Abstract
The possession of some HLA class I molecules is associated with delayed progression to AIDS. The mechanism behind this beneficial effect is unclear. We tested the idea that cytotoxic T-cell responses restricted by advantageous HLA class I molecules impose stronger selection pressures than those restricted by other HLA class I alleles. As a measure of the selection pressure imposed by HLA class I alleles, we determined the extent of HLA class I-associated epitope variation in a cohort of European human immunodeficiency virus (HIV)-positive individuals (n = 84). We validated our findings in a second, distinct cohort of African patients (n = 516). We found that key HIV epitopes restricted by advantageous HLA molecules (B27, B57, and B51 in European patients and B5703, B5801, and B8101 in African patients) were more frequently mutated in individuals bearing the restricting HLA than in those who lacked the restricting HLA class I molecule. HLA alleles associated with clinical benefit restricted certain epitopes for which the consensus peptides were frequently recognized by the immune response despite the circulating virus's being highly polymorphic. We found a significant inverse correlation between the HLA-associated hazard of disease progression and the mean HLA-associated prevalence of mutations within epitopes (P = 0.028; R2 = 0.34). We conclude that beneficial HLA class I alleles impose strong selection at key epitopes. This is revealed by the frequent association between effective T-cell responses and circulating viral escape mutants and the rarity of these variants in patients who lack these favorable HLA class I molecules, suggesting a significant pressure to revert.
Human immunodeficiency virus (HIV) is controlled by an immune response of variable efficacy. Crucial protection is conferred by the CD8+ T-cell response as dictated by human leukocyte antigen (HLA) class I alleles (16). However, not all HLA class I molecules are the same. In Caucasians, HLA-B27 and -B57 delay disease progression (27); others, such as some HLA-B35 alleles, are associated with early onset of AIDS (12). In African patients HLA-B5703, -B5801, and -B8101 are associated with lower viral loads (19). The mechanism behind this differential HLA class I effect is elusive.
Immune responses to different epitopes vary in their degree of efficacy and impact on plasma viral loads (20). In acute (3) and chronic (31) infection, epitopes restricted by beneficial HLA class I alleles are more frequently recognized than those restricted by other HLA class I alleles. Responses to some epitopes are associated with controlled viremia (18), but immune responses can also be detected despite ongoing viral replication and disease progression (17). These have been described, respectively, as “driver” and “passenger'” cytotoxic T lymphocytes in which the former are effective and influence viral evolution while the latter, although detectable by gamma interferon enzyme-linked immunospot (ELISPOT) assay, are less effective and impose weaker selection pressure (21).
Viral mutations allow “escape” from the HLA class I-mediated immune response (16). Associations between individual HLA class I molecules and specific viral polymorphisms are well documented (25), suggesting a widespread impact of HLA class I on the HIV genome (34). However, the implications of immune escape for populations remain unclear. In individual case studies, immune escape is linked to a rise in viral load (9, 15). The simian model also implicates escape from certain well-studied epitopes to be part of disease progression (7). There is good evidence which contradicts the simplistic hypothesis that immune escape from CD8+ T-cell responses is inevitably followed by elevation in viral load and disease progression. Viral sequences from a group of HLA-B57-positive patients with preserved CD4 cell counts—“elite” controllers—reveal a high degree of immune escape in the presence of controlled viremia (4). Reversion of escape mutants upon transfer to a non-major histocompatibility complex class I-matched host (22) and other evidence of fitness costs attributable to these mutants (10, 23) indicate that the outcome of adaptation to immune selection may not be wholly disadvantageous to the host.
Here, we present data from a cohort of HIV-positive individuals from Switzerland (n = 84) and confirm our findings in a second, distinct cohort of patients from southern Africa (n = 516). We show how HLA class I alleles linked with slow disease progression are associated with very strong selection pressures at key epitopes, which results in a high degree of variation in HLA-matched individuals compared with their HLA-unmatched controls. This finding holds for the slow progressor alleles HLA-B27, -B57, -B51, and -A11 in Caucasians and for a distinct set of molecules (HLA-B5703, -B5801, and -B8101) linked to lower viral loads in Africans. We hypothesize that for the protective HLA class I alleles, viral escape variants evolve that carry heavy mutation costs, implying that the selecting immune responses are particularly efficacious.
MATERIALS AND METHODS
Cohorts.
The Swiss-Spanish Intermittent Therapy Trial (SSITT) was a study of structured treatment interruptions in chronically HIV-infected patients, described in detail elsewhere (8). The 516 African patients were recruited in southern Africa, as described previously (19).
Immune responses.
From the SSITT cohort, we identified 133 patients for whom HLA class I immune responses were measured by gamma interferon ELISPOT assays using optimal peptides for the HLA class I molecules each patient expressed. The epitopes were described in the Los Alamos Database and are listed in Table S1 in the supplemental material. The patients were tested for these responses repeatedly (mean number of times tested, 16; range, 3 to 26) over a mean of 14 months (range, 3 to 19). A positive response to an epitope was defined as two or more responses greater than 50 spots/million cells (after subtraction of background). Full details of the immunological responses in this cohort are described elsewhere (28). Immune responses for the African patients were tested using a panel of 410 18-mer peptides based on the HIV subtype C consensus sequence spanning the HIV genome (1).
DNA amplification and sequencing.
Analysis of the HIV-1 Gag, Pol, and Nef genes encoding 54 HLA class I optimal epitopes (see Table S1 in the supplemental material) was carried out in 84 Swiss patients. Viral DNA was extracted from peripheral blood mononuclear cells collected while patients were off therapy. Cells were stimulated in culture for 3 days with phytohemagglutinin and interleukin-2 as described previously (15). HIV-1 Gag, Pol, and Nef genes were amplified by nested PCR and sequenced using ABI Big Dye terminator sequencing kits (Applied Biosystems). Sequences were aligned manually using Se-Al software. For the African patients, sequencing was carried out from DNA using primers specific for HIV-1 subtype C, as described previously (22). Variation was defined in relation to the sequences of the peptides with which they were tested in the ELISPOT assays. For the Swiss study these were the same sequence as the laboratory reference strain HIV-1 HXB2, except for three peptides which varied at one amino acid site each. For the African study, the reference strain was the cohort subtype C consensus sequence from 2001, as previously described (19).
Statistics and analysis.
Sequences of 54 and 70 “optimal” epitopes were identified within the HIV Gag, Pol, and Nef genes for the Caucasian and African cohorts, respectively. Epitope sequences were compared with the sequence of the optimal peptide and scored in a binary manner as polymorphic or consensus. For each epitope the cohort was divided into two groups: patients who carried the restricting HLA class I molecule and a control group who lacked the HLA class I molecule under study. The difference in proportions of polymorphic epitopes in the HLA-matched and -unmatched individuals was scored as a measure of HLA-driven variation. The proportions of epitopes containing polymorphisms were compared using the nonparametric Wilcoxon signed-rank test for paired values and the Mann-Whitney test for independent values. The significance of sites within epitopes which varied from the original optimal peptide sequence was tested using Fisher's exact test. Statistical analyses were carried out using Prism 4 for Macintosh (GraphPad Software).
RESULTS
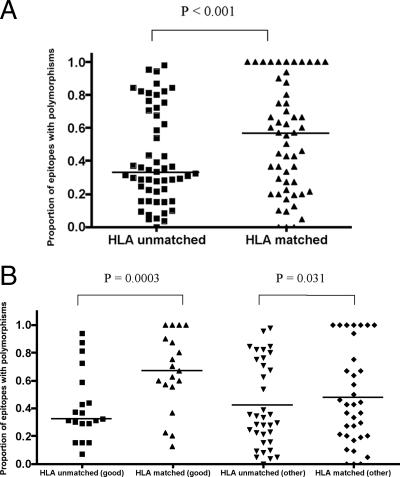
To investigate the relationship between epitope sequence variation and HLA class I molecules in our Swiss cohort (n = 84), we studied three immunogenic genes (HIV gag, pol, and nef) encoding 54 epitopes, defined from the Los Alamos Database. A comparison of the proportion of epitopes with polymorphisms within HLA-matched and -unmatched patients shows a statistically significant difference (median values for HLA-unmatched and -matched epitopes, 0.328 and 0.564, respectively; P < 0.0001, Wilcoxon signed-rank test) (Fig. 1A) in favor of HLA-matched epitopes. Statistical significance remained after removal of epitopes with 100% variation from the analysis (P = 0.0029) (data not shown).
FIG. 1.
Proportion of optimal epitopes containing polymorphisms in HLA-matched and HLA-unmatched (control) patients. (A) For each of the 54 HLA class I-restricted optimal epitopes in the Swiss cohort, the proportion of variant sequences was calculated in patients expressing the relevant HLA class I allele (matched) and in a control group of patients not carrying that allele (unmatched). The figure shows the distribution of variation in the matched and unmatched groups (Wilcoxon signed-rank test). Statistical significance remained after removal of epitopes with 100% variation from the analysis (P = 0.0007) (data not shown). (B) Epitopes were divided according to whether they were restricted by good (HLA-A*11, -B*27, -B*51, -B*57, and -B*58, which are associated with clinical advantage) or other (not associated with clinical advantage) HLA class I alleles. The proportion of variants in each group was compared using the Wilcoxon signed-rank test.
Epitopes were then segregated according to whether they were restricted by beneficial, or “good,” HLA class I alleles A*11, B*27, B*51, B*57, and B*58 and the “other” HLA class I molecules. The good HLA alleles restricted epitopes which were significantly more polymorphic in the HLA-matched patients than in HLA-unmatched controls (median values, 0.67 versus 0.32, respectively; P = 0.0003, Wilcoxon signed-rank test). Epitopes restricted by the nonadvantageous, or other, HLA class I alleles showed a much smaller, although statistically significant, difference in the variation in matched and unmatched patients (median values, 0.43 versus 0.33, respectively; P = 0.03, Wilcoxon signed-rank test). However, this finding for the other HLA alleles was driven by seven epitopes which had reached 100% variation, and their removal from the analysis resulted in the loss of statistical significance (P = 0.34). This was not the case for the good alleles (P = 0.0006).
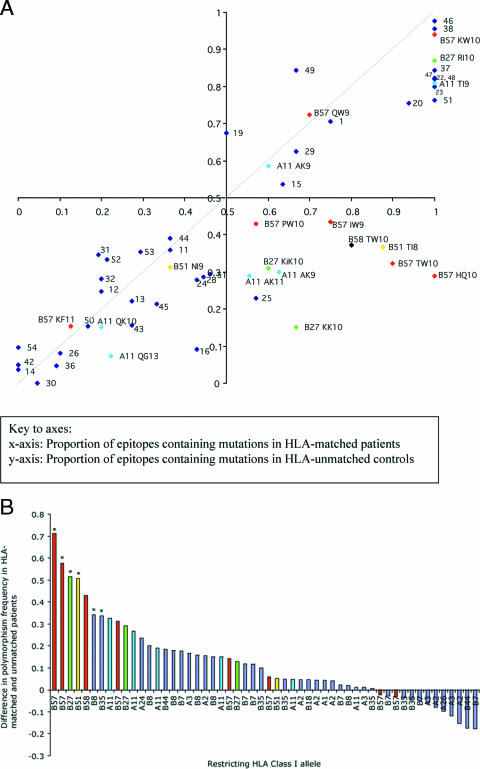
To avoid the subjectivity of nominating good and other HLA class I alleles, sequence variation among all 54 optimal epitopes was analyzed. Figure 2A plots each optimal epitope according to the prevalence of polymorphisms in HLA-matched (x axis) and HLA-unmatched (y axis) patients. The axes intersect at (0.5,0.5) rather than (0,0) in order to define four equal quadrants. If the selection pressure imposed by HLA class I had minimal effect, we would expect all points to lie near the line x = y (hashed). Any epitope lying below the line x = y has more mutations in the HLA-matched patients than in the HLA-unmatched control group. In the bottom right quadrant of the plot (furthest from the line x = y) are the epitopes that are highly polymorphic in individuals bearing the restricting HLA and also comparatively conserved in individuals who do not. It is interesting that these epitopes are almost entirely restricted by protective HLA class I alleles, B*27, B*57, B*51, and A*11.
FIG. 2.
Prevalence of mutations within epitopes in HLA-matched and HLA-unmatched control patients in a Swiss cohort. (A) Patients are from the Swiss HIV cohort (n = 84). For 54 HLA class I-restricted optimal epitopes, the proportion of variant sequences in patients expressing the relevant HLA class I allele (matched) is plotted on the x axis, and the proportion of variant sequences in a control group of patients not carrying that allele (unmatched) is plotted on the y axis. The axes intersect at (0.5,0.5) rather than (0, 0) in order to define four equal quadrants within the figure. The dashed line represents x = y. Epitopes restricted by HLA-B27, -B51, -B57, -B58, and -A11 are color coded and labeled. All other epitopes are represented as numbered blue dots (see Table S1 in the supplemental material for a key to this labeling). (B) The difference between the proportions of polymorphic epitopes in the HLA-matched and -unmatched Swiss patients was calculated and plotted according to rank. To highlight the dominant contribution of the protective HLA class I alleles HLA-B57 and -B58, HLA-B27, HLA-B51, and HLA-A11, these epitopes are color coded; all other epitopes are represented in blue. An asterisk above a column indicates a P value of <0.05 for the distribution of variant epitopes in HLA-matched and -unmatched populations for each epitope using Fisher's exact test.
Variation across the HIV genome may arise for many reasons. To control for polymorphisms that result from forces other than HLA-induced selection, we subtracted the frequency of mutations in the control group of HLA-unmatched patients from the frequency in HLA-matched patients. This difference reflects the additional variation attributable to HLA class I selection. Figure 2B ranks the distribution of this parameter and reveals that the majority of epitopes that show HLA-driven variation (i.e., those that are much more common in HLA-matched than -unmatched patients) are HLA-B alleles associated with slow disease progression (27). Of the 10 most polymorphic epitopes, 7, including the top 5, were restricted by either HLA-B*27, -B*51, or -B*57. The most polymorphic epitopes restricted by an HLA-A allele were restricted by HLA-A*11, also associated with slow disease progression (27).
In the European cohort the epitopes that showed the greatest degree of HLA-attributable variation were the Nef epitope HTQGYFPDWQ (abbreviated HQ10 for the initial and final residues and the number of residues in the epitope; Nef amino acids [aa] 116 to 125), the HLA-B*57- and HLA-B*58-restricted Gag epitope TSTLQEQIGW (TW10; p24 aa 108 to 117), the HLA-B*27-restricted Gag epitope KRWIILGLNK (KK10; p24 aa 131 to 140), and the HLA-B*51-restricted epitope TAFTIPSI (TI8). We found mutations similar to those which have been reported previously (13, 15, 18, 24, 25). Specifically, the Nef HQ10 epitope was predominantly polymorphic at position 1 (histidine to asparagine) and was present in only 28.8% of 45 HLA-unmatched patients but in all HLA-B57-positive patients (odds ratio [OR], ∞; P = 0.0015). The Gag TW10 epitope had mutations at positions 3 (threonine to asparagine) and 9 (predominantly glycine to alanine) contributing to mutations in 13 out of 15 (86.7%) of the HLA-matched patients compared with 16 out of 57 (28.3%) of the HLA-unmatched controls (OR, 16; P < 0.0001). For the Gag KK10 epitope, the predominant polymorphisms were at position 2 (all arginine to lysine) and position 6 (all leucine to methionine, except for one patient with isoleucine) (OR, 12.7; P = 0.001). The HLA-B*51-restricted TI8, epitope which escapes very soon after seroconversion (11), was polymorphic (isoleucine to threonine) at the carboxyl terminus in 7 out of 8 HLA-B*51-positive patients compared with 21 out of 57 HLA-B51-negative patients (OR, 12; P = 0.0087). Phylogenetic analysis (CodeML in the PAML software package) identified the threonine at position 3 in TW10, the leucine at position 6 of KK10, and the isoleucine at position 8 of TI8 to be under strong positive selection pressure. All mutations are detailed in Table S3 of the supplemental material.
It is interesting that although some epitopes restricted by beneficial HLA class I alleles show a high degree of HLA-attributable variation, other epitopes restricted by the same beneficial HLA alleles do not. Some epitopes are highly polymorphic in both HLA-matched and HLA-unmatched patients such as Pol KW10 (B*57), Nef RI10 (B*27), and Gag TI9 (A*11) (see Table S1 in the supplemental material for sequences). Other epitopes restricted by beneficial HLA alleles are highly conserved in both groups, including Gag KF11 (B*57), Pol QG13 (A*11), Nef QK10 (A*11), and Gag NI9 (B*51). These data suggest that the benefit associated with certain HLA class I alleles may be epitope specific rather than an HLA class effect exerted across all restricted sites. It also suggests that there may be protective epitopes that are restricted by HLA alleles which otherwise are not associated with a generalized clinical advantage.
To confirm the observation that epitopes with the greatest degree of HLA-attributable variation are restricted by beneficial class I alleles, we turned to a distinct cohort of African patients (n = 516). Aspects of this population have been published previously (19). The cohort provides an opportunity to test whether our results are sustained in a population infected with a different HIV viral subtype (subtype C) and with a different distribution of HLA class I alleles. The clinical benefit associated with different HLA class I alleles has not been as thoroughly characterized in Africa as it has in Caucasian cohorts, so we used the distribution of HLA-associated mean viral loads in this cohort (19) as a measure of HLA advantage. In this population only HLA class I alleles B5703, B5801, and B8101 have been associated, with statistical significance, with higher CD4 cell counts and lower viral loads (19).
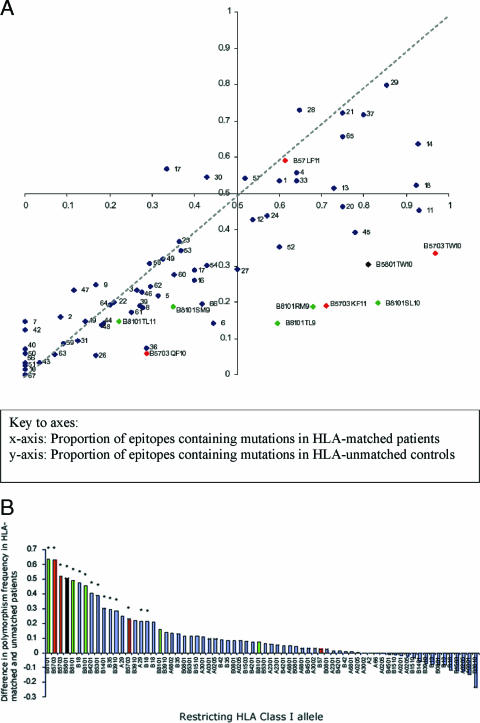
Sequences were obtained, as for the Swiss cohort, from the immunogenic HIV Gag, Pol, and Nef genes. Figure 3A shows the prevalence of polymorphisms within 70 optimal epitopes in HLA-matched (x axis) and HLA-unmatched (y axis) African individuals. As with Fig. 2A, if HLA class I had little effect on polymorphism prevalence, one would expect all points to lie on or near the line x = y (hatched). As for the Swiss data, we find a cluster of epitopes in the lower right quadrant lying furthest from the line x = y that are highly polymorphic in hosts bearing the restricting HLA but conserved in unmatched controls. These were Gag TSTLQEQIAW (Gag TW10; B*5703 and B*5801), Pol SPIETVPVKL (Pol SL10; B*8101), Gag TPQDLNTML (Gag TL9; B*8101), Nef RPQVPLRPM (Nef RM9; B*8101), and Gag KAFSPEVIPMF (Gag KF11; B*5703).
FIG. 3.
Prevalence of mutations within epitopes in HLA-matched and HLA-unmatched control patients in an African cohort. (A) Patients are from the African HIV cohort (n = 512). For 70 HLA class I-restricted optimal epitopes, the proportion of variant sequences in patients expressing the relevant HLA class I allele (matched) is plotted on the x axis, and the proportion of variant sequences in a control group of patients not carrying that allele (unmatched) is plotted on the y axis. The axes intersect at (0.5,0.5) rather than (0,0) in order to define four equal quadrants within the figure. The dashed line represents x = y. The epitopes restricted by HLA-B5703, -B5801, and -B8101 are color coded and labeled. All other epitopes are represented as numbered blue dots (see Table S2 in the supplemental material for a key to the numbering). (B) The difference between the proportions of polymorphic epitopes in the HLA-matched and -unmatched African patients was calculated and displayed according to rank. To highlight the dominant contribution of the protective HLA class I alleles HLA-B5703, HLA-B5801, and HLA-B8101, these epitopes are color coded as shown; all other epitopes are represented in blue. An asterisk above a column indicates a P value of <0.05 for each epitope using Fisher's exact test.
For Gag TW10, patients who carried HLA-B*5703 or -B*5801 were more likely to have a polymorphism at position 3 from threonine to asparagine, often in conjunction with a second polymorphism at positions 5, 8, or 9. (P < 0.00001; OR of 51.4 for B*5703 and a P value of <0.0001; OR of 8.12 for B*5801). In patients with B*8101, the Pol SL10 epitope was more likely to be polymorphic at positions 2 (predominantly proline to serine) and 4 (predominantly glutamic acid to lysine) (P < 0.00001; OR, 20.4), the Gag TL9 epitope was variant at positions 3 (predominantly glutamine to serine) and 7 (predominantly threonine to serine) (P < 0.00001; OR, 8.86), and the Nef RM9 epitope was mostly variant at position 6 (predominantly leucine to valine) (P < 0.00001; OR, 9.1). All mutations are detailed in Table S3 in the supplemental material. As previously described (33) the Gag epitope KF11 is highly variant in the African cohort when restricted by B*5703 but highly conserved in the European B*5701 positive cohort.
As with the European cohort, it is striking that these epitopes are also predominantly restricted by the HLA class I molecules associated with clinical benefit. Figure 3B ranks epitopes according to HLA-attributable variation (the difference in proportions of variability in HLA-matched and -unmatched individuals) and shows that of the eight most variable epitopes, seven are restricted by HLA class I alleles associated with benefit in this African cohort.
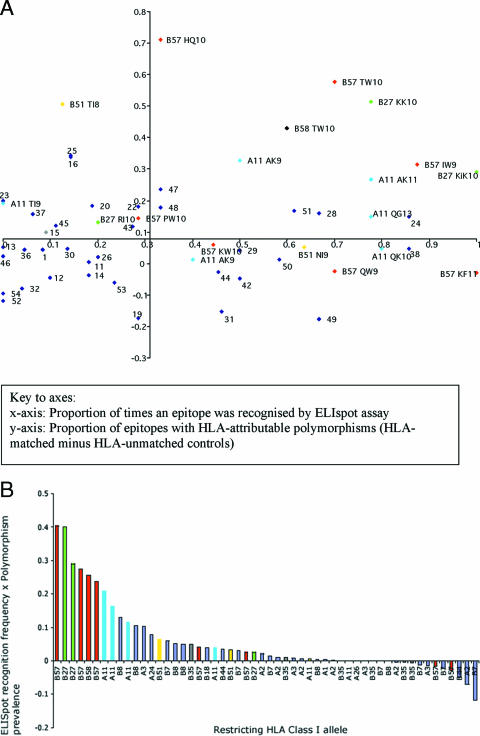
We next sought to determine whether there was also a differential relationship between the immune response to an epitope and its degree of polymorphism. Turning back to the European cohort, each of the 54 epitopes was plotted according to its frequency of recognition in a gamma interferon ELISPOT assay (x axis) and its HLA-attributable degree of genetic variability (the difference in the proportion of variants in the HLA-matched and HLA-unmatched patients) (y axis) (Fig. 4A). The axes of the plot intersect at the median value for each distribution. In this figure epitopes which were frequently recognized in a gamma interferon ELISPOT assay are found on the right-hand side. Epitopes that were more polymorphic in HLA-matched hosts are found in the upper half of the plot. Accordingly, the upper-right quadrant of the plot shows epitopes with both high degrees of polymorphism and high frequency of ELISPOT recognition. This quadrant predominantly contains epitopes restricted by the protective HLA class I alleles such as HLA-B27, -B57, and -B58 and HLA-A11.
FIG. 4.
Frequency of epitope recognition by ELISPOT and prevalence of intraepitopic polymorphisms. (A) Each optimal epitope was represented on a plot according to the proportion of patients who recognized it in a gamma interferon ELISPOT assay (x axis) and the difference in proportions of polymorphic sites in the HLA-matched and -unmatched patients (y axis). The axes intersect at the median value for each distribution. Epitopes restricted by protective (HLA-B27, -B57, -B58, and -A11) HLA class I molecules are labeled (restricting HLA class I allele and epitope abbreviation) and color coded. All other alleles are represented in blue and are numbered (a key to the numbering is available in Table S1 of the supplemental material). (B) A score was calculated for each epitope based on the product of the proportion of times it was recognized in an ELISPOT assay and the difference in proportion of variant sequences in HLA-matched and -unmatched patients. This quality score is presented plotted in order of rank. The epitopes restricted by HLA-B57 and -B58, HLA-B27, HLA-B51, and HLA-A11 are color coded; all other epitopes are represented in blue.
These data can be also be presented as a score for each epitope which uses the mathematical product of the difference in proportions of intraepitopic mutations and the frequency with which each epitope was recognized by patients carrying the restricting HLA class I molecule. This allowed us to rank epitopes which are well recognized in conjunction with high levels of polymorphism (Fig. 4B). It is interesting that of the top 10 epitopes in Fig. 4B, 9 are restricted by HLA-B27, -B57, or -B58 or HLA-A11. These are protective HLA class I alleles.
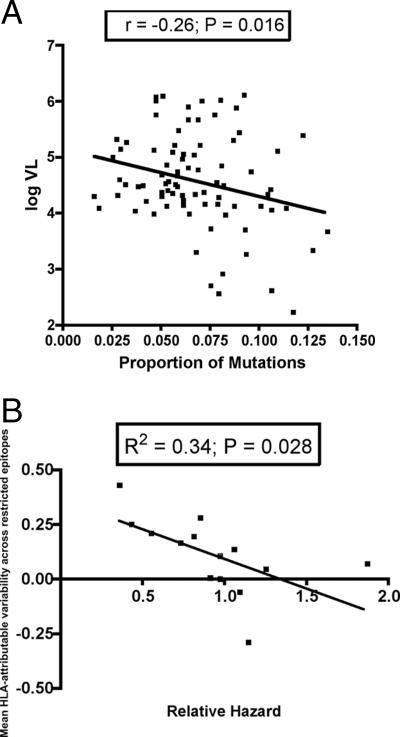
For each patient in the Swiss cohort, the proportion of all polymorphic epitope sequences in the Gag, Pol, and Nef genes (regardless of HLA type) was calculated and correlated with the baseline pretherapy viral load (Fig. 5A). We find a statistically significant correlation between lower plasma viral loads and higher proportions of mutated epitope sequences (P = 0.016; r = −0.26, Pearson's correlation). Next, we calculated the mean value of epitope variability for each HLA class I molecule by averaging the differences between the frequencies of polymorphisms (defined as variant from the population consensus sequence) in epitopes in the HLA-matched and -unmatched patients. Again, we found a statistically significant negative correlation between the mean prevalence of intraepitopic polymorphisms and mean baseline plasma HIV-1 viral load for each HLA class I molecule (P = 0.023; R2 = 0.36) (data not shown). This result supports the assertion that strong selection pressures produce viral escape variants with reduced replicative capacity, although this latter analysis did not survive correction for pseudo-replication. We also found a significant inverse correlation between the mean epitope variability for each HLA class I allele and the HLA-associated relative hazard of disease progression (27) (P = 0.028; R2 = 0.34) (Fig. 5B), confirming that beneficial HLA class I alleles are associated with a greater prevalence of epitope variation.
FIG. 5.
Epitope variability predicts plasma viral load and relative hazard of disease progression. (A) For each patient the proportion of sequence variants in 54 optimal epitopes in the Gag, Pol, and Nef genes was calculated and correlated with baseline plasma viral load, using Pearson's correlation coefficient. (B) For each epitope the difference in the proportion of variant epitopes in HLA-matched and -unmatched patients was calculated. A mean value for each HLA class I molecule was then calculated from all the epitopes restricted by each HLA class I allele. These values were used as a measure of polymorphic variability associated with each HLA class I molecule and correlated negatively with the associated relative hazard of disease progression for each HLA class I allele.
DISCUSSION
Why HLA class I molecules confer distinct rates of progression to AIDS is unclear. Here, we show that some epitopes restricted by HLA alleles associated with clinical protection are frequently recognized in gamma interferon ELISPOT assays and are highly polymorphic in patients bearing the restricting HLA class I but more conserved in individuals who do not. In the Swiss cohort these epitopes were predominantly restricted by HLA-B27, -B51, -B57, and -B58, and in a much larger African cohort with a different HLA class I distribution we found these epitopes to be restricted by B5703, B5801, and B8101. The epitopes identified using the plots in Fig. 2, 3, and 4 are a subset of those targeted by beneficial HLA class I alleles and fulfill the criteria of driver responses (21).
Why are the epitopes that are associated with protective immune responses more polymorphic in HLA-positive hosts than in the HLA-negative controls? We suggest that beneficial HLA class I molecules impose stronger selection pressure on the virus so that immune escape variants with high viral fitness costs are selected. Fitness costs associated with escape have recently been shown for the HLA-B57- and -B5801-restricted Gag TW10 epitope (23). We hypothesize that immune escape associated with a substantial fitness cost results in lower viral loads than escape from a weaker immune pressure which exacts less cost to the virus. There is a precedent for this in the field of antiviral drug resistance. Some patients who develop multidrug resistance mutations do not experience extensive viral load rebound and clinical disease progression but, rather, maintain stable CD4 cell counts in combination with a low-grade viremia (5, 6). This is directly comparable with our hypothesis, except that the HLA class I-restricted immune response rather than drug therapy is the positive selection pressure and the outcome is immune escape rather than drug resistance. We along with others (31, 32) have previously reported frequency-dependent selection in relationship to HLA class I advantage. The finding here that beneficial alleles are associated with a higher degree of viral mutation suggests that the advantage conferred by rare alleles is not simply related to the low frequency of escape variant transmission.
Why is it that, despite being more variant in HLA-matched hosts, epitopes restricted by beneficial HLA alleles are recognized with greater frequency than those restricted by less advantageous HLA alleles? Firstly, even when polymorphisms that confer immune escape approach or reach fixation in peripheral blood, wild-type forms persist at low levels and the CD8+ T-cell capacity to recognize the nonmutant epitope will be sustained (2, 4, 7, 24, 29). This repeated antigenic stimulus would explain the persistence of cytotoxic T cells that can recognize the wild-type optimal peptide and, in turn, maintain selection pressure for HIV escape variants. A second explanation for sustained immunity is T-cell memory. Despite the appearance and evolution of viral escape, T-cell clones that recognize the original sequence persist in the host and can be identified by T-cell receptor motifs (26, 30). These clones may decay in frequency but will be maintained through intermittent exposure to the original wild-type viral variants (29, 30), as they are in other chronic viral infections (14).
We also found that not all the epitopes restricted by beneficial HLA class I alleles showed this pattern of discordant variation in the HLA-matched and -unmatched groups. This suggests that the clinical benefit conferred by specific immune responses is not just HLA specific but also dependent on the gene being targeted and possibly even the position of the epitope within the gene. This would fit with previous data showing an HLA-independent advantage conferred by immune responses to Gag epitopes (20). Another explanation could be that the variant epitopes had escaped immune recognition and that subdominant conserved epitopes had become the focus of the immune response. The interplay between responses to different epitopes within and across HLA alleles is clearly complex. Although subdominant responses are likely to become important as escape develops, we did not see in the longitudinal ELISPOT analyses from the SSITT cohort any clear patterns of sequential epitope selection (data not shown). It seems more likely that at some epitopes restricted by beneficial HLA class I alleles, the sequence conservation reflects a weak selection pressure, and so escape variants are not driven out.
We conclude that the clinical advantage conferred by favorable HLA class I molecules is a reflection of the potent selection pressure exerted at key, dominant epitopes. We suggest that these are the epitopes which elicit effective driver responses, compared with passenger responses—epitopes to which a strong ELISPOT response does not correlate with protection. These protective epitopes are associated with a high prevalence of variation in combination with a strong propensity to revert. Circulating escape mutants persist in patients with good HLA class I molecules but are not associated with the rebound viremia and the clinical progression associated with less costly escape. This has implications for vaccine design as the emergence of immune escape is not necessarily a harbinger of failure of viral control.
Supplementary Material
Acknowledgments
We express our gratitude to the physicians and patients involved in the SSITT (10, 11) and the patients and staff at the Cato Manor Clinic and Sinikithemba Clinic, Durban, South Africa, without whom this study would not have been possible. We thank Paul Klenerman for helpful comments on the manuscript.
This work was supported by the Medical Research Council (United Kingdom) (A.J.F.) and the Wellcome Trust (R.E.P. and P.J.R.G.). B.D.W. is a Doris Duke Distinguished Professor. A.M. and R.E.P. are foundation fellows of the James Martin 21st Century School. The SSITT study was financed in the framework of the Swiss HIV Cohort Study, supported by the Swiss National Science Foundation (grant no. 3347C0-108787).
The members of the Swiss HIV Cohort Study are M. Battegay, E. Bernasconi, J. Böni, H. Bucher, P. Bürgisser, S. Cattacin, M. Cavassini, R. Dubs, M. Egger, L. Elzi, P. Erb, M. Fischer, M. Flepp, A. Fontana, P. Francioli (President of the SHCS, Centre Hospitalier Universitaire Vaudois, CH-1011 Lausanne, Switzerland), H. Furrer (Chairman of the Clinical and Laboratory Committee), M. Gorgievski, H. Günthard, H. Hirsch, B. Hirschel, I. Hösli, C. Kahlert, L. Kaiser, U. Karrer, C. Kind, T. Klimkait, B. Ledergerber, G. Martinetti, B. Martinez, N. Müller, D. Nadal, M. Opravil, F. Paccaud, G. Pantaleo, M. Rickenbach (Head of Data Center), C. Rudin (Chairman of the Mother and Child Substudy), P. Schmid, D. Schultze, J. Schüpbach, R. Speck, P. Taffé, P. Tarr, A. Telenti, A. Trkola, P. Vernazza (Chairman of the Scientific Board), R. Weber, and S. Yerly.
Footnotes
Published ahead of print on 4 April 2007.
Supplemental material for this article is available at http://jvi.asm.org/.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 3.Altfeld, M., E. T. Kalife, Y. Qi, H. Streeck, M. Lichterfeld, M. N. Johnston, N. Burgett, M. E. Swartz, A. Yang, G. Alter, X. G. Yu, A. Meier, J. K. Rockstroh, T. M. Allen, H. Jessen, E. S. Rosenberg, M. Carrington, and B. D. Walker. 2006. HLA alleles associated with delayed progression to AIDS contribute strongly to the initial CD8(+) T cell response against HIV-1. PLOS Med. 3:e403. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, J. R., T. M. Williams, R. F. Siliciano, and J. N. Blankson. 2006. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J. Exp. Med. 203:1357-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deeks, S. G., J. D. Barbour, J. N. Martin, M. S. Swanson, and R. M. Grant. 2000. Sustained CD4+ T cell response after virologic failure of protease inhibitor-based regimens in patients with human immunodeficiency virus infection. J. Infect. Dis. 181:946-953. [DOI] [PubMed] [Google Scholar]
- 6.Deeks, S. G., and R. M. Grant. 1999. Sustained CD4 responses after virological failure of protease inhibitor-containing therapy. Antivir. Ther. 4(Suppl. 3):7-11. [PubMed] [Google Scholar]
- 7.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 8.Fagard, C., A. Oxenius, H. Gunthard, F. Garcia, M. Le Braz, G. Mestre, M. Battegay, H. Furrer, P. Vernazza, E. Bernasconi, A. Telenti, R. Weber, D. Leduc, S. Yerly, D. Price, S. J. Dawson, T. Klimkait, T. V. Perneger, A. McLean, B. Clotet, J. M. Gatell, L. Perrin, M. Plana, R. Phillips, and B. Hirschel. 2003. A prospective trial of structured treatment interruptions in human immunodeficiency virus infection. Arch. Intern. Med. 163:1220-1226. [DOI] [PubMed] [Google Scholar]
- 9.Feeney, M. E., Y. Tang, K. A. Roosevelt, A. J. Leslie, K. McIntosh, N. Karthas, B. D. Walker, and P. J. Goulder. 2004. Immune escape precedes breakthrough human immunodeficiency virus type 1 viremia and broadening of the cytotoxic T-lymphocyte response in an HLA-B27-positive long-term-nonprogressing child. J. Virol. 78:8927-8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez, C. S., I. Stratov, R. De Rose, K. Walsh, C. J. Dale, M. Z. Smith, M. B. Agy, S. L. Hu, K. Krebs, D. I. Watkins, H. O'Connor, D. M. P. Davenport, and S. J. Kent. 2005. Rapid viral escape at an immunodominant simian-human immunodeficiency virus cytotoxic T-lymphocyte epitope exacts a dramatic fitness cost. J. Virol. 79:5721-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frater, A. J., C. T. Edwards, N. McCarthy, J. Fox, H. Brown, A. Milicic, N. Mackie, T. Pillay, J. W. Drijfhout, S. Dustan, J. R. Clarke, E. C. Holmes, H. T. Zhang, K. Pfafferott, P. J. Goulder, M. O. McClure, J. Weber, R. E. Phillips, and S. Fidler. 2006. Passive sexual transmission of human immunodeficiency virus type 1 variants and adaptation in new hosts. J. Virol. 80:7226-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao, X., G. W. Nelson, P. Karacki, M. P. Martin, J. Phair, R. Kaslow, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, S. J. O'Brien, and M. Carrington. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344:1668-1675. [DOI] [PubMed] [Google Scholar]
- 13.Geels, M. J., M. Cornelissen, H. Schuitemaker, K. Anderson, D. Kwa, J. Maas, J. T. Dekker, E. Baan, F. Zorgdrager, R. van den Burg, M. van Beelen, V. V. Lukashov, T. M. Fu, W. A. Paxton, L. van der Hoek, S. A. Dubey, J. W. Shiver, and J. Goudsmit. 2003. Identification of sequential viral escape mutants associated with altered T-cell responses in a human immunodeficiency virus type 1-infected individual. J. Virol. 77:12430-12440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillespie, G. M., M. R. Wills, V. Appay, C. O'Callaghan, M. Murphy, N. Smith, P. Sissons, S. Rowland-Jones, J. I. Bell, and P. A. Moss. 2000. Functional heterogeneity and high frequencies of cytomegalovirus-specific CD8+ T lymphocytes in healthy seropositive donors. J. Virol. 74:8140-8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 16.Goulder, P. J., and D. I. Watkins. 2004. HIV and SIV CTL escape: implications for vaccine design. Nat. Rev. Immunol. 4:630-640. [DOI] [PubMed] [Google Scholar]
- 17.Islam, S. A., C. M. Hay, K. E. Hartman, S. He, A. K. Shea, A. K. Trocha, M. J. Dynan, N. Reshamwala, S. P. Buchbinder, N. O. Basgoz, and S. A. Kalams. 2001. Persistence of human immunodeficiency virus type 1-specific cytotoxic T-lymphocyte clones in a subject with rapid disease progression. J. Virol. 75:4907-4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiepiela, P., A. J. Leslie, I. Honeyborne, D. Ramduth, C. Thobakgale, S. Chetty, P. Rathnavalu, C. B. Moore, K. J. Pfafferott, L. Hilton, P. Zimbwa, S. Moore, T. Allen, C. Brander, M. M. Addo, M. Altfeld, I. R. James, S. A. Mallal, M. Bunce, L. D. Barber, J. Szinger, C. Day, P. Klenerman, J. Mullins, B. T. Korber, H. M. Coovadia, B. D. Walker, and P. J. Goulder. 2004. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature 432:769-774. [DOI] [PubMed] [Google Scholar]
- 20.Kiepiela, P., K. Ngumbela, C. Thobakgale, D. Ramduth, I. Honeyborne, E. Moodley, S. Reddy, C. de Pierres, Z. Mncube, N. Mkhwanazi, K. Bishop, M. van der Stok, K. Nair, N. Khan, H. Crawford, R. Payne, A. Leslie, J. Prado, A. Prendergast, J. Frater, N. McCarthy, C. Brander, G. H. Learn, D. Nickle, C. Rousseau, H. Coovadia, J. I. Mullins, D. Heckerman, B. D. Walker, and P. Goulder. 2007. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat. Med. 13:46-53. [DOI] [PubMed] [Google Scholar]
- 21.Klenerman, P., Y. Wu, and R. Phillips. 2002. HIV: current opinion in escapology. Curr. Opin. Microbiol. 5:408-413. [DOI] [PubMed] [Google Scholar]
- 22.Leslie, A. J., K. J. Pfafferott, P. Chetty, R. Draenert, M. M. Addo, M. Feeney, Y. Tang, E. C. Holmes, T. Allen, J. G. Prado, M. Altfeld, C. Brander, C. Dixon, D. Ramduth, P. Jeena, S. A. Thomas, A. S. John, T. A. Roach, B. Kupfer, G. Luzzi, A. Edwards, G. Taylor, H. Lyall, G. Tudor-Williams, V. Novelli, J. Martinez-Picado, P. Kiepiela, B. D. Walker, and P. J. Goulder. 2004. HIV evolution: CTL escape mutation and reversion after transmission. Nat. Med. 10:282-289. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Picado, J., J. G. Prado, E. E. Fry, K. Pfafferott, A. Leslie, S. Chetty, C. Thobakgale, I. Honeyborne, H. Crawford, P. Matthews, T. Pillay, C. Rousseau, J. I. Mullins, C. Brander, B. D. Walker, D. I. Stuart, P. Kiepiela, and P. Goulder. 2006. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80:3617-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Migueles, S. A., A. C. Laborico, H. Imamichi, W. L. Shupert, C. Royce, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, C. W. Hallahan, and M. Connors. 2003. The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences. J. Virol. 77:6889-6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 26.Moss, P. A., S. L. Rowland-Jones, P. M. Frodsham, S. McAdam, P. Giangrande, A. J. McMichael, and J. I. Bell. 1995. Persistent high frequency of human immunodeficiency virus-specific cytotoxic T cells in peripheral blood of infected donors. Proc. Natl. Acad. Sci. USA 92:5773-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Brien, S. J., X. Gao, and M. Carrington. 2001. HLA and AIDS: a cautionary tale. Trends Mol. Med. 7:379-381. [DOI] [PubMed] [Google Scholar]
- 28.Oxenius, A., D. A. Price, H. F. Gunthard, S. J. Dawson, C. Fagard, L. Perrin, M. Fischer, R. Weber, M. Plana, F. Garcia, B. Hirschel, A. McLean, and R. E. Phillips. 2002. Stimulation of HIV-specific cellular immunity by structured treatment interruption fails to enhance viral control in chronic HIV infection. Proc. Natl. Acad. Sci. USA 99:13747-13752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price, D. A., S. M. West, M. R. Betts, L. E. Ruff, J. M. Brenchley, D. R. Ambrozak, Y. Edghill-Smith, M. J. Kuroda, D. Bogdan, K. Kunstman, N. L. Letvin, G. Franchini, S. M. Wolinsky, R. A. Koup, and D. C. Douek. 2004. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity 21:793-803. [DOI] [PubMed] [Google Scholar]
- 31.Scherer, A., J. Frater, A. Oxenius, J. Agudelo, D. A. Price, H. F. Gunthard, M. Barnardo, L. Perrin, B. Hirschel, R. E. Phillips, and A. R. McLean. 2004. Quantifiable cytotoxic T lymphocyte responses and HLA-related risk of progression to AIDS. Proc. Natl. Acad. Sci. USA 101:12266-12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trachtenberg, E., B. Korber, C. Sollars, T. B. Kepler, P. T. Hraber, E. Hayes, R. Funkhouser, M. Fugate, J. Theiler, Y. S. Hsu, K. Kunstman, S. Wu, J. Phair, H. Erlich, and S. Wolinsky. 2003. Advantage of rare HLA supertype in HIV disease progression. Nat. Med. 9:928-935. [DOI] [PubMed] [Google Scholar]
- 33.Yu, X. G., M. Lichterfeld, S. Chetty, K. L. Williams, S. K. Mui, T. Miura, N. Frahm, M. E. Feeney, Y. Tang, F. Pereyra, M. X. Labute, K. Pfafferott, A. Leslie, H. Crawford, R. Allgaier, W. Hildebrand, R. Kaslow, C. Brander, T. M. Allen, E. S. Rosenberg, P. Kiepiela, M. Vajpayee, P. A. Goepfert, M. Altfeld, P. J. Goulder, and B. D. Walker. 2007. Mutually exclusive T-cell receptor induction and differential susceptibility to human immunodeficiency virus type 1 mutational escape associated with a two-amino-acid difference between HLA class I subtypes. J. Virol. 81:1619-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yusim, K., C. Kesmir, B. Gaschen, M. M. Addo, M. Altfeld, S. Brunak, A. Chigaev, V. Detours, and B. T. Korber. 2002. Clustering patterns of cytotoxic T-lymphocyte epitopes in human immunodeficiency virus type 1 (HIV-1) proteins reveal imprints of immune evasion on HIV-1 global variation. J. Virol. 76:8757-8768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.