Abstract
Human rhinovirus 14 (HRV14) is a member of the rhinovirus genus, which belongs to the picornavirus family, which includes clinically and economically important members, such as poliovirus, foot-and-mouth disease virus, and endomyocarditis virus. Capsid stability plays an important role in the viral infection process, in that it needs to be stable enough to move from cell to cell and yet be able to release its genetic material upon the appropriate environmental cues from the host cell. It has been suggested that certain host cell molecules, “pocket factors,” bind to the WIN drug-binding cavity beneath the canyon floor and provide transient stability to a number of the picornaviruses. To directly test this hypothesis, HRV14 was mutated in (V1188M, C1199W, and V1188M/C1199W) and around (S1223G) the drug-binding pocket. Infectivity, limited proteolysis, and matrix-assisted laser desorption ionization analyses indicate that filling the drug-binding pocket with bulky side chains is not deleterious to the viral life cycle and lends some stabilization to the capsid. In contrast, studies with the S1223G mutant suggest that this mutation at least partially overcomes WIN drug-mediated inhibition of cell attachment and capsid breathing. Finally, HRV16, which is inherently more stable than HRV14 in a number of respects, was found to “breathe” only at 37°C and did not tolerate stabilizing mutations in the drug-binding cavity. These results suggest that it is the drug-binding cavity itself and not the putative pocket factor that is crucial for the capsid dynamics, which is, in turn, necessary for infection.
Human rhinovirus 14 (HRV14) belongs to the rhinovirus genus of the picornavirus family of viruses (28). This family of viruses is characterized by small, icosahedral pseudo-T = 3 protein shells, which encapsidate a positive-sense RNA genome. The rhinovirus genus is further divided into major and minor groups of serotypes based on which cellular receptor they use for viral entry. Members of the largest group, the major group of HRV serotypes, recognize intercellular adhesion molecule 1 (ICAM-I) (10), while members of the minor group of serotypes enter the cell via very low density lipoprotein receptors (11, 12). The capsid consists of four viral proteins, VP1 to VP4 (Fig. 1), with VP1 to VP3 forming the bulk of the outer surface of the capsid and VP4 lying buried at the capsid/RNA interface in an extended conformation (27). A prominent depression, called the canyon, encircles each of the fivefold axes on the capsid surface and has been shown to be the site for receptor (ICAM-I) attachment (4, 23). A hydrophobic pocket lies beneath the canyon floor and is the binding site for antiviral compounds (33). Once bound to this pocket, these hydrophobic compounds stabilize the capsid and inhibit uncoating (5, 8). In the case of some serotypes, viral attachment to the host cell is also inhibited (24); however, this effect on attachment may be directly related to capsid stabilization (25, 31).
FIG. 1.
Stereo ribbon diagram of a protomeric unit of HRV14 with drug bound. The structure is oriented such that the RNA interior is towards the bottom of the figure, a fivefold axis is on the right side, and the exterior of the capsid is at the top of the figure. VP1 to -4 are colored blue, green, red, and mauve, respectively. A WIN compound is shown as a gray ball-and-stick model, with its molecular surface represented by a transparent cloud. Similarly, the side chains of the mutation sites described here are labeled and colored orange. Note that S1223 is exterior to the drug-binding cavity and points towards the floor of the canyon, while the other two mutations significantly fill the drug-binding cavity and sterically interfere with drug binding.
The role of a virus capsid is to transfer the genetic material from one host cell to another while protecting it from the extracellular milieu. To this end, the capsid has to be stable outside of the host cell when it is moving from one cell to another and at the same time readily uncoat upon its interaction with the host cell to deliver its genetic material. A number of nonenveloped viruses have been shown to undergo a dynamic “breathing” that is likely relevant to the eventual triggering of genomic release (2, 3, 9, 17, 18, 25). In the case of rhinovirus, the capsid is highly dynamic (17) and is affected by antibodies binding in the canyon and drugs binding in the hydrophobic pocket beneath the canyon (25). In both cases, it is likely that capsid stabilization is afforded by preventing conformational changes in the canyon region. In the case of the monoclonal antibody 17-IA, the stabilization is likely steric, since it binds deep in the canyon and contacts both the north and south sides of the receptor-binding region (25, 32). In contrast, the antiviral compounds likely stabilize the capsid by filling the hydrophobic pocket (33). Stabilization of proteins by filling internal cavities has been previously observed in many other systems (e.g., see references 19 and 20), and in the case of these compounds binding to poliovirus, stabilization has been shown, not surprisingly, to be entropy driven (35).
The drug-binding cavity has also been suggested to be the binding site for host cell-derived factors called “pocket factors” (PFs) (22). These putative factors have not been isolated or identified, and their existence has been wholly implied from the observation of small islands of electron density in the hydrophobic pocket in the 3.2-Å-resolution crystal structure of HRV1A (15) and in the 3.5-Å-resolution crystal structure of HRV16 (22). Similar electron density has been found in several other picornaviruses (13, 34, 36). The PFs have been suggested (22) to play an important role in the viral life cycle by providing transient stability to the capsid during its movement from one host cell to another. The interaction of the virus capsid with the host cell receptor in the canyon region has been suggested to be competitive with the binding of PFs (22). This was based on the observation that the conformation of the bottom of the canyon slightly changes upon drug binding (33) and that the antiviral compounds block receptor binding in the case of some HRV serotypes. To explain the interplay between WIN drugs and PFs, it has been hypothesized that WIN drugs have greater affinity than PFs and hence they compete PFs out of the hydrophobic pocket. Thus, according to the PF hypothesis, the affinities of the ICAM-I receptor, WIN compounds, and PFs are as follows: WIN has greater affinity than ICAM-I, which has much greater affinity than PF (22).
The purpose of the current study is to directly test the tenets of the PF hypothesis. If PFs are essential for the viral life cycle, then complete abrogation of their binding through mutagenesis should affect viral replication. In this study, four drug-resistant mutants of HRV14 (HRV14/V1188M, HRV14/C1199W, HRV14/V1188M/C1199W, and HRV14/S1223G) and the native HRV16 were analyzed using limited proteolysis followed by matrix-assisted laser desorption ionization (MALDI) analysis to study the effect of mutations on capsid “breathing.” Single-step growth curves and thermostability assays in the presence or absence of WIN drugs demonstrate the effects of various mutations on capsid dynamics and replication. The results presented here are entirely consistent with the hypothesis that the drug-binding cavity itself is essential to maintaining capsid dynamics and that the putative PFs are unlikely to play a major role in the viral life cycle.
MATERIALS AND METHODS
Mutagenesis.
HRV14 cDNA that produces infectious RNA was used as a template for mutagenesis by the PCR overlap method. The two mutation sites, V1188 and C1199, are in the cDNA region that is enclosed by the two unique restriction sites DraIII and AvrII. Two oligonucleotides were synthesized, one in 5′-3′ orientation, covering the DraIII site, and one in 3′-5′ orientation, covering the AvrII site. For each mutation, fragments of the two portions of the cDNA were made by PCR using one primer containing the mutation. These fragments were used as primers for subsequent PCRs to make the full-length DraIII/AvrII fragment. For the double mutation, the V1188M PCR product was used as a template for amplification with the C1199W primers. These fragments were inserted into the HRV14 cDNA.
A similar method was followed for HRV16 mutagenesis. The infectious HRV16 cDNA was used as a template for mutagenesis by PCR overlap method. Two mutation sites, L1181 and M1192, are located between the two unique restriction sites AvrII and ClaI. Using oligonucleotides covering the two restriction sites, two single mutations, L1181M and M1192W, and a double mutation, L118M/M1192W, were made and inserted back into the HRV16 cDNA.
In vitro transcription.
cDNA clones of HRV14 and HRV16 were linearized using MluI and SacI, respectively, followed by purification using the QIAGEN gel elution kit. The linearized cDNA clones were used as a template for RNA synthesis using in vitro transcription. Approximately 2 μg of linearized cDNA was mixed with the following components: ribonucleoside triphosphates to a final concentration of 1 mM, 1× T7 RNA polymerase buffer from Promega, 10 mM dithiothreitol, 20 U of RNase inhibitor, 20 U of T7 RNA polymerase enzyme from Promega, and nuclease-free water to a final volume of 20 μl. The reaction mixture was then incubated at 37°C for 60 min for RNA synthesis. RNA was stored at −20°C prior to transfection.
Transfection and amplification of the mutants.
As described previously (16), transfection of mutant RNA transcripts into HeLa cells was carried out using DEAE-dextran. RNA transcripts were diluted in HEPES-buffered saline containing 200 μg DEAE-dextran/ml. The solutions were added to HeLa cell monolayers and incubated at room temperature for 60 min. The cells were washed to remove DEAE-dextran and supplemented with 4 ml of medium A supplemented with 37.5 mM HEPES, pH 7.2, in a liquid overlay method. The plates were incubated for 48 h before the cells were scrapped and the virus particles released with repeated freezing and thawing. Plaque assays were performed to quantify the infectious particles. Virus titer amplification was accomplished using monolayers of HeLa cells on T75 and T150 flasks followed by roller bottles until the titer was ∼107 PFU/ml. Virus was purified and RNA extracted as described elsewhere (37).
Sequencing of viral RNA corresponding to coat proteins.
Viral RNA was extracted from purified virions, and cDNA was generated by reverse transcription-PCR using avian myeloblastosis virus reverse transcriptase. The regions enclosing the point mutations were amplified using the primers used for mutagenesis (AvrII, DraIII, and ClaI). The fragments were sequenced using the automated DNA sequencing facility at Iowa State. In the case of the HRV14 double mutant, the complete P1 region corresponding to the viral proteins VP1 to VP4 was sequenced to check for possible secondary site mutations. Six different primers were made at intervals of 500 bp and were used to read the entire 2.5-kbp fragment that encompasses the complete P1 region that encodes VP1-VP4.
Plaque assays.
All plaque assays were performed in 60- by 15-mm culture plates treated with vacuum gas plasma. HeLa cells grown in suspension cultures were spun down and resuspended in AH medium to a final density of 1.41 × 106 cells/ml. Five milliters of the resuspended cells were added to each plate and incubated in a 37°C CO2 incubator for 6 to 8 h to form a monolayer. The AH medium was suctioned off the plate, and the monolayers were washed with 5 ml phosphate-buffered saline. Serial dilutions of the virus were added to the middle of the plates and distributed uniformly over the monolayers. For assaying the effect of the WIN drugs, the serial dilutions of the virus were mixed with the specified amount of WIN drugs and incubated at room temperature for 1 h before infecting the monolayers. The virus dilutions were allowed to attach for 1 h, and then the unattached virus particles were removed by washing with phosphate-buffered saline (PBS). The infected monolayers were then overlaid with 2.5 ml of P6 medium (2×) and 1.6% agar in a 1:1 ratio. After allowing 10 min for P6-agar to form a semisolid gel, 2.5 ml P6 (1×) was added. The plates were then incubated at 37°C in a CO2 incubator for 48 h before staining with crystal violet.
Thermostability assays.
The virus stocks were incubated in the presence and absence of antiviral (WIN) drugs for 1 h prior to thermal inactivation. The virus samples were transferred to a 52°C water bath, and aliquots were removed at various time points and immediately transferred to ice. The samples containing WIN drugs were diluted 100-fold before the plaque assays were performed. This was sufficient to eliminate the inhibitory effect of the WIN compounds.
Single-step growth curves.
For single-step growth analysis, HeLa cells were maintained in a suspension culture and pelleted down at 600 × g for 10 min to a final concentration of ∼4 × 107 cells per ml. The cells were resuspended in 100 ml PBS at 4°C and pelleted as above to remove the calf serum from the growth medium. The cells were transferred to 15-ml Falcon tubes and infected with virus at an MOI of 10 to 15. For the experiments where the effect of WIN drugs on the single-step growth curve was studied, the inoculum was incubated with appropriate amounts of WIN 52084 for 1 h before infection. After allowing 1 h for attachment, the unattached virus particles were removed by washing with PBS. The infected cells were then resuspended in 20 ml prewarmed medium B (35°C). Aliquots of 0.5 ml were taken at every hour for 8 h and frozen immediately in a dry-ice ethanol bath. Virus titer at various time points were determined by plaque assays after the cells were lysed with repeated freeze-thaw cycles.
Limited proteolysis and MALDI analysis.
MALDI was performed as described previously (25). Briefly, purified virus samples (1 mg/ml) were used for limited proteolysis, and the digested fragments were analyzed using MALDI. Modified trypsin (Promega) was dissolved in sterile Milli Q water and mixed with the virus sample in 20 mM Tris (pH 7.6). The virus/trypsin reaction mixture was 10 μl in volume, with a virus:trypsin ratio of 100:1. To study the effect of WIN compounds, the purified virus sample was incubated with an appropriate amount of WIN 52084 for 1 h before it was subjected to proteolysis. An aliquot of virus with or without WIN drug before addition of trypsin was denoted the zero-time-point sample. Aliquots of 0.5 μl from the reaction mixture were taken at various time points and mixed with 0.5 μl of the matrix solution (3,5-dimethyl-4-hydroxy cinnamic acid in a saturated solution of acetonitrile-water containing 0.25% trifluoroacetic acid) on the MALDI plate. A Voyager-DESTER MALDI time-of-flight reflectron mass spectrometer was used for mass analysis. MALDI-generated ions were accelerated into the time-of-flight mass analyzer by a 20-kV pulse after a 100-ns delayed extraction period.
RESULTS
Stability of drug resistance mutations.
As one measure of the viability of the various mutations, the stabilities of the RNA genomes of the single (HRV14/V1188M, HRV14/C1199W, and HRV14/S1223G) and double (HRV14/V1188M/C1199W) mutants were monitored. RNA was extracted from the purified virions obtained after passaging six times in the presence of WIN 52084 (2.0 μg/ml). cDNA was generated using oligo(dT) from the RNA, and the entire P1 region was sequenced by amplifying the 2.5 kbp encompassing the entire structural protein coding region. In all cases, neither secondary site mutations nor revertants were observed. This suggests that these mutations are stable and therefore are likely beneficial to the virus in the presence of WIN drug. However, such mutations may have a deleterious effect on viral replication in the absence of drug. To test for this, the double mutant was passaged three times in the absence of WIN 52084, RNA was extracted from the purified virions, and the entire P1 region was sequenced. Neither secondary site compensation mutations nor site revertants were observed. This suggests that the double mutation does not have any adverse effect on the growth and amplification of HRV14 even in the absence of the selection pressure of the WIN compounds. Similar results were obtained in the cases of the V1188M, C1199W, and S1223G mutants.
Infectivity studies in the presence and absence of WIN 52084.
Infectivity studies were performed to determine the effect of WIN 52084 on the double and single mutants relative to that on the wild type. As expected, the wild-type virus was completely inhibited at 10 μg/ml of WIN compound (Fig. 2). Also as expected, the mutations within the drug-binding cavity, V1188M, C1199W, and the double mutation V1188M/C1199W, all demonstrated resistance to this high concentration of drug. It is important to note that there were differences in the numbers of PFU added to the plates shown in Fig. 2 in order to exemplify the effects of the drugs on wild-type virus in contrast to that on the drug-resistant forms. In fact, the particle/PFU ratio for the double mutant was slightly lower than that for the wild type, suggesting that the double mutant is actually more infectious (data not shown). Finally, the S1223G mutant, where the mutation lies outside the hydrophobic pocket and near the canyon floor, was also resistant to antiviral compounds when tested at 5 μg/ml WIN 52084 (Fig. 2).
FIG. 2.
Plaque assays showing drug resistance of the V1188M/C1199W HRV14 double mutant and V1188M and S1223G single mutants. The virus samples were incubated in the presence or absence of WIN compound for 1 h at room temperature before addition to the HeLa cell monolayers. A concentration of 10 μg/ml WIN 52084 was used, except in the case of the S1223G mutant, where 5 μg/ml was used because this mutation confers slightly lower resistance to the drug.
Thermostability assays demonstrate the ability of the HRV14 mutants to bind WIN drugs.
It has been shown that these antiviral compounds offer significant protection against heat inactivation (29). Therefore, this type of analysis provides a good measure of the ability of the virus particles to bind WIN compounds and confer capsid stabilization. In this study, the thermostability of the mutants was measured in the presence and absence of 10 μg/ml and 5 μg/ml WIN 52084 at 52°C. The infectivity of wild-type virus decreased by approximately three logs in the first 2 min (Fig. 3A) in the absence of drug. However, in the presence of 5 or 10 μg/ml WIN 52084, HRV14 was nearly completely protected against thermal inactivation (Fig. 3A). Even in the absence of drug, the V1188M and V1188M/C1199W mutants showed modest resistance to heat inactivation compared to the wild type (Fig. 3A and B). In stark contrast to the case with the wild type, WIN 52084 did not protect the mutants against thermal denaturation to nearly the same degree (Fig. 3A and B). Only the C1199W mutant shows modest resistance to heat inactivation at 10 μg/ml (Fig. 3C).
FIG. 3.
Thermostability assays showing ability of WIN compounds to bind and stabilize the various forms of HRV14 against heat shock at 52°C in the presence or absence of WIN 52084. A concentration of 10 μg/ml of drug was used for the wild-type, V1188M, and V1188M/C1199W forms of the virus. A concentration of 5 μg/ml was used in for the analysis of the S1223G mutant, and both concentrations were used in the case of the C1199W mutant, as noted on the graph.
As mentioned above, the S1223G mutation in HRV14 does not lie in the drug-binding cavity but rather near the canyon floor (Fig. 1). Unlike the case with the other drug-resistant mutants, WIN compounds protected the virus against heat inactivation to approximately the same extent as wild-type virus (Fig. 3D). This clearly shows that WIN compounds are able to bind to this mutant but that the mode of resistance to the drug is quite different than that with mutations that lie in the drug-binding cavity. However, it should be noted that at least with regard to thermal stability, the S1223G mutation does not cause an apparent destabilization of the capsid in the absence of the receptor. In fact, this mutation apparently affords modest protection against thermal denaturation.
Single-step growth curve experiments.
To determine whether the mutations had any effect on any stage of the virus life cycle, single-step growth curves for all the mutants were studied in the presence and absence of WIN 52084. The growth curves were determined by measuring the virus titer in aliquots taken from the cell suspension at various time points. With wild-type HRV14 in the absence of drug, there is typically a decrease in the number of infectious particles during the first 2 to 3 h of incubation. This has been ascribed to the particles undergoing the “eclipse” phase whereby they interact with the viral receptor and lose infectivity. This phenomenon is also observed in these studies (Fig. 4), albeit not being quite as pronounced as in other published work (29). In the presence of drug, previous studies with the major group of HRV serotypes have shown that this phase of the viral life cycle is muted by the drug, and it is therefore assumed that the drugs affect the initial attachment phase of the reaction (29). In contrast, the drugs do not apparently affect the early attachment events with the minor serotypes (24). As shown in Fig. 4, the yield of HRV14 in subsequent phases of viral growth is greatly impacted in the presence of drug. While for the major group of HRV serotypes, this has been entirely ascribed to the inhibition of attachment, it seems logical that it could be, at least in part, also due to the stabilizing effect that the drugs have on the viral particle. In the cases of the V1188M (Fig. 4A), C1199W (Fig. 4C), V1188M/C1199W (Fig. 4B), and S1223G (Fig. 4D) mutants, the addition of drug does not affect the growth curve profile, and this suggests that all of the mutants overcome the effect of the WIN drugs on cell attachment. Only in the case of the S1223G mutant is there a delay in the rise period, suggesting the inhibitory effect of WIN compounds on uncoating. Studies with a similar mutant (V1153I) demonstrated that WIN compounds inhibit the release of VP4, leading to a delay in the rise period after 4 h postinoculation (29). It is important to note that based on the thermal inactivation analysis, the V1188M and C1199W mutants likely act by blocking drug binding, whereas the S1233G mutant clearly binds drug and yet cell attachment is unaffected by the drug.
FIG. 4.
Single-step growth curves of wild type and mutants in the presence or absence of WIN 52084 (2 μg/ml).
Analysis of capsid breathing by MALDI studies.
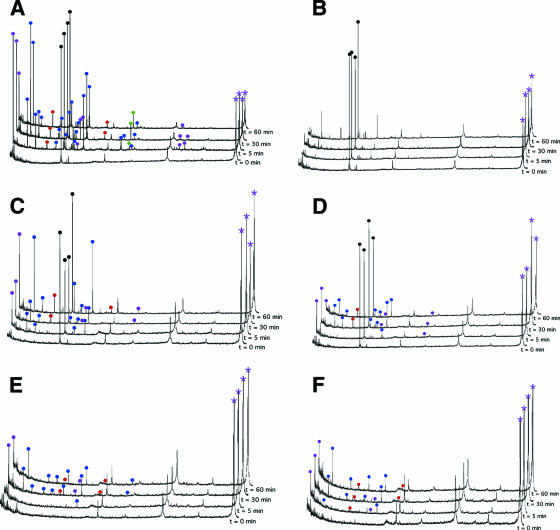
HRV undergoes a dynamic “breathing” process whereby the buried N termini of VP1 and VP4 are transiently exposed (17). As shown in these studies, this process can be monitored using limited proteolysis and MALDI analysis of the resulting proteolytic fragments. Also, as was previously described, WIN compounds completely inhibit this breathing process. Using this procedure, the effects of the mutations on capsid dynamics were measured. The MALDI spectra of the drug-binding cavity mutants (V1188M, C1199W, and V1188M/C1199W) are unaffected by WIN 52084 (Fig. 5A to F). However, in comparison to the wild type, the mutants show a relative decrease in capsid dynamics. This suggests that filling the pocket region with bulkier, hydrophobic residues reduces capsid “breathing.” In contrast, the S1223G mutant shows decreased capsid dynamics even at high WIN 52084 concentrations (Fig. 6), demonstrating that the mutation serves to abrogate the drug-induced inhibition of capsid breathing. It is important to note that the S1223G mutation did not cause an apparent increase in capsid dynamics, nor did it decrease the thermal stability of the capsid (Fig. 3). Together, this suggests that the effects of the S1223G mutation are not global (e.g., destabilizing the entire capsid) but rather are targeting the way in which the drug is blocking breathing and infectivity.
FIG. 5.
Limited trypsinolysis followed by MALDI analysis of wild-type (A and B), V1188M (C and D), or C1199W (E and F) form of HRV14 in the absence (A, C, and E) or presence (B, D, and F) of 10 μg/ml of WIN 52084. Fragments belonging to various capsid proteins are denoted by colored balls at the tops of the peaks in the MALDI spectrum. VP1, VP2, VP3, and VP4 are denoted by blue, green, red, and mauve, respectively. Black balls denote an internal MALDI standard. The mauve asterisk identifies the peak representing full-length VP4.
FIG. 6.
Capsid breathing of the S1223G mutant in the presence of various amounts of WIN 52084 (0, 2, 5, or 10 μg/ml). Fragments belonging to various capsid proteins are denoted by colored balls at the tops of the peaks in the MALDI spectrum. VP1, VP2, VP3, and VP4 are denoted by blue, green, red, and mauve, respectively. Black represents an internal MALDI standard. The mauve asterisk denotes full-length VP4.
MALDI analysis of HRV16.
HRV16 has been suggested to be more stable than HRV14 under certain circumstances (14). If, as suggested by the analysis of the drug pocket mutants, capsid dynamics is somehow correlated with overall capsid stability, then it follows that HRV16 should have capsid dynamics different from those of HRV14. Limited proteolysis followed by MALDI analysis of the resulting capsid fragments showed that HRV16 was resistant to digestion at room temperature (Fig. 7). Digested fragments belonging to VP4 and VP1 N termini appear only by the end of a 1-h exposure to trypsin (Fig. 7, left). At 37°C, digested fragments belonging to VP4 and VP1 N termini begin to appear within the first 5 min of exposure to trypsin (Fig. 7, right). The N termini of VP1 and VP4 of HRV14 are extremely sensitive to proteolysis at room temperature, and this is not appreciably increased at 37°C. This is entirely consistent with the previous observation that while the HRV16-ICAM-I complex is stable at room temperature (23), the HRV14-ICAM-I complex is stable only at 4°C.
FIG. 7.
Capsid breathing of HRV16 shows temperature dependence of capsid breathing. Fragments belonging to various capsid proteins are denoted by colored balls at the tops of the peaks in the MALDI spectrum. VP1, VP2, VP3, and VP4 are denoted by blue, green, red, and mauve, respectively. Black balls denote an internal MALDI standard. The mauve asterisk denotes the peak representing full-length VP4. The figure on the left represents digestion at room temperature, with that at 37°C shown on the right.
DISCUSSION
The PF hypothesis is essentially based on the fact that the antiviral WIN compounds were found to bind in a hydrophobic cavity beneath the canyon floor (33), followed by the observation that electron density was also found in this region of the crystal structures of poliovirus (13), HRV1A (15), HRV16 (22), and bovine enterovirus (34). It was initially suggested that the WIN compounds might be mimicking a “natural factor” that might play some role in the viral life cycle (33). Since the other viral structures were found to have electron density associated within the drug-binding cavity, it was thought that the density represented such “natural factors” from the cell and that these compounds facilitated the extracellular spread of the virus by transiently stabilizing the virions. This led to a model whereby “pocket factors” bind in a competitive manner with regard to receptor and WIN compounds, with the WIN drugs binding with a higher affinities than the cellular receptors, which, in turn, bind better than PFs (22). Again, the main tenet of this hypothesis is that the PFs, and not the pocket itself, play the crucial role in viral infectivity. One immediate contradiction to this model was the fact that the structure of native HRV14 did not have “pocket factor” bound within the cavity and yet was highly infectious and stable in this purified form.
There are additional findings that call into question the existence and role of the proposed cell-derived PFs. These cell-derived factors must have very high affinity for the capsid in order to remain bound to the capsid in spite of lengthy purification processes. However, according to the proposed model, PFs necessarily have a lower binding affinity than WIN compounds (22). As shown in the thermostability assays presented here, diluting the virus-WIN reaction mix by a factor of 100 is sufficient to diffuse the WIN compounds from the pocket in rapid order, as made evident by the retention of infectivity. Further, direct measurements of the dissociation constants have shown even the best WIN compounds have off rates of a few hours (7). According to the model (22), the PFs bind much more weakly than WIN compounds and would therefore have off rates much faster than this. These results are therefore not consistent with the suggestion that the cell-derived PFs, whose binding affinity is lower that that of WIN drugs, remain bound to the virions even after lengthy purification processes (e.g., see reference 15) that include multiple dilutions and buffer exchanges.
A direct challenge to the notion that the density observed in the drug-binding pocket represents host cell compounds comes from the fact that the drug-binding cavity is empty in the native HRV14 crystal structure (27) but has apparent “pocket factors” bound in the Fab17-HRV14 (32) and HRV14 chimera structures (6). In the case of the Fab17-HRV14 structure, not only is density observed in the drug-binding cavity, but the conformational changes associated with drug binding were also observed (30). The only difference among these three structures is that the cryoprotectant, polyethylene glycol 400, was used while collecting the crystallographic data for the Fab17 complex and the HRV14 chimera. Therefore, it logically follows that the density observed in drug-binding pockets of Fab17-HRV14 and the HRV14 chimera actually consists of molecules found in the polyethylene glycol 400 solution. Further, it is also possible that the molecules observed in the other viral structures may not be from the host cell but rather from the crystallization and/or purification milieu.
Results on the minor group of serotypes of rhinovirus also suggest that at least some of the details in the PF model are unlikely to be broadly applicable even among the rhinoviruses. The cell receptor for the minor-group serotypes of rhinovirus, the very low density lipoprotein receptor, interacts with the virus at the dome of the fivefold axes (12, 21, 26) rather than the canyon region (23). However, as with the major HRV serotypes, PF has been observed in the hydrophobic pocket of HRV2 (36). Due to the relative locations of the receptor and the PF, it seems unlikely that the two ligands bind in an antagonistic manner. This is substantiated by the fact that the WIN drugs do not have any effect on virus/receptor interactions and therefore act by stabilizing the capsid and preventing uncoating. Therefore, it seems unlikely that the minor HRV serotypes can have the same interplay between receptor and PF as described previously (22).
The drug-resistant mutations of HRV14 against WIN compounds also call into question the PF hypothesis. There are two main types of mutants: those that block WIN drug binding in the pocket region, referred to as the drug exclusion mutants, and the drug compensation mutants, which block drug efficacy without affecting binding. In the case of the drug exclusion mutants, smaller hydrophobic compounds might still bind, but previous studies have shown that smaller molecules have significantly weaker binding constants (1). These smaller WIN compounds have been shown to cause conformational changes in the virus capsid, similar to the longer WIN compounds. However, their extremely high 50%-effective-dose concentrations make it unlikely that host cell-derived PFs exist at effective levels in nature. The double pocket mutant has the drug-binding cavity filled to such an extent that it is unlikely that anything other than the smallest of molecules, and most certainly not PFs, is likely to bind. In spite of this, these variants exhibit the same growth and replication phenotype as wild-type virus. While it might be argued that these hydrophobic residues mimic the PFs, this cannot be the case, since they do not associate with the drug-binding pocket in a reversible manner. This strongly suggests that even if cell-derived molecules do bind to the drug-binding pocket, they apparently do not play a demonstrable role in the viral life cycle.
The drug compensation mutants seem to overcome the inhibitory effect of the drug without abrogating binding. When the drug binds under the canyon floor, there is a relatively small conformational change in the peptide strands between it and the canyon floor. The S1223G drug compensation mutant lies on the strand that undergoes the largest of these movements (33). There is no structural evidence that this particular residue directly contacts the bound receptor, and therefore it is unlikely that this mutation acts by eliminating some sort of steric clash between the canyon floor and the receptor. Instead, as shown by thermostability and infectivity assays, it is apparent that this mutation partially abrogates drug inhibition by overcoming some of the stabilizing effects of the drug. As is made evident by the fact that the antiviral compound is still able to protect the virus from thermal denaturation, it is apparent that the drug can still bind to this mutant but no longer inhibits replication or cellular attachment. This is entirely consistent with the fact that this mutant partially abrogates the WIN drug inhibition of viral capsid breathing. As we proposed previously (31), the model that is entirely consistent with all of the above results is that the drug-binding cavity, and not the putative PFs, is crucial for the viral life cycle by affording capsid dynamics at the canyon region. From this model, the drug mainly acts by stabilizing the capsid, which, in the case of the major serotypes, in turn affects interactions with the receptor. When the drug-binding pocket is partially filled with hydrophobic residues (HRV14/V1188M/C1199W), the stabilizing effect of the drug is partially mimicked, as shown by the decrease in viral breathing of the mutants. The S1223G mutation circumvents some of the stabilizing effects by increasing the flexibility of the canyon region, as made evident by infectivity and capsid breathing in the presence of the drug. It should be noted, however, that the effects of the S1223G mutation are very much targeted towards abrogating the effects of the drug. The mutation does not decrease the thermal stability of the capsid (Fig. 3), nor does it increase the overall capsid dynamics (Fig. 6). Instead, it appears to break some conduit between the drug and its ability to inhibit breathing and infectivity.
In the case of HRV16, filling the hydrophobic cavity (L1181M and M1192W mutations) with bulkier residues did not result in viable mutants. This is not wholly surprising, since HRV16 is more stable than HRV14 and filling the cavity probably increases the stability to a level beyond the ability of the receptor to trigger uncoating. Further, HRV16 WIN drug-resistant mutants have mutations outside the hydrophobic pocket at the protomer-protomer interface (37). These mutants fail to assemble in the absence of the WIN compounds (16) and can be rescued by adding drugs late in the life cycle. These studies suggest that the mutants are able to interact with ICAM-I and undergo uncoating even while the WIN drugs are bound to the hydrophobic pocket. The HRV16 drug-resistant mutants, similar to the S1223G HRV14 mutant, counteract the stabilizing effect of WIN compounds, thereby leading to host cell infection. Unlike the S1223G mutant, these mutations likely cause global destabilization of the capsid that is countermanded by the WIN compounds. Nevertheless, these results are directly contradictory to the PF model, where there is a direct competition between drug and receptor binding.
From the results presented here, together with those from previous studies, it seems very unlikely that PFs play a significant role in the life cycle of picornaviruses. Instead, the results suggest that the empty pocket affords necessary flexibility to the capsid. In the case of the major serotypes, this flexibility is also important in enhancing receptor interactions with the canyon, which are then followed by uncoating. Overall, the results presented here support our previous model for uncoating (Fig. 8) (31). In order for uncoating to occur, the capsid must overcome an activation energy barrier. The virus capsid is necessarily dynamic and samples various conformational states up to the precipice of uncoating, as made evident by the capsid “breathing.” In the case of the major serotypes, the magnitude of the activation energy is diminished by the receptor and perhaps the low pH of the endosome. This process is akin to the action of enzymes that serve to decrease the energy of the activation barrier. In contrast, the WIN drugs decrease the initial energy state, presumably through entropic changes, and hence, the energy of activation increases to the point where even the breathing and receptor-mediated changes are inhibited. The most prominent change during this breathing is the transient externalization of VP1 and VP4 N termini in a process that is likely the normal prelude to capsid uncoating. Key to this process, we propose, is the empty drug-binding cavity. Therefore, it is the drug-binding cavity itself, common to all HRV serotypes, that is crucial for viral uncoating and not the previously observed PFs.
FIG. 8.
Thermodynamic model describing the capsid dynamics of HRV14. Ea represents the activation energy barrier the virus has to overcome in order to uncoat and release its RNA and VP4. The black line represents the normal process of HRV14 in the absence of drug and receptor, the green line in the presence of the receptor, and the blue line in the presence of WIN compounds, with the drug-dependent mutants in red.
Acknowledgments
This work was supported by an NIH grant to T.J.S. (GM10704).
Footnotes
Published ahead of print on 11 April 2007.
REFERENCES
- 1.Bibler-Muckelbauer, J. K., M. J. Kremer, M. G. Rossmann, G. D. Diana, F. J. Dutko, D. C. Pevear, and M. A. McKinlay. 1994. Human rhinovirus 14 complexed with fragments of active antiviral compounds. Virology 202:360-369. [DOI] [PubMed] [Google Scholar]
- 2.Bothner, B., X. F. Dong, L. Bibbs, J. E. Johnson, and G. Siuzdak. 1998. Evidence of viral capsid dynamics using limited proteolysis and mass spectrometry. J. Biol. Chem. 273:673-676. [DOI] [PubMed] [Google Scholar]
- 3.Broo, K., J. Wei, D. Marshall, F. Brown, T. J. Smith, J. E. Johnson, A. Schneemann, and G. Siuzdak. 2001. Viral capsid mobility: a dynamic conduit for inactivation. Proc. Natl. Acad. Sci. USA 98:2274-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colonno, R. J., J. H. Condra, S. Mizutani, P. L. Callahan, M. E. Davies, and M. A. Murcko. 1988. Evidence for the direct involvement of the rhinovirus canyon in receptor binding. Proc. Natl. Acad. Sci. USA 85:5449-5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Diana, G. D., D. C. Pevear, M. J. Otto, M. A. McKinlay, M. G. Rossmann, T. J. Smith, and J. Badger. 1989. Inhibitors of viral uncoating. Pharmacol. Ther. 42:289-305. [DOI] [PubMed] [Google Scholar]
- 6.Ding, J., A. D. Smith, S. C. Geisler, X. Ma, G. F. Arnold, and E. Arnold. 2002. Crystal structure of a human rhinovirus that displays part of the HIV-1 V3 loop and induces neutralizing antibodies. Structure 10:999-1011. [DOI] [PubMed] [Google Scholar]
- 7.Fox, M. P., M. A. McKinlay, G. D. Diana, and F. J. Dutko. 1991. Binding affinities of structurally related human rhinovirus capsid-binding compounds are related to their activities against human rhinovirus type 14. Antimicrob. Agents Chemother. 35:1040-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fox, M. P., M. J. Otto, and M. A. McKinlay. 1986. Prevention of rhinovirus and poliovirus uncoating by WIN 51,711, a new antiviral drug. Antimicrob. Agents Chemother. 30:110-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fricks, C. E., and J. M. Hogle. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greve, J. M., G. Davis, A. M. Meyer, C. P. Forte, S. C. Yost, C. W. Marlor, M. E. Kamarck, and A. McClelland. 1989. The major human rhinovirus receptor is ICAM-I. Cell 56:839-847. [DOI] [PubMed] [Google Scholar]
- 11.Hewat, E. A., and D. Blaas. 1996. Structure of a neutralizing antibody bound bivalently to human rhinovirus 2. EMBO J. 15:1515-1523. [PMC free article] [PubMed] [Google Scholar]
- 12.Hewat, E. A., E. Neumann, J. Conway, R. Moser, B. Ronacher, T. C. Marlovits, and D. Blaas. 2000. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. EMBO J. 19:6317-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 14.Hoover-Litty, H., and J. M. Greve. 1993. Formation of rhinovirus-soluble ICAM-I complexes and conformational changes in the virion. J. Virol. 67:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, S., T. J. Smith, M. S. Chapman, M. G. Rossmann, D. C. Peavear, F. J. Dutko, P. J. Felock, G. D. Diana, and M. A. McKinlay. 1989. The crystal structure of human rhinovirus serotype 1A (HRV1A). J. Mol. Biol. 210:91-111. [DOI] [PubMed] [Google Scholar]
- 16.Lee, W. M. 1992. Human rhinovirus 14: synthesis and characterization of a molecular cDNA clone which makes highly infectious transcripts. Ph.D. dissertation. University of Wisconsin, Madison.
- 17.Lewis, J. K., B. Bothner, T. J. Smith, and G. Siuzdak. 1998. Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. USA 95:6774-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, Q., A. G. Yafal, Y. M. H. Lee, J. Hogle, and M. Chow. 1994. Poliovirus neutralization by antibodies to internal epitopes of VP4 and VP1 results from reversible exposure of the sequences at physiological temperatures. J. Virol. 68:3965-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews, B. W. 1995. Studies on protein stability with T4 lysozyme. Adv. Protein Chem. 46:249-278. [DOI] [PubMed] [Google Scholar]
- 20.Morton, A., and B. W. Matthews. 1995. Specificity of ligand binding in a buried nonpolar cavity of T4 lysozyme: linkage of dynamics and structural plasticity. Biochemistry 34:8576-8588. [DOI] [PubMed] [Google Scholar]
- 21.Okun, V., R. Moser, B. Ronacher, E. Kenndler, and D. Blaas. 2001. VLDL receptor fragments of different lengths bind to human rhinovirus HRV2 with different stoichiometry. An analysis of virus-receptor complexes by capillary electrophoresis. J. Biol. Chem. 276:1057-1062. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira, M. A., R. Zhao, W.-M. Lee, M. J. Kremer, I. Minor, R. R. Rueckert, G. D. Diana, D. C. Pevear, F. J. Dutko, M. A. McKinlay, and M. G. Rossmann. 1993. The structure of human rhinovirus 16. Structure 1:51-68. [DOI] [PubMed] [Google Scholar]
- 23.Olson, N. H., P. R. Kolatkar, M. A. Oliveira, R. H. Cheng, J. M. Greve, A. McClelland, T. S. Baker, and M. G. Rossmann. 1993. Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. USA 90:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pevear, D. C., F. J. Fancher, P. J. Feloc, M. G. Rossmann, M. S. Miller, G. Diana, A. M. Treasurywala, M. A. McKinlay, and F. J. Dutko. 1989. Conformational change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J. Virol. 63:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reisdorph, N., J. Thomas, U. Katpally, E. Chase, K. Harris, G. Siuzdak, and T. J. Smith. 2003. Human rhinovirus capsid dynamics is controlled by canyon flexibility. Virology 314:34-44. [DOI] [PubMed] [Google Scholar]
- 26.Ronacher, B., T. C. Marlovits, R. Moser, and D. Blaas. 2000. Expression and folding of human very-low-density lipoprotein receptor fragments: neutralization capacity toward human rhinovirus HRV2. Virology 278:541-550. [DOI] [PubMed] [Google Scholar]
- 27.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, R. R. Rueckert, B. Sherry, and G. Vriend. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature (London) 317:145-153. [DOI] [PubMed] [Google Scholar]
- 28.Rueckert, R. R. 1996. Picornaviridae and their replication. Raven Press, New York, NY.
- 29.Shepard, D. A., B. A. Heinz, and R. R. Rueckert. 1993. WIN 52035-2 inhibits both attachment and eclipse of human rhinovirus 14. J. Virol. 67:2245-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, T. J. 2002. Antibody interactions with rhinovirus, p. 39-49. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 31.Smith, T. J., and T. S. Baker. 1999. Picornaviruses: epitopes, canyons, and pockets. Adv. Virus Res. 52:1-23. [DOI] [PubMed] [Google Scholar]
- 32.Smith, T. J., E. S. Chase, T. J. Schmidt, N. H. Olson, and T. S. Baker. 1996. Neutralizing antibody to human rhinovirus 14 penetrates the receptor-binding canyon. Nature (London) 383:350-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, T. J., M. J. Kremer, M. Luo, G. Vriend, E. Arnold, G. Kamer, M. G. Rossmann, M. A. McKinlay, G. D. Diana, and M. J. Otto. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233:1286-1293. [DOI] [PubMed] [Google Scholar]
- 34.Smyth, M., J. Tate, E. Hoey, C. Lyons, S. Martin, and D. Stuart. 1995. Implications for viral uncoating from the structure of bovine enterovirus. Nat. Struct. Biol. 2:224-231. [DOI] [PubMed] [Google Scholar]
- 35.Tsang, S. K., P. Danthi, M. Chow, and J. M. Hogle. 2000. Stabilization of poliovirus by capsid-binding antiviral drugs is due to entropic effects. J. Mol. Biol. 296:335-340. [DOI] [PubMed] [Google Scholar]
- 36.Verdaguer, N., D. Blaas, and I. Fita. 2000. Structure of human rhinovirus serotype 2 (HRV2). J. Mol. Biol. 300:1179-1194. [DOI] [PubMed] [Google Scholar]
- 37.Wang, W., W. M. Lee, A. G. Mosser, and R. R. Rueckert. 1998. WIN 52035-dependent human rhinovirus 16: assembly deficiency caused by mutations near the canyon surface. J. Virol. 72:1210-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]