Abstract
The release of human immunodeficiency virus type 1 (HIV-1) and of other retroviruses from certain cells requires the presence of distinct regions in Gag that have been termed late assembly (L) domains. HIV-1 harbors a PTAP-type L domain in the p6 region of Gag that engages an endosomal budding machinery through Tsg101. In addition, an auxiliary L domain near the C terminus of p6 binds to ALIX/AIP1, which functions in the same endosomal sorting pathway as Tsg101. In the present study, we show that the profound release defect of HIV-1 L domain mutants can be completely rescued by increasing the cellular expression levels of ALIX and that this rescue depends on an intact ALIX binding site in p6. Furthermore, the ability of ALIX to rescue viral budding in this system depended on two putative surface-exposed hydrophobic patches on its N-terminal Bro1 domain. One of these patches mediates the interaction between ALIX and the ESCRT-III component CHMP4B, and mutations which disrupt the interaction also abolish the activity of ALIX in viral budding. The ability of ALIX to rescue a PTAP mutant also depends on its C-terminal proline-rich domain (PRD), but not on the binding sites for Tsg101, endophilin, CIN85, or for the newly identified binding partner, CMS, within the PRD. Our data establish that ALIX can have a dramatic effect on HIV-1 release and suggest that the ability to use ALIX may allow HIV-1 to replicate in cells that express only low levels of Tsg101.
The C-terminal p6 domain of the human immunodeficiency virus type 1 (HIV-1) Gag polyprotein harbors a highly conserved P(T/S)APP motif that is critical for virus replication in most cell types (10). In adherent cells such as macrophages, PTAPP mutants exhibit a defect at the level of virus release from the cell surface, whereas in T-cell lines and primary lymphocytes a defect in virion-virion detachment has been noted (10, 14, 17). Functionally equivalent short sequence motifs that facilitate virus budding and release, collectively referred to as late assembly (L) domains, have also been identified in other retroviruses and in several other enveloped RNA viruses. While most lentiviruses have L domains of the PTAP type, the L domain of equine infectious anemia virus (EIAV) maps to a conserved YPxL sequence, and other retroviruses harbor a conserved PPxY motif in Gag that is required for efficient budding (12). Remarkably, L domains do not depend on a particular location within Gag to function and can even be moved between unrelated viruses (37), an observation that suggested that L domains may act as docking sites for cellular factors.
Indeed, it is now clear that the PTAP-type L domain in HIV-1 p6 recruits the host protein Tsg101, which is essential for HIV-1 budding from adherent cells (9, 13, 31, 44). Tsg101 is a component of the ESCRT-I complex, which mediates the sorting of cargo into vesicles that bud form the limiting membrane into the lumen of multivesicular bodies (MVBs) (2, 15, 22). Virus budding at the cell membrane and vesicle formation at the MVB are topologically related, and ESCRT-I could thus have similar roles in both budding events. In yeast, the absence of any of three of the four components of ESCRT-I leads to the accumulation of an aberrant endosome-derived structure called the class E compartment (21). Altogether, at least 17 distinct class E vacuolar protein sorting (Vps) proteins appear to be required for MVB biogenesis and are conserved from yeast to humans (2). Apart from ESCRT-I, class E Vps proteins are the constituents of two additional heteromeric endosomal sorting complexes called ESCRT-II and ESCRT-III. ESCRT-II is activated by ESCRT-I and in turn stimulates the assembly of ESCRT-III on endosomal membranes (3, 4). In humans, ESCRT-III is formed by the charged MVB proteins (CHMPs), which belong to a large family of distantly related coiled-coil proteins (16, 45). At least some of the CHMPs interact with the ATPase Vps4 (30, 45), a class E Vps protein that disassembles the ESCRT complexes for further rounds of sorting (3, 5). Retroviral budding in general exhibits a remarkable sensitivity to mutant versions of Vps4 or of the CHMPs (13, 30, 43, 45, 51), indicating that all types of L domains ultimately engage the MVB sorting machinery.
In addition to the Tsg101 binding site, HIV-1 p6 contains a region that has an auxiliary role in virus release and binds to ALIX/AIP1 (43). ALIX also binds avidly to the YPxL-type L domain of EIAV and is required for EIAV budding (30, 43, 45). Intriguingly, ALIX interacts with Tsg101 through a PSAP motif in its C-terminal proline-rich domain (PRD) (43, 45), which also binds to endophilin, CIN85, and ALG-2 (35). ALIX has been linked to MVB biogenesis on the basis of its affinity for the unconventional phospholipid lysobisphosphatidic acid, which is abundant in the internal membranes of MVBs (32). A role in the MVB pathway is also suggested by the observation that ALIX binds to CHMP4 proteins via its N-terminal Bro1 domain (19, 30, 43, 45), which is further reported to interact with a Rab GTPase-activating protein (18). Bro1, one of the two yeast homologs of ALIX, functions in the MVB pathway and associates with endosomes in a manner that depends specifically on SNF7, the single yeast homolog of the human CHMP4 proteins (36). The other yeast homolog of ALIX is an essential component of a signal transduction pathway that also depends on SNF7 and on Vps23, the Saccharomyces cerevisiae ortholog of Tsg101 (27, 48, 49).
HIV-1 mutants that lack the Tsg101 binding site in p6 exhibit a severe release defect in adherent cells, which implies that the ALIX binding site alone is not sufficient for efficient virus budding. However, we now report that Tsg101 binding site mutants can be efficiently rescued by increasing the cellular expression levels of ALIX. We also show that the binding site for CHMP4B on the Bro1 domain of ALIX is required for this activity, as is a second putative interaction surface on the Bro1 domain. Interestingly, while the binding sites for Tsg101, endophilin, CIN85, and CMS in the PRD are all dispensable, the very C terminus of the PRD is essential and may connect ALIX to an as-yet-unidentified cofactor that is required for its function in virus budding.
MATERIALS AND METHODS
Proviral constructs.
HXBH10, the parental proviral plasmid used in this study, is a vpu-positive version of the infectious HXB2 proviral clone of HIV-1. The ΔPTAPP mutant is a variant of HXBH10 with an in-frame deletion of codons 7 through 11 of p6 (25). The PTAP/LIRL mutant of HXBH10 has a 4-amino-acid substitution of the PTAP L domain in p6 that does not change the coding sequence of the overlapping pol reading frame (17). Similarly, the T8I mutation targets the PTAP motif, and the previously described Y36s mutation (14) introduces a premature termination codon in place of Tyr-36 of p6 without altering the pol frame.
Expression vectors.
The expression vectors for HA-ALIX, HA-ALIXΔPRD, CHMP31-150FLAG, and FLAG-CMS have been described previously (24, 43, 51). An EagI-SmaI fragment encoding ALIX residues 1 to 847 with an N-terminal hemagglutinin (HA) tag was excised from the HA-ALIX vector and subcloned between the NotI and EcoRI sites (filled in with Klenow enzyme) of pBJ5. This yielded an expression vector for HA-ALIX1-847 followed by two unrelated amino acids. An EcoRV site near the 3′ end of the ALIX coding sequence was used to insert premature termination codons directly after ALIX codons 853, 858, and 863 using a PCR-based cloning strategy. ALIX point mutants were made using the QuikChange mutagenesis strategy (Stratagene). The coding sequence for CIN85 with a C-terminal HA tag was amplified from BC015806 (Open Biosystems) and that of endophilin 2 with an N-terminal HA tag from BC001270. The primers incorporated restriction sites that were used to clone the PCR fragments between the XhoI and EcoRI sites of pBJ5. For the expression of glutathione S-transferase (GST)-ALIX PRD fusion proteins, PCR fragments encompassing ALIX residues 714 through 868 or mutant versions of the same region were inserted in frame between the EcoRI and XhoI sites of pCAGGS/GST (30).
Analysis of viral particle production.
293T cells (1.2 × 106) were seeded into T25 flasks and transfected 24 h later using a calcium phosphate precipitation technique. The cultures were transfected with 1 μg of HIV-1 proviral DNA together with expression vectors for wild-type (WT) or mutant HA-ALIX (between 1 and 3 μg) and, where indicated, for CHMP31-150FLAG (0.5 μg) or the appropriate empty vectors. The total amount of transfected DNA was brought to 8 μg with carrier DNA (pTZ18U). Twenty-four hours posttransfection, the cells were lysed in radioimmunoprecipitation assay buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% Nonidet P-40 [NP-40], 0.5% sodium deoxycholate, 0.05% sodium dodecyl sulfate [SDS]), and the culture supernatants were clarified by low-speed centrifugation and passaged through 0.45-μm filters. Virions released into the medium were then pelleted through 20% sucrose cushions and analyzed as described elsewhere (1) by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting with a rabbit anti-HIV CA serum (Advanced Biotechnologies). Proteins in the cell lysates were detected by Western blotting with rabbit anti-HIV CA serum or with anti-HA antibody HA.11 (Covance).
Coimmunoprecipitations.
293T cells (3.5 × 106) were seeded into T80 flasks and cotransfected with expression vectors for FLAG-tagged WT or mutant ALIX and HA-tagged CHMP4B (8 μg each). Twenty-four hours posttransfection, the cells were lysed in NP-40 buffer (0.5% NP-40, 20 mM Tris HCl [pH 7.4], 150 mM NaCl, and protease inhibitor cocktail [Complete; Roche Molecular Biochemicals]). The lysates were centrifuged at 16,000 × g and immunoprecipitated for 2.5 h at 4°C with anti-FLAG antibody M2 (Sigma). Immunoprecipitates and the cell lysates were analyzed by immunoblotting with anti-HA or anti-FLAG antibody as indicated.
GST pull-down assay.
293T cells seeded into T80 flasks were cotransfected with expression vectors for GST- and either HA- or FLAG-tagged proteins (5 μg each) together with 8 μg carrier DNA. Twenty-four hours later the cells were lysed in 0.5% NP-40 buffer, and clarified lysates were incubated with glutathione-Sepharose beads (GE Healthcare) for 2.5 h at 4°C. After extensive washing in NP-40 buffer, bound proteins were eluted by boiling in SDS-PAGE sample buffer and resolved by SDS-PAGE. Epitope-tagged proteins were detected by Western blotting with anti-HA or anti-FLAG M2 antibody, and GST fusion proteins were visualized with colloidal Coomassie brilliant blue G-250.
Protein identification.
A 70-kDa protein pulled down by WT GST-PRD expressed in 293T cells, but not by the PR744AG mutant, was excised from a Coomassie-stained gel and subjected to in-gel tryptic digestion. Extracted peptides were analyzed on a Shimadzu Biotech Axima TOF2 (Shimadzu Instruments) matrix-assisted laser desorption ionization-time-of-flight mass spectrometer. Peptide mass fingerprinting yielded 31 peptides that matched CMS/CD2AP, and collisionally induced dissociation analysis of the six most abundant peptides verified the peptide mass fingerprint identification.
RESULTS
Exogenous ALIX rescues particle production by Tsg101 binding site mutants.
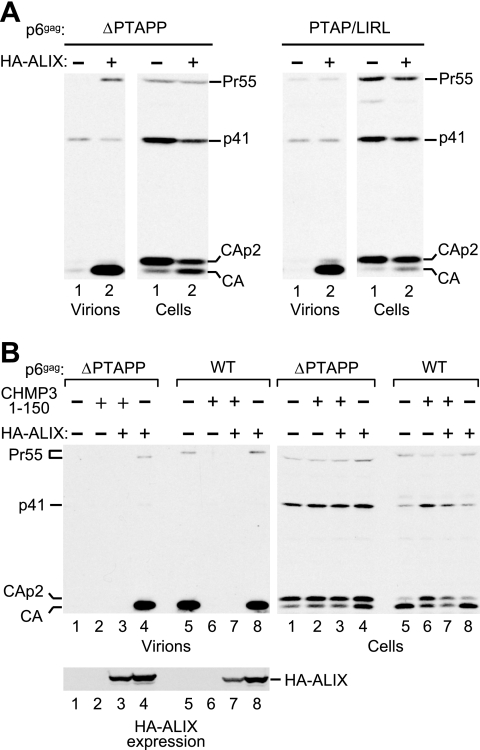
While examining the ability of HIV-1 p6 mutants to replicate in T-cell lines, we noticed that Jurkat cells are partially permissive for viruses that have only the Tsg101 or only the ALIX binding site mutated but are much less permissive for a virus that harbors both mutations simultaneously (data not shown). This observation prompted us to explore the possibility that ALIX, if expressed at sufficiently high levels, can complement viruses defective for Tsg101 binding. To examine this possibility, we first used the previously described ΔPTAPP mutant of the infectious HXBH10 molecular clone, which lacks the entire Tsg101 binding site and exhibits a severe defect in particle release in adherent cells (1). The ΔPTAPP provirus was transfected into 293T cells along with an expression vector for HA-ALIX or the empty vector. Remarkably, the coexpression of HA-ALIX led to a profound increase in viral particle production by the ΔPTAPP mutant (Fig. 1A, left panel). Furthermore, exogenous HA-ALIX at least partially corrected the Gag processing defect of the ΔPTAPP mutant, which is characterized by the accumulation of the cleavage intermediates CA-p2 and p41 and is considered a hallmark of late assembly defects. Exogenous HA-ALIX also profoundly improved particle production by a mutant that had the Tsg101 binding site replaced without changing the overlapping pol reading frame (Fig. 1A, right panel). As previously reported (17), this mutant released essentially no mature CA when cotransfected with the vector control.
FIG. 1.
Rescue of Tsg101 binding site mutants by ALIX. (A) Exogenous ALIX profoundly increases the release of virions by HIV-1 mutants that have the PTAP L domain either deleted or replaced without changing the overlapping pol frame. 293T cells were transfected with mutant HIV-1 proviral DNA (1 μg) and either empty pBJ5 or a version expressing HA-ALIX (3 μg). Virion production and Gag expression levels were compared by Western blotting with anti-CA serum. (B) ALIX increases virion production by ΔPTAPP HIV-1 to WT levels, and its effect is blocked by dominant-negative CHMP3. 293T cells were transfected with HIV-1 proviral DNA (1 μg) together with expression vectors for HA-ALIX (2 μg), and/or CHMP31-150FLAG (0.5 μg), as indicated. Virion production and Gag expression were compared as above, and the expression of tagged ALIX was determined by Western blotting with anti-HA.
A side-by-side comparison of the ΔPTAPP mutant and the WT virus confirmed the dramatic particle release defect of the mutant (Fig. 1B, lanes 1 and 5). Nevertheless, HA-ALIX rescued particle production by the ΔPTAPP mutant to WT levels (lanes 4 and 5). In contrast, HA-ALIX had no effect on the WT virus (lane 8). We also examined whether HA-ALIX suppresses the effect of CHMP31-150FLAG, a truncated version of the ESCRT-III component CHMP3 that blocks HIV-1 budding (Fig. 1B, lane 6), as previously reported (51). However, HA-ALIX rescued neither the WT virus nor the ΔPTAPP mutant in the presence of CHMP31-150FLAG (lanes 3 and 7). We conclude that the dominant-negative effect of CHMP31-150FLAG does not depend on the Tsg101 binding site in p6.
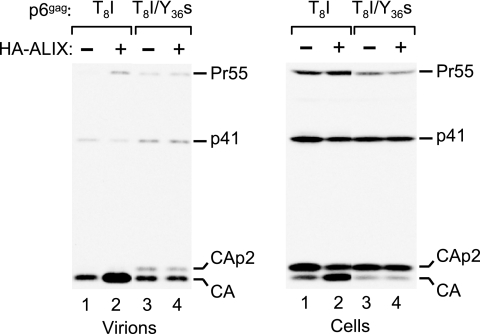
We previously showed that an LYP and an LxxLF motif in a C-terminal region of p6 are required for the interaction with ALIX (43). To determine whether this region is also required for the rescue of a Tsg101 binding site mutant by HA-ALIX, we used a provirus that harbors both a point mutation in the PTAP L domain (T8I) and a premature termination codon (Y36s) near the C terminus of p6 that removes the LYP and LxxLF motifs. While the T8I mutant exhibited a defect in virus release as reported previously (17), at least in 293T cells particle production was somewhat easier to detect than with the ΔPTAPP mutant (Fig. 2), perhaps because the T8I mutation disrupts only part of the Tsg101 binding site (13). In the context of the T8I mutant, the Y36s truncation did not further reduce particle production but caused a moderate accumulation of the CA-p2 cleavage intermediate in virions. Interestingly, while the coexpression of HA-ALIX clearly increased particle production by the T8I mutant and improved the processing of cell-associated Gag, both of these effects were abolished by the Y36s truncation (Fig. 2) and by a 2-amino-acid substitution in the LxxLF motif (LF45PS) that prevents the interaction with ALIX (43) (data not shown). We conclude that the rescue of Tsg101 binding site mutants by HA-ALIX requires the presence of an intact ALIX binding site in p6.
FIG. 2.
The rescue of a Tsg101 binding site mutant by ALIX depends on the ALIX binding site in p6. 293T cells were transfected with the indicated HIV-1 p6 mutants (1 μg) and either empty pBJ5 or a version expressing HA-ALIX (3 μg). Virion production and Gag expression levels were compared by Western blotting with anti-CA serum. The T8I substitution targets the Tsg101 binding site, and the Y36s truncation removes the ALIX binding site.
CHMP4B binding site mutants of ALIX fail to rescue HIV-1 budding.
We and others have shown that ALIX interacts with all three human CHMP4 isoforms (19, 30, 43, 45). However, the interaction with CHMP4B is particularly robust (19). Vps32/SNF7, the single yeast ortholog of the human CHMP4 proteins, binds to the N-terminal Bro1 domain of the ALIX homolog Bro1 (23). The crystal structure of the Bro1 domain of the yeast protein reveals a boomerang-shaped folded core of 367 residues with two exposed hydrophobic patches (23). Interestingly, certain point mutations in patch 1 disrupted the interaction with SNF7, indicating that patch 1 constitutes at least part of the binding site (23).
ALIX shares an N-terminal Bro1 domain with its yeast homolog, and at least some of the residues that make up hydrophobic patch 1 on the Bro1 domain are conserved among the yeast and human proteins. This is particularly evident for Phe-199 and Leu-216 of ALIX, which correspond to patch 1 residues Phe-189 and Leu-209 of Bro1. Additionally, Ile-148, Ala-149, and Leu-213 of ALIX appear to correspond to patch 1 residues Ile-144, Ala-145, and Leu-206 of its yeast homolog.
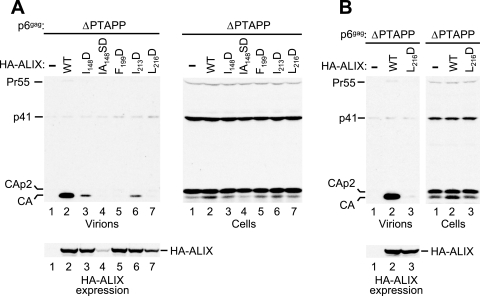
To examine the importance of these putative patch 1 residues for the activity of ALIX in virus budding, we changed each of these residues to Asp. This substitution was chosen because of previous work showing that the replacement of Bro1 residues Ile-144 and Leu-336 by Asp disrupts the interaction with SNF7 (23). All mutations were introduced into N-terminally HA-tagged ALIX to allow a comparison of the expression levels of the WT and mutant proteins. As shown in Fig. 3A, one mutant (IA148SD) was poorly expressed and therefore not analyzed further. The I148D and I213D mutants were expressed at WT levels and retained the ability to rescue particle production by the p6 ΔPTAPP mutant (Fig. 3A, lanes 3 and 6), albeit at a reduced level compared to WT ALIX (lane 2). In contrast, the F199D and L216D mutants showed no activity in this assay (lanes 5 and 7). Whereas the F199D mutation did not affect the expression of ALIX, the L216D mutant was expressed at a slightly lower level than the WT protein. Therefore, we performed an additional experiment in which the amount of vector encoding the L216D mutant was doubled. Although this led to expression levels for the mutant protein that approached those obtained for WT ALIX in the same experiment, the L216D mutant again proved unable to rescue particle production by the ΔPTAPP HIV-1 provirus (Fig. 3B).
FIG. 3.
Effects of mutations targeting surface patch 1 of the ALIX Bro1 domain on the ability of ALIX to rescue virion release by ΔPTAPP HIV-1. (A) 293T cells were transfected with the ΔPTAPP mutant (1 μg) and vectors expressing WT HA-ALIX or the indicated mutants, or empty vector (2 μg). Virion production and the expression levels of Gag and HA-ALIX were monitored by Western blotting. (B) Comparison of the abilities of WT and L216D ALIX to rescue virion release when expressed at similar levels. 293T cells were transfected with the ΔPTAPP mutant (1 μg) and vectors expressing WT HA-ALIX (2 μg), L216D HA-ALIX (4 μg), or empty vector (2 μg).
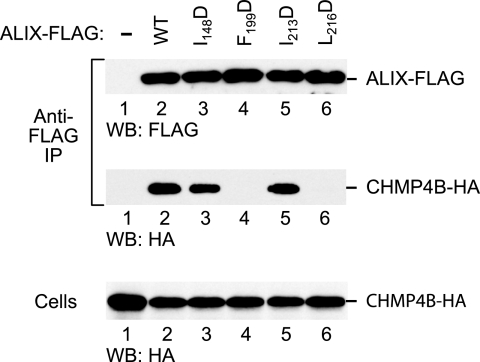
To determine the effects of the patch 1 mutations on the interaction between ALIX and CHMP4B in human cells, we used a previously described coprecipitation assay (43). The I148D, F199D, I213D, and L216D mutations were individually introduced into FLAG-tagged ALIX, and the mutants were coexpressed in 293T cells together with HA-tagged CHMP4B. The cells were then lysed in 0.5% NP-40 buffer, and proteins precipitated from the lysates with anti-FLAG antibody were analyzed by Western blotting. As expected, CHMP4B-HA coprecipitated specifically with WT ALIX-FLAG (Fig. 4, lanes 1 and 2). Interestingly, whereas the I148D and I213D mutations had little effect on the coprecipitation of CHMP4B-HA (lanes 3 and 5), the F199D and L216D mutations abolished the interaction between ALIX and CHMP4B (lanes 4 and 6). Since the F199D and L216D mutations also abolished the rescue of ΔPTAPP HIV-1 by ALIX, these data indicate that the ability to interact with CHMP4B is required for the function of ALIX in viral budding.
FIG. 4.
Effects of mutations targeting surface patch 1 of the ALIX Bro1 domain on the interaction with CHMP4B. 293T cells were cotransfected with vectors expressing WT ALIX-FLAG or the indicated mutants together with CHMP4B-HA. The cell lysates and proteins immunoprecipitated (IP) with anti-FLAG antibody were analyzed by Western blotting (WB) as indicated.
Surface patch 2 is crucial for the function of ALIX in HIV-1 budding.
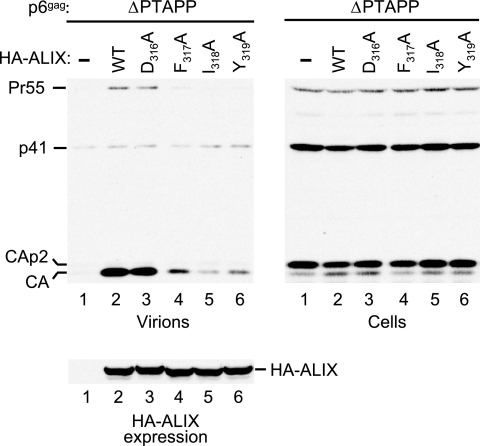
In addition to the binding site for SNF7, the crystal structure of the Bro1 domain revealed a second hydrophobic patch at one tip of the boomerang (23). Because of the striking exposure of several hydrophobic residues in this patch to solvent, it has been proposed that patch 2 is an interaction site for an unknown ligand (23). Interestingly, the three hydrophobic side chains in the patch that are hyperexposed (Phe-318, Ile-319, and Tyr-320) are conserved between yeast Bro1 and human ALIX, where the equivalent residues are Phe-317, Ile-318, and Tyr-319. Furthermore, these three residues are located within a highly conserved sequence context that also includes several charged or polar residues and which represents the most conserved region of ALIX.
To examine the role of this conserved region in the virus release function of ALIX, we individually replaced the three consecutive hydrophobic residues, Phe-317, Ile-318, and Tyr-319, with alanine in the context of HA-tagged ALIX. In addition, one of the polar residues in the conserved region (Asp-316) was changed to alanine. In the crystal structure of yeast Bro1, the side chain of the corresponding residue (Asp-317) is directly adjacent to the hydrophobic side chains that form surface patch 2 (23). As shown in Fig. 5, neither of these mutations affected the expression levels of HA-ALIX in 293T cells. However, only the D316A mutant remained as active as WT ALIX in rescuing the budding defect of ΔPTAPP HIV-1 (Fig. 5, lanes 2 and 3). While one of the hydrophobic patch mutants (F317A) also retained some activity, the other two (I318A and Y319A) were poorly active in this assay, particularly the I318A mutant (Fig. 5, lanes 4 to 6). We thus conclude that hydrophobic patch 2 is critical for the role of ALIX in virus release.
FIG. 5.
Effects of mutations targeting surface patch 2 of the ALIX Bro1 domain on the ability of ALIX to rescue virion release by ΔPTAPP HIV-1. 293T cells were transfected with the ΔPTAPP mutant (1 μg) and vectors expressing WT HA-ALIX or the indicated mutants, or empty vector (1 μg). Virion production and the expression levels of Gag and HA-ALIX were monitored by Western blotting.
Essential role of the very C terminus of ALIX.
In addition to the N-terminal Bro1 domain, all ALIX homologs share a C-terminal PRD which, in the case of human ALIX, spans 152 amino acids. The PRD is thought to be largely unstructured and to contain multiple docking sites for other cellular proteins. Interaction partners that have been identified include Tsg101 (43, 45), CIN85 (8), endophilin (7), and ALG2 (33).
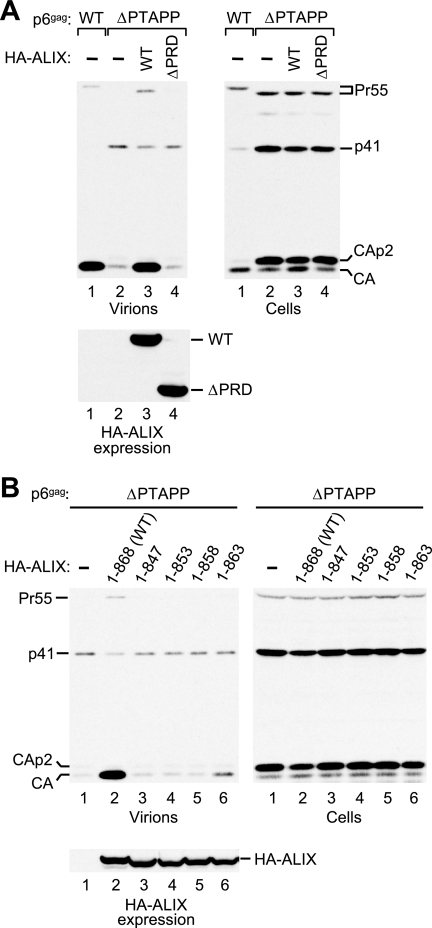
To determine whether the PRD is required for the rescue of ΔPTAPP HIV-1 by ALIX, we first used a previously described C-terminal truncation mutant called HA-ALIXΔPRD, which retains the first 702 residues of ALIX (43). As shown in Fig. 6A, the coexpression of ALIXΔPRD did not at all improve particle production by the ΔPTAPP mutant (lane 4). In marked contrast, WT ALIX expressed at similar levels again rescued particle production to the level obtained with WT HIV-1 (lane 3). Since ALIXΔPRD lacks 14 residues upstream of the PRD, we next tested a series of truncation mutants that lacked only PRD residues. The results of these experiments showed that ALIX1-847 as well as mutants with more extensive deletions into the PRD all had completely lost the ability to rescue the particle release defect of ΔPTAPP HIV-1 (Fig. 6B and data not shown). ALIX1-847 retains the binding site for ALG-2, because it has been shown that ALIX796-847 is sufficient for full binding activity (41). Furthermore, ALIX1-847 retains the reported binding sites for Tsg101, CIN85, and endophilin (7, 26, 40, 45). We thus infer that the presence of binding sites for all of the known PRD interaction partners is not sufficient for the function of the PRD in HIV-1 budding.
FIG. 6.
Effects of ALIX PRD truncations on the rescue of ΔPTAPP HIV-1. (A) The PRD is essential for the rescue of virus release by ALIX. 293T cells were transfected with HIV-1 proviral DNAs (1 μg) and either empty pBJ5 or versions expressing HA-ALIX or HA-ALIXΔPRD (3 μg). Virion production and the expression levels of Gag and HA-ALIX were monitored by Western blotting. (B) The very C terminus of the PRD is required for the activity of ALIX in virus release. 293T cells were transfected with the ΔPTAPP mutant (1 μg) and vectors expressing WT HA-ALIX or the indicated truncation mutants, or empty vector (1 μg).
The inactive ALIX1-847 mutant lacks the 21 C-terminal residues of ALIX. To map the essential C-terminal region in more detail, we generated three additional truncation mutants that lacked only the 15 (ALIX1-853), 10 (ALIX1-858), or 5 (ALIX1-863) C-terminal residues of the PRD. When tested for their ability to rescue virion release by ΔPTAPP HIV-1, ALIX1-853 and ALIX1-858 showed no activity at all in repeated experiments (Fig. 6B). ALIX1-863, which lacks only the sequence YYPQQ at the C terminus of ALIX, increased virus particle production only slightly (Fig. 6B). These data indicate that the very C terminus of the PRD is essential for the ability of ALIX to promote virus budding.
Role of specific protein interaction sites in the ALIX PRD.
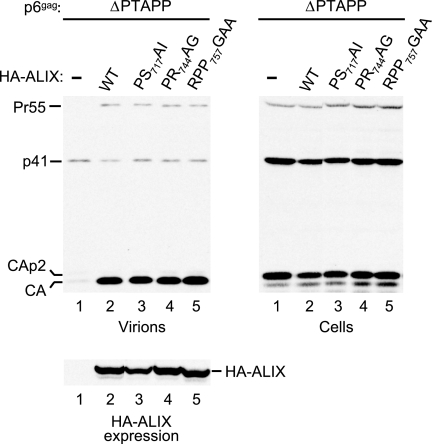
We specifically targeted the interaction sites for Tsg101, CIN85, and endophilin to determine whether these are required to rescue HIV-1 budding in our assay. The PS717AI mutation was designed to disrupt the 717PSAP720 motif at the very beginning of the PRD, which constitutes the Tsg101 binding site of ALIX (45). The PR744AG mutation targets 740PTPAPR745, which conforms to the atypical PxxxPR polyproline-arginine motif recognized by the SH3 domains of CIN85 (26). Lastly, the RPP757GAA mutation disrupts the sequence 754PPARPP759, which matches the consensus binding site for endophilin (28). As shown in Fig. 7, none of these mutations compromised the ability of HA-ALIX to rescue virus particle production by ΔPTAPP HIV-1.
FIG. 7.
The binding sites for Tsg101, CIN85, CMS/CD2AP, and endophilin are dispensable for the ability of ALIX to rescue virus release by ΔPTAPP HIV-1. 293T cells were transfected with the ΔPTAPP mutant (1 μg) and vectors expressing WT HA-ALIX or the indicated PRD mutants (1 μg). Virion production and the expression levels of Gag and HA-ALIX were monitored by Western blotting. PS717AI targets the binding site for Tsg101, PR744AG targets the binding site for CIN85 and CMS/CD2AP, and RPP757GAA targets the binding site for endophilin.
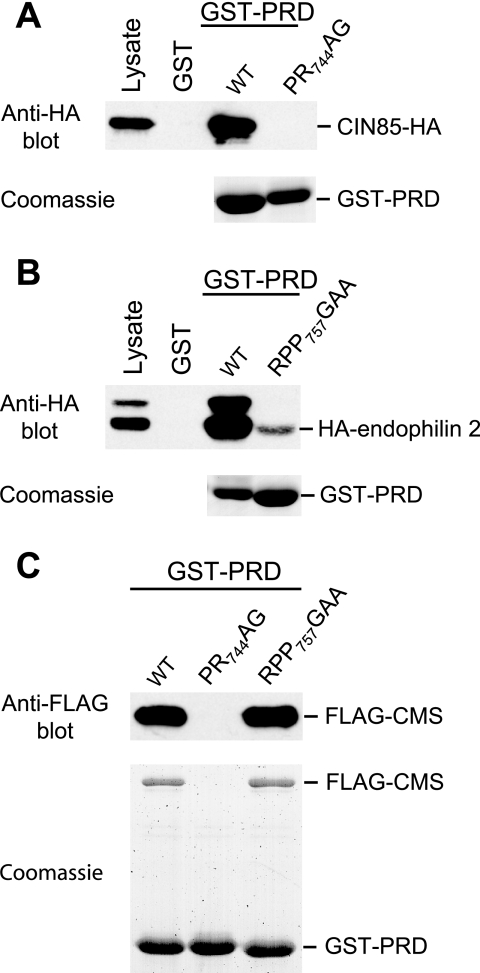
To confirm that the mutations in the PRD affected the interaction with CIN85 or endophilin, we expressed GST-PRD fusion proteins in human 293T cells together with epitope-tagged binding partners and performed GST pull-down assays. As can been seen in Fig. 8A, the PR744AG mutation abolished the interaction with CIN85, as expected. Furthermore, we observed that the RPP757GAA mutation drastically impaired the interaction with endophilin 2 (Fig. 8B), the ubiquitously expressed member of the endophilin family (38).
FIG. 8.
Effects of mutations in the ALIX PRD on the interactions with CIN85, endophilin, and CMS/CD2AP in human cells. (A) The PR744AG mutation abolishes the interaction with CIN85-HA. GST-PRD fusion proteins were expressed in 293T cells together with CIN85-HA, and proteins precipitated from the cell lysates by glutathione-Sepharose beads were analyzed by Western blotting with anti-HA or by Coomassie staining to detect the GST-PRD fusion proteins. (B) GST pull-down assays performed as for panel A, showing that the RPP757GAA mutation severely impairs the interaction with endophilin. (C) GST pull-down assays performed as for panel A, showing that the PR744AG mutation also abolishes the interaction with FLAG-CMS, whereas the RPP757GAA mutation does not affect this interaction.
During the course of these experiments, we noticed that the WT GST-PRD fusion protein specifically coprecipitated a protein of about 70 kDa, and microsequencing unequivocally identified this protein as CMS/CD2AP (data not shown; see Materials and Methods). CMS belongs to the same family of endocytic adaptor proteins as CIN85, and its SH3 domains recognize the same proline-arginine motif as those of CIN85 (34). However, it has been proposed that, in addition to 740PTPAPR745, ALIX may contain a second binding site for CMS that overlaps with the endophilin binding site (752PQPPARP758). As shown in Fig. 8C, the interaction between the ALIX PRD and CMS was abolished by the PR744AG mutation but unaffected by the RPP757GAA mutation, indicating that the PRD contains only one binding site for CMS. Taken together, our results imply that the interactions with Tsg101, CIN85, CMS, and endophilin are all dispensable for the rescue of HIV-1 budding by ALIX.
DISCUSSION
In this study, we have shown that exogenous ALIX can fully correct the profound budding defect of HIV-1 mutants that lack the PTAP-type primary L domain near the N terminus of p6. Exogenous ALIX also alleviated the characteristic Gag processing defect of PTAP mutants, but this effect was less robust and more variable between experiments than the effect on virus release. While the reason for this is not known, it may be that impaired processing of mature CA in particular is a more sensitive indicator of an L domain defect than impaired virus release and, thus, more difficult to correct.
The rescue of virus budding by ALIX was fully dependent on the integrity of the ALIX binding site near the C terminus of p6, confirming the functional relevance of the interaction between ALIX and p6. While in established cell lines the ALIX binding site is less critical for virus release and/or infectivity than the Tsg101 binding site, the contribution of the ALIX binding site would be expected to be more evident in cells that express relatively low levels of Tsg101. Indeed, HIV-1 may have evolved the ability to use both Tsg101 and ALIX to cope with situations in which the expression level of one of these cellular factors alone is insufficient to support efficient virus replication.
Several lines of evidence suggest that HIV-1 budding depends on an intact class E Vps vesicle formation pathway. For instance, HIV-1 budding requires Tsg101 and is strongly inhibited by a defective version of Vps4, the ATPase which recycles class E Vps proteins (13). Furthermore, HIV-1 budding is potently blocked by various dominant-negative versions of the CHMPs, the components of ESCRT-III (30, 45, 51). Nevertheless, it has recently become clear that HIV-1 budding is unaffected by the depletion of a subunit of ESCRT-II (29), which at least in yeast is an essential component of the MVB biogenesis pathway (4). Perhaps even more surprisingly, given the strong inhibitory effect of mutant CHMPs on retrovirus budding, HIV-1 release was also unaffected by the efficient depletion of CHMP5 or CHMP6 (29, 47). The yeast ortholog of CHMP6 is myristylated and, together with the yeast ortholog of CHMP4, forms a subcomplex that is required for the membrane association of ESCRT-III (3). Human CHMP6 is also myristylated and interacts directly with CHMP4B (50), suggesting that it has a similar function in ESCRT-III assembly as its yeast ortholog. The observation that CHMP6 is apparently dispensable for HIV-1 budding thus argues against a requirement for the full ESCRT-III complex.
However, our results now provide evidence for a role of at least one of the components of ESCRT-III in HIV-1 release. ALIX interacts with the ESCRT-III component CHMP4B (30, 43, 45), and this interaction is mediated by the N-terminal Bro1 domain (20). Based on the structure of the Bro1 domain of yeast Bro1, we have mutated the putative binding site and found that point mutations that abolish the interaction with CHMP4B also abolish the function of ALIX in HIV-1 budding. In yeast, the endosomal association of Bro1 specifically depends on the CHMP4 homolog SNF7 (36), and there is also evidence that the recruitment of ALIX to endosomes is mediated by CHMP4 proteins (20). Thus, one possible explanation for our findings is that ALIX must be targeted to endosomes in order to function in viral budding. However, a recent study suggests that ALIX is also found in discrete endosome-like domains of the plasma membrane that are competent for outward vesicle budding and that ALIX and HIV-1 Gag colocalize in these domains (6). Another possibility is that the association of CHMP4B allows ALIX to recruit yet another factor that is required for its function. This scenario is not unprecedented, because an ALIX homolog that acts in the fungal pH response pathway recruits a downstream effector through SNF7 (48).
The structure of the yeast Bro1 domain points to the existence of a second hydrophobic interaction surface (23), and the exceptionally high degree of conservation of the residues that contribute to this surface among ALIX homologs implies functional importance. Our results establish that the three hydrophobic residues at the core of this surface are indeed important for the ability of ALIX to promote HIV-1 budding. It has been reported that one of these residues (Tyr-319) is phosphorylated by the Src tyrosine kinase, which antagonizes the activity of ALIX as a regulator of receptor endocytosis (39). However, we consider it unlikely that a requirement for Tyr-319 phosphorylation explains the effects we observed. First, Ile-318 is at least as important as Tyr-319 for the activity of ALIX in HIV-1 budding but is not part of the consensus sequence for a tyrosine kinase phosphorylation site. Second, one of the residues that does match the consensus (Asp-316) was dispensable for the activity of ALIX in our assay.
ALIX connects to interaction partners not only through its Bro1 domain but also through its C-terminal PRD. Although dispensable for the interaction between ALIX and HIV-1 p6 (43), the PRD nevertheless proved to be essential for the rescue of a Tsg101 binding site mutant in 293T cells. This observation contrasts with our previous finding that the overexpression of ALIXΔPRD in HeLa cells significantly increased virus-like particle production by minimal HIV-1 Gag constructs, even though these lacked the ALIX binding site in p6 (43). However, no rescue was observed if p6 was deleted entirely (43). Thus, consistent with the present study, ALIXΔPRD was unable to rescue a mutant that lacked the Tsg101 binding site.
Endophilin, one of the known interaction partners of the ALIX PRD, is constitutively associated with the adaptor protein CIN85, which independently binds to the ALIX PRD (42). CIN85 also associates with the ubiquitin ligase Cbl and plays a role in the delivery of activated tyrosine kinase receptors to late endosomes (42). In the present study, we additionally identified CMS, an endocytic adaptor protein related to CIN85, as a binding partner for the PRD of ALIX. Intriguingly, endophilin has a BAR domain through which it can generate membrane curvature (11) and is thus a plausible candidate for a factor involved in retrovirus release. Indeed, it has been reported that endophilin 2 specifically interacts with the Gag protein of Moloney murine leukemia virus (46). Furthermore, the overexpression of endophilin selectively inhibited virion production by Moloney murine leukemia virus, and it was therefore suggested that endophilin is hijacked by retroviruses to promote virion production (46). Nevertheless, our results indicate that the interactions of ALIX with endophilin, CIN85, or CMS are all dispensable for the activity of ALIX in HIV-1 budding, as is the interaction with Tsg101. In contrast, the very C terminus of the PRD is essential for the activity, and the identification of a binding partner for this region may shed light on the mechanism by which ALIX promotes virus budding.
Acknowledgments
We thank Paul D. Bieniasz for the pCAGGS/GST vector, Kathrin H. Kirsch for the FLAG-CMS expression vector, Bettina Strack for making the HA-ALIX1-847 expression vector, and John Leszyk for performing the matrix-assisted laser desorption ionization-time-of-flight mass spectrometry.
This work was supported by National Institutes of Health grant AI29873.
Footnotes
Published ahead of print on 11 April 2007.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babst, M. 2005. A protein's final ESCRT. Traffic 6:2-9. [DOI] [PubMed] [Google Scholar]
- 3.Babst, M., D. J. Katzmann, E. J. Estepa-Sabal, T. Meerloo, and S. D. Emr. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell 3:271-282. [DOI] [PubMed] [Google Scholar]
- 4.Babst, M., D. J. Katzmann, W. B. Snyder, B. Wendland, and S. D. Emr. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3:283-289. [DOI] [PubMed] [Google Scholar]
- 5.Babst, M., B. Wendland, E. J. Estepa, and S. D. Emr. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Booth, A. M., Y. Fang, J. K. Fallon, J. M. Yang, J. E. Hildreth, and S. J. Gould. 2006. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J. Cell Biol. 172:923-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatellard-Causse, C., B. Blot, N. Cristina, S. Torch, M. Missotten, and R. Sadoul. 2002. Alix (ALG-2-interacting protein X), a protein involved in apoptosis, binds to endophilins and induces cytoplasmic vacuolization. J. Biol. Chem. 277:29108-29115. [DOI] [PubMed] [Google Scholar]
- 8.Chen, B., S. C. Borinstein, J. Gillis, V. W. Sykes, and O. Bogler. 2000. The glioma-associated protein SETA interacts with AIP1/Alix and ALG-2 and modulates apoptosis in astrocytes. J. Biol. Chem. 275:19275-19281. [DOI] [PubMed] [Google Scholar]
- 9.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farsad, K., N. Ringstad, K. Takei, S. R. Floyd, K. Rose, and P. De Camilli. 2001. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell Biol. 155:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 14.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruenberg, J., and H. Stenmark. 2004. The biogenesis of multivesicular endosomes. Nat. Rev. Mol. Cell Biol. 5:317-323. [DOI] [PubMed] [Google Scholar]
- 16.Howard, T. L., D. R. Stauffer, C. R. Degnin, and S. M. Hollenberg. 2001. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell Sci. 114:2395-2404. [DOI] [PubMed] [Google Scholar]
- 17.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ichioka, F., M. Horii, K. Katoh, Y. Terasawa, H. Shibata, and M. Maki. 2005. Identification of Rab GTPase-activating protein-like protein (RabGAPLP) as a novel Alix/AIP1-interacting protein. Biosci. Biotechnol. Biochem. 69:861-865. [DOI] [PubMed] [Google Scholar]
- 19.Katoh, K., H. Shibata, K. Hatta, and M. Maki. 2004. CHMP4b is a major binding partner of the ALG-2-interacting protein Alix among the three CHMP4 isoforms. Arch. Biochem. Biophys. 421:159-165. [DOI] [PubMed] [Google Scholar]
- 20.Katoh, K., H. Shibata, H. Suzuki, A. Nara, K. Ishidoh, E. Kominami, T. Yoshimori, and M. Maki. 2003. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. J. Biol. Chem. 278:39104-39113. [DOI] [PubMed] [Google Scholar]
- 21.Katzmann, D. J., M. Babst, and S. D. Emr. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106:145-155. [DOI] [PubMed] [Google Scholar]
- 22.Katzmann, D. J., G. Odorizzi, and S. D. Emr. 2002. Receptor downregulation and multivesicular-body sorting. Nat. Rev. Mol. Cell Biol. 3:893-905. [DOI] [PubMed] [Google Scholar]
- 23.Kim, J., S. Sitaraman, A. Hierro, B. M. Beach, G. Odorizzi, and J. H. Hurley. 2005. Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell 8:937-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirsch, K. H., M. M. Georgescu, S. Ishimaru, and H. Hanafusa. 1999. CMS: an adapter molecule involved in cytoskeletal rearrangements. Proc. Natl. Acad. Sci. USA 96:6211-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo, E., and H. G. Gottlinger. 1996. A conserved LXXLF sequence is the major determinant in p6gag required for the incorporation of human immunodeficiency virus type 1 Vpr. J. Virol. 70:159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kowanetz, K., I. Szymkiewicz, K. Haglund, M. Kowanetz, K. Husnjak, J. D. Taylor, P. Soubeyran, U. Engstrom, J. E. Ladbury, and I. Dikic. 2003. Identification of a novel proline-arginine motif involved in CIN85-dependent clustering of Cbl and down-regulation of epidermal growth factor receptors. J. Biol. Chem. 278:39735-39746. [DOI] [PubMed] [Google Scholar]
- 27.Kullas, A. L., M. Li, and D. A. Davis. 2004. Snf7p, a component of the ESCRT-III protein complex, is an upstream member of the RIM101 pathway in Candida albicans. Eukaryot. Cell 3:1609-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landgraf, C., S. Panni, L. Montecchi-Palazzi, L. Castagnoli, J. Schneider-Mergener, R. Volkmer-Engert, and G. Cesareni. 2004. Protein interaction networks by proteome peptide scanning. PLoS Biol. 2:E14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langelier, C., U. K. von Schwedler, R. D. Fisher, I. De Domenico, P. L. White, C. P. Hill, J. Kaplan, D. Ward, and W. I. Sundquist. 2006. Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J. Virol. 80:9465-9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin-Serrano, J., A. Yarovoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 32.Matsuo, H., J. Chevallier, N. Mayran, I. Le Blanc, C. Ferguson, J. Faure, N. S. Blanc, S. Matile, J. Dubochet, R. Sadoul, R. G. Parton, F. Vilbois, and J. Gruenberg. 2004. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303:531-534. [DOI] [PubMed] [Google Scholar]
- 33.Missotten, M., A. Nichols, K. Rieger, and R. Sadoul. 1999. Alix, a novel mouse protein undergoing calcium-dependent interaction with the apoptosis-linked-gene 2 (ALG-2) protein. Cell Death Differ. 6:124-129. [DOI] [PubMed] [Google Scholar]
- 34.Moncalian, G., N. Cardenes, Y. L. Deribe, M. Spinola-Amilibia, I. Dikic, and J. Bravo. 2006. Atypical polyproline recognition by the CMS N-terminal Src homology 3 domain. J. Biol. Chem. 281:38845-38853. [DOI] [PubMed] [Google Scholar]
- 35.Odorizzi, G. 2006. The multiple personalities of Alix. J. Cell Sci. 119:3025-3032. [DOI] [PubMed] [Google Scholar]
- 36.Odorizzi, G., D. J. Katzmann, M. Babst, A. Audhya, and S. D. Emr. 2003. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 116:1893-1903. [DOI] [PubMed] [Google Scholar]
- 37.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ringstad, N., Y. Nemoto, and P. De Camilli. 1997. The SH3p4/Sh3p8/SH3p13 protein family: binding partners for synaptojanin and dynamin via a Grb2-like Src homology 3 domain. Proc. Natl. Acad. Sci. USA 94:8569-8574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt, M. H., I. Dikic, and O. Bogler. 2005. Src phosphorylation of Alix/AIP1 modulates its interaction with binding partners and antagonizes its activities. J. Biol. Chem. 280:3414-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmidt, M. H., D. Hoeller, J. Yu, F. B. Furnari, W. K. Cavenee, I. Dikic, and O. Bogler. 2004. Alix/AIP1 antagonizes epidermal growth factor receptor downregulation by the Cbl-SETA/CIN85 complex. Mol. Cell. Biol. 24:8981-8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shibata, H., K. Yamada, T. Mizuno, C. Yorikawa, H. Takahashi, H. Satoh, Y. Kitaura, and M. Maki. 2004. The penta-EF-hand protein ALG-2 interacts with a region containing PxY repeats in Alix/AIP1, which is required for the subcellular punctate distribution of the amino-terminal truncation form of Alix/AIP1. J. Biochem. (Tokyo) 135:117-128. [DOI] [PubMed] [Google Scholar]
- 42.Soubeyran, P., K. Kowanetz, I. Szymkiewicz, W. Y. Langdon, and I. Dikic. 2002. Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature 416:183-187. [DOI] [PubMed] [Google Scholar]
- 43.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 44.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 46.Wang, M. Q., W. Kim, G. Gao, T. A. Torrey, H. C. Morse III, P. De Camilli, and S. P. Goff. 2003. Endophilins interact with Moloney murine leukemia virus Gag and modulate virion production. J. Biol. 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward, D. M., M. B. Vaughn, S. L. Shiflett, P. L. White, A. L. Pollock, J. Hill, R. Schnegelberger, W. I. Sundquist, and J. Kaplan. 2005. The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J. Biol. Chem. 280:10548-10555. [DOI] [PubMed] [Google Scholar]
- 48.Xu, W., and A. P. Mitchell. 2001. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J. Bacteriol. 183:6917-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, W., F. J. Smith, Jr., R. Subaran, and A. P. Mitchell. 2004. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol. Biol. Cell 15:5528-5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yorikawa, C., H. Shibata, S. Waguri, K. Hatta, M. Horii, K. Katoh, T. Kobayashi, Y. Uchiyama, and M. Maki. 2005. Human CHMP6, a myristoylated ESCRT-III protein, interacts directly with an ESCRT-II component EAP20 and regulates endosomal cargo sorting. Biochem. J. 387:17-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zamborlini, A., Y. Usami, S. R. Radoshitzky, E. Popova, G. Palu, and H. Gottlinger. 2006. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl. Acad. Sci. USA 103:19140-19145. [DOI] [PMC free article] [PubMed] [Google Scholar]