1-213aa) (lane 9). Further analysis showed that Bcl-XL(1-223aa) did not interact significantly with 7a (Fig. 5B, lane 6). However, Bcl-XL(1-224aa) interacted weakly and Bcl-XL(1-225aa) interacted as strongly as full-length Bcl-XL (Fig. 5B, lanes 4 and 5). Thus, the results show that aa 224 and 225 of Bcl-XL are essential for the interaction between Bcl-XL and 7a (Fig. 5B and C). Interestingly, aa 225 to 227 of Bcl-XL were previously shown to be important for its interaction with the proapoptotic Bax protein (13). Although 7a and Bax bind to similar regions within the C-terminal transmembrane domain of Bcl-XL, the exact binding mechanisms appear to be different, because Bcl-XL(1-224aa) could bind 7a but not Bax (Fig. 5C) (13).
7a is partially localized in the mitochondria.
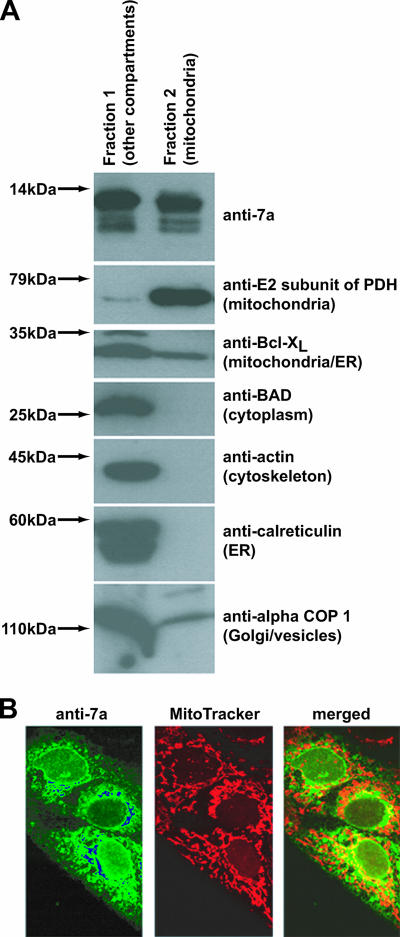
Previously, 7a was found predominantly in the ER-Golgi intermediate compartments (10). Here, 293T cells expressing 7a were fractionated into two fractions in order to determine if 7a can translocate to the mitochondria (Fig. 6A). One of the fractions contained the mitochondria (fraction 2), while the other contained all other cellular compartments (fraction 1). Western blot analyses showed that 7a was found in both fractions (Fig. 6A, first panel) whereas a mitochondrion-resident protein, the E2 subunit of PDH, was only found in fraction 2 (second panel). Endogenous Bcl-XL protein, which has been reported to localize mainly at the ER and mitochondria, was also detected in both fractions (Fig. 6, third panel). On the other hand, nonmitochondrial proteins, like Bad, actin, calreticulin, and alpha COP I, were found only in fraction 1 (fourth to seventh panels). Similar results were obtained when Vero E6 cells were used instead (data not shown).
FIG. 6.
Cellular distribution of the SARS-CoV 7a protein, expressed in mammalian cells through the transfection of a 7a-expression cDNA construct, as determined by fractionation and indirect immunofluorescence experiments. (A) 293T cells expressing 7a were fractionated into two fractions, one that contained mitochondria (fraction 2) and another that contained all other cellular compartments (fraction 1). Western blot analyses were performed to determine the amounts of 7a and cellular proteins in these two fractions. Molecular mass markers are shown on the left. (B) Vero E6 cells expressing 7a were probed with anti-7a monoclonal antibody and a fluorescein isothiocyanate-conjugated anti-mouse secondary antibody or MitoTracker red dye which labeled the mitochondria of the cell. In the merged image, the fraction of 7a protein localized in the mitochondria is indicated by the yellow staining.
Immunofluorescence experiments were not performed on 293T cells because of their poor morphology. Instead, the experiments were performed on 7a-expressing Vero E6 cells to determine if 7a can be found in the mitochondria. Consistent with the results of the fractionation experiments, the results with MitroTracker red dye showed that 7a partially localized in the mitochondria (Fig. 6B). Hence, these results show that 7a is found in the mitochondria as well as the ER-Golgi intermediate compartments.
The fractionation experiment was repeated using SARS-CoV-infected Vero E6 cells harvested at 0, 16, and 24 h postinfection. As shown in Fig. 7, a portion of the 7a protein is found in the mitochondria in cells harvested at 16 h and 24 h postinfection (fraction 2, lanes 4 and 6). Consistent with the results obtained from cells transfected with a 7a-expression cDNA construct (Fig. 6), a portion of the 7a protein is also found in the nonmitochondrial fraction (fraction 1, lanes 3 and 5). The monoclonal antibodies against the E2 subunit of PDH could not recognize the protein in Vero E6 cells (derived from African green monkey) probably due to species differences (data not shown). However, endogenous calreticulin (ER), actin (cytoskeleton), and alpha-COP 1 (Golgi/vesicles) proteins are not found in the mitochondrial fraction, indicating that the fractionation has successfully removed all nonmitochondrial proteins from fraction 2. It is interesting to note that the higher-molecular-mass forms of 7a, which were detected in cells transfected with a 7a-expressing cDNA construct (Fig. 4), are also detected in infected cells.
The SARS-CoV 7a protein also interacts with other prosurvival members of the Bcl-2 family.
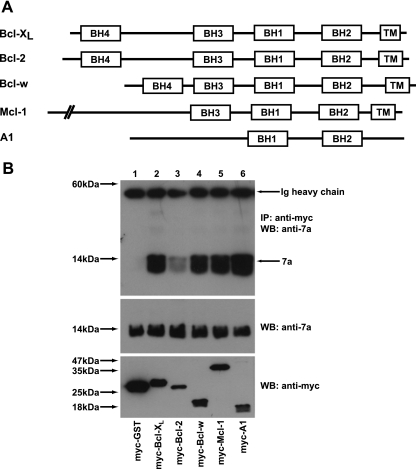
As most of the prosurvival members of the Bcl-2 family contain conserved BH1 to BH4 domains, as well as a transmembrane domain near their C termini (Fig. 8A), and have similar apoptosis regulatory functions (2, 34), coimmunoprecipitation experiments were also performed to examine the interaction of 7a with other prosurvival members of the Bcl-2 family. As shown in Fig. 8B, 7a interacted strongly with Bcl-XL (lane 2), Bcl-w (lane 4), Mcl-1 (lane 5), and A1 (lane 6) but weakly with Bcl-2 (lane 3).
FIG. 8.
Interaction of the SARS-CoV 7a protein with different prosurvival members of the Bcl-2 family. (A) A schematic diagram showing the positions of the BH1 to BH4 domains and transmembrane domains (TM) found in the prosurvival Bcl-2 family members (Bcl-XL, Bcl-2, Bcl-w, Mcl-1, and A1). (B) 293T cells were transfected with cDNA constructs expressing full-length 7a and myc-tagged GST (negative control; myc-GST) or myc-tagged prosurvival members of the Bcl-2 family (myc-Bcl-XL, myc-Bcl-2, myc-Bcl-w, myc-Mcl-1, and myc-A1). Coimmunoprecipitation experiments were performed as described in the legend for Fig. 2A, and the amounts of 7a that coimmunoprecipitated with the myc-tagged proteins (IP: anti-myc) were determined by using an anti-7a mouse monoclonal antibody (WB: anti-7a). Ig, immunoglobulin. The amounts of 7a (WB: anti-7a) and myc-tagged (WB: anti-myc) proteins in the lysates before IP were determined by subjecting aliquots of the lysates to Western blot analysis (middle and bottom panels). Molecular mass markers are shown on the left.
DISCUSSION
The SARS-CoV genome encodes eight putative accessory proteins (i.e., ORFs 3a, 3b, 6, 7a, 7b, 8a, 8b, and 9b) which have no homologues in other known coronaviruses. Although most of these viral proteins were expressed during SARS-CoV infection in vitro and in vivo (29), they appeared to be dispensable for viral replication in cell culture and the mouse model (38). Nevertheless, the expression of some of these accessory proteins has been shown to affect the physiological state of the cell, suggesting that they may be important for virus-host interactions and thus contribute to viral stability and/or pathogenesis in vivo (29). Previously, we showed that the overexpression of the SARS-CoV 7a accessory protein induces apoptosis in a caspase-3-dependent manner (28). Here, we show that the induction of apoptosis by 7a can be blocked by the overexpression of the Bcl-XL protein (Fig. 1), which is a prosurvival member of the Bcl-2 family, suggesting that 7a is acting at or upstream of the Bcl-2 family.
Using coimmmunoprecipitation experiments, we further demonstrated that 7a can interact with Bcl-XL in SARS-CoV-infected cells (Fig. 2). Mutagenesis experiments showed that the transmembrane domain of 7a is essential for both apoptosis induction and interaction with Bcl-XL (Fig. 3), suggesting that 7a induces apoptosis via its interaction with Bcl-XL. Our results also showed that 7a interacts predominantly with aa 224 and 225 within the C-terminal transmembrane domain of Bcl-XL (Fig. 5). However, the BH3 domain of Bcl-XL also contributes to this interaction, because the replacement of a critical residue within this domain (Y101→K) reduces the strength of the interaction significantly (Fig. 5). As the transmembrane domain of Bcl-XL is important for its interaction with Bax (13) and association with intracellular membranes (25), the binding of 7a to Bcl-XL during SARS-CoV infection may interfere with the prosurvival function of Bcl-XL. Like Bcl-XL, 7a can be found in the mitochondria as well as the ER (Fig. 6 and 7); hence it is possible that complexes of 7a and Bcl-XL are found in different membranous compartments. The efficient cleavage of the signal peptide of 7a (presumably by ER signal peptidases) suggests that newly synthesized 7a translocates first to the ER. Subsequently, the mature 7a protein is transported to the ER-Golgi intermediate compartments (10) as well as the mitochondria (Fig. 6 and 7). Future studies are needed to understand how the distribution of 7a is regulated during infection and the function of 7a in the different membranous compartments.
Interestingly, 7a does not interact with the proapoptotic members of the Bcl-2 family, but it interacts with several other prosurvival members of the Bcl-2 family, namely, Bcl-2, Bcl-w, A1, and Mcl-1 (Fig. 2 and 8). Despite the sequence variation between these prosurvival members of the Bcl-2 family, 7a still has the ability to form complexes with these proteins in vitro. Unlike the other prosurvival proteins, the A1 protein does not have a transmembrane domain at its C terminus (Fig. 8), which suggests that it is likely to interact with 7a by a mechanism that is different from that of Bcl-XL or indirectly through another prosurvival protein. Therefore, further studies are needed to determine if these interactions occur simultaneously and how they are modulated during different stages of the SARS-CoV infection.
The occurrence of caspase-dependent apoptosis in SARS-CoV-infected cells is well documented (1, 24, 36). Similar to our observations on the SARS-CoV 7a protein, Yang and coworkers also showed that the induction of apoptosis by the SARS-CoV envelope protein can be inhibited by the overexpression of Bcl-XL (37). As it has been reported that inhibition of apoptosis does not affect SARS-CoV replication in Vero cells (1), the induction of apoptosis by SARS-CoV proteins could be more important for virus-induced pathogenesis. The observation of apoptosis (and necrosis) in various infected tissues obtained during autopsy studies on SARS casualties also suggested that induction of apoptosis could be important for viral pathogenesis. Although the 7a protein is not essential for SARS-CoV replication in cell culture and young mice (38), it is still unclear if 7a contributes to viral pathogenesis in other animal models, such as aged BALB/c mice, hamsters, ferrets, and nonhuman primates. Unlike young mice, these animals showed some histopathologic damage upon SARS-CoV infection (26). Even then, such studies may still not give the full picture, as the severe disease seen in SARS-CoV-infected humans is not completely reproduced in any of the currently available animal models (26). Yount and coworkers (38) also reported that a mutant virus without the 7a/7b gene still induced extensive cytopathic effects in the Vero cell line, suggesting that 7a is not a dominant contributor to virus-induced cell death, at least in this cell culture system. However, there is currently no data on the effects of 7a-induced apoptosis in the mouse or other animal models.
Besides the 7a and envelope proteins, numerous other SARS-CoV proteins have also been shown to induce apoptosis. They are the 3C-like protease (19); the major structural proteins, spike, nucleocapsid, and membrane (7, 27, 40); and two more accessory proteins, ORFs 3a and 3b (18, 35, 39). The mechanisms for induction of apoptosis by these viral proteins and their relative contributions to SARS-CoV-induced apoptosis have not been investigated in detail. For these viral proteins, it has also not yet been determined if the apoptosis can be inhibited by prosurvival members of the Bcl-2 family. In addition, the SARS-CoV 3b protein induces both necrosis and apoptosis in Vero E6 cells (15). It is interesting to note that some of these viral proteins (envelope, nucleocapsid, and membrane) induce apoptosis only in the absence of growth factors, while others (3C-like protease, spike, and ORFs 3a, 3b, and 7a) induce apoptosis under normal growth conditions. This may imply that the induction of apoptosis by these different proteins occurs at different stages of the infection cycle. In future studies, it will be crucial to determine the interplay between these SARS-CoV proteins and the manner in which they modulate cell death during the virus life cycle.
Acknowledgments
We thank personnel at the Biological Resource Centre (Agency for Science, Technology and Research [A*STAR], Singapore), Monoclonal Antibody Unit (Institute of Molecular and Cell Biology, Singapore), and physical containment level 4 laboratory (Victorian Infectious Diseases Reference Laboratory, Australia.) for technical assistance.
This work was supported by grants from A*STAR.
Y.-J.T. and V.C.Y. are adjunct staff of the Department of Microbiology and Department of Pharmacology, respectively, at the National University of Singapore.
Footnotes
Published ahead of print on 11 April 2007.
REFERENCES
- 1.Bordi, L., C. Castilletti, L. Falasca, F. Ciccosanti, S. Calcaterra, G. Rozera, A. Di Caro, S. Zaniratti, A. Rinaldi, G. Ippolito, M. Piacentini, and M. R. Capobianchi. 2006. Bcl-2 inhibits the caspase-dependent apoptosis induced by SARS-CoV without affecting virus replication kinetics. Arch. Virol. 151:369-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borner, C. 2003. The Bcl-2 protein family: sensors and checkpoints for life-or-death decisions. Mol. Immunol. 39:615-647. [DOI] [PubMed] [Google Scholar]
- 3.Chau, T. N., K. C. Lee, H. Yao, T. Y. Tsang, T. C. Chow, Y. C. Yeung, K. W. Choi, Y. K. Tso, T. Lau, S. T. Lai, and C. L. Lai. 2004. SARS-associated viral hepatitis caused by a novel coronavirus: report of three cases. Hepatology 39:302-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, R. F., J. C. Chang, W. T. Yeh, C. H. Lee, J. W. Liu, H. L. Eng, and K. D. Yang. 2006. Role of vascular cell adhesion molecules and leukocyte apoptosis in the lymphopenia and thrombocytopenia of patients with severe acute respiratory syndrome (SARS). Microbes Infect. 8:122-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chng, W. J., H. C. Lai, A. Earnest, and P. Kuperan. 2005. Haematological parameters in severe acute respiratory syndrome. Clin. Lab. Haematol. 27:15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chong, P. Y., P. Chui, A. E. Ling, T. J. Franks, D. Y. Tai, Y. S. Leo, G. J. Kaw, G. Wansaicheong, K. P. Chan, L. L. Ean Oon, E. S. Teo, K. B. Tan, N. Nakajima, T. Sata, and W. D. Travis. 2004. Analysis of deaths during the severe acute respiratory syndrome (SARS) epidemic in Singapore: challenges in determining a SARS diagnosis. Arch. Pathol. Lab. Med. 128:195-204. [DOI] [PubMed] [Google Scholar]
- 7.Chow, K. Y., Y. S. Yeung, C. C. Hon, F. Zeng, K. M. Law, and F. C. Leung. 2005. Adenovirus-mediated expression of the C-terminal domain of SARS-CoV spike protein is sufficient to induce apoptosis in Vero E6 cells. FEBS Lett. 579:6699-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuconati, A., and E. White. 2002. Viral homologs of BCL-2: role of apoptosis in the regulation of virus infection. Genes Dev. 16:2465-2478. [DOI] [PubMed] [Google Scholar]
- 9.Ding, Y., H. Wang, H. Shen, Z. Li, J. Geng, H. Han, J. Cai, X. Li, W. Kang, D. Weng, Y. Lu, D. Wu, L. He, and K. Yao. 2003. The clinical pathology of severe acute respiratory syndrome (SARS): a report from China. J. Pathol. 200:282-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fielding, B. C., Y. J. Tan, S. Shuo, T. H. Tan, E. E. Ooi, S. G. Lim, W. Hong, and P. Y. Goh. 2004. Characterization of a unique group-specific protein (U122) of the severe acute respiratory syndrome (SARS) coronavirus. J. Virol. 78:7311-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guan, M., H. Y. Chen, P. H. Tan, S. Shen, P. Y. Goh, Y. J. Tan, P. H. Pang, Y. Lu, P. Y. Fong, and D. Chin. 2004. Use of viral lysate antigen combined with recombinant protein in Western immunoblot assay as confirmatory test for serodiagnosis of severe acute respiratory syndrome. Clin. Diagn. Lab. Immunol. 11:1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardwick, J. M., and D. S. Bellows. 2003. Viral versus cellular BCL-2 proteins. Cell Death Differ. 10:S68-S76. [DOI] [PubMed] [Google Scholar]
- 13.Jeong, S. Y., B. Gaume, Y. J. Lee, Y. T. Hsu, S. W. Ryu, S. H. Yoon, and R. J. Youle. 2004. Bcl-x(L) sequesters its C-terminal membrane anchor in soluble, cytosolic homodimers. EMBO J. 23:2146-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaye, M., J. Druce, T. Tran, R. Kostecki, D. Chibo, J. Morris, M. Catton, and C. Birch. 2006. SARS-associated coronavirus replication in cell lines. Emerg. Infect. Dis. 12:128-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan, S., B. C. Fielding, T. H. Tan, C. F. Chou, S. Shen, S. G. Lim, W. Hong, and Y. J. Tan. 2006. Over-expression of severe acute respiratory syndrome coronavirus 3b protein induces both apoptosis and necrosis in Vero E6 cells. Virus Res. 122:20-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, and P. Palese. 2006. 7a protein of severe acute respiratory syndrome coronavirus inhibits cellular protein synthesis and activates p38 mitogen-activated protein kinase. J. Virol. 80:785-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lang, Z. W., L. J. Zhang, S. J. Zhang, X. Meng, J. Q. Li, C. Z. Song, L. Sun, Y. S. Zhou, and D. E. Dwyer. 2003. A clinicopathological study of three cases of severe acute respiratory syndrome (SARS). Pathology 35:526-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Law, P. T., C. H. Wong, T. C. Au, C. P. Chuck, S. K. Kong, P. K. Chan, K. F. To, A. W. Lo, J. Y. Chan, Y. K. Suen, H. Y. Chan, K. P. Fung, M. M. Waye, J. J. Sung, Y. M. Lo, and S. K. Tsui. 2005. The 3a protein of severe acute respiratory syndrome-associated coronavirus induces apoptosis in Vero E6 cells. J. Gen. Virol. 86:1921-1930. [DOI] [PubMed] [Google Scholar]
- 19.Lin, C. W., K. H. Lin, T. H. Hsieh, S. Y. Shiu, and J. Y. Li. 2006. Severe acute respiratory syndrome coronavirus 3C-like protease-induced apoptosis. FEMS Immunol. Med. Microbiol. 46:375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lip, K. M., S. Shen, X. Yang, C. T. Keng, A. Zhang, H. L. Oh, Z. H. Li, L. A. Hwang, C. F. Chou, B. C. Fielding, T. H. Tan, J. Mayrhofer, F. G. Falkner, J. Fu, S. G. Lim, W. Hong, and Y. J. Tan. 2006. Monoclonal antibodies targeting the HR2 domain and the region immediately upstream of the HR2 of the S protein neutralize in vitro infection of severe acute respiratory syndrome coronavirus. J. Virol. 80:941-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minn, A. J., C. S. Kettlun, H. Liang, A. Kelekar, M. G. Vander Heiden, B. S. Chang, S. W. Fesik, M. Fill, and C. B. Thompson. 1999. Bcl-xL regulates apoptosis by heterodimerization-dependent and -independent mechanisms. EMBO J. 18:632-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peiris, J. S., K. Y. Yuen, A. D. Osterhaus, and K. Stohr. 2003. The severe acute respiratory syndrome. N. Engl. J. Med. 349:2431-2441. [DOI] [PubMed] [Google Scholar]
- 23.Polster, B. M., J. Pevsner, and J. M. Hardwick. 2004. Viral Bcl-2 homologs and their role in virus replication and associated diseases. Biochim. Biophys. Acta 1644:211-227. [DOI] [PubMed] [Google Scholar]
- 24.Ren, L., R. Yang, L. Guo, J. Qu, J. Wang, and T. Hung. 2005. Apoptosis induced by the SARS-associated coronavirus in Vero cells is replication-dependent and involves caspase. DNA Cell Biol. 24:496-502. [DOI] [PubMed] [Google Scholar]
- 25.Schinzel, A., T. Kaufmann, and C. Borner. 2004. Bcl-2 family members: integrators of survival and death signals in physiology and pathology. Biochim. Biophys. Acta 1644:95-105. [DOI] [PubMed] [Google Scholar]
- 26.Subbarao, K., and A. Roberts. 2006. Is there an ideal animal model for SARS? Trends Microbiol. 14:299-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surjit, M., B. Liu, S. Jameel, V. T. Chow, and S. K. Lal. 2004. The SARS coronavirus nucleocapsid protein induces actin reorganization and apoptosis in COS-1 cells in the absence of growth factors. Biochem. J. 383:13-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan, Y. J., B. C. Fielding, P. Y. Goh, S. Shen, T. H. Tan, S. G. Lim, and W. Hong. 2004. Overexpression of 7a, a protein specifically encoded by the severe acute respiratory syndrome coronavirus, induces apoptosis via a caspase-dependent pathway. J. Virol. 78:14043-14047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan, Y. J., S. G. Lim, and W. Hong. 2006. Understanding the accessory viral proteins unique to the severe acute respiratory syndrome coronavirus. Antiviral Res. 72:78-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan, Y. J., S. P. Lim, A. E. Ting, P. Y. Goh, Y. H. Tan, S. G. Lim, and W. Hong. 2003. An anti-HIV-1 gp120 antibody expressed as an endocytotic transmembrane protein mediates internalization of HIV-1. Virology 315:80-92. [DOI] [PubMed] [Google Scholar]
- 31.Tan, Y. J., E. Teng, S. Shen, T. H. Tan, P. Y. Goh, B. C. Fielding, E. E. Ooi, H. C. Tan, S. G. Lim, and W. Hong. 2004. A novel SARS coronavirus protein, U274, is transported to the cell surface and undergoes endocytosis. J. Virol. 78:6723-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei, L., S. Sun, C. H. Xu, J. Zhang, Y. Xu, H. Zhu, S. C. Peh, C. Korteweg, M. A. McNutt, and J. Gu. 2007. Pathology of the thyroid in severe acute respiratory syndrome. Hum. Pathol. 38:95-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White, E. 2006. Mechanisms of apoptosis regulation by viral oncogenes in infection and tumorigenesis. Cell Death Differ. 13:1371-1377. [DOI] [PubMed] [Google Scholar]
- 34.Willis, S., C. L. Day, M. G. Hinds, and D. C. Huang. 2003. The Bcl-2-regulated apoptotic pathway. J. Cell Sci. 116:4053-4056. [DOI] [PubMed] [Google Scholar]
- 35.Wong, S. L., Y. Chen, C. M. Chan, C. S. Chan, P. K. Chan, Y. L. Chui, K. P. Fung, M. M. Waye, S. K. Tsui, and H. Y. Chan. 2005. In vivo functional characterization of the SARS-Coronavirus 3a protein in Drosophila. Biochem. Biophys. Res. Commun. 337:720-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan, H., G. Xiao, J. Zhang, Y. Hu, F. Yuan, D. K. Cole, C. Zheng, and G. F. Gao. 2004. SARS coronavirus induces apoptosis in Vero E6 cells. J. Med. Virol. 73:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang, Y., Z. Xiong, S. Zhang, Y. Yan, J. Nguyen, B. Ng, H. Lu, J. Brendese, F. Yang, H. Wang, and X. F. Yang. 2005. Bcl-xL inhibits T cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem. J. 392:135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yount, B., R. S. Roberts, A. C. Sims, D. Deming, M. B. Frieman, J. Sparks, M. R. Denison, N. Davis, and R. S. Baric. 2005. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 79:14909-14922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan, X., Y. Shan, Z. Zhao, J. Chen, and Y. Cong. 2005. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol. J. 2:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao, G., S. Q. Shi, Y. Yang, and J. P. Peng. 2006. M and N proteins of SARS coronavirus induce apoptosis in HPF cells. Cell Biol. Toxicol. 22:313-322. [DOI] [PMC free article] [PubMed] [Google Scholar]