Abstract
Deletion of the terminal repeats (TR) from herpesvirus saimiri (HVS) renders it unable to produce infectious virus or generate plaques. However, a TR-deleted HVS bacterial artificial chromosome can form replication compartments. Complementation of this mutant shows that one copy of the TR, plus the right junction of the genome with the TR, is sufficient for efficient plaque formation and generation of infectious virus. Within the TR unit, the region around the cleavage site of the genome appears both necessary and sufficient for virus production. Analysis of episomes from productive cells indicates a propensity to amplify TR numbers during the lytic cycle.
Herpesvirus saimiri (HVS) is the prototype gamma-2-herpesvirus whose primary host is the squirrel monkey. The linear viral genome of HVS is composed of an AT-rich unique region (L-DNA) encoding 75 open reading frames (ORFs) (1), flanked by a noncoding GC-rich repetitive region (H-DNA) (6). This H-DNA is composed of about 30 copies of the 1.44-kb terminal repeat (TR) unit, randomly distributed between termini by the cleavage that creates the linear genome of the virion (16). This cleavage occurs next to the ApaI restriction site of one of the TR units and leaves a blunt terminus (4, 16). This genome structure is common to other gamma-2-herpesviruses, such as human herpesvirus 8 (Kaposi's sarcoma herpesvirus) and murine herpesvirus 68.
Like other gammaherpesviruses, the HVS genome is a circular episome in latently infected cells (18), replicating and segregating efficiently within a dividing cell population. Maintenance of the gammaherpesvirus episome is mediated by the binding of the latency-associated nuclear antigen (LANA, or ORF73) to the terminal repeats, with three to four repeat units required for episome persistence of a plasmid (2, 3, 7, 8, 11, 17). In order to characterize the TR requirements of a gammaherpesvirus genome, we have used the bacterial artificial chromosome (BAC) containing the A11-S4 strain of HVS (pHVS15+) and its derivative pHVS15+ΔTR, from which the TRs and part of ORF75 have been deleted (19). pHVS15+ efficiently forms plaques upon transfection of permissive owl monkey kidney (OMK) cells, while pHVS15+ΔTR fails to do so (19).
pHVS15+ΔTR is able to form replication compartments.
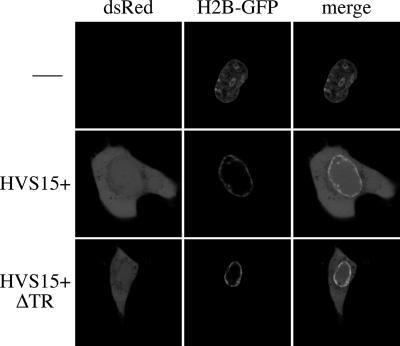
The formation of viral replication compartments in herpes simplex virus forces the host chromatin to the margins of the cell. This redistribution of host chromosomes can be followed by a histone 2B-green fluorescent protein (H2B-GFP) fusion (15). We have observed this phenomenon within OMK cells infected by HVS (not shown) and therefore used H2B-GFP as a surrogate marker for the formation of viral replication compartments. Confocal microscopy of OMK cells cotransfected with either pHVS15+ or pHVS15+ΔTR and an expression plasmid for H2B-GFP (pCLNR-H2BGFP [13]) identified cells that were positive for both peripheral H2B-GFP and the DsRed expressed by the HVS-BAC in both cases (Fig. 1). This indicates that the lytic cycle is essentially functional in pHVS15+ΔTR and that its defect is likely to be at a later stage of virion production, such as packaging, as seen for a TR-deleted Epstein-Barr virus (EBV), which is competent for trans packaging of amplicons (9).
FIG. 1.
pHVS15+ΔTR forms replication compartments. Confocal images of OMK cells cotransfected with pHVS15+, pHVS15+ΔTR, or nothing alongside pCLNR-H2BGFP (an expression vector for the H2B-GFP fusion protein). Two days posttransfection, a subset of cells expressed DsRed (from the viral DNA). Of these, some (typical examples are shown) have relocalized the H2B-GFP to the periphery of the nucleus, indicative of replication compartment formation. For contrast, the cell transfected with H2B-GFP alone was the one that most closely resembled the peripheral localization seen in infected cells; most uninfected cells exhibit a more-diffuse H2B-GFP distribution within the nucleus.
Complementation of pHVS15+ΔTR identifies requirements for packaging.
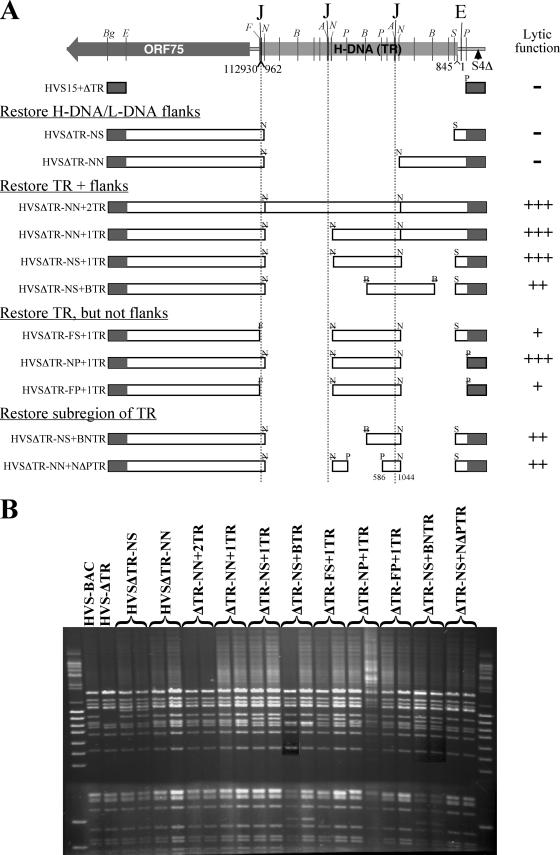
In order to identify the regions responsible for this defect, we have constructed recombinant BACs in order to complement the deletion of the TRs. Recombinants were made by introducing regions of the TRs into pHVS15+ΔTR, using a RecA-based markerless recombination system described previously (14, 19). To control for second-site mutations during the recombination process, it would be ideal to reconstruct wild-type virus for each mutant; however, the large size of the TR makes it impossible to reconstruct these viruses within the current recombination system. For each complementing virus, we have therefore made and tested two independent recombinants, as the best alternative to revertants. The recombinant BACs are shown schematically in Fig. 2A. The nomenclature of the BACs is based on the restriction sites used to subclone the right and then left ends of the HVS genome, along with the number and nature of TR units added. The DNA for subsequent experiments was prepared with a QIAGEN Maxiprep kit and checked for integrity by pulsed-field gel electrophoresis (Fig. 2B).
FIG. 2.
The recombinant BACs used in this study. (A) Schematic representation of the HVS-A11 TR region and the recombinant BACs. Nearly three TR units are shown, although the parental wild-type HVS-BAC has around 30 repeat units. Homology regions through which recombination occurs are indicated by shaded boxes. Enzyme sites used to subclone regions are as follows: Bg, BglII; E, EcoRV; F, FspI; N, NotI; B, BssHII; P, PstI; S, SmaI; and A, ApaI. Where letters are struck through, the enzyme site was destroyed in the cloning process. Where noncompatible enzymes are adjacent in the scheme, or enzyme sites are lost, a short artificial linking sequence is present. Positions within the terminal repeat sequence (GenBank accession no. K03361) are shown for the junctions with the ends of the unique region of the genome (16) and for the smallest region sufficient to mediate packaging. The nucleotide positions of the ends of the L-DNA (GenBank accession no. X64346) are also indicated. “J” represents the sequence to the right of the terminal cleavage site. The left of the cleavage site is indicated by ApaI sites. “E” is the position at which TR-derived sequences cease. “S4Δ” is the site of the ORF1-3 deletion of the A11-S4 strain that was used to make the parental BAC. Nomenclature of the recombinant BACs uses the restriction enzymes employed to subclone regions and the number and nature of TR units where added. The relative lytic activities of the BACs are shown for ease of reference. The ability of the construct to produce virus is indicated by − (unable), +++ (near that of wild type), ++ (slightly retarded), or + (substantially retarded), as described for Fig. 3A. (B) Pulsed-field gel of the BAC DNA preparation used in the study, digested with NheI. For each construct, two independently generated recombinants were made and are shown here to be identical.
To assess the phenotype of each recombinant, 1 μg of DNA for each BAC was transfected in duplicate into a well of a six-well dish of 80% confluent OMK cells using a Lipofectin-peptide transfection reagent (12, 20). The cells were incubated in Dulbecco's modified Eagle medium supplemented with l-glutamine, penicillin and streptomycin, and 2% fetal calf serum (10% fetal calf serum was used to culture OMK cells). The mean times taken to destroy the cell sheets are presented in Fig. 3A.
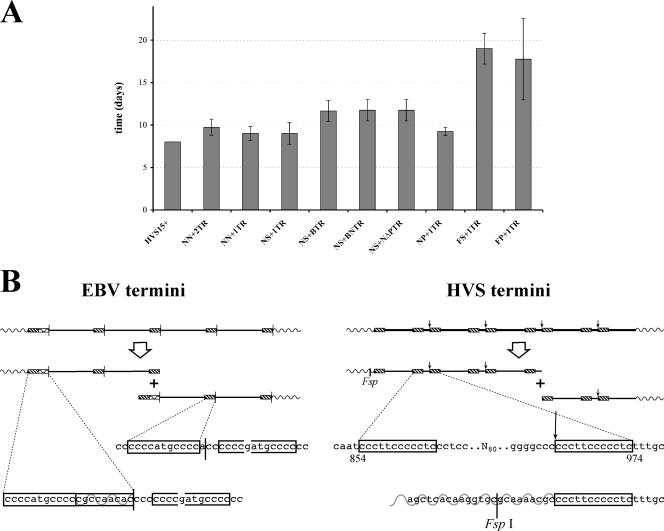
FIG. 3.
(A) Lytic efficiency of recombinant BACs. For each recombinant BAC, one microgram of two independently generated recombinant BACs was transfected in duplicate into OMK cells. The mean time taken to lyse the cell sheet of a well of a six-well plate is shown. Error bars represent one standard deviation above and below the mean. Three recombinant BACs (pHVS15+ΔTR, pHVSΔTR-NS, and pHVSΔTR-NN) failed to form plaques or lyse the cells, so they are not shown here. (B) Schematic and nucleotide resolution scheme comparing HVS and EBV TRs and their cleavage. The EBV genome has been reversed from its conventional orientation so that the EBV and HVS genomes are colinear. Boxes indicate C-rich regions repeated within the TR unit that are adjacent to the cleavage site (vertical arrow/line). The broken box indicates a near repeat of this C-rich region. Wavy lines indicate the unique genome region; solid lines indicate TRs. The FspI restriction site used to delete the right end of the genome is indicated.
Three of the BACs failed to generate plaques. Of these, pHVSΔTR-NN contains almost a full copy of a TR unit, lacking only a 117-bp region immediately upstream of the terminal cleavage site (positions 845 to 961 in the TR as sequenced by Bankier et al. [4]; GenBank accession no. K03361). All of the BACs containing a whole TR unit (or more) were able to produce virus efficiently.
The functional recombinant BACs fall into three groups, taking 9, 12, or 18+ days from transfection to destruction of the cell sheet (Fig. 2A and 3A). The minimum requirement of the BACS closest to wild type in efficiency (pHVSΔTR-NP+1TR) is a single TR (cloned using NotI) and the right end of the L-DNA plus the first 80 bp of the TR. When the TR is subcloned with BssHII (pHVSΔTR-NS+BTR), the productivity of the BAC is slightly impaired. This disrupts the 14-bp element of a set of three tandem sequence duplications (4). Since the BACs containing smaller parts of the TR (pHVSΔTR-NS+BNTR and pHVSΔTR-NS+NΔPTR) have the same functionality, a single functional element that contributes to virus production must lie across the BssHII site. Since pHVSΔTR-NN fails to make virus, we can conclude that the region shared by both of these partial TRs (position 588 to 1044) is both necessary and sufficient for packaging. It contains the site of genome linearization (962/3) but lacks the tandem duplications and the ORF73 binding sites (1118 to 1166) previously identified within the TR (17).
Deletion of the junction at the right end of the L-DNA (pHVSΔTR-FS+1TR and pHVSΔTR-FP+1TR) initially resulted in extremely small and slow-growing plaques, but after a while, some plaques spread much more rapidly and dominated the destruction of the cell sheet. The FspI site used to make this construct is just 8 bp from the H/L-DNA junction (Fig. 3B), so the critical element must be very close to the flank. The leftmost bases of the TR are those that immediately follow the cleavage site within the TR (Fig. 3B). It is tempting to speculate that this half-site plays a role in the cleavage process, but invoking a requirement for a second half-site overlooks the fact that another copy of this sequence exists 100 bp 5′ of the cleavage site itself. If the role of the flank were simply to provide an additional copy of the cleavage half-site, then duplication of the TR would account for the escape from the impairment that is observed. This is consistent with observations for murine gammaherpesvirus 68 amplicons, where a single TR is sufficient for packaging, while two TRs give a higher packaging efficiency (10).
The possible requirement for the right end of the genome offers an interesting comparison with the generation of EBV termini (21). There, an element from the junction of unique sequence and TR at the equivalent end of the genome is duplicated on one terminus of the linear viral DNA. Figure 3B compares the structures of the TRs of HVS and EBV. In both cases, two copies of the C-rich elements at the ‘left’ terminus of the linearized genome are duplicated elsewhere within the repeat unit, albeit with a single base insertion for EBV. Also, the sequence from the junction at the left flank of the TR is important for packaging/linearization, and the C-rich element is repeated here. In EBV, the addition of the element from the TR's left flank onto the terminus invokes a recombination-based model for conversion from a circular genome to a linear one. The termini of HVS appear to have undergone a simple cleavage. However, the parallels between EBV and HVS suggest that there may be some mechanistic similarity between the TR cleavage processes of the two viruses. Using flanking regions might also represent a common mechanism that prevents genome cleavage at multiple sites within the TR.
Multimerization of TRs is a common phenomenon.
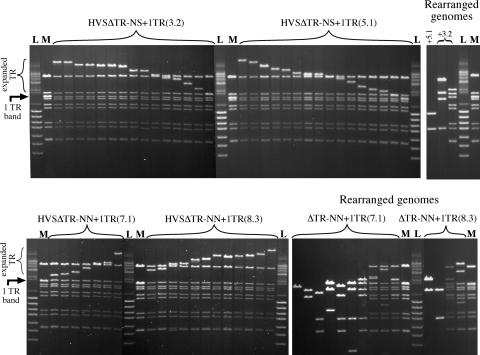
In order to assess the nature of the virus produced by the transfected BACs, equal titers of the duplicates of the two most productive viruses were used to infect OMK cells. After 40 h, the cells were lysed and genomic DNA extracted. One percent of this DNA was electroporated into Electromax DH10B E. coli (Invitrogen), to recover circular BACs. For each virus, eighteen individual colonies were picked, and plasmid DNA was isolated using standard small-scale methods and analyzed by digestion and pulsed-field gel electrophoresis (Fig. 4). Most of the rescued BACs were observed to be intact, with the exception of variations in the size of the TR band—the rescues have been ordered in Fig. 4 to make this more apparent. A minority of BACs have undergone deletions or rearrangements that affect the digestion pattern of the unique region of the genome, as shown to the right of the intact BACs in Fig. 4. The extensive nature of the deletions in some of these BACs suggest that they are nonproductive, although only an oriLyt and a TR would be required for packaging in trans.
FIG. 4.
Analysis of DNA from BACs recovered from OMK cells infected with recombinant virus. DNA was isolated from 18 bacterial colonies for two independent BAC clones of two viruses and digested with EcoRV. The pulsed-field gel of the digests is organized with intact genomes (arranged in order of TR size for clarity) to the left, and BACs that have undergone deletion/rearrangement(s) in regions outside the TR are separated to the right. The original size of the TR band is indicated by an arrow, and the variable sized band containing the TR array is indicated by a bracket. “M” indicates lanes containing the parental BAC. “L” is a DNA size standard, which is a mixture of the ‘lambda mono-cut mix’ and ‘BstEII lambda’ (both from New England Biolabs). The numbers in parentheses after the virus names indicate the identities of the specific BAC clones from which the viruses originated.
Presumably, the tendency to increase TR numbers is a consequence of single-TR BACs being less efficient at packaging, being well below the packaging capacity of the virion. The distribution of repeat numbers at each end of the genome suggests a ‘head-full’ packaging mechanism for the virus, with a fixed size observed for the genome in the virion (16), consistent with studies on EBV-based amplicons, which were packaged most efficiently at the size closest to an apparent maximum of 180 kb (5). This contrasts with rescued episomes, which have no apparent optimal number of repeats. Indeed, rescue of pHVS15+ episomes from infected cells showed a similar variation in TR size. The recircularization of HVS presumably occurs by recombination between TR units on opposite flanks. Unless some mechanism favors repeats nearer the termini, this will naturally result in a variable number of TR units.
The study of the latency requirements of HVS is both complicated and simplified by these observations. The tendency of HVS to change TR numbers makes the establishment of latent episomes with specific TR numbers difficult, preventing us from comparing the viral context with observations that three to four repeat units are sufficient to replicate plasmid vectors (2, 8). However, this also means that a single modified TR unit should produce an equivalent array, simplifying the construction of HVS with a mutant TR array, allowing the analysis of precisely modified TRs in the future.
This study reveals that the HVS terminal repeats are not required for the formation of replication compartments and that the region around the cleavage site of the HVS TRs is both necessary and sufficient for the packaging of the virus genome. Furthermore, the junction between the right end of the unique region of the genome and the left end of the TR also plays an important role in packaging. Comparison of the HVS TR with sequences within the EBV TR leads us to propose some mechanistic similarities between the cleavage of the two virus genomes, despite the different natures of the termini. In addition, we have shown that in the course of lytic replication and genome circularization, single TRs are amplified into multimers.
Acknowledgments
We thank the Wellcome Trust for financial support of this research.
We also thank Teru Kanda for providing pCLNR-H2BGFP and Paul Farrell for helpful comments on the manuscript.
Footnotes
Published ahead of print on 11 April 2007.
REFERENCES
- 1.Albrecht, J. C., J. Nicholas, D. Biller, K. R. Cameron, B. Biesinger, C. Newman, S. Wittmann, M. A. Craxton, H. Coleman, B. Fleckenstein, et al. 1992. Primary structure of the herpesvirus saimiri genome. J. Virol. 66:5047-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., and K. M. A. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 4.Bankier, A. T., W. Dietrich, R. Baer, B. G. Barrell, F. Colbere-Garapin, B. Fleckenstein, and W. Bodemer. 1985. Terminal repetitive sequences in herpesvirus saimiri virion DNA. J. Virol. 55:133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bloss, T. A., and B. Sugden. 1994. Optimal lengths for DNAs encapsidated by Epstein-Barr virus. J. Virol. 68:8217-8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bornkamm, G. W., H. Delius, B. Fleckenstein, F. J. Werner, and C. Mulder. 1976. Structure of herpesvirus saimiri genomes: arrangement of heavy and light sequences in the M genome. J. Virol. 19:154-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calderwood, M., R. E. White, R. A. Griffiths, and A. Whitehouse. 2005. Open reading frame 73 is required for herpesvirus saimiri A11-S4 episomal persistence. J. Gen. Virol. 86:2703-2708. [DOI] [PubMed] [Google Scholar]
- 8.Collins, C. M., M. M. Medveczky, T. Lund, and P. G. Medveczky. 2002. The terminal repeats and latency-associated nuclear antigen of herpesvirus saimiri are essential for episomal persistence of the viral genome. J. Gen. Virol. 83:2269-2278. [DOI] [PubMed] [Google Scholar]
- 9.Delecluse, H. J., D. Pich, T. Hilsendegen, C. Baum, and W. Hammerschmidt. 1999. A first-generation packaging cell line for Epstein-Barr virus-derived vectors. Proc. Natl. Acad. Sci. USA 96:5188-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng, H., J. T. Chu, N. H. Park, and R. Sun. 2004. Identification of cis sequences required for lytic DNA replication and packaging of murine gammaherpesvirus 68. J. Virol. 78:9123-9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fowler, P., S. Marques, J. P. Simas, and S. Efstathiou. 2003. ORF73 of murine herpesvirus-68 is critical for the establishment and maintenance of latency. J. Gen. Virol. 84:3405-3416. [DOI] [PubMed] [Google Scholar]
- 12.Hart, S. L., C. V. Arancibia-Carcamo, M. A. Wolfert, C. Mailhos, N. J. O'Reilly, R. R. Ali, C. Coutelle, A. J. George, R. P. Harbottle, A. M. Knight, D. F. Larkin, R. J. Levinsky, L. W. Seymour, A. J. Thrasher, and C. Kinnon. 1998. Lipid-mediated enhancement of transfection by a nonviral integrin-targeting vector. Hum. Gene Ther. 9:575-585. [DOI] [PubMed] [Google Scholar]
- 13.Kanda, T., K. F. Sullivan, and G. M. Wahl. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8:377-385. [DOI] [PubMed] [Google Scholar]
- 14.Lalioti, M., and J. Heath. 2001. A new method for generating point mutations in bacterial artificial chromosomes by homologous recombination in Escherichia coli. Nucleic Acids Res. 29:E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monier, K., J. C. Armas, S. Etteldorf, P. Ghazal, and K. F. Sullivan. 2000. Annexation of the interchromosomal space during viral infection. Nat. Cell Biol. 2:661-665. [DOI] [PubMed] [Google Scholar]
- 16.Stamminger, T., R. W. Honess, D. F. Young, W. Bodemer, E. D. Blair, and B. Fleckenstein. 1987. Organization of terminal reiterations in the virion DNA of herpesvirus saimiri. J. Gen. Virol. 68:1049-1066. [DOI] [PubMed] [Google Scholar]
- 17.Verma, S. C., and E. S. Robertson. 2003. ORF73 of herpesvirus saimiri strain C488 tethers the viral genome to metaphase chromosomes and binds to cis-acting DNA sequences in the terminal repeats. J. Virol. 77:12494-12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werner, F. J., G. W. Bornkamm, and B. Fleckenstein. 1977. Episomal viral DNA in a herpesvirus saimiri-transformed lymphoid cell line. J. Virol. 22:794-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.White, R. E., M. A. Calderwood, and A. Whitehouse. 2003. Generation and precise modification of a herpesvirus saimiri bacterial artificial chromosome demonstrates that the terminal repeats are required for both virus production and episomal persistence. J. Gen. Virol. 84:3393-3403. [DOI] [PubMed] [Google Scholar]
- 20.White, R. E., R. Wade-Martins, S. L. Hart, J. Frampton, B. Huey, A. Desai-Mehta, K. M. Cerosaletti, P. Concannon, and M. R. James. 2003. Functional delivery of large genomic DNA to human cells with a peptide-lipid vector. J. Gene Med. 5:883-892. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann, J., and W. Hammerschmidt. 1995. Structure and role of the terminal repeats of Epstein-Barr virus in processing and packaging of virion DNA. J. Virol. 69:3147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]