Abstract
Hepatic fibrosis is the primary mediator of disease due to chronic infection with hepatitis C virus (HCV). HCV exists as a quasispecies in each infected individual, and longitudinal viral sequence changes may reveal viral dynamics and the selection pressures applied by the host immune system. Thus, we hypothesized that patterns of sequence change might reveal the immunopathogenesis of fibrosis progression. We tested this hypothesis by studying individuals enrolled in a prospective study of chronic HCV-related hepatic fibrosis with little or no fibrosis at first biopsy (stage 0 or 1) and a second planned liver biopsy sample obtained 4 years later. Serum was obtained from five individuals with fast progression (FP; defined as a >2-stage change between visits) and 10 carefully matched individuals with slow progression (SP; defined as a <2-stage change between visits). We sequenced multiple cloned hemigenomic cDNAs from each person spanning six genes (core through NS3). Phylogenetic analysis revealed temporal shifts in phylogenetic clustering over time, suggesting frequent quasispecies replacement rather than simple diversification. In addition, mixed infections were detected in three subjects, with coexistence in two subjects (one FP, one SP) of subtypes 1a and 1b throughout the 4-year biopsy interval. Subjects with FP had a higher rate of evolution than subjects with SP, with a preponderance of synonymous changes, suggesting purifying selection, except in hypervariable region 1, where positive selection pressure is frequently detected. Thus, in a small but carefully matched cohort we found evidence for rapid neutral evolution of HCV in persons with rapid progression of hepatic fibrosis, suggesting higher turnover of infected cells.
In the United States, approximately 3.9 million persons have chronic hepatitis C virus (HCV) infection, which may progress to cirrhosis or hepatocellular carcinoma (3, 17, 43). HCV infection is the leading indication for liver transplantation in the United States. Complications of HCV infection result in approximately 10,000 deaths annually, and this rate is expected to rise sharply in the next 10 to 20 years, since most infections were acquired between 1970 and 1990 and cirrhosis generally manifests 20 to 30 years after exposure (52).
The frequency at which HCV infection progresses to cirrhosis or end-stage liver disease (ESLD) is controversial. Di Bisceglie and coworkers found that 20% of persons with posttransfusion HCV infection had cirrhosis 1 to 16 years later (6). At another extreme, of 232 Irish women infected 17 years earlier through HCV-contaminated immunoglobulin, only 2.4% had cirrhosis (16). This variability underscores the importance of determining the biological basis and risk factors for disease progression.
Mechanisms and correlates of fibrosis are largely unknown and cannot be investigated directly using in vitro or animal models. To date, the most important model system for HCV infection has been the chimpanzee, but HCV-infected chimpanzees rarely develop significant hepatic fibrosis (46). Therefore, carefully matched humans remain the only direct means to study HCV-related liver fibrosis. Cross-sectional and longitudinal observational studies have shown that accelerated liver fibrosis may be associated with older age at HCV infection, coinfection with human immunodeficiency virus (HIV) or HBV, alcohol use, gender, and possibly race (6, 16, 40, 41, 49).
Evolution of the HCV genome has proven to be an informative marker of host-virus interaction. Each HCV-infected person harbors a viral quasispecies, or swarm of genetically distinct but related variants. The genetic diversity and evolution rate of HCV are highest in the envelope genes, particularly hypervariable region 1 (HVR1) at the N terminus of the E2 region, but other regions are informative as well. Analyses of envelope gene sequences obtained during acute infection suggest that greater diversity in HVR1 is associated with viral persistence, rather than spontaneous clearance, during acute HCV infection (8, 37). In addition, sequences obtained during acute (4, 42) and chronic (35) infection reveal strong evidence of selection pressure on nonstructural genes exerted by CD8-positive T cells, as well as significant constraints on sequence evolution. Therefore, HCV sequences recoverable from serum have the potential to reveal important aspects of viral pathogenesis.
Studies of the relationship between HCV-related liver disease and the viral quasispecies have generated conflicting results. Using single-stranded conformational polymorphism, investigators have found higher viral complexity associated with higher alanine aminotransferase (ALT) (54) and advanced fibrosis (18) in some studies but not in others (10, 22, 27, 29, 38). Rather than use a gel shift assay, some studies have used nucleotide sequencing to study the viral quasispecies. More advanced fibrosis has been associated with higher nucleotide diversity in the structural core, E1, and E2 genes in some studies (12, 39). These studies are typically cross-sectional, which is problematic because the evolution of liver disease might distort the viral quasispecies, potentially obscuring important phenomena that might be present during the earlier stages of progressive liver fibrosis. In a longitudinal study of HCV quasispecies variation in the setting of renal transplantation in persons with chronic HCV infection, lower rates of amino acid replacements in HVR1 were strongly associated with more rapid progression of fibrosis (13). In a longitudinal study of HCV sequence variation in 20 persons with or without ESLD in the setting of inherited bleeding disorders (80% of whom had HIV infection), HVR1 complexity and diversity were higher early in disease and narrowed with disease progression (33). The extent to which these findings obtained with immunosuppressed patients can be extrapolated to the general population is unclear.
We sought to address the limitations of prior studies, including the retrospective approach, lack of paired biopsies, prospectively collected data on other risk factors (e.g., alcohol intake), and limitation to single genomic regions (preventing comparison of structural and nonstructural genes, for example). Nested within a longitudinal study of HCV infection and fibrosis progression (51), we identified case subjects (fast progressors [FP]) with little or no fibrosis on initial liver biopsy followed by rapid progression of fibrosis 4 years later and carefully matched control subjects (slow progressors [SP]) with little or no fibrosis at baseline and no significant increase in fibrosis 4 years later. Serum obtained within 6 months of each biopsy was used to obtain cDNA clones of the HCV hemigenome spanning the core, E1, E2, p7, NS2, and NS3 regions. Principal findings associated with FP included higher overall genetic diversity at baseline and higher rate of change across all genes, without strong evidence of greater immune selection pressure. These findings have potential implications for mechanistic understanding of HCV pathogenesis, and modeling of disease pathogenesis.
MATERIALS AND METHODS
Study subjects.
We studied chronically HCV-infected persons with paired biopsies obtained approximately 4 years apart in a prospective study of determinants of liver disease progression (51). Between 1996 and 1998, 210 subjects were randomly selected from 1,667 HCV-infected injection drug users (IDUs) who were members of the community-based ALIVE study cohort in Baltimore, MD, which was described previously (34, 41, 44). Four years later, the 210 subjects originally biopsied were offered a second liver biopsy to determine rate and predictors of liver fibrosis progression. Exclusion criteria included decompensated liver disease or HIV infection at first biopsy, severe medical comorbidity, increased bleeding risk, and advanced immunosuppression. Levels of serum transaminases (aspartate aminotransferase [AST] and ALT) were determined in a clinical laboratory, and HCV RNA was measured using the COBAS Amplicor HCV monitor test, version 2.0 (Roche Molecular Systems), in accordance with the manufacturer's instructions.
Five subjects who had stage 0 or 1 fibrosis at baseline and an increase of ≥3 in stage number at the second biopsy were defined as case subjects. Controls were also required to have stage 0 or 1 fibrosis at baseline but an increase of ≤1 at the second biopsy. To enhance separation of the two groups, no subject with a 2-stage change in fibrosis was included. Controls were matched 2:1 with case subjects, based on age ± 10 years, HBsAg seropositivity, HIV status, and alcohol use. All subjects were HIV and HBsAg negative. All study subjects in this sample identified themselves as African Americans, reflecting the study population, which is more than 90% African-American (34). None of these subjects had been treated for HCV infection at the time of the second biopsy.
Liver biopsies and histologic examination.
A transcutaneous liver biopsy was performed as previously described (34, 51). Liver tissue was fixed in 10% formalin, and paraffin-embedded sections were stained with hematoxylin and eosin and Masson's trichrome stains. The degree of inflammation was graded and the amount of fibrosis staged using the Ishak modified hepatic activity index by a single experienced hepatopathologist (M.S.T.), who was blinded to clinical markers other than HCV antibody status.
Amplification of a 5.2-kb fragment of the entire genome.
The region encoding core, E1, E2, p7, NS2, and NS3 was amplified and cloned from 140 μl of serum as previously described (24), except that the reverse transcription (RT) primer was 6080G1R-16 (5′-CCGGTTCATCCAYTGC-3′; positions 6080 to 6095 in strain H77C; GenBank accession number AF009606), used at a concentration of 0.2 μM. Appropriate measures to prevent contamination were routinely followed, including segregation of specimen handling, PCR, and plasmid manipulations into separate rooms (20).
To ensure that template resampling (23) was unlikely, five randomly selected specimens were subjected to additional testing. HCV RNA was extracted in the same manner from serum diluted 1:10 and 1:100, with positive results, indicating that the clones we obtained from undiluted serum were not obtained by repetitive sampling of a limited number of templates.
Cloning of 5.2-kb cDNA.
In order to facilitate the cloning of the 5.2-kb fragment, PCR products were further extended to add an A base at the terminus of the fragment at 68°C for 10 min with dATP and Taq polymerase (Invitrogen) before cloning. After that, the products were gel purified with SeaKem GTG agarose (FMC Bioproducts, Rockland, ME), visualized by crystal violet, and cloned by utilizing a TOPO XL PCR cloning kit (Invitrogen, Carlsbad, CA) according to the manufacturer's protocols. Forty colonies were randomly picked for each specimen and cultured in 300 μl of Luria-Bertani broth with 50 μg/ml kanamycin. Ten of them were randomly selected for further study. All 40 cDNA clones were stored in 20% glycerol at −80°C.
Sequencing of clones.
Rolling circle amplification was performed utilizing a bacteriophage DNA polymerase (TempliPhi; Invitrogen) for the preparation of sequencing templates according to the protocol of the manufacturer. Briefly, the mixture, containing 5 μl sample buffer and 1 μl liquid bacterial culture in a 0.2-ml reaction tube, was heated at 95°C for 3 min and cooled to 4°C. A premix solution containing 5 μl reaction buffer and 0.2 μl enzyme mix was added to each reaction mixture. After incubation at 30°C for 6 h, the polymerase was inactivated at 65°C for 10 min, and 40 μl of sterile water was added and mixed by vortexing. Finally, 10 μl of the diluted solution mixed with 10 μl of 1 μM sequencing primer was applied to a Prism 3100 automated sequencer (ABI, Foster City, CA). Primer sequences were removed prior to analysis.
Identification of cDNA clones for 5-kb sequencing.
Sequencing of the 5.2-kb hemigenome was performed for all five case subjects and the first five control subjects (SP1 through SP5), two cDNA clones per specimen (four clones each). For each specimen, envelope sequences from 10 random randomly selected clones were determined using primer H77-1868a21 (24). Phylogenetic analysis with outgroup sequences AF009606 (strain H77, subtype 1a), D90208 (strain J, subtype 1b), D14853 (strain G9, subtype 1c), and Y11604 (strain ED43, subtype 4a) was used to determine which 2 of the 10 clones for each subject were closest to their most recent common ancestor, based on patristic distance. These were sequenced across the 5.2-kb length of the cDNA clone, and sequences were assembled into contigs using Aligner (version 1.5.2; CodonCode Corporation, Dedham, MA).
Phylogenetic analysis.
Sequences were assembled and analyzed by using BioEdit (version 7.0.5; http://www.mbio.ncsu.edu/BioEdit/bioedit.html), with alignment performed using ClustalX (version 1.8.3; http://bips.u-strasbg.fr/fr/Documentation/ClustalX/) (14). Complexity and normalized Shannon entropy were calculated as previously described (47). The best-fit phylogenetic models (general time reversible with invariant sites and variable rates modeled by a gamma distribution [GTR+I+G]) was selected by Akaike's information criterion, as implemented in ModelTest (version 3.7; http://darwin.uvigo.es/software/modeltest.html) (32). Phylogenetic trees were inferred using PAUP (version 4b10; Sinauer Associates) and visualized using Mega (version 3.1; http://www.megasoftware.net). Mega was also used to calculate nonsynonymous and synonymous distance (Nei-Gojobori method with the Jukes-Cantor correction). To detect evidence of selection at individual codons, fixed-effects likelihood analyses was used as implemented in DataMonkey (http://www.datamonkey.org) (19), using default parameters and the server-selected Hasegawa-Kishino-Yano evolutionary model. Sliding window analyses were performed using VarPlot (version 1.5; http://sray.med.som.jhmi.edu/SCRoftware/VarPlot) (37), with genetic distance calculated using maximum likelihood distance (F84 as implemented in Phylip) and nonsynonymous and synonymous distances calculated using the method of Nei and Gojobori with the Jukes-Cantor correction. Signature pattern analysis was performed using VisSPA (version 1.6; http://sray.med.som.jhmi.edu/SCRoftware/VisSPA) (36). To detect evidence for recombination, similarity scanning, bootscanning, and informative sites analysis were performed using SimPlot (version 3.5.1; http://sray.med.som.jhmi.edu/SCRoftware/SimPlot) (26).
Statistical analysis.
To avoid the assumption of normal distribution, median values were compared using the nonparametric Mann-Whitney U test. For binomial distributions (as in Fig. 5), the Poisson distribution was used to calculate P values.
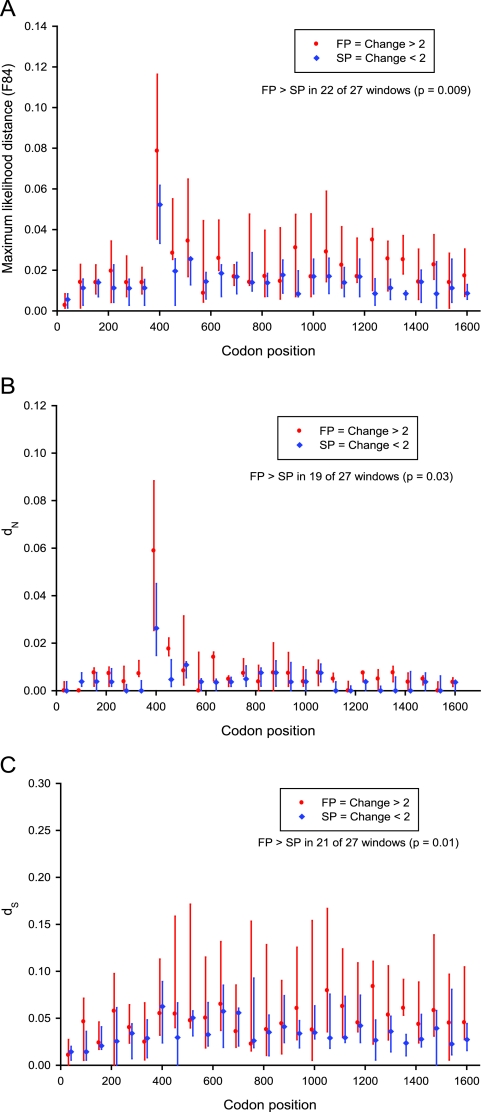
FIG. 5.
Divergence between visits 1 and 2 in nonoverlapping windows of 60 codons. Polyprotein position is indicated along the horizontal axis, relative to the gene positions indicated above the plot. For FP (in red; n = 5) and SP (in blue; n = 5) at each window midpoint, a vertical line indicates the interquartile range, and a marker indicates the median. For each of the 10 study subjects examined, four hemigenomic sequences were analyzed, two for each biopsy visit. Distances were calculated in VarPlot using maximum likelihood distance (F84 model from Phylip) (A) and nonsynonymous (B) and synonymous (C) distance (method of Nei and Gojobori with the Jukes-Cantor correction).
Nucleotide sequence accession numbers.
All newly determined sequences analyzed in this report have been deposited in GenBank, with accession numbers EF560218 through EF560558.
RESULTS
Study cohort.
The clinical characteristics of study subjects with rapid and slow progression of liver fibrosis are shown in Table 1. As designed, cases (subjects FP1 to FP5) and two sets of controls (subjects SP1 to SP5 and SP6 to SP10) were matched with respect to variables previously shown to have an effect on fibrosis progression and/or sequence evolution in persons with hepatitis C, including age, HIV seropositivity (all negative), HBsAg seropositivity (all negative), and alcohol use. Duration of HCV infection was also similar.
TABLE 1.
Clinical characteristics of the study subjects
| Subject | Age (yr) at first biopsy | Gender | HCV duration (yr)a | Alcohol useb | IDU frequencyc
|
Fibrosis stage
|
Fibrosis change (no. of stages) | Biopsy interval (mo) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Biopsy 1 | Biopsy 2 | Biopsy 1 | Biopsy 2 | |||||||
| FP1 | 55 | M | 27 | 0 | 0 | 0 | 1 | 4 | 3 | 51 |
| FP2 | 40 | M | 23 | ++ | + | + | 1 | 6 | 5 | 53 |
| FP3 | 34 | M | 18 | + | ++ | +++ | 0 | 3 | 3 | 44 |
| FP4 | 42 | M | 25 | + | + | 0 | 1 | 4 | 3 | 47 |
| FP5 | 43 | M | 30 | + | 0 | ++ | 0 | 3 | 3 | 53 |
| SP1 | 43 | M | 22 | + | + | +++ | 0 | 0 | 0 | 51 |
| SP2 | 46 | M | 30 | 0 | 0 | 0 | 0 | 0 | 0 | 44 |
| SP3 | 47 | M | 34 | + | +++ | ++ | 1 | 2 | 1 | 44 |
| SP4 | 42 | M | 29 | ++ | + | 0 | 0 | 0 | 0 | 48 |
| SP5 | 40 | M | 23 | + | 0 | 0 | 1 | 0 | −1 | 44 |
| SP6 | 36 | M | 10 | ++ | + | ++ | 0 | 0 | 0 | 53 |
| SP7 | 47 | M | 30 | 0 | 0 | 0 | 0 | 1 | 1 | 49 |
| SP8 | 42 | M | 23 | + | 0 | 0 | 0 | 1 | 1 | 46 |
| SP9 | 43 | F | 16 | + | ++ | 0 | 1 | 2 | 1 | 48 |
| SP10 | 51 | F | 17 | + | + | + | 1 | 2 | 1 | 48 |
Duration of HCV infection, estimated using time from first injection drug use to first biopsy (9).
Proportion of semiannual visits at which the behavior was acknowledged: 0, none of visits; +, some but not all visits; ++, all visits.
Self-reported number of injection drug use (IDU) episodes during the preceding 6 months: 0, none; +, 1 to 100; ++, 101 to 500; +++, more than 500.
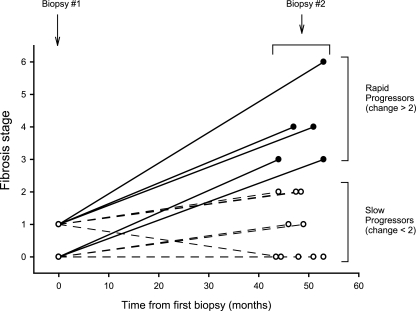
Figure 1 shows the histological results from liver biopsies at two visits separated by 44 to 53 months (medians, 51 for FP and 48 for SP; P = 0.4). As intended in the study design, case subjects FP1 to FP5 had much more rapid progression of fibrosis (change in modified Ishak fibrosis score median, 3; range, 3 to 5) than the controls (SP1 to SP10; median, 0.5; range, −1 to 1).
FIG. 1.
Timing and stage of liver biopsies. From a prospective cohort of 210 subjects who had undergone serial biopsies approximately 4 years apart, 5 case subjects with fast progression of fibrosis and 10 matched controls with slow progression were identified.
Quasispecies complexity and entropy of HVR1.
From each serum specimen, the sequence of 10 cDNA clones was determined for a 753-nucleotide (nt) region spanning the E1-E2 junction, including HVR1. Analysis of HVR1 nucleotide sequences allowed comparison to other studies that focused on this region. Complexity, or the number of distinct sequences divided by the number of sequences examined, was higher (P = 0.02) at the first biopsy visit in FP (median, 0.7; range, 0.7 to 0.9) than in SP (median, 0.4; range, 0.2 to 0.9). Similarly, the normalized Shannon entropy was twofold higher (P = 0.01) at the first biopsy visit in FP (median, 0.82; range, 0.76 to 0.94) than in SP (median, 0.41; range, 0.14 to 0.94).
At the second biopsy visit, the complexity and entropy values for SP (0.4 and 0.46, respectively) were highly consistent with the values from the first visit. In contrast, complexity and entropy for FP at the time of the second biopsy (when advanced fibrosis was detected) had fallen to 0.6 and 0.65, similar to those for SP (P > 0.05).
Evolutionary patterns of envelope sequences.
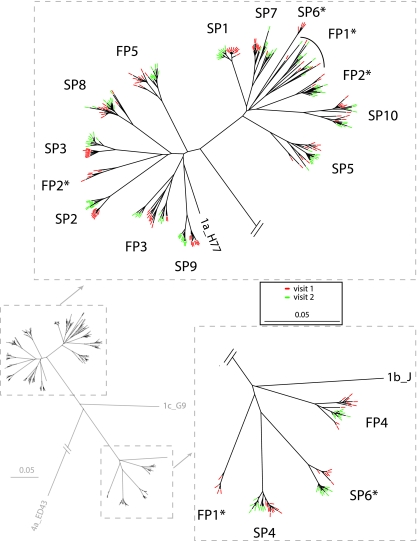
Phylogenetic analysis of envelope sequences from baseline and the second biopsy revealed monophyletic subject-specific clustering for 12 of 15 subjects (Fig. 2). In contrast, three subjects’ quasispecies were divided into more than one clade, and two of these indicated coexistence of subtypes 1a and 1b, whereas the third revealed two distinct clades within subtype 1a.
FIG. 2.
Phylogenetic analysis of HCV envelope sequences (753 nt) from subjects with fast (FP1 to FP5) and slow (SP1 to SP10) progression of liver fibrosis. Asterisks indicate the three subjects found to have mixed infections (more than one clade). Sequences from visit 1 are indicated in red, and those from visit 2 (4 years later) are indicated in green. The phylogenetic model was HKY85 with invariant sites, and rate variation was modeled using a gamma distribution. An initial neighbor-joining tree was used in a heuristic search, with tree bisection-reconnection branch swapping performed 10 times.
The subjects for whom more than one clade was found were examined in more detail. In each case, a primary clade was present, containing most of the sequences, and at least one sequence from each biopsy visit. For subject SP6, the primary clade was subtype 1b, whereas the secondary clade was subtype 1a and contained four sequences from biopsy visit 1. For subject FP1, the primary clade clustered with subtype 1a, with a secondary clade containing four sequences from biopsy visit 1. A tertiary clade, containing one sequence from each visit, also clustered with subtype 1a. For subject FP2, both the primary clade and secondary clades clustered with subtype 1a but were distinct. The primary FP2 clade contained three sequences from visit 1 and all 10 sequences from visit 2, whereas the secondary clade contained seven sequences from visit 1. Taken together, these findings strongly support the presence of mixed infections in three subjects and strongly support the persistent presence of two distinct quasispecies for 4 years in two subjects (one from each study group).
In no case did primary or secondary clades intermingle with another subject's sequences. Subsequent analyses of diversity and divergence used only sequences from primary clades (i.e., sequences from the same quasispecies as discussed below), because inclusion of secondary clades would have distorted results by including the distance between widely separated clades. Had secondary clades been included, estimates of diversity at visit 1 and of divergence between visits 1 and 2 would have been three- to fivefold larger for subjects FP1, FP2, and SP6.
Hemigenomic sequence evolution.
Phylogenetic analysis of 5.2-kb hemigenomic sequences from 10 subjects (FP1 to FP5 and SP1 to SP5) supported relationships observed in the envelope region. Phylogenetic analysis of each gene separately (core, E1, E2, p7, NS2, and NS3) was also similar in structure, with no shifts in subtype or consistency in the subject-specific clades seen in the envelope analysis (data not shown). These similarities indicate proportional (though not uniform) rates and no evidence for recombination within clade or between clades for subjects FP1 and FP2, who had hemigenomic sequencing performed and had secondary clades at visit 1. Because only two 5-kb cDNA clones/visit were examined (three for subject FP2 visit 1), and we did not examine the NS4 and NS5 regions, recombinants may have been missed if the breakpoint was located in the NS4-NS5 region. However, the region examined is relevant because it includes the postulated recombination “hot spot” in NS2 observed in a natural recombinant and also generated efficiently in vitro (15, 31). We conclude that recombinants involving exchanges within the 5.2-kb core-NS3 region we examined had not completely replaced the parental strains during at least 4 years of concurrent viremia in subjects FP1 and FP2, though we cannot exclude the presence of lower-frequency recombinant variants.
Quasispecies diversity at separate biopsy visits.
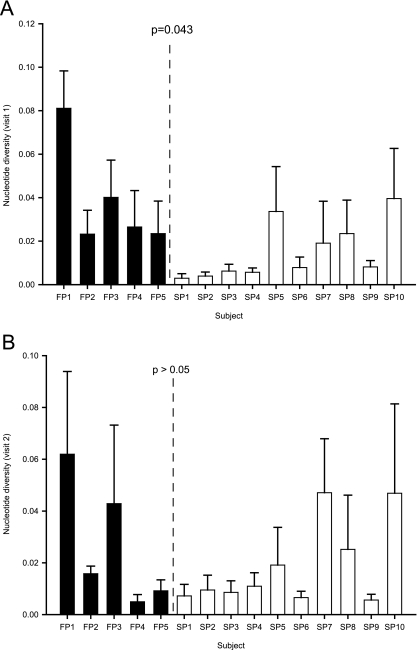
Diversity in the envelope genes was calculated as the mean pairwise distance for sequences obtained for each subject at each visit. Diversity was higher at the first visit (when the fibrosis stage was 0 or 1 for all subjects) in subjects with FP versus SP (P = 0.043), but there was overlap (Fig. 3A). At the second visit, FP diversity was not higher than SP diversity (Fig. 3B).
FIG. 3.
Diversity, or the average pairwise genetic distance among clones, of envelope gene sequences (753 nt) obtained from FP and SP at the first (A) and second (B) biopsy visits. Diversity was calculated from the 10 clones obtained per specimen using the GTR+I+G model (model and parameters determined by ModelTest). Error bars indicate standard deviations. P values were calculated using a nonparametric comparison of medians.
Quasispecies divergence between biopsy visits in envelope genes.
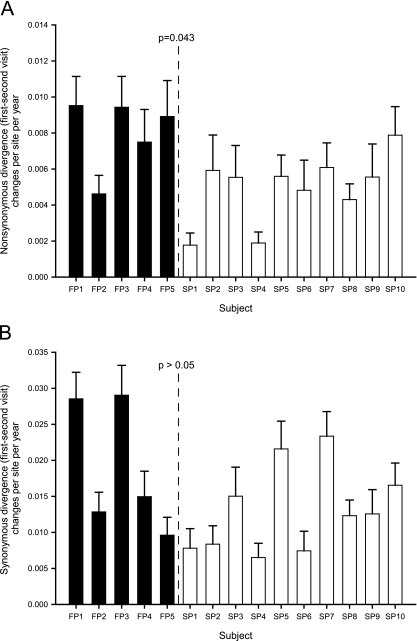
The rate of divergence was estimated by averaging pairwise distances among envelope gene sequences obtained at the two visits for each study subject. As noted above, outlier clades were excluded. The median divergence at all sites was greater for FP (0.046) than for SP (0.029), though this difference did not reach statistical significance (P = 0.058). To determine whether these changes resulted in amino acid changes, we examined nonsynonymous (dN) and synonymous (dS) envelope gene divergence. Median intervisit dN was higher for FP than SP (P = 0.043) (Fig. 4A), whereas there was no significant difference in either dS (Fig. 4B) or the ratio of these measures (dN/dS) (data not shown).
FIG. 4.
Nonsynonymous (A) and synonymous (B) divergence, or mean pairwise distance between visits, of envelope gene sequences (753 nt; 10 sequences per study subject per visit, for a total of 20 sequences per subject) was calculated using the method of Nei and Gojobori with the Jukes-Cantor correction. Error bars indicate standard deviations. P values were calculated using a nonparametric comparison of medians.
Differences in evidence of selection averaged over multiple sites may be due to changes in single codons or clusters of adjacent sites. A higher-resolution analysis was performed by applying the fixed-effects likelihood tool in DataMonkey to envelope codons 235 to 484. For each study subject, 20 sequences were analyzed, except for the three subjects with multiclade infections (FP1, FP2, and SP6) (Fig. 2), from whom only major-clade sequences were used. Individual analyses of the 15 subjects (5 FP and 10 SP) at a threshold P value of <0.05 showed no difference in the number of positively selected (dN > dS) sites (one each: codon 395, with P = 0.02 in FP1, and codon 404, with P = 0.04 in SP5). Relaxing the threshold to P < 0.1 provided additional support for positive selection at codons 399 (SP5), 400 (FP5), 404 (FP1, FP4, and FP5), 405 (FP5), and 476 (SP9) but no significant difference between FP and SP (P > 0.05 for difference in median number of positively selected codons).
Comparative rates of change across the hemigenome.
Hemigenomic sequencing was performed for five FP and five SP. Sliding-window analysis was used to examine nonoverlapping windows of 60 codons for overall divergence (maximum likelihood distance using the F84 model in Phylip), dN, and dS. As an unbiased estimator, we examined the number of windows in which the median distance measure was higher for FP than SP, testing the null hypothesis that only 13 or 14 (half) of the 27 windows would be higher for FP. Instead, divergence was significantly greater in FP than SP in terms of overall divergence (Fig. 5A) (22 of 27 windows; P < 0.01), more consistently for synonymous divergence (Fig. 5C) (21 of 27 windows; P = 0.01) than for nonsynonymous divergence (Fig. 5B) (19 of 27 windows, P = 0.03). There were too few hemigenomic sequences per study subject for individual codon-based analyses.
Analysis using VisSPA did not reveal any sites (among the 1,644 examined) bearing signature patterns that distinguished FP from SP, i.e., where an amino acid was uniquely (or predominantly, at a threshold of 60%) shared by FP. In a separate analysis, sequences obtained from FP at the time of the second biopsy (when disease had progressed) were not distinct from sequences obtained at the first visit (when little or no liver disease was present). At one site (position 1200 in the polyprotein), the most commonly observed residue had changed: three of five FP had an N1200S substitution, whereas two consistently had S1200. This difference was not significantly associated with disease progression (P > 0.05) in these five FP; moreover, examination of the SP showed that four of five had S1200 while one had N1200, with no substitution during the 4-year interval. We did not detect strong evidence for a signature sequence associated with disease progression.
Hepatocyte injury and level of HCV viremia.
The higher synonymous divergence rate for FP than SP suggested that higher levels of hepatocyte turnover and/or higher levels of HCV replication might be involved. We examined transaminase levels as a well-established surrogate marker of hepatocyte injury (5, 11). Median levels of AST and ALT were higher in FP than SP (Table 2) . In addition, we examined HCV RNA levels and found that they were also higher, but only at the first visit (Table 2).
TABLE 2.
Serum transaminase and HCV RNA levels
| Subject | Biopsy 1
|
Biopsy 2
|
||||
|---|---|---|---|---|---|---|
| AST (IU/liter) | ALT (IU/liter) | Log10[HCV RNA (IU/ml)] | AST (IU/liter) | ALT (IU/liter) | Log10[HCV RNA (IU/ml)] | |
| FP1 | 72 | 80 | 6.9 | 51 | 66 | 6.8 |
| FP2 | 56 | 55 | 7.2 | 100 | 108 | 6.7 |
| FP3 | 31 | 33 | 6.6 | 171 | 169 | 7.3 |
| FP4 | 62 | 30 | 5.8 | 46 | 60 | 4.4 |
| FP5 | 108 | 135 | 6.5 | 47 | 42 | 5.6 |
| SP1 | 64 | 63 | 6.4 | 56 | 55 | 6.1 |
| SP2 | 62 | 51 | 5.3 | 26 | 29 | 5.2 |
| SP3 | 34 | 29 | 4.5 | 25 | 24 | 4.7 |
| SP4 | 32 | 37 | 6.6 | 38 | 52 | 6.5 |
| SP5 | 29 | 33 | 6.1 | 39 | 52 | 6.3 |
| SP6 | 25 | 28 | 5.1 | 23 | 23 | 5.2 |
| SP7 | 39 | 15 | 5.6 | 131 | 64 | 5.4 |
| SP8 | 22 | 26 | 5.5 | 26 | 31 | 5.6 |
| SP9 | 28 | 37 | 6.6 | 42 | 73 | 7.2 |
| SP10 | 28 | 22 | 5.9 | 33 | 30 | 6.3 |
| P value (FP vs SP)a | 0.05 | 0.09 | 0.03 | 0.03 | 0.04 | 0.4 |
Boldface indicates statistical significance.
DISCUSSION
In this longitudinal study involving hemigenomic sequencing of HCV and sequential liver biopsies, we found evidence of greater HCV sequence divergence in persons who developed more progressive hepatic fibrosis. This finding was consistent across all six genes examined, structural and nonstructural. Detailed analysis of HVR1, the most variable portion of the genome, also revealed greater diversity and divergence, with an excess of nonsynonymous changes consistent with immune selection. We also found evidence of mixed infections with persistence of the same mixtures for years, suggesting that concurrent infection is not rare.
This study focused on the evolution of HCV sequence during the 4-year interval between serial biopsies performed in a prospective cohort. The nested case-control design allowed us to focus on individuals at the extremes of fibrosis progression, with FP (case subjects) having an increase in fibrosis of more than two stages, whereas matched SP (controls) had no significant change in fibrosis over the same period. We were also able to use matching to reduce the influence of nonviral factors, a measure that is very important in research on human subjects, which cannot be experimentally controlled. In addition, we used long-amplicon RT-PCR to clone a 5.2-kb region spanning all of the structural genes plus p7, NS2, and NS3, permitting comparisons of evolutionary rates in genes that are rarely examined. To select hemigenomic clones for sequencing for each specimen (two specimens per study subject), we sequenced a 700-nt region spanning HVR1 from 10 randomly selected clones, allowing us to assess quasispecies diversity.
We found that quasispecies complexity and diversity were higher in FP than SP at the first visit (prior to disease progression), but not at the second visit (once fibrosis had progressed). Greater complexity and diversity in FP at the first visit, when fibrosis stage was matched between FP and SP, suggests that these measures might be predictive of disease progression. That this difference was no longer significant at the time of the second biopsy 4 years later might simply be due to the small size of this study (type 2 error), but it agrees with a recent longitudinal study of persons with and without ESLD (33). This shift may reflect changing interactions between host and virus as the disease progresses and also might explain inconsistent results from cross-sectional studies.
Diverse viral sequences might be a marker of a more intense immune response, more rapid viral turnover, and infection of a larger proportion of hepatocytes. The first of these should be associated with evidence of positive selection pressure (high dN/dS), which was not demonstrated in this study. In contrast, weak selective pressure, combined with greater replication due to the short life span of infected hepatocytes with replacement and reinfection, and/or infection of a large proportion of the liver, could be associated with increased diversity in the absence of evidence for strong positive selection pressure (50).
We also examined multiple genes for evidence of selective pressures during the interval between biopsies and found that FP had a consistently higher evolutionary rate than SP for all genes examined (Fig. 5). This difference was more consistent for dS than for dN, suggesting that it was not due to strong selection at most sites but rather to greater viral population turnover. Use of codon-based models did reveal a small number of sites with a greater dN than dS, particularly in HVR1, as others have noted (28). While these results do not exclude the possibility that higher evolutionary rates might have been driven by selection at a small number of sites (particularly HVR1), they do argue against widespread positive selection explaining differences between FP and SP.
We and others recently showed that persons with chronic, but not acute, HCV infection produce high titers of broadly neutralizing antibodies, using HCV-lentivirus pseudotyping systems and HCV cell culture (25, 30, 53). Even though late-appearing neutralizing antibodies are insufficient to clear HCV infection, the present data suggest that they may affect fitness of HCV. Taken together, these studies suggest that immune selection in envelope genes is a marker of (i) high neutralizing antibody titers and (ii) high turnover of HCV-infected cells (i.e., destruction of HCV-infected cells, coupled with hepatic regeneration and subsequent HCV infection). It is also possible that immune pressure on envelope genes could be mediated by cytolytic T lymphocytes; however, the lack of a similar pressure in nonstructural genes, in the same amplicons, suggests dominance of envelope-specific (i.e., humoral) selection.
We also found evidence for mixed infections, with two subjects showing persistent codominance of two distinct strains, and no evidence to suggest that recombination had occurred despite 4 years of mixed infection. The persistence of such mixed infection has been reported previously, though most longitudinal studies have found that only one strain persists, with exclusion of the other (7, 21, 48). Another recent study examining IDUs with HCV superinfection also found no evidence for recombination (1). The absence of evidence for recombination in these studies, combined with published evidence that intersubtype recombination has happened in the past (15), suggests that this phenomenon is real but rare. Our results also strongly suggest that our hemigenomic RT-PCR assay is unlikely to generate artifactual evidence of recombination, which must always be considered (2). Our study does not address the relative timing of these multiple infections, nor does it exclude the possibility that the other subjects might harbor low-frequency viral populations that are similarly divergent. The occurrence of such stable (persistent over 4 years) mixed infections indicates that either these viral populations do not compete with each other or their fitness is very closely matched.
The presence of mixed infection also has analytical implications. Amplification could be selective, and the most accurate means to ascertain the “true” distribution of variants is probably digital PCR, but that procedure is extremely expensive (45). In addition, unrecognized mixed infection could distort estimates of viral diversity and divergence. Such estimates generally assume that the diversity and divergence have occurred in the current host, whereas this is not the case in mixed infections. This can be rectified, as in the current study, through phylogenetic analysis and exclusion of heterologous sequences. In contrast, gel shift assays do not provide enough information to distinguish divergent from heterologous variants. It is possible that conflicting results from prior studies of HCV diversity might be due, in part, to the presence of mixed infections.
The possibility of RT-PCR contamination was carefully evaluated. Negative control samples were used in every reaction and were consistently negative. Specimens obtained from the same study subject at different visits were processed on different days, interspersed with specimens from other study subjects, so that contamination would be readily recognizable; no evidence of contamination was observed. On important limitation of this study is the relatively small number of subjects. If many factors affect HCV sequence evolution (e.g., distinct epitopes in different individuals), it is possible that we would fail to detect their effects; because we did not assess immune responses in this study, we could not correlate epitopes with sequence changes.
Taken together, our findings suggest that rapid progression of liver fibrosis during chronic HCV infection is associated with greater quasispecies diversity at an early stage in disease and more rapid viral population turnover, with evidence of selective pressure in HVR1 but not in other regions of the genome.
Acknowledgments
This study was supported by NIH grant R01 DA16078 (D.L.T.). The ALIVE cohort is supported by NIH grants R01 DA04334 and R01 DA12568.
Footnotes
Published ahead of print on 28 February 2007.
REFERENCES
- 1.Bernardin, F., B. Herring, K. Page-Shafer, C. Kuiken, and E. Delwart. 2006. Absence of HCV viral recombination following superinfection. J. Viral Hepatol. 13:532-537. [DOI] [PubMed] [Google Scholar]
- 2.Bradley, R. D., and D. M. Hillis. 1997. Recombinant DNA sequences generated by PCR amplification. Mol. Biol. Evol. 14:592-593. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. 1998. Recommendations for prevention and control of hepatitis C virus (HCV) infection and HCV-related chronic disease. Morb. Mortal. Wkly. Rep. 47(RR-19):1-39. [Google Scholar]
- 4.Cox, A. L., T. Mosbruger, Q. Mao, Z. Liu, X. H. Wang, H. C. Yang, J. Sidney, A. Sette, D. Pardoll, D. L. Thomas, and S. C. Ray. 2005. Cellular immune selection with hepatitis C virus persistence in humans. J. Exp. Med. 201:1741-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Ritis, F., M. Coltorti, and G. Giusti. 1972. Serum-transaminase activities in liver disease. Lancet i:685-687. [DOI] [PubMed] [Google Scholar]
- 6.Di Bisceglie, A. M., Z. D. Goodman, K. G. Ishak, J. H. Hoofnagle, J. J. Melpolder, and H. J. Alter. 1991. Long-term clinical and histopathological follow-up of chronic posttransfusion hepatitis. Hepatology 14:969-974. [DOI] [PubMed] [Google Scholar]
- 7.Fan, X., D. M. Lang, Y. Xu, A. C. Lyra, K. Yusim, J. E. Everhart, B. T. Korber, A. S. Perelson, and A. M. Di Bisceglie. 2003. Liver transplantation with hepatitis C virus-infected graft: interaction between donor and recipient viral strains. Hepatology 38:25-33. [DOI] [PubMed] [Google Scholar]
- 8.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 9.Garfein, R. S., M. C. Doherty, E. R. Monterroso, D. L. Thomas, K. E. Nelson, and D. Vlahov. 1998. Prevalence and incidence of hepatitis C virus infection among young adult injection drug users. J. Acquir. Immune. Defic. Syndr. Hum. Retrovirol. 18(Suppl. 1):S11-S19. [DOI] [PubMed] [Google Scholar]
- 10.González-Peralta, R. P., J. Y. She, G. L. Davis, T. Ohno, M. Mizokami, and J. Y. N. Lau. 1996. Clinical implications of viral quasispecies heterogeneity in chronic hepatitis C. J. Med. Virol. 49:242-247. [DOI] [PubMed] [Google Scholar]
- 11.Guidotti, L. G., K. Ando, M. V. Hobbs, T. Ishikawa, L. Runkel, R. D. Schreiber, and F. V. Chisari. 1994. Cytotoxic T lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc. Natl. Acad. Sci. USA 91:3764-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Honda, M., S. Kaneko, A. Sakai, M. Unoura, S. Murakami, and K. Kobayashi. 1994. Degree of diversity of hepatitis C virus quasispecies and progression of liver disease. Hepatology 20:1144-1151. [PubMed] [Google Scholar]
- 13.Izopet, J., L. Rostaing, K. Sandres, J. M. Cisterne, C. Pasquier, J. L. Rumeau, M. Duffaut, D. Durand, and J. Puel. 2000. Longitudinal analysis of hepatitis C virus replication and liver fibrosis progression in renal transplant recipients. J. Infect. Dis. 181:852-858. [DOI] [PubMed] [Google Scholar]
- 14.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 15.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kenny-Walsh, E., et al. 1999. Clinical outcomes after hepatitis C infection from contaminated anti-D immune globulin. N. Engl. J. Med. 340:1228-1233. [DOI] [PubMed] [Google Scholar]
- 17.Kiyosawa, K., T. Sodeyama, E. Tanaka, Y. Gibo, K. Yoshizawa, Y. Nakano, S. Furuta, Y. Akahane, K. Nishioka, and R. H. Purcell. 1990. Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology 12:671-675. [DOI] [PubMed] [Google Scholar]
- 18.Koizumi, K., N. Enomoto, M. Kurosaki, T. Murakami, N. Izumi, F. Marumo, and C. Sato. 1995. Diversity of quasispecies in various disease stages of chronic hepatitis C virus infection and its significance in interferon treatment. Hepatology 22:30-35. [PubMed] [Google Scholar]
- 19.Kosakovsky Pond, S. L., and S. D. Frost. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208-1222. [DOI] [PubMed] [Google Scholar]
- 20.Kwok, S., and R. Higuchi. 1989. Avoiding false positives with PCR. Nature 339:237-238. [DOI] [PubMed] [Google Scholar]
- 21.Laskus, T., L. F. Wang, M. Radkowski, H. Vargas, M. Nowicki, J. Wilkinson, and J. Rakela. 2001. Exposure of hepatitis C virus (HCV) RNA-positive recipients to HCV RNA-positive blood donors results in rapid predominance of a single donor strain and exclusion and/or suppression of the recipient strain. J. Virol. 75:2059-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leone, F., H. Zylberberg, G. Squadrito, B. Le Guen, P. Berthelot, S. Pol, and C. Brechot. 1998. Hepatitis C virus (HCV) hypervariable region 1 complexity does not correlate with severity of liver disease, HCV type, viral load or duration of infection. J. Hepatol. 29:689-694. [DOI] [PubMed] [Google Scholar]
- 23.Liu, S. L., A. G. Rodrigo, R. Shankarappa, G. H. Learn, L. Hsu, O. Davidov, L. P. Zhao, and J. I. Mullins. 1996. HIV quasispecies and resampling. Science 273:415-416. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Z., D. M. Netski, Q. Mao, O. Laeyendecker, J. R. Ticehurst, X. H. Wang, D. L. Thomas, and S. C. Ray. 2004. Accurate representation of the hepatitis C virus quasispecies in 5.2-kilobase amplicons. J. Clin. Microbiol. 42:4223-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Logvinoff, C., M. E. Major, D. Oldach, S. Heyward, A. Talal, P. Balfe, S. M. Feinstone, H. Alter, C. M. Rice, and J. A. McKeating. 2004. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc. Natl. Acad. Sci. USA 101:10149-10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez-Labrador, F. X., S. Ampurdanes, M. Gimenez-Barcons, M. Guilera, J. Costa, M. T. Jimenez de Anta, J. M. Sanchez-Tapias, J. Rodes, and J. C. Saiz. 1999. Relationship of the genomic complexity of hepatitis C virus with liver disease severity and response to interferon in patients with chronic HCV genotype 1b interferon. Hepatology 29:897-903. (Erratum, 29:1915.) [DOI] [PubMed] [Google Scholar]
- 28.McAllister, J., C. Casino, F. Davidson, J. Power, E. Lawlor, P. L. Yap, P. Simmonds, and D. B. Smith. 1998. Long-term evolution of the hypervariable region of hepatitis C virus in a common-source-infected cohort. J. Virol. 72:4893-4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naito, M., N. Hayashi, T. Moribe, H. Hagiwara, E. Mita, Y. Kanazawa, A. Kasahara, H. Fusamoto, and T. Kamada. 1995. Hepatitis C viral quasispecies in hepatitis C virus carriers with normal liver enzymes and patients with type C chronic liver disease. Hepatology 22:407-412. [PubMed] [Google Scholar]
- 30.Netski, D. M., J. A. McKeating, E. Depla, K. A. Dowd, D. L. Thomas, and S. C. Ray. 2004. The development of neutralizing antibodies during acute hepatitis C virus infection, abstr. P-201. 11th Int. Symp. Hepatitis C Virus Related Viruses, Heidelberg, Germany.
- 31.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 103:7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 33.Qin, H., N. J. Shire, E. D. Keenan, S. D. Rouster, M. E. Eyster, J. J. Goedert, M. J. Koziel, and K. E. Sherman. 2005. HCV quasispecies evolution: association with progression to end-stage liver disease in hemophiliacs infected with HCV or HCV/HIV. Blood 105:533-541. [DOI] [PubMed] [Google Scholar]
- 34.Rai, R., L. E. Wilson, J. Astemborski, F. Anania, M. Torbenson, C. Spoler, D. Vlahov, S. A. Strathdee, J. Boitnott, K. E. Nelson, and D. L. Thomas. 2002. Severity and correlates of liver disease in hepatitis C virus-infected injection drug users. Hepatology 35:1247-1255. [DOI] [PubMed] [Google Scholar]
- 35.Ray, S. C., L. Fanning, X.-H. Wang, D. M. Netski, E. Kenny-Walsh, and D. L. Thomas. 2004. Hemigenomic analysis of hepatitis C sequence variation in a common-source outbreak reveals evidence of selection and reversion, abstr. P-200. 11th Int. Symp. Hepatitis C Virus Related Viruses, Heidelberg, Germany.
- 36.Ray, S. C., L. Fanning, X. H. Wang, D. M. Netski, E. Kenny-Walsh, and D. L. Thomas. 2005. Divergent and convergent evolution after a common-source outbreak of hepatitis C virus. J Exp. Med. 201:1753-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ray, S. C., Y. M. Wang, O. Laeyendecker, J. Ticehurst, S. A. Villano, and D. L. Thomas. 1998. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as decoy. J. Virol. 73:2938-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rothman, A. L., C. Morishima, H. L. Bonkovsky, S. J. Polyak, R. Ray, A. M. Di Bisceglie, K. L. Lindsay, P. F. Malet, M. Chang, D. R. Gretch, D. G. Sullivan, A. K. Bhan, E. C. Wright, and M. J. Koziel. 2005. Associations among clinical, immunological, and viral quasispecies measurements in advanced chronic hepatitis C. Hepatology 41:617-625. [DOI] [PubMed] [Google Scholar]
- 39.Sakai, A., S. Kaneko, M. Honda, E. Matsushita, and K. Kobayashi. 1999. Quasispecies of hepatitis C virus in serum and in three different parts of the liver of patients with chronic hepatitis. Hepatology 30:556-561. [DOI] [PubMed] [Google Scholar]
- 40.Seeff, L. B., F. B. Hollinger, H. J. Alter, E. C. Wright, C. M. Cain, Z. J. Buskell, K. G. Ishak, F. L. Iber, D. Toro, A. Samanta, R. L. Koretz, and R. P. Perrillo. 2001. Long-term mortality and morbidity of transfusion-associated non-A, non-B, and type C hepatitis: a National Heart, Lung, and Blood Institute collaborative study. Hepatology 33:455-463. [DOI] [PubMed] [Google Scholar]
- 41.Thomas, D. L., J. Astemborski, R. M. Rai, F. A. Anania, M. Schaeffer, N. Galai, K. Nolt, K. E. Nelson, S. A. Strathdee, L. Johnson, O. Laeyendecker, J. Boitnott, L. E. Wilson, and D. Vlahov. 2000. The natural history of hepatitis C virus infection: host, viral, and environmental factors. JAMA 284:450-456. [DOI] [PubMed] [Google Scholar]
- 42.Timm, J., G. M. Lauer, D. G. Kavanagh, I. Sheridan, A. Y. Kim, M. Lucas, T. Pillay, K. Ouchi, L. L. Reyor, J. S. Zur Wiesch, R. T. Gandhi, R. T. Chung, N. Bhardwaj, P. Klenerman, B. D. Walker, and T. M. Allen. 2004. CD8 epitope escape and reversion in acute HCV infection. J. Exp. Med. 200:1593-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tong, M. J., N. S. El-Farra, A. R. Reikes, and R. L. Co. 1995. Clinical outcomes after transfusion-associated hepatitis C. N. Engl. J. Med. 332:1463-1466. [DOI] [PubMed] [Google Scholar]
- 44.Vlahov, D., J. C. Anthony, A. Muñoz, J. Margolik, D. D. Celentano, L. Solomon, and B. F. Polk. 1991. The ALIVE Study: a longitudinal study of HIV-1 infection in intravenous drug users: description of methods. J. Drug Issues 21:759-776. [PubMed] [Google Scholar]
- 45.Vogelstein, B., and K. W. Kinzler. 1999. Digital PCR. Proc. Natl. Acad. Sci. USA 96:9236-9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker, C. M. 1997. Comparative features of hepatitis C virus infection in humans and chimpanzees. Springer Semin. Immunopathol. 19:85-98. [DOI] [PubMed] [Google Scholar]
- 47.Wang, Y. M., S. C. Ray, O. Laeyendecker, J. R. Ticehurst, and D. L. Thomas. 1998. Assessment of hepatitis C virus sequence complexity by electrophoretic mobilities of both single- and double-stranded DNAs. J. Clin. Microbiol. 36:2982-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Widell, A., S. Månsson, N. H. Persson, H. Thysell, S. Hermodsson, and I. Blohme. 1995. Hepatitis C superinfection in hepatitis C virus (HCV)-infected patients transplanted with an HCV-infected kidney. Transplantation 60:642-647. [DOI] [PubMed] [Google Scholar]
- 49.Wiley, T. E., J. Brown, and J. Chan. 2002. Hepatitis C infection in African Americans: its natural history and histological progression. Am. J. Gastroenterol. 97:700-706. [DOI] [PubMed] [Google Scholar]
- 50.Williamson, S., S. M. Perry, C. D. Bustamante, M. E. Orive, M. N. Stearns, and J. K. Kelly. 2005. A statistical characterization of consistent patterns of human immunodeficiency virus evolution within infected patients. Mol. Biol. Evol. 22:456-468. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, L. E., M. Torbenson, J. Astemborski, H. Faruki, C. Spoler, R. Rai, S. Mehta, G. D. Kirk, K. Nelson, N. Afdhal, and D. L. Thomas. 2006. Progression of liver fibrosis among injection drug users with chronic hepatitis C. Hepatology 43:788-795. [DOI] [PubMed] [Google Scholar]
- 52.Wong, J. B., G. M. McQuillan, J. G. McHutchison, and T. Poynard. 2000. Estimating future hepatitis C morbidity, mortality, and costs in the United States. Am. J. Public Health 90:1562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. USA 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuki, N., N. Hayashi, T. Moribe, Y. Matsushita, T. Tabata, T. Inoue, Y. Kanazawa, K. Ohkawa, A. Kasahara, H. Fusamoto, and T. Kamada. 1997. Relation of disease activity during chronic hepatitis C infection to complexity of hypervariable region 1 quasispecies. Hepatology 25:439-444. [DOI] [PubMed] [Google Scholar]