Abstract
The herpes simplex virus type 1 (HSV-1) US3 gene encodes a serine/threonine kinase that, when inactivated, causes capsids to aggregate aberrantly between the inner and outer nuclear membranes (INM and ONM, respectively) within evaginations/extensions of the perinuclear space. In both Hep2 cells and an engineered cell line derived from Hep2 cells expressing lamin A/C fused to enhanced green fluorescent protein (eGFP-lamin A/C), lamin A/C localized mostly in a reticular pattern with small regions of the INM devoid of eGFP-lamin A/C when they were either mock infected or infected with wild-type HSV-1(F). Cells infected with HSV-1(F) also contained some larger diffuse regions lacking lamin A/C. Proteins UL31 and UL34, markers of potential envelopment sites at the INM and perinuclear virions, localized within the regions devoid of lamin A/C and also in regions containing lamin A/C. Similar to previous observations with Vero cells (S. L. Bjerke and R. J. Roller, Virology 347:261-276, 2006), the proteins UL34 and UL31 localized exclusively in very discrete regions of the nuclear lamina lacking lamin A/C in the absence of US3 kinase activity. To determine how US3 alters lamin A/C distribution, US3 was purified and shown to phosphorylate lamin A/C at multiple sites in vitro, despite the presence of only one putative US3 kinase consensus site in the lamin A/C sequence. US3 kinase activity was also sufficient to invoke partial solubilization of lamin A/C from permeabilized Hep2 cell nuclei in an ATP-dependent manner. Two-dimensional electrophoretic analyses of lamin A/C revealed that lamin A/C is phosphorylated in HSV-infected cells, and the full spectrum of phosphorylation requires US3 kinase activity. These data suggest that US3 kinase activity regulates HSV-1 capsid nuclear egress at least in part by phosphorylation of lamin A/C.
Like orthologues in other members of the subfamily Alphaherpesvirinae, the US3 gene of herpes simplex virus type 1 (HSV-1) encodes a serine/threonine kinase (13, 37). These proteins have been implicated in nuclear egress, prevention of apoptosis, and modulation of the actin cytoskeleton to promote the cell-to-cell spread of virions (11, 14, 19, 22, 25, 42, 44, 46, 52). The current study focuses on the role of the US3 gene-encoded kinase activity in nuclear egress of nucleocapsids and virions.
Although models of HSV virion egress differ as to the extent of its contribution, all models propose that at some point during the course of infection with wild-type herpesviruses, nucleocapsids assemble in the nucleoplasm and bud through the inner nuclear membrane (INM) and into the perinuclear space (20, 51, 53). This compartment is delimited by the INM and outer nuclear membrane (ONM) and is continuous with the lumen of the endoplasmic reticulum. To become enveloped, capsids must bypass the nuclear lamina, a fibrous meshwork lining the nucleoplasmic face of the INM. The nuclear lamina provides structural rigidity to the nucleus and is essential for transcription and DNA replication (15, 48). The lamina contains a series of type 5 intermediate filaments composed of lamin types A, B1, B2, and C; types A and C are products of RNA splice variants from the LmnA transcript, whereas types B1 and B2 are derived from other genes (12, 16, 17). Like all intermediate filaments, lamins comprise globular head and tail domains that flank a rod domain (12). The rod domains of two lamins intertwine to form protomers, whereas regions bordering the rod/head and rod/tail domains likely interact with other lamin protomers to form longer filaments (40). The globular domains interact with a variety of proteins in the lamina and INM.
One remarkable feature of the lamina is its dynamic nature. The lamina expands by the addition of protomers during interphase, is completely disassembled prior to mitosis, and is partially disrupted during apoptosis. Phosphorylation likely plays a role in lamin dynamics in all phases of the cell cycle. The disassembly of the lamina during mitosis is associated with phosphorylation of lamin A/C by cdc2 kinase at Ser390 and Ser392 and during apoptosis by protein kinase C (PKC) delta (5, 7, 8, 32, 33). Protein kinase C can phosphorylate lamin A/C at Ser572 in vitro (8).
The architecture of the nuclear lamina is altered from its normal state during HSV-1 infection (3, 31, 40, 47, 49). Depending on the cell line and time after infection, these changes include (i) limited displacement and conformational changes of lamin A/C (3, 40, 49), (ii) redistribution of lamin B to a perinuclear region (31, 47), and (iii) increased mobility and mislocalization of lamin B receptor, one of several integral membrane proteins that anchor the nuclear lamina to the INM (47, 48). In attempts to understand the mechanism(s) by which the lamina becomes displaced, it was found that PKC of the alpha and delta subfamilies are recruited to the nuclear rim of HSV-1-infected cells and that lamin B becomes hyperphosphorylated during HSV infection, in part due to phosphorylation by PKC (31). Given these observations, it was postulated that HSV coopts cellular mechanisms to disrupt the nuclear lamina and thereby promotes egress of nucleocapsids (31). The state of phosphorylation of lamin A/C in HSV-infected cells is unknown, in contrast to that of lamin B. Previous analyses using one-dimensional gel electrophoresis indicated that HSV infection did not drastically alter the electrophoretic mobility of lamin A/C from human foreskin fibroblasts or from Hep2 cells, suggesting that phosphorylation is not extensive (39, 40).
As detected by immunoelectron microscopy, nascent virions located in the perinuclear space contain the US3 kinase and at least two of its substrates, the proteins encoded by the UL31 and UL34 genes (as products pUL31 and pUL34, respectively) (21, 34, 38, 42). The proteins pUL31 and pUL34 are required for efficient envelopment of nucleocapsids at the INM (4, 27, 41, 43). The observations that proteins pUs3, pUL31, and pUL34 are located within perinuclear virions and at the INM suggest that these proteins are incorporated into the virion during budding at the INM (42). Interestingly, all of the HSV-1-induced alterations in lamin A and B distribution/conformation require expression of pUL31 and pUL34 (3, 31, 40, 49). How pUL31 and pUL34 alter the structure of the nuclear lamina is not known, but the alteration likely involves multiple mechanisms. For example, both pUL31 and pUL34 are required for the recruitment of protein kinase C alpha and delta to the nuclear rim, and both can interact with lamin A/C in vitro (31, 40). Thus, UL31/UL34 might interrupt the nuclear lamina by mechanical interference with lamin-lamin interactions, by a disruption of lamin folding, by recruitment of lamina-disrupting kinases, or by all three mechanisms.
Deletion of US3 delays the onset of production of infectious virus and reduces peak infectious titers by approximately 10- to 30-fold with Hep2 cells (42, 44). The absence of US3 or its kinase activity does not preclude budding at the INM, nor incorporation of the pUL31/pUL34 into perinuclear virions, but causes these virions to aggregate aberrantly within the perinuclear space (22, 42, 44, 46, 52). These observations with both the HSV and the pseudorabies herpesvirus (PRV) systems have led to the conclusion that US3 is involved in the exit of virions from the perinuclear space into the cytoplasm (22, 42). On the other hand, if the exit from the perinuclear space was the sole contribution of US3 to nuclear egress, one might expect that pUS3(−) perinuclear virions would distribute evenly throughout the perinuclear space, but this is not the case. Rather, virions are tightly packed together within discrete invaginations/extensions of the nuclear membrane (NM). How US3 kinase activity precludes this aberrant aggregation of perinuclear virions is unknown.
In vitro biochemical studies characterized a US3 kinase minimal consensus sequence as (R)n-X-(S/T)-Y-Y, where n is >2, S/T is the target site where either serine or threonine is phosphorylated, X can be absent or any amino acid but preferably Arg, Ala, Val, Pro, or Ser, and Y is similar to X except that it cannot be an absent amino acid, proline, or an acidic residue (24, 35, 36). The optimal consensus sequence is similar except that X is not absent and n is ≥3. Both pUL34 and pUL31 have one or more sequences matching this consensus motif (28). Of the known US3 kinase substrates localizing at the NM, the lack of phosphorylation of pUL34 is not likely responsible for the aberrant NM morphology in US3 mutant-infected cells (44), whereas the role of pUL31 phosphorylation has not yet been tested. The current studies were initiated under the hypothesis that substrates in addition to pUL31 and pUL34 may be involved in capsid and virion egress from the NM.
MATERIALS AND METHODS
Cells and viruses.
Wild-type HSV-1(F) virus and a US3 deletion mutant virus, R7041, have been described and were obtained from B. Roizman (10, 37). Virus vRR1204, containing a mutation at US3 codon 220 changing lysine to alanine (K220A) in an HSV-1(F) background, was a kind gift of R. J. Roller (44). The viruses were grown and titers were determined on Vero cells as described previously (29).
An enhanced green fluorescent protein (eGFP)-lamin A/C-expressing cell line was made by transfection of DNA containing a cDNA of LmnA fused to eGFP-C1 and encoding neomycin resistance (a kind gift from D. Gilbert, Upstate Medical Center) into human Hep2 cells (18). Individual eGFP-positive cells were sorted based on the intensity of fluorescence determined using an ExCalibur fluorescence-activated cell sorter (FACS), into individual wells of a 96-well plate. The cells were grown in growth medium (Dulbecco's minimal essential medium supplemented with 10% fetal bovine serum and antibiotics) supplemented with 250 μg/ml Geneticin (Invitrogen). The highest-expression cells failed to expand further, whereas some intermediate expressers and all low expressers expanded into colonies (not shown). A single clone of several, containing intermediate levels of nuclear fluorescence, was further amplified in growth medium supplemented with 50 to 250 μg/ml Geneticin and was used for further studies.
Construction of recombinant baculoviruses.
Full-length US3 and US3(K220A) were PCR amplified from viral DNA of HSV-1(F) or the US3 mutant virus vRR1204, respectively. The PCRs used primers with a BglII restriction endonuclease site incorporated into the forward primer (5′-TAA TAG ATC TAT GGC CTG TCG TAA GTT TTG-3′) and an EcoRI site in the reverse primer (5′-ATA TGA ATT CTC ATT TCT GTT GAA ACA GCG-3′; restriction sites are in italics). PCR products were then cloned into the BamHI and EcoRI sites of the pGEX4T-1 vector such that they were in frame with the gene encoding glutathione-S-transferase (GST). DNA in the plasmid clones was sequenced, and those clones with the proper GST-US3 fusions were used as templates for a second PCR to amplify GST-US3 and GST-US3(K220A). This second PCR used primer 5′-AGG CAG ATC TAT TCA TGT CCC CTA TAC TAG-3′ containing another BglII site (italicized) and the US3 reverse primer. Amplicons were then subcloned into the BglII/EcoRI sites of the pBacPAK8 vector (Clontech). This construct was then transfected into insect Sf9 cells along with BacPAK6 baculoviral DNA (Clontech) according to the instructions of the BacPAK baculovirus expression system, and recombinant baculoviruses expressing the target proteins were plaque purified and amplified to produce viral stocks.
Antibodies and immunofluorescence.
Polyclonal chicken antibody against pUL34 was a kind gift from R. J. Roller (41). Chicken anti-lamin A/C polyclonal and rabbit anti-pUL31 antisera were prepared in our laboratory and were described previously (40, 41). Lamin B-specific goat polyclonal antibody was purchased from Santa Cruz Biotechnology (catalog number sc-6218).
To characterize the localization of pUL31 and lamin A/C in infected cells, 200 μl of rabbit anti-pUL31 antisera was adsorbed against an acetone powder (45) prepared from approximately 2 × 108 Hep2 cells that were infected 24 h previously with a UL31 deletion virus (27). The powder and antisera were added to 4 ml phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA) and were mixed overnight. After centrifugation, the supernatant was passed through a 0.2-μm-pore-sized filter.
Hep2 cells growing on glass coverslips were mock infected or infected with wild-type HSV-1(F), US3 null, or US3(K220A) HSV-1 at a multiplicity of infection (MOI) of 5.0 for 16 h. Cells were fixed with 3% paraformaldehyde for 15 min, followed by treatment for 5 min at −20°C in methanol. The cells were then permeabilized with 0.1% Triton X-100 and reacted with 10% human serum in PBS to block nonspecific immunoreactivity and were subsequently probed with the preadsorbed pUL31 rabbit polyclonal antibody diluted 1:50 in PBS supplemented with 1% BSA. Ten percent BlockHen II (Aves Lab) was used for a second round of blocking before probing with a 1:200 dilution of chicken anti-lamin A/C polyclonal antibody. Bound primary antibodies were recognized by Texas Red-conjugated donkey anti-rabbit and fluorescein isothiocyanate (FITC)-conjugated donkey anti-chicken immunoglobulin (Jackson ImmunoResearch).
In some experiments, the cell line expressing eGFP-lamin A/C was mock infected or infected as described above, fixed in 3% paraformaldehyde, and permeabilized with 0.1% Triton X-100. Epitopes were then blocked by incubation with 10% human serum in PBS for 1 h, followed by blocking with 10% BlockHen II and reaction with pUL34-specific antisera diluted 1:200. After extensive washing, primary antibody was recognized by Texas Red-conjugated donkey anti-chicken immunoglobulin (Jackson ImmunoResearch).
Images were visualized and recorded using either an Olympus or a Zeiss laser scanning confocal microscope equipped with a 488-nm argon or 568-nm krypton laser, respectively, and associated software. Digital images were exported in a tagged image file format (TIFF) to Adobe Photoshop for processing.
Purification of wild-type and mutant HSV-1 pUS3.
Sf9 cells were infected with recombinant baculoviruses expressing GST-pUS3 or GST-pUS3(K220A) and were lysed 42 h postinfection by brief sonication in ice-cold lysis buffer (50 mM Tris [pH 7.5], 100 mM NaCl, 5 mM MgCl2, 0.1% NP-40, 10% glycerol) containing 1× Complete protease inhibitor cocktail (Roche). The US3 proteins were purified from the lysates by affinity chromatography using glutathione Sepharose beads (Amersham) as previously described (21). Proteins were eluted in 100 mM reduced glutathione according to the manufacturer's protocol and dialyzed into storage buffer (20 mM Tris-HCl [pH 7.5], 25 mM KCl, 0.1 mM EDTA, 2 mM dithiothreitol [DTT], 50% glycerol). Protein concentration was determined by an RC DC protein assay kit (Bio-Rad), and aliquots were frozen at −80°C.
GST fusion proteins were partially purified from Escherichia coli lysates, as described previously (40), and retained on the Sepharose beads without elution for reactions. Protein concentration was estimated by elution and separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by comparison with known amounts of BSA standards stained with Coomassie brilliant blue.
In vitro kinase assay.
A kinase assay was performed using purified GST-pUS3 or GST-pUS3(K220A) essentially as described previously, with minor modifications (21). Briefly, 0.1 μg GST-pUS3 or at least an equivalent amount of GST-pUS3(K220A) was incubated separately with 1 to 3 μg of partially purified E. coli-expressed GST fusion proteins for 30 min at 30°C in 50 μl US3-specific kinase buffer (50 mM Tris [pH 9.0], 20 mM MgCl2, 0.1% NP-40, 1 mM DTT) containing 10 μM ATP, and 10 μCi [γ-32P]ATP (Amersham). The Sepharose beads bearing the fusion proteins were then washed three times with TNE buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA) and in some cases were resuspended in bacteriophage lambda pyrophosphatase (λ-Ppase) buffer containing 200 U of enzyme (New England Biolabs) and incubated for another 30 min at 30°C. The proteins on the beads were again washed three times in TNE. All the samples were dissolved in SDS-PAGE sample buffer (10 mM Tris-HCl [pH 8.0], 10 mM β-mercaptoethanol, 20% glycerol, 5% SDS, trace amounts of bromophenol blue) and subjected to electrophoresis in a 12% polyacrylamide gel in the presence of 0.1% SDS. Gels were then stained with Coomassie brilliant blue, dried, and autoradiographed using X-ray film (Pierce).
Lamin disassembly assay.
A previously described lamin disassembly assay was modified for the current studies (6). Approximately 1 × 107 Hep2 cells were homogenized in 5.0 ml nuclear buffer (250 mM sucrose, 5 mM MgCl2, 25 mM KCl, 10 mM Tris [pH 7.4], 1 mM DTT, 1× Complete protease inhibitor cocktail [Roche]) by three strokes of a Dounce homogenizer. Nuclei were pelleted at 800 × g for 10 min in a refrigerated tabletop centrifuge (Eppendorf). The supernatant was discarded, and pelleted nuclei were resuspended in nuclear buffer and then permeabilized by the addition of 0.1% Triton X-100, followed by incubation on ice for 20 min. Nuclei were again pelleted at 800 × g for 10 min, and the supernatant was removed. The pelleted permeabilized nuclei were then washed twice with an excess volume of US3-specific kinase buffer and finally resuspended in 400 μl of this buffer.
For the in vitro disassembly assay, three 80-μl aliquots of resuspended nuclei were incubated with 1 μg GST-pUS3 or GST-pUS3(K220A) in 120 μl kinase buffer in the presence or absence of 1 mM ATP. After 30 min of incubation at 30°C, the reaction mixture tubes were gently vortexed, and 10 μl of the reaction mixtures (samples designated L) were set aside. All subsequent steps were performed at 0 to 4°C. After centrifugation at 2,000 × g for 5 min, supernatants of each sample (30 μl) were collected and were designated S1. The remaining nuclei were centrifuged again at 10,000 × g for 2 min, and equal volumes of the supernatants were collected and designated S2. Proteins in the L, S1, and S2, samples were denatured in SDS-PAGE sample buffer, electrophoretically separated in denaturing polyacrylamide gels, and transferred to nitrocellulose for immunoblotting with lamin-specific antibodies.
Two-dimensional gel electrophoresis.
To solubilize lamins, 2 × 106 infected Hep2 cells were lysed directly on culture dishes into 250 μl of urea rehydration buffer {9 M urea, 4% CHAPS [3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate], 50 mM DTT, 0.2% ampholytes [Bio-Rad]} containing 1× Complete protease inhibitor cocktail (Roche) and a serine/threonine phosphatase inhibitor (10 mM NaF) (23). Approximately 100 μg of protein was subjected to isoelectric focusing in a Bio-Rad Protean isoelectric focusing cell using an 11-cm-long pH 3.0 to 10.0 nonlinear or a pH 5.0 to 8.0 linear immobilized pH gradient ReadyStrip (Bio-Rad). For separation in the second dimension, the strips were loaded on precast Criterion 10% bis-Tris polyacrylamide gels (Bio-Rad), followed by electrophoresis. The separated proteins were then transferred electrically to nitrocellulose for immunoblotting.
In some experiments, cells were first permeabilized in 0.5% CHAPS on ice for 5 min in the presence of Complete protease inhibitor, clarified by centrifugation at 800 × g for 5 min at 4°C, and incubated with 400 U of λ-Ppase in a supplied buffer (New England Biolabs) at 30°C for 30 min. After this reaction, cells were repelleted and lysed in urea rehydration buffer, followed by two-dimensional electrophoresis and immunoblotting.
Immunoblotting.
Nitrocellulose sheets bearing proteins of interest were blocked in 5% nonfat milk plus 0.2% Tween 20 for at least 2 h. Membranes were then probed with lamin A/C chicken polyclonal antibody and lamin B goat polyclonal antibody if needed. Primary antibodies were detected by horseradish peroxidase-conjugated rabbit anti-chicken (Jackson ImmunoResearch) and/or bovine anti-goat secondary antibodies (Santa Cruz Biotechnology). Bound immunoglobulin was visualized by enhanced chemiluminescence (Pierce) followed by exposure to X-ray film. Signal intensities were quantified and calibrated using a Syngene Chemi-Genius imaging system and associated GeneTools software as needed.
RESULTS
US3 kinase activity modifies nuclear lamin A/C organization and localization of pUL31 and pUL34 during HSV-1 infection.
Because Hep2 cells are generally resistant to HSV-induced cytoskeletal changes, one goal of these studies was to investigate whether the nuclear lamina of Hep2 cells was affected by US3, as was previously reported with Vero cells (3). In considering the design of such experiments, we noted in previous studies that some commercial lamin A/C antibodies are rendered less immunoreactive upon infection with HSV-1 (40). To avoid the potentially misleading effects of antibody staining, we attempted to use eGFP-lamin A/C in transient-expression assays but found that overexpression of such constructs disrupted the nuclear lamina in many cells, even in the absence of infection (not shown). To avoid these overexpression artifacts, and under the expectation that cells experiencing associated cytotoxic effects should be outgrown by healthy cells expressing the construct, we sought to produce a cell line stably expressing eGFP-lamin A/C. Hep2 cells were therefore transfected with a plasmid conferring G418 resistance and encoding eGFP fused to lamin A/C. Individual cells expressing eGFP were cloned by FACS, followed by amplification in G418-containing medium. Cell lines expressing fluorescence in association with the nuclear rim were selected for further study.
One such cell line was either mock infected or infected with wild-type HSV-1(F) or virus vRR1204 bearing a mutation inactivating US3 kinase activity [designated US3(K220A) in Fig. 1 ] (44). At 16 h postinfection, cells were fixed, permeabilized, and reacted with chicken antibody directed against UL34 protein. Bound chicken antibody was revealed by reaction with Texas Red-conjugated donkey anti-chicken antibody, followed by confocal microscopy.
FIG. 1.
Confocal analyses of a Hep2-derived cell line stably expressing eGFP-lamin A/C. Cells were either mock infected (Mock) or infected with 5.0 PFU/cell of wild-type HSV-1(F) (designated F) or mutant US3(K220A), lacking pUS3 kinase activity. Cells were fixed and permeabilized at 16 h postinfection and stained with polyclonal anti-pUL34 chicken polyclonal antibody, followed by Texas Red-conjugated secondary antibody, and examined with confocal microscopy. (A) Analysis of optical sections taken through the middle of cells. (B) Optical sections taken at the bottom of cells. Regions indicated in white rectangles in panels D, E, F, J, K, and L are digitally magnified in the panel immediately below them (panels G, H, I, L, M and N, respectively). Coincident signals in the merged images (rightmost column) are indicated by a yellow color. An arrow indicates one region of the infected cell containing less lamin A/C than surrounding areas.
In general, the shape of nuclei infected with HSV-1(F) was more irregular than that of uninfected cells. As revealed upon collection of confocal images at high magnification, followed by digital enlargement, eGFP-lamin A/C appeared mostly as a series of filaments and dots linking larger solid areas of fluorescence. Overall, the fluorescence appeared in a reticular pattern in the nuclear rim of both the mock-infected and the HSV-1(F)-infected cells. The reticular pattern was most readily noticeable in optical sections of the tops and bottoms of the cells in which a planar view of the nuclear lamina was most readily apparent (Fig. 1B). In both mock-infected and HSV-1(F)-infected cells, the reticular filaments were bordered by spaces that lacked eGFP fluorescence. Although the reticular pattern of NM fluorescence in mock-infected and HSV-1-infected cells did not differ significantly, most cells infected with HSV-1(F) contained larger regions near the bottom of the cell that were devoid of fluorescence (Fig. 1B, arrow). In general, the appearance of these cells is in contrast to the appearance reported for HSV-infected Hep2 cells expressing eGFP-lamin A/C transiently, in which large discrete holes in the eGFP-specific fluorescence were seen (50). Differences in the two studies include the use of transient expression of eGFP-lamin A/C, followed by infection with 50 PFU/cell in the previous study, versus the use of a stably expressing cell line and an MOI of 5 in the current study.
In contrast to the results obtained upon infection with wild-type virus, the pattern of eGFP-lamin A/C fluorescence was markedly different after infection with the kinase-inactivated US3 mutant virus. Specifically, sites within the reticular pattern lacking eGFP fluorescence were substantially larger in diameter and more likely oval to round. In addition, the larger regions of HSV-1(F)-infected cells lacking the most eGFP in the nuclear rim displaying a paucity of lamin A/C were absent. These results indicated that the kinase activity of pUS3 is necessary for the distribution of lamin A/C normally seen in cells infected with wild-type HSV.
As noted previously, pUL34-specific immunostaining was distributed throughout the nuclear rim in Hep2 cells infected with the wild-type virus HSV-1(F) (41). This was readily apparent in optical cross-sections taken through the middle of cells (Fig. 1A). Superficially, eGFP-lamin A/C fluorescence and pUL34-specific immunostaining mostly colocalized in these cross-sections. However, high magnification of planar sections near the bottom of the cells revealed regions containing only pUL34 immunostaining or only eGFP-lamin A/C, as well as regions where eGFP-lamin A/C and pUL34 colocalized (Fig. 1B). In the larger regions lacking lamin A/C, seen only in cells infected with HSV-1(F), pUL34 immunostaining could also be detected.
In striking contrast to the localization of pUL34 in HSV-1(F)-infected cells, cells infected with the US3 kinase mutant contained pUL34 almost exclusively within punctate foci at the nuclear rim in optical cross-sections taken near the centers of cells (Fig. 1A) (3, 42, 44). These regions invariably corresponded to the well-defined breaches in the reticular distribution of eGPF-lamin A/C noted above. This dramatic change in the distribution of lamin A/C became even more striking in optical sections that focused on the bottom of infected cell nuclei. In such sections (Fig. 1B, lower two rows), foci staining intensely with pUL34-specific antibody coincided with regions that entirely lacked detectable eGPF-lamin A/C. We conclude that the kinase activity of pUS3 is necessary for partial colocalization of pUL34 and eGFP-lamin A/C at the nuclear rim and for the wide distribution of pUL34 throughout the nuclear rim. These data are consistent with previous immunostaining studies indicating that US3 kinase activity is necessary for the normal distribution of pUL34 with Vero cells (3, 42, 44).
To ensure that the above-described observations were not idiosyncratic to the eGFP-lamin A/C cell line, Hep2 cells were also mock infected or were infected with the wild-type HSV-1(F), the US3 kinase-dead viral mutant, or a viral mutant containing a deletion of US3. The cells were fixed at 16 h after infection and were immunostained with antibody against pUL31 and a chicken polyclonal antibody directed against lamin A/C that was insensitive to HSV-induced epitope masking (40).
As shown in Fig. 2, lamin A/C localized in a reticular pattern in mock-infected cells that superficially resembled that of cells infected with HSV-1(F), except that some regions near the nuclear rim of infected cells did not stain with lamin A/C antibody (e.g., as shown by the arrow in Fig. 2D). pUL31 immunostaining also appeared in a reticular pattern, but this pattern did not completely overlap that of lamin A/C immunostaining. Specifically, regions containing only pUL31 immunostaining or only lamin A/C immunostaining and colocalization of these were observed. Thus, the patterns of pUL31 immunostaining were similar to those of pUL34, as indicated above.
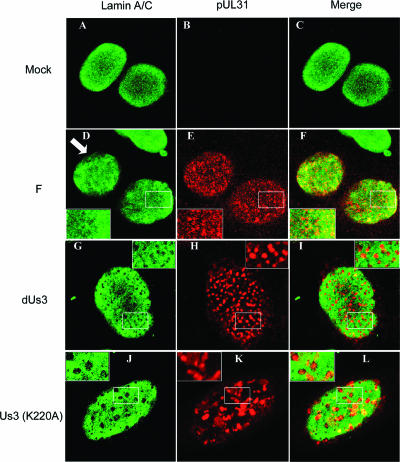
FIG. 2.
Digital confocal images of Hep2 cells immunostained with antibodies to lamin A/C and pUL31. Hep2 cells were mock infected (Mock) or infected with a US3 deletion mutant (dUs3), a mutant lacking US3 kinase activity [US3 (K220A)], or wild-type virus HSV-1(F) (designated F). Sixteen hours after infection, the cells were fixed, permeabilized, and reacted with preadsorbed rabbit polyclonal antiserum against pUL31 or chicken immunoglobulin Y directed against lamin A/C. Bound antibodies were revealed with Texas Red-conjugated anti-rabbit antibody or FITC-conjugated anti-chicken antibodies. Optical sections closest to the glass coverslips are shown. An arrow indicates regions of the nuclear rim that are mostly devoid of lamin A/C immunostaining. White boxes indicate areas of interest that are magnified in an inset in the same panel.
More importantly for the purposes of subsequent experiments herein, planar sections of cells infected with either the US3 deletion virus or the kinase-dead mutant virus revealed well-circumscribed, roughly circular regions that completely lacked lamin A/C immunostaining. These regions invariable contained most of the pUL31 immunostaining in the cell, i.e., there were very few regions in which pUL31 and lamin A/C colocalized except on the edges of these “holes” in the nuclear lamina. Taken together, the results are reminiscent of the above-described study using the eGFP-lamin A/C-expressing cell line and indicate that US3 kinase and its activity are required for (i) partial colocalization of pUL31/pUL34 with lamin A/C, (ii) for the reticular distribution of lamin A/C and pUL31/pUL34 seen in cells infected with wild-type viruses, and (iii) for the large regions devoid of lamin A/C immunostaining as seen in cells infected with HSV-1(F).
Adaptation of a US3 in vitro kinase assay and identification of lamin A as a substrate.
The above-described observations indicated that the kinase activity encoded by the US3 gene altered lamin A/C distribution. The simplest mechanism to explain this phenomenon would be that US3 phosphorylates lamin A/C directly, thereby altering lamin A/C's interaction with other proteins in the nuclear lamina. Further studies were undertaken to test this possibility.
Inasmuch as preliminary observations indicated that the Thr-10 residue of lamin A/C (within the sequence Arg7-Arg8-Ala9-Thr10-Arg11-Ser12, from GenBank accession number NP_733821) matched the optimal consensus target, experiments were designed to determine whether lamin A/C could be phosphorylated by the US3 kinase. Thus, US3 fused to GST was expressed in insect SF9 cells and was purified on glutathione beads as previously described (21). As a control, the kinase-inactivated mutant US3(K220A) was purified in the same manner. Coomassie blue staining of the proteins indicated at least 90% purity (Fig. 3A).
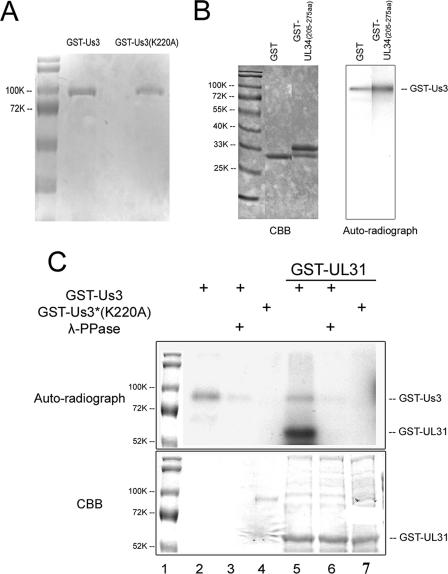
FIG. 3.
Analysis of in vitro kinase activities of purified GST-pUS3 and the mutant GST-pUS3(K220A). (A) Denaturing polyacrylamide gel containing purified GST-pUS3 and GST-pUS3(K220A) and stained with Coomassie brilliant blue. (B) In vitro kinase reaction. The fusion proteins indicated at the top of the left panel were mixed with 100 ng GST-US3. After 30 min at 30°C in the presence of [γ-32P]ATP, the reaction mixtures were resolved on a denaturing polyacrylamide gel and visualized by Coomassie (CBB) staining (left panel) and autoradiography (right panel). The position of the approximately 90,000 apparent Mr GST-US3 fusion protein is shown in the right panel. GST-UL34 (amino acids 205 to 275) is a negative control inasmuch as it does not contain the phosphorylation consensus site in pUL34. (C) In vitro kinase activities of GST-pUS3 and GST-pUS3(K220A). Purified GST-pUS3 (0.1 μg) or 0.5 μg GST-pUS3(K220A) was incubated with [γ-32P]ATP in the presence or absence of partially purified GST-pUL31 as a potential substrate, followed by subsequent incubation in the presence or absence of λ-Ppase. The reaction components were denatured, electrophoretically separated, and stained with Coomassie brilliant blue (CBB, bottom panel) or dried and autoradiographed (upper panel). For comparative purposes, the molecular weight standards from the bottom panel were copied and aligned with the top panel, using Adobe Photoshop. Sizes of the standards are indicated to the left of the figure in thousands [K]).
To test their enzymatic activities, 0.1 μg of the purified proteins was reacted with purified GST, or with pUL31 fused to GST, in the presence of [γ-32P]ATP. As an additional negative control, the pUL34 amino acids 205 to 275 region, comprising a region of pUL34 separate from that of the known US3 phosphorylation site, was also subjected to the kinase assay. The reaction mixtures were then electrophoretically separated on a denaturing polyacrylamide gel and subjected to autoradiography (Fig. 3B and C). Consistent with a previous report (21), GST-pUS3 phosphorylated itself (Fig. 3B and C, lanes 2 and 5), whereas GST-pUS3(K220A) had no such activity (Fig. 3C, lane 4). GST-pUS3 also readily phosphorylated the partially purified pUL31-GST fusion protein (Fig. 3C, lane 5). In contrast, GST-pUS3 did not phosphorylate GST: GST fused to the UL34 amino acids 205 to 275 or other E. coli proteins within the GST-pUL31 preparation that were readily visible on the Coomassie blue-stained gel (Fig. 3B and C, lane 5). GST-pUs3(K220A) did not phosphorylate GST-pUL31 or other proteins in the E. coli lysate (Fig. 3C, lane 7). As expected, addition of bacteriophage λ-Ppase significantly reduced the phosphorylation of GST-pUS3 (Fig. 3C, lane 3) and eliminated the phosphorylation of GST-pUL31 that was mediated by GST-pUS3 (Fig. 3C, lane 6).
Collectively, these data indicate that the US3 gene-encoded kinase activity was functional and specific inasmuch as it induced phosphorylation of two known substrates (pUL31 and pUS3) but not GST or other tested proteins. In addition, the data indicate that the K220A mutation inactivated all detectable US3 kinase activity. Thus, an efficient and specific in vitro kinase assay system was established for testing of additional substrates.
To determine whether lamin A was a target of the Us3 kinase, GST fused to lamin A was purified from E. coli and subjected to the in vitro kinase assay. The results, shown in Fig. 4, indicated that the lamin A-GST fusion protein was heavily phosphorylated in the presence of GST-pUS3 (Fig. 4, lane 1) but not in the presence of GST-pUS3(K220A) (Fig. 4, lane 3). The phosphorylation of lamin A-GST was eliminated upon incubation with λ-Ppase (Fig. 4, lane 2). These data indicate that lamin A is an in vitro substrate of the US3 kinase.
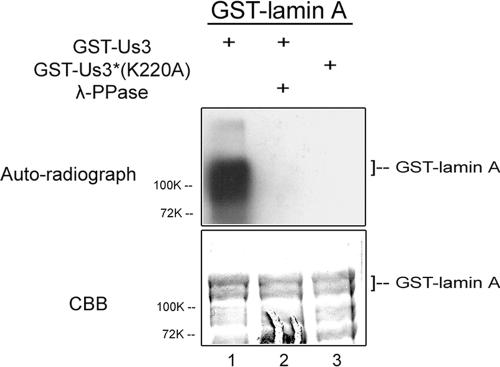
FIG. 4.
Phosphorylation of lamin A by US3 kinase in vitro. Full-length lamin A fused to GST was partially purified and reacted with either purified GST-pUS3 or GST-pUS3(K220A) in the presence of [γ-32P]ATP, followed by incubation in the presence or absence of λ-Ppase. The reaction components were separated on a polyacrylamide gel and stained with Coomassie brilliant blue (CBB, lower panel) and autoradiographed (upper panel).
US3 has broader kinase activity than predicted and phosphorylates lamin A/C at multiple sites in vitro.
Because lamin A/C is phosphorylated at different sites depending on the physiological mechanism of lamin disassembly (see the introduction), it was of interest to identify which region of lamin A was phosphorylated by the US3 kinase. To identify the region targeted by the US3 kinase, five subfragments of lamin A fused to GST were purified from lysates of E. coli and incubated with GST-pUS3 in the presence of [γ-32P]ATP, followed by denaturing gel electrophoresis, Coomassie blue staining, and autoradiography. The amount of radioactive phosphate in a given protein band was normalized to the amount of protein present in the reaction mixtures and expressed as a percentage of the degree to which full-length lamin A was phosphorylated. The results are shown in Fig. 5.
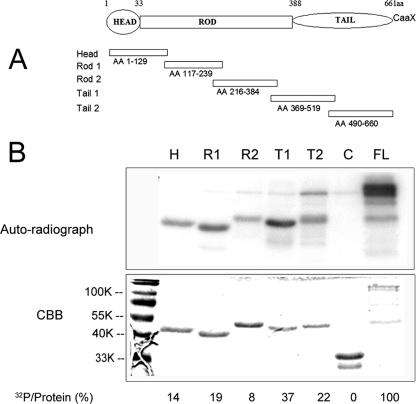
FIG. 5.
(A) Schematic diagram of lamin A that primary structure and five subdomains fused to GST. The amino acids (AA) in each peptide subdomain are indicated, assuming that the start methionine codon is 1. (Reprinted from reference 40.) (B) Full-length (FL) lamin A or lamin A fragments fused to GST and reacted with GST-pUS3 kinase. The five GST fusion proteins bearing fragments of lamin A as detailed for panel A were purified, mixed with US3-GST and [γ-32P]ATP, and electrophoretically separated and stained with Coomassie brilliant blue (lower panel) and autoradiographed (upper panel). H, head; R1, rod1; R2, rod2; T1, tail 1; T2, tail2; lane C, GST-pUL34 (amino acids 205 to 275).
Comparing the radiolabeling of full-length protein with that of truncated lamin A proteins normalized to the amount of protein in individual reaction mixtures revealed that full-length lamin A was phosphorylated to a higher degree than any single subfragment. While only Thr-10 matched the optimal US3 kinase consensus sequence, it was surprising to find that all five subfragments of lamin A were phosphorylated to various extents when reacted with purified GST-pUS3. Although the head (Fig. 5, lane H) domain (amino acids 1 to 129) containing the US3 consensus site was phosphorylated to a level that was 14% of that of full-length lamin A/C, the tail 1 (Fig. 5, lane T1) domain (amino acids 369 to 519) was even more heavily phosphorylated (to 37% of that of full-length lamin A/C), despite the lack of a pUS3 phosphorylation consensus motif. As expected from previous results, GST-pUL34 (amino acids 205 to 275) lacking the US3 kinase consensus motif was not phosphorylated by pUS3 (Fig. 5, lane C).
Taken together, these observations indicate that the full-length lamin A-GST was phosphorylated at multiple sites within lamin A. It follows that, at least in vitro, the previously derived US3 kinase consensus site is overly restrictive and other sites can also be phosphorylated.
US3 induces partial lamin A/C solubilization in purified Hep2 cell nuclei.
Because the phosphorylation of lamins is well known to regulate lamin disassembly, it was of interest to determine whether phosphorylation by US3 kinase could increase the solubility of lamin A/C. This was of particular interest because the complex structure of the nuclear lamina might preclude the access of pUs3 to lamin A/C or lamin A/C phosphorylation sites. To test whether US3 kinase activity could modify the nuclear lamina, an in vitro lamina disassembly assay was developed.
Briefly, nuclei from Hep2 cells were isolated, permeabilized, and incubated in US3 kinase buffer with GST-pUs3 or GST-pUs3(K220A) in the presence or absence of ATP. After the reaction, nuclei were pelleted by centrifugation, and equal amounts of supernatant were collected for analysis. To ensure minimal contamination of the supernatants with insoluble material, nuclei were subjected to a second high-speed centrifugation, and a portion of the supernatants were again collected. Protein in the supernatants and pellets were denatured in SDS, electrophoretically separated, transferred to nitrocellulose, and probed with a chicken polyclonal antibody to lamin A/C.
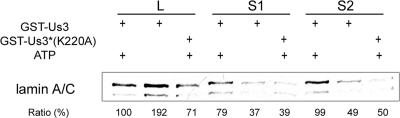
As shown in Fig. 6, the presence of GST-pUs3 and ATP increased the amount of lamin A/C solubilized into the supernatants by approximately twofold compared to that in reaction mixtures containing either GST-pUS3(K220A) plus ATP or GST-pUS3 without ATP. This reaction was repeated three times with similar results. We conclude that US3 kinase activity and ATP are sufficient to increase the solubility of lamin A/C from an endogenous preexisting nuclear lamina in vitro.
FIG. 6.
Immunoblots of total and solubilized lamin A/C from permeabilized Hep2 cell nuclei reacted with wild-type and mutant US3-encoded kinases. Purified and permeabilized Hep2 cell nuclei were incubated for 30 min in the presence or absence of the indicated fusion proteins and ATP. As a loading control, a sample of the total material (L) was collected immediately after the reactions. S1 was collected after centrifugation at low speed. A second supernatant fraction (S2) was collected after high-speed centrifugation. Proteins in the various fractions were denatured in SDS, electrophoretically separated, transferred to nitrocellulose, and probed with lamin A/C-specific antibody. Bound immunoglobulin was detected by a horseradish peroxidase-conjugated secondary antibody and chemiluminescence. The intensity of the chemiluminescence signals of the lamin A and C bands were quantified using a Syngene Chemi-Genius imaging system and associated GeneTools software, and the amount of chemiluminescence was pooled. The reported percentages represent the intensity of chemiluminescence in that lane relative to that detected in the first lane (sample L of GST-pUs3 plus ATP).
Lamin A/C is phosphorylated in HSV-infected cells, and the full spectrum of phosphorylation requires pUs3 kinase activity.
The studies described above were performed in vitro. In order to determine whether US3 phosphorylates lamin A/C in infected cells, two-dimensional electrophoretic analysis of lamin A/C was performed under various conditions. Specifically, Hep2 cells were mock infected or infected with 5.0 PFU/cell of wild-type HSV-1(F) or the US3(K220A) mutant virus and were lysed 16 h postinfection in 9 M urea to solubilize nuclear lamins (23). The proteins were separated by isoelectric focusing in the first dimension and by size in the second dimension. The electrophoretically separated proteins were transferred to nitrocellulose, and the positions of various isoforms of lamin A/C were revealed by immunoblotting with lamin A/C-specific chicken polyclonal antibody. The immunoblots were also probed with a goat antibody directed against lamin B1 and B2 with predicted isoelectric points of 5.11 and 5.29, respectively.
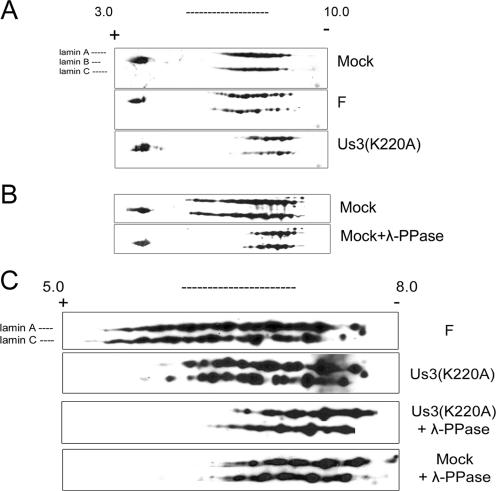
The results (Fig. 7) showed that unlike lamin A/C, neither lamin B isoform showed consistent or obvious shifts in isoelectric points under the various conditions tested. The lamin B-specific spots were therefore used as an internal control for alignment of the lamin A/C immunoblots. Upon separation in the first dimension in the pH 3.0 to 10.0 range, lamin A/C from mock-infected cells and cells infected with wild-type HSV-1(F) were broadly distributed throughout a wide pH range. Superficially, the patterns of lamin A/C-specific spots were similar in distribution, but mock-infected cells contained lamin A/C isoforms that were evenly distributed throughout the middle of the range, whereas some individual spots were more defined and easier to discern in lamin A/C taken from cells infected with HSV-1(F). This suggested that lamin A/C in HSV-1(F)-infected cells are modified differently than in mock-infected cells, but these differences are subtle, at least as viewed by two-dimensional gel electrophoresis. In contrast, however, US3(K220A)-infected cells lacked many of the more acidic lamin A/C species present within the populations of lamin A/C isoforms in HSV-1(F)-infected or mock-infected cells. The predicted isoelectric points of lamin A and C are 6.57 or 6.40, respectively. Separation in the first dimension using a narrower pH range, closer to the lamin A/C isoelectric point (pH 5.0 to 8.0) (Fig. 7C), confirmed that numerous acidic lamin A/C species detected in HSV-1(F)-infected cells were absent from cells infected with the US3 kinase mutant.
FIG. 7.
Two-dimensional gel electrophoresis and immunoblots of lamin A/C isoforms from mock-infected and HSV-infected cells. Hep2 cells were mock infected or infected with wild-type HSV-1(F) (panels labeled F) or mutant Us3(K220A). Proteins were extracted in buffer containing 9 M urea to solubilize lamins and separated by isoelectric focusing in the first dimension using nonlinear pH gradients from pH 3.0 to 10.0 (A and B) or linear pH gradients 5.0 to 8.0 (C) and by size on denaturing polyacrylamide gels in the second dimension. The proteins were then transferred to nitrocellulose and probed with polyclonal chicken antibody directed against lamin A/C or a rabbit antibody against lamin B1 and lamin B2. Bound immunoglobulin was detected by appropriate horseradish peroxidase-conjugated secondary antibodies. Chemiluminescence generated by the addition of appropriate substrates was recorded on X-ray film. In some experiments (λ-Ppase), proteins were reacted with λ-Ppase before electrophoretic separation. For comparative purposes, the three blots in panels A and B were aligned by position of lamin B-specific spots and by distance from the cationic and anionic poles. The positions of the terminal poles on the pH strip of the four immunoblots shown in panel C are also aligned. For orientation, the positions of the lamin A-, B-, and C-specific spots are indicated to the left of the top immunoblot shown in panels A and C.
Because the wide range of distribution of lamin A/C on first dimension might be due to posttranslational modifications other than phosphorylation (such as polyadenylylation), samples were treated with λ-Ppase, followed by two-dimensional electrophoretic separation and immunoblotting with lamin A/C antibody. As shown in Fig. 7B and C, phosphatase treatment of lamins from mock-infected cells resulted in a dramatic loss of the most acidic species of lamin A/C, indicating that the acidity of these species is a consequence of phosphorylation. Moreover, λ-Ppase treatment of lamin A/C also eliminated some acidic species detected in cells infected with the US3(K220A) mutant virus. Thus, US3 kinase activity cannot account for all of the lamin A/C phosphorylation in HSV-infected cells.
Taken together, these data indicate that many of the acidic lamin A/C isoforms are phosphorylated species whose acidity in infected cells depends on US3 kinase activity. We conclude that endogenous lamin A/C is phosphorylated during HSV-1 infection and that the full spectrum of phosphorylation requires US3 kinase activity.
DISCUSSION
The effect of US3 on the distribution of lamin A/C in HSV-1-infected cells.
Using two different methods, we have shown that US3 kinase activity is necessary for the pattern of lamin A/C distribution in HSV-1-infected Hep2 cells. Whereas lamin A/C and pUL31 and pUL34 immunostaining partially colocalized in a reticular pattern in Hep2 cells infected with wild-type virus, the lack of kinase activity in infected cells caused pUL31 and pUL34 to localize solely within circular regions lacking lamin A/C. These regions were wider in diameter and more circumscribed than most areas of cells infected with wild-type viruses containing pUL31/pUL34 but lacking lamin A/C. Similar conclusions were reached in studies of Vero cells, with the exception that the disruption of the lamina induced by both wild-type and US3(−) viruses were considerably more dramatic than that of uninfected cells, probably as a result of using different cell lines in the two studies (3). Our results are not a consequence of epitope masking because we used an eGFP-lamin A/C-expressing cell line and a polyclonal antibody insensitive to this HSV-induced effect with similar results.
We also noted subtle differences in the distribution of lamin A/C between the lamina of uninfected and HSV-1(F)-infected Hep2 cells. In both cases, the lamina contained small regions that lacked eGFP-lamin A/C. In infected cells, many, but not all, of these contained pUL34 and pUL31, proteins that are incorporated into nascent virions and are required for envelopment of nucleocapsids (41-43). Unlike the regions in uninfected Hep2 cells, larger regions of the nuclear rim, as viewed in planar sections, seemed to be devoid of lamin A/C in infected cells. It is unclear if these regions also lack lamin B or correspond to regions that communicate with the HSV DNA replication compartment (49). In any event, the concentration of pUL31 and pUL34 in both of the larger regions lacking lamin A/C and small gaps in the reticular pattern of lamin A/C argue that they both represent potential sites of virion egress. In contrast, the absence of US3 kinase or its activity restricted potential envelopment sites to fewer, very discrete, and round foci that also lacked lamin A/C.
These data suggest that during infection with wild-type virus, some potential envelopment sites (i.e., in the reticular pattern) may be very small or a consequence of localized thinning of the lamina, whereas others, while larger, remain somewhat diffuse. These observations suggest a sophisticated manipulation of the dynamics of nuclear lamins by HSV to promote virus egress while maintaining most laminar structure. This balance depends on US3 kinase activity, inasmuch as the absence of this activity causes very discrete breaches in the lamina as viewed by the distribution of eGFP-lamin A/C. It seems likely that capsids pass through these exaggerated breaches in the lamina and accumulate in the perinuclear space adjacent to these sites. This is supported by the observation that in the absence of US3, perinuclear virions accumulate in evaginations of the NM (42, 44). It would be useful to confirm this by live cell microscopy.
How does US3 kinase activity preclude the presence of discrete “holes” in the nuclear lamina and aggregation of virions in the perinuclear space? We speculate that there are three possibilities. The first possibility is that US3 kinase activity serves to increase lamin turnover and dynamics. Thus, phosphorylation by US3 would remove lamin A/C from some regions where US3 accumulates. Subsequent dephosphorylation of these lamins (not accounted for in these studies) would allow their reincorporation into gaps in the lamina, similar to mechanisms by which the lamina expands during interphase (30). The result is a more dynamic and flexible lamina to facilitate nuclear egress at multiple sites and the absence of most obvious gaps, since these are subsequently filled in. A second, related possibility is that US3's effects on lamin A/C distribution are unrelated to the direct phosphorylation of lamin A/C by US3. In this scenario, US3 kinase would phosphorylate other proteins that would then serve to increase lamin A/C phosphorylation in infected cells, leading to fewer discrete breaches in the lamina. A third possibility is related to the effect of US3 kinase on the localization of pUL31 and pUL34. At least pUL31 likely encodes its own depolymerizing effects on the lamina (40, 49), and its concentration in discrete areas of the nuclear rim in the absence of US3 kinase activity might promote highly localized lamina disruption. The converse of this hypothesis is that US3's kinase activity serves to redistribute the lamina depolymerizing, envelopment machinery (specifically the pUL34/pUL31 complex) more widely throughout the NM. Clearly, further studies are necessary to test specific aspects of these models and to determine whether US3's phosphorylation and displacement of lamin A/C are related to the localization of pUL34/pUL31.
Lamin A/C phosphorylation in infected cells.
Two-dimensional gel electrophoresis was chosen to characterize lamin A/C in infected and uninfected cells because of its ability to rapidly characterize differences between soluble and insoluble isoforms of lamin A/C. While it might seem surprising that the migration of lamin A/C in uninfected and HSV-1(F)-infected cells was only subtly different, we speculate that some of the phosphorylation of lamin A/C in mock-infected cells is consequential to phosphorylation by cellular kinases like cdc2 kinase in some mitotic cells (32, 33), whereas in HSV-infected cells, lamin A/C is primarily phosphorylated by the US3 kinase (Fig. 7). Thus, while collectively the electrophoretic profiles under the two conditions are similar, the mechanisms responsible for lamin A/C phosphorylation in the two circumstances are very different. These interpretations are supported by the observations that (i) HSV-infected cells do not undergo mitosis late in infection (9), (ii) US3 can directly phosphorylate lamin A/C at multiple sites in vitro (Fig. 5), and (iii) most highly phosphorylated lamin A/C species in HSV-1(F)-infected cells are not detected upon infection with the US3 kinase mutant (Fig. 7). On the other hand, some phosphorylated species of lamin A/C were detected even in the absence of US3 kinase activity, suggesting that other kinases also phosphorylate lamin A/C in HSV-1(F)-infected cells.
Substrates of the US3 kinase.
It was surprising that multiple regions of lamin A could be phosphorylated by purified GST-pUS3, despite the fact that only codon 10 matched the consensus sequence derived from previous analyses of peptide substrates (24, 35, 36). Indeed, some lamin A peptides lacking a bona fide pUS3 phosphorylation consensus sequence (e.g., amino acids 369 to 519 in the tail domain) were phosphorylated more heavily than the lamin A head domain bearing this motif (Fig. 5). In the same reactions, the US3 kinase did not phosphorylate a variety of E. coli proteins, nor GST, suggesting kinase specificity for lamin A/C. Parenthetically, the consensus phosphorylation site of cyclic AMP-dependent PKA phosphorylation is R-X-S/T or R-R/K-X-S/T, i.e., very similar to and partially overlapping that of pUS3, although the Km values of the respective enzymes differ (35). Although it has been speculated that pUS3 kinase activity functionally overlaps that of PKA in HSV-infected cells (1), the spectrum of US3 kinase substrates is likely broader inasmuch as protein kinase A phosphorylates lamin A/C only at N-terminal amino acids 1 to 32 in vitro (8).
These data indicate that the US3 kinase is more promiscuous than previously thought and suggest the existence of more substrates than previously predicted. Like other promiscuous kinases, it seems likely that the most important factor limiting phosphorylation by pUS3 would be access to the substrate in the living cell or virion, rather than the presence or absence of the previously reported phosphorylation motif. Given this consideration, it is reasonable to postulate that US3 kinase substrates other than lamin A/C could also contribute to alterations in the nuclear lamina of HSV-infected cells. Candidates include lamin B, various lamin receptors, and the HSV-1 UL31 protein.
Acknowledgments
We thank Kaihua Sun, Teresa Monique Gunn, and Ted Clark for help with two-dimensional electrophoresis; David Gilbert for the eGFP-lamin A/C expression construct; Richard Roller and Bernard Roizman for antisera and recombinant viruses; Carol Duffy for help with confocal microscopy; James Lee Smith for help with FACs analysis and sorting; Klaus Osterrieder for comments on the manuscript; and members of the Baines laboratory for interesting discussions.
These studies were supported by grants R01AI 52341 and S10 RR020981 from the National Institutes of Health.
Footnotes
Published ahead of print on 11 April 2007.
REFERENCES
- 1.Benetti, L., and B. Roizman. 2004. Herpes simplex virus protein kinase US3 activates and functionally overlaps protein kinase A to block apoptosis. Proc. Natl. Acad. Sci. USA 101:9411-9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjerke, S. L., J. M. Cowan, J. K. Kerr, A. E. Reynolds, J. D. Baines, and R. J. Roller. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J. Virol. 77:7601-7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjerke, S. L., and R. J. Roller. 2006. Roles for herpes simplex virus type 1 U(L)34 and U(S)3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347:261-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, Y. E., C. Van Sant, P. W. Krug, A. E. Sears, and B. Roizman. 1997. The null mutant of the UL31 gene of herpes simplex virus 1: construction and phenotype of infected cells. J. Virol. 71:8307-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collas, P. 1999. Sequential PKC- and Cdc2-mediated phosphorylation events elicit zebrafish nuclear envelope disassembly. J. Cell Sci. 112:977-987. [DOI] [PubMed] [Google Scholar]
- 6.Collas, P., L. Thompson, A. P. Fields, D. L. Poccia, and J. C. Courvalin. 1997. Protein kinase C-mediated interphase lamin B phosphorylation and solubilization. J. Biol. Chem. 272:21274-21280. [DOI] [PubMed] [Google Scholar]
- 7.Cross, T., G. Griffiths, E. Deacon, R. Sallis, M. Gough, D. Watters, and J. M. Lord. 2000. PKC-delta is an apoptotic lamin kinase. Oncogene 19:2331-2337. [DOI] [PubMed] [Google Scholar]
- 8.Eggert, M., N. Radomski, D. Tripier, P. Traub, and E. Jost. 1991. Identification of phosphorylation sites on murine nuclear lamin C by RP-HPLC and microsequencing. FEBS Lett. 292:205-209. [DOI] [PubMed] [Google Scholar]
- 9.Ehmann, G. L., T. I. McLean, and S. L. Bachenheimer. 2000. Herpes simplex virus type 1 infection imposes a G1/S block in asynchronously growing cells and prevents G1 entry in quiescent cells. Virology 267:335-349. [DOI] [PubMed] [Google Scholar]
- 10.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 11.Favoreel, H. W., G. Van Minnebruggen, D. Adriaensen, and H. J. Nauwynck. 2005. Cytoskeletal rearrangements and cell extensions induced by the US3 kinase of an alphaherpesvirus are associated with enhanced spread. Proc. Natl. Acad. Sci. USA 102:8990-8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher, D. Z., N. Chaudhary, and G. Blobel. 1986. cDNA sequencing of nuclear lamins A and C reveals primary and secondary structural homology to intermediate filament proteins. Proc. Natl. Acad. Sci. USA 83:6450-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frame, M. C., F. C. Purves, D. J. McGeoch, H. S. Marsden, and D. P. Leader. 1987. Identification of the herpes simplex virus protein kinase as the product of virus gene US3. J. Gen. Virol. 68:2699-2704. [DOI] [PubMed] [Google Scholar]
- 14.Galvan, V., and B. Roizman. 1998. Herpes simplex virus 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell-type-dependent manner. Proc. Natl. Acad. Sci. USA 95:3931-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruenbaum, Y., A. Margalit, R. D. Goldman, D. K. Shumaker, and K. L. Wilson. 2005. The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6:21-31. [DOI] [PubMed] [Google Scholar]
- 16.Hoger, T. H., G. Krohne, and W. W. Franke. 1988. Amino acid sequence and molecular characterization of murine lamin B as deduced from cDNA clones. Eur. J. Cell Biol. 47:283-290. [PubMed] [Google Scholar]
- 17.Hoger, T. H., K. Zatloukal, I. Waizenegger, and G. Krohne. 1990. Characterization of a second highly conserved B-type lamin present in cells previously thought to contain only a single B-type lamin. Chromosoma 100:67-69. [DOI] [PubMed] [Google Scholar]
- 18.Izumi, M., O. A. Vaughan, C. J. Hutchison, and D. M. Gilbert. 2000. Head and/or CaaX domain deletions of lamin proteins disrupt preformed lamin A and C but not lamin B structure in mammalian cells. Mol. Biol. Cell 11:4323-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jerome, K. R., R. Fox, Z. Chen, A. E. Sears, H.-Y. Lee, and L. Corey. 1999. Herpes simplex virus inhibits apoptosis through the action of two genes, Us5 and Us3. J. Virol. 73:8950-8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson, D. C., and P. G. Spear. 1982. Monensin inhibits the processing of herpes simplex virus glycoproteins, their transport to the cell surface, and the egress of virions from infected cells. J. Virol. 43:1102-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato, A., M. Yamamoto, T. Ohno, H. Kodaira, Y. Nishiyama, and Y. Kawaguchi. 2005. Identification of proteins phosphorylated directly by the Us3 protein kinase encoded by herpes simplex virus 1. J. Virol. 79:9325-9331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 23.Krohne, G. 2004. Lamins. Methods Cell Biol. 78:573-596. [DOI] [PubMed] [Google Scholar]
- 24.Leader, D. P., A. D. Deana, F. Marchiori, F. C. Purves, and L. A. Pinna. 1991. Further definition of the substrate specificity of the alpha-herpesvirus protein kinase and comparison with protein kinases A and C. Biochim. Biophys. Acta 1091:426-431. [DOI] [PubMed] [Google Scholar]
- 25.Leopardi, R., C. Van Sant, and B. Roizman. 1997. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. USA 94:7891-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang, L., and J. D. Baines. 2005. Identification of an essential domain in the herpes simplex virus 1 UL34 protein that is necessary and sufficient to interact with UL31 protein. J. Virol. 79:3797-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang, L., M. Tanaka, Y. Kawaguchi, and J. D. Baines. 2004. Cell lines that support replication of a novel herpes simplex 1 UL31 deletion mutant can properly target UL34 protein to the nuclear rim in the absence of UL31. Virology 329:68-76. [DOI] [PubMed] [Google Scholar]
- 28.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 29.Nalwanga, D., S. Rempel, B. Roizman, and J. D. Baines. 1996. The UL16 gene product of herpes simplex virus is a virion protein that colocalizes with intranuclear capsid proteins. Virology 226:236-242. [DOI] [PubMed] [Google Scholar]
- 30.Ottaviano, Y., and L. Gerace. 1985. Phosphorylation of the nuclear lamins during interphase and mitosis. J. Biol. Chem. 260:624-632. [PubMed] [Google Scholar]
- 31.Park, R., and J. D. Baines. 2006. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J. Virol. 80:494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peter, M., E. Heitlinger, M. Haner, U. Aebi, and E. A. Nigg. 1991. Disassembly of in vitro formed lamin head-to-tail polymers by CDC2 kinase. EMBO J. 10:1535-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peter, M., J. Nakagawa, M. Doree, J. C. Labbe, and E. A. Nigg. 1990. In vitro disassembly of the nuclear lamina and M phase-specific phosphorylation of lamins by cdc2 kinase. Cell 61:591-602. [DOI] [PubMed] [Google Scholar]
- 34.Poon, A. P., and B. Roizman. 2005. Herpes simplex virus 1 ICP22 regulates the accumulation of a shorter mRNA and of a truncated US3 protein kinase that exhibits altered functions. J. Virol. 79:8470-8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purves, F. C., A. D. Deana, F. Marchiori, D. P. Leader, and L. A. Pinna. 1986. The substrate specificity of the protein kinase induced in cells infected with herpesviruses: studies with synthetic substrates [corrected] indicate structural requirements distinct from other protein kinases. Biochim. Biophys. Acta 889:208-215. [DOI] [PubMed] [Google Scholar]
- 36.Purves, F. C., M. Katan, W. S. Stevely, and D. P. Leader. 1986. Characteristics of the induction of a new protein kinase in cells infected with herpesviruses. J. Gen. Virol. 67:1049-1057. [DOI] [PubMed] [Google Scholar]
- 37.Purves, F. C., R. M. Longnecker, D. P. Leader, and B. Roizman. 1987. The herpes simplex virus 1 protein kinase is encoded by open reading frame US3 which is not essential for virus growth in cell culture. J. Virol. 61:2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Purves, F. C., D. Spector, and B. Roizman. 1991. The herpes simplex virus protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J. Virol. 65:5757-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radsak, K. D., K. H. Brucher, and S. D. Georgatos. 1991. Focal nuclear envelope lesions and specific nuclear lamin A/C dephosphorylation during infection with human cytomegalovirus. Eur. J. Cell Biol. 54:299-304. [PubMed] [Google Scholar]
- 40.Reynolds, A. E., L. Liang, and J. D. Baines. 2004. Conformational changes in the nuclear lamina of cells infected with herpes simplex virus 1 requires genes UL31 and UL34. J. Virol. 78:5564-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds, A. E., B. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the HSV-1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roller, R., Y. Zhou, R. Schnetzer, J. Ferguson, and D. Desalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryckman, B. J., and R. J. Roller. 2004. Herpes simplex virus type 1 primary envelopment: UL34 protein modification and the US3-UL34 catalytic relationship. J. Virol. 78:399-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 46.Schumacher, D., B. K. Tischer, S. Trapp, and N. Osterrieder. 2005. The protein encoded by the US3 orthologue of Marek's disease virus is required for efficient de-envelopment of perinuclear virions and involved in actin stress fiber breakdown. J. Virol. 79:3987-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott, E. S., and P. O'Hare. 2001. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 75:8818-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shumaker, D. K., E. R. Kuczmarski, and R. D. Goldman. 2003. The nucleoskeleton: lamins and actin are major players in essential nuclear functions. Curr. Opin. Cell Biol. 15:358-366. [DOI] [PubMed] [Google Scholar]
- 49.Simpson-Holley, M., J. Baines, R. Roller, and D. M. Knipe. 2004. Herpes simplex virus 1 UL31 and UL34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 78:5591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simpson-Holley, M., R. C. Colgrove, G. Nalepa, J. W. Harper, and D. M. Knipe. 2005. Identification and functional evaluation of cellular and viral factors involved in the alteration of nuclear architecture during herpes simplex virus 1 infection. J. Virol. 79:12840-12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stackpole, C. W. 1969. Herpes-type virus of the frog renal adenocarcinoma: I. Virus development in tumor transplants maintained at low temperature. J. Virol. 4:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagenaar, F., J. M. Pol, B. Peeters, A. L. Gielkens, N. de Wind, and T. G. Kimman. 1995. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J. Gen. Virol. 76:1851-1859. [DOI] [PubMed] [Google Scholar]
- 53.Wild, P., M. Engels, C. Senn, K. Tobler, U. Ziegler, E. M. Schraner, E. Loepfe, M. Ackermann, M. Mueller, and P. Walther. 2005. Impairment of nuclear pores in bovine herpesvirus 1-infected MDBK cells. J. Virol. 79:1071-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]