Abstract
Sindbis virus is a single-stranded positive-sense RNA virus. It is composed of 240 copies of three structural proteins: E1, E2, and capsid. These proteins form a mature virus particle composed of two nested T=4 icosahedral shells. A complex network of disulfide bonds in the E1 and E2 glycoproteins is developed through a series of structural intermediates as virus maturation occurs (M. Mulvey and D. T. Brown, J. Virol. 68:805-812, 1994; M. Carleton et al., J. Virol. 71:1558-1566, 1997). To better understand the nature of this disulfide network, E1 and E2 cysteinyl residues were labeled with iodoacetamide in the native virus particle and analyzed by liquid chromatography-tandem mass spectrometry. This analysis identified cysteinyl residues of E1 and E2, which were found to be label accessible in the native virus particle, as well as those that were either label inaccessible or blocked by their involvement in disulfide bonds. Native virus particles alkylated with iodoacetamide demonstrated a 4-log decrease in viral infectivity. This suggests that the modification of free cysteinyl residues results in the loss of infectivity by destabilizing the virus particle or that a rearrangement of disulfide bonds, which is required for infectivity, is blocked by the modification. Although modification of these residues prevented infectivity, it did not alter the ability of virus to fuse cells after exposure to acidic pH; thus, modification of free cysteinyl residues biochemically separated the process of infection from the process of membrane fusion.
Sindbis virus (SV) is a positive-sense, single-stranded RNA virus of the Alphavirus genus in the Togaviridae family and is propagated in nature by both insect and mammalian hosts (3). The mature SV particle contains three structural proteins—E1, E2, and capsid (C)—in a 1:1:1 stoichiometric ratio. The SV particle is approximately 70 nm in diameter and serves as an excellent model for structural studies. Each virus particle is composed of 240 copies of each structural protein producing 80 equivalent units, which are organized into two geometrically identical T=4 icosahedral shells (31). An outer protein shell, composed of the E1 and E2 glycoproteins, is organized into trimers of E1/E2 heterodimers (1, 5, 36). The outer protein shell surrounds an inner shell, which is composed entirely of the capsid protein. The 11.7-kb genomic RNA is packed inside the inner capsid shell. Positioned between the two protein shells is a host-derived membrane bilayer that is penetrated by the transmembrane (TM) domain anchors of E1 and E2 (12, 18, 38, 43, 52). The endodomain of E2 interacts with the capsid protein linking the two shells together (17, 29, 48, 49).
SV produces a subgenomic mRNA that encodes a polyprotein which is posttranslationally cleaved into capsid, E1, E2, E3, and 6K (45). The capsid protein is processed by an autoproteolytic activity after translation on cytoplasmic ribosomes. Removal of the capsid protein exposes the polyprotein signal sequences, which directs insertion of the multipass polyprotein that contains six membrane-spanning domains (18). Signalase cleaves the polyprotein into the structural components PE2, 6K, and E1 (18). The 6K protein plays a role in the correct integration of E1 into the membrane and is necessary for efficient particle budding (18). 6K is found only at very low levels in mature virus (9). The E1 glycoprotein, containing a functional domain necessary for viral entry, is folded into a stable compact high-energy conformation as it is assembled into heterotrimers with the precursor to E2, PE2 (5, 22-24). PE2 is transported to the cell surface through the trans-Golgi network, where it is processed to E2 and E3 by furin (20). Evidence suggests that E2 is responsible for receptor recognition. E3 is not present in the mature virus. During transport of the trimeric complexes to the plasma membrane, a second membrane-spanning region of E2 is retracted from the membrane, and a 33-amino-acid endodomain is exposed to the cytoplasm (19). At the plasma membrane the E1-E2 heterotrimers assemble into the outer protein shell by association of the E2 endodomain with a hydrophobic cleft in the capsid protein (15, 17, 27, 29, 30, 48, 49). This association of the E2 endodomain with the capsid protein gives stability to the structure of the virus (16) and plays a critical role in the formation of the viral outer protein shell around the preformed inner protein shell as the process of envelopment takes place (8).
The SV E1 protein contains 439 amino acids with a predicted ectodomain consisting of the first 408 residues. E2 contains 423 amino acids and has an ectodomain consisting of the first 362 residues. Each of these membrane proteins contains 17 Cys residues (SV capsid does not contain any Cys residues), which are completely conserved across the Alphavirus genus with the exception of E1 C430 and E2 C388 and C390. These nonconserved Cys residues are located in the transmembrane domains. Residues C396, C416, and C417 of the E2 protein are located in the 33-amino-acid endodomain. Of the remaining Cys residues, there is the possibility of having eight disulfide bonds in the E1 protein and six in the E2 protein.
Relatively little is known about the E2 disulfide network. Gidwitz et al. suggested that all Cys residues located in the E2 protein were either involved in disulfide bonds or were not accessible when treated with [14C]iodoacetamide at a 1.1 molar excess of iodoacetamide to total cysteinyl content (10). The E1 protein of Semliki Forest virus, also a member of the Alphavirus genus, has a crystal structure solved at a 3-Å resolution (40). The authors of that study reported disulfide bonds located at E1 residues 49 to 114, 62 to 94, 63 to 96, 259 to 271, 328 to 370, 301 to 376, and 306 to 380. Thus, they report that all Cys residues in the ectodomain are involved in disulfide bridges.
The E1 protein proceeds through a complicated set of interactions as folding occurs in the endoplasmic reticulum (ER). Once translated, E1 quickly adopts an E1 alpha configuration, proceeds through a series of disulfide bond-stabilized intermediates (beta and gamma), and ultimately adopts a compact high-energy E1 epsilon form (4, 5, 23). The energy stored in E1 may be used to breach a cell membrane as penetration takes place (32). E1 epsilon associates with PE2 in a heterotrimeric spike and leaves the ER for the trans-Golgi network. It was concluded from these studies (26) that the E1 protein acquires a meta-stable conformation through the sequential formation of disulfide intermediates. Attempts to extract the E1 protein from mature virus particles with detergent resulted in the generation of a number of disulfide stabilized conformations which could be separated on nondenaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (23). The propensity of E1 to adopt alternative conformations complicates the isolation of its native form. The Semliki Forest virus E1 crystal structure presented by Roussel et al. was extracted by detergent-based methods (40) and thus may not represent the native protein structure.
We identify here the location of label-accessible Cys residues in the SV E1 and E2 proteins by incubation of native virus particles in the presence of iodoacetamide. We determined the effect of alkylation on virus infectivity and membrane fusion and also examined the role of several identified label-accessible Cys residues in virus assembly and function by mutagenesis. These data suggest that reduced Cys residues are present in the infectious virus particle and that disulfide bond rearrangements may occur during the progression of virus maturation and infection.
MATERIALS AND METHODS
Cell maintenance, virus growth, purification, and labeling.
BHK-21 cells were grown and maintained in supplemented minimum essential medium as described previously (37, 52). The SV heat-resistant strain was used to infect BHK cells at a multiplicity of infection of 10 (12). Virus was harvested from the cell supernatant at 24 h postinfection and was purified over a 15% to 35% step potassium tartrate gradient by centrifugation at 24,000 rpm at 4°C in a Beckman SW28 rotor overnight. The visible band of virus was extracted with a needle, diluted twofold, and placed over a continuous 15 to 35% potassium tartrate gradient. This gradient was centrifuged in a Beckman SW28 rotor at 26,000 rpm for 5 h at 4°C. The virus band was collected with a needle and dialyzed against phosphate-buffered saline-Dulbecco medium (PBS-D; pH 8.0) overnight. The amount of viral protein was quantified by using the MicroBCA protein assay kit (Pierce, Rockford, IL).
Purified virus, along with an equimolar amounts of RNase B (Sigma) based on cysteinyl content to serve as a control, was either maintained at pH 8.0 or was adjusted to pH 5.3, placed at 37°C for 10 min, and then returned to pH 8.0. Samples that were switched to pH 5.3 were diluted to approximately 50 μg/ml due to precipitation at the normal working concentration of approximately 200 μg/ml. All samples were then incubated with either 100- or 500-fold molar excess iodoacetamide (Sigma, St. Louis MO) added to the total cysteinyl content and kept at 37°C for 2 h. Our choice of iodoacetamide over other labels such as N-ethylmaleimide was based on several considerations. First, in general, only the most reactive sulfhydryl groups of proteins are capable of reacting with Michael acceptors such as maleimides. Thus, iodoacetamide was more suitable due to its higher reactivity but lower Cys residue specificity. This nonspecificity was not an issue because the product ion spectra we obtained for our experiments were searched using all possible mass modifications due to iodoacetamide labeling on Cys, His, Lys, Met, and Tyr residues in order to detect all possible residue modifications. Second, iodoacetamide is a smaller molecule than N-ethylmaleimide, so it is a better topological probe and thus has a better chance of reacting with a partially exposed Cys residue. Third, iodoacetamide treatment of a Cys residue produces the S-carboxamidomethyl modification, a thioether adduct that is very stable and well suited for liquid chromatography-tandem mass spectrometry (LC/MS/MS) analysis. For Cys residues modified by N-ethylmaleimide, the S-ethylsuccinimido modification undergoes partial ring hydrolysis to form an isomeric mixture of maleamic acid adducts (13, 28). This complicates MS analysis and makes identifying Cys modifications more difficult, while severely impacting the ability to quantify the percentage of Cys residue exposure due to differential labeling.
Gidwitz et al. (10) reported that all Cys residues located in the E2 protein were either involved in disulfide bonds or were not accessible when treated with [14C]iodoacetamide at a 1.1 molar excess of total cysteinyl content. We report (below) that Cys residues 105, 152, and 220 are not present as part of a disulfide bond. The reason for this discrepancy is most likely due to the concentration of iodoacetamide used in the labeling reaction. We had difficulty identifying labeled peptides at a molar excess of less than 100, and therefore labeling at 1.1 molar excesses might yield undetectable amounts of labeled protein.
A portion of the iodoacetamide-treated virus, using our labeling approach, was removed for titration (37), and the corresponding titer was compared to a nonlabeled control processed in the same fashion. Iodoacetamide-treated virus was purified away from unreacted reagent by dialysis or size-exclusion chromatography (for initial analysis only). After several experiments, excess reagent was removed by gel purification only to conserve protein concentration. Peptides verified with or without this step were in agreement. The virus proteins were purified by SDS-electrophoresis (see below) under nonreducing conditions to further separate them from unreacted iodoacetamide prior to reduction and reaction with iodomethane. It should be noted that only a single protein band was detected for E1 and E2 under nondenaturing SDS-electrophoresis, indicating that disulfide bond rearrangements did not occur prior to iodomethane treatment.
SDS-PAGE and preparation of samples for LC/MS/MS analysis.
Samples were boiled in 5× SDS sample buffer devoid of any reductants and were run on a 10.8% polyacrylamide gel. The gel was fixed twice with 50% methanol, and 10% acetic acid for 15 min at room temperature. The gel was stained overnight with Ruby Red (Invitrogen, Carlsbad, CA) and washed twice for 30 min each time with 10% methanol and 7% acetic acid. Bands were then visualized with UV illumination, excised, and placed in 1.5-ml Eppendorf tubes.
Gel slices were washed twice with 50 mM ammonium bicarbonate (pH 8.0) for 10 min and destained three times for 20 min with 40% acetonitrile-60% 50 mM ammonium bicarbonate (pH 8.0) (vol/vol). Samples were dehydrated with 95% acetonitrile for 5 min, and solvent was removed by vacuum centrifugation. Samples were rehydrated in 50 mM ammonium bicarbonate (pH 8.0) containing 5 mM dithiothreitol (DTT) and then kept at 37°C for 1 h. The samples were dried and then rehydrated with 20 mM iodomethane (Sigma) in 50 mM ammonium bicarbonate and incubated at 37°C for 1 h. Gel slices were washed three times for 10 min each time with 50 mM ammonium bicarbonate (pH 8.0) and then dehydrated with 95% acetonitrile for 10 min. After vacuum centrifugation, the samples were rehydrated with 50 mM ammonium bicarbonate (pH 8.0), and the gel slices were crushed into pieces and then dried. The gel pieces were rehydrated with 50 mM ammonium bicarbonate (pH 8.0) containing protease (either trypsin, chymotrypsin, Glu-C, or Lys-C) in a 1:5 enzyme/protein ratio. Samples were then incubated overnight at the recommended temperature of the protease used for in-gel digestion. Peptides were then extracted by sonication three times for 20 min each time with 60% acetonitrile-35% 50 mM ammonium bicarbonate-5% formic acid (vol/vol/vol). Samples were dried and stored at −80°C until LC/MS/MS analysis was performed.
Microcapillary reversed-phase LC/MS/MS analysis.
Peptides were analyzed by microcapillary reversed-phase LC/MS/MS using an Agilent 1100 series capillary LC system (Agilent Technologies, Inc., Palo Alto, CA) coupled to an LCQ Deca ion-trap mass spectrometer (Thermo Electron, San Jose, CA) with an in-house manufactured electrospray interface operating in the positive ion mode at 2.2 kV. A reversed-phase capillary column was manufactured in-house by slurry packing a 5-μm 300 Ε Jupiter C18 stationary-phase apparatus (Phenomenex, Torrance, CA) into a 55-cm-by-360 μm (outer diameter)-by-150 μm (inner diameter) capillary (Polymicro Technologies, Inc., Phoenix, AZ) incorporating a 2-μm-pore-size retaining mesh in a high-pressure liquid chromatography stainless steel union (Valco Instruments Co., Houston, TX) containing a flame-pulled capillary tip. The mobile phases consisted of 0.1% formic acid in water (phase A) and 0.1% formic acid in acetonitrile (phase B). Peptide fractions were solubilized with 20 μl of 5% phase B-95% phase A. After a volume of 8 μl was loaded onto the reversed-phase column, a gradient program held the flow rate of 1.5 μl min−1 at 5% phase B for 20 min and then initiated a linear gradient to 95% phase B over 90 min. After the column was washed with 95% phase B for 20 min, the column was equilibrated with 5% phase B for 60 min prior to the next injection. The LCQ Deca was operated in the datum-dependent MS/MS mode in which the four most intense ions detected in the precursor MS scan were selected for collision-induced dissociation (CID). A 2-min dynamic m/z exclusion list for selected precursor ions was utilized to increase the detection of lower abundant peptides during gradient elution. The data were acquired in an m/z range of 400 to 2,000 using a normalized collision energy setting of 45%.
Peptide identification.
Peptides were identified by searching the product ion spectra against a SV structural protein database (which also contained the sequence of the protease used for in-gel digestion) using TurboSEQUEST (BioWorks 3.1; Thermo Electron, San Jose, CA). In the present study, the initial SEQUEST scoring criteria used for peptide identification were peptides that contained appropriate cleavage sites based on the protease used in the protein digestion and that displayed a cross-correlation score (Xcorr) of greater than 2.0, 1.5, and 3.3, for +1, +2, and +3 charged precursor ions, respectively, and a delta-correlation score (ΔCn) of >0.08 (33). The Xcorr of a peptide is based on the “fit” of the MS/MS data to the theoretical distribution of ions produced for the peptide, and the ΔCn is the “difference” between the top two Xcorrs for a given product ion spectrum. The product ion spectra selected by SEQUEST were then manually inspected to verify acceptable ion coverage for identification and the site(s) of modification, including those identified with an Xcorr above the threshold and an ΔCn value below 0.08 for a labeled peptide. To determine the extent of protein labeling using the alkylating reagents, all possible residue mass modifications were considered dynamic when analyzing the data. SEQUEST searches included a mass modification of 57.0 u (unified atomic mass unit) on Cys, His, Lys, and Tyr and 58.0 u on Met, corresponding to the net additional mass due to the alkylation reaction with iodoacetamide. Values of 114 and 115 u were used to search for double carboxamidomethylation on Lys and His, respectively, and 43.0 u for carboxamidomethylation and demethylation on Met. For methylation due to iodomethane treatment, 14.0 u was used to search for modifications on Cys, His, Lys, and Tyr. A dynamic mass modification of 16.0 u on Met residues was used to search for oxidation (methionine sulfoxide). Area calculations from peaks produced in the extracted-ion chromatograms (EICs) of the m/z values corresponding to the alkylated and nonalkylated precursor ions for all detected charge states were used to estimate the percentage of modified versus nonmodified cysteinyl peptides.
Mutants, transfection, and virus titration.
Mutants were constructed in the virus strain Toto 1101 (39) by using the QuikChange mutagenesis protocol (Stratagene, La Jolla, CA) as described previously (12). Mutants were named for the specific amino acid mutated from the amino acid terminus of the E1 protein. In each case a Cys residue was mutated to a Ser residue. The PCR primers utilized were C49S coding (5′-GAGTACATTACATCTAAATTCACCACTG-3′), C49S noncoding (5′-CAGTGGTGAATTTAGATGTAATGTACTC-3′), C259S coding (5′-CCGCACCTTTCGGATCCAAGATTGCAG-3′), C259S noncoding (5′-CTGCAATCTTGGATCCGAAAGGTGCGG-3′, C271S coding (5′-CCGAGCGGTGGACAGTTCATACGGG-3′), and C271S noncoding (5′-CCCGTATGAACTGTCCACCGCTCGG-3′). The PCR conditions were as follows: 94°C for 2 min, 95°C for 12 s, 50°C for 30 s, and 68°C for 30 min for 30 cycles. After the first 10 cycles the 30-min extension time was increased by 10 s/cycle. A final extension at 72°C for 30 min was performed, and the products were held at 4°C. Mutations were confirmed by sequencing and, after large-scale DNA growth and purification, plasmids were linearized with XhoI (New England Biolabs, Ipswich, MA).
Mutant cDNA constructs were prepared for transcription and transcribed by using Sp6 reverse transcriptase (New England Biolabs, Ipswich, MA) as described previously (11, 12). Electroporation was performed on BHK cells as described by Lilijestrom and Garoff (18). Virus was harvested after 24 h, and titers were determined on BHK cells (37).
FFWO assays.
Cell monolayers prepared for fusion from without (FFWO) were prepared in 24-well plates as described above. Monolayers that were just subconfluent were placed on ice for 15 min. While still on ice, gradient purified virus (treated or untreated with iodoacetamide) was added to the appropriate wells. The cells were then treated at pH 5.3 for 5 min and returned to pH 7.4 for 1 h. The fusion process was initiated by the addition of fusion medium (1× minimal essential medium composed of fusion salts [1.8 mM CaCl2, 5.3 mM KCl, 0.1 μM MgSO4·6H2O, 116 mM NaCl, and 2.9 mM sucrose], 1× MEM-E amino acids [Invitrogen], 1× MEM-E vitamins [Invitrogen], 2 mM l-glutamine, and 0.2% fetal bovine serum). Fusion medium was adjusted to pH 5.3 in medium containing 10 mM MES buffer or to pH 7.4 in medium containing 10 mM HEPES buffer. The cell monolayers were then scored for the amount of fusion seen relative to mock treated samples as a percentage of a non-virus-containing control. Cell fusion into polykaryons was defined as cells that contained three or more nuclei per cell.
RESULTS
Location of the free Cys residues in SV E1 and E2 proteins.
Mature virus particles were collected from the media of infected BHK cells and purified over potassium tartrate gradients (as described in Materials and Methods). Purified virus was then dialyzed against PBS-D and alkylated by the addition of a 500-fold molar excess iodoacetamide to total cysteinyl content. After nonreducing SDS-PAGE to separate the viral proteins and remove excess iodoacetamide (as described in Materials and Methods), the gel bands corresponding to the virus proteins were excised, reduced with DTT, and treated with 20 mM iodomethane. The presence of each label is detected by its unique mass as determined by LC/MS/MS analysis. Labeling with iodoacetamide results in carboxamidomethylation and a net increase of 57.0 u per Cys residue (Cam-Cys), while labeling with iodomethane results in methylation and a net increase of 14.0 u per Cys residue (m-Cys). Final alkylation with iodomethane is a necessary step in order to prevent disulfide rearrangement from occurring before LC/MS/MS analysis. With this labeling technique, all native Cys residues containing a free sulfhydryl group (not involved in a disulfide bond) that are accessible to solvent would be labeled as Cam-Cys, and all others would be labeled as m-Cys after reduction with DTT. Several representative product ion spectra of differentially mass labeled Cys peptides identified by this approach are presented in Fig. 1 with their corresponding SEQUEST scores listed in Table 1. Although the conditions were optimized to maximize the labeling of the thiolate side chains of Cys residues, a few His and Lys residues were detected as carboxyamidomethylated. These modifications were at levels that barely permitted detection and were not consistently detected from sample to sample, as was observed for the Cys modifications. Interestingly, these non-Cys modifications were within several residues of the primary sequence to the detected Cam-Cys modifications or very close to oxidized Met residues. These other modifications were distinguishable from the Cys modifications due to peptide fragmenting. With iodomethane labeling, only Cys residues were detected as being methylated (m-Cys). Together, these data indicate that native E1 protein cysteines are carboxamidomethylated. Figure 2, Fig. 3, and Table 2 represent a compilation of the data collected for the SV E1 and E2 proteins.
FIG. 1.
MS/MS identification of Cys-labeled peptide pairs obtained from differential alkylation of E1. The product ion spectra of several modified Cys peptides obtained by labeling native E1 at pH 8.0 using iodoacetamide (A and B) or denatured E1 using iodomethane (C and D) are shown. Each product ion spectrum (MS/MS spectrum) is a collection of ions produced by fragmentation of the intact peptide. Under the collision-induced dissociation conditions implemented in these measurements, fragmentation preferentially occurs along the backbone at peptide bonds to generate N-terminal fragments (b ions) and C-terminal fragments (y ions) at specific mass-to-charge (m/z) ratios and intensities which provide information regarding amino acid sequence and sites of modification. The predominant singly charged b and y product ion peaks are labeled accordingly, with the subscripts denoting their position within the identified peptide. Product ions eliciting neutral mass losses of H2O and NH3 are also indicated. Identified Cys residues are denoted as C* and C# for modifications corresponding to carboxamidomethylation and methylation, respectively. Analysis of the fragmentation patterns was performed by using SEQUEST with the corresponding scores presented in Table 1. The peptides were generated by in-gel proteolytic digestion of E1 using trypsin (A and C) or chymotrypsin (B and D) prior to extraction and LC/MS/MS analysis.
TABLE 1.
Representative list of Cys-labeled peptide pairs obtained from E1 at pH 8.0 as identified by LC/MS/MS analysis
| Peptidea | [M+H]+b
|
Charge statec | Xcorrd | ΔCne | Labeled residuef | |
|---|---|---|---|---|---|---|
| Calculated | Measured | |||||
| F.GC#KIAVNPL.R | 928.5 | 928.9 | 2 | 2.577 | 0.105 | C259 |
| F.GC*KIAVNPL.R | 971.5 | 972.0 | 2 | 2.510 | 0.088 | C259 |
| R.AVDC#SYGNIPISIDIPNAAFIR.T | 2,364.7 | 2,363.7 | 2 | 4.494 | 0.975 | C271 |
| R.AVDC*SYGNIPISIDIPNAAFIR.T | 2,407.7 | 2,406.5 | 2 | 3.292 | NCg | C271 |
Residue modifications: *, 57.0 u (carboxamidomethylation); and #, 14.0 u (methylation). The amino acid residues appearing before and after the periods correspond to the residues proceeding and following the peptide in the protein sequence.
Average mass calculated based on the peptide sequence (calculated) and the deconvoluted mass-to-charge (m/z) ratio based on the measured centroid mass (measured).
Charge state of the precursor ion selected for CID.
The SEQUEST cross-correlation score (Xcorr) of the peptide is based on the match of the obtained product ion spectra to the theoretical ion distribution for the corresponding peptide contained in the database.
The SEQUEST difference of cross-correlation scores (ΔCn) is the “difference” between the top two Xcorr values for a given product ion spectrum.
Labeled residues correspond to their position in the primary sequence of the E1 protein as determined by CID peptide fragmentation analysis.
NC, not calculated. Since SEQUEST could not match another peptide with the corresponding precursor ion mass and generated fragmentation pattern, the ΔCn could not be calculated.
FIG. 2.
Representation of labeled cysteine residues in the amino acid sequence of the SV E1 and E2 proteins as determined by MS at pH 8.0. Boxed Cys residues were carboxamidomethylated by iodoacetamide. Underlined Cys residues were methylated by iodomethane. Boxed and underlined Cys residues represent alkylation by both reagents. RNase B served as a control.
FIG. 3.
Representation of labeled cysteine residues in the amino acid sequence of the SV E1 and E2 proteins as determined by S after a pH shift from 8.0 to 5.3 to 8.0. Boxed Cys residues were carboxamidomethylated by iodoacetamide. Underlined Cys residues were methylated by iodomethane. Boxed and underlined Cys residues represent alkylation by both reagents.
TABLE 2.
Label accessibility of Cys residues for native SV E1 and E2 structural proteins determined by differential mass labeling and LC/MS/MS analysis
| Protein | Residue(s)
|
|||||
|---|---|---|---|---|---|---|
| Constant pH (pH 8.0)
|
pH shift (pH 7.4-5.3-8.0)
|
|||||
| Iodoacetamide inaccessible | Cam-Cys modification | Relative iodoacetamide accessibility | Iodoacetamide inaccessible | Cam-Cys modification | Relative iodoacetamide accessibility | |
| E1 | 62, 63, 68, 78, 301, 306, 328, 370, 376, 380 | 49, 114, 259, 271, 430 | 49 (both forms detected but not quantifiable) | 62, 301, 306, 328 | 259, 271 | 259 = 29% 271 = 100% (only the Cam form detected) |
| 114 = 90% | ||||||
| 259 = 17% | ||||||
| 271 = 73% | ||||||
| 430 = 100% (only the Cam form was detected) | ||||||
| E2 | 201, 411 | 105, 152, 220 | 105 = 100% (only the Cam form was detected) | 152, 411 | 220 | 220 = 10% |
| 152 (the Cam form was detected but not quantifiable | ||||||
| 220 (the Cam form was detected but not quantifiable | ||||||
aIodoacetamide inaccessible, Cys residues that were only detected as methylated (m) due to iodomethane treatment; Cam-Cys modification, Cys residues that were only detected as carboxamdiomethylated (Cam) due to iodoacetamide treatment. For relative iodoacetamide accessibility determinations, the measured relative percentage of the Cam-Cys peptide was calculated by integrating the peaks corresponding to the Cam-Cys and m-Cys modified peptides within the same LC/MS/MS analysis. For this type of measurement, a measurable difference of 10% was required to determine the relative iodoacetamide accessibility. Residues detected by MS/MS analysis but not quantifiable due to their lower abundance are indicated.
Using our approach for virus at neutral pH, 14 of the 17 Cys residues of E1 were labeled (Fig. 2). E1 Cys residues 49, 114, 259, 271, and 430 were found to be labeled by both iodoacetamide and iodomethane. This suggests that these Cys residues contained free sulfhydryl groups that were partially labeled by iodoacetamide. The remainder of the population was labeled by iodomethane after E1 had been reduced. E1 Cys residues 62, 63, 68, 78, 301, 306, 328, 370, 376, and 380 were only identified as m-Cys, suggesting that these residues were either label inaccessible or that their sulfhydryl groups were blocked due to their participation in disulfide bonds. E1 C430 was only detected as Cam-Cys. This Cys residue is within the transmembrane domain and thus is not expected to be in a disulfide bond and is labeled due to the membrane permeability of iodoacetamide. In Table 2, the percent labeled values represent a measure of the relative percentage of the modified Cys residue alkylated by iodoacetamide (i.e., the population that was label-accessible and quantified as Cam-Cys). EICs of the corresponding precursor ions were utilized to obtain these measurements. The percentages of the identified Cam-Cys modification for E1 Cys residues 114, 259, 271, and 430 were 90, 17, 73, and 100%, respectively, with the remaining percentage being the m-Cys modification. These values indicate that some Cys residues in the native virus particle are more accessible to iodoacetamide, while some are less accessible and are presumed to be buried deeper within the virus particle.
Five of the 17 Cys residues were labeled in the E2 protein of the virus at neutral pH (Fig. 2). C105, C152, and C220 were all detected as Cam-Cys, indicating that these Cys residues have free sulfhydryl groups, are accessible to iodoacetamide, and therefore are not involved in disulfide bonds in the native particle. E2 C201 and C411 were only detected as m-Cys, and thus these residues may participate in disulfide bonds. Analysis of the EICs of the precursor ions revealed that only detectable amounts of E2 C152 and C220 were present as the Cam-Cys modification, whereas E2 C105 was only detected as the Cam-Cys form (Table 2).
As a control, RNase B was examined under the same cysteinyl concentration and labeling conditions as the purified virus samples at neutral pH. Five of the eight Cys residues were labeled and detected as m-Cys by LC/MS/MS (Fig. 2) with no Cam-Cys modification detected for any Cys residue. This is in agreement with the published disulfide network present in RNase B in which all eight Cys residues participate in disulfide bonds and are thus inaccessible to solvent (42). This validates our alkylation labeling strategy to probe solvent-accessible Cys residues using iodoacetamide, since no Cys residues of RNase B were found to have been carboxamidomethylated. At very high concentrations of iodoacetamide (>1,000-fold excess), some Cam-Cys modifications of RNase B were detected, so the use of iodoacetamide at these concentrations was avoided for viral particle labeling. Interestingly, when these extreme labeling conditions were used, the viral proteins tended to precipitate.
Effect of iodoacetamide labeling on virus infectivity.
Purified virus was incubated at 100- or 500-fold excess iodoacetamide to total cysteinyl content at 37°C and pH 8.0 for 2 h, and the titers were determined on BHK cells. The infectivity of the purified virus alkylated with iodoacetamide was determined by plaque assay on BHK cell monolayers. The titer of the purified virus alone was 2.0 × 1010 PFU/ml; the titers for purified virus treated with 100- and 500-fold excesses of iodoacetamide were 1.5 × 108 and 9.0 × 106 PFU/ml, respectively. Purified virus treated with a 100-fold excess of iodoacetamide demonstrated a 2-log decrease in virus infectivity, whereas treatment of the virus with 500-fold excess iodoacetamide resulted in a 3.5-log decrease in infectivity compared to wild-type purified virus. These data reveal that iodoacetamide is alkylating exposed Cys residues in the native virus particle and that the presence of the carboxamidomethyl group has a detrimental effect on virus infectivity. This modification alone may either destabilize the virus structure or prevent the Cys residues that are in the reduced form in the native virus particle from participating in disulfide bond reshuffling to facilitate the reorganization of the virus surface during an early step of virus infection. It contrast, purified virus remained infectious when stored at either pH 7.4 or 8.0 (data not shown), indicating that the pH 8.0 environment had no effect on infectivity. Since iodoacetamide alkylation of Cys residues is most selective at pH 8.0 and no differences in infectious virus titer were observed at pH 8.0, this pH was adopted for our labeling experiments.
Effect of iodoacetamide labeling on virus mediated low-pH-induced cell-cell fusion.
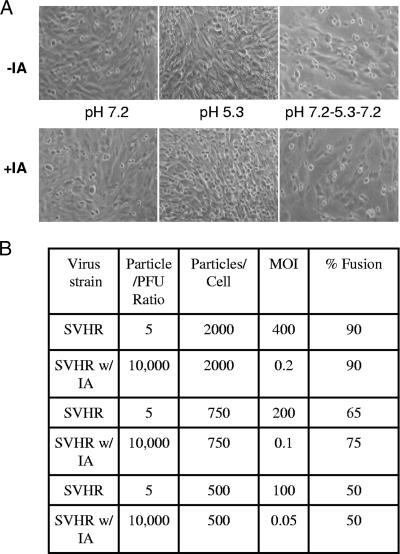
The attachment of large numbers of virus particles to cells followed by brief exposure to acidic pH and a subsequent return to neutral pH results in cell-cell fusion (6, 32). This observation has served as evidence that exposure to acidic pH and membrane fusion are the events that lead to infection of cells by alphaviruses (50, 51). The finding that the process of cell infection could be inhibited by alkylation of free envelope protein Cys residues suggested that if fusion is an integral part of infection, fusion might also be blocked by alkylation. To test this hypothesis, we determined the infectivity of a preparation of SV prior to and after alkylation as described previously (12). We then exposed cells to decreasing numbers of virus particles per cell (Fig. 4). After we lowered the pH to 5.3 and returned to pH 7.2, we estimated the percentage of cells fused by determining the relative amount of syncytium formation in addition to the number of cells containing one nucleus before and after low-pH treatment. The results of this experiment are presented in Fig. 4. We found that the ability to fuse cells was determined by the number of virus particles and not by the number of PFU associated with the cells. Thus, iodoacetamide-treated virus with a particle/PFU ratio of 10,000 to 1 could fuse cells with 750 cell-associated virus particles, which represents less than one infectious unit per cell (Fig. 4). The decreased ability of iodoacetamide-treated or untreated virus to fuse cells followed a reduction in particle number, not a decrease in the quantity of infectious units (PFU/ml). This result suggests that modification of free sulfhydryls in the virus glycoproteins blocks infectivity but not the ability to fuse cells.
FIG. 4.
(A) Fusion of BHK cell monolayers by iodoacetamide-treated SV. In this example, 500 particles of iodoacetamide-treated (+IA) or untreated (−IA) SVHR/cell were placed on BHK monolayers and were examined for fusion at different pH values (see Materials and Methods). (B) Table relating the amounts of fusion observed at various concentrations of virus particles applied to BHK cells.
Conformational changes induced with low pH do not alter the location of free Cys residues.
As described above, exposure of SV to acidic pH (5.3) and a subsequent return to neutral pH (pH 7.4) induces cell-to-cell FFWO of cell monolayers (6, 32). Using electron cryomicroscopy, we have described dramatic conformational changes in the surface of the virion as the virus is shifted into acidic pH and returned to neutral pH (32). These pH-induced conformational changes may correlate to a structural change that occurs on the virus surface during entry and may be induced by interactions with the host receptor (32). Electron cryomicroscopy analysis has revealed that as the pH is reduced from 7.4 to 5.3 a new spike structure emerges at the strict fivefold axis of the icosahedral protein lattice (32). It was suggested that this results from a reconfiguration of the E1 protein, which normally lays flat along the surface of the virus (32, 35). This reconfiguration of E1 may be necessary for delivery of the viral genome into the host cell (32). Once the virus is returned to neutral pH, the virus structure partially reverts to a more native structure where this new spike structure is withdrawn into the virus surface. Since this low-pH-mediated fusion event may mimic entry, the location(s) of the free Cys residues was investigated to determine whether disulfide rearrangement occurred after induction of these conformational changes. The intermediate low-pH form (5.3) could not be alkylated due to the kinetics of Cys residue labeling with iodoacetamide at low pH, and thus the viral structure was examined by alkylating after exposure to low pH (5.3) and the return to neutral pH.
The locations of the free Cys residues after the pH shift described above are presented in Fig. 3 and Table 2. Six of the seventeen Cys residues of E1 were identified by using our labeling approach. The reduction in the amount of coverage for the pH shift experiments compared to the nonshifted sample was probably due to the sample concentration used in each labeling reaction. If the initial concentration of virus was too high, the virus precipitated when the pH was shifted to 5.3 (32). Therefore, the virus concentration was adjusted to an amount where no precipitation occurred (see Materials and Methods). Both C259 and C271 of E1 were detected as the Cam-Cys form (Fig. 3 and Table 2), suggesting that they are not involved in disulfide bonds and are accessible to iodoacetamide labeling. Cys residues at positions 62, 301, 306, and 328 were only identified as the m-Cys form, suggesting that these residues are not accessible to iodoacetamide and are involved in disulfide bonds. The relative percentages of the identified Cam-Cys modifications for E1 Cys residues C259 and C271 in the pH-shifted sample were determined to be 29 and 100% (i.e., only Cam-Cys form detected), respectively. This shows a general increase in accessibility to iodoacetamide for these Cys residues in the pH-shifted sample compared to the sample maintained at pH 8.0. In fact, no Cys residues that were determined to have label-accessible sulfhydryl groups at pH 8.0 were found to be inaccessible or blocked after the pH shift or vice versa. In addition, no detectable rearrangement of disulfide bonds was observed after low-pH exposure.
As indicated in Fig. 3, three of the seventeen Cys residues of E2 were alkylated after the pH shift (7.4-5.3-8.0). E2 C220 was determined to have a free sulfhydryl group, whereas C152 and C411 were only detected as methylated, indicating that they were inaccessible to iodoacetamide or participating in disulfide bonds after the pH shift. After the pH shift, a quantitative increase in the carboxamidomethylation of E2 C220 was observed where nearly 10% was detected as Cam-Cys form, whereas 90% was identified as m-Cam. Thus, there is an increase in the amount of iodoacetamide-labeled C220 when the pH is shifted from 7.4 to 5.3 to 8.0 compared to when it is held constant at pH 8.0. The low-pH shift likely induces a conformation where C220 of E2 is more accessible to iodoacetamide. Once again, no detectable rearrangement of disulfide bonds was observed after low-pH exposure.
The lack of disulfide bond rearrangement after the pH shift of both E1 and E2 suggests that the conformational changes observed during the pH shift (26) may not be involved in viral infection since labeling virus with iodoacetamide reduces infectivity and yet promotes no differences in disulfide bond rearrangement. The conformational changes that occur during the low-pH shift may correlate to fusion events that are not involved in viral infection. It is also possible that a two-step process is necessary for infectivity where disulfide bond rearrangement may not occur as the first step. Alternatively, a disulfide bond rearrangement may have occurred and was not detected using our labeling method and LC/MS/MS measurements.
The effect of low pH exposure on virus infectivity was also examined. Virus was maintained at pH 8.0 or was reduced to pH 5.3 for 10 min at 37°C and then returned to pH 8.0. Virus held constant at pH 8.0 had a titer of 3.0 × 1010 PFU/ml, while virus exposed to low pH resulted in a 3-log decrease in virus infectivity to 2.0 × 107 PFU/ml (data not shown). This finding is in agreement with previously published research (7).
Effects of E1 Cys residue mutations on infectious virus production.
The effects on virus assembly and function of the E1 Cys residues that were determined to be labeled with iodoacetamide were examined. Cys-to Ser residue substitution mutants were constructed for C49, C114, C259, and C271. These four residues were all identified as possessing reactive sulfhydryl groups according to LC/MS/MS analysis. Very little virus was produced by any of these point mutations (ca. 104 PFU/ml), and analysis of the virus produced (by reverse transcription-PCR) indicated that reversion to wild-type sequence had occurred in all cases (data not shown). Thus, all of the point mutations of Cys to Ser resulted in a complete failure to assemble virus. These data indicate that while these residues may not be involved in disulfide bonds in the native virus particle they are critical for normal virus production. We have previously shown that the E1 glycoprotein is folded sequentially though several disulfide stabilized intermediates as it matures in the ER (5, 23). The presence of free sulfhydryl groups may be required to mediate this assembly process.
DISCUSSION
Disulfide bonds are essential to SV assembly and infectivity (2, 4, 22, 23). SV E1 and E2 proteins each contain 17 Cys residues. In order to gain insight into the nature of the disulfide network a differential labeling strategy, in combination with MS, was used. This strategy would permit the identification of reduced Cys residues present in the mature virus particle.
At pH 8.0, E1 Cys residues 49, 114, 259, and 271 were alkylated by both iodoacetamide and iodomethane reagents (Fig. 2). Since these E1 residues were labeled, at least in part, by iodoacetamide they must be present as reduced sulfhydryl groups and therefore are not part of a disulfide bond. E1 C114 and C 271 were primarily detected as Cam-Cys (90 and 73% labeled, respectively), suggesting that these Cys residues were more accessible to the iodoacetamide reagent than C259 (17% labeled). E1 C259 may be buried deeper within the virus particle than C114 and C271, which would account for its reduced labeling efficiency. C430, which lies within the transmembrane domain, was accessible to iodoacetamide, and only the Cam-Cys form was detected. E1 Cys residues 62, 63, 68, 78, 301, 306, 328, 370, 376, and 380 were only detected in their m-Cys form. This suggests that these Cys residues are involved in disulfide bonds or are inaccessible to iodoacetamide in the native virus particle but are labeled by iodomethane once the protein is denatured and reduced.
Much of these data are in agreement with the reported distribution of disulfide bonds described by Roussel et al. based on the crystallographic structure of the Semliki Forest virus E1 protein (40). Roussel et al. indicated that all E1 Cys residues with the exception of 430 are involved in disulfide bonds (40). Here, we report that Cys residues 49, 114, 259, 271, and 430 have label-accessible sulfhydryl groups for which Roussel et al. reported disulfide bonds for C49-C114 and C259-C271. One reason for the discrepancy may be the utilization of detergent-based extraction methods used during the preparation of the Semliki Forest virus E1 protein for crystallography. It was previously shown that SV E1 adopts many disulfide stabilized configurations upon extraction from the native virus particle with detergent (23). The crystal structure of E1 from Semliki Forest virus may be of one of these non-native conformations. Recently, a crystal structure of the E protein of West Nile virus has been produced (26). The structure is similar to that produced for the E1 proteins of alphaviruses and other E protein structures produced for flaviviruses. Nybakken et al. (26) point out that it is paradoxical that the fusion loop domain of the E protein is accessible for specific neutralizing antibody binding in the mature virion but is inaccessible in the virion structure as determined by cryo-EM (21). The authors of that study explain this by raising the possibility that the crystal structure does not represent the structure in the mature virion but may represent a transition structure through which the protein passes during maturation or infection. Likewise in the case of the crystal structures produced for the alphavirus E1 glycoprotein, studies of the configuration of this protein in the intact virion do not agree with the crystal structures produced from the individual protein (34, 41). As in the case of the West Nile E protein structure the structures produced for alphavirus E1 may be of an intermediate in the assembly or penetration process.
At pH 8.0, 5 of the 17 E2 Cys residues were detected as labeled (Fig. 2). Of these, C105, C152, and C220 were partially labeled by iodoacetamide, indicating that they contain reduced sulfhydryl groups and are not involved in disulfide bonds. E2 residues C201 and C411 were only detected as the m-Cys form and therefore are either participating in disulfide bonds or are inaccessible to iodoacetamide. E1 C152 and C220 were detected in the Cam-Cys form but were of low abundance and could not be quantified. In fact, the vast majority of the E2 Cys residues were only labeled by the iodomethane reagent after the virus was reduced by DTT, as indicated by the enhanced detection of each m-Cys modification. These data suggest that these Cys residues are deeply buried within the native virus particle and are largely unavailable to the iodoacetamide reagent. In any case, the determination of E2 C152 and C220 as labeled by iodoacetamide suggests that they are not involved in disulfide bonds in the native protein structure.
After the virus was treated with low pH and then returned to neutral pH, our labeling approach indicated that no E1 or E2 disulfide bond rearrangement occurred. These correlating data imply that a disulfide bond rearrangement does not exist, based on the Cys residues detected, between the native state and the state occupied after the pH shift to 5.3 and back to pH 8.0. The conformational changes induced by the pH shift may be necessary for virus-induced cell fusion but may be independent of infectivity.
Labeling of Cys residues by iodoacetamide in the SV E1 and E2 proteins also provides information regarding the regions of the native virus particle that are solvent accessible (Table 3). SV E1 Cys residues 49, 114, 259, 271, and 430 and E2 Cys residues 105,152, and 220 were found to be accessible to iodoacetamide at neutral pH. E1 C259 and C271 and E2 C220 were found to be accessible at pH 5.3. It should also be mentioned that some of the non-Cys modifications that were detected in our study correlate with the observations presented in Table 3. These data, along with others (25, 44, 47) (Table 3), provide insight regarding the surface-accessible regions of the virus particle under various conditions.
TABLE 3.
Accessible regions of the SV E1 and E2 proteinsa
| E1 | E2 | Assayb | Source or reference |
|---|---|---|---|
| K16 | K70, K76, K97, K131, K149, K202, K235 | Accessible to biotin | 41 |
| 1-21, 161-176, 212-220 | 31-84, 134-148, 158-186, 231-260, 299-314, 324-337 | Accessible to protease at neutral pH | 34 |
| 1-29, 86-91, 110-119, 121-128, 145-158, 162-170, 177-200, 206-217, 273-287 | 42-66, 99-102, 262-309, 168-179, 237-257, 330-338 | Accessible to protease at pH 4.5 | 34 |
| Y1, Y15, Y24 | Y53, Y140, Y155, Y165, Y188, Y328, Y339 | Accessible to iodination | 34 |
| 181-216 | Interacts with neutralizing antibody | 44 | |
| 173-220 | Epitope recognized by antibody | 47 | |
| 130, 133, 392, 393 | Accessible to furin | 25 | |
| C49, C114, C259, C271, C430 | C105, C152, C220 | Accessible to iodoacetamide at neutral pH | This study |
| C259, C271 | C220 | Accessible to iodoacetamide at pH 5.3 | This study |
| K211, K223 | K70, K76, K235, K254 | Accessible to cross-linker | Unpublished data |
This table reflects a brief summary of the literature in reference to accessible regions located on the SV E1 and E2 proteins as determined by various methods (see last column).
All assays were carried out at neutral pH unless stated otherwise.
Labeling the native virus particles with iodoacetamide was found to greatly reduce infectivity (see above). The loss of infectivity was dependent on the concentration of iodoacetamide used in the labeling reaction. The addition of the alkyl group to the sulfhydryl of an exposed Cys residue may be responsible for the loss of virus infectivity by destabilizing the structure of the native viral particle. Alternatively, early steps in virus infectivity may be inhibited by the presence of the alkyl group on the modified Cys residues. It is possible that a reduced sulfhydryl group is necessary to serve as a donor in a disulfide reshuffling reaction that correctly configures the virus for cell penetration. Alternatively, the creation of modified Cys residues may block critical events in disassembly required for release of the virus RNA into the cell interior. Interestingly, the carboxamidomethylation of Cys residues did not appear to effect the process of virus-mediated cell-cell fusion induced by transient exposure to acidic pH. This fact mechanistically separates the process of infection from the process of low-pH-mediated membrane fusion and supports the hypothesis that fusion itself cannot lead to infection (32). This observation is in support of an increasing body of evidence suggesting that membrane fusion and endocytosis are not involved in the infection of cells by alphaviruses (14, 32, 46).
Since several Cys residues of E1 were determined not to be part of disulfide bonds in the native structure, four mutants were constructed to evaluate the effect of E1 Cys residues C49, C114, C259, and C271 on infectious virus assembly. In each case the Cys residue was converted to a Ser residue. Interestingly, all of these mutants had a marked effect on the amount of infectious virus particles produced, as may be expected since all of these Cys residues are completely conserved across the Alphavirus genus. All four of these mutants yielded at least a 4-log decrease in infectious virus production compared to the wild type. This finding reinforces the idea that disulfide bond rearrangement occurs as virus maturation proceeds (5, 22-24). If these Cys residues remain free (i.e., not participating in a disulfide bond) during the complete SV life cycle, such a large decrease in infectious virus production would not be expected. These residues may contain free sulfhydryl groups in the native infectious virion but may be involved in a non-native disulfide bridge intermediate that is resolved at some point as virus maturation progresses.
Acknowledgments
This research was supported by a grant from the Foundation for Research, Carson City, NV, and by the North Carolina Agricultural Research Service.
We thank the research agencies of North Carolina State University and the North Carolina Agricultural Research Service for continued support of biological mass spectrometry research.
Footnotes
Published ahead of print on 4 April 2007.
REFERENCES
- 1.Anthony, R. P., and D. T. Brown. 1991. Protein-protein interactions in an alphavirus membrane. J. Virol. 65:1187-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anthony, R. P., A. M. Paredes, and D. T. Brown. 1992. Disulfide bonds are essential for the stability of the Sindbis virus envelope. Virology 190:330-336. [DOI] [PubMed] [Google Scholar]
- 3.Brown, D. T., and L. D. Condreay. 1986. Replication of alphaviruses in mosquito cells, p. 171-207. In M. J. Schlesinger (ed.), The Togaviridae and Flaviviridae. Plenum Publishing Corp., Inc., New York, NY.
- 4.Carleton, M., and D. T. Brown. 1996. Disulfide bridge-mediated folding of Sindbis virus glycoproteins. J. Virol. 70:5541-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carleton, M., H. Lee, M. Mulvey, and D. T. Brown. 1997. Role of glycoprotein PE2 in formation and maturation of the Sindbis virus spike. J. Virol. 71:1558-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards, J., and D. T. Brown. 1986. Sindbis virus-mediated cell fusion from without is a two-step event. J. Gen. Virol. 67:377-380. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, J., E. Mann, and D. T. Brown. 1983. Conformational changes in Sindbis virus envelope proteins accompanying exposure to low pH. J. Virol. 45:1090-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreira, D. F., R. Hernandez, M. Horton, and D. T. Brown. 2003. Morphological variants of Sindbis virus produced by a mutation in the capsid protein. Virology 307:54-66. [DOI] [PubMed] [Google Scholar]
- 9.Gaedigk-Nitschko, K., and M. J. Schlesinger. 1990. The Sindbis virus 6K protein can be detected in virions and is acylated with fatty acids. Virology 175:274-281. [DOI] [PubMed] [Google Scholar]
- 10.Gidwitz, S., J. M. Polo, N. L. Davis, and R. E. Johnston. 1988. Differences in virion stability among Sindbis virus pathogenesis mutants. Virus Res. 10:225-239. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez, R., H. Lee, C. Nelson, and D. T. Brown. 2000. A single deletion in the membrane-proximal region of the Sindbis virus glycoprotein E2 endodomain blocks virus assembly. J. Virol. 74:4220-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez, R., C. Sinodis, M. Horton, D. Ferreira, C. Yang, and D. T. Brown. 2003. Deletions in the transmembrane domain of a Sindbis virus glycoprotein alter virus infectivity, stability, and host range. J. Virol. 77:12710-12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishii, Y., and S. S. Lehrer. 1986. Effects of the state of the succinimido-ring on the fluorescence and structural properties of pyrene maleimide-labeled alpha alpha-tropomyosin. Biophys. J. 50:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koschinski, A., G. Wengler, G. Wengler, and H. Repp. 2005. Rare earth ions block the ion pores generated by the class II fusion proteins of alphaviruses and allow analysis of the biological functions of these pores. J. Gen. Virol. 86:3311-3320. [DOI] [PubMed] [Google Scholar]
- 15.Lee, H., and D. T. Brown. 1994. Mutations in an exposed domain of Sindbis virus capsid protein result in the production of noninfectious virions and morphological variants. Virology 202:390-400. [DOI] [PubMed] [Google Scholar]
- 16.Lee, H., P. D. Ricker, and D. T. Brown. 1994. The configuration of Sindbis virus envelope proteins is stabilized by the nucleocapsid protein. Virology 204:471-474. [DOI] [PubMed] [Google Scholar]
- 17.Lee, S., K. E. Owen, H. K. Choi, H. Lee, G. Lu, G. Wengler, D. T. Brown, M. G. Rossmann, and R. J. Kuhn. 1996. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure 4:531-541. [DOI] [PubMed] [Google Scholar]
- 18.Liljestrom, P., and H. Garoff. 1991. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J. Virol. 65:147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, N., and D. T. Brown. 1993. Transient translocation of the cytoplasmic (endo) domain of a type I membrane glycoprotein into cellular membranes. J. Cell Biol. 120:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moehring, J. M., N. M. Inocencio, B. J. Robertson, and T. J. Moehring. 1993. Expression of mouse furin in a Chinese hamster cell resistant to Pseudomonas exotoxin A and viruses complements the genetic lesion. J. Biol. Chem. 268:2590-2594. [PubMed] [Google Scholar]
- 21.Mukhopadhyay, S., B. S. Kim, P. R. Chipman, M. G. Rossmann, and R. J. Kuhn. 2003. Structure of West Nile virus. Science 302:248. [DOI] [PubMed] [Google Scholar]
- 22.Mulvey, M., and D. T. Brown. 1996. Assembly of the Sindbis virus spike protein complex. Virology 219:125-132. [DOI] [PubMed] [Google Scholar]
- 23.Mulvey, M., and D. T. Brown. 1994. Formation and rearrangement of disulfide bonds during maturation of the Sindbis virus E1 glycoprotein. J. Virol. 68:805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mulvey, M., and D. T. Brown. 1995. Involvement of the molecular chaperone BiP in maturation of Sindbis virus envelope glycoproteins. J. Virol. 69:1621-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson, S., R. Hernandez, D. Ferreira, and D. T. Brown. 2005. In vivo processing and isolation of furin protease-sensitive alphavirus glycoproteins: a new technique for producing mutations in virus assembly. Virology 332:629-639. [DOI] [PubMed] [Google Scholar]
- 26.Nybakken, G. E., C. A. Nelson, B. R. Chen, M. S. Diamond, and D. H. Fremont. 2006. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 80:11467-11474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oates, A., E. Polmear, R. Herrington, A. Farrugia, S. Sykes, G. Raines, H. Aumann, and A. Street. 1992. von Willebrand factor characterization of a severe dry-heat treated factor VIII concentrate, AHF (high purity). Thromb. Res. 65:389-399. [DOI] [PubMed] [Google Scholar]
- 28.Oda, Y., T. Nagasu, and B. T. Chait. 2001. Enrichment analysis of phosphorylated proteins as a tool for probing the phosphoproteome. Nat. Biotechnol. 19:379-382. [DOI] [PubMed] [Google Scholar]
- 29.Owen, K. E., and R. J. Kuhn. 1997. Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology 230:187-196. [DOI] [PubMed] [Google Scholar]
- 30.Owen, K. E., and R. J. Kuhn. 1996. Identification of a region in the Sindbis virus nucleocapsid protein that is involved in specificity of RNA encapsidation. J. Virol. 70:2757-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paredes, A. M., D. T. Brown, R. Rothnagel, W. Chiu, R. J. Schoepp, R. E. Johnston, and B. V. Prasad. 1993. Three-dimensional structure of a membrane-containing virus. Proc. Natl. Acad. Sci. USA 90:9095-9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paredes, A. M., D. Ferreira, M. Horton, A. Saad, H. Tsuruta, R. Johnston, W. Klimstra, K. Ryman, R. Hernandez, W. Chiu, and D. T. Brown. 2004. Conformational changes in Sindbis virions resulting from exposure to low pH and interactions with cells suggest that cell penetration may occur at the cell surface in the absence of membrane fusion. Virology 324:373-386. [DOI] [PubMed] [Google Scholar]
- 33.Peng, J., J. E. Elias, C. C. Thoreen, L. J. Licklider, and S. P. Gygi. 2003. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2:43-50. [DOI] [PubMed] [Google Scholar]
- 34.Phinney, B. S., K. Blackburn, and D. T. Brown. 2000. The surface conformation of Sindbis virus glycoproteins E1 and E2 at neutral and low pH, as determined by mass spectrometry-based mapping. J. Virol. 74:5667-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pletnev, A. G., R. Putnak, J. Speicher, E. J. Wagar, and D. W. Vaughn. 2002. West Nile virus/dengue type 4 virus chimeras that are reduced in neurovirulence and peripheral virulence without loss of immunogenicity or protective efficacy. Proc. Natl. Acad. Sci. USA 99:3036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renz, D., and D. T. Brown. 1976. Characteristics of Sindbis virus temperature-sensitive mutants in cultured BHK-21 and Aedes albopictus (mosquito) cells. J. Virol. 19:775-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice, C. M., J. R. Bell, M. W. Hunkapiller, E. G. Strauss, and J. H. Strauss. 1982. Isolation and characterization of the hydrophobic COOH-terminal domains of the Sindbis virion glycoproteins. J. Mol. Biol. 154:355-378. [DOI] [PubMed] [Google Scholar]
- 39.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roussel, A., J. Lescar, M. C. Vaney, G. Wengler, G. Wengler, and F. A. Rey. 2006. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure 14:75-86. [DOI] [PubMed] [Google Scholar]
- 41.Sharp, J. S., S. Nelson, D. Brown, and K. B. Tomer. 2006. Structural characterization of the E2 glycoprotein from Sindbis by lysine biotinylation and LC-MS/MS. Virology 348:216-223. [DOI] [PubMed] [Google Scholar]
- 42.Shin, H. C., M. Narayan, M. C. Song, and H. A. Scheraga. 2003. Role of the [65-72] disulfide bond in oxidative folding of bovine pancreatic ribonuclease A. Biochemistry 42:11514-11519. [DOI] [PubMed] [Google Scholar]
- 43.Strauss, E. G., E. M. Lenches, and J. H. Strauss. 2002. Molecular genetic evidence that the hydrophobic anchors of glycoproteins E2 and E1 interact during assembly of alphaviruses. J. Virol. 76:10188-10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strauss, E. G., D. S. Stec, A. L. Schmaljohn, and J. H. Strauss. 1991. Identification of antigenically important domains in the glycoproteins of Sindbis virus by analysis of antibody escape variants. J. Virol. 65:4654-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, G., R. Hernandez, K. Weninger, and D. T. Brown. 5 February 2007. Infection of cells by Sindbis virus at low temperature. Virology [Epub ahead of print.] [DOI] [PubMed]
- 47.Wang, K. S., and J. H. Strauss. 1991. Use of a lambda gt11 expression library to localize a neutralizing antibody-binding site in glycoprotein E2 of Sindbis virus. J. Virol. 65:7037-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.West, J., and D. T. Brown. 2006. Role of a conserved tripeptide in the endodomain of Sindbis virus glycoprotein E2 in virus assembly and function. J. Gen. Virol. 87:657-664. [DOI] [PubMed] [Google Scholar]
- 49.West, J., R. Hernandez, D. Ferreira, and D. T. Brown. 2006. Mutations in the endodomain of Sindbis virus glycoprotein E2 define sequences critical for virus assembly. J. Virol. 80:4458-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White, J., and A. Helenius. 1980. pH-dependent fusion between the Semliki Forest virus membrane and liposomes. Proc. Natl. Acad. Sci. 77:3273-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White, J., J. Kartenbeck, and A. Helenius. 1980. Fusion of Semliki Forest virus with the plasma membrane can be induced by low pH. J. Cell Biol. 87:264-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitehurst, C. B., J. H. Willis, C. N. Sinodis, R. Hernandez, and D. T. Brown. 2006. Single and multiple deletions in the transmembrane domain of the Sindbis virus E2 glycoprotein identify a region critical for normal virus growth. Virology 347:199-207. [DOI] [PubMed] [Google Scholar]