Abstract
Infectious pancreatic necrosis virus (IPNV) is a bisegmented, double-stranded RNA (dsRNA) virus of the Birnaviridae family that causes widespread disease in salmonids. Its two genomic segments are encapsulated together with the viral RNA-dependent RNA polymerase, VP1, and the assumed internal protein, VP3, in a single-shell capsid composed of VP2. Major aspects of the molecular biology of IPNV, such as particle assembly and interference with host macromolecules, are as yet poorly understood. To understand the infection process, analysis of viral protein interactions is of crucial importance. In this study, we focus on the interaction properties of VP3, the suggested key organizer of particle assembly in birnaviruses. By applying the yeast two-hybrid system in combination with coimmunoprecipitation, VP3 was proven to bind to VP1 and to self-associate strongly. In addition, VP3 was shown to specifically bind to dsRNA in a sequence-independent manner by in vitro pull-down experiments. The binding between VP3 and VP1 was not dependent on the presence of dsRNA. Deletion analyses mapped the VP3 self-interaction domain within the 101 N-terminal amino acids and the VP1 interaction domain within the 62 C-terminal amino acids of VP3. The C-terminal end was also crucial but not sufficient for the dsRNA binding capacity of VP3. For VP1, the 90 C-terminal amino acids constituted the only dispensable part for maintaining VP3-binding ability. Kinetic analysis revealed the presence of VP1-VP3 complexes prior to the formation of mature virions in IPNV-infected CHSE-214 cells, which indicates a role in promoting the assembly process.
Infectious pancreatic necrosis virus (IPNV) is the prototype species of the Aquabirnavirus genus and the most common pathogenic microorganism in the aquatic fauna. IPNV causes infectious pancreatic necrosis, an acute and serious disease in juvenile salmonid fish worldwide (19). Control of this disease is of major importance for the aquaculture industry. In addition to high mortality, survivors of the disease may become lifelong carriers, maintaining the virus in the population (34). Due to the ecological importance of IPNV and to the expansion and diversification of aquaculture, much effort has been put into studies of both IPNV biology and the pathogenesis of the disease in recent years (44).
Aquabirnavirus belongs to the Birnaviridae family, which also includes the Infectious bursal disease virus (IBDV), an Avibirnavirus that infects young chickens (11). Most knowledge about the birnaviruses is based on studies of IBDV and IPNV (35, 44). Members of the Birnaviridae family have several unique features in common. Their bisegmented, double-stranded RNA (dsRNA) genome (designated segments A and B) is enclosed in a nonenveloped single-shelled icosahedral particle of 60 to 70 nm in diameter. The major components of the birnavirus particle arise from the proteolytic processing of the 106-kDa polyprotein pVP2-VP4-VP3 encoded by segment A (13, 14). The polyprotein is cotranslationally cleaved by the virus-encoded serine-lysine protease (VP4), releasing proteins pVP2 and VP3 within the infected cell (3, 14). Segment A contains a second, smaller open reading frame in the 5′ end, which encodes the nonstructural protein, VP5 (30). VP1, the viral RNA-dependent RNA polymerase (RdRp) encoded by segment B, is present in the virion in two forms, as a free protein and linked to the 5′ ends of the two genomic segments by covalent binding, the so-called VPg (5, 10, 36). VP1 has been shown to be guanylated in vitro, and this VP1pGpG complex in turn serves as a primer for RNA synthesis (50).
The structures associated with IPNV replication and genome and particle assembly in infected cells have only started to be elucidated. In a recent paper, two types of viral particles of different sizes were identified during the IPNV infective cycle (49). Immediately after synthesis, noninfectious, immature particles 66 nm in diameter appeared. These provirion particles were detected simultaneously with the viral dsRNA in infected cells, suggesting that viral assembly occurs as soon as dsRNA replication has begun. Subsequently, through proteolytic cleavage of the viral precursors within the capsid, the smaller (60 nm in diameter) and mature infectious virions are made. For IBDV, the particle morphogenesis appears to be controlled by VP3, which interacts both with the C-terminal end of the precursor pVP2 and with VP1 (6, 38). Additionally, VP3 of IBDV has been shown to be an RNA-binding protein (27). An early study of IPNV also suggested VP3 to be associated with viral RNA. VP3-containing ribonucleoprotein core structures were identified by electron microscopy studies, and the basic C-terminal end of the protein was proposed to be associated with the viral genome (20).
In this study of IPNV, we focus on the interaction properties of VP3 by employing the yeast two-hybrid system, as well as coimmunoprecipitation (co-IP) and an in vitro dsRNA pull-down assay. In addition to being a self-interacting protein, VP3 was also shown to interact with both VP1 and dsRNA in a mutually independent manner. The domains responsible for these three binding properties of VP3 and for the VP3-binding property of VP1 were further characterized by deletion mapping. In IPNV-infected CHSE-214 cells, VP1-VP3 complexes were detected prior to the formation of mature virus progeny, which indicates a role in the virion assembly process.
MATERIALS AND METHODS
Cell cultures and virus.
Chinook salmon embryo cells (CHSE-214) were grown as monolayers at 20°C, 5.0% CO2 in Eagle minimal essential medium (EMEM) (Invitrogen) supplemented with 100 μg/ml streptomycin, 60 μg/ml penicillin, 2 mM l-glutamine, 1% nonessential amino acids, and 7.5% fetal bovine serum (FBS) (Euroclone). For immunoprecipitation (IP) and Western analyses, CHSE-214 cells were seeded into six-well plates (106 cells/well) or 24-well plates (2 × 105 cells/well), respectively. Prior to infection experiments, cells were grown to 80% confluence.
HEK-293 cells (GP-293; Clonetech) were maintained at 37°C, 5.0% CO2, in EMEM supplemented with 100 μg/ml streptomycin, 60 μg/ml penicillin, 4 mM l-glutamine, and 10% FBS. For transfection, cells were seeded into six-well plates with a density of 106 cells/well and grown to 80 to 90% confluence. Prior to addition of DNA, the cell medium was removed and replaced with 1 ml of serum-free EMEM. Transfection was performed by using the Lipofectamine 2000 (Invitrogen) transfection reagent according to the manufacturer's protocol. For each well, 4 μg of plasmid DNA was incubated with 10 μl Lipofectamine 2000 in 500 μl serum-free EMEM for 20 min at room temperature before being added to the cells. Three hours posttransfection, FBS was added to a total concentration of 2%. The cells were harvested for IP or sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) 18 to 24 h after transfection.
IPNV of the N1 strain, serotype Sp (8, 18), was used in this study. The experiments were performed with a multiplicity of infection (MOI) of 1 or 2 infectious particles/cell in CHSE-214 cells. After absorption of the virus for 2 h in serum-free culture medium, the medium containing virus was carefully removed from the cells. The infection was then carried out at 17.5°C in the presence of 2% FBS and cells harvested at different time points. For propagation of virus, cells were grown in 240-ml culture flasks and infected with virus at a MOI of about 0.001. After 1 week, when a massive cytopathic effect had occurred, the cells were detached and the virus released by freezing and thawing the cell flasks twice. The supernatants were cleared by centrifugation at 1,500 × g for 20 min, divided into aliquots, and stored at −80°C. Virus titers were determined by end-point titration and calculated by the 50% tissue culture infective dose method (42).
Construction of expression vectors for two-hybrid, co-IP, and pull-down analyses.
All PCR primers, cloning vectors, and resulting plasmid constructs are listed in Table 1. The primers were designed for directional insertion of amplified fragments into the pENTR/D-TOPO vector (Invitrogen), with a 5′ CACC sequence in addition to an ATG initiation codon in the forward primer and a stop codon in the reverse primer. The VP2 and VP3 encoding sequences were amplified by PCR from a plasmid containing segment A of IPNV N1 (pETSegA; kindly provided by E. Biering, Intervet Norbio) by using Platinum pfx DNA polymerase (Invitrogen) or Pfu DNA polymerase (Stratagene). cDNA synthesized from IPNV-infected cells served as a template for PCR amplification of VP1. Constructs were verified by DNA sequencing using BigDye chemistry and a 3100 gene analyzer (Applied Biosciences). Once in pENTR/D-TOPO, the VP1, VP2, and VP3 inserts and their truncated derivatives were transferred to eukaryotic expression vectors (as shown in Table 1) by Gateway recombination using LR Clonase II enzyme mix (Invitrogen), following the manufacturer's instructions. For yeast two-hybrid analysis, modified Clontech vectors (pGADT7 and pGBKT7) were used. The vectors (kindly provided by O. M. Seternes, University of Tromsø) were made Gateway compatible by insertion of the Gateway polylinker region as described previously (28) and are named pGal4AD and pGal4DBD, respectively, in Table 1. Recombination into pGal4DBD required an intermediate cloning step with the pDONOR207 vector (Invitrogen). Control plasmids pTD1-1, pGBKT7-53, and pGBKT7-Lam were purchased from Clontech. For IP or dsRNA binding analysis, inserts were transferred to Gateway-compatible vectors introducing N-terminal tags, the 4-kDa hemagglutinin (HA) epitope, the 4-kDa c-Myc tag (both kindly provided by T. Lamark, University of Tromsø), or the 25-kDa glutathione S-transferase (GST) protein (pDEST27; Invitrogen), to the expressed proteins.
TABLE 1.
Primers and plasmid constructs used for expression of IPNV fusion proteins
| Construct | Presence of expression vectora
|
Cloning primers | Primer sequences 5′-3′ | ||||
|---|---|---|---|---|---|---|---|
| pGal4AD | pGal4DBD | pDEST-HA | pDEST-myc | pDEST27 | |||
| pVP1 | x | x | x | x | VP1fw | CACCATGTCGGACATCTTCAATTCACCTCAG | |
| VP1rev1 | TCAGTTTCTTCTCTGCTTCTCCCGACG | ||||||
| pPreVP2 | x | x | x | x | VP2fw | CACCATGAACACAAACAAGGCAACCGCAACT | |
| VP2rev1 | CTATGGAAACAGCGTGGACAGTACAGGAGC | ||||||
| pVP3 | x | x | x | x | VP3fw | CACCATGGACGAGGAACTGCAACGCCTC | |
| VP3rev1 | TTACACCTCAGCGTTGTCTCCGCTGGGGG | ||||||
| pVP1ΔN40 | x | VP1ΔN40F | CACCATGCCGTTAGATAGCCCCCAGGCTG | ||||
| VP1rev1 | TCAGTTTCTTCTCTGCTTCTCCCGACG | ||||||
| pVP1ΔN120 | x | VP1ΔN120F | CACCATGGGGGCTCCAGACATAACACTA | ||||
| VP1rev1 | TCAGTTTCTTCTCTGCTTCTCCCGACG | ||||||
| pVP1ΔN220 | x | VP1ΔN220F | CACCATGCCAAGCTGGTTGATACAGTACAC | ||||
| VP1rev1 | TCAGTTTCTTCTCTGCTTCTCCCGACG | ||||||
| pVP1ΔC90 | x | VP1fw | CACCATGTCGGACATCTTCAATTCACCTCAG | ||||
| VP1ΔC90R | CTACTAGGATGTTGACGCTGCGGGTTTTA | ||||||
| pVP1ΔC135 | x | VP1fw | CACCATGTCGGACATCTTCAATTCACCTCAG | ||||
| VP1ΔC135R | CTATTATACCTCGGCGAGGGTTTTGCCTG | ||||||
| pVP1ΔC179 | x | VP1fw | CACCATGTCGGACATCTTCAATTCACCTCAG | ||||
| VP1ΔC179R | TTATTATCCGAACTCCGACATGGTGTTCC | ||||||
| pVP1ΔC234 | x | VP1fw | CACCATGTCGGACATCTTCAATTCACCTCAG | ||||
| VP1ΔC234R | TTATTATATTTGGTATGCGATCTCGGCC | ||||||
| pVP1ΔN40,C90 | x | VP1ΔN40F | CACCATGCCGTTAGATAGCCCCCAGGCTG | ||||
| VP1ΔC90R | CTACTAGGATGTTGACGCTGCGGGTTTTA | ||||||
| pVP3ΔC56 | x | x | x | VP3fw | CACCATGGACGAGGAACTGCAACGCCTC | ||
| VP3ΔC56R | TTATTAGTTCTGTGCGAATACAGCTGCA | ||||||
| pVP3ΔC135 | x | x | VP3fw | CACCATGGACGAGGAACTGCAACGCCTC | |||
| VP3ΔC135R | TTATTATTCCGGTGTGGCGAAGTCCGCC | ||||||
| pVP3ΔN42 | x | x | VP3ΔN42F | CACCATGTACGAGTGGTCAAAAGAG | |||
| VP3rev1 | TTACACCTCAGCGTTGTCTCCGCTGGGGG | ||||||
| pVP3ΔN81 | x | VP3ΔN81F | CACCATGAGAGCCACCCGCATATCACTGGA | ||||
| VP3rev1 | TTACACCTCAGCGTTGTCTCCGCTGGGGG | ||||||
| pVP3ΔN107 | x | VP3ΔN107F | CACCATGTACCGCGGCCCATCTCCCGG | ||||
| VP3rev1 | TTACACCTCAGCGTTGTCTCCGCTGGGGG | ||||||
| pVP3ΔN174 | x | x | x | VP3ΔN174F | CACCATGGTATTCGCACAGAACGGAGGCAGA | ||
| VP3rev1 | TTACACCTCAGCGTTGTCTCCGCTGGGGG | ||||||
| pVP3ΔN225 | x | VP3ΔN225F | CACCATGTTCACCCCCAGCGGAGACAACGCTGAGGTGTAA | ||||
| VP3ΔN225R | TTACACCTCAGCGTTGTCTCCGCTGGGGGTGAACAT | ||||||
| pVP3ΔN42,C43 | x | VP3ΔN42F | CACCATGTACGAGTGGTCAAAAGAG | ||||
| VP3ΔC43R | TTACTAGAGGTCTTGCATTTGGTCCTG | ||||||
x, vector is present.
Two-hybrid analysis.
Both rich and selective yeast growth media were made from commercially available powders (BD Biosciences Clonetech). Yeast cells were grown at 30°C for 2 to 4 days. Plasmid constructs based on the pGal4DBD vector or the pGal4AD vector were transformed, using the Frozen-EZ Yeast Transformation II kit (Zymo Research), into competent yeast cells of strain Saccharomyces cerevisiae Y187 (MATα) or PJ69-2A (MATa) by selection for growth on medium lacking leucine or tryptophan, respectively. At least 10 (each) transformants carrying VP1, VP2, or VP3 were mated to each other in pairwise combinations, while truncated mutants of VP1 were mated with VP3 and truncated mutants of VP3 were mated with VP3 or VP1 as shown in Fig. 5. Ten diploid yeast cells from each mating were plated and scored for growth on a triple dropout medium (TDO) lacking leucine, tryptophan, and histidine or a quadruple dropout medium (QDO) lacking leucine, tryptophan, histidine, and adenine. Growth on TDO plates indicates a weak interaction, whereas growth on QDO plates indicates a stronger interaction. Simian virus 40 (SV40) T-antigen (pTD1-1) and p53 (pGBKT7-53) served as positive controls, Lamin C (pGBKT7-Lam) and empty vectors as negative controls. Growth was quantified with the Oxygen BioSensor assay (BD Biosciences Clontech) after 50 h in liquid TDO medium. Growth was recorded as fluorescence intensity. The maximum fluorescence intensity was established with Na2SO3, a strong reducing agent, and set to equal a fivefold increase. The n-fold increase of the interactions was then estimated by dividing the final florescence readout by the number that equalled onefold. Additionally, for some of the interactions, a β-galactosidase assay was carried out with liquid cultures using an o-nitrophenyl β-galactopyranoside substrate according to BD Biosciences Clontech's protocol.
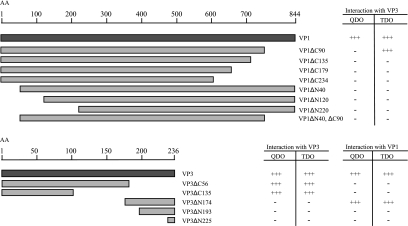
FIG. 5.
Deletion mapping of the VP1-VP3 and VP3-VP3 interaction domains using the yeast two-hybrid system. The figure shows an overview of the VP1 and VP3 deletion mutants cloned into the pGAl4AD yeast expression vector. All VP1 deletion mutants were tested for interaction with full-length VP3. All VP3 deletion mutants were tested for their ability to interact with both full-length VP1 and full-length VP3. Interactions were assayed for histidine autotrophy (TDO) and for histidine and adenine autotrophy (QDO). +++, massive growth, i.e., a strong interaction; −, no growth (no interaction). All results shown are representative of at least 10 independent transformants. AA, amino acids.
Gel electrophoresis, Western blotting, and antibodies.
Protein samples were boiled in SDS sample buffer (160 mM Tris-HCl [pH 6.8], 10% β-mercaptoethanol, 2% SDS, 20% glycerol, 0.1% bromophenol blue) for 5 min, quickly centrifuged, and then subjected to SDS-PAGE and Western blotting using the Invitrogen NuPAGE system. Precast 4 to 12% gradient NuPAGE Novex Bis-Tris gels were used with morpholineethanesulfonic acid buffer. Gel electrophoresis, blotting, blocking, and antibody incubation were performed as described by the manufacturer. Primary antibodies are described below. Goat anti-rabbit horseradish peroxidase antibody or goat anti-mouse horseradish peroxidase antibody (Santa Cruz Biotechnology) diluted 1:25,000 was used as a secondary antibody. Detection was performed by using the SuperSignal West Pico chemiluminescent substrate (Pierce Biotechnology, Inc.). Stripping of the membranes was performed in 0.2 M NaOH for 10 min, followed by washing, blocking, and new antibody incubation.
Polyclonal rabbit antibodies against VP1 (named αVP1) and VP3 (named αVP3*) and monoclonal antibodies against VP3 (named αVP3) and VP2 (named αVP2) (both kindly provided by K. E. Christie, Intervet Norbio), HA (named αHA), and GST (named αGST), (Santa Cruz Biotechnology) were used for IP and Western blot analysis. Polyclonal αVP3* antiserum was generated against a C-terminal fragment (amino acids 102 to 234) of N1 VP3. The recombinant VP3 fragment was expressed in the Escherichia coli BL21 strain with an N-terminal six-His tag and purified by affinity chromatography to a nickel-nitrilotriacetic acid column, following the QIAexpress protocol for large-scale purification of insoluble proteins (QIAGEN). The purified protein fraction was dialyzed against phosphate-buffered saline, identified on gel by Coomassie staining and Western blot analysis, and then submitted for production of polyclonal antibody following the Eurogentec Rabbit Standard Protocol. The resulting antiserum showed specificity in Western blot analysis against tagged VP3 proteins expressed in transfected cells. The specificity was also tested by IP of IPNV-infected cell lysates, followed by gel purification and liquid chromatography-tandem mass spectrometry, confirming that the antibody specifically precipitated VP3. Polyclonal antibodies against VP1 were prepared by Eurogentec (Belgium) using the peptides RGWTNEDGSPRYNPT and TQKKNAKRREKQRRN. The two conjugated peptides were pooled and injected into two rabbits according to Eurogentec's Double XP procedure. The resulting antisera were purified by affinity chromatography using the respective peptides. The specificities of the purified immunoglobulin Gs was tested by Western blot analysis against tagged VP1 proteins expressed in transfected cells.
Radiolabeling and autoradiography.
For in vivo labeling of proteins, confluent monolayers of CHSE-214 cells were infected with IPNV strain N1 at a MOI of 1 or 2 or mock infected. At different time points (8, 12, 14, 16, 18, 24, 36, or 48 h) postinfection (p.i.), cells were starved of methionine and cysteine for 1 h in EMEM medium lacking these amino acids (Gibco/BRL) and pulse-labeled for 3 h with 20 to 50 μCi/ml l-[35S]methionine/cysteine (Pro-mix L-[35S] in vitro cell labeling mix; Amersham) in methionine- and cysteine-free EMEM medium. For in vitro translation of proteins, T7 polymerase-driven expression was carried out by using the TNT coupled reticulocyte lysate system (Promega) in the presence of 1 μCi/ml l-[35S]methionine/cysteine as described by the manufacturer. pDEST-HA vectors (0.4 μg of each) expressing VP1, VP2, VP3, or truncated versions of VP3 (VP3ΔN42, VP3ΔN81, VP3ΔN107, VP3ΔN174, VP3ΔC56, or VP3ΔN42,ΔC43) and plasmid pEQ843 (0.4 μg) expressing E3L (kindly provided by A. P. Geballe, Fred Hutchinson Cancer Research Center, Seattle, WA) were used as templates. Radiolabeled protein samples were analyzed by SDS-PAGE and visualized by autoradiography. Gels were dried in a gel dryer prior to exposure on Kodak BioMax MS film overnight.
Co-IP and GST pull-down analyses.
Cells were washed two times with ice-cold phosphate-buffered saline and harvested in HA lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2 mM EDTA, 1 mM EGTA, 1% Triton X-100) with a protease inhibitor cocktail added (Complete EDTA-free; Roche). Cell lysates were incubated on ice for 15 min and cleared by centrifugation for 15 min at 18,000 × g in a microcentrofuge. Lysates were then subjected to IP with either αVP1 or αVP3 together with preblocked Protein A/G PLUS-Agarose beads (Santa Cruz biotechnology) or with an αHA affinity matrix (Roche). In a similar manner, gluthatione Sepharose 4B (GE Healthcare) was used to pull down GST fusion proteins from the transfected cell lysates. The Sepharose beads were then washed four times with HA lysis buffer and all traces of buffer removed with a pipette tip before elution in 50 μl 2× SDS sample buffer. Eluted proteins were subjected to SDS-PAGE and visualized by Western blotting or by autoradiography.
dsRNA-agarose binding assay.
dsRNA-agarose beads were prepared by covalent binding of polyinosinic poly(C) [poly(I-C); Sigma] to adipic acid dihydrazide agarose (Sigma). Poly(I-C) (2.5 mg/ml) in 0.1 M sodium acetate (NaOAc) (pH 5.0) was incubated with 0.1 M NaIO4 for 1 h in the dark, precipitated with ethyl alcohol, and resuspended in 0.1 M NaOAc at 2.5 mg/ml. Agarose [0.6 ml/mg poly(I-C)], preequilibrated in NaOAc, was incubated with poly(I-C) for 3 h at 4°C with rotation. Binding was terminated by addition of NaCl (final concentration, 2 M), followed by incubation for another 30 min. The agarose beads were then washed several times in buffer A (100 mM KCl, 20 mM HEPES [pH 7.5], 10% glycerol [vol/vol], 5 mM magnesium acetate, 1 mM dithiothreitol [Sigma], and 1 mM benzamidine [Sigma]) and resuspended in an equal volume of buffer A for storage at 4°C. For each binding assay, 200 μl of the 50% slurry was used. The dsRNA pull-down assay was performed as described by Hakki and Geballe (17). Radiolabeled protein samples were added to dsRNA agarose beads resuspended in 200 μl buffer A and 50 μl buffer B (120 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 10% glycerol, 1 mM benzamidine, and 1% NP-40 [Calbiochem]), followed by incubation on a rotator at 4°C for 1 h. The beads were then washed twice in buffer C (identical to buffer A, except 200 mM KCl) and twice in buffer A. Each wash was followed by centrifugation at 200 × g at 4°C for 2 min. Traces of buffer were removed by a pipette tip after the last washing step. Bound proteins were eluted by boiling the agarose beads in 25 μl 2× SDS sample buffer for 4 min and analyzed by SDS-PAGE gel electrophoresis followed by autoradiography.
RESULTS
Examination of VP3 interactions with VP1, VP2, and VP3 in the yeast two-hybrid system.
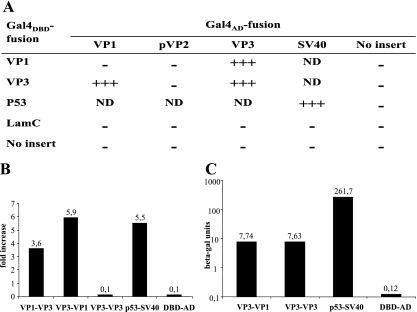
VP3 has been suggested to be the key organizer of the birnavirus assembly process, since it maintains critical interactions with the other viral components (47). The interaction between VP3 and VP1 has been assigned a crucial role in IBDV morphogenesis (31) and in viral replication (47). To explore the interaction properties of IPNV VP3, sequences encoding full-length VP3, VP1, and pVP2 (shown in Fig. 1) were cloned into yeast two-hybrid vectors to assay protein-protein interactions by coexpressing the fusion proteins in pairwise combinations in diploid Saccharomyces cerevisiae (15). Interacting proteins were assessed by the ability for growth on complete minimal medium deficient in histidine (TDO) or histidine and adenine (QDO), and the results are shown in Fig. 2A. Strong interactions were found between VP3 and VP1 and also between VP3 and VP3. Although VP2 and VP3 are the constituents of the IPNV capsid, no interaction was detected between them. Growth was recorded at day 4, when the positive control expressing p53 fused to Gal4DBD and SV40 T antigen fused to Gal4AD showed massive growth. The specificity of all interactions was tested and confirmed by coexpressing the Gal4AD fusion proteins both with Gal4DBD alone and with Gal4DBD fused to the human Lamin C protein, which is known to be a noninteracting protein (1, 23). The positive interactions were further evaluated by fluorometric quantification of yeast growth (MATCHMAKER Oxygen Biosensor kit; Clontech). The results are presented in Fig. 2B as n-fold increases in fluorescence resulting from yeast growth in liquid TDO medium. The fluorescence was recorded when the positive control conferred a more than fivefold increase and the negative controls, the Gal4DBD protein coexpressed with empty Gal4AD vector, gave a 0.1-fold increase. The interaction between VP1 and VP3 resulted in a more than threefold increase in fluorescence independent of whether VP1 or VP3 was expressed as a Gal4DBD or Gal4AD fusion protein. Surprisingly, no VP3-VP3 interaction was detected by the fluorescence assay (0.1-fold increase). But the strong interaction between VP3 and VP3 was confirmed when a β-galactosidase assay utilizing the lacZ reporter gene was employed, as shown in Fig. 2C. Expression of all fusion proteins was verified in yeast by Western blot analysis (data not shown).
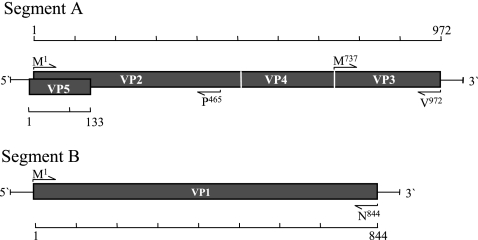
FIG. 1.
Schematic presentation of proteins encoded by the two genomic segments of IPNV. Segment A contains two partially overlapping open reading frames encoding the polyprotein (pVP2-VP4-VP3) and the smaller VP5 protein. Segment B encodes VP1, the RNA-dependent polymerase. Sequences of primers used for generating the different cloned constructs are listed in Table 1. Positions of the first and the last amino acids encoded by each cloned fragment are indicated next to the PCR primers (arrows).
FIG. 2.
IPNV protein interactions in yeast two-hybrid system. (A) Growth was scored after 4 days, when the positive control showed massive growth on medium lacking histidine and adenine (QDO). +++, massive growth (strong interaction); −, no growth (no interaction); ND, not determined. All results shown are representative for at least 2 independent experiments, and at least 10 independent transformants were tested in each experiment. The strength of the positive interactions was measured by (B) quantifying the yeast growth as n-fold increase in fluorescence, where fluorescence was measured after 50 h of growth in liquid medium lacking histidine (TDO) by using the Oxygen BioSensor assay (BD Biosciences Clontech) or by (C) determining expression of the lacZ reporter gene as β-galactosidase (beta-gal) units. As a positive control for interaction in all assays, SV40 T antigen fused to DNA-binding domain (DBD) and P53 fused to activation domain (AD) were applied, while DBD expressed together with AD served as a negative control. In addition, Lamin C fused to DBD expressed together with AD served as a negative control in panel A.
Verification of VP3-VP3 and VP1-VP3 protein interactions by IP and pull-down assays.
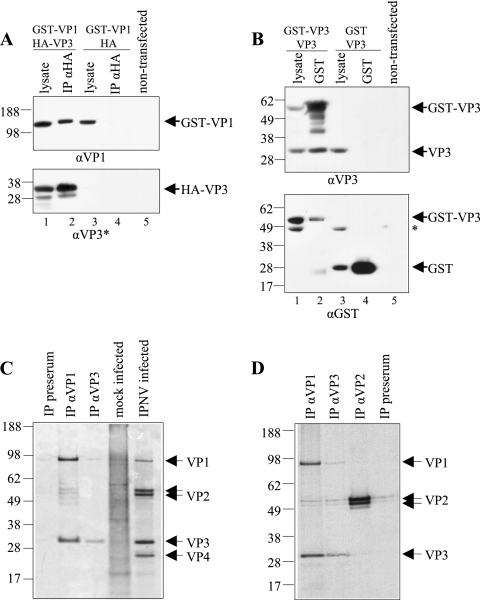
As shown in Fig. 3A, GST-VP1 was coimmunoprecipitated with HA-VP3 using an anti-HA affinity matrix when coexpressed in HEK-293 cells. The precipitates were analyzed by Western blotting, first using αVP1 as the primary antibody (upper panel), followed by stripping and detection with a VP3-antibody (αVP3*) (lower panel). GST-VP1 was detected at the expected 110 kDa. HA-VP3 was detected as a double band around 30 to 34 kDa, which may represent a full-length form and a degraded form of the protein. A reverse experiment using the tagged VP1 and VP3 constructs was performed and gave the same results when using the anti-VP1 antibody for IP (results not shown). A control was included in order to exclude the possibility of cross-reactivity of the tags. When lysate from cells coexpressing GST-VP1 and HA was subjected to IP with anti-HA, no VP1 reactive protein was precipitated (lane 4). These results verify that the interaction between VP1 and VP3 is specific and independent of the tags.
FIG. 3.
Co-IP and pull-down analyses of the VP1-VP3 and VP3-VP3 interactions. (A) GST-VP1 was coexpressed with HA-VP3 or unfused HA (negative control) in HEK-293 cells and the lysed cells subjected to IP with an anti-HA affinity matrix. Samples were analyzed along with the total cell lysate by SDS-PAGE followed by Western blotting using a VP1 antibody (αVP1) and subsequently, after stripping, a polyclonal VP3 antiserum (αVP3*). (B) GST-VP3 was coexpressed with VP3 or unfused GST (negative control) in HEK-293 cells and the lysed cells subjected to gluthathione Sepharose pull-down. Samples were analyzed along with the total cell lysate by SDS-PAGE followed by Western blotting using a monoclonal VP3 antibody (αVP3) and subsequently, after stripping, αGST. The GST-VP3 and GST proteins were detected in cell lysates (lanes 1 and 3, respectively) and correspondingly after GST pull-down (lanes 2 and 4, respectively). An additional band (asterisk) is observed in the cell lysates. (C) Radioimmunoprecipitation analysis of the VP1-VP3 interaction in IPNV-infected CHSE-214 cells. At 48 h p.i., cells were metabolically labeled with [35S]methionine/cysteine for 3 h prior to harvesting. The cell lysates were subjected to IP with monoclonal VP3 (αVP3) or polyclonal VP1 (αVP1) antibody or with rabbit preserum as indicated. The precipitated proteins were analyzed along with lysates of mock-infected and IPNV-infected cells by SDS-PAGE and autoradiography. (D) IP was performed with a monoclonal VP2 antibody (αVP2) and the resulting immunoprecipitates analyzed along with immunoprecipitates from αVP1, αVP3, or preserum as indicated. Positions of the viral proteins and sizes of marker proteins (in kDa) are indicated.
Figure 3B shows that VP3 was pulled down together with GST-VP3 from lysate of cotransfected HEK-293 cells by gluthatione Sepharose. The protein complexes were analyzed by Western blotting, first applying monoclonal αVP3 (upper panel). Two protein bands, corresponding to GST-VP3 (54 kDa) and VP3 (30 kDa), were detected in the cell lysate (lane 1) and after GST pull-down (lane 2). Additional bands reacting with the VP3 antibody were detected in lane 2 which may represent degraded forms of GST-VP3. When VP3 was coexpressed with GST (in the absence of GST-VP3), no VP3 reactive bands were pulled down by gluthatione Sepharose, showing that GST-VP3 specifically interacts with VP3 through VP3 and that the interaction is not mediated by the protein tags. To show that GST is expressed in the transfected cells, the membrane was stripped and reincubated with αGST (lower panel). The GST-VP3 and GST proteins were detected in cell lysates (lanes 1 and 3, respectively) and correspondingly after GST pull-down (lanes 2 and 4, respectively). An additional band was observed in the cell lysates. This protein may represent an endogenous protein with cross-reactivity to the GST antibody, but it is not pulled down by the glutathione Sepharose.
In vivo verification of the VP1-VP3 interaction in IPNV-infected cells.
To obtain evidence that the VP1-VP3 interaction also takes place in vivo, IPNV-infected CHSE-214 cells were analyzed by a radioimmunoprecipitation assay. IPNV-infected and mock-infected cells were metabolically labeled at 48 h p.i. with [35S]methionine/cysteine. The proteins in the cleared cell lysates were subjected to IP and analyzed by SDS-PAGE, followed by autoradiography, along with total lysates from mock-infected and IPNV-infected cells, as shown in Fig. 3C. Metabolically labeled proteins corresponding to the sizes of VP1, VP2 (two bands), VP3, and VP4 present in the IPNV-infected cells are indicated in the figure. VP3 and VP1 were detected in the precipitates independently of whether IP was performed with αVP1 or αVP3 (as indicated above the lanes), clearly showing that both antibodies coimmunoprecipitated the other protein. When using αVP1 for IP, two additional protein bands, corresponding to the sizes of the two VP2 forms, appeared. To test whether VP2 was pulled down along with the VP3-VP1 complex as a result of specific complex formation, an additional IP using αVP2 was performed. In the resulting autoradiography, presented in Fig. 3D, the two bands corresponding to the size of pVP2/VP2 could be detected. VP3 and VP1 were detected in the precipitates solely when αVP1 or αVP3 was used for IP. The IP performed with rabbit preserum resulted in two weak bands, corresponding to VP2 (Fig. 3D), which indicates that VP2 does bind to other proteins or to the agarose beads. Such unspecific binding properties can be explained by the abundance of VP2, constituting 62% of the IPNV particle (12). No other viral proteins were precipitated by the preserum negative control (Fig. 3C and D). The identities of the precipitated proteins were confirmed by the presence of corresponding protein bands in the total lysates of IPNV-infected cells, as indicated in Fig. 3C.
Kinetic analysis of the in vivo VP1-VP3 protein interactions in IPNV-infected cells.
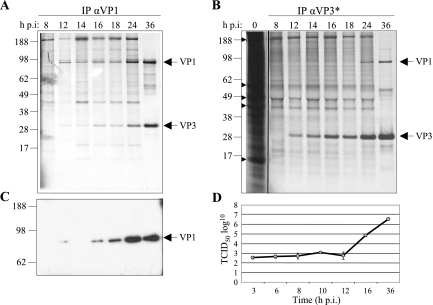
The kinetics of the VP1-VP3 protein association was studied in lysates of IPNV-infected, metabolically labeled CHSE-214 cells. Antibodies specific for either VP1 or VP3 were used to coimmunoprecipitate the two proteins at 8, 12, 14, 16, 18, 24, and 36 h p.i. When analyzed by SDS-PAGE followed by autoradiography (Fig. 4A), bands corresponding to both VP1 and VP3 appeared at 12 h p.i. and increased in strength until 36 h p.i. when precipitating with αVP1. When precipitating with αVP3 (Fig. 4B), the complexes were detectable somewhat later, at 24 h p.i. To establish the time point for the appearance of mature virus progeny, supernatants derived from the virus-infected cells at 3, 6, 8, 10, 12, 16, and 36 h p.i. were harvested and their viral titers estimated. The virus titration curve in Fig. 4D shows that no viral progenies were detectable until 12 h p.i., while at 16 h p.i., there was an increase in viral titers. Accordingly, we detect the VP1-VP3 interaction before the first infectious IPNV particles are produced, although it cannot be ruled out that intracellular infectious virions exist at earlier time points.
FIG. 4.
Kinetics of in vivo VP1-VP3 interaction. CHSE-214 cells infected with IPNV were metabolically labeled with [35S]methionine/cysteine for 3 h and harvested at 8, 12, 14, 16, 18, and 36 h p.i. IP was performed with a VP1 antibody (αVP1) (A) or with polyclonal VP3 antiserum (αVP3*) (B). Arrowheads point to highly abundant proteins in mock-infected cell lysates. (C) Western blot analysis of immunoprecipitates using αVP1 as the primary antibody. IP was preformed on cell lysates harvested in parallel to the experiments in A and B by using αVP1. Positions of viral proteins and sizes of marker proteins (in kDa) are indicated. (D) Virus titers were determined in cell supernatants from infected CHSE-214 cells at 3, 6, 8, 10, 12, 16, and 36 h p.i. The estimated 50% tissue culture infective dose is shown as the average for three parallel wells of infected cells. Three independent experiments were performed with similar results.
Earlier studies have shown that IPNV possesses two forms of VP1, a free polypeptide and a genome-linked protein (VPg) (5). In our study we observed that although some proteins in the high-molecular-mass region (>95 kDa) were immunoprecipitated by use of αVP1 at the early time points (Fig. 4A), a single VP1-specific band of approximately 95 kDa was confirmed by Western blot analysis with αVP1 (Fig. 4C). The specificity of the VP3-precipitated bands was also confirmed in a corresponding manner (data not shown). The additional bands observed in the co-IP experiment (Fig. 4A and B) may correspond to highly abundant cellular proteins, since these are also seen in the mock-infected cells (Fig. 4B) and probably do not represent proteins that interact with VP1 or VP3, since they disappear later in the infection.
Deletion mapping of the VP3- and VP1-interacting domains by yeast two-hybrid analysis.
To map the domains involved in the interactions between VP3 and VP1 and between VP3 and VP3, eight different deletion mutants were generated from the VP1 protein and five from the VP3 protein (Fig. 5). All of the deletion mutants were expressed by the Gal4AD fusion vector. The VP1 deletion mutants were tested for their capability to interact with VP3 expressed from pGal4DBD, while the VP3 deletion mutants were tested both for their capability to interact with Gal4DBD-VP1 and Gal4DBD-VP3 by scoring of growth on TDO and QDO. As indicated in Fig. 5, a VP3 deletion mutant (VP3ΔC135) lacking the 135 C-terminal amino acids was fully capable of binding to VP3, which maps the VP3 self-interaction domain within the 101 N-terminal amino acids. No growth on TDO or QDO medium demonstrates that neither mutant VP3ΔN225 nor VP3ΔN193, consisting of an 11- or a 43-amino-acid C-terminal peptide, respectively, was able to interact with VP1. The VP3ΔN174 mutant and VP1 showed a strong interaction and thus mapped the interaction domain to be within the 62 C-terminal amino acids of VP3. All VP1 mutants, except for VP1ΔC90, scored negative in both the QDO and TDO growth tests when mated with VP3. This mutant, having a C-terminal deletion of 90 amino acids, scored positive on TDO, which indicates a weak interaction with VP3. The results for VP1 imply that the overall three-dimensional structure of the VP1 protein is crucial for the VP1-VP3 interaction. The possibility of transactivation of the reporter genes was ruled out by testing all clones for reporter gene activation when expressed either alone or together with the control plasmids Gal4DBD-Lamin C or Gal4DBD-empty as described above (data not shown). Consistent results were obtained when using the oxygen biosensor assay described above (data not shown).
VP3 expressed in vitro or in IPNV infections binds dsRNA.
In a previous study carried out by Hjalmarsson et al. (20), ribonucleoprotein complexes were isolated from disrupted IPNV virions. By serological methods and electron microscopy, these complexes were shown to contain VP3 and RNA. However, it was not determined whether VP3 was directly bound to RNA molecules or to VP1 in complex with dsRNA, since VP1 is known to bind both genomic segments to form VPg (5). VP3-RNA complexes were also purified from supernatants of IPNV-infected cell cultures, although the nature of the RNA binding was not determined (21).
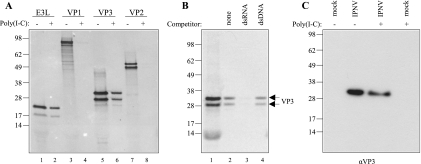
Here we analyzed the dsRNA binding properties of VP3 by using an in vitro dsRNA pull-down assay previously used to characterize dsRNA binding proteins (17). Correspondingly, the E3L protein from the vaccinia virus, which is known as a dsRNA-binding protein, was used as a positive control (24). Along with VP3, we also tested the dsRNA binding properties of VP1 and included VP2, which is reported not to bind RNA, as a negative control. The dsRNA pull-down assay was performed by incubating radiolabeled proteins with poly(I-C) agarose beads. As seen in Fig. 6A, in vitro translation of VP1, VP3, and VP2 yielded proteins of approximately 95 kDa, 30/34 kDa, and 52/56 kDa, respectively, consistent with their predicted sizes (lanes 3, 5, and 7). Double bands appeared, probably due to initiation of translation from two in-frame AUG codons (in the N-terminal tag and in the virus-encoded mRNA). For E3L, two proteins of about 20 and 25 kDa (lane 1) were detected, which is in accordance with previous observations (17). Both forms of the VP3 protein were able to bind to poly(I-C) agarose (lane 6), as did the positive control (lane 2). No binding to dsRNA was detected for VP1 (lane 4), nor for VP2 (lane 8), which validates that the conditions used in the dsRNA binding experiments did not favor nonspecific bindings.
FIG. 6.
dsRNA binding activity of VP3. The binding capacity of in vitro- or in vivo-synthesized, [35S]methionine/cysteine-labeled VP3 was tested by pull-down analysis. (A) In vitro-translated E3L, VP1, VP3, and VP2 pulled down with poly(I-C) agarose beads (+) were analyzed by SDS-PAGE and autoradiography along with the corresponding translation reaction products (−). (B) The specificity of VP3 binding to dsRNA was tested by incubating the translated protein with 200 μg of free dsRNA [poly(I-C)] or free dsDNA (salmon sperm DNA) for 30 min prior to dsRNA pull-down (lane 3 and 4, respectively). In lane 2, no competitor was added to the poly(I-C) beads. Samples were run along with the VP3 translation reaction products (lane 1). (C) Cell lysates of IPNV-infected (24 h p.i.) or mock-infected CHSE-214 cells were subjected to dsRNA pull-down. Precipitated proteins (+) were analyzed along with the total lysates (−) by SDS-PAGE followed by Western blotting using the monoclonal VP3 antibody (αVP3). Molecular size marker is indicated (in kDa).
Next, the specificity of the observed dsRNA binding capacity of VP3 was assessed by performing a competition experiment. In vitro-translated VP3 was incubated with 200 μg of free dsRNA [poly(I-C)]) (approximately the same amount as covalently bound to agarose) or free double-stranded DNA (dsDNA) (calf thymus DNA) for 30 min prior to incubation with dsRNA conjugated to agarose beads. As shown in Fig. 6B, binding of VP3 to dsRNA-agarose (lane 2) was prevented by preincubation with an excess amount of dsRNA (lane 3) but not when a corresponding amount of dsDNA was added as a competitor (lane 4). These results show that in vitro-translated VP3 binds dsRNA but not double-stranded DNA, indicating that VP3 binds preferentially to dsRNA in a sequence-independent manner. Furthermore, the binding took place in the absence of other viral proteins.
In order to determine whether VP3 expressed in vivo by viral infection also could bind to dsRNA, the dsRNA pull-down assay was applied on cleared cell lysates of CHSE-214 cells infected for 24 h with IPNV- or mock-infected cells. As seen in Fig. 6C, VP3 was detected in lysates from infected CHSE-214 cells but not from mock-infected cells by Western blot analysis using a polyclonal anti-VP3 antiserum. After a dsRNA pull-down assay, VP3 appeared in the agarose fraction, demonstrating the dsRNA binding capacity of in vivo-synthesized VP3. No other protein bands were detected in the cell lysates or in agarose-bound fractions, which indicates a specific detection of VP3. The same result was obtained by using a monoclonal antibody against VP3 (data not shown). Thus, VP3 expressed either by in vitro translation or in virus-infected cells binds to dsRNA in vitro.
Deletion mapping of the dsRNA binding domain (dsRBD) of VP3.
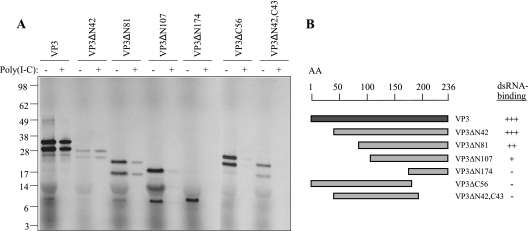
Inspection of the VP3 amino acid sequence did not reveal regions of homology to known dsRNA binding domains of other dsRNA binding proteins. Previously it was suggested that the positively charged C-terminal part of VP3 is the part of the protein that associates with the genomic segments of IPNV (20). For IBDV VP3, on the other hand, an internal N-terminal domain has been reported to be sufficient for dsRNA binding (27). In order to define the regions of IPNV VP3 responsible for dsRNA binding, we made different VP3 deletion mutants lacking one or both of the suggested amino acid regions and assessed the dsRNA binding properties of each in vitro-translated mutant by using the dsRNA pull-down assay. As shown in Fig. 7A, the presence of proteins in the RNA-bound fractions was analyzed, along with the corresponding in vitro-translated truncated VP3 protein samples. As noted earlier, double bands appeared, probably due to initiation of translation at AUG codons both in the N-terminal tag and in the VP3 mRNA. The results of this experiment are summarized in Fig. 7B. When truncated at the N-terminal end, a mutant lacking 42 amino acids was capable of binding dsRNA, while increasing the truncation to 81, 107, or 174 amino acids N terminally resulted in a successive reduction and loss in the dsRNA binding capacity. Truncation of 56 amino acids at the C-terminal end resulted in complete loss of dsRNA binding capacity. The same result was obtained with a mutant lacking 43 C-terminal amino acids, in addition to the nonessential 42 N-terminal amino acids. In the absence of the 43 C-terminal amino acids, no binding take place, nor is a peptide consisting of the 62 C-terminal amino acids (VP3ΔN174) capable of binding. Taken together, these results show that neither the N-terminal nor the C-terminal part of VP3 is sufficient for dsRNA binding. Furthermore, the influence of the N-terminal domain appears to be dependent on the size of the truncation, where increased truncation results in gradual loss of binding capacity.
FIG. 7.
Mapping of the dsRNA binding domain of VP3: dsRNA pull-down analysis of truncated VP3 proteins. (A) In vitro-translated, [35S]methionine/cysteine-labeled VP3 mutants were pulled down with poly(I-C) agarose beads (+) and analyzed by SDS-PAGE and autoradiography along with 1 μl of the corresponding translation reaction product (−) as indicated. (B) Schematic drawing of truncated VP3 proteins that were tested for their dsRNA binding capacity. +++, strong binding; ++, intermediate binding; +, weak binding; −, no binding.
DISCUSSION
Knowledge about the nature and significance of the interactions between the viral components during the IPNV life cycle is still limited. Studies of IBDV have revealed that VP3 is a multifunctional viral component, exhibiting several activities throughout the viral life cycle (6, 7, 31, 32, 38). Although VP3 probably possesses an equally important role for IPNV, little is yet known about the interactions between VP3 and other viral components of IPNV. The elucidation of such interactions was therefore the main focus in this study. Here we report a strong interaction between the viral RdRp VP1 and VP3, detected by several approaches. In accordance with the results for IBDV (47), our data propose that the interaction between VP3 and VP1 was independent of the presence of dsRNA. Furthermore, the VP3 protein was also shown to interact with itself and with dsRNA, which together implies that VP3 is an internal protein with several roles in organizing the IPNV replication cycle.
The C terminus of VP3 interacts with VP1, while the overall structure of VP1 is important for binding to VP3.
The VP1-interacting domain of VP3, which encompasses the 62 C-terminal residues, is characterized by a sequence rich in proline and charged amino acids. Due to its hydrophilic features, this region is predicted to be well exposed, hence having the potential to bind proteins and nucleic acids. For IBDV, Tacken et al. (47) showed that the 10 C-terminal residues were sufficient for binding to VP1. Maraver et al. (31) reported in a later study that the 5 C-terminal residues of IBDV VP3 did not bind VP1, while the last 16 amino acids were sufficient for binding. In our work, not even 43 C-terminal amino acids were sufficient for interaction with VP1. The divergence between the two viruses in this region of the protein (11 of 43 amino acids are different) may offer an explanation for this.
Also, for VP1, the binding domain responsible for VP3 interaction appeared to differ between the two viruses. For IBDV, an internal core domain of VP1 was reported to be the region responsible for VP3 binding (47). In our work, all of the N-terminal and C-terminal VP1 deletion mutants, except for VP1ΔC90, which showed a weak interaction with VP3, tested negative, indicating that neither the N-terminal end nor the C-terminal end of VP1 is dispensable for VP3 binding. The deletions introduced in VP1 may have affected the folding of the molecule, which indeed will have an impact on the ability of a protein to interact with other proteins.
VP1 and VP3 form complexes in the absence of dsRNA.
As reported earlier for IBDV (31, 47), both VP1 and VP3 interact with viral dsRNA independently of each other, hence raising the possibility that the interaction between the proteins is mediated by dsRNA. Our data show that the VP1-VP3 interaction takes place in the absence of dsRNA, both when the two proteins are expressed during transient cotransfection and in the yeast-two hybrid system. RNase treatments of in vivo-derived VP1-VP3 immune complexes strengthened this interpretation by not causing degradation of these complexes (data not shown). These results strongly suggest that VP1-VP3 complexes are formed by direct protein-protein interactions.
VP1-VP3 complexes are detected before the formation of mature virus particles, which suggests a role in the assembly process.
The presence of cytoplasmic VP1-VP3 complexes was detected 12 h p.i. These proteins were found to associate shortly after their translation. At 16 h p.i., viral progeny was detectable, indicating that complex formation takes place before viral particles appear and thus plays a role in viral assembly. In a recent study of IPNV, assembly was shown to occur simultaneously with RNA replication, implying a temporal and spatial coordination between these events (49). These authors propose a continuous model where the morphogenesis of IPNV is completed when viral precursor proteins have been cleaved within the immature capsid, leading to particle maturation and formation of infectious icosahedral virions.
The observation that only a small amount of VP1 was coprecipitated by the antibodies against VP3 (Fig. 3C and D and 4B) may be a consequence of the efficiency of antibody binding or radioactive amino acid incorporation or may be due to an unequal stoichiometric relationship between VP1 and VP3. VP3 is reported to constitute 30% of the IPNV particle, whereas VP1 constitutes only 4% (12).
The N-terminal domain of VP3 is responsible for the VP3-VP3 interactions, while no VP3-VP2 binding was found.
Data from IBDV suggest that VP3 is a scaffolding protein that contributes to the bending of the pVP2/VP2 capsomers into the icosahedral capsid (33). As shown here, IPNV VP3 also shares a common feature of scaffolding proteins in its ability to establish self-interactions. No interaction was detected between VP3 and VP2 either by applying the yeast two-hybrid analysis or by co-IP analysis of both transfected (data not shown) and infected cells. This is in accordance with results reported for IBDV (47). Still, a possible weak interaction between VP3 and the C-terminal domain of pVP2, based on coexpression in insect cells that leads to colocalization, has been pointed out (38). The crystal structure of IBDV VP2 was recently determined (9). Fitting the atomic model of the capsid protein into the virion electron density map revealed that VP2 is the only component of the virion capsid. Surprisingly, no VP3 was detected in the outer shell (40). The fact that birnavirus VP2 and VP3 proteins have a different structural arrangement in the capsid suggests only a weak affinity and no direct interaction between them.
VP3 binds to dsRNA in a sequence-independent manner in the absence of other factors.
We demonstrate that VP3 binds specifically to dsRNA in the absence of VP1 or other viral proteins. For IBDV, a direct interaction between VP3 and dsRNA has been reported. Tacken et al. (47) showed that VP3 interacts with both viral genome segments. In competition experiments, we showed that VP3 favors binding to dsRNA over dsDNA, which is a common feature for RNA binding proteins. Although differing in their dsRBD motifs (48), they typically interact with the ribose 2′-OH groups of RNA (2), thereby conferring specificity for RNA over DNA. In the study by Tacken et al. (47), formation of VP3-dsRNA complexes took place in the absence of the VP1-interacting domain of VP3. The ability of IBDV VP3 to bind RNA has been further confirmed by Kochan et al. (27), who demonstrated that VP3 binds RNA in the absence of other virus-encoded proteins in vitro.
The ability of binding to synthetic dsRNA, poly(I-C), reveals that VP3 binds dsRNA in a sequence-independent manner. In contrast, VP1 was not capable of binding to poly(I-C). This is in accordance with the presumption that VP1 recognizes specific sequences at the ends of the two genomic sequences (5). The lack of sequence specificity has also been reported for IBDV VP3 when single-stranded RNA probes were applied for in vitro binding assays (27). This property is proven for several virus-encoded nucleoproteins (16, 37, 46), where other factor(s) are believed to be necessary to confer specificity.
Both C- and N-terminal domains of VP3 contribute to RNA binding.
Except for the positively charged C-terminal end, suggested to play a role in dsRNA binding (20, 22), analysis of the primary amino acid sequence did not reveal known dsRBD motifs within VP3. Deletion mapping of the dsRBD of VP3 showed that both the N-terminal and the C-terminal domains contribute to binding. Accordingly, our result partly contrasts the reported finding for IBDV, where the VP3 RNA binding domain was mapped to a highly conserved 69-amino-acid stretch close to the N terminus of the protein (27), although these authors also suggest that VP3 could contain more than one RNA-binding domain. This specific region of the protein, however, differs between the two viruses, which may entail differences in their binding properties as well. This also applies for the C-terminal ends of the two VP3s.
VP3 has three partly overlapping but distinct interaction domains.
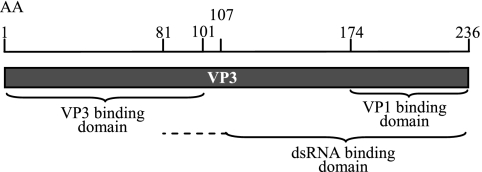
Although IPNV is closely related to IBDV and these viruses have many features in common, the binding domains of both VP3 and VP1 differ slightly between the two viruses. A model presented in Fig. 8 summarizes the domains of VP3 responsible for different interactions. The C-terminal domain, binding VP1, differs from the dsRNA binding part, since a larger polypeptide in the C terminus is needed to establish RNA binding. Finally, the N-terminal end of the VP3 molecule is involved in the VP3 self-interactions, possibly giving rise to VP3 trimers similar to those described as making up the inner surface of IBDV particles (4). These multiple interaction properties suggest the involvement of VP3 in replication and packaging of the IPNV genome. Studies of IBDV support this assumption, whereas virion morphogenesis is controlled by interactions of VP3 with both pVP2 and VP1 (6, 7, 31, 32, 38).
FIG. 8.
Model postulating VP3 binding domains mapped in this work. The binding site for VP3 self-interaction is within the 101 N-terminal amino acids. The C-terminal end of VP3 contains two functional domains: the VP1 binding site is within the 62 C-terminal amino acids, while the dsRNA binding ability also depends on a region extending into the N terminus of the protein. AA, amino acids.
There are several interesting possibilities regarding the biological significance of the interaction between VP1 and VP3. This complex may be involved in the regulation of viral RNA synthesis or may be a part of the viral replication apparatus, as has been proposed for the interaction between the RdRp and the virus coat protein of tobacco vein mottling virus (41). Interactions between viral structural proteins may affect their polymerase activity. In rotavirus, an interaction between the inner capsid protein, VP6, and the inner core protein, VP3, is necessary to recover RNA polymerase activity (45). Future studies aimed at mapping the different steps during IPNV replication and morphogenesis will be facilitated by introduction of specific mutations into the viral genome according to the mapped binding domains of VP3 and VP1. A reverse genetics system has been described for IPNV (51) and can be used to further characterize the in vivo roles of these domains.
VP3 binding to dsRNA—possible mechanism for avoiding antiviral host responses?
Throughout the replication cycle of dsRNA viruses, their genome remains encased within the capsid, and they are transcribed and replicated within viral capsids by the RdRp (39). This property may have evolved in part to prevent the genomic dsRNA from inducing a comprehensive innate immune response. It is therefore tempting to propose that the dsRNA-binding properties of VP3 described here make the genomic dsRNA less accessible for the interferon (IFN) system. In fact, recent studies report the absence of type I IFN responses in salmonid cell lines during IPNV infections (25, 29), while treatment by IFN stimulatory agents prior to infection strongly inhibits IPNV (25, 26, 43). The possibility that IPNV VP3 plays an antagonist role in antiviral host cell responses through its RNA binding properties is an interesting and so far unexplored question.
Acknowledgments
This work was supported by the National Program for Research in Functional Genomics in Norway (FUGE) of the Research Council of Norway (grant no. 159326/S10).
The excellent technical assistance of Marit N. Hegseth and Toril Aamo is highly appreciated. We thank Karen E. Christie and Eirik Biering, Intervet Norbio, for providing the IPNV N1 strain, monoclonal antibodies specific for VP2 and VP3, and the IPNV plasmid pETSegA. Further, we thank Adam P. Geballe, Fred Hutchinson Cancer Research Center, Seattle, WA, for providing the plasmid pEQ843 expressing E3L and Jack Bruun, University of Tromsø, for liquid chromatography-tandem mass spectrometry analysis. We very much acknowledge Ole M. Seternes and Trond Lamark, University of Tromsø, for providing several Gateway-compatible vectors and for all technical advice concerning yeast two-hybrid screening and co-IP.
Footnotes
Published ahead of print on 11 April 2007.
REFERENCES
- 1.Bartel, P., C. T. Chien, R. Sternglanz, and S. Fields. 1993. Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14:920-924. [PubMed] [Google Scholar]
- 2.Bevilacqua, P. C., and T. R. Cech. 1996. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry 35:9983-9994. [DOI] [PubMed] [Google Scholar]
- 3.Birghan, C., E. Mundt, and A. E. Gorbalenya. 2000. A non-canonical lon proteinase lacking the ATPase domain employs the Ser-Lys catalytic dyad to exercise broad control over the life cycle of a double-stranded RNA virus. EMBO J. 19:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bottcher, B., N. A. Kiselev, V. Y. Stel'Mashchuk, N. A. Perevozchikova, A. V. Borisov, and R. A. Crowther. 1997. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J. Virol. 71:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvert, J. G., E. Nagy, M. Soler, and P. Dobos. 1991. Characterization of the VPg-dsRNA linkage of infectious pancreatic necrosis virus. J. Gen. Virol. 72:2563-2567. [DOI] [PubMed] [Google Scholar]
- 6.Chevalier, C., J. Lepault, B. Da Costa, and B. Delmas. 2004. The last C-terminal residue of VP3, glutamic acid 257, controls capsid assembly of infectious bursal disease virus. J. Virol. 78:3296-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chevalier, C., J. Lepault, I. Erk, B. Da Costa, and B. Delmas. 2002. The maturation process of pVP2 requires assembly of infectious bursal disease virus capsids. J. Virol. 76:2384-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie, K. E., L. S. Havarstein, H. O. Djupvik, S. Ness, and C. Endresen. 1988. Characterization of a new serotype of infectious pancreatic necrosis virus isolated from Atlantic salmon. Arch. Virol. 103:167-177. [DOI] [PubMed] [Google Scholar]
- 9.Coulibaly, F., C. Chevalier, I. Gutsche, J. Pous, J. Navaza, S. Bressanelli, B. Delmas, and F. A. Rey. 2005. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120:761-772. [DOI] [PubMed] [Google Scholar]
- 10.Dobos, P. 1993. In vitro guanylylation of infectious pancreatic necrosis virus polypeptide VP1. Virology 193:403-413. [DOI] [PubMed] [Google Scholar]
- 11.Dobos, P. 1995. The molecular biology of infectious pancreatic necrosis virus (IPNV). Annu. Rev. Fish Dis. 5:25-54. [Google Scholar]
- 12.Dobos, P. 1977. Virus-specific protein synthesis in cells infected by infectious pancreatic necrosis virus. J. Virol. 21:242-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duncan, R., and P. Dobos. 1986. The nucleotide sequence of infectious pancreatic necrosis virus (IPNV) dsRNA segment A reveals one large ORF encoding a precursor polyprotein. Nucleic Acids Res. 14:5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan, R., E. Nagy, P. J. Krell, and P. Dobos. 1987. Synthesis of the infectious pancreatic necrosis virus polyprotein, detection of a virus-encoded protease, and fine structure mapping of genome segment A coding regions. J. Virol. 61:3655-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 16.Gott, P., R. Stohwasser, P. Schnitzler, G. Darai, and E. K. Bautz. 1993. RNA binding of recombinant nucleocapsid proteins of hantaviruses. Virology 194:332-337. [DOI] [PubMed] [Google Scholar]
- 17.Hakki, M., and A. P. Geballe. 2005. Double-stranded RNA binding by human cytomegalovirus pTRS1. J. Virol. 79:7311-7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Havarstein, L. S., K. H. Kalland, K. E. Christie, and C. Endresen. 1990. Sequence of the large double-stranded RNA segment of the N1 strain of infectious pancreatic necrosis virus: a comparison with other Birnaviridae. J. Gen. Virol. 71:299-308. [DOI] [PubMed] [Google Scholar]
- 19.Hill, B. J., and K. Way. 1995. Serological classification of infectious pancreatic necrosis (IPN) virus and other aquatic birnaviruses. Annu. Rev. Fish Dis. 5:55-77. [Google Scholar]
- 20.Hjalmarsson, A., E. Carlemalm, and E. Everitt. 1999. Infectious pancreatic necrosis virus: identification of a VP3-containing ribonucleoprotein core structure and evidence for O-linked glycosylation of the capsid protein VP2. J. Virol. 73:3484-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hjalmarsson, A., and E. Everitt. 1999. Identification of IPNV-specified components released from productively infected RTG-2 cells following massive cytopathic effect. Arch. Virol. 144:1487-1501. [DOI] [PubMed] [Google Scholar]
- 22.Hudson, P. J., N. M. McKern, B. E. Power, and A. A. Azad. 1986. Genomic structure of the large RNA segment of infectious bursal disease virus. Nucleic Acids Res. 14:5001-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes, S. R., S. Goyal, J. E. Sun, P. Gonzalez-DeWhitt, M. A. Fortes, N. G. Riedel, and S. R. Sahasrabudhe. 1996. Two-hybrid system as a model to study the interaction of beta-amyloid peptide monomers. Proc. Natl. Acad. Sci. USA 93:2065-2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs, B. 2000. Translational control in poxvirus-infected cells, p. 951-971. In J. Sonenberg and M. Mathewa (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Jensen, I., and B. Robertsen. 2002. Effect of double-stranded RNA and interferon on the antiviral activity of Atlantic salmon cells against infectious salmon anemia virus and infectious pancreatic necrosis virus. Fish Shellfish Immunol. 13:221-241. [DOI] [PubMed] [Google Scholar]
- 26.Jørgensen, J. B., A. Johansen, B. Stenersen, and A. I. Sommer. 2001. CpG oligodeoxynucleotides and plasmid DNA stimulate Atlantic salmon (Salmo salar L.) leucocytes to produce supernatants with antiviral activity. Dev. Comp. Immunol. 25:313-321. [DOI] [PubMed] [Google Scholar]
- 27.Kochan, G., D. Gonzalez, and J. F. Rodriguez. 2003. Characterization of the RNA-binding activity of VP3, a major structural protein of Infectious bursal disease virus. Arch. Virol. 148:723-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamark, T., M. Perander, H. Outzen, K. Kristiansen, A. Overvatn, E. Michaelsen, G. Bjorkoy, and T. Johansen. 2003. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J. Biol. Chem. 278:34568-34581. [DOI] [PubMed] [Google Scholar]
- 29.Larsen, R., T. P. Rokenes, and B. Robertsen. 2004. Inhibition of infectious pancreatic necrosis virus replication by Atlantic salmon Mx1 protein. J. Virol. 78:7938-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magyar, G., and P. Dobos. 1994. Evidence for the detection of the infectious pancreatic necrosis virus polyprotein and the 17-kDa polypeptide in infected cells and of the NS protease in purified virus. Virology 204:580-589. [DOI] [PubMed] [Google Scholar]
- 31.Maraver, A., R. Clemente, J. F. Rodriguez, and E. Lombardo. 2003. Identification and molecular characterization of the RNA polymerase-binding motif of infectious bursal disease virus inner capsid protein VP3. J. Virol. 77:2459-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maraver, A., A. Ona, F. Abaitua, D. Gonzalez, R. Clemente, J. A. Ruiz-Diaz, J. R. Caston, F. Pazos, and J. F. Rodriguez. 2003. The oligomerization domain of VP3, the scaffolding protein of infectious bursal disease virus, plays a critical role in capsid assembly. J. Virol. 77:6438-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Torrecuadrada, J. L., J. R. Caston, M. Castro, J. L. Carrascosa, J. F. Rodriguez, and J. I. Casal. 2000. Different architectures in the assembly of infectious bursal disease virus capsid proteins expressed in insect cells. Virology 278:322-331. [DOI] [PubMed] [Google Scholar]
- 34.McAllister, P. E., W. J. Owens, and T. M. Ruppenthal. 1987. Detection of infectious pancreatic necrosis virus in pelleted cell and particulate components from ovarian fluid of brook trout Salvelinus fontinalis. Dis. Aquat. Org. 2:235-237. [Google Scholar]
- 35.Muller, H., M. R. Islam, and R. Raue. 2003. Research on infectious bursal disease—the past, the present and the future. Vet. Microbiol. 97:153-165. [DOI] [PubMed] [Google Scholar]
- 36.Muller, H., and R. Nitschke. 1987. The two segments of the infectious bursal disease virus genome are circularized by a 90,000-Da protein. Virology 159:174-177. [DOI] [PubMed] [Google Scholar]
- 37.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ona, A., D. Luque, F. Abaitua, A. Maraver, J. R. Caston, and J. F. Rodriguez. 2004. The C-terminal domain of the pVP2 precursor is essential for the interaction between VP2 and VP3, the capsid polypeptides of infectious bursal disease virus. Virology 322:135-142. [DOI] [PubMed] [Google Scholar]
- 39.Patton, J. T., and E. Spencer. 2000. Genome replication and packaging of segmented double-stranded RNA viruses. Virology 277:217-225. [DOI] [PubMed] [Google Scholar]
- 40.Pous, J., C. Chevalier, M. Ouldali, J. Navaza, B. Delmas, and J. Lepault. 2005. Structure of birnavirus-like particles determined by combined electron cryomicroscopy and X-ray crystallography. J. Gen. Virol. 86:2339-2346. [DOI] [PubMed] [Google Scholar]
- 41.Quadt, R., H. J. M. Rosdorff, T. W. Hunt, and E. M. J. Jaspars. 1991. Analysis of the protein composition of alfalfa mosai virus RNA-dependent RNA polymerase. Virology 182:309-315. [DOI] [PubMed] [Google Scholar]
- 42.Reed, L., and H. Muench. 1938. A simple method for estimating fifty percent endpoints. Am. J. Hygiene 27:493-497. [Google Scholar]
- 43.Robertsen, B., V. Bergan, T. Rokenes, R. Larsen, and A. Albuquerque. 2003. Atlantic salmon interferon genes: cloning, sequence analysis, expression, and biological activity. J. Interferon Cytokine Res. 23:601-612. [DOI] [PubMed] [Google Scholar]
- 44.Rodriguez Saint-Jean, S., J. J. Borrego, and S. I. Perez-Prieto. 2003. Infectious pancreatic necrosis virus: biology, pathogenesis, and diagnostic methods. Adv. Virus Res. 62:113-165. [DOI] [PubMed] [Google Scholar]
- 45.Sandino, A. M., J. Fernandez, J. Pizarro, M. Vasquez, and E. Spencer. 1994. Structure of rotavirus particle: interaction of the inner capsid protein VP6 with the core polypeptide VP3. Biol. Res. 27:39-48. [PubMed] [Google Scholar]
- 46.Skuzeski, J. M., and T. J. Morris. 1995. Quantitative analysis of the binding of turnip crinkle virus coat protein to RNA fails to demonstrate binding specificity but reveals a highly cooperative assembly interaction. Virology 210:82-90. [DOI] [PubMed] [Google Scholar]
- 47.Tacken, M. G., B. P. Peeters, A. A. Thomas, P. J. Rottier, and H. J. Boot. 2002. Infectious bursal disease virus capsid protein VP3 interacts both with VP1, the RNA-dependent RNA polymerase, and with viral double-stranded RNA. J. Virol. 76:11301-11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian, B., P. C. Bevilacqua, A. Diegelman-Parente, and M. B. Mathews. 2004. The double-stranded-RNA-binding motif: interference and much more. Nat. Rev. Mol. Cell Biol. 5:1013-1023. [DOI] [PubMed] [Google Scholar]
- 49.Villanueva, R. A., J. L. Galaz, J. A. Valdes, M. M. Jashes, and A. M. Sandino. 2004. Genome assembly and particle maturation of the birnavirus infectious pancreatic necrosis virus. J. Virol. 78:13829-13838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu, H. T., W. D. Si, and P. Dobos. 2004. Mapping the site of guanylylation on VP1, the protein primer for infectious pancreatic necrosis virus RNA synthesis. Virology 322:199-210. [DOI] [PubMed] [Google Scholar]
- 51.Yao, K., and V. N. Vakharia. 1998. Generation of infectious pancreatic necrosis virus from cloned cDNA. J. Virol. 72:8913-8920. [DOI] [PMC free article] [PubMed] [Google Scholar]