Abstract
Human immunodeficiency virus type 1 (HIV-1) fusion inhibitors blocking viral entry by binding the gp41 heptad repeat 1 (HR1) region offer great promise for antiretroviral therapy, and the first of these inhibitors, T20 (Fuzeon; enfuvirtide), is successfully used in the clinic. It has been reported previously that changes in the 3-amino-acid GIV motif at positions 36 to 38 of gp41 HR1 mediate resistance to T20 but usually not to second-version fusion inhibitors, such as T1249, which target an overlapping but distinct region in HR1 including a conserved hydrophobic pocket (HP). Based on the common lack of cross-resistance and the difficulty of selecting T1249-resistant HIV-1 variants, it has been suggested that the determinants of resistance to first- and second-version fusion inhibitors may be different. To further assess HIV-1 resistance to fusion inhibitors and to analyze where changes in HR1 are tolerated, we randomized 16 codons in the HR1 region, including those making contact with HR2 codons and/or encoding residues in the GIV motif and the HP. We found that changes only at positions 37I, 38V, and 40Q near the N terminus of HR1 were tolerated. The propagation of randomly gp41-mutated HIV-1 variants in the presence of T1249 allowed the effective selection of highly resistant forms, all containing changes in the IV residues. Overall, the extent of T1249 resistance was inversely correlated to viral fitness and cytopathicity. Notably, one HIV-1 mutant showing ∼10-fold-reduced susceptibility to T1249 inhibition replicated with wild type-like kinetics and caused substantial CD4+-T-cell depletion in ex vivo-infected human lymphoid tissue in the presence and absence of an inhibitor. Taken together, our results show that the GIV motif also plays a key role in resistance to second-version fusion inhibitors and suggest that some resistant HIV-1 variants may be pathogenic in vivo.
Presently, more than 20 drugs have been approved by the Food and Drug Administration for the treatment of human immunodeficiency virus (HIV) infection and AIDS, and highly active antiretroviral therapy has greatly increased the life expectancy for individuals with HIV type 1 (HIV-1) infection, at least in industrialized countries (46). However, with a single exception described below, all these drugs belong to one of only three classes targeting the viral reverse transcriptase (RT) or protease (1, 25), and resistance to one drug often results in cross-resistance to others in the same class (11). Some patients die from AIDS caused by multiresistant viruses (39), and the incidence of primary HIV resistance is increasing in various parts of the world (9, 12, 34, 60). Thus, there is an urgent need for antiretrovirals with novel mechanisms of action that can be used for salvage therapy.
One highly promising approach to improve AIDS therapy is the inhibition of virus entry. The knowledge of the mechanisms involved in this process allowed the development of a variety of agents inhibiting the process at different steps: the attachment of gp120 to the CD4 cell receptor, the binding of gp120 to CCR5 or CXCR4, and the gp41-mediated fusion of viral and cellular membranes (4, 43, 48). Several entry inhibitors are presently in clinical development and will hopefully soon complement the present therapeutic armamentarium against HIV infection and AIDS. To date, however, only one of them, T20 (Fuzeon; envufirtide), has been approved for clinical use (30, 40). T20 is a 36-amino-acid (aa) peptide which corresponds to heptad repeat 2 (HR2) of gp41 (58, 59). It binds to the HR1 domain in gp41, which becomes exposed after CD4 binding (20, 26), thereby preventing the formation of the six-helix bundle essential for membrane fusion and virus entry (4, 47). T20 is a good option for rescue therapy in combination with other antiretrovirals. One problem is, however, that HIV-1 easily becomes resistant to this inhibitor. The main mechanism of resistance to T20 is the selection of changes within a 36- to 45-aa domain of the HR1 region, particularly within a conserved 3-aa sequence (GIV) of gp41 (2, 23, 35, 49, 52, 53, 55).
To overcome this problem, second-version fusion inhibitors with greater antiviral potency than T20, such as T1249, composed of gp41 HR2 sequences derived from HIV-1, HIV-2, and simian immunodeficiency virus, have been generated (17, 24). Like other second-version fusion inhibitors, T1249 targets a region within HR1 overlapping but distinct from that targeted by T20. The nonoverlapping residues in T1249 include three highly conserved hydrophobic residues predicted to project into the deep hydrophobic pocket (HP) of the HR1 trimer; these residues are important for HR2 binding and hence six-helix bundle stability (6, 15, 42, 54). Although T1249 is more effective than T20, its clinical development is presently on hold due to formulation problems. Nonetheless, modified, more stable and potent forms of T1249 or related peptides that require fewer injections and are more appropriate for chronic administration may become the next generation of HIV-1 fusion inhibitors. Therefore, further evaluation of HIV-1 resistance to both T20 and T1249 remains of great interest. Some changes in the GIV motif can reduce the susceptibility of HIV-1 to T1249 or to another fusion inhibitor targeting the HP, C34 (3, 32, 44). It has been clearly demonstrated, however, that T1249 and other second-iteration fusion inhibitors are usually active against T20-resistant HIV-1 isolates (14, 41, 51) and decrease the viral load in patients harboring T20-resistant viruses (29, 32). Based on the common lack of cross-resistance and the difficulty of generating T1249-resistant HIV-1 variants in vitro, it has been suggested that the resistance profiles for T20 and T1249 might be clearly distinct and that the compounds can be used in combination (51).
In the present study, we utilized a site-specific random PCR mutagenesis approach to select T1249-resistant forms of HIV-1 and to assess where in the gp41 HR1 changes may or may not be tolerated. We found that changes in the amino acids of the hydrophobic cavity and other residues predicted to be important for the formation of the six-helix bundle are hardly tolerated, and thus, these residues are excellent targets for antiretroviral drugs. However, our approach also allowed the rapid selection of HIV-1 mutants resistant to both T20 and T1249. All mutations conferring cross-resistance to both fusion inhibitors corresponded to the IV residues of the GIV motif, suggesting that the resistance profiles for these drugs are highly similar. Overall, the level of resistance was inversely correlated with viral fitness. Not all mutations reducing the susceptibility of HIV-1 to T1249 and T20, however, impaired the ability of the virus to replicate efficiently and to cause CD4+-T-cell depletion in ex vivo human lymphoid tissue (HLT), suggesting that some HIV-1 variants resistant against fusion inhibitors may be pathogenic.
MATERIALS AND METHODS
Cells and virus stocks.
TZM-bl cells were obtained from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program (ARRRP) and were contributed by John Kappes and Xiaoyun Wu. This cell line expresses large amounts of CD4, CCR5, and CXCR4 and contains Tat-responsive reporter genes for firefly luciferase and β-galactosidase under the control of an HIV-1 long terminal repeat promoter (55). Adherent TZM-bl and 293T cells were maintained in Dulbecco's modified Eagle's medium (GIBCO BRL Life Technologies) containing 10% heat-inactivated fetal bovine serum. Cell monolayers were split 1:10 at confluence by treatment with 0.25% trypsin-1 mM EDTA (Invitrogen). The nonadherent cell lines CEMx174 5.25M7 (kindly provided by Nat Landau, La Jolla, CA) and PM1 (38), provided by M. Reitz through the NIH ARRRP, were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum. CEMx174 5.25M7 cells contain the gene encoding the enhanced version of the green fluorescent protein under the control of the HIV-1 long terminal repeat promoter. Human peripheral blood mononuclear cells (PBMC) were isolated by using lymphocyte separation medium (Organon Teknika Corporation, Durham, NC) and stimulated with phytohemagglutinin (4 μg/ml; Sigma) for 3 days in RPMI 1640 medium supplemented with 20% fetal calf serum and 100 U of interleukin 2 per ml. Virus stocks were generated by the transient transfection of 293T cells with the CXCR4-tropic HIV-1 NL4-3 molecular clone or specific gp41-mutated variants of NL4-3 by using the calcium phosphate method as described elsewhere (5). The p24 antigen levels were quantitated with an HIV p24 enzyme-linked immunosorbent assay (ELISA) kit obtained through the NIH ARRRP.
Generation of random gp41-mutated HIV-1 variants.
Site-specific random mutagenesis of the HIV-1 NL4-3 molecular clone was performed by splice overlap extension PCR using wobble primers. PCR fragments were gel purified and ligated into the full-length NL4-3 molecular clone by using the NheI and BamHI restriction sites flanking the HR1 region, and supercompetent Escherichia coli XL2-Blue cells (Stratagene, La Jolla, CA) were transformed with the constructs. Eighty percent of the transformation mixture was inoculated directly into Luria-Bertani medium for expansion while the remaining 20% of the mixture was plated onto agar plates to check the number of transformants. High cloning efficiencies (>1,000 transformants per HR1 variant pool) and the presence of wobble codons at the desired positions were verified by restriction and sequence analyses of the obtained plasmid populations.
Selection and genotyping of T1249-resistant viruses.
CEMx174 5.25M7 cells were infected in the presence of 50 nM T1249 with virus containing 10 ng of the p24 core antigen derived from transfected 293T cells. At 12 h postinfection, the cells were washed to remove the inoculum, and they were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum and an inhibitor. The cells were split regularly, fresh cells were added when required, and supernatant aliquots were taken periodically. Green fluorescent protein expression was monitored to detect the emergence of drug-resistant variants. Finally, cells were centrifuged and the supernatant was used to extract viral RNA (QIAGEN). RT-PCR was performed by using the Invitrogen RT-PCR system as recommended by the manufacturer, and the gel-purified RT-PCR fragments were sequenced directly for genotypic analysis. Individual RT-PCR clones were also analyzed, and full-length gp41 HR1-mutated NL4-3 variants were generated as described above.
Infectivity and inhibition assays.
TZM-bl cells were seeded into flat-bottomed 96-well dishes and cultured overnight. Infections were performed in triplicate in the absence of an inhibitor (control) or in the presence of 20, 100, and 500 nM T20 or 1, 10, and 100 nM T1249 with virus containing 1 ng of p24. Infectivity was measured in a luminometer at 2 days postinfection by using the β-galactosidase screening kit from TROPIX as recommended by the manufacturer. Virus infectivity was calculated by dividing the β-galactosidase activity (expressed as relative light units per second) produced at each concentration of the inhibitor by the relative light units measured in the absence of the inhibitor. The mean 50% inhibitory concentrations (IC50s) were calculated as described previously (53) and compared by using Student's t test to determine whether the observed differences were statistically significant.
Viral fitness.
To analyze the fitness of resistant viruses, phytohemagglutinin-stimulated PBMC and PM1 cells were challenged with viruses containing 2 ng of the p24 core antigen. Supernatants were collected at regular intervals (1, 3, 6, 9, 12, and 15 days postinfection), and virus production was quantified by the p24 ELISA.
Env expression analysis.
Viral particles containing normalized p24 antigen produced by transfected 293T cells were pelleted and analyzed for gp120, gp41, and p24 expression by immunoblotting using HIV-1 gp120 rabbit antiserum (Advanced Biotechnologies), hmAb5F3, and rabbit anti-HIV-1 p24 provided by the NIH ARRRP.
HIV infection of HLT ex vivo.
Human tonsillar tissue removed during a routine tonsillectomy was received within 5 h of excision. The tonsils were washed thoroughly with medium containing antibiotics, sectioned into 2- to 3-mm blocks, placed on top of collagen sponge gels in the culture medium at the air-liquid interface, and infected as described previously (21, 22). Briefly, for the testing of tissues from one donor, p24-normalized viruses were inoculated into each of 18 tissue blocks. HIV-1 production was assessed by using the HIV-1 p24 ELISA. To evaluate CD4+-T-cell depletion, flow cytometry was performed on cells mechanically isolated from control and infected tissue blocks (22). The cells were stained for the cell surface antigens CD3, CD4, and CD8 by using anti-CD3-fluorescein isothiocyanate, anti-CD4-allophycocyanin, and anti-CD8-Tri color; fixed and permeabilized with Cytofix-Cytoperm (Caltag); and stained for the intracellular marker by using anti-p24 antibody KC57 RD1 (Coulter Clone). The depletion of CD4+ T cells was assessed as described previously (22) and expressed as a ratio of CD4+ T cells to CD8+ T cells. For the experiments with T1249, tonsillar tissue blocks were incubated in medium containing 50 nM T1249 in a rotating vessel for 5 h before infection and maintained in medium containing the inhibitor.
RESULTS
Many residues in the HR1 region do not tolerate changes.
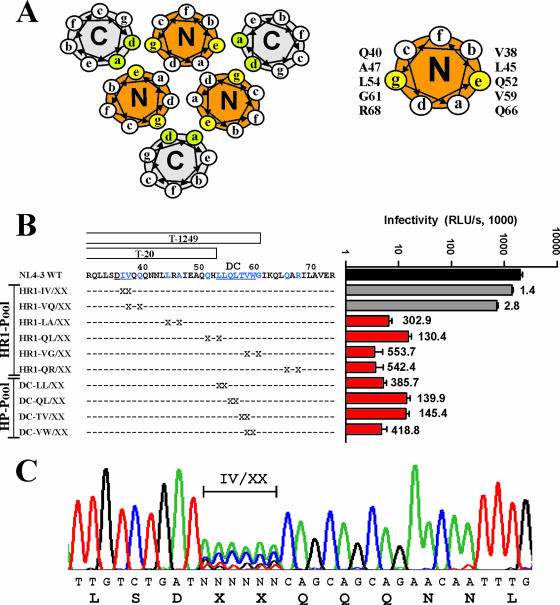
To analyze at which amino acid positions changes may be tolerated in the gp41 HR1 regions previously implicated in resistance to T20 (2, 24, 35, 49, 52, 53, 55) and the formation and stability of the six-helix bundle (6, 15, 28, 37, 42, 54, 57), we used a random PCR mutagenesis approach to generate a comprehensive set of gp41-mutated variants of the replication-competent and well-characterized HIV-1 NL4-3 molecular clone. Notably, the GIV motif is changed to DIV in the NL4-3 gp41, and this alteration slightly reduces the sensitivity of this clone to T20 (31) but not to T1249 (Table 1). One set of mutations targeted residues at the e and g positions of the N helices of the trimeric coiled coil that pack in an antiparallel orientation against residues at positions a and d of the C helices (Fig. 1A) and are important for the formation of the six-helix bundle and hence viral infectivity (7, 16, 19, 56). The remaining positions targeted were either those of the IV residues in the GIV motif, where the majority of known mutations mediating T20 resistance are located (2, 24, 35, 49, 52, 53, 55), or those of residues involved in the formation of the conserved hydrophobic coiled-coil cavity in the gp41 core. It has been proposed previously that the deep hydrophobic cavity formed by the stretch of hydrophobic amino acids in the N helix trimer is important for six-helix bundle stability (15) and provides an excellent antiviral drug target (6, 16, 28). In fact, second-version fusion inhibitors containing this cavity binding region, such as T1249 (Fig. 1B), are thought to be much less susceptible to the evolution of resistant virus than the T20 fusion inhibitor lacking this region (14, 29, 41, 51). We targeted only two amino acid residues per mutant (Fig. 1B) and cloned the randomly mutagenized env alleles as a pool into the proviral constructs to ensure that all mutant HIV-1 stocks contained a complex mixture of amino acid changes at the desired locations. Control experiments confirmed that >95% of the proviral constructs contained an insert of the expected size and that each plasmid preparation represented at least 1,000 independent transformants (data not shown).
TABLE 1.
Properties of randomly mutagenized HIV-1 variants selected in the presence of T1249
| NL4-3 clone | Amino acid sequence or change(s)a | Nucleotide sequence or change(s)b | Frequencyc | Pool of origin | Infectivityd | T20 IC50 (nM)e | T1249 IC50 (nM)e | Reduction in T1249 susceptibility (n-fold) | Zidovudine IC50 (nM)e |
|---|---|---|---|---|---|---|---|---|---|
| wt | DIV | ATAGTG | 0/15 | NA | 100 ± 18 | 57 ± 37 | 1.2 ± 0.1 | 25.8 ± 8.0 | |
| IV/SN mutant | DSN | TC-AAC | 4/15 | HR1 | 19 ± 4 | >500 | >50 | >50 | 23.1 ± 4.9 |
| IV/QQ mutant | DQQ | CAGCAA | 1/15 | HR1 | 35 ± 8 | >500 | 16.8 ± 6.2 | 14.0 | 28.1 ± 10.5 |
| IV/TA mutant | DTA | -C-CA | 1/15 | HR1 | 15 ± 2 | >500 | 41.3 ± 20.0 | 34.4 | 32.9 ± 6.0 |
| IV/SQ mutant | DSQ | TC-CAA | 1/15 | HR1 | 18 ± 4 | >500 | >50 | >50 | 23.8 ± 11.6 |
| IV/HE mutant | DHE | CAC-A- | 5/15 | HR1 | 17 ± 5 | >500 | >50 | >50 | 23.4 ± 11.9 |
| IV/QM mutant | DQM | CAGA- | 3/15 | HR1 | 44 ± 3 | >500 | 30.5 ± 18.2 | 25.4 | 14.8 ± 4.1 |
| IV/VT mutant | DVT | G-AC- | 2/6 | IV/XX-VQ/XX | 69 ± 13 | >500 | 9.3 ± 3.6 | 7.7 | 26.2 ± 11.1 |
| IV/QH mutant | DQH | CAGCAC | 2/6 | IV/XX-VQ/XX | 15 ± 4 | >500 | 20.5 ± 13.8 | 17.1 | 21.3 ± 1.6 |
| IV/AT mutant | DAT | GCCAC- | 2/6 | IV/XX-VQ/XX | 22 ± 2 | >500 | >50 | >50 | 22.2 ± 10.4 |
| L33V mutant | L33V | TTA to GTA | NA | NA | 76 ± 18 | 341 + 63 | 1.3 ± 0.6 | 1.1 | 35.7 ± 18.4 |
| L33S mutant | L33S | TTA to TCA | NA | NA | 54 ± 15 | >500 | 18.8 ± 11.8 | 15.7 | 23.1 ± 4.9 |
The positions of randomly mutated amino acids in the HIV-1 HR1 and IV/XX-VQ/XX viral stocks are indicated in Fig. 1B.
Dashes in nucleotide sequences represent nucleotide identity.
Number of clones containing the indicated sequence changes per total number of clones analyzed. NA, not applicable.
Infectivity was measured using TZM-bl cells, and the data shown are mean values ± standard deviations derived from triple infections with each of three different virus stocks. Results are expressed as percentages of the infectivity of the NL4-3 wt.
Results shown are the means ± standard deviations of results from three independent experiments.
FIG. 1.
Random mutagenesis of the HIV-1 gp41 HR1 region. (A) Helical wheel diagram of the six-helix bundle (modified from reference 37 with permission). The left panel shows the positions of heptad repeat residues in a cross-section of the helices in the gp41 core. The residues at the g and e positions in the N helices (N) and the a and d positions in the C helices (C) that are important for the interaction between the coiled-coil domain and the antiparallel external helices are highlighted in yellow and green, respectively. The right panel shows the amino acid residues at positions g and e of the N helix subjected to random mutagenesis. (B) Levels of infectivity of gp41-mutated HIV-1 NL4-3 variants containing randomized codons corresponding to the HR1 region. The amino acid sequences of the N-helical region and the positions of the randomized residues (denoted by X's) are indicated. Viral infectivity was measured by infecting TZM-bl cells with virus stocks containing normalized amounts (1 ng) of the p24 core antigen, and the data represent average values ± standard deviations derived from triplicate infections. The numbers to the right of the bars indicate the reductions (n-fold) in infectivity compared to that of the NL4-3 wt virus. Similar results were obtained in an independent experiment. RLU/s, relative light units per second. (C) Sequence analysis of the NL4-3 HR1 IV/XX-VQ/XX mutant mix. Proviral DNA encompassing the mutated positions was subjected to direct sequence analysis. The nucleotide and deduced amino acid sequences of the mutated region are shown. Similar results were obtained for the remaining nine HR1 and HP mutants except that complex nucleotide mixtures were observed at different positions.
The direct sequencing of the proviral mixture showed a complex array of nucleotide changes in the codons corresponding to the targeted amino acid positions in the gp41 HR1 (an example is shown in Fig. 1C). Virus stocks were generated by the transient transfection of 293T cells with the gp41-mutated HIV-1 constructs. To assess the impact of the random substitutions on HIV-1 infectivity, we infected TZM-bl indicator cells with virus stocks containing normalized quantities of the p24 core antigen. The results demonstrated that HIV-1 variants containing random changes at positions 37 or 38 and 38 or 40 in or near the GIV motif were highly infectious, suggesting that changes in these amino acid residues are frequently tolerated (Fig. 1B). In contrast, the levels of infectivity of all remaining virus stocks were >100-fold lower than that of the HIV-1 NL4-3 wild type (wt). In several cases, the infectivity levels of the virus stocks were reduced to the same extent as the expected frequencies of the two wt codons in the proviral pools (Fig. 1B). Thus, in agreement with the high conservation of these residues among HIV-1 and simian immunodeficiency virus strains (33) and an important role for gp41 function, alterations were hardly or not at all tolerated at the e and g positions of the N helices and in the residues forming the HP.
Effective selection of T1249-resistant HIV-1 mutants.
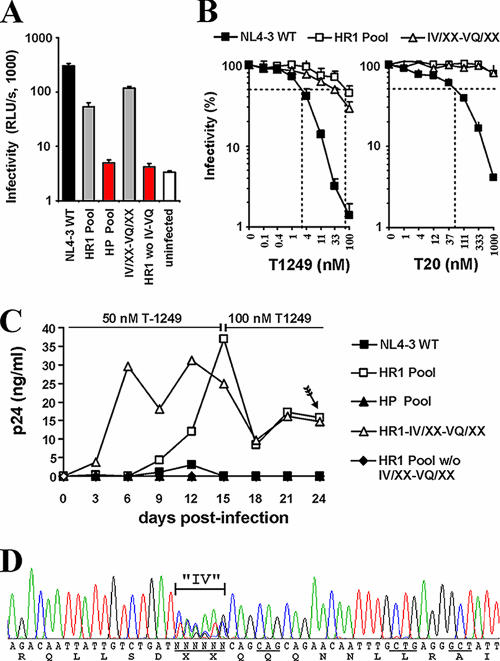
To test whether random changes in the HR1 region altered the sensitivity of HIV-1 to fusion inhibitors, we infected TZM-bl cells with wt and mutant virus stocks in the presence and absence of T20 and T1249. To further evaluate the role of the IV residues in HIV-1 resistance to fusion inhibitors, we used four mutant virus stocks: (i) the HR1 pool, with variants containing alterations at a total of 11 positions in the N helix, including those in and around the GIV motif; (ii) the HP pool, with variants containing alterations at a total of eight positions in the residues forming the deep cavity; (iii) the IV/XX-VQ/XX pool, with variants harboring mutations in residues in or near the GIV motif; and (iv) a pool of variants carrying mutations in HR1 other than those in or near the GIV motif (the HR1 pool without the IV/XX-VQ/XX virus stock) (Fig. 1B). As expected from the results obtained using the individual virus stocks (Fig. 1B), the levels of infectivity of the HP pool and the HR1 pool without the IV/XX-VQ/XX virus stock were very low (Fig. 2A). In comparison, the levels of infectivity of the HIV-1 HR1 pool and the IV/XX-VQ/XX virus stock were only moderately reduced compared to that of wt NL4-3. Most importantly, infection with HIV-1 mutants containing random changes in the HR1 region was only moderately inhibited by T1249 and hardly at all affected by T20, whereas infection with wt NL4-3 was efficiently blocked (Fig. 2B). These results strongly suggested that a significant proportion of the gp41-mutated HIV-1 variants present in the HR1 pool and in the IV/XX-VQ/XX virus stock showed reduced sensitivity to T20 and T1249 inhibition.
FIG. 2.
Selection of T1249-resistant HIV-1 variants. (A) Levels of infectivity of the indicated HIV-1 NL4-3 stocks (see Fig. 1B for details). Viral infectivity was measured by infecting TZM-bl cells with virus stocks containing normalized amounts (1 ng) of the p24 core antigen, and data represent average values ± standard deviations derived from triplicate infections. HR1 wo IV-VQ, HR1 pool without the IV/XX-VQ/XX viral stock. (B) Changes in the HR1 region reduce the susceptibility of HIV-1 to fusion inhibitors. TZM-bl indicator cells were infected with wt NL4-3, the HR1 pool, or the IV/XX-VQ/XX viral stock in the presence of the indicated concentrations of T1249 or T20. Shown are average values derived from triplicate infections with virus stocks containing 20 ng of the p24 antigen. (C) Changes in the gp41 HR1 region allow efficient viral replication in the presence of T1249. CEMx174 5.25M7 cells were infected in triplicate with the indicated virus stocks in the presence of 50 nM T1249. At day 15, the concentration of the inhibitor was increased to 100 nM. The arrow indicates the point at which the gp41 region was amplified by RT-PCR from the cell-free culture supernatants. The replication curves indicate average levels of p24 derived from triplicate infections. (D) The selected T1249-resistant HIV-1 mutants contain alterations mainly or exclusively in the IV residues. Shown is the result of direct sequence analysis of the RT-PCR product obtained from the cell-free supernatant of CEMx174 5.25M7 cells infected with the HR1 pool mentioned in the legend to panel B. Codons that were subjected to random PCR mutagenesis are underlined in the nucleotide sequence.
Previous studies have suggested that it is difficult to select HIV-1 variants resistant to second-version fusion inhibitors targeting the HP in vitro (14, 29, 32, 41, 51). To evaluate whether our random mutagenesis approach may be more successful than standard viral passage in the presence of an inhibitor, we infected CEMx174 5.25M7 cells with NL4-3 wt and mutant virus stocks in the presence of 50 nM T1249. Efficient viral replication was observed already at 6 days after infection with the HIV-1 IV/XX-VQ/XX pool (Fig. 2C). In comparison, infection with the HR1 pool of mutants containing changes at a total of 11 positions, including those of the IV residues, also resulted in effective albeit substantially delayed replication (Fig. 2C). In contrast, in the presence of T1249, no effective viral spread was observed after inoculation with wt HIV-1 NL4-3, the HP pool, or the HR1 pool of variants without changes in the IV residues. These data suggested that HIV-1 mutants containing changes in the IV residues were capable of replicating in the presence of T1249, whereas changes at the remaining positions, including those in the HP domain, either impaired viral replication or did not confer T1249 resistance.
To select for the most resistant HIV-1 variants, we increased the dose of T1249 to 100 nM after 15 days of culture. At day 24 after infection, viral RNA was extracted from the cell-free supernatants of the cultures infected with the HR1 and IV/XX-VQ/XX pools (Fig. 2C) and the gp41 coding region was amplified by RT-PCR. Direct sequencing of the PCR products showed that HIV-1 variants selected in the presence of T1249 contained changes exclusively in the IV residues (an example is shown in Fig. 2D). To further assess which gp41-mutated HIV-1 variants emerged in the presence of the inhibitor, we analyzed a total of 21 individual PCR clones (summarized in Table 1). As expected from the results of the bulk sequence analysis, all of these clones contained changes in the IV residues but no changes elsewhere in the gp41 region. Notably, the selected HIV-1 mutants differed from wt NL4-3 by an average of five and a minimum of three nucleotide changes that always changed both codons (Table 1). This finding may explain the difficulty in selecting T1249-resistant forms by cell culture passage of NL4-3 wt virus.
Fitness of T1249-resistant HIV-1 mutants.
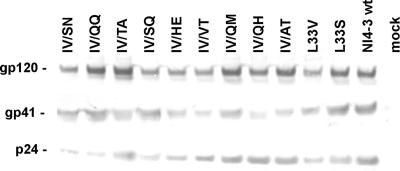
Since reduced viral fitness can contribute to the continued benefit of antiretroviral therapy despite the presence of high-level drug resistance (11), we next analyzed the effect of the gp41 HR1 sequence variations on viral infectivity, replication, and susceptibility to fusion inhibitors. First, all changes observed in the presence of T1249 (Table 1) were introduced into the replication-competent HIV-1 NL4-3 proviral clone. We found that all nine gp41-mutated HIV-1 variants were less infectious than wt NL4-3 (Table 1). Most substitutions in the IV residues reduced viral infectivity by more than fivefold, and only one mutant virus, containing a change from IV to VT (the IV/VT mutant), was almost as infectious as the parental NL4-3 clone. Western blot analysis of viral particles pelleted from the supernatant of transfected 293T cells showed that all gp41 mutant forms were efficiently expressed (Fig. 3). In addition to the gp41-mutated variants selected by our random PCR mutagenesis approach, we analyzed two gp41-mutated clones, one containing the L33V change mediating T20 resistance and the other carrying the L33S mutation previously shown to reduce the susceptibility of HIV-1 to T1249 and C34 inhibition (3, 8). The external gp120 Env glycoprotein was readily detected in the cell-free supernatants of all cultures transduced with the proviral HIV-1 constructs (Fig. 3). Thus, the greatly reduced infectivity of the majority of mutant HIV-1 clones was obviously not due to inefficient gp41 expression or gp120 virion incorporation.
FIG. 3.
Expression of wt and gp41 mutant Env proteins. Cell-free supernatants from transfected 293T cells were centrifuged to pellet viral particles, and the pellets were probed with antibodies against gp120, gp41, and p24. Similar results were obtained in an independent experiment.
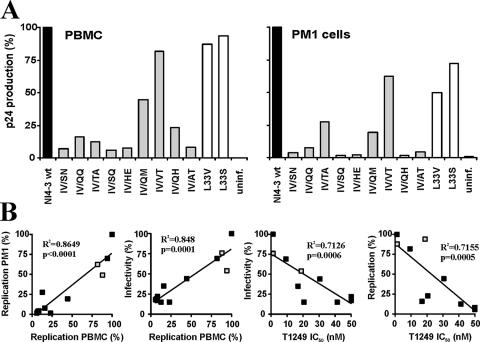
Next, we analyzed the susceptibility of the nine gp41-mutated HIV-1 variants to fusion inhibitors. We found that all but one of them were entirely resistant to T20 (Table 1). Moreover, they showed greatly reduced susceptibility to inhibition by T1249. The only exception was the L33V mutant virus, which showed only partial resistance to T20 and remained fully susceptible to T1249 inhibition. Notably, changes from IV to SN, SQ, HE, and AT rendered HIV-1 entirely insensitive to this second-version fusion inhibitor (Table 1). To further assess the fitness of the gp41-mutated HIV-1 variants, we tested their capacities for replication in human PBMC and PM1 cells. In agreement with the results of the infectivity assays (Table 1), only the IV/VT, L33V, and L33S mutant viruses replicated almost as efficiently as the NL4-3 wt in the absence of an inhibitor (Fig. 4A). In the presence of T1249, however, higher levels of mutant virus production were observed (data not shown). Statistical analysis demonstrated that the efficiencies of replication in PBMC and those in PM1 cells correlated with each other and with the levels of infectivity of the gp41-mutated HIV-1 variants (Fig. 4B, left panels). Notably, we also found an inverse correlation between the replicative capacities and the levels of infectivity of the mutant viruses and their degrees of resistance against T1249 (Fig. 4B, right panels). Thus, mutations that confer resistance to T1249 are usually associated with substantially reduced viral fitness.
FIG. 4.
Replication of T1249-resistant HIV-1 mutants. (A) Cumulative production of p24 by infected PBMC or PM1 cells. Values were measured at 3, 6, 9, 12, and 15 days after infection. Shown are representative levels of p24 production expressed as percentages of those measured in cultures infected with the wild-type virus. Similar results were obtained in independent experiments. uninf., uninfected. (B) Correlation between the replicative capacities and the levels of infectivity and T1249 resistance of gp41-mutated HIV-1 NL4-3 variants. Shown are the correlations between levels of p24 production in PBMC and PM1 cells (left panel), virus infectivity for TZM-bl cells and virus production by PBMC (left middle panel), virus infectivity and the IC50 of T1249 (right middle panel), and viral replication in PBMC and the IC50 (right panel). The values obtained for the L33V and L33S mutants are indicated by gray squares.
Replication and cytopathicity in ex vivo HLT.
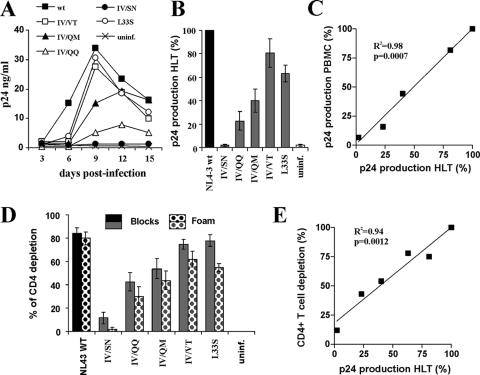
In vivo, HIV-1 replicates in infected individuals mainly in the lymphoid tissues (18). It has been shown previously that HIV-1 also spreads efficiently and causes CD4+-T-cell depletion in ex vivo-infected HLT (21, 22). This experimental system is most likely of high physiological relevance, because HLT maintains its original complexity of cell populations and cytoarchitecture and does not require exogenous activation to support productive HIV infection and viral spread (21, 22). It has been reported previously that in a significant proportion of patients in whom highly active antiretroviral therapy is failing, the CD4+-T-cell counts remain significantly above the pretreatment levels, despite poor control of viral replication (2, 9), and it is presently largely unclear whether HIV-1 mutants resistant to second-version fusion inhibitors may be pathogenic in vivo. To address this question, we analyzed the replicative capacities of five T20- and T1249-resistant gp41-mutated HIV-1 variants in ex vivo lymphoid tissue. We found that with the exception of the inactive IV/SN mutant, all gp41-mutated HIV-1 variants replicated in ex vivo HLT, albeit with differential efficiencies (Fig. 5A and B). Overall, the abilities of the gp41-mutated HIV-1 variants to replicate in ex vivo HLT correlated with their capacities for replication in PBMC (Fig. 5C). The IV/VT mutant replicated almost as effectively as the NL4-3 wt, whereas changes from IV to QM or QQ clearly reduced the replicative capacity (Fig. 5).
FIG. 5.
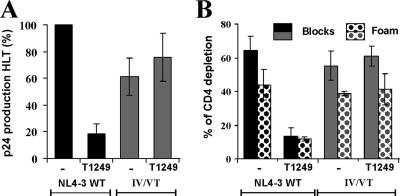
Replication and cytopathicity of gp41-mutated HIV-1 variants in lymphoid tissue ex vivo. (A) Representative replication kinetics of wt NL4-3 and gp41-mutated variants. uninf., unifected. (B) Average virus production. Matched tissues from seven donors were inoculated with the wt virus or with gp41-mutated variants as indicated, and for each condition, the cumulative production of p24 over 15 days was measured. Presented are means ± standard errors of the means of these measurements expressed as percentages of p24 production in cultures infected with the wt virus. (C) Correlation between virus production by infected PBMC and that by HLT ex vivo. (D) CD4+-T-cell depletion in HLT infected ex vivo with gp41-mutated HIV-1 variants. To evaluate CD4+-T-cell depletion, cells were mechanically isolated from matched control and infected tissues (18 pooled blocks for each variant) on day 15 postinfection, stained for CD3, CD4, CD8, and p24, and analyzed with flow cytometry as described previously (21, 22). Both T-cell populations in the tissue blocks (Blocks) and those that migrated in the gel foam (Foam) on which the tissues were incubated were analyzed. Presented are average depletion values ± standard errors of the means for tissues from seven donors. (E) Correlation between CD4+-T-cell depletion and p24 production in ex vivo-infected HLT cultures.
Since CD4+-T-cell depletion is a hallmark of the pathogenesis of AIDS, we also determined the levels of cytopathicity of the gp41-mutated HIV-1 variants in ex vivo HLT. In agreement with their differential replicative capacities, they also caused different levels of CD4+-T-cell depletion. The IV/VT and L33S mutants were almost as cytopathic as the NL4-3 wt, whereas the IV/QQ and IV/QM mutants caused only moderate and the IV/SN mutant caused no loss of CD4+ T cells (Fig. 5D). The levels of cytopathicity of the gp41-mutated HIV-1 variants correlated with their replicative capacities, indicating that the mutations in gp41 did not alter viral cytopathicity per se (Fig. 5E). Finally, we analyzed HIV-1 replication and cytopathicity in ex vivo HLT in the presence of a fusion inhibitor. The replication of the HIV-1 NL4-3 wt was inhibited and cytopathic effects were blocked by 50 nM T1249 (Fig. 6A and B). In contrast, this inhibitor did not reduce the levels of viral replication and CD4+-T-cell depletion in tissues infected with the IV/VT mutant virus (Fig. 6). The levels of p24 production and CD4+-T-cell loss were only marginally reduced compared to those in tissues infected with the NL4-3 wt in the absence of an inhibitor. Taken together, our results show that some mutations in gp41 conferring resistance to both T20 and T1249 have only marginal effects on HIV-1 replication and cytopathicity in ex vivo HLT.
FIG. 6.
T1249-resistant HIV-1 causes CD4+-T-cell depletion in the presence of an inhibitor. (A) Cumulative p24 production over 15 days of HLT infection with the HIV-1 NL4-3 wt or the IV/VT mutant in the absence of an inhibitor (−) or in the presence of 50 nM T1249. Cumulative virus production was monitored at 3, 6, 9, 12, and 15 days postinfection. Results shown in panels A and B are average values ± standard errors of the means for tissues from three independent donors. (B) CD4+-T-cell depletion in the tissue blocks (Blocks) and among the cells that migrated in the gel foam (Foam) at the end of culture at 15 days postinfection. Tissue culture, infections, and fluorescence-activated cell sorter analysis were performed as described previously (21, 22).
DISCUSSION
HIV-1 entry is a highly attractive target for antiretroviral therapy because it can be blocked at several steps and because HIV-1 does not show cross-resistance to many of the entry inhibitors and to other presently available antiretrovirals because of the drugs’ distinct mechanisms of action (4, 43, 47, 48). It has been suggested that even T20 and second-version fusion inhibitors blocking the same step during viral entry and targeting an overlapping site in the HR1 gp41 region can be employed in combination because most alterations in HR1 conferring resistance to T20 have little, if any, effect on virus sensitivity to T1249 (32, 50, 51). It is well documented that single-nucleotide changes altering the GIV motif may render HIV-1 resistant to T20 (2, 24, 35, 49, 52, 53, 55). In comparison, all T1249-resistant viruses identified by the random PCR mutagenesis approach in our study contained at least three mutations resulting in 2-aa changes (Table 1). Thus, a greater number of changes in the gp41 HR1 region seems to be required for T1249 resistance, which largely explains why it has proven difficult to select T1249-resistant HIV-1 variants by standard cell culture passage. It remains elusive, however, why the L33S mutation selected in the presence of C34 (3) has not also been observed in previous studies of T1249 resistance, since this amino acid substitution requires only a single-nucleotide change (TTA to TCA). One possible reason is that the NL4-3 L33S mutant shows only partial resistance to T1249 (Table 1) and may not replicate efficiently under stringent selective conditions. More importantly, our results clearly suggest that the mechanisms of HIV-1 resistance to first- and second-version fusion inhibitors are not fundamentally different and involve alterations in the GIV motif. Notably, some of the changes contributing to T1249 resistance, such as V38 to A, M, or E (Table 1), were the most commonly detected resistance mutations in HIV-1-infected patients receiving T20 treatment (2, 35). Thus, while these alterations alone seem to be insufficient to confer significant resistance to T1249 (50), they may prime the virus for easier development of resistance to second-iteration fusion inhibitors targeting the HP. To challenge this hypothesis, it will be interesting to determine whether T20-resistant HIV-1 variants containing changes at V38 may allow the selection of T1249-resistant forms by using the standard passage approach. It is conceivable, however, that individual amino acid changes can be more easily selected under treatment than multiple changes. Thus, our results argue against the usage of second-iteration fusion inhibitors for salvage therapy of HIV-infected patients harboring T20-resistant viral strains. Moreover, we found that all mutations in the IV residues identified conferred resistance to both T20 and T1249, suggesting that these drugs should not be used in combination. Further studies are required, however, to clarify whether our results obtained using the molecular NL4-3 clone are also valid for the majority of primary HIV-1 isolates and to assess the impact of the G36D variation on the susceptibilities of different gp41-mutated variants to fusion inhibitors.
Although changes in the GIV motif clearly play a key role in resistance to T20, alterations at other positions in HR1 or elsewhere in gp41 modulating viral susceptibility to fusion inhibitors have been reported previously (2, 13, 24, 27, 35, 49, 52, 53, 55). Of the 16 amino acid positions targeted by our random mutagenesis approach, only positions I37 and V38 tolerated changes well and allowed the selection of mutants in the presence of T1249. The results of the infectivity assays suggest that the vast majority of changes at the remaining positions in the HP and in the e or g residues involved in six-helix formation were hardly tolerated and disrupted Env function (Fig. 1B). These findings agree with those of previous studies, in which several of these residues were analyzed by alanine-scanning mutagenesis (37, 42, 57). Our results showing that the randomization of all amino acid residues in the hydrophobic cavity dramatically reduced HIV-1 infectivity further emphasize that this region is an excellent target for novel antiretroviral drugs (6). Further development of small-molecule inhibitors targeting this region (10) seems of high interest because any resistance-conferring mutations in these residues may severely impair viral fitness and hence lead to an attenuated phenotype in vivo in HIV-1-infected patients.
By using growth competition assays, it has been shown previously that the replicative fitness of recombinant viruses carrying T20 resistance mutations is highly correlated with the susceptibility to this inhibitor (36, 51). Another study reported, however, that T20-insensitive HIV-1 isolates from naive patients exhibit high levels of viral fitness (45). It has also been shown previously that some mutations in gp41 are associated with virus rebound or a nonresponder status of T1249-treated individuals (32), but it remained largely elusive whether mutations mediating T20 or T1249 resistance might result in a virological or immunological benefit and attenuate viral pathogenicity in vivo. To address this question in an adequate model, we investigated the replicative capacities and levels of cytopathicity of HIV-1 clones containing mutations conferring resistance to both T20 and T1249 in ex vivo-infected HLT. We found that the levels of replicative fitness and cytopathicity of T1249-resistant HIV-1 mutants in ex vivo HLT were inversely correlated to viral resistance. The majority of gp41-mutated HIV-1 variants, including all those showing a >50-fold increase in resistance to T1249, also caused much lower levels of CD4+-T-cell depletion than the NL4-3 wt (Fig. 5 and data not shown), suggesting an attenuated phenotype in vivo. The IV-to-VT mutation, however, hardly attenuated HIV-1 replication and virulence in ex vivo HLT, although this change was associated with complete resistance to T20 and about 10-fold-reduced susceptibility to T1249 (Table 1). Thus, resistance to fusion inhibitors comes at the cost of reduced viral fitness, but partially resistant virus variants may be almost as replication competent and cytopathic as wt HIV-1. It will be of interest to clarify whether variants with HR1 mutations conferring T20 and T1249 resistance without reducing viral fitness in vitro are commonly more susceptible to neutralizing antibodies targeting fusion intermediates, as previously suggested (51). It also remains to be determined whether compensatory changes restoring viral fitness and pathogenicity may be selected in HIV-1-infected patients treated with T20 or second-version fusion inhibitors.
Our results demonstrate that the site-specific random PCR mutagenesis approach is a much faster and more effective method to select viruses resistant to fusion inhibitors in vitro than the standard cell culture passage of wt virus in the presence of an inhibitor. In agreement with previous results (52), we were able to efficiently select HIV-1 variants resistant to T20 by using the standard approach (data not shown). Our attempts to select T1249-resistant HIV-1 mutants in this manner, however, consistently failed. In contrast, gp41-mutated HIV-1 variants containing randomized codons at positions corresponding to the IV residues replicated with wild type-like replication efficiencies in the presence of high concentrations of T1249 (Fig. 2C). Taken together, these results suggest that individual changes in gp41 may already reduce viral susceptibility to T20, whereas usually several changes are required for resistance to T1249. The selection of resistant HIV-1 variants will depend on the levels of viral replication, the numbers of target cells, and the length of time. Given that HIV-1-infected individuals contain an enormous number of target cells and need to be treated for many years, it is conceivable that whatever is observed in vitro will most likely also occur in vivo. Thus, we feel that our results are relevant and that similar HIV-1 variants would most likely also emerge during long-term treatment with second-version fusion inhibitors. Our collection of gp41-mutated HIV-1 variants will allow rapid assessments of whether changes in HR1 also mediate resistance to other fusion inhibitors and may be used to predict possible resistance profiles in vivo.
In conclusion, our data indicate that changes in the GIV motif at positions 36 to 38 of gp41 play a key role in HIV-1 resistance to both first- and second-version fusion inhibitors. Individual changes at position 38, which are commonly observed in patients treated with T20 (2, 35, 49, 53, 55), do not confer cross-resistance to T1249. If this change is combined with a second mutation at position 37, however, the virus can become highly resistant to T1249. Our data suggest that second-version fusion inhibitors should not be used in combination with T20 or in salvage therapy in pretreated patients, although clearly more clinical data are required to make definitive treatment recommendations. Some parts of HR1, particularly the HP, represent excellent targets for HIV-1 fusion inhibitors because changes are hardly tolerated. Finally, our collection of gp41 HR1-mutated HIV-1 variants should be useful to readily assess the possible mechanisms of resistance to such novel entry inhibitors in future studies.
Acknowledgments
We thank Nicola Bailer and Daniela Krnavek for excellent technical assistance and Thomas Mertens for support and encouragement.
This work was supported by the Landesstiftung BW and the Deutsche Forschungsgemeinschaft and by NIH grant 1R01AI067057-01A2.
Footnotes
Published ahead of print on 11 April 2007.
REFERENCES
- 1.Agrawal, L., X. Lu, Q. Jin, and G. G. Alkhatib. 2006. Anti-HIV therapy: current and future directions. Curr. Pharm. Des. 12:2031-2055. [DOI] [PubMed] [Google Scholar]
- 2.Aquaro, S., R. D'Arrigo, V. Svicher, G. Perri, S. L. Caputo, U. Visco-Comandini, M. Santoro, A. Bertoli, F. Mazzotta, S. Bonora, V. Tozzi, R. Bellagamba, M. Zaccarelli, P. Narciso, and C. F. Antinori. 2006. Specific mutations in HIV-1 gp41 are associated with immunological success in HIV-1-infected patients receiving enfuvirtide treatment. J. Antimicrob. Chemother. 58:714-722. [DOI] [PubMed] [Google Scholar]
- 3.Armand-Ugon, M., A. Gutierrez, B. Clotet, and E. A. Este. 2003. HIV-1 resistance to the gp41-dependent fusion inhibitor C-34. Antivir. Res. 59:137-142. [DOI] [PubMed] [Google Scholar]
- 4.Briz, V., E. Poveda, and V. Soriano. 2006. HIV entry inhibitors: mechanisms of action and resistance pathways. J. Antimicrob. Chemother. 57:619-627. [DOI] [PubMed] [Google Scholar]
- 5.Carl, S., R. Daniels, A. J. Iafrate, P. Easterbrook, T. C. Greenough, J. Skowronski, and F. Kirchhoff. 2000. Partial “repair” of defective nef genes in a long-term nonprogressor with human immunodeficiency virus type 1 infection. J. Infect. Dis. 181:132-140. [DOI] [PubMed] [Google Scholar]
- 6.Chan, D. C., C. T. Chutkowski, and P. S. Kim. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 95:15613-15637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 8.Chinnadurai, R., J. Munch, and F. Kirchhoff. 2005. Effect of naturally-occurring gp41 HR1 variations on susceptibility of HIV-1 to fusion inhibitors. AIDS 19:1401-1405. [DOI] [PubMed] [Google Scholar]
- 9.Clavel, F., and A. W. Hance. 2004. HIV drug resistance. N. Engl. J. Med. 350:1023-1035. [DOI] [PubMed] [Google Scholar]
- 10.Debnath, A., K., L. Radigan, and S. Jiang. 1999. Structure-based identification of small molecule antiviral compounds targeted to the gp41 core structure of the human immunodeficiency virus type 1. J. Med. Chem. 42:3203-3209. [DOI] [PubMed] [Google Scholar]
- 11.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 12.Deeks, S. G. 2003. Treatment of antiretroviral-drug-resistant HIV-1 infection. Lancet 362:2002-2011. [DOI] [PubMed] [Google Scholar]
- 13.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, Z. Zhang, W. A. O'Brien, L. Ratner, G. M. Shaw, and E. Hunter. 2001. Sensitivity of human immunodeficiency virus type 1 to fusion inhibitors targeted to the gp41 first heptad repeat involves distinct regions of gp41 and is consistently modulated by gp120 interactions with the coreceptor. J. Virol. 75:8605-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dwyer, J. J., A. Hasan, K. L. Wilson, J. M. White, T. J. Matthews, and M. K. Delmedico. 2003. The hydrophobic pocket contributes to the structural stability of the N-terminal coiled coil of HIV gp41 but is not required for six-helix bundle formation. Biochemistry 42:4945-4953. [DOI] [PubMed] [Google Scholar]
- 16.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 17.Eron, J. J., R. M. Gulick, J. A. Bartlett, T. Merigan, R. Arduino, J. M. Kilby, B. Yangco, A. Diers, C. Drobnes, R. DeMasi, M. Greenberg, T. Melby, C. Raskino, P. Rusnak, Y. Zhang, R. Spence, and G. D. Miralles. 2004. Short-term safety and antiviral activity of T-1249, a second generation fusion inhibitor. J. Infect. Dis. 189:1075-1083. [DOI] [PubMed] [Google Scholar]
- 18.Fauci, A. S. 1996. Host factors and the pathogenesis of HIV-induced disease. Nature 384:529-534. [DOI] [PubMed] [Google Scholar]
- 19.Follis, K. E., S. J. Larson, M. Lu, and J. H. Nunberg. 2002. Genetic evidence that interhelical packing interactions in the gp41 core are critical for transition of the human immunodeficiency virus type 1 envelope glycoprotein to the fusion-active state. J. Virol. 76:7356-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallo, S. A., A. Puri, and R. Blumenthal. 2001. HIV-1 gp41 six-helix bundle formation occurs rapidly after the engagement of gp120 by CXCR4 in the HIV-1 Env-mediated fusion process. Biochemistry 40:12231-12236. [DOI] [PubMed] [Google Scholar]
- 21.Glushakova, S., B. Baibakov, L. B. Margolis, and J. Zimmerberg. 1995. Infection of human tonsil histocultures: a model for HIV pathogenesis. Nat. Med. 1:1320-1322. [DOI] [PubMed] [Google Scholar]
- 22.Glushakova, S., B. Baibakov, J. Zimmerberg, and L. B. Margolis. 1997. Experimental HIV infection of human lymphoid tissue: correlation of CD4+ T cell depletion and virus syncytium-inducing/non-syncytium-inducing phenotype in histoculture inoculated with laboratory strains and patient isolates of HIV type 1. AIDS Res. Hum. Retrovir. 13:461-471. [DOI] [PubMed] [Google Scholar]
- 23.Greenberg, M., and L. N. Cammack. 2004. Resistance to enfuvirtide, the first HIV fusion inhibitor. J. Antimicrob. Chemother. 54:333-340. [DOI] [PubMed] [Google Scholar]
- 24.Greenberg, M. L., D. Davison, and L. Jin. 2002. In vitro antiviral activity of T-1249, a second generation fusion inhibitor. Antivir. Ther. 7(Suppl. 1):S10. [Google Scholar]
- 25.Hammer, S. M., M. S. Saag, M. Schechter, J. S. Montaner, R. T. Schooley, D. M. Jacobsen, M. A. Thompson, C. C. Carpenter, M. A. Fischl, B. G. Gazzard, J. M. Gatell, M. S. Hirsch, D. A. Katzenstein, D. D. Richman, S. Vella, P. G. Yeni, P. A. Volberding, and the International AIDS Society—USA Panel. 2006. Treatment for adult HIV infection: 2006 recommendations of the International AIDS Society—USA Panel. JAMA 296:827-843. [DOI] [PubMed] [Google Scholar]
- 26.He, Y., R. Vassell, M. Zaitseva, N. Nguyen, Z. Yang, Y. Weng, and C. D. Weiss. 2003. Peptides trap the human immunodeficiency virus type 1 envelope glycoprotein fusion intermediate at two sites. J. Virol. 77:1666-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heil, M. L., J. M. Decker, J. N. Sfakianos, G. M. Shaw, E. Hunter, and C. A. Derdeyn. 2004. Determinants of human immunodeficiency virus type 1 baseline susceptibility to the fusion inhibitors enfuvirtide and T-649 reside outside the peptide interaction site. J. Virol. 78:7582-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji, H., W. Shu, E. T. Burling, S. Jiang, and M. Lu. 1999. Inhibition of human immunodeficiency virus type 1 infectivity by the gp41 core: role of a conserved hydrophobic cavity in membrane fusion. J. Virol. 73:8578-8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kilby, J. M., and J. J. Eron. 2003. Novel therapies based on mechanisms of HIV-1 cell entry. N. Engl. J. Med. 348:2228-2238. [DOI] [PubMed] [Google Scholar]
- 30.Kilby, J. M., S. Hopkins, T. Venetta, B. DiMassimo, G. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 31.Kinomoto, M., M. Yokoyama, H. Sato, A. Kojima, T. Kurata, K. Ikuta, T. Saga, and K. Tokunaga. 2005. Amino acid 36 in the human immunodeficiency virus type 1 gp41 ectodomain controls fusogenic activity: implications for the molecular mechanism of viral escape from a fusion inhibitor. J. Virol. 79:5996-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lalezari, J. P., N. C. Bellos, K. Sathasivam, G. J. Richmond, C. J. Cohen, R. A. Myers, Jr., D. H. Henry, C. Raskino, T. Melby, H. Murchison, Y. Zhang, R. Spence, M. L. Greenberg, R. A. Demasi, G. D. Miralles, and the T1249-102 Study Group. 2005. T-1249 retains potent antiretroviral activity in patients who had experienced virological failure while on an enfuvirtide-containing treatment regimen. J. Infect. Dis. 191:1155-1163. [DOI] [PubMed] [Google Scholar]
- 33.Leitner, T., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber. 2003. HIV sequence compendium. LA-UR 04-7420. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, NM.
- 34.Little, S. J., S. Holte, J. P. Routy, E. S. Daar, M. Markowitz, A. C. Collier, R. A. Koup, J. W. Mellors, E. Connick, B. Conway, M. Kilby, L. Wang, J. M. Whitcomb, N. S. Hellmann, and D. D. Richman. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347:385-394. [DOI] [PubMed] [Google Scholar]
- 35.Lu, J., S. G. Deeks, R. Hoh, G. Beatty, B. A. Kuritzkes, J. N. Martin, and D. R. Kuritzkes. 2006. Rapid emergence of enfuvirtide resistance in HIV-1-infected patients: results of a clonal analysis. J. Acquir. Immune Defic. Syndr. 43:60-64. [DOI] [PubMed] [Google Scholar]
- 36.Lu, J., P. Sista, F. Giguel, M. Greenberg, and D. R. Kuritzkes. 2004. Relative replicative fitness of human immunodeficiency virus type 1 mutants resistant to enfuvirtide (T-20). J. Virol. 78:4628-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu, M., M. O. Stoller, S. Wang, J. Liu, M. B. Fagan, and J. H. Nunberg. 2001. Structural and functional analysis of interhelical interactions in the human immunodeficiency virus type 1 gp41 envelope glycoprotein by alanine-scanning mutagenesis. J. Virol. 75:11146-11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lusso, P., F. Cocchi, C. Balotta, P. D. Markham, A. Louie, P. Farci, R. Pal, R. C. Gallo, and M. S. Reitz, Jr. 1995. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 69:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markowitz, M., H. Mohri, S. Mehandru, A. Shet, L. Berry, R. Kalyanaraman, A. Kim, C. Chung, P. Jean-Pierre, A. Horowitz, M. La Mar, T. Wrin, N. Parkin, M. Poles, C. Petropoulos, M. Mullen, D. Boden, and D. D. Ho. 2005. Infection with multidrug resistant, dual-tropic HIV-1 and rapid progression to AIDS: a case report. Lancet 365:1031-1038. [DOI] [PubMed] [Google Scholar]
- 40.Matthews, T., M. Salgo, M. Greenberg, J. Chung, R. DeMasi, and D. Bolognesi. 2004. Enfuvirtide: the first therapy to inhibit the entry of HIV-1 into host CD4 lymphocytes. Nat. Rev. Drug Discov. 3:215-225. [DOI] [PubMed] [Google Scholar]
- 41.Menzo, S., A. Castagna, A. Monachetti, H. Hasson, A. Danise, E. Carini, P. Bagnarelli, A. Lazzarin, and M. Clementi. 2004. Genotype and phenotype patterns of human immunodeficiency virus type 1 resistance to enfuvirtide during long-term treatment. Antimicrob. Agents Chemother. 48:3253-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mo, H., A. K. Konstantinidis, K. D. Stewart, T. Dekhtyar, T. Ng, K. Swift, E. D. Matayoshi, W. Kati, W. Kohlbrenner, and A. Molla. 2004. Conserved residues in the coiled-coil pocket of human immunodeficiency virus type 1 gp41 are essential for viral replication and interhelical interaction. Virology 329:319-327. [DOI] [PubMed] [Google Scholar]
- 43.Moore, J. P., and R. W. Doms. 2003. The entry of entry inhibitors: a fusion of science and medicine. Proc. Natl. Acad. Sci. USA 100:10598-10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nameki, D., E. Kodama, M. Ikeuchi, N. Mabuchi, A. Otaka, H. Tamamura, M. Ohno, N. Fujii, and M. Matsuoka. 2005. Mutations conferring resistance to HIV-1 fusion inhibitors are restricted by gp41 and Rev-responsive element functions. J. Virol. 79:764-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neumann, T., I. Hagmann, S. Lohrengel, M. L. Heil, C. A. Derdeyn, H.-G. Kräusslich, and M. T. Dittmar. 2005. T-20-insensitive HIV-1 from naive patients exhibits high viral fitness in a novel dual colour competition assay on primary cells. Virology 332:251-262. [DOI] [PubMed] [Google Scholar]
- 46.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, S. D. Holmberg, et al. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 47.Pierson, T. C., R. W. Doms, and S. Pohlmann. 2004. Prospects of HIV-1 entry inhibitors as novel therapeutics. Rev. Med. Virol. 14:255-270. [DOI] [PubMed] [Google Scholar]
- 48.Pohlmann, S., and J. D. Reeves. 2006. Cellular entry of HIV: evaluation of therapeutic targets. Curr. Pharm. Des. 12:1963-1973. [DOI] [PubMed] [Google Scholar]
- 49.Poveda, E., B. Rodes, C. Toro, L. Martin-Carbonero, J. Gonzalez-Lahoz, and V. Soriano. 2002. Evolution of the gp41 env region in HIV-infected patients receiving T-20, a fusion inhibitor. AIDS 16:1959-1961. [DOI] [PubMed] [Google Scholar]
- 50.Reeves, J. D., S. A. Gallo, N. Ahmad, J. L. Miamidian, P. E. Harvey, M. Sharron, S. Pohlmann, J. N. Sfakianos, C. A. Derdeyn, R. Blumenthal, E. Hunter, and R. W. Doms. 2002. Sensitivity of HIV-1 to entry inhibitors correlates with envelope/coreceptor affinity, receptor density, and fusion kinetics. Proc. Natl. Acad. Sci. USA 99:16249-16254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reeves, J. D., F. H. Lee, J. L. Miamidian, C. B. Jabara, M. M. Juntilla, and R. W. Doms. 2005. Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J. Virol. 79:4991-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sista, P. R., T. Melby, D. Davison, L. Jin, S. Mosier, M. Mink, E. L. Nelson, R. DeMasi, N. Cammack, M. P. Salgo, T. J. Matthews, and M. L. Greenberg. 2004. Characterization of determinants of genotypic and phenotypic resistance to enfuvirtide in baseline and on-treatment HIV-1 isolates. AIDS 18:1787-1794. [DOI] [PubMed] [Google Scholar]
- 54.Wang, S., J. York, W. Shu, M. O. Stoller, J. H. Nunberg, and M. Lu. 2002. Interhelical interactions in the gp41 core: implications for activation of HIV-1 membrane fusion. Biochemistry 41:7283-7292. [DOI] [PubMed] [Google Scholar]
- 55.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 57.Weng, Y., and C. D. Weiss. 1998. Mutational analysis of residues in the coiled-coil domain of human immunodeficiency virus type 1 transmembrane protein gp41. J. Virol. 72:9676-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wild, C., T. Greenwell, and T. Matthews. 1993. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell-cell fusion. AIDS Res. Hum. Retrovir. 9:1051-1053. [DOI] [PubMed] [Google Scholar]
- 59.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeni, P. 2006. Update on HAART in HIV. J. Hepatol. 44:100-103. [DOI] [PubMed] [Google Scholar]